- 1Environmental Biotechnology Group, Biotechnology Department, Iranian Research Organization for Science and Technology, Tehran, Iran
- 2Maryland Pathogen Research Institute, University of Maryland, College Park, MD, USA
- 3The Geneva Foundation, Frederick, MD, USA
Helicobacter pylori is recognized as the most common pathogen to cause gastritis, peptic and duodenal ulcers, and gastric cancer. The organisms are found in two forms: (1) spiral-shaped bacillus and (2) coccoid. H. pylori coccoid form, generally found in the environment, is the transformed form of the normal spiral-shaped bacillus after exposed to water or adverse environmental conditions such as exposure to sub-inhibitory concentrations of antimicrobial agents. The putative infectious capability and the viability of H. pylori under environmental conditions are controversial. This disagreement is partially due to the fact of lack in detecting the coccoid form of H. pylori in the environment. Accurate and effective detection methods of H. pylori will lead to rapid treatment and disinfection, and less human health damages and reduction in health care costs. In this review, we provide a brief introduction to H. pylori environmental coccoid forms, their transmission, and detection methods. We further discuss the use of these detection methods including their accuracy and efficiency.
Introduction
Helicobacter pylori is recognized as the most common cause of gastritis, peptic and duodenal ulcers, and gastric cancer (1, 2). For many years, the transmission dynamics of H. pylori largely remained unknown and has thus gained the interest of many researchers around the world. In many studies, contaminated water is implicated as a source of transmission of this pathogen that colonizes more than 50% of humans (3). Water supplies contaminated by sewage with bodily fluids or feces from infected people have been considered as a potential source of H. pylori infection (4, 5). The transmission of H. pylori may occur from person to person both via the oral-to-oral and fecal-to-oral routes (6). Some previous studies showed a positive correlation between H. pylori infection and consumption of untreated or low-quality drinking water suggesting the waterborne transmission of H. pylori (7–9). H. pylori transforms from the normal spiral-shaped bacillary form into the coccoid form when it is exposed to water in adverse conditions (5, 10). Like other Gram-negative bacteria, the coccoid forms of H. pylori are also usually in viable but non-culturable (VBNC), less virulent, and less likely to colonize and induce inflammation than the spiral forms. It has been demonstrated that bacteria in the VBNC state are able to maintain their metabolic activity and pathogenicity (11) as well as may revert to active re-growth conditions (12, 13). It is well known that the detection of H. pylori in coccoid forms is difficult using traditional methods (14). It was long assumed that the bacterial cells were dead when they were no longer able to form colonies on routine culture media. We now know this assumption is too simplistic and there are many situations where bacterial cells lose culturability but remain viable and are potentially able to regrow. Recently, investigators have demonstrated that the coccoid forms of H. pylori can be cultured on enrichment culture (15). Epidemiological studies suggest that the level of sanitation, particularly, water sanitation influences the probability of infection with H. pylori. The risk of H. pylori infection was suggested to be 2–13 times higher in people who drink untreated river or well water and swim in rivers, streams, or pools than those who drink municipal tap water and do not swim in such environments (8, 9, 16, 17).
Helicobacter pylori has been detected from drinking water (4, 18–20) as well as sea water (5). One H. pylori strain stored in deep ground water or in natural seawater at 4°C was observed to survive significantly longer than the same strain stored in nutrient-rich media (21). Several studies indicate that H. pylori may survive as culturable forms for weeks in water and may survive longer in natural systems than in artificial nutrient-rich systems (14, 22). Only a few studies reported the detection of H. pylori coccoid form in environmental water samples. In one study, the bacterium was found in a municipal wastewater canal on the U.S.–Mexico border, which was suggestive to be a fecal–oral route of contamination (4). In another study, H. pylori coccoid form was identified from a seawater sample (23). Furthermore, Samra et al. (24) examined 600 drinking water samples collected by water and sanitation agencies from ground-drilled water in different localities. In this review, we summarize the current approaches to detect the environmental coccoid form of H. pylori and discuss their sensitivity, specificity, and accuracy.
VBNC State and Environmental H. pylori Coccoid Forms
Viable but non-culturable state is a bacterial response to some forms of natural adverse conditions such as nutrient starvation (25), extreme temperatures (26), incubation outside of the permissive pH or saltiness ranges for cell growth (27, 28), high- or low-osmotic concentrations (29), variable oxygen concentrations (30), exposure to food preservatives (31), and exposure to visible light and UV irradiation (32). Shahamat et al. demonstrated the entrance of H. pylori into the VBNC state for the first time during laboratory studies in which cells were observed to become non-culturable in freshwater microcosms (33). Cells in the VBNC state typically demonstrate very low levels of metabolic activity, but on resuscitation become culturable again (5, 34–36). Many suggest that H. pylori persists in the environment in a VBNC form (21, 34, 37, 38) and there is only scattered evidence for reversion to the actively dividing form (39, 40). H. pylori is mostly found in a spiral shape within the human host, but it converts into a coccoid shape when is exposed to unfavorable environments (41). It has been suggested that based on evidence gathered over the last few years, the VBNC cells of human pathogens should be viewed as a potential hazard to public health rather than considered as dead cells (42). In addition, pathogens in a VBNC state may remain virulent or produce enterotoxins (43). An issue of much significance is to detect VBNC and viable-culturable (VC) cells by novel and more efficient methods. There is an urgent need for a method, which lowers selectivity, reduces bias from sample storage and incubation, and decreases assay time (44).
Culture
There are no established culture methods for the detection of H. pylori in the drinking water supplies (45). Despite efforts to produce a culture-specific, media-culturing H. pylori from drinking water has not been successful (46, 47). A simple plating medium was suggested to detect H. pylori in the environment (48). Several studies have reported that H. pylori enters into the coccoid form when exposed to a nutrient deficient environment (49), drug supplementation (50), pH change (39), abnormal temperature (51), or prolonged culture (52). It is believed that the spiral form (Figure 1A) is transformed immediately and rapidly into the coccoid form (Figure 1B) (34). The first successful isolation of H. pylori from environmental water using the enriched culture was from a municipal wastewater canal heavily contaminated with untreated raw sewage at the U.S.–Mexico border where H. pylori infection was reported frequently (4). However, the history of unsuccessful attempts to culture H. pylori from environmental waters led investigators to explore the use of molecular methods to detect and identify this organism.
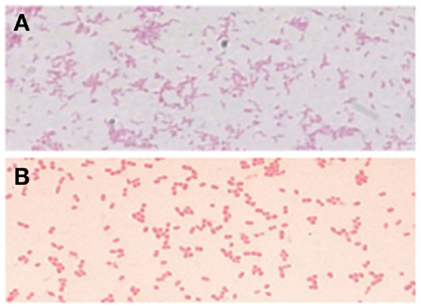
Figure 1. Conversion from the H. pylori bacillary form (A) to the coccoid form (B) (15).
Autoradiography
Autoradiography was optimized and employed to detect metabolic activity of VBNC cells of H. pylori in water. Tritium-labeled cells of H. pylori showed the aggregations of silver grains associated with uptake by H. pylori of radiolabeled substrate. Temperature is a significant environmental factor for the viability of the organism in water. Autoradiography revealed that H. pylori remain viable at 4°C for 26 months. However, sterile water does not reflect the natural environment in which competition with naturally occurring populations of microorganisms can occur. Findings based on an autoradiography approach provided evidence supporting the hypothesis that there is a waterborne route of infection for H. pylori (51).
Electron Microscopy
Coccoid forms have been divided into two types, a and b, by electron microscopy although the function of the two different coccoid forms of H. pylori is unclear. One possibility is that coccoid form in general represents a degenerating state of the organism (53). Kusters et al. (54) indicated that the coccoid cells of H. pylori were the morphological manifestation of bacterial cell death, observing the transformation process by electron microscopy. However, others suggested this form to be VBNC (55). Benaissa et al. (56) asserted that coccoid H. pylori was devoid of degenerative change. Willén et al. (57) studied morphologic conversion of H. pylori from spiral (Figure 2A) to coccoid (Figure 2B) form where scanning electron microscopy and transmission electron microscopy were employed.
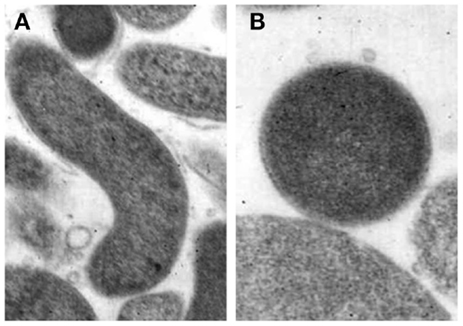
Figure 2. Morphological appearances of Helicobacter pylori. (A) Rod-shaped (B) Coccoid form. Original magnification × 20,000 (58).
Fluorescent In Situ Hybridization
Fluorescent in situ hybridization (FISH) with ribosomal RNA oligonucleotide probes has been used successfully for the detection and identification of VBNC forms of bacteria (59). FISH was validated as a quick and sensitive method for the detection of H. pylori in environmental samples (60). In the U.S., actively respiring H. pylori from surface and well water has been detected using fluorescent antibody-tetrazolium reduction (FACTC) microscopy (18) and confirmed using species-specific polymerase chain reaction (PCR) (61). These findings helped to determine the presence of H. pylori in the natural environment and a possible waterborne route of transmission. Use of FISH provides an alternative to PCR detection of H. pylori in water (60) and raw bovine milk (62). These findings imply that at some point in time helicobacters have entered the water source but it is not possible for PCR or hybridization methods to establish if viable organisms are present although coccoids in VBNC forms may be transmitted via water (63, 64).
DNA-Based Techniques
Among the molecular methods, PCR has been widely used for the diagnosis of H. pylori infection as well as the analysis of diversity, virulence, persistence, and resistance patterns of these bacteria (65) including detection of the organisms in environmental samples (66, 67). Specific target genes are selected to avoid cross-reactivity between H. pylori and other bacteria. For example, PCR targeting the 16S rRNA gene, random chromosome sequences, the 26-kDa species specific antigen gene, the urease A (ureA) gene, and the urease C (ureC) gene or glmM gene have been used (68–70). Among these gene targets, PCR-based detection of the ureC gene appears to be the most promising for the detection of H. pylori (69).
The presence of H. pylori in drinking water, which was detected by PCR, has been reported from many different countries (71, 72). Despite the requirement for a microaerobic atmosphere, helicobacters can possibly survive for short periods in water in a VBNC coccoid form (40, 49), which would allow them to be transmitted via the water distribution system while remain undetectable by culture techniques. Moreover, recent findings suggest that H. pylori cells may be able to tolerate the levels of disinfectant normally used in water purification plants. Results from one study showed the presence of H. pylori from U.S. surface water (18). H. pylori DNA has also been amplified from drinking water samples in Japan (73), Mexico (74), and Peru (75), untreated well water in the U.S. (76), from water samples taken from a water delivery truck and two lakes near Repulse Bay in the Canadian arctic (77), and from drinking water storage pots in Gambia (70).
Clearly, PCR is the only way to demonstrate the presence of H. pylori in water supplies and seawater (5, 78). H. pylori could not be cultured and the cell membrane was disintegrated but nucleic acid was still detected by PCR (47, 61). Furthermore, because of its high sensitivity, PCR was suggested to be an appropriate method to detect organisms when they are in low numbers, slow growing, or non-culturable form (79).
Despite the findings of much research to identify H. pylori in water, it is important to consider the fact that the use of PCR and other molecular methods for the detection of pathogens in environmental samples suffers from a number of limitations. The most serious limitation is that PCR does not enable us to distinguish between live and dead cells. It also suffers from the fact that it is biased and time consuming (44).
Nayak and Rose (80) demonstrated that quantitative PCR (qPCR) could determine H. pylori concentrations in water. In this study, qPCR was shown to be a specific, sensitive, and rapid method to quantify H. pylori in sewage. Another study showed that coccoid forms, regardless of viability, are readily detected in small numbers by qPCR assays (81). Nayak and Rose (80) investigated the detection of H. pylori in sewage and water using a new qPCR method with SYBR green. Janzon et al. (81) detected H. pylori DNA in drinking and environmental water in Dhaka, Bangladesh, using highly sensitive real-time PCR. Sen et al. (82) developed an internal control for evaluation and standardization of a qPCR assay for the detection of H. pylori in drinking water. There are also reports of the failure to identify H. pylori in drinking water in the U.S. (83) and in drinking water or reclaimed wastewater in low-endemic developed countries such as Belgium, Spain, and Italy (84).
The H. pylori qPCR test has several advantages. First, it does not rely on culturing. Second, many samples can be analyzed quickly, since real-time qPCR instruments are easily available and it can analyze up to 384 samples in 2 h. Third, real-time qPCR removes many of the sources of human error from the analysis process and lessens the potential of contamination. The results of McDaniels et al. (83) support the idea that a rapid real-time qPCR may be useful for the screening of large numbers of drinking water samples for the presence of H. pylori at low concentrations. Due to metabolic and morphological changes that can prevent H. pylori cells in water from growing on conventional media, an H. pylori-specific TaqMan qPCR assay that uses a 6-carboxyfluorescein-labeled probe has been developed (83).
In addition, there are a number of studies reporting traces of H. pylori in various water sources, mainly using PCR-based methods, although the successful isolation of live H. pylori from river water (4) or marine zooplankton (5) has been reported. However, some of these studies were performed with river water, lake water, or seawater (20, 85, 86) rather than drinking water, and some used nested PCR (20, 78, 87), which may increase detection sensitivity but is more prone to contamination.
Flow Cytometry
Flow cytometry is an analytical technique, which has the potential to make a distinction among the four physiological states of bacteria: reproductively viable, metabolically active, intact, and permeabilized. It can determine the proportions of VBNC and VC states and dead cells, based on membrane integrity of Gram-negative bacteria (44). The application of this technique makes rapid in situ analysis of single cells possible. In addition, using this technique along with staining techniques such as live/dead staining, it is possible to obtain qualitative data (88). Although the use of flow cytometry has revealed four physiological states, rapid approaches to distinguish between VBNC and VC cells are not yet available (44).
Loop-Mediated Isothermal Amplification Method
Loop-mediated isothermal amplification (LAMP) is a promising technique that can overcome some of the technical shortcomings of PCR. LAMP is a novel gene amplification strategy in which all reactions are conducted under isothermal conditions (i.e., no need for thermocycling) using a single type of enzyme. This method has high-amplification efficiency and provides faster amplification times than PCR (65, 89). LAMP amplifies targeted DNA producing magnesium pyrophosphate as a by-product DNA amplification can be detected by turbidity measured via photometry due to the increase of magnesium pyrophosphate in solution (65, 90) or by SYBR green addition, which can change the color detectable with naked eyes without the need for expensive equipment. Also, the detection of DNA amplification can use manganese loaded calcein, which starts fluorescing upon mixing with manganese by pyrophosphate during in vitro DNA synthesis (91).
It is, therefore, possible to detect the amplification of the products without gel electrophoresis using the white precipitate of magnesium pyrophosphate in the reaction mixture. This can be achieved due to high specificity and amplification efficiency of LAMP (65, 89). The simple operation of the LAMP assay offers advantages over currently available DNA probe and PCR methods (65). Although PCR methods are rapid and accurate compared to other detection techniques to detect coccoid forms of H. pylori, LAMP was found to be even more efficient detecting H. pylori coccoid forms in water samples (Figure 3) in terms of accuracy, rapidity, and sensitivity based on laboratory microcosm experiments (15), similar level of H. pylori detection was archived in the stomach biopsy samples, employing LAMP method (92).
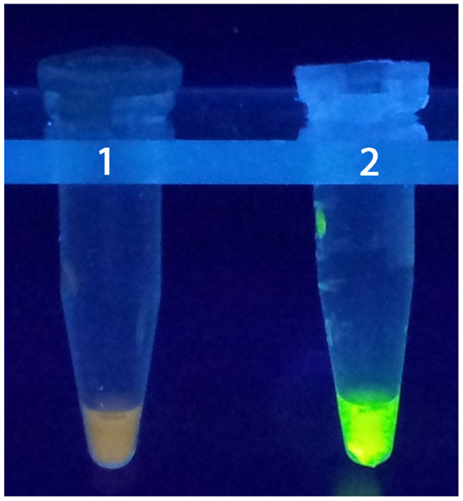
Figure 3. LAMP test. 1: negative control, 2: positive control (15).
Next-Generation DNA Sequencing and Metagenomics
Metagenomics is the application of modern genomics techniques for studying microbial community directly in their natural environments (93). Importantly, metagenomics bypasses the need for laboratory cultivation of individual bacterial species, thereby allowing for the study of unculturable microorganisms (94). The presence of unculturable bacteria in the environment has been known for more than a century and has often been referred to as the “Great Plate Count Anomaly” (95). This concept is the condition in which there is a discrepancy – often by several orders of magnitude – between the sizes of a bacterial population estimated by culture compared to that observed under microscope. It may be fair to mention that the demonstration that H. pylori causing gastric ulcers and cancer helped to draw attention on the importance of the unculturable microbial world. Although spiral bacteria were observed in the gastric mucosa of dogs in 1893 and in humans in 1906 (96), and correlations between the occurrence of the bacteria and peptic ulcers were noted in 1938 (97), it was not until H. pylori was cultured when its role as the disease causing agent was accepted (98, 99).
By the mid-1980s, microbiologists began describing the phylogenetic diversity of microorganisms in “exotic” environments, such as oceans, deep sea vents, hot springs, soil, and others using molecular methods alone. Much of these culture-independent methods were based on isolating total DNA from an environmental sample, cloning the DNA into a suitable vector, transforming the clones into a host bacterium (i.e., producing a clone library), and screening the clones for a phylogenetic marker (e.g., 16S rRNA). Clones were then sequenced and 16S rRNA gene sequences cataloged to reveal the diverse taxa present in the sample (94). This technique has been used widely to identify bacteria and archaea from a variety of environments (100–103). Today, high-throughput, next-generation DNA sequencing has made this process vastly more efficient. Advancements in next-generation sequencing (and reduced costs) now provide a technical means by which to not only monitor environmental microbial communities but also to study the occurrence of pathogens in the natural environmental, especially those that are no longer culturable, such as VBNC forms of bacterial pathogens. Metagenomics coupled to next-generation sequencing has been used to study microbial communities in many natural environments, including coastal areas of Thailand (104), waters of the Puget Sound in the U.S. (105), freshwater and marine sediments along the Pearl River in China (106), among others. While none of these studies have specifically focused on non-culturable bacteria or the coccoid form of H. pylori, Zheng et al. developed methods for metagenomic analysis of H. pylori from old formalin-fixed and paraffin-embedded gastrointestinal biopsies using Roche 454 high-throughput pyrosequencing (107). It is reasonable to suggest that these approaches can be used for the detection and characterization of H. pylori in the natural aquatic environment.
Conclusion
Helicobacter pylori is a significant human pathogen that is estimated to infect the gastric mucosa of half of the world’s population (108). The transmission dynamics of H. pylori are poorly understood; however, epidemiological data and the detection of H. pylori in a wide variety of natural aquatic environments points to waterborne transmission. Although, most attempts to culture H. pylori from water samples have proved unsuccessful, likely due to the presence of VBNC coccoid form, great tools to detect H. pylori in water samples, most commonly, PCR and qPCR are now available. As a powerful and accurate detection method for H. pylori, the qPCR technique provides diagnostic microbiology laboratories with a capacity to quantify and achieve a high degree of sensitivity and specificity of targets as compared to standard PCR. Other promising techniques for the detection of environmental H. pylori include the LAMP assay, which can be performed without a thermocycler and results can be visualized by eye (Figure 3). Perhaps one of the most exciting areas in microbiology, in the past decade, is the increasing use of next-generation DNA sequencing and metagenomics. As the instruments for DNA sequencing become more widespread and conveniently portable while the cost of sequencing continues to decrease, metagenomics will likely become the mainstream technology used in environmental microbiology and microbial ecology, including research into the transmission dynamics and potential reservoirs of environmental H. pylori. This awareness eventually will help public health official to take necessary action to protect people from H. pylori infection.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Malfertheiner P. The intriguing relationship of Helicobacter pylori infection and acid secretion in peptic ulcer disease and gastric cancer. Dig Dis (2011) 29:459–64. doi:10.1159/000332213
2. Calvet X, Ramírez Lázaro MJ, Lehours P, Mégraud F. Diagnosis and epidemiology of Helicobacter pylori infection. Helicobacter (2013) 18(1):5–11. doi:10.1111/hel.12071
3. Aziz RK, Khalifa MM, Sharaf RR. Contaminated water as a source of Helicobacter pylori infection. J Adv Res (2013). doi:10.1016/j.jare.2013.07.007
4. Lu Y, Redlinger TE, Avitia R, Galindo A, Goodman K. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl Environ Microbiol (2002) 68(3):1436–9. doi:10.1128/AEM.68.3.1436-1439.2002
5. Cellini L, Del Vecchio A, Di Candia M, Di Campli E, Favaro M, Donelli G. Detection of free and plankton associated Helicobacter pylori in seawater. J Appl Microbiol (2004) 97:285–92. doi:10.1111/j.1365-2672.2004.02307.x
6. Cellini L, Robuffo I, Maraldi NM, Donelli G. Searching the point of no return in Helicobacter pylori life: necrosis and/or programmed death? J Appl Micr (2001) 90(5):727–32. doi:10.1046/j.1365-2672.2001.01300.x
7. McCallion WA, Murray LJ, Bailie AG, Dalzell AM, O’Reilly DP, Bamford KB. Helicobacter pylori infection in children: relation with current household living conditions. Gut (1996) 39:18–21. doi:10.1136/gut.39.1.18
8. Nurgalieva ZZ, Malaty HM, Graham DY, Almuchambetova R, Machmudova A, Kapsultanova D, et al. Helicobacter pylori infection in Kazakhstan: effect of water source and household hygiene. Am J Trop Med Hyg (2002) 67(2):201–6.
9. Karita M, Teramukai S, Matsumoto S. Risk of Helicobacter pylori transmission from drinking well water is higher than that from infected intrafamilial members in Japan. Dig Dis Sci (2003) 48:1062–7. doi:10.1023/A:1023752326137
10. Al-Sulami AA, Al-Edani TAA, Al-Abdula AA. Culture method and PCR for the detection of Helicobacter pylori in drinking water in Basrah governorate Iraq. Gastroenterol Res Pract (2012) 2012(5):245167. doi:10.1155/2012/245167
11. Colwell RR, Brayton P, Herrington D, Tall B, Huq A, Levine MM. Viable but non-culturable Vibrio cholerae O1 revert to a cultivable state in the human intestine. World J Microbiol Biotechnol (1996) 12(1):28–31. doi:10.1007/BF00327795
12. Su X, Chen X, Hu J, Shen C, Ding L. Exploring the potential environmental functions of viable but non-culturable bacteria. World J Microbiol Biotechnol (2013) 29:2213–8. doi:10.1007/s11274-013-1390-5
13. Senoh M, Ghosh-Banerjee J, Ramamurthy T, Colwell RR, Miyoshi S, Nair GB, et al. Conversion of viable but nonculturable enteric bacteria to culturable by co-culture with eukaryotic cells. Microbiol Immunol (2012) 56:342–5. doi:10.1111/j.1348-0421.2012.00440.x
14. Andersen LP, Rasmussen L. Helicobacter pylori coccoid forms and biofilm formation. FEMS Immunol Med Microbiol (2009) 56:112–5. doi:10.1111/j.1574-695X.2009.00556.x
15. Chamanrokh P, Shahhosseiny MH, Mazaheri Assadi M, Nejadsattari T, Esmaili D. Evaluation of three tests used to detect non-culturable forms of Helicobacter pylori in water samples. Jundishapur J Microbiol (2014).
16. Goodman KJ, Correa P, Aux Tengana HJ, Ramirez H, DeLany JP, Guerrero Pepinosa G, et al. Helicobacter pylori infection in the Colombian Andes: a population-based study in transmission pathways. Am J Epidemiol (1996) 144:290–9. doi:10.1093/oxfordjournals.aje.a008924
17. Ahmed KS, Khan AA, Ahmed I, Tiwari SK, Habeeb A, Ahi JD, et al. Impact of household hygiene and water source on the prevalence and transmission of Helicobacter pylori: a South Indian perspective. Singapore Med J (2007) 48:543–9.
18. Hegarty J, Dowd M, Baker KH. Occurrence of Helicobacter pylori in surface water in the United States. J Appl Microbiol (1999) 87:697–701. doi:10.1046/j.1365-2672.1999.00912.x
19. Glynn MK, Friedman CR, Gold BD, Khanna B, Hutwagner L, Iihoshi N, et al. Seroincidence of Helicobacter pylori infection in a cohort of rural Bolivian children: acquisition and analysis of possible risk factors. Clin Infect Dis (2002) 35:1059–65. doi:10.1086/342910
20. Queralt N, Bartolomé R, Araujo R. Detection of Helicobacter pylori DNA in human faeces and water with different levels of faecal pollution in the north-east of Spain. J Appl Microbiol (2005) 98:889–95. doi:10.1111/j.1365-2672.2004.02523.x
21. Konishi K, Saito N, Shoji E, Takeda H, Kato M, Asaka M, et al. Helicobacter pylori: longer survival in deep ground water and sea water than in a nutrient-rich environment. APMIS (2007) 115:1285–91. doi:10.1111/j.1600-0643.2007.00594.x
22. Adams BL, Bates TC, Oliver JD. Survival of Helicobacter pylori in a natural freshwater environment. Appl Environ Microbiol (2003) 69(12):7462–6. doi:10.1128/AEM.69.12.7462-7466.2003
23. Cellini L, Grand R, Prenna M, Pasquantonio M, Pane L. Detection of Helicobacter pylori associated with zooplankton. Aquat Microb Ecol (2005) 40:115–20. doi:10.3354/ame040115
24. Samra ZQ, Javaid U, Ghafoor S, Batool A, Dar N, Athar MA. PCR assay targeting virulence genes of Helicobacter pylori isolated from drinking water and clinical samples in Lahore metropolitan, Pakistan. J Water Health (2011) 9(1):208–16. doi:10.2166/wh.2010.169
25. Cook KL, Bolster CH. Survival of Campylobacter jejuni and Escherichia coli in ground water during prolonged starvation at low temperatures. J Appl Microbiol (2007) 103:573–83. doi:10.1111/j.1365-2672.2006.03285.x
26. Besnard V, Federighi M, Declerq E, Jugiau F, Cappelier JM. Environmental and physico-chemical factors induce VBNC state in Listeria monocytogenes. Vet Res (2002) 33(4):359–70. doi:10.1051/vetres:2002022
27. Cunningham E, O’Byrne C, Oliver JD. Effect of weak acids on Listeria monocytogenes survival: evidence for available but nonculturable state in response to low pH. Food Control (2009) 20:1141–4. doi:10.1016/j.foodcont.2009.03.005
28. Del Campo R, Russi P, Mara P, Mara H, Peyrou M, deLeón IP, et al. Xanthomonas axonopodis pv. Citri enters the VBNC state after copper treatment and retains its virulence. FEMS Microbiol Lett (2009) 298:143–8. doi:10.1111/j.1574-6968.2009.01709.x
29. Asakura H, Kawamoto K, Haishima Y, Igimi S, Yamamoto S, Makino S-I. Differential expression of the outermembrane protein W(OmpW) stress response in enterohemorrhagic Escherichia coli O157:H7 corresponds to the viable but non-culturable state. Res Microbiol (2008) 159:709–17. doi:10.1016/j.resmic.2008.08.005
30. Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, et al. There resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol (2008) 67:672–84. doi:10.1111/j.1365-2958.2007.06078.x
31. Quirós C, Herrero M, García LA, Díaz M. Quantitative approach to determining the contribution of viable-but-nonculturable subpopulations to malolactic fermentation processes. Appl Environ Microbiol (2009) 75(9):2977–81. doi:10.1128/AEM.01707-08
32. Gourmelon M, Cillard J, Pommepuy M. Visible light damage to Escherichia coli in sea water: oxidative stress hypothesis. J Appl Bacteriol (1994) 77:105–12. doi:10.1111/j.1365-2672.1994.tb03051.x
33. Shahamat M, Paszko-Kolva C, Yamamoto H, Colwell R. Ecological studies of Campylobacter pylori. Klin Wochenschr (1989) 67(Suppl XVII):62–3.
34. Saito N, Konishi K, Sato F, Kato M, Takeda H, Sugiyama T, et al. Plural transformation-processes from spiral to coccoid Helicobacter pylori and its viability. J Infect (2003) 46:49–55. doi:10.1053/jinf.2002.1047
35. Azevedo NF, Pacheco AP, Vieira MJ, Keevil CW. Nutrient shock and incubation atmosphere influence recovery of culturable Helicobacter pylori from water. Appl Environ Microbiol (2004) 70:490–3. doi:10.1128/AEM.70.1.490-493.2004
36. Chaput C, Ecobichon C, Cayet N, Girardin SE, Werts C, Guadagnini S, et al. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog (2006) 2:e97. doi:10.1371/journal.ppat.0020097
37. Queralt N, Araujo R. Analysis of the survival of H. pylori within a laboratory-based aquatic model system using molecular and classical techniques. Microb Ecol (2007) 54:771–7. doi:10.1007/s00248-007-9242-1
38. Azevedo C, Almeida L, Cerqueira S, Dias C, Keevil W, Vieira MJ. Coccoid form of Helicobacter pylori as a morphological manifestation of cell adaptation to the environment. Appl Environ Microbiol (2007) 73:3423–7. doi:10.1128/AEM.00047-07
39. Cellini L, Allocati N, Angelucci D, Iezzi T, Di CE, Marzio L, et al. Coccoid Helicobacter pylori not culturable in vitro reverts in mice. Microbiol Immunol (1994) 38:843–50. doi:10.1111/j.1348-0421.1994.tb02136.x
40. She FF, Lin JY, Liu JY, Huang C, Su DH. Virulence of water-induced coccoid Helicobacter pylori and its experimental infection in mice. World J Gastroenterol (2003) 9(3):516–20.
41. Andersen LP, Wadstrom T. Basic bacteriology and culture. In: Mobley HLT, Mendz GL, Hazell SL, editors. Helicobacter Pylori: Physiology and Genetics. Washington, DC: ASM Press (2001). p. 27–38.
42. Huq A, Rivera I, Colwell R. Epidemiological significance of viable but nonculturable microorganisms. In: Colwell R, Grimes D, editors. Nonculturable Microorganisms in the Environment. Washington, DC: American Society for Microbiology Press (2000). p. 301–23.
43. Pommepuy M, Butin M, Derrien A, Gourmelon M, Colwell RR, Cormier M. Retention of enteropathogenicity by viable but nonculturable Escherichia coli exposed to seawater and sunlight. Appl Environ Microbiol (1996) 62:4621–6.
44. Khan MMT, Pyle BH, Camper AK. Specific and rapid enumeration of viable but nonculturable and viable-culturable gram-negative bacteria by using flow cytometry. Appl Environ Microbiol (2010) 76(15):5088–96. doi:10.1128/AEM.02932-09
45. Percival SL, Thomas JG. Transmission of Helicobacter pylori and the role of water and biofilms. J Water Health (2009) 7(3):469–77. doi:10.2166/wh.2009.070
46. Azevedo NF, Almeida C, Fernandes I, Cerqueira L, Dias S, Keevil CW, et al. Survival of gastric and enterohepatic Helicobacter spp. in water: implications for transmission. Appl Environ Microbiol (2008) 74(6):1805–11. doi:10.1128/AEM.02241-07
47. Giao MS, Azevedo NF, Wilks SA, Vieira MJ, Keevil CW. Persistence of Helicobacter pylori in heterotrophic drinking-water biofilms. Appl Environ Microbiol (2008) 74:5898–904. doi:10.1128/AEM.00827-08
48. Stoodley P, Wilson S, Hall-Stoodley L, Boyle JD, Lappin-Scott HM, Costerton JW. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl Environ Microbiol (2001) 67:5608–13. doi:10.1128/AEM.67.12.5608-5613.2001
49. Mizoguchi H, Fujioka T, Nasu M. Evidence for viability of coccoid forms of Helicobacter pylori. J Gastroenterol (1999) 34:32–6.
50. Bode G, Mauch F, Malfertheiner P. The coccoid forms of Helicobacter pylori. Criteria for their viability. Epidemiol Infect (1993) 111:483–90. doi:10.1017/S0950268800057216
51. Shahamat M, Mai U, Paszko-Kolva C, Kessel M, Colwell RR. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl Environ Microbiol (1993) 59:1231–5.
52. Sato F. Helicobacter pylori in culture: an ultrastructural study. Hokkaido Igaku Zasshi (2000) 75(3):187–96.
54. Kusters JG, Gerrits MM, Van Strijp JA, Vandenbroucke-Grauls CM. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect Immun (1997) 65:3672–9.
55. Eaton KA, Catrenich CE, Makin KM, Krakowka S. Virulence of coccoid and bacillary forms of Helicobacter pylori in gnotobiotic piglets. J Infect Dis (1995) 171:459–62. doi:10.1093/infdis/171.2.459
56. Benaissa M, Babin P, Quellard N, Pezennec L, Cenatiempo Y, Fauchere JL. Changes in Helicobacter pylori ultrastructure and antigens during conversion from the bacillary to the coccoid form. Infect Immun (1996) 64(6):2331–5.
57. Willén R, Carlén B, Wang X, Papadogiannakis N, Odselius R, Wadstrom T. Morphologic conversion of Helicobacter pylori from spiral to coccoid form. Scanning (SEM) and transmission electron microscopy (TEM) suggest viability. Ups J Med Sci (2000) 105(1):31–40.
58. Cellini L. Helicobacter pylori: a chameleon-like approach to life. World J Gastroenterol (2014) 20(19):5575–82. doi:10.3748/wjg.v20.i19.5575
59. Rowan NJ. Viable but non-culturable forms of food and waterborne bacteria: quo vadis? Trends Food Sci Technol (2004) 15:462–7. doi:10.1016/j.tifs.2004.02.009
60. Moreno Y, Ferrus MA, Alonso JL, Jimenez A, Herandez J. Use of fluorescent in situ hybridization to evidence the presence of Helicobacter pylori in water. Water Res (2003) 37:2251–6. doi:10.1016/S0043-1354(02)00624-3
61. Azevedo NF, Pacheco AP, Keevil CW, Vieira MJ. Adhesion of water stressed Helicobacter pylori to abiotic surfaces. J Appl Microbiol (2006) 101:718–24. doi:10.1111/j.1365-2672.2006.03029.x
62. Angelidis AS, Tirodimos I, Bobos M. Detection of Helicobacter pylori in raw bovine milk by fluorescence in situ hybridization (FISH). Int J Food Microbiol (2011) 151:252–6. doi:10.1016/j.ijfoodmicro.2011.09.007
63. Sato F, Saito N, Shouji E, Rani A, Takeda H, Sugiyama T, et al. The maintenance of viability and spiral morphology of Helicobacter pylori in mineral water. J Med Microbiol (1999) 48:971. doi:10.1099/00222615-48-10-971
64. Nilsson HO, Blom J, Abu-Al-Soud W, Ljungh AA, Andersen LP, Wadstrom T. Effect of cold starvation, acid stress, and nutrients on metabolic activity of Helicobacter pylori. Appl Environ Microbiol (2002) 68:11–9. doi:10.1128/AEM.68.1.11-19.2002
65. Minami M, Ohta M, Ohkura T, Ando T, Torii K, Hasegawa T, et al. Use of a combination of brushing technique and the loop-mediated isothermal amplification method as a novel, rapid, and safe system for detection of Helicobacter pylori. J Clin Microbiol (2006) 44:4032–7. doi:10.1128/JCM.00898-06
66. Mapstone NP, Lynch DA, Lewis FA, Axon AT, Tompkins DS, Dixon MF, et al. Identification of Helicobacter pylori DNA in the mouths and stomachs of patients with gastritis using PCR. J Clin Pathol (1993) 46:540–3. doi:10.1136/jcp.46.6.540
67. Enroth H, Engstrand L. Immunomagnetic separation and PCR for detection of Helicobacter pylori in water and stool specimens. J Clin Microbiol (1995) 33:2162–5.
68. De Reuse H, Labigne A, Mengin-Lecreulx D. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J Bacteriol (1997) 179:3488–93.
69. Lu JJ, Perng CL, Shyu RY, Chen CH, Lou Q, Chang SKF, et al. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. J Clin Microbiol (1999) 37:772–4.
70. Bunn JE, MacKay WG, Thomas JE, Reid DC, Weaver LT. Detection of Helicobacter pylori DNA in drinking water biofilms: implications for transmission in early life. Lett Appl Microbiol (2002) 34:450–4. doi:10.1046/j.1472-765X.2002.01122.x
71. Shahamat M, Alavi M, Watts JE, Gonzalez JM, Sowers KR, Maeder DW, et al. Development of two PCR-based techniques for detecting helical and coccoid forms of Helicobacter pylori. J Clin Microbiol (2004) 42:3613–9. doi:10.1128/JCM.42.8.3613-3619.2004
72. Watson CL, Owen RJ, Said B, Lai S, Lee JV, Surman-Lee S, et al. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J Appl Microbiol (2004) 97:690–8. doi:10.1111/j.1365-2672.2004.02360.x
73. Sasaki K, Tajiri Y, Sata M, Fujii Y, Matsubara F, Zhao M, et al. Helicobacter pylori in the natural environment. Scand J Infect Dis (1999) 31:275–9. doi:10.1080/00365549950163572
74. Mazari-Hiriart M, Lopez-Vidal Y, Calva JJ. Helicobacter pylori in water systems for human use in Mexico City. Water Sci Technol (2001) 43(12):93–8.
75. Hulten K, Han SW, Enroth H, Klein PD, Opekum AR, Gilman RH, et al. Helicobacter pylori in the drinking water in Peru. Gastroenterology (1996) 110:1031–5. doi:10.1053/gast.1996.v110.pm8612990
76. Baker KH, Hegarty JP. Presence of Helicobacter pylori in drinking water is associated with clinical infection. Scand J Infect Dis (2001) 33:744–6. doi:10.1080/003655401317074536
77. McKeown I, Orr P, McDonald S, Kabani A, Brown R, Coghlan G, et al. Helicobacter pylori prevalence in the Canadian arctic: seroprevalence and detection in community water samples. Am J Gastroenterol (1999) 94:1823–9. doi:10.1111/j.1572-0241.1999.01212.x
78. Park SR, Mackay WG, Reid DC. Helicobacter sp recovered from drinking water biofilm sampled from a water distribution system. Water Res (2001) 35:1624–6. doi:10.1016/S0043-1354(00)00582-0
79. Kabir S. Detection of Helicobacter pylori in faeces by culture, PCR and enzyme immunoassay. J Med Microbiol (2001) 50(12):1021–9.
80. Nayak AK, Rose JB. Detection of Helicobacter pylori in sewage and water using a new quantitative PCR method with SYBR green. J Appl Microbiol (2007) 103:1931–41. doi:10.1111/j.1365-2672.2007.03435.x
81. Janzon A, Sjoling Å, Lothigius Å, Ahmed D, Qadri F, Svennerholm AM. Failure to detect Helicobacter pylori DNA in drinking and environmental water in Dhaka, Bangladesh, using highly sensitive real-time PCR assays. Appl Environ Microbiol (2009) 75:3039–44. doi:10.1128/AEM.02779-08
82. Sen K, Schable NA, Lye DJ. Development of an internal control for evaluation and standardization of a quantitative PCR assay for detection of Helicobacter pylori in drinking water. Appl Environ Microbiol (2007) 73:7380–7. doi:10.1128/AEM.00687-07
83. McDaniels AE, Wymer L, Rankin C, Haugland R. Evaluation of quantitative real time PCR for the measurement of Helicobacter pylori at low concentrations in drinking water. Water Res (2005) 39:4808–16. doi:10.1016/j.watres.2005.09.030
84. Böckelmann U, Dörries HH, Ayuso-Gabella MN, Salgot de Marcay M, Tandoi V, Levantesi C, et al. Quantitative PCR monitoring of antibiotic resistance genes and bacterial pathogens in three European artificial groundwater recharge systems. Appl Environ Microbiol (2009) 75:154–63. doi:10.1128/AEM.01649-08
85. Fujimura S, Kato S, Kawamura T. Helicobacter pylori in Japanese river water and its prevalence in Japanese children. Lett Appl Microbiol (2004) 38:517–21. doi:10.1111/j.1472-765X.2004.01529.x
86. Voytek MA, Ashen JB, Fogarty LR, Kirshtein JD, Landa ER. Detection of Helicobacter pylori and fecal indicator bacteria in five North American rivers. J Water Health (2005) 3(4):405–22.
87. Mazari-Hiriart M, Lopez-Vidal Y, Castillo-Rojas G, Ponce de Leon S, Cravioto A. Helicobacter pylori and other enteric bacteria in freshwater environments in Mexico City. Arch Med Res (2001) 32:458–67. doi:10.1016/S0188-4409(01)00304-6
88. Verthé K, Verstraete W. Use of flow cytometry for analysis of phage-mediated killing of Enterobacter aerogenes. Res Microbiol (2006) 157:613–8. doi:10.1016/j.resmic.2006.02.007
89. Mori Y, Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J Infect Chemother (2009) 15:62–9. doi:10.1007/s10156-009-0669-9
90. Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun (2001) 289(1):150–4. doi:10.1006/bbrc.2001.5921
91. Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc (2008) 3(5):877–82. doi:10.1038/nprot.2008.57
92. Chamanrokh P, Shahhosseiny MH, Mazaheri Assadi M, Nejadsattari T, Esmaili D. A comparison of loop-mediated isothermal amplification (LAMP) with PCR and rapid urease test (RUT) to detect Helicobacter pylori in biopsy samples. J Pure Appl Microbiol (2014) 8(2):1079–86.
93. Chen K, Pachter L. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput Biol (2005) 1(2):106–12. doi:10.1371/journal.pcbi.0010024
94. Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev (2004) 68(4):669–85. doi:10.1128/MMBR.68.4.669-685.2004
95. Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol (1985) 39:321–46. doi:10.1146/annurev.mi.39.100185.001541
96. Buckley MJ, O’Morain CA. Helicobacter biology-discovery. Br Med Bull (1998) 54(1):7–16. doi:10.1093/oxfordjournals.bmb.a011681
97. Doenges JL. Spirochaetes in the gastric glands of Macacus rhesus and humans without definite history of related disease. Proc Soc Exp Biol Med (1938) 38:536–8. doi:10.3181/00379727-38-9924P
98. Marshall BJ, Armstrong JA, McGechie DB, Glancy RJ. Attempt to fulfil Koch’s postulates for pyloric Campylobacter. Med J Aust (1985) 142(8):436–9.
99. Marshall BJ, McGechie DB, Rogers PA, Glancy RJ. Pyloric Campylobacter infection and gastroduodenal disease. Med J Aust (1985) 142(8):439–44.
100. Relman DA. The identification of uncultured microbial pathogens. J Infect Dis (1993) 168(1):1–8. doi:10.1093/infdis/168.1.1
101. Hugenholtz P, Pace NR. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol (1996) 14(6):190–7. doi:10.1016/0167-7799(96)10025-1
102. Pace NR. A molecular view of microbial diversity and the biosphere. Science (1997) 276(5313):734–40. doi:10.1126/science.276.5313.734
103. DeLong EF, Pace NR. Environmental diversity of bacteria and archaea. Syst Biol (2001) 50(4):470–8. doi:10.1080/106351501750435040
104. Somboonna N, Assawamakin A, Wilantho A, Tangphatsornruang S, Tongsima S. Metagenomic profiles of free-living archaea, bacteria and small eukaryotes in coastal areas of Sichang Island, Thailand. BMC Genomics (2012) 13(Suppl 7):S29. doi:10.1186/1471-2164-13-S7-S29
105. Port JA, Wallace JC, Griffith WC, Faustman EM. Metagenomic profiling of microbial composition and antibiotic resistance determinants in puget sound. PLoS One (2012) 7(10):e48000. doi:10.1371/journal.pone.0048000
106. Wang Y, Sheng HF, He Y, Wu JY, Jiang YX, Tam NF, et al. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl Environ Microbiol (2012) 78(23):8264–71. doi:10.1128/AEM.01821-12
107. Zheng Z, Andersson AF, Ye W, Nyren O, Normark S, Engstrand L. A method for metagenomics of Helicobacter pylori from archived formalin-fixed gastric biopsies permitting longitudinal studies of carcinogenic risk. PLoS One (2011) 6(10):e26442. doi:10.1371/journal.pone.0026442
Keywords: Helicobacter pylori, environmental coccoid form, detection methods, LAMP, PCR
Citation: Mazaheri Assadi M, Chamanrokh P, Whitehouse CA and Huq A (2015) Methods for detecting the environmental coccoid form of Helicobacter pylori. Front. Public Health 3:147. doi: 10.3389/fpubh.2015.00147
Received: 18 February 2015; Accepted: 08 May 2015;
Published: 28 May 2015
Edited by:
Mohiuddin Md. Taimur Khan, Washington State University, USAReviewed by:
M. Jahangir Alam, University of Houston College of Pharmacy, USAM. A. Karim, Kennesaw State University, USA
Copyright: © 2015 Mazaheri Assadi, Chamanrokh, Whitehouse and Huq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahnaz Mazaheri Assadi, Iranian Research Organization for Science and Technology (IROST), Sh. Ehsani Rad St., Enqelab St., Parsa Sq., Ahmadabad Mostoufi Rd., Azadegan Highway, P. O. Box 3353-5111, Tehran 3353136846, I. R., Iran, mxmazaheriassadi@yahoo.com
 Mahnaz Mazaheri Assadi
Mahnaz Mazaheri Assadi Parastoo Chamanrokh
Parastoo Chamanrokh Chris A. Whitehouse
Chris A. Whitehouse Anwar Huq
Anwar Huq