Longitudinal characterization of Escherichia coli in healthy captive non-human primates
- 1Department of Veterinary and Biomedical Sciences, College of Veterinary Medicine, University of Minnesota, Saint Paul, MN, USA
- 2Department of Veterinary Population Medicine, College of Veterinary Medicine, University of Minnesota, Saint Paul, MN, USA
- 3Como Park Zoo & Conservatory, Saint Paul, MN, USA
The gastrointestinal (GI) tracts of non-human primates (NHPs) are well known to harbor Escherichia coli, a known commensal of human beings and animals. While E. coli is a normal inhabitant of the mammalian gut, it also exists in a number of pathogenic forms or pathotypes, including those with predisposition for the GI tract as well as the urogenital tract. Diarrhea in captive NHPs has long been a problem in both zoo settings and research colonies, including the Como Zoo. It is an animal welfare concern, as well as a public health concern. E. coli has not been extensively studied; therefore, a study was performed during the summer of 2009 in collaboration with a zoo in Saint Paul, MN, which was previously experiencing an increased incidence and severity of diarrhea among their NHP collection. Fresh fecal samples were collected weekly from each member of the primate collection, between June and August of 2009, and E. coli were isolated. A total of 33 individuals were included in the study, representing eight species. E. coli isolates were examined for their genetic relatedness, phylogenetic relationships, plasmid replicon types, virulence gene profiles, and antimicrobial susceptibility profiles. A number of isolates were identified containing virulence genes commonly found in several different E. coli pathotypes, and there was evidence of clonal transmission of isolates between animals and over time. Overall, the manifestation of chronic diarrhea in the Como Zoo primate collection is a complex problem whose solution will require regular screening for microbial agents and consideration of environmental causes. This study provides some insight toward the sharing of enteric bacteria between such animals.
Introduction
Escherichia coli is a Gram-negative bacteria and known gut commensal of animals, including non-human primates (NHPs) (1–11). This diverse organism not only plays a role in the maintenance of gut health by helping to prevent the establishment of pathogenic bacteria in the gastrointestinal (GI) tract, but can also exist in a number of pathogenic forms (5, 12). In fact, pathogenic forms of E. coli are a major cause of diarrheal illness worldwide (13, 14). In addition, E. coli is a model laboratory organism, which has been used to examine basic evolutionary processes, bacterial pathogenesis, and epidemiological transmission (14–17). The ubiquity and ease of isolation of E. coli have enabled ecological studies to investigate how anthropogenic change (e.g., forest fragmentation) affects bacterial transmission among wildlife, including primates, human beings, and livestock (18–21). Information generated from these types of studies provides insight into the disease transmission process, enabling for the calculation of potential risk factors associated with human encroachment on wildlife populations and vice versa (18–21). Such studies also aid in the development and implementation of conservation strategies to protect endangered wildlife (18–21).
Diarrhea in captive primates has long been a problem in both zoo settings and research colonies (22–28). In fact, since at least 1998, the primate collection at the Como Zoo in Saint Paul, MN, has experienced issues with diarrhea. Diarrhea is a medical concern not only because it is physiologically taxing to the animals, but also because it is an ideal mechanism for disease transmission, making it both a public health concern and an animal welfare concern. Management of intermittent diarrhea is confounded by its complexities, including the lack of a consistently identifiable causative agent.
In many animals, including primates, some pathotypes of E. coli such as enteropathogenic E. coli (EPEC) are associated with the development of persistent diarrhea (3, 7, 29). Although the E. coli pathotypes have been clearly defined, they often differ in virulence between hosts, even between closely related mammals (21, 30, 31). For example, one of the most pathogenic forms of E. coli toward human beings is Shiga toxin-producing E. coli, or STEC, which includes serotype 0157:H7. STEC are commensal organisms in ruminants, but are highly virulent pathogens in human beings that can cause severe hemorrhagic colitis (32–34). In NHPs, it is unclear whether certain pathotypes exist as commensals or pathogens.
In this study, we examined E. coli isolates from 33 NHPs within a single zoo over a 7-week period, surveying genetic relatedness using a sequence-based approach, phylogenetic types, plasmid replicon types, virulence gene profiles, and antimicrobial susceptibility profiles. E. coli was the organism most suited for the purposes of this study, as it is easily culturable, has been previously implicated in episodes of diarrheal illness in captive primate populations, and is an ideal organism to assess genetic relatedness and microbial gene flow. The goal of this work was to determine whether primates are in fact a reservoir for E. coli harboring virulence genes and plasmid types typical of different E. coli pathotypes, to determine the genetic relatedness of E. coli over time and between co-habitating (i.e., same primate complex) animal groups, to assess the antimicrobial susceptibilities of E. coli isolates, and to evaluate the potential link between the zoo’s history of diarrhea with pathogenic E. coli presence.
Materials and Methods
Primate Study Population and Fecal Sample Collection
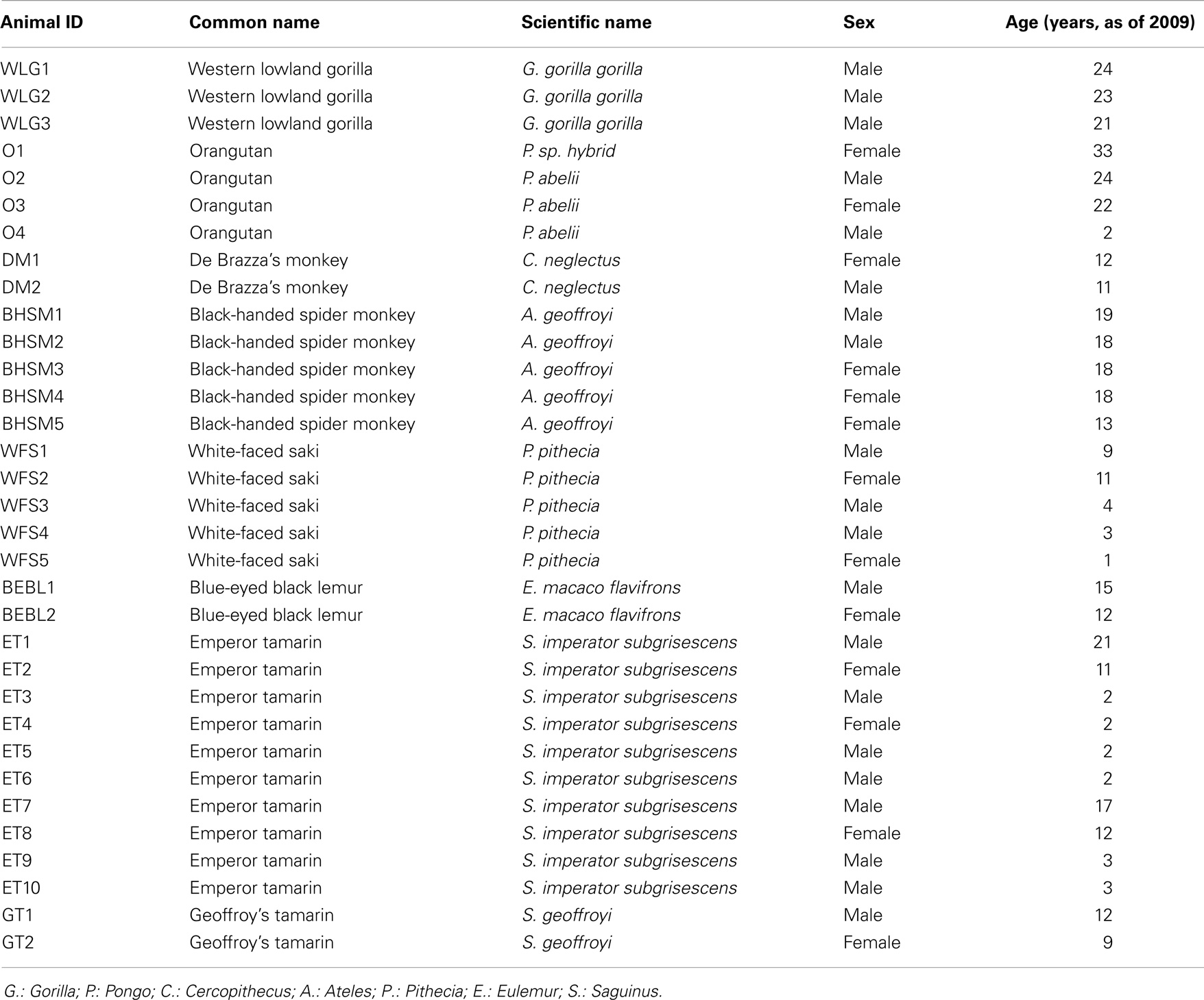
The research in this study complied with protocols approved through the University of Minnesota Institutional Animal Care and Use Committee. The primate collection at the Como Zoo in Saint Paul, MN, an American Zoological Association (AZA) accredited zoo, served as the study population, which included 33 healthy primates representing eight primate species (Table 1). Fresh fecal samples were collected weekly from each primate, between June and August of 2009. Samples were collected from freshly voided feces using a sterile spatula, with care to collect from the top of the sample to avoid ground contamination. After collection, samples were immediately placed on ice for transport from the zoo to the laboratory and subsequently processed.
Bacterial Isolates
E. coli isolates (N = 162) were included in this study. E. coli was isolated from fecal samples as follows: 1 g of fresh fecal material was homogenized in 10 mL of buffered peptone water (BD, Sparks, MD, USA). Ten-fold serial dilutions were performed on each sample, and dilutions were plated on MacConkey agar (BD). Plates with isolated colonies were used to pick one presumptive colony per fecal sample. Suspect E. coli colonies were then re-plated on Levine eosin methylene blue (EMB) agar (BD) as a secondary confirmation of E. coli. Finally, the suspect E. coli colonies from the EMB agar were confirmed using PCR primers specific for E. coli (35).
DNA Extraction and PCR Analysis
DNA templates for PCR were prepared using boiled lysates of overnight cultures from each individual sample being examined (36). A total of 22 E. coli control strains were used in this study (Table S1 in Supplementary Material). For plasmid replicon typing and virulence gene screening, amplifications were performed in a 25 μL reaction mixture, containing 5 μL 5 × PCR buffer (Promega, Madison, WI, USA), 0.25 mM dNTP mixture, 2.5 U Taq DNA polymerase (Promega), 0.15 μmol of each primer, and 2 μL template DNA. Following PCR (described below), the samples were subjected to horizontal gel electrophoresis in a 2.0% (w/v) Tris–acetate–EDTA (TAE) agarose, and the size of the amplicons was determined by comparison to the 100 bp DNA marker and known positive controls (New England Biolabs, Beverly, MA, USA).
Phylogenetic Group Typing
Isolates were assigned to one of the four phylogenetic groups (A, B1, B2, or D) using the interpretive approach of Clermont et al. (37). As determined by PCR, isolates were assigned to groups based on their possession of two genes (chuA and yjaA) and a DNA fragment (TSPE4.C2).
Sequence-Based Typing
A subset of isolates was examined for genetic relatedness using fumC gene sequencing, as previously described (38). For each isolate, PCR amplicons were generated and subjected to bidirectional sequencing using the same PCR primers used for amplification. Resulting sequences were quality trimmed, assembled, and concatenated to a similar size. Phylogenetic relationships between the isolates were examined using MEGA5 (39). Data from this project are freely available at Figshare (http://figshare.com/articles/Zoo_E_coli_fumC_dataset/956237).
Virulence Genotyping
Test and control organisms were examined for the presence of 10 genes known for their association with Extraintestinal pathogenic E. coli (ExPEC) virulence: cvaC, etsB, hlyA, hlyF, ireA, iroN, iss, iutA, papC, and sitA (40, 41); three genes associated with EPEC virulence: eae, espC, and tir; three genes associated with Enterotoxigenic E. coli (ETEC) virulence: eltA, sta, and stb; three genes associated with Enterohemorrhagic E. coli (EHEC) virulence: etpD, stx1A, and stx2A; three genes associated with Enteroaggregative E. coli (EAEC) virulence: aatP, aggR, and pet; and two genes associated with Enteroinvasive E. coli (EIEC) virulence: sepA and shET2. The multiplex procedures for the majority of ExPEC-associated genes screened for in this study are previously described (40). Three additional multiplex PCR assays were designed in this study to screen for the remainder of genes (Table S2 and Figure S1 in Supplementary Material). Reaction mixtures associated with the three multiplex PCR assays designed in this study were subjected to the following cycling parameters: initial denaturation (95°C, 10 min), followed by 30 cycles of denaturation (94°C, 30 s), annealing (55°C, 30 s), and extension (72°C, 60 s), and a final extension (72°C, 7 min). Specificity of the primers was confirmed using known positive and negative bacterial controls.
Plasmid Replicon Typing
Isolates were examined for the presence of 15 sequences specific for different plasmid incompatibility types using a combination of multiplex PCR assays previously described (42). The following plasmid incompatibility groups were sought: IncP-1α, IncA/C, IncFIC, IncB/O, IncK/B, IncW, IncFIA, IncFIB, IncY, IncI1, IncX, IncHI1, IncN, IncHI2, and IncL/M.
Antimicrobial Susceptibility Testing
E. coli isolates (N = 162) were tested for susceptibility to seven antimicrobial agents by Kirby-Bauer disk diffusion, according to the Clinical Laboratory Standards Institute (CLSI) MIC Interpretative Standards using Mueller–Hinton agar plates (43, 44). Disks containing the following antimicrobial agents were used: ampicillin (AM, 10 μg), gentamicin (GM, 10 μg), nalidixic acid (NA, 30 μg), streptomycin (S, 10 μg), tetracycline (TE, 30 μg), sulfisoxazole (G, 0.25 mg) and trimethoprim (TMPS or W5, 5 μg) (BD). The following E. coli strains were used as positive controls: ATCC 25922 and APEC 01 (43–45).
Results
Over the course of this seven-week study, a total of 229 fecal samples were collected from the Como Zoo’s primate collection. Of the 229 fecal samples collected, E. coli was successfully isolated from 162 samples (70.7%). Unsuccessful isolations typically involved overgrowth with Proteus spp. which was unable to be resolved. For the E. coli isolates (N = 162) used in this study, one colony per sample was selected. The majority of the individuals were sampled weekly throughout the seven-week study period (i.e., seven total samples collected per individual). However, there were two instances where an individual was not sampled due to unavailable sample material during the designated sample collection period.
Phylogenetic Group Typing
Phylogenetic groups A, B1, B2, and D were all found to be present in the captive population of NHPs tested in this study. Of the four phylogenetic groups tested for in this study, B1 was the most commonly identified group, with 53.1% (N = 86) of the isolates testing positive. In addition, 22.8% (N = 37) of the isolates tested positive for group A and 19.8% (N = 32) of the isolates tested positive for group B2. The least prevalent phylogenetic group in this study was group D, which tested positive in 4.3% (N = 7) of the isolates.
Sequence-Based Typing
Sequence-based typing was performed on a subset (N = 91) of the E. coli isolates, selected to be representative of the total population based upon date of isolation and animal source, using the fumC gene (Figure 1). A total of 18 fumC sequence types were identified using the Achtman E. coli MLST database (46). Sequence types spanned multiple isolates, species, and collection time points. Some common patterns were observable, though. For example, in many cases isolates were identified that originated from the same animal at different time points, and possessed identical sequence types as well as identical genotyping profiles, suggesting common clones persisting over time. There was also evidence of transfer of common E. coli clones between animals of the same species, and between animals of differing species over time. In particular, the following identical genotyping patterns were observed: fumC65 (two sets of two isolates), fumC29 (five isolates), fumC23 (two sets of two isolates), fumC24 (four isolates), fumC43 (eight isolates), fumC260 (three sets of two isolates), and fumC11 (two sets of seven and five isolates).
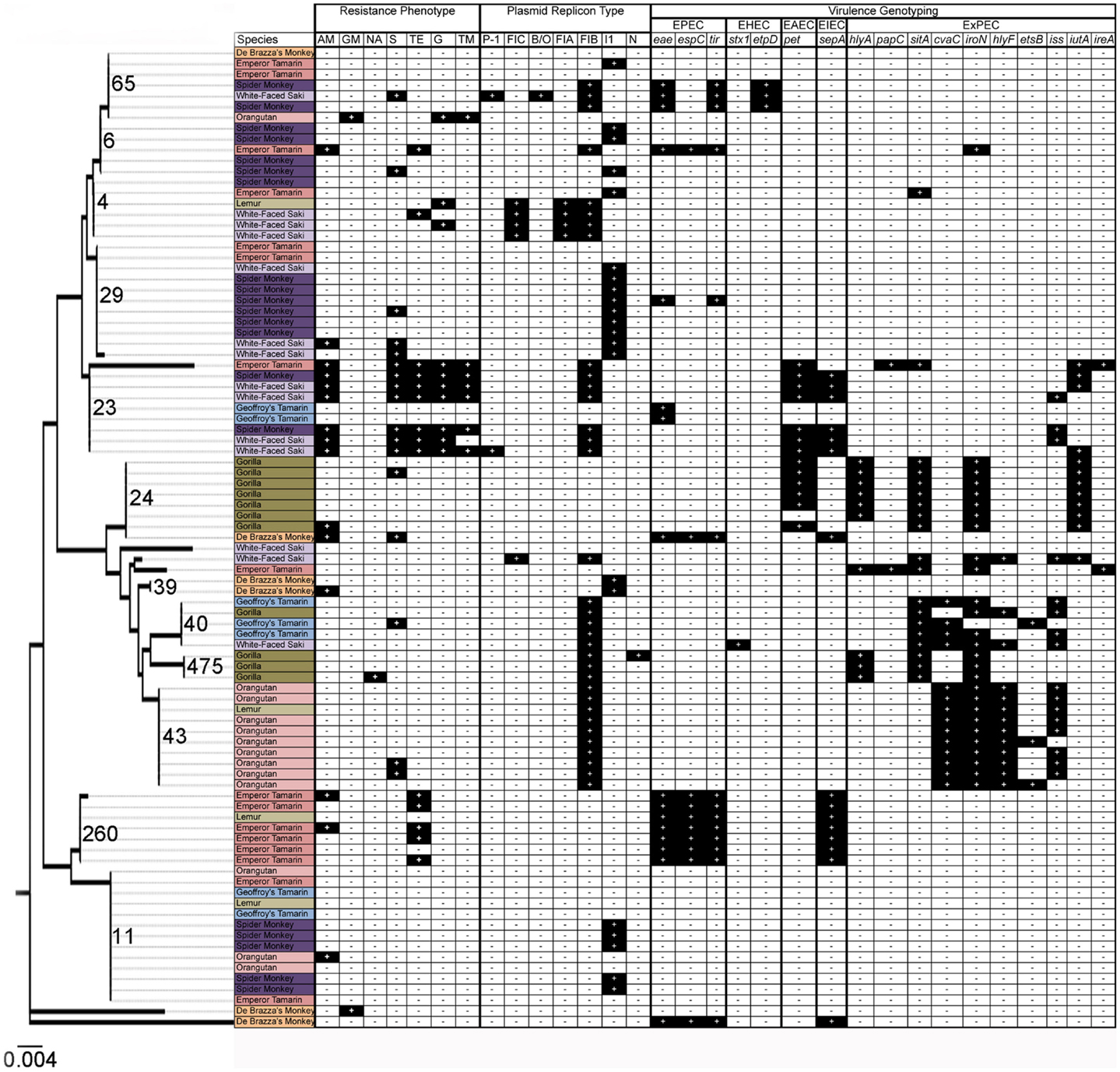
Figure 1. Results of resistance phenotyping, plasmid replicon analysis, virulence genotyping, and fumC sequence analysis among 91 isolates in this study. Species are colored as follows: orange: De Brazza’s monkey; light pink: orangutan; pink: emperor tamarin; dark purple: spider monkey; light purple: white-faced saki; light green: lemur; dark green: gorilla; and light blue: Geoffroy’s tamarin. Dendrogram depicts relationships of isolates using fumC sequence analysis.
Plasmid Replicon Typing
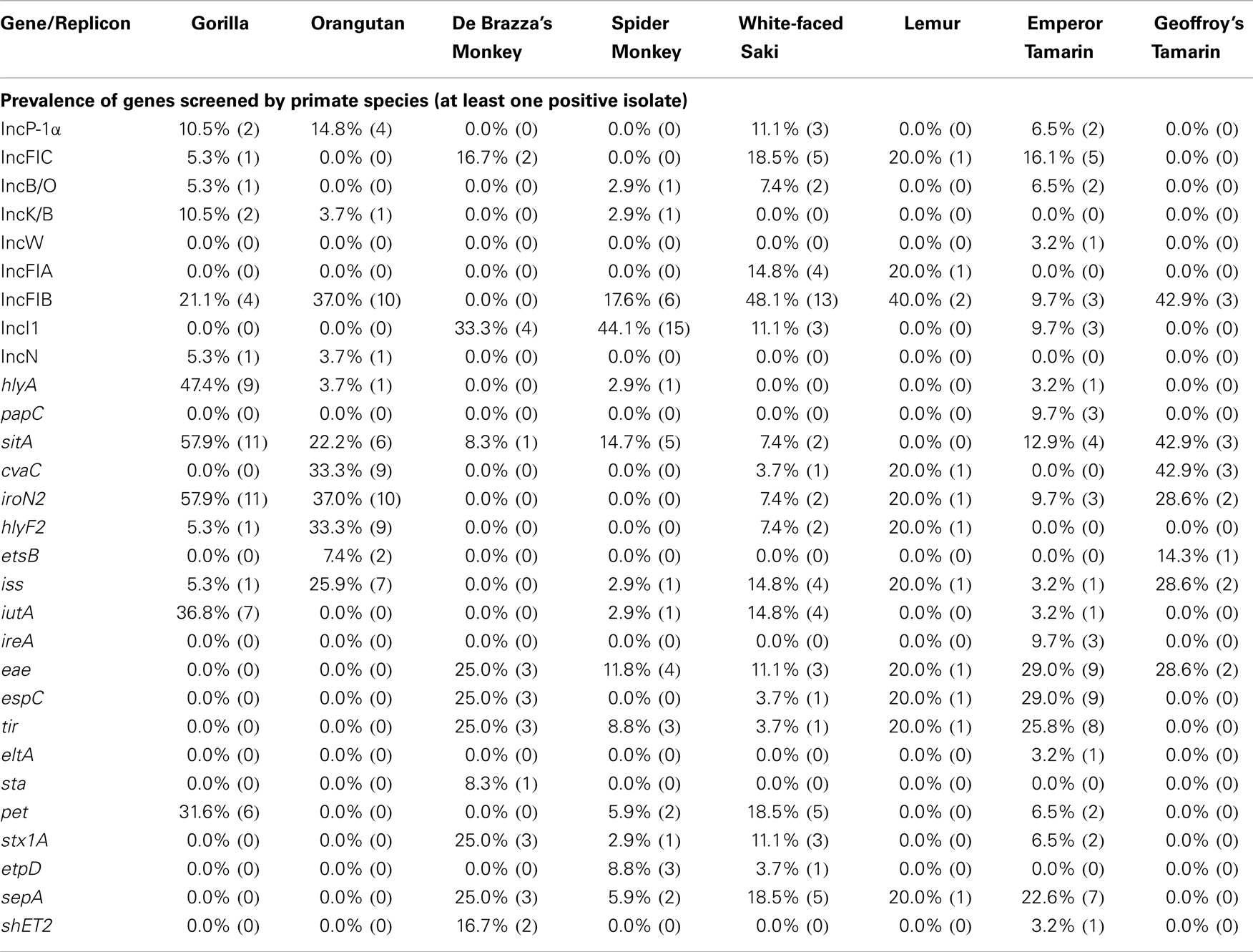
All eight NHP species examined were found to harbor E. coli with DNA sequences associated with at least one of the 15 plasmid incompatibility groups sought in this study. Of the 15 plasmid incompatibility groups examined, 9 were found to be present in the isolates, including IncFIB, IncI1, IncFIC, IncP-1α, IncB/O, IncFIA, IncK/B, IncN, and IncW (Table 2). However, only two plasmid types were prevalent among isolates at rates greater than 10% (RepFIB and RepI1).

Table 2. Overall prevalence of each gene and plasmid replicon studied (at least one positive isolate).
Virulence Genotyping
Screening for 24 virulence factors associated with ExPEC, EPEC, ETEC, EAEC, EHEC, and EIEC, all eight NHP species tested were found to contain E. coli possessing known virulence factors. Of these 24 virulence factors, 20 were identified in the E. coli isolates from 31 of the 33 primates in this study.
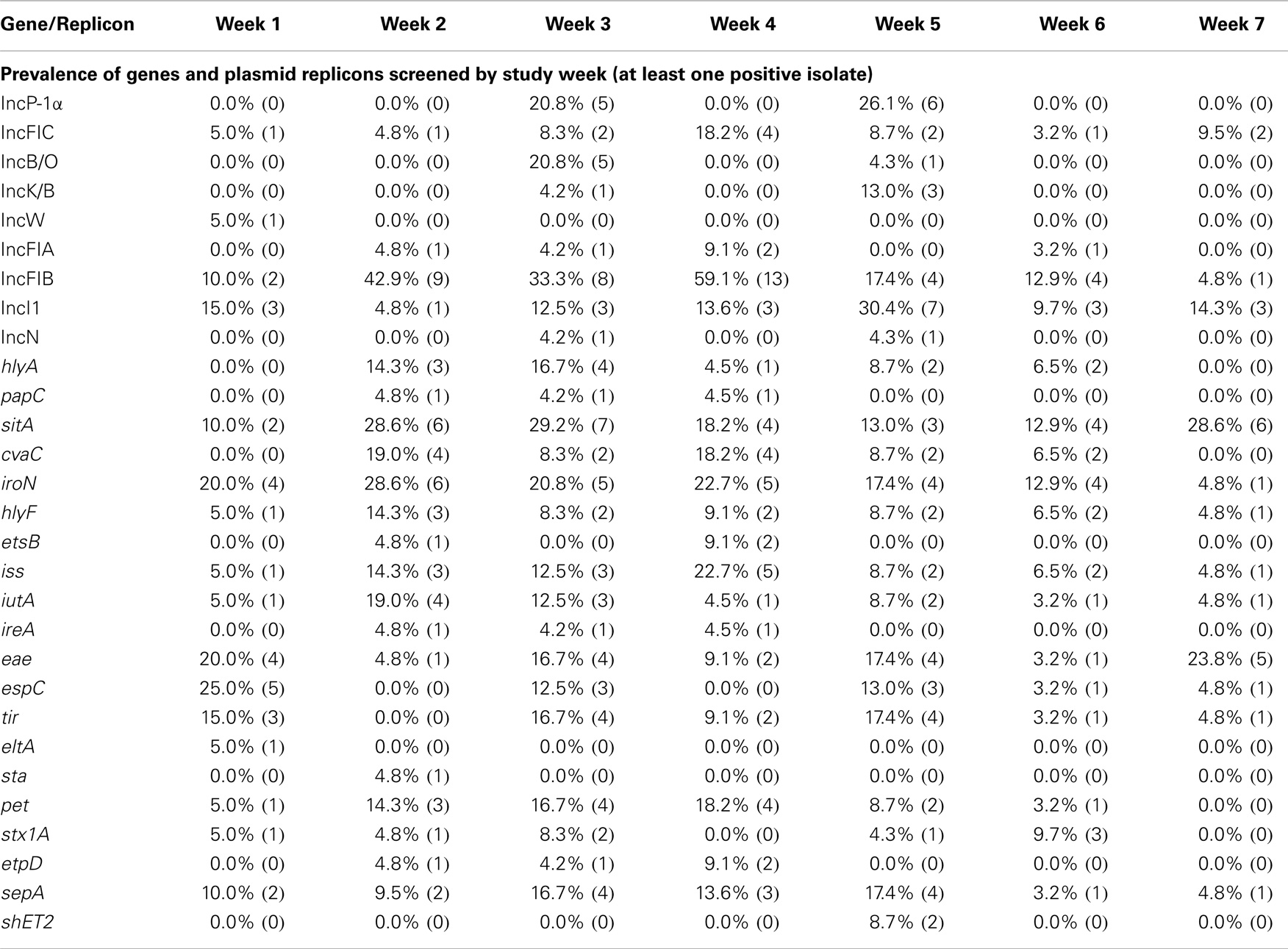
Of the 10 ExPEC-associated genes sought, all were found to be present in at least one NHP, including sitA, iroN, iss, cvaC, hlyF, iutA, hlyA, etsB, ireA, and papC (Table 2). Furthermore, ExPEC-associated genes were found among E. coli isolated from every NHP species examined (Table 3). In terms of gene function, ExPEC genes sought included those associated with iron acquisition (sitA, iroN, iutA, ireA, and etsB), serum survival (iss), colicin production (cvaC), hemolysin production (hlyA and hlyF), and fimbriae production (papC) (40, 41, 47–50). Three ExPEC-associated genes were present in greater than 10% of the isolates examined: sitA (19.8%), iroN (17.3%), and iss (10.5%). These genes often occurred together in the same isolates. At weeks 6 and 7 of sampling, ExPEC virulence genes had the highest prevalence of all genes screened (Table 4).
Of the three EPEC-associated genes screened, all three were present in at least one primate, including eae (N = 21), tir (N = 15), and espC (N = 13) (Table 2). EPEC-associated genes were present in six of eight primate species examined in this study, including Old World monkeys, New World monkeys, and lemurs. Isolates from only two species, gorillas, and orangutans, did not harbor EPEC-associated genes. Both primate species are great apes and belong to the same taxonomic family as humans (i.e., Hominidae) (Table 3). The most prevalent EPEC-associated gene, eae (13.0%), which is required for attaching/effacing adherence in E. coli (51, 52), was identified in isolates from the following primate species: De Brazza’s monkeys, spider monkeys, white-faced sakis, lemurs, emperor tamarins, and Geoffroy’s tamarins (Table 4). The EPEC-associated gene, tir (9.3%), which plays a role in adherence to small intestinal epithelial cells (51), was identified in isolates from the following primate species: De Brazza’s monkeys, spider monkeys, white-faced sakis, lemurs, and emperor tamarins (Table 4). The EPEC-associated gene, espC (8.0%), which is responsible for the production of an autotransporter protein/enterotoxin (53), was identified in isolates from the following primate species: De Brazza’s monkeys, white-faced sakis, lemurs, and emperor tamarins (Table 3). The espC-encoded protein is unique as it is the only one of the five proteins secreted extracellularly that is secreted independently of the type III secretion system encoded on the enterocyte effacement pathogenicity island (53).
Of the three ETEC-associated genes screened, sta and eltA were present in at least one NHP albeit at very low prevalence (Table 2), while stb was not found in any isolates examined. sta and eltA are both enterotoxin-encoding genes (54). sta encoding heat-stable toxin type A and eltA encoding heat-labile enterotoxin A prepeptide gene (54).
Of the three EAEC-associated genes screened, only pet was present in at least one primate, with an overall prevalence of 9.3% (Table 2). Among the isolates analyzed via fumC sequencing, two lineages of strains possessed pet, with or without co-carriage of ExPEC virulence genes and/or sepA (Figure 1). Some of these isolates possessed an IncI1 plasmid, which is characteristic of EAEC strains possessing a virulence plasmid (55). The EAEC-associated gene pet is an autotransporter enterotoxin gene that contributes to diarrheal illness, which is predominantly secretory in nature, seen in patients with EAEC (56). Specifically, pet was found in the gorillas, spider monkeys, white-faced sakis, and emperor tamarins (Table 3).
Of the three EHEC-associated genes screened, stx1A and etpD were present in at least one primate (Table 2). stx2a was not found in any isolates. The most prevalent EHEC-associated gene, stx1A (4.9%), was identified in isolates from the following primate species: De Brazza’s monkeys, spider monkeys, white-faced sakis, and emperor tamarins (Table 3).
Of the two EIEC-associated genes screened, both were present in at least one primate, including sepA and shET2 (Table 2). The most prevalent EIEC-associated gene, sepA (10.5%), was identified in isolates from the following primate species: De Brazza’s monkeys, spider monkeys, white-faced sakis, lemurs, and emperor tamarins (Table 3). The EIEC-associated gene sepA encodes a serine protease (57). The remaining EIEC-associated gene shET2 (1.2%) was identified in the De Brazza’s monkeys and emperor tamarins only. The EIEC-associated gene shET2 encodes an enterotoxin (58). E. coli isolated from the gorillas, orangutans, and Geoffroy’s tamarins did not harbor any EIEC-associated genes (Table 3).
Antimicrobial Susceptibility Testing
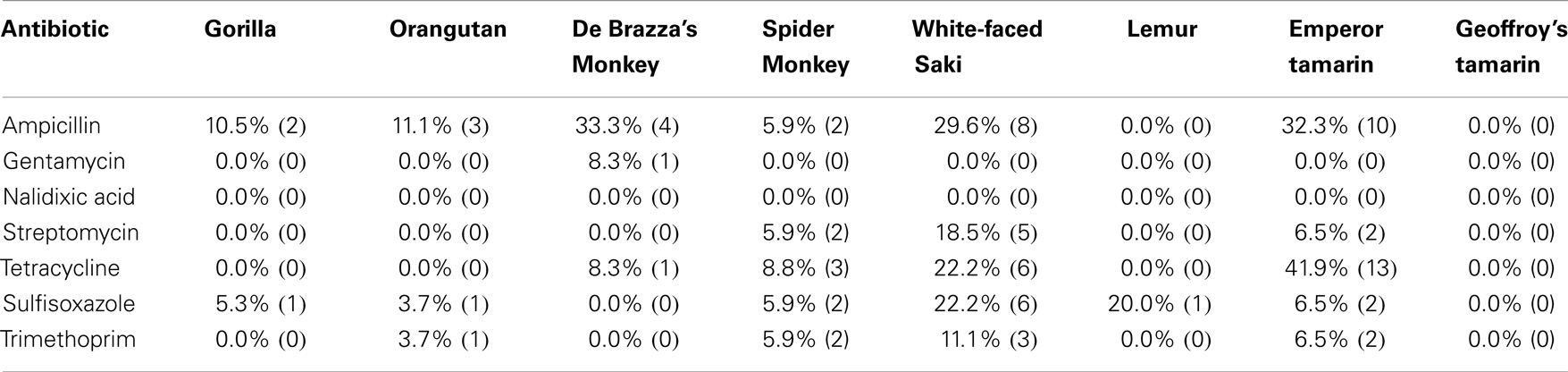
Based on the disk diffusion results, the majority of isolates were susceptible to the seven antibiotics tested (Table 5). However, there were isolates displaying individual or combined decreased antibiotic susceptibility to six of the seven antibiotics tested. Of the antibiotics tested in this study, isolates were most commonly resistant to ampicillin (17.9%) and tetracycline (14.2%). The remaining antibiotics tested had lower occurrence of resistance among the isolates tested (Table 5). Of the eight primate species screened in this study, E. coli isolated from feces of De Brazza’s monkeys showed the highest prevalence of ampicillin resistance (33.3%) and E. coli isolated from feces of emperor tamarins showed the highest prevalence of tetracycline resistance (41.9%) (Table 5). In contrast, the Geoffroy’s tamarins were fully susceptible to all seven antibiotics tested for the duration of the study. There were nine isolates displaying multidrug resistance, defined as decreased antibiotic susceptibility to >3 classes of antibiotics (59). Five of 27 isolates from white-faced sakis tested were multidrug resistant, which was the most of any primate species tested in this study.
Discussion
For this study, we aimed to determine the prevalence of pathogenic E. coli in a captive population of NHPs, to determine the genetic relatedness of E. coli over time and between co-habitating (i.e., same primate complex) animal groups, to examine the antimicrobial susceptibility profiles of E. coli isolates, and to assess the potential link between the zoo’s history of diarrhea with pathogenic E. coli presence. E. coli was the organism most suited for the purposes of this study, as it is easily cultured, has been previously associated with diarrheal illness in captive primate populations, and enables assessment of genetic relatedness and microbial gene flow. Initially, we used a phylogenetic typing method to classify the E. coli isolates into four groups (A, B1, B2, and D) (37). Of the four phylogenetic groups, B1 is the group most commonly associated with intestinal disease, while A is the group often associated with commensal strains (37). Additionally, previous research indicates that the majority of virulent extraintestinal strains belong to group B2 (37). Virulent extraintestinal strains also belong to phylogenetic group D, but to a lesser degree (37). Our findings suggest that NHPs carry E. coli belonging to multiple phylogenetic types, and possibly multiple pathotypes, but whether or not these pathotypes are associated with disease in these animals is unknown at this point.
Genes associated with all aforementioned pathotypes were present among isolates in this NHP collection, but ExPEC-associated genes were the most prevalent pathotype-associated genes identified. ExPEC is best known for its role in causing urinary tract infections (UTIs) and sepsis in human beings (60, 61). However, it is unknown whether or not ExPEC causes disease in NHPs. In terms of UTI pathogenesis, some evidence suggests that monkeys are not suitable hosts for experimental infection models, as they do not appear to develop UTIs naturally from human ExPEC strains (60). Throughout the study period, none of the subjects exhibited any clinical signs suggesting the presence of UTIs or associated sepsis amongst the primate collection and therefore it is more likely that these isolates were a component of the normal GI flora of these animals.
Over the course of this study, all primate species examined, aside from the orangutans, contained at least one isolate testing positive for at least one of the diarrheagenic E. coli pathotypes screened, including EPEC, ETEC, EAEC, EHEC, and EIEC. Of these, EPEC-associated genes were the most prevalent. EPEC is a major problem in developing countries because it is a major cause of infant diarrhea, which is sometimes fatal (62). While no causation was established, it is certainly plausible that EPEC could play a role in the intermittent diarrhea seen in this primate population. It is likely that EPEC exhibits different effects on different animals, with some animals acting as asymptomatic carriers and others susceptible to diarrhea (3, 6, 7, 29).
As previously mentioned, we tested each E. coli isolate for susceptibility toward the following seven antibiotics: ampicillin, gentamicin, nalidixic acid, streptomycin, tetracycline, sulfisoxazole, and trimethoprim. Past antibiotic treatments attempted at the zoo to treat diarrheal illness have included ampicillin, tetracycline, erythromycin, azithromycin, enrofloxacin, and sulfasalazine. Recently, antibiotics were rarely used in the white-faced saki and emperor tamarin populations, yet we observed a higher percentage of resistance among the E. coli isolates obtained from these two primate species (Table 5). In fact, neither the emperor tamarins nor the white-faced sakis were treated with ampicillin, yet E. coli isolated from the emperor tamarins showed the highest prevalence of ampicillin resistance (32.3%), and E. coli isolated from the white-faced sakis showed the third highest prevalence of antibiotic resistance (29.6%). Along these same lines, the emperor tamarins were never treated with tetracycline, yet E. coli isolated from the emperor tamarins showed the highest prevalence of tetracycline resistance (41.9%) (Table 5). The fact that tetracycline resistance and ampicillin resistance were high in the white-faced sakis and emperor tamarins, while antibiotics were used infrequently, could suggest that resistance genes carried by the obtained E. coli isolates were not acquired in response to selective pressures. Another plausible explanation is unknown co-selection may be occurring through use of another antibiotic or antimicrobial agent. Also, ampicillin has been used historically at the zoo to treat physical injuries, which are often times incurred by the males. Specifically, ampicillin has been the treatment of choice for the gorillas, orangutans, and spider monkeys. The proximity of the spider monkey holding enclosure to the white-faced sakis and emperor tamarins could be a factor in bacterial transmission, including isolates resistant to ampicillin, as the same zoo staff member typically cleans these three enclosures on a given day (Figure 2). Overall, the rates of resistance observed in this study were much lower than those observed in a previous study examining Snub-nosed monkeys at zoos in China (63). This could be attributed to numerous factors, including geographical location, zoo practices, and primate species examined.
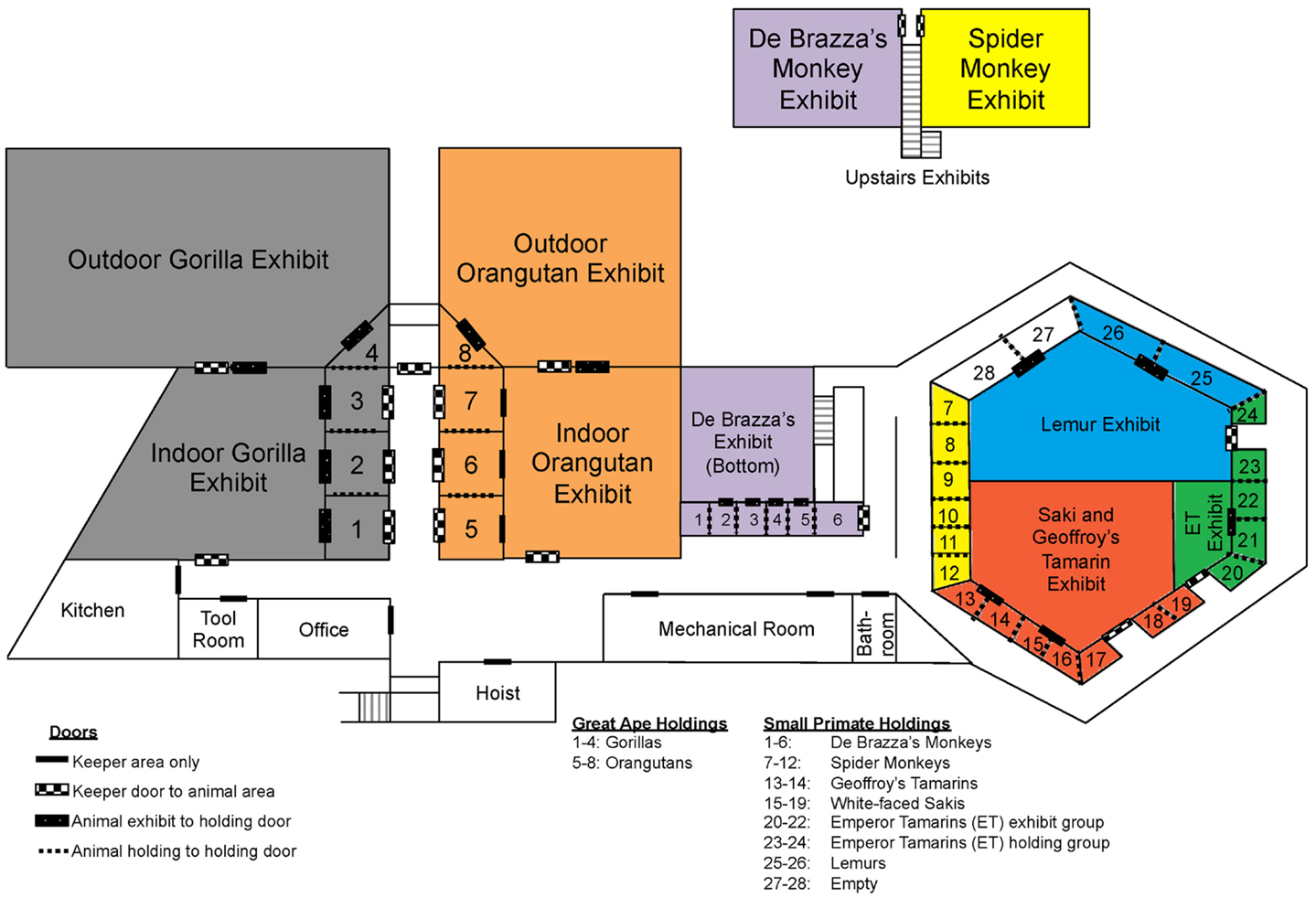
Figure 2. Map of Como Zoo’s primate enclosure locations, including those in holding and on-exhibit. Species are colored as follows: gray = gorilla, orange = orangutan, purple = De Brazza’s monkey, yellow = spider monkey, blue = lemur, green = Emperor tamarin, and red = white-faced saki and Geoffroy’s tamarin.
In order to determine the genetic relatedness of E. coli over time and between co-habitating (i.e., same primate complex) animal groups, sequence-based typing was performed on a subset of the isolates using the fumC gene. The results of the sequence-based typing indicated that E. coli strains of similar phylogenetic backgrounds, harboring similar sets of virulence factors, are found in multiple animals and species throughout the primate complex at this zoo (Figure 1). This is reflected in the fumC sequence typing, where isolates often belonged to the same sequence types and possessed similar virulence gene repertoires and plasmid types (Figure 1). Specifically, common isolates were found between spider monkeys and white-faced sakis, and between gorillas and orangutans, among others.
One important factor to consider when considering potential routes of bacterial transmission among the primates housed at the Como Zoo’s primate complex is the layout of primate holding and the system being employed at the time of this study for washing enclosures, feeding, among other keeper-related tasks. In an effort to visualize the potential routes of bacterial transmission from different animal groups (i.e., primate species), we constructed a detailed map of the primate complex showing the housing locations of all animal groups (i.e., primate species), as well as the locations of doors and primate keeper areas (Figure 2). The map provides perspective, as it allows for the visualization of where each primate species is housed in relation to the others, including those housed together, in close proximity, and at opposite sides of the complex.
As previously mentioned, results of the sequence-based typing showed that isolates with similar genetic backgrounds were found between spider monkeys and white-faced sakis, and between gorillas and orangutans, among others. As is illustrated in Figure 2, the spider monkeys and white-faced sakis enclosures were physically separated, as were the gorillas and orangutans enclosures. However, the gorillas and orangutans shared the same cleaning equipment, as they were housed in close proximity to each other. The same is true for the white-faced sakis and emperor tamarins. In addition, the cleaning procedure at the zoo was designed so that enclosures were located in relation to each other throughout the complex, as well as the placement of cleaning equipment (e.g., hoses). This allows for one person to clean all enclosures located in the great ape holdings section of the complex while another person cleans the enclosures located in the small primate holdings section of the complex simultaneously (Figure 2). Thus, on any given day, the same keeper would be responsible for cleaning both the spider monkeys and white-faced sakis enclosures (Figure 2). Due to the cleaning system employed by the zoo, bacterial transmission between animals likely occurred. One probable route of bacterial transmission that could have taken place is via fomites; in this case, water hoses, brushes, brooms, clothing, footwear, as well as other primate keeper accessories and cleaning equipment could have served as fomites. Overall, shared usage of cleaning equipment, along with designating one keeper responsible for cleaning the enclosures of multiple primate species simultaneously could have resulted in the sharing of bacteria between physically separated primates species.
Diarrhea in captive primates has long been a problem in both zoo settings and research colonies, including the Como Zoo (22–28). Diarrhea is a medical concern not only because it is physiologically taxing to the animals, but also because it is an ideal mechanism for disease transmission. This makes it both a public health concern and an animal welfare concern. In this study, we identified a number of E. coli isolates containing virulence genes from various pathotypes, and there was evidence of clonal transmission of isolates between animals and over time in the complex. Overall, the manifestation of chronic diarrhea in the Como Zoo’s primate collection is a complex problem whose solution will require regular screening for microbial agents and consideration of environmental causes. The use of E. coli as a surrogate for pathogen transmission, in this setting, aids in the identification of approaches that can be used in captive NHP populations to reduce such transmission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The research in this study complied with protocols approved through the University of Minnesota Institutional Animal Care and Use Committee (IACUC) and adhered to the legal requirements of the country in which it was conducted. The authors would like to thank the Como Zoo primate keepers for their assistance with fecal collection, sample identification, and providing all background information necessary to conduct this study. We thank current and past members of the Johnson Laboratory, Kevin Lang and Claudia Fernandez for technical assistance, as well as Bonnie Youmans for providing critical reading of the manuscript. We thank the University of Minnesota College of Veterinary Medicine’s Veterinary Diagnostic Lab (VDL), especially Arno Wunschmann, for providing information related to the diagnostic testing history at Como Zoo. We thank Dr. Edward Dudley (Penn State University) and Dr. Richard Isaacson (University of Minnesota) for providing positive controls for PCR genotyping. This research was funded by Morris Animal Foundation through a Veterinary Student Scholars Grant, by Merial/NIH through a Summers Scholars Grant, and by the University of Minnesota.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fvets.2014.00024/abstract
References
1. Bailey C, Mansfield K. Review paper: emerging and reemerging infectious diseases of nonhuman primates in the laboratory setting. Vet Pathol (2010) 47(3):462–81. doi: 10.1177/0300985810363719
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Blanco M, Blanco JE, Blanco J, de Carvalho VM, Onuma DL, de Castro AFP. Typing of intimin (eae) genes in attaching and effacing Escherichia coli strains from monkeys. J Clin Microbiol (2004) 42(3):1382–3. doi:10.1128/JCM.42.3.1382-1383.2004
3. Carvalho VM, Gyles CL, Ziebell K, Ribeiro MA, Catao-Dias JL, Sinhorini IL, et al. Characterization of monkey enteropathogenic Escherichia coli (EPEC) and human typical and atypical EPEC serotype isolates from neotropical nonhuman primates. J Clin Microbiol (2003) 41(3):1225–34. doi:10.1128/JCM.41.3.1225-1234.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Carvalho VM, Irino K, Onuma D, de Castro AFP. Random amplification of polymorphic DNA reveals clonal relationships among enteropathogenic Escherichia coli isolated from non-human primates and humans. Braz J Med Biol Res (2007) 40(2):237–41. doi:10.1590/S0100-879X2007000200010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Micro (2004) 2(2):123–40. doi:10.1038/nrmicro818
6. Mansfield KG, Lin KC, Newman J, Schauer D, MacKey J, Lackner AA, et al. Identification of enteropathogenic Escherichia coli in simian immunodeficiency virus-infected infant and adult Rhesus Macaques. J Clin Microbiol (2001) 39(3):971–6. doi:10.1128/JCM.39.3.971-976.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Mansfield KG, Lin KC, Xia D, Newman JV, Schauer DB, MacKey J, et al. Enteropathogenic Escherichia coli and ulcerative colitis in cotton-top Tamarins (Saguinus oedipus). J Infect Dis (2001) 184:803–7. doi:10.1086/322990
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Mansfield KGKG, Kemnitz JWJW. Introduction: challenges in microbial quality control for nonhuman primate. ILAR J (2008) 49(2):133–6. doi:10.1093/ilar.49.2.133
9. Martin HR, Taylor NS, Buckley EM, Marini RP, Patterson MM, Fox JG. Characterization of cytotoxic necrotizing factor 1-producing Escherichia coli strains from faeces of healthy macaques. J Med Microbiol (2009) 58(10):1354–8. doi:10.1099/jmm.0.012088-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. McClure HM, Strozier LM, Keeling ME. Enteropathogenic Escherichia coli infection in anthropoid apes. J Am Vet Med Assoc (1972) 161(6):687–9. doi:10.1128/IAI.69.1.315-324.2001
11. Thomson JA, Scheffler JJ. Hemorrhagic typhlocolitis associated with attaching and effacing Escherichia coli in common marmosets. Lab Anim Sci (1996) 46(3):275–9.
12. Bentley R, Meganathan R. Biosynthesis of vitamin K (Menaquinone) in bacteria. Microbiol Rev (1982) 46(3):241–280
13. DuPont HL, Formal SB, Hornick RB, Snyder MJ, Libonati JP, Sheahan DG, et al. Pathogenesis of Escherichia coli diarrhea. New Engl J Med (1971) 285(1):1–9. doi:10.1056/NEJM197107012850101
14. Levine MM. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis (1987) 155(3):377–89. doi:10.1093/infdis/155.3.377
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature (2009) 461(7268):1243–7. doi:10.1038/nature08480
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Lee PS, Lee KH. Escherichia coli – a model system that benefits from and contributes to the evolution of proteomics. Biotechnol Bioeng (2003) 84(7):801–14. doi:10.1002/bit.10848
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol (2005) 3(2):e45. doi:10.1371/journal.pbio.0030045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Goldberg TL, Gillespie TR, Rwego IB, Estoff EL, Chapman CA. Forest fragmentation and bacterial transmission among nonhuman primates, humans, and livestock, Uganda. Emerging Infect Dis (2008) 14(9):1375–82. doi:10.3201/eid1409.071196
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Goldberg TL, Gillespie TR, Rwego IB, Wheeler E, Estoff EL, Chapman CA. Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biol Conserv (2007) 135(4):511–7. doi:10.1016/j.biocon.2006.10.048
20. Rwego IB, Gillespie TR, Isabirye-Basuta G, Goldberg TL. High rates of Escherichia coli transmission between livestock and humans in rural Uganda. J Clin Microbiol (2008) 46(10):3187–91. doi:10.1128/JCM.00285-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Rwego IB, Isabirye-Basuta G, Gillespie TR, Goldberg TL. Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi impenetrable National Park. Uganda. Conserv Biol (2008) 22(6):1600–7. doi:10.1111/j.1523-1739.2008.01018.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Bethune MT, Borda JT, Ribka E, Liu M-X, Phillippi-Falkenstein K, Jandacek RJ, et al. A non-human primate model for gluten sensitivity. PLoS One (2008) 3(2):e1614. doi:10.1371/journal.pone.0001614
23. Blackwood RS, Tarara RP, Christe KL, Spinner A, Lerche NW. Effects of the macrolide drug tylosin on chronic diarrhea in rhesus macaques (Macaca mulatta). Comp Med (2008) 58(1):81–7.
24. Johnson LD, Ausman LM, Rolland RM, Chalifoux LV, Russell RG. Campylobacter-induced enteritis and diarrhea in captive cotton-top tamarins (Saguinus oedipus) during the first year of life. Comp Med (2001) 51(3):257–61.
25. Ludlage E, Mansfield K. Clinical care and diseases of the common marmoset (Callithrix jacchus). Comp Med (2003) 53(4):369–82.
26. Muñoz-Zanzi CA, Thurmond MC, Hird DW, Lerche NW. Effect of weaning time and associated management practices on postweaning chronic diarrhea in captive rhesus monkeys (Macaca mulatta). Comp Med (1999) 49(6):617–21.
27. Prongay K, Park B, Murphy SJ. Risk factor analysis may provide clues to diarrhea prevention in outdoor-housed Rhesus macaques (Macaca mulatta). Am J Primatol (2013) 75(8):872–82. doi:10.1002/ajp.22150
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Wilk JL, Maginnis GM, Coleman K, Lewis A, Ogden B. Evaluation of the use of coconut to treat chronic diarrhea in Rhesus macaques (Macaca mulatta). J Med Primatol (2008) 37(6):271–6. doi:10.1111/j.1600-0684.2008.00313.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. Chen HD, Frankel G. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol Rev (2005) 29(1):83–98. doi:10.1016/j.femsre.2004.07.002
30. Bueris V, Sircili MP, Taddei CR, dos Santos MF, Franzolin MR, Martinez MB, et al. Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador. Bahia, Brazil. Mem Inst Oswaldo Cruz (2007) 102(7):839–44. doi:10.1590/S0074-02762007005000116
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Trabulsi LR, Keller R, Gomes TAT. Typical and atypical enteropathogenic Escherichia coli. Emerging Infect Dis (2002) 8(5):508–13. doi:10.3201/eid0805.010385
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Callaway TR, Anderson RC, Edrington TS, Genovese KJ, Bischoff KM, Poole TL, et al. What are we doing about Escherichia coli O157:H7 in cattle? J Anim Sci (2004) 82:E93–9.
33. Ferens WA, Hovde CJ. Escherichia coli O157: H7: animal reservoir and sources of human infection. Foodborne Pathog Dis (2011) 8(4):465–87. doi:10.1089/fpd.2010.0673
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Greig JD, Waddell L, Wilhelm B, Wilkins W, Bucher O, Parker S, et al. The efficacy of interventions applied during primary processing on contamination of beef carcasses with Escherichia coli: A systematic review-meta-analysis of the published research. Food Control (2012) 27(2):385–97. doi:10.1016/j.foodcont.2012.03.019
35. Maheux AF, Picard FJ, Boissinot M, Bissonnette L, Paradis S, Bergeron MG. Analytical comparison of nine PCR primer sets designed to detect the presence of Escherichia coli/Shigella in water samples. Water Res (2009) 43(12):3019–28. doi:10.1016/j.watres.2009.04.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Johnson JR, Brown JJ. A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(α1–4)Gal-binding PapG Adhesins of Escherichia coli. J Infect Dis (1996) 173(4):920–6. doi:10.1093/infdis/173.4.920
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol (2000) 66(10):4555–8. doi:10.1128/AEM.66.10.4555-4558.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, et al. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol (2012) 78(5):1353–60. doi:10.1128/AEM.06663-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol (2011) 28(10):2731–9. doi:10.1093/molbev/msr121
40. Johnson TJ, Wannemuehler Y, Doetkott C, Johnson SJ, Rosenberger SC, Nolan LK. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J Clin Microbiol (2008) 46(12):3987–96. doi:10.1128/JCM.00816-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Nolan LK. Characterizing the APEC pathotype. Vet Res (2005) 36(2):241–56. doi:10.1051/vetres:2004057
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
42. Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, et al. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol (2007) 73(6):1976–83. doi:10.1128/AEM.00760-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Clinical Laboratory Standards Institute. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals: Approved standard M31-A2. 2nd ed. Wayne, PA: National Committee for Clinical Laboratory Standards (2002).
44. Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 12th International Supplement. Wayne, PA: National Committee for Clinical Laboratory Standards (2002).
45. Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, et al. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol (2007) 189(8):3228–36. doi:10.1128/JB.00537-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
46. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol (2006) 60(5):1136–51. doi:10.1111/j.1365-2958.2006.05172.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Johnson TJ, Nolan LK. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol Mol Biol Rev (2009) 73(4):750–74. doi:10.1128/MMBR.00015-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Kemmett K, Humphrey T, Rushton S, Close A, Wigley P, Williams NJ. A longitudinal study simultaneously exploring the carriage of APEC virulence associated genes and the molecular epidemiology of faecal and systemic E. coli in commercial broiler chickens. PLoS One (2013) 8(6):e67749. doi:10.1371/journal.pone.0067749
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Oswald E, Schmidt H, Morabito S, Karch H, Marches O, Caprioli A. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Immun (2000) 68(1):64–71. doi:10.1128/IAI.68.1.64-71.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Russo TA, Carlino UB, Johnson JR. Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect Immun (2001) 69(10):6209–16. doi:10.1128/IAI.69.10.6209-6216.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Batchelor M, Prasannan S, Daniell S, Reece S, Connerton I, Bloomberg G, et al. Structural basis for recognition of the translocated intimin receptor (Tir) by intimin from enteropathogenic Escherichia coli. EMBO J (2000) 19(11):2452–64. doi:10.1093/emboj/19.11.2452
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Cantey JR, Moseley SL. HeLa cell adherence, actin aggregation, and invasion by nonenteropathogenic Escherichia coli possessing the eae gene. Infect Immun (1991) 59(11):3924–9.
53. Mellies JL, Navarro-Garcia F, Okeke I, Frederickson J, Nataro JP, Kaper JB. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect Immun (2001) 69(1):315–24. doi:10.1128/IAI.69.1.315-324.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Zhang W, Francis DH. Genetic fusions of heat-labile toxoid (LT) and heat-stable toxin b (STb) of porcine enterotoxigenic Escherichia coli elicit protective anti-LT and anti-STb antibodies. Clin Vaccine Immunol (2010) 17(8):1223–31. doi:10.1128/CVI.00095-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Dudley EG, Abe C, Ghigo JM, Latour-Lambert P, Hormazabal JC, Nataro JP. An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces. Infect Immun (2006) 74(4):2102–14. doi:10.1128/IAI.74.4.2102-2114.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Eslava C, Navarro-Garcia F, Czeczulin JR, Henderson IR, Cravioto A, Nataro JP. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect Immun (1998) 66(7):3155–63.
57. Lan R, Stevenson G, Reeves PR. Comparison of two major forms of the Shigella virulence plasmid pINV: positive selection is a major force driving the divergence. Infect Immun (2003) 71(11):6298–306. doi:10.1128/IAI.71.11.6298-6306.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Yavzori M, Cohen D, Orr N. Prevalence of the genes for Shigella enterotoxins 1 and 2 among clinical isolates of Shigella in Israel. Epidemiol Infect (2002) 128(3):533–5. doi:10.2307/3864756
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
59. Sahm DF, Thornsberry C, Mayfield DC, Jones ME, Karlowsky JA. Multidrug-resistant urinary tract isolates of Escherichia coli: prevalence and patient demographics in the United States in 2000. Antimicrob Agents Chemother (2001) 45(5):1402–6. doi:10.1128/AAC.45.5.1402-1406.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
60. Johnson JR, Stell AL, Delavari P, Murray AC, Kuskowski M, Gaastra W. Phylogenetic and pathotypic similarities between Escherichia coli isolates from urinary tract infections in dogs and extraintestinal infections in humans. J Infect Dis (2001) 183(6):897–906. doi:10.1086/319263
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Kucheria R. Urinary tract infections: new insights into a common problem. Postgrad Med J (2005) 81(952):83–6. doi:10.1136/pgmj.2004.023036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Abreu AG, Bueris V, Porangaba TM, Sircili MP, Navarro-Garcia F, Elias WP. Autotransporter protein-encoding genes of diarrheagenic Escherichia coli are found in both typical and atypical enteropathogenic E. coli strains. Appl Environ Microbiol (2012) 79(1):411–4. doi:10.1128/AEM.02635-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Wang Y, He T, Han J, Wang J, Foley SL, Yang G, et al. Prevalence of ESBLs and PMQR genes in fecal Escherichia coli isolated from the non-human primates in six zoos in China. Vet Microbiol (2012) 159(1–2):53–9. doi:10.1016/j.vetmic.2012.03.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Escherichia coli, virulence, plasmid, antibiotic resistance, non-human primate
Citation: Clayton JB, Danzeisen JL, Trent AM, Murphy T and Johnson TJ (2014) Longitudinal characterization of Escherichia coli in healthy captive non-human primates. Front. Vet. Sci. 1:24. doi: 10.3389/fvets.2014.00024
Received: 26 September 2014; Accepted: 31 October 2014;
Published online: 17 November 2014.
Edited by:
Paul Wigley, University of Liverpool, UKReviewed by:
Md.Tanvir Rahman, Bangladesh Agricultural University, BangladeshSherry Layon, Vetanco, Argentina
Copyright: © 2014 Clayton, Danzeisen, Trent, Murphy and Johnson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Timothy J. Johnson, Department of Veterinary and Biomedical Sciences, University of Minnesota, 1971 Commonwealth Avenue, 205 Veterinary Science, Saint Paul, MN 55025, USA e-mail: joh04207@umn.edu
 Jonathan B. Clayton
Jonathan B. Clayton Jessica L. Danzeisen
Jessica L. Danzeisen Ava M. Trent2
Ava M. Trent2
 Tami Murphy
Tami Murphy Timothy J. Johnson
Timothy J. Johnson