- Department of Pharmaceutics, Division of Microbiology, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana
A review of the literature was undertaken to delineate the current level and mechanisms of resistance to carbapenems, colistin, and tigecycline in South Africa. Thirty-two English publications and 32 National Institute of Communicable Diseases communiqués identified between early January 2000 and 20 May, 2016 showed substantial reports of NDM (n = 860), OXA-48 (n = 584), VIM (n = 131), and IMP (n = 45) carbapenemases within this period, mainly in Klebsiella pneumoniae (n = 1138), Acinetobacter baumannii (n = 332), Enterobacter cloacae (n = 201), and Serratia marcescens (n = 108). Colistin and tigecycline resistance was prevalent among K. pneumoniae, A. baumannii, S. marcescens, and E. cloacae. The first mcr-1 colistin resistance gene to be detected in South Africa was reported in Escherichia coli from livestock as well as from hospitalized and outpatients. There are increasing reports of NDM and OXA-48 carbapenemases among Enterobacteriaceae and A. baumannii in South Africa. Mcr-1 is now present in South African patients and livestock. Resistance to carbapenems, colistin, and tigecycline restricts infection management options for clinicians.
Bullet Points/Highlights
• Resistance to last-resort antibiotics, such as carbapenems, tigecycline, and colistin, is increasing among Gram-negative bacteria in South Africa, restricting infection management options for clinicians and posing a threat to food safety.
• NDM-1 and OXA-48-like carbapenemases are the most common carbapenemases found among Gram-negative bacteria in South Africa and have been implicated in clinical outbreaks.
• The mcr-1 colistin resistance gene has been reported among Escherichia coli in both clinical and poultry samples in two provinces in South Africa. This is a threat to all Africa and Europe as many patients from several countries in these continents seek health care in South Africa.
Introduction
The inexorable adaptations of bacteria to the therapeutic effects of antibiotics, with its grave implications, are dawning upon the world. This is witnessed in the increasing awareness being created by the WHO and EU about antibiotic resistance, specifically through the formers’ global/regional antibiotic surveillance reports (1, 2). Moreover, antibiotic resistance awareness days and weeks have been instituted by both organizations (3), while efforts to engage policy makers and governments (4), clinicians, and patients in antibiotic stewardship have increased concomitantly (1, 2). Until recently, the effect of antibiotic use in livestock production on clinical medicine was a subject of debate (5). However, the detection of mcr-1 colistin resistance gene in swine, pork, imported chicken (in Denmark), and hospitalized patients has helped settle the argument that antibiotics use in veterinary medicine, specifically as growth promoters, can be a source of resistance genes in human pathogenic bacteria (6, 7). Subsequently, the concept of “One Health” that triangulates clinical, environmental, and veterinary antibiotic resistance surveillance and molecular epidemiological studies as a means to containing resistance is gaining grounds (8).
A recent study in the UK detected large amounts of resistance genes to clinically useful antibiotics, such as sulfonamides, trimethoprim, and tetracyclines, in rivers that were fed with runoff effluents from farms on which antibiotics were used, as well as from sewage processing plants (9). In India and Bangladesh, substantial amounts of broad-spectrum antibiotic resistance enzymes, such as NDM-1 and CTX-M-15, which, respectively, confer resistance to most β-lactams (except aztreonam) and cephalosporins (except cephamycins), were detected in the environment, as well as in tap water (10–12). The implications of this escalating preponderance of resistance genes in the environment, and subsequently in human pathogens, cannot be gainsaid, but briefly it portends the end of antibiotics as useful therapeutic agents in combatting bacterial infections (7, 13).
The emergence of antibiotics quickly and significantly reduced the prevalence and mortality of several hitherto fatal infections, such as sepsis, meningitis, tuberculosis, gangrenes, dysentery, food-borne diarrhea, salmonellosis, and pneumonia (1). Increased life-expectancy, better quality of life, and increased wealth were the palpable results of antibiotics use; however, antibiotic resistance remains a threat to these gains. Expansions in resistance among bacteria have been blamed on increased unrestricted antibiotic use in clinical and veterinary medicines (7, 13). Currently, the carbapenems, colistin, and tigecycline, alone or in combinations, are used as reserve antibiotics to treat fatal bacterial infections (14). Unfortunately, reports of resistance to these last-resort antibiotics have been increasing with concerning frequency (15).
Detection of resistance to these last-resort antibiotics in South Africa are increasingly, but worryingly, being reported. To draw attention to this menace, this article provides a comprehensive review of the current burden of resistance to reserve antibiotics, as reported in published literature, as well as discusses their public health implications. This review thus aims to create an awareness of the precarious state of South Africa’s public health as a means of stimulating government action toward implementing policies that will address the situation.
Last-Resort Antibiotics: From Carbapenems to Colistin and Tigecycline
Until recently, carbapenems, such as meropenem, imipenem, ertapenem, and doripenem, were the last-resort antibiotics used for managing multidrug-resistant bacterial infections (14, 16). However, their use selected for resistant strains, necessitating a change to colistin and tigecycline as last-resort antibiotics (14). It has been established that bacteria develops resistance to carbapenems through the expression/production of enzymes called carbapenemases, and/or the reduction in outer membrane permeability resulting from porin mutations (17). Carbapenemases remain the most clinically important mechanism of carbapenem resistance (18) and are categorized into three classes, namely class A, B, and D; class A includes the clinically important KPC and GES enzymes, while VIM, IMP, and NDM are the most described types under class B. OXA-48-like enzymes remain the commonly reported types under class D (17, 19). Resistance to colistin and tigecycline are, respectively, due to lipid A mutations and efflux hyper expression (14). Recently, Liu et al. (7) found a new plasmid-borne colistin resistance mcr-1 gene that is quickly spreading worldwide (20). The increasing but worrying emergence of these carbapenemases and carbapenem-, colistin- (particularly mcr-1), and tigecycline-resistant Gram-negative bacteria in South Africa are presented herein.
Resistance to Carbapenems in South Africa
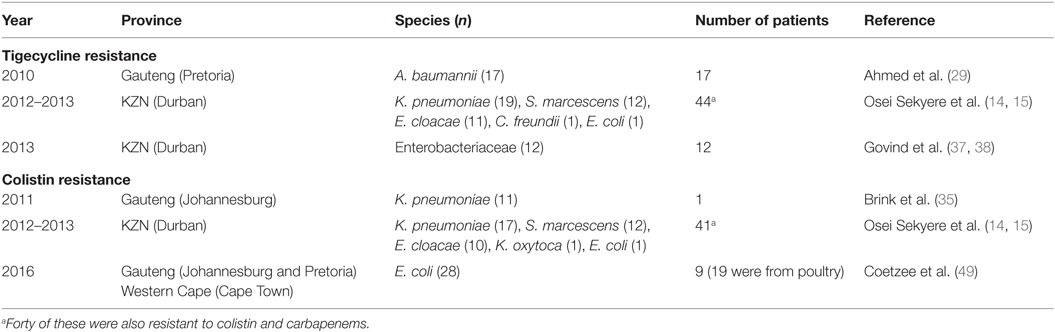
Analysis of carbapenem resistance-reporting publications showed that an estimated 2315 carbapenem-resistant cases/infections occurred between January 2000 and May 20, 2016 (Tables 1 and 2 and Table S1 in Supplementary Material). The majority of these estimated cases (n = 1220) were from the Gauteng province, followed by a substantial number from KwaZulu-Natal (n = 515) province (Figure 1). Klebsiella pneumoniae (n = 1138), Acinetobacter baumannii (n = 332), Enterobacter cloacae (n = 201), and Serratia marcescens (n = 108) were the most common carbapenem-resistant isolates. The most described carbapenemases were NDMs (n = 860) and OXA-48 (n = 584), which were increasingly detected after 2012, subsequent to its detection in 2011 (Tables 1 and 2 and Table S1 in Supplementary Material; Figure 1). The National Institute of Communicable Diseases (NICD) currently only reports carbapenem-resistant Enterobacteriaceae (CRE) from the public and private health sectors, and not carbapenem-resistant, non-fermenting Gram-negative bacteria or colistin and tigecycline resistance; however, it is on alert to report on the mcr-1 gene (as at the time of writing this paper). As a reference laboratory, the NICD receives and confirms CRE from selected hospitals from South Africa and publishes them in monthly communiques.
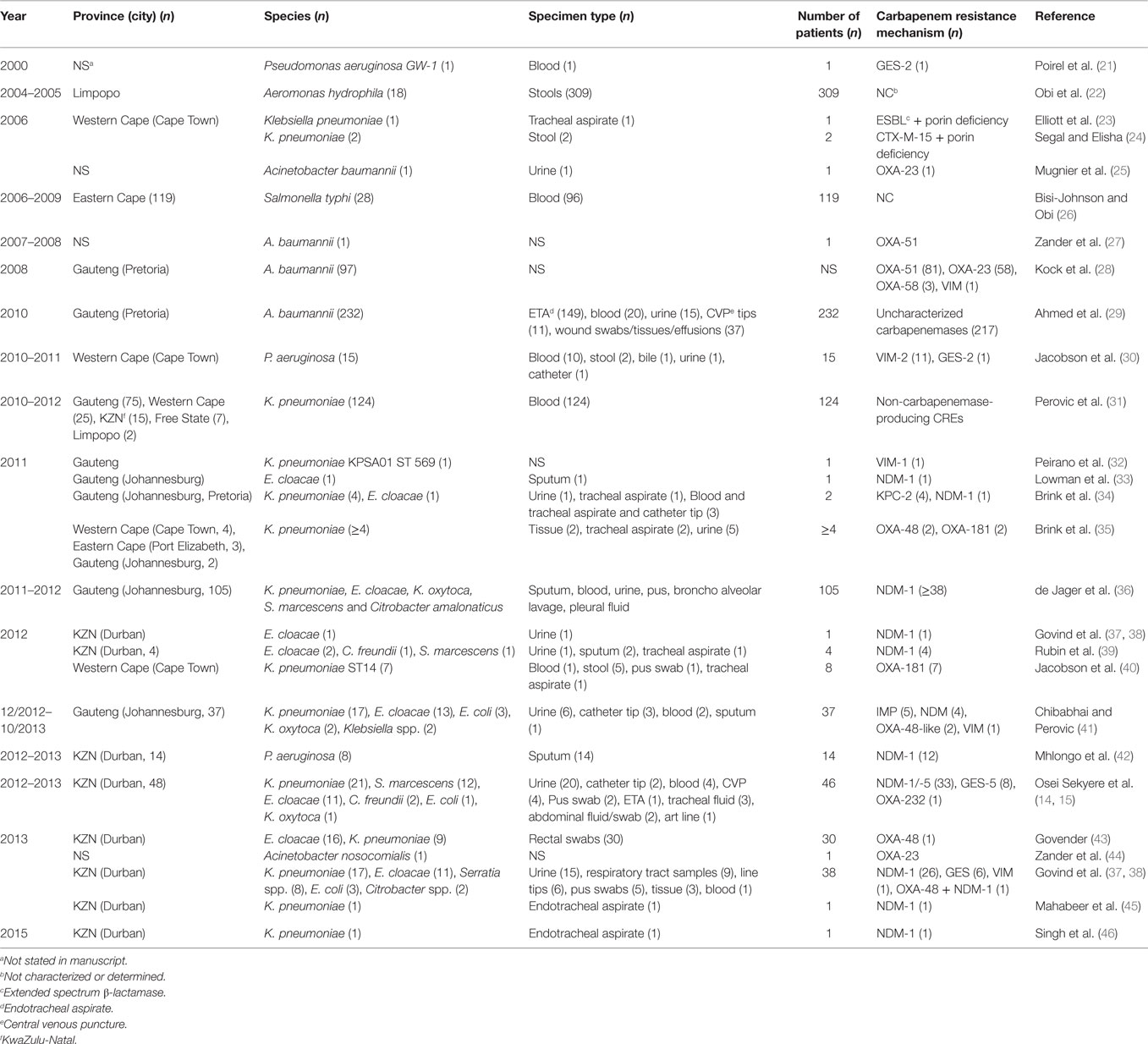
Table 1. Timeline of published carbapenem-resistant Enterobacteriaceae (CREs) and carbapenemases detected in Gram-negative bacteria in South Africa.
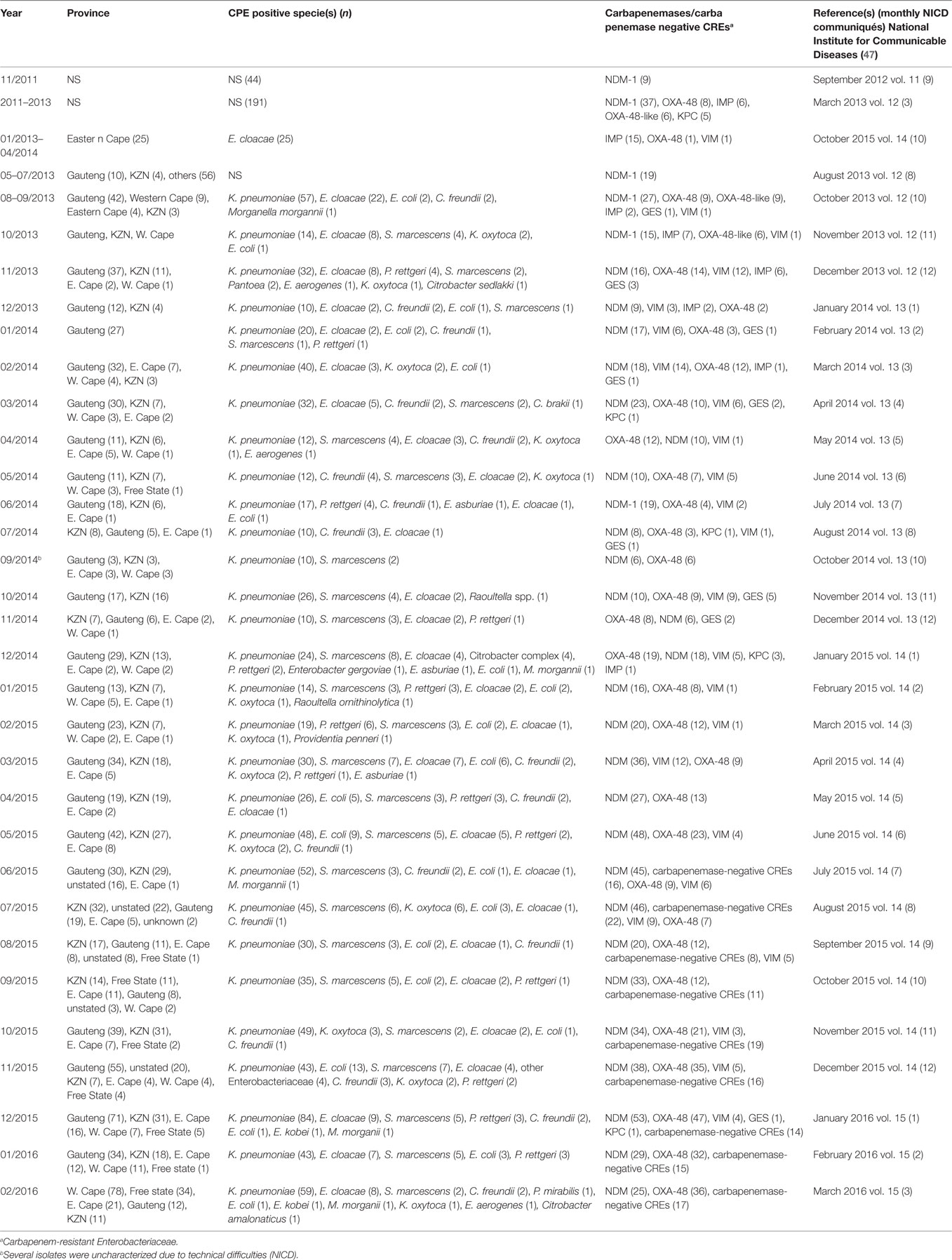
Table 2. Prevalence of carbapenemases and CRE per province as detected by NICD between 2011 and 2015.
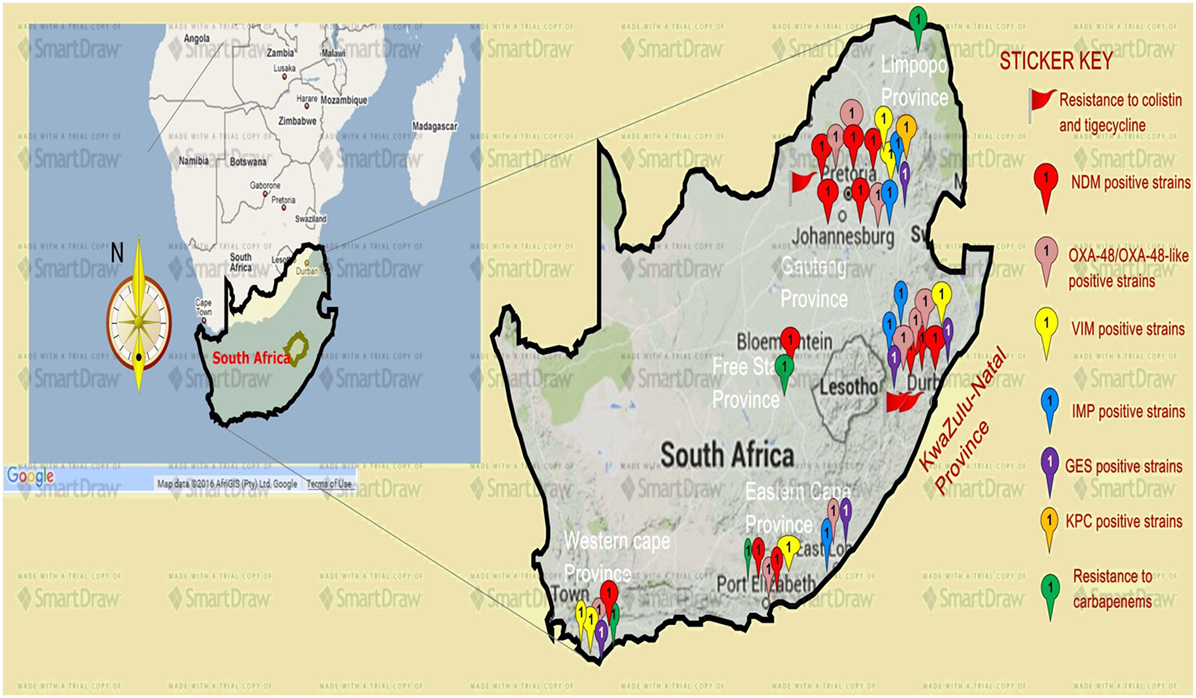
Figure 1. A map showing the distribution, resistance mechanisms, and density of carbapenem, colistin, and tigecycline resistance described so far in South Africa. Resistance to carbapenems is highest in the Gauteng and KwaZulu-Natal provinces, followed by the Eastern and Western Capes, respectively. Resistance to colistin (mediated by the mcr-1 gene in some cases) and tigecycline are increasingly also being reported in Gauteng and KwaZulu-Natal provinces. NDM-1 (red ribbons) and OXA-48 (pink ribbons) variants are the commonly reported carbapenem resistance mechanism described in South Africa, particularly in Enterobacteriaceae such as K. pneumoniae, Enterobacter cloacae, Serratia marcescens, and Acinetobacter baumannii. Relatively minor resistance cases were reported from the Free state and Limpopo provinces, while VIM, IMP, GES, and other carbapenemases are less described in South Africa. NB: a free version of smartdraw software was used to design this map, hence the watermark “smartdraw” in the background.
These observations suggest the substantial dissemination of carbapenem resistance, mediated mostly by NDM-1 or OXA-48 carbapenemases in K. pneumoniae, A. baumannii, E. cloacae, S. marcescens, and other Gram-negative bacteria. After 2000 A.D., the number of carbapenem resistance reports increased in South Africa, specifically in Gauteng and KwaZulu-Natal provinces (Tables 1 and 2). Reports of NDM and OXA-48-like carbapenemases increased after 2011 in Gauteng, KwaZulu-Natal, Eastern, and Western Capes provinces (Tables 1 and 2; Figure 1). Most of these carbapenem resistance cases had no travel history outside South Africa, suggesting that these strains were selected from increased carbapenem use within the country (48). Published articles and the NICD communiqués tend to agree in a large measure with regards to the detection patterns and distribution of carbapenemases and CREs among the provinces (Table S1 in Supplementary Material). Carbapenem resistance has been described in at least 10 Gram-negative bacterial hosts/species, which have been isolated from various clinical specimens (Tables 1 and 2); but with the exception of mcr-1 that was found in Escherichia coli in poultry (49), no reports of carbapenem- or tigecycline-resistant isolates were found from animals.
Reports of carbapenem-resistant Enterobacteriaceae from animals or the environment were not seen, and the NICD does not survey or collect CRE from livestock or environmental sources. Given the detection of mcr-1 among poultry and humans (50) in two provinces in South Africa, the need for a “One Health” approach to antimicrobial resistance surveillance and genomic epidemiology is dire (8). It is particularly concerning that these data are not representative of the whole burden of carbapenem resistance in South Africa. Subsequently, the burden of carbapenem resistance could therefore be more than reported, should all hospitals and laboratories, veterinarians, and environmentalists/ecologists report their carbapenem-resistant cases. In addition, the distribution, resistance mechanisms, and bacterial hosts could change with a holistic “One Health” approach.
Given the high attributable mortalities resulting from carbapenem-resistant, and especially NDM-positive bacterial infections (51, 52), their public health impacts cannot be over emphasized, particularly as studies indicate that many CRE-infected South African patients demised (35, 40, 53). Given the persistent and undetected nature of these carbapenem-resistant strains, specifically in many South African hospitals, the possibility of outbreaks, interhospital and intrahospital circulation (30, 40), and their transmission via hospital staff and patients to homes and communities is very high. Due to the association of CREs with intensive care units and invasive medical procedures, all patients who patronize these services are highly at risk (34, 54). With the detection of carbapenem resistance among Salmonella typhi in a hospital in Port Elizabeth, CREs can easily be transferred via contaminated food and water, a route that will expose many to these germs (26). This is further evidence of the need for a “One Health” approach to the antibiotic resistance problem in South Africa as resistant bacteria are not only reserved in clinical settings.
The escalating prevalence of these resistant strains is not only significant for South Africa but also to Africa and Europe as a whole, as published evidence shows that many patients from across Africa and Europe are transferred to South Africa for better and cheaper medical attention. For instance, it is established that many Europeans troop to South Africa as a destination for cosmetic surgeries, such as hip replacements, rhinoplasty, breast augmentation, liposuction, facelifts, and tummy tucks (55); the possibility of South Africa thus spreading these resistant strains to other African and European nations cannot be ruled out as medical tourism in South Africa increased from 3.9 to 5.0% of all entries between 2006 and 2010 (55). This should alert the government, the health-care industry, and health workers to the economic costs associated with the increasing prevalence of CREs as it could deter patients from across Africa and Europe from seeking health care in the country.
The main challenge regarding the combatting of these resistant strains lies in the limited antibiotics available for their treatment. In KwaZulu-Natal, CREs have been isolated from neonates, including a 24-h neonate (45, 46) for whom antibiotic therapy are further limited due to their age. Clinicians are then forced to resort to more expensive but toxic combination therapies that include the nephrotoxic and neurotoxic colistin and/or tigecycline (56, 57), and to extend the patient’s stay in the hospital. The cost arising from the extended hospital stay to the patient, their family, and the government, as shown by the WHO in the US, EU, and Thailand, are considerable (1). Moreover, hospitals with outbreaks of CREs need to spend considerable resources sterilizing all wards and instituting stricter infection management controls, with some being forced to close down until the outbreaks are traced and eradicated (40). It is therefore in the interest of hospitals to institute periodic surveillance and effective infection control measures to preempt full-scale outbreaks, as well as screen all incoming patients to quickly isolate carriers.
Increasing Resistance to Tigecycline and Colistin
The exponential consumption of carbapenems in South Africa between January 2009 and June 2011, due to rising extended-spectrum β-lactamases among invasive Enterobacteriaceae, was early signaled as a potential factor in selecting CREs (14, 49, 54). It has been shown that colistin use in South Africa is increasing rapidly, and the failure of the carbapenems has been fingered as a cause (48, 49). Therefore, the increasing use of colistin and tigecycline due to an ever-expanding prevalence of CREs will result in colistin resistance (14, 58) as has been already observed among 45 CREs, 40 of which were resistant to both colistin and tigecycline (15), and also reported by Brink et al. (35) in K. pneumoniae (35). With the emergence of colistin resistance among CREs, the pre-antibiotic age, due to pandrug resistance, is not far off. While reports of colistin- and tigecycline-resistant CREs in South Africa are relatively few, rather than suggesting their low prevalence, it indicates the absence of surveillance to detect them.
For instance, surveillance for mcr-1 in poultry and patients led to the first detection of this gene in multiclonal E. coli strains in Gauteng and Western Cape provinces (49). This is an unfortunate finding, from a public health perspective, as consumption of the infected poultry can quickly spread mcr-1 among consumers and further compromise colistin therapy in CRE-infected patients in South Africa. The selection of mcr-1 among the poultry was suggested as due to the high use of colistin among South African poultry as growth promoters, prophylactic, and therapeutic agents, underscoring the need for stricter regulations on antibiotic use in the veterinary sector. Being found in E. coli, it is not surprising that the affected patients included outpatients; this is due to the common presence of E. coli in both community and hospital settings. This further portrays an ominous future as E. coli and the plasmid-borne mcr-1 gene can easily find their way into environmental sources through humans and animals waste. Moreover, other bacterial species can easily obtain this gene from E. coli through the mcr-1 plasmid, leading to further spread (7). The higher prevalence of colistin and tigecycline resistance recorded in Durban (15) suggests that it remains largely undetected and underscores the need for further surveillance studies in hospitals, farms, and environments. Without colistin and tigecycline (there are no other reserve antibiotics), the prognosis of bacterial infections in South Africa would be bleak.
Colistin and tigecycline resistance cases were higher in KwaZulu-Natal province (Figure 1) than in Gauteng province and were common among K. pneumoniae, A. baumannii, S. marcescens, and E. cloacae (Table 3), albeit mcr-1 has been detected in Gauteng (Johannesburg) and Western Cape (Cape Town) provinces so far (49). In the abovementioned study, 44 CREs were resistant to tigecycline; 41 of which were resistant to colistin, with 40 being resistant to both colistin and tigecycline (15) (Table 3). The first mcr-1 gene to be detected in South Africa was hosted in polyclonal E. coli strains isolated from poultry (n = 19) and from outpatients and inpatients in Gauteng and Western Cape provinces (49).
The geographical location of the carbapenem-, colistin-, and tigecycline-resistant strains may not necessarily suggest their higher prevalence in Gauteng, KwaZulu-Natal, and the Eastern, and Western Cape provinces, but rather a higher surveillance effort in these areas. However, the higher risk posed to the citizens of these provinces cannot be downplayed, as these pandrug-resistant strains could be easily spread into the environment and community (12, 13). The broad species range (Tables 1–3 and Table S1 in Supplementary Material) with which these carbapenem resistances are associated are also worrying, as these bacteria are commonly implicated in many human illnesses. Thus, the absence of reserve antibiotics in treating common infections from these resistant human pathogens will be precarious to public health.
Necessary Remedial and Policy Interventions
The escalating reports of NDMs, estimated at 34% in 2014 to 59% in 2015 (59), and other carbapenemases in addition to the inadequate national data, belie the porous infection control measures and surveillance programs in South Africa. This must change if the resistance menace is to be contained. A stricter enforcement of antibiotic use regulations among prescribers, dispensers, and livestock farmers is warranted to reduce the amount consumed in South Africa (48). It is incumbent on government and health regulators to enforce antibiotic stewardship programs as well as institute weekly or fortnight surveillance in all hospitals in addition to well-established livestock and environmental surveillance. Anal swab screening of incoming and hospitalized patients, as well as screening of hospital staff, wards, and all invasive medical instruments should be enforced in all hospitals. Food animals must be periodically sampled and tested for resistant bacteria. Contact precautions and patient isolation protocols, as advised by the Centers for Disease Control and Prevention (CDC), should be taught to health workers and strictly implemented for infected patients (60). In addition, patients returning from high-risk countries (with higher prevalence of carbapenem, colistin, or tigecycline resistance) should be screened before admission (60).
Currently, the NICD monitors and receives CRE from selected hospitals in South Africa, which it uses for its monthly communiques (Table 2). This is a good step toward describing the trends in CRE prevalence in South Africa, albeit it falls short of establishing their molecular epidemiology that is pivotal for any meaningful intervention. Moreover, efforts should be made to include all hospitals, in both private and public sectors, livestock farms, and environments, to obtain a true picture of the situation in the country. To further enhance epidemiological analysis, isolates included in published manuscripts should be removed from the NICD communiques or flagged therein to avoid their being double counted. In addition, the NICD should collaborate with universities and hospitals to establish “Centers for Genomic Epidemiology of Resistant Infections” to train and equip students, clinicians (physicians and veterinarians), and environmentalists/ecologists to undertake periodic antibiotic resistance surveillance and genomic epidemiology studies in hospitals, the environment, and livestock farms under a “One Health” (8) approach using next-generation sequencing (NGS) technology. This will help trace the routes and describe the evolutionary biology of resistant infections over time, which will provide adequate data to inform intervention policies.
There should be periodic in-service training for all hospital staff to increase awareness about carbapenem, colistin, and tigecycline resistance and the need to reduce their use. Such training must also involve the imparting of CRE diagnosis or detection skills and education in the available detection tools and methods as well as results interpretation. Hence, skills in undertaking and interpreting antimicrobial sensitivity tests using “interpretative reading” (61), the Carba NP test, multiplex real-time PCR, and/or whole genome sequencing will be useful.
Furthermore, prescribers should be encouraged to adopt evidence-based therapies to reduce antibiotic prescriptions, specifically for non-bacterial-based infections. Dispensers and community pharmacies must only serve antibiotics under prescription, with regulations to punish offending practitioners (1, 2). Government, the media, and schools must complement the efforts of the WHO by broadcasting antibiotic awareness days and weeks throughout South Africa, to alert the inhabitants of the need to finish their antibiotic courses, avoid buying antibiotics without prescriptions, avoid sharing their unfinished antibiotics with family and friends, and/or avoid advising friends or family to buy antibiotics when they develop symptoms similar to that of a known disease treated with a particular antibiotic (1, 2).
Conclusion
There is an escalating prevalence and possible endemicity of NDM and OXA-48 among Enterobacteriaceae and A. baumannii in South Africa, which is largely under-detected. Resistance to colistin, mediated by the plasmid-borne mcr-1 gene and other chromosomal mutations, and tigecycline is burgeoning among Gram-negative bacteria, leaving clinicians with no reserve antibiotics for treating fatal bacterial infections. Stricter infection control and antibiotic stewardship, “One Health” surveillance and genomic epidemiology studies, education, and awareness creation are warranted.
Author Contributions
This paper was designed, prepared, and finalized by John Osei Sekyere, Ph.D.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author is grateful to Carrin Martin, College of Health Sciences, University of KwaZulu-Natal, Durban, South Africa for her criticisms and proof reading of the text as well as cross-checking the data, methods, and results for accuracy.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fpubh.2016.00209
References
1. WHO. Antimicrobial Resistance: Global Report on Surveillance. France: World Health Organization (2014).
2. WHO. Tackling Antibiotic Resistance from a Food Safety Perspective in Europe, Word Health Organization. Denmark: WHO-Europe (2011).
3. European Union. Antibiot. Aware. Day. (2015). Available from: http://ecdc.europa.eu/en/EAAD/Pages/Home.aspx
4. White House. Pledge to Love, Prot. Cherish Antibiot. (2015). Available from: https://www.whitehouse.gov/blog/2015/06/05/pledge-love-protect-and-cherish-antibiotics
5. Bester LA, Essack SY. Antibiotic resistance via the food chain: fact or fiction? S Afr J Sci (2010) 106:1–5. doi:10.4102/sajs.v106i9/10.281
6. Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, et al. Detection of MCR-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill (2015) 20:30085. doi:10.2807/1560-7917.ES.2015.20.49.30085
7. Liu Y-Y, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis (2016) 16:161–8. doi:10.1016/S1473-3099(15)00424-7
8. Hueston W, Appert J, Denny T, King L, Umber J, Valeri L. Assessing global adoption of one health approaches. Ecohealth (2013) 10:228–33. doi:10.1007/s10393-013-0851-5
9. Rowe W, Verner-Jeffreys DW, Baker-Austin C, Ryan JJ, Maskell DJ, Pearce GP. Comparative metagenomics reveals a diverse range of antimicrobial resistance genes in effluents entering a river catchment. Water Sci Technol (2016) 73(7):1541–9. doi:10.2166/wst.2015.634
10. Rashid M, Rakib MM, Hasan B. Antimicrobial-resistant and ESBL-producing Escherichia coli in different ecological niches in Bangladesh. Infect Ecol Epidemiol (2015) 5:26712. doi:10.3402/iee.v5.26712
11. Toleman MA, Bugert JJ, Nizam SA. Extensively drug-resistant New Delhi metallo-β-lactamase-encoding bacteria in the environment, Dhaka, Bangladesh, 2012. Emerg Infect Dis (2015) 21:1027–30. doi:10.3201/eid2106.141578
12. Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis (2011) 11:355–62. doi:10.1016/S1473-3099(11)70059-7
13. Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, et al. Antibiotic resistance – the need for global solutions. Lancet Infect Dis (2013) 13:1057–98. doi:10.1016/S1473-3099(13)70318-9
14. Osei Sekyere J, Govinden U, Bester LA, Essack SY. Colistin and tigecycline resistance in carbapenemase-producing Gram negative bacteria: emerging resistance mechanisms and detection methods. J Appl Microbiol (2016) 121(3):601–17. doi:10.1111/jam.13169
15. Osei Sekyere J, Pedersen T, Sivertsen A, Govinden U, Essack SY, Moodley K, et al. Molecular epidemiology of carbapenem, colistin and tigecycline resistant Enterobacteriaceae in Durban, South Africa. ECCMID Poster Presentation, Amsterdam (2016). doi:10.13140/RG.2.1.1049.0003
16. Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect (2014) 20:821–30. doi:10.1111/1469-0691.12719
17. Queenan AM, Bush K. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev (2007) 20:440–58. doi:10.1128/CMR.00001-07
18. Sekyere JO, Govinden U, Essack S. The molecular epidemiology and genetic environment of carbapenemases detected in Africa. Microb Drug Resist (2015) 22:59–68. doi:10.1089/mdr.2015.0053
19. Bush K, Fisher JF. Epidemiological expansion, structural studies, and clinical challenges of new β-lactamases from gram-negative bacteria. Annu Rev Microbiol (2011) 65:455–78. doi:10.1146/annurev-micro-090110-102911
20. Schwarz S, Johnson AP. Transferable resistance to colistin: a new but old threat: table 1. J Antimicrob Chemother (2016) 71(8):2066–70. doi:10.1093/jac/dkw274
21. Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. GES-2, a class A beta-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother (2001) 45:2598–603. doi:10.1128/AAC.45.9.2598-2603.2001
22. Obi CL, Ramalivhana J, Samie A, Igumbor EO. Prevalence, pathogenesis, antibiotic susceptibility profiles, and in-vitro activity of selected medicinal plants against Aeromonas isolates from stool samples of patients in the Venda region of South Africa. J Health Popul Nutr (2007) 25(4):428–35.
23. Elliott E, Brink AJ, van Greune J, Els Z, Woodford N, Turton J, et al. In vivo development of ertapenem resistance in a patient with pneumonia caused by Klebsiella pneumoniae with an extended-spectrum beta-lactamase. Clin Infect Dis (2006) 42:e95–8. doi:10.1086/503264
24. Segal H, Elisha BG. Resistance to β-lactams, and reduced susceptibility to carbapenems, in clinical isolates of Klebsiella pneumoniae due to interplay between CTX-M-15 and altered outer membrane permeability. South Afr J Epidemiol Infect (2006) 21(2):41–4.
25. Mugnier PD, Poirel L, Naas T, Nordmann P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii1. Emerg Infect Dis (2010) 16:35. doi:10.3201/eid1601.090852
26. Bisi-Johnson M, Obi C. Detection of carbapenem resistance in Salmonella species from a tertiary hospital in Eastern Cape, South Africa. Br Microbiol Res J (2015) 10:1–6. doi:10.9734/BMRJ/2015/18586
27. Zander E, Higgins PG, Fernández-González A, Seifert H. Detection of intrinsic blaOXA-51-like by multiplex PCR on its own is not reliable for the identification of Acinetobacter baumannii. Int J Med Microbiol (2013) 303:88–9. doi:10.1016/j.ijmm.2012.12.007
28. Kock MM, Bellomo AN, Storm N, Ehlers MM. Prevalence of carbapenem resistance genes in Acinetobacter baumanii isolated from clinical specimens obtained from an academic hospital in South Africa. South Afr J Epidemiol Infect (2013) 23:28–32.
29. Ahmed NH, Baba K, Clay C, Lekalakala R, Hoosen AA. In vitro activity of tigecycline against clinical isolates of carbapenem resistant Acinetobacter baumannii complex in Pretoria, South Africa. BMC Res Notes (2012) 5:215. doi:10.1186/1756-0500-5-215
30. Jacobson RK, Minenza N, Nicol M, Bamford C. VIM-2 metallo-β-lactamase-producing Pseudomonas aeruginosa causing an outbreak in South Africa. J Antimicrob Chemother (2012) 67:1797–8. doi:10.1093/jac/dks100
31. Perovic O, Singh-Moodley A, Dusé A, Bamford C, Elliott G, Swe-Han K, et al. National sentinel site surveillance for antimicrobial resistance in Klebsiella pneumoniae isolates in South Africa, 2010 – 2012. S Afr Med J (2014) 104:563–8. doi:10.7196/samj.7617
32. Peirano G, Moolman J, Pitondo-Silva A, Pitout JDD. The characteristics of VIM-1-producing Klebsiella pneumoniae from South Africa. Scand J Infect Dis (2012) 44:74–8. doi:10.3109/00365548.2011.614276
33. Lowman W, Sriruttan C, Nana T, Bosman N, Duse A, Venturas J, et al. NDM-1 has arrived: first report of a carbapenem resistance mechanism in South Africa. S Afr Med J (2011) 101:873–5.
34. Brink AJ, Coetzee J, Clay C, Corcoran C, van Greune J, Deetlefs JD, et al. The spread of carbapenem-resistant Enterobacteriaceae in South Africa: risk factors for acquisition and prevention. S Afr Med J (2012) 102:599–601. doi:10.7196/SAMJ.5789
35. Brink AJ, Coetzee J, Corcoran C, Clay CG, Hari-Makkan D, Jacobson RK, et al. Emergence of OXA-48 and OXA-181 carbapenemases among Enterobacteriaceae in South Africa and evidence of in vivo selection of colistin resistance as a consequence of selective decontamination of the gastrointestinal tract. J Clin Microbiol (2013) 51:369–72. doi:10.1128/JCM.02234-12
36. de Jager P, Chirwa T, Naidoo S, Perovic O, Thomas J. Nosocomial outbreak of New Delhi metallo-β-lactamase-1-producing Gram-negative bacteria in South Africa: a case-control study. PLoS One (2015) 10:e0123337. doi:10.1371/journal.pone.0123337
37. Govind CN, Moodley K, Mahabeer P, Khan S. The emergence of carbapenem resistant Enterobacteriaceae in KwaZulu Natal, South Africa. 5th FIDSSA Conference 2013. Drakensberg, KwaZulu Natal, South Africa: South Afr J Epidemiol Infect (2013). 217 p.
38. Govind CN, Moodley K, Peer AK, Pillay N, Maske C, Wallis C, et al. NDM-1 imported from India – first reported case in South Africa. S Afr Med J (2013) 103:476–8. doi:10.7196/SAMJ.6593
39. Rubin JE, Peirano G, Peer AK, Govind CN, Pitout JDD. NDM-1–producing Enterobacteriaceae from South Africa: moving towards endemicity? Diagn Microbiol Infect Dis (2014) 79:378–80. doi:10.1016/j.diagmicrobio.2014.04.003
40. Jacobson RK, Manesen MR, Moodley C, Smith M, Williams S, Nicol M, et al. Molecular characterisation and epidemiological investigation of an outbreak of blaOXA-181 carbapenemase-producing isolates of Klebsiella pneumoniae in South Africa. S Afr Med J (2015) 105:1030–5. doi:10.7196/SAMJ.2015.v105i12.9926
41. Chibabhai V, Perovic O. Epidemiology of carbapenem resistant Enterobacteriaceae at Charlotte Maxeke Johannesburg Academic Hospital. Int J Infect Dis (2014) 21:410. doi:10.1016/j.ijid.2014.03.1265
42. Mhlongo N, Essack S, Govinden U. NDM-1, novel TEM-205, novel TEM-213 and other extended-spectrum β-lactamases co-expressed in isolates from cystic fibrosis patients from South Africa. S Afr J Infect Dis (2015) 30:103–7. doi:10.1080/23120053.2015.1074441
43. Govender K. Detection of Carbapenem-Resistant Enterobacteriaceae amongst Neonates in a Regional Hospital in KwaZulu-Natal: Screening for Carbapenemase Production and MIC Correlation. Durban: University of KwaZulu-Natal (2014). Available from: http://researchspace.ukzn.ac.za/handle/10413/12030
44. Zander E, Fernández-González A, Schleicher X, Dammhayn C, Kamolvit W, Seifert H, et al. Worldwide dissemination of acquired carbapenem-hydrolysing class D β-lactamases in Acinetobacter spp. other than Acinetobacter baumannii. Int J Antimicrob Agents (2014) 43:375–7. doi:10.1016/j.ijantimicag.2014.01.012
45. Mahabeer P, Singh R, Moodley P. NDM-1 producing Klebsiella pneumoniae in a neonate. 5th FIDSSA Conference. Drakensberg, KwaZulu Natal: Southern African Journal of Epidemiology and Infection (2013) 28(3):218.
46. Singh R, Mahabeer P, Mlisana K, Naidoo K, Moodley P. Isolation of New Delhi metallo-ßeta-lactamase-1(NDM-1) producing Klebsiella pneumoniae in a neonate in KwaZulu-Natal, South Africa. J Pediatr Neonatal Care (2015) 2:1–2. doi:10.15406/jpnc.2015.02.00082
47. National Institute for Communicable Diseases. Update on carbapenemase-producing Enterobacteriaceae. NICD Commun Dis Commun. (2016). Available from: http://www.nicd.ac.za/?page=archives&id=134
48. Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis (2014) 14(8):742–50. doi:10.1016/S1473-3099(14)70780-7
49. Coetzee J, Corcoran C, Prentice E, Moodley M, Mendelson M, Poirel L, et al. Emergence of plasmid-mediated colistin resistance (MCR-1) among Escherichia coli isolated from South African patients. S Afr Med J (2016) 106:449–50. doi:10.7196/SAMJ.2016.v106i5.10710
50. Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates, South Africa. Antimicrob Agents Chemother (2016) 60(7):4394–7. doi:10.1128/AAC.00444-16
51. Doi Y, Paterson DL. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med (2015) 36:74–84. doi:10.1055/s-0035-1544208
52. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis (2011) 17:1791–8. doi:10.3201/eid1710.110655
53. Brink AJ, Coetzee J, Clay CG, Sithole S, Richards GA, Poirel L, et al. Emergence of New Delhi metallo-beta-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa. J Clin Microbiol (2012) 50:525–7. doi:10.1128/JCM.05956-11
54. Coetzee J, Brink AJ. The emergence of carbapenem resistance in Enterobacteriaceae in South Africa. South Afr J Epidemiol Infect (2011) 26:239–40.
55. Crush J, Chikanda A, Maswikwa B. Patients without Borders: Medical Tourism and Medical Migration in Southern Africa. Cape Town and Toronto: Southern African Migration Programme (2012).
56. Stein C, Makarewicz O, Bohnert JA, Pfeifer Y, Kesselmeier M, Hagel S, et al. Three dimensional checkerboard synergy analysis of colistin, meropenem, tigecycline against multidrug-resistant clinical Klebsiella pneumonia isolates. PLoS One (2015) 10:e0126479. doi:10.1371/journal.pone.0126479
57. Tascini C, Tagliaferri E, Giani T, Leonildi A, Flammini S, Casini B, et al. Synergistic activity of colistin plus rifampin against colistin-resistant KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother (2013) 57:3990–3. doi:10.1128/AAC.00179-13
58. Giani T, Arena F, Vaggelli G, Conte V, Chiarelli A, Henrici De Angelis L, et al. Large nosocomial outbreak of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae traced to clonal expansion of an mgrB deletion mutant. J Clin Microbiol (2015) 53:3341–4. doi:10.1128/JCM.01017-15
59. Perovic O. FIDSSA Congress 2015 abstract submissions oral presentations. S Afr J Infect Dis (2015) 30:177–90.
60. Centers for Disease Control and Prevention (CDC). Risk of CRE Infections. CDC Vital Signs Rep. Atlanta, Georgia: Centers for Disease Control and Prevention (2013) 62(09):165–170.
Keywords: carbapenem, tigecycline, colistin, carbapenemase, mcr-1, South Africa, antibiotic resistance
Citation: Osei Sekyere J (2016) Current State of Resistance to Antibiotics of Last-Resort in South Africa: A Review from a Public Health Perspective. Front. Public Health 4:209. doi: 10.3389/fpubh.2016.00209
Received: 07 August 2016; Accepted: 12 September 2016;
Published: 30 September 2016
Edited by:
Fumito Maruyama, Kyoto University, JapanReviewed by:
Diamantis Plachouras, European Centre for Disease Prevention and Control, SwedenTorahiko Okubo, Hokkaido University, Japan
Copyright: © 2016 Osei Sekyere. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Osei Sekyere, jod14139@yahoo.com
†Present address: John Osei Sekyere, Antimicrobial Research Unit, Discipline of Pharmaceutical Sciences, University of KwaZulu-Natal, Durban, South Africa
 John Osei Sekyere
John Osei Sekyere