A Review of Selected Genes with Known Effects on Performance and Health of Cattle
- National Animal Disease Center, USDA, ARS, Ames, IA, USA
There are genetic conditions that influence production in dairy and beef cattle. The objective of this review was to describe relevant genetic conditions that have been associated with productivity and health in cattle. Genes or genomic regions that have been identified as a candidate for the condition will be included, and the genetic basis of the condition will be defined. Genes and genetic conditions included in this review are bovine leukocyte adhesion deficiency, deficiency of the uridine monophosphate synthase, bovine chronic interstitial nephritis, horn development, myostatin, complex vertebral malformation, leptin, osteopetrosis, apoptosis peptide activating factor 1, chondrodysplastic dwarfism, caseins, calpastatin, umbilical hernia, lactoglobulin, citrullinemia, cholesterol deficiency, prions, thyroglobulin, diacylglycerol acyltransferase, syndactyly, maple syrup urine disease, slick hair, Factor XI deficiency, and μ-Calpain. This review is not meant to be comprehensive, and relevant information is provided to ascertain genetic markers associated with the conditions.
Introduction
Some traits or genetic conditions are controlled by a single gene (monogenic or qualitative traits), while others are controlled by many genes (polygenic or quantitative traits). Eighty-seven percent of qualitative traits in cattle are recessively inherited (1). It is not surprising that genetic conditions are breed specific, given that cattle breeds were developed in relative genetic isolation and independently of each other (1). Until the advent of modern molecular biology methods, the technology was unavailable to identify genes associated with quantitative traits and the variants within the gene that produce differences in productivity or its expression.
Genome-wide association studies (GWASs) are possible due to the availability of technology that allows high-throughput genotyping of single-nucleotide polymorphisms (SNPs). These SNPs are variants, or alleles, in the DNA sequence that may be associated with the expression of a trait or characteristic in cattle. The technology allows deciphering the genetics behind the expression of economically important traits. Genomic regions associated with productive traits have been identified in dairy (2–4) and beef cattle (5–8). It has also been possible to identify genomic regions associated with infectious and genetic diseases that affect performance in cattle (9, 10).
High-throughput sequencing offers the opportunity to identify causative genetic variants, which was previously unavailable (11). Accessibility of this technology has allowed the identification of unknown variants that could possibly be responsible for the conditions. This is particularly important when a condition has been reported in several breeds, and the putative causative mutation has been identified in one breed but not in another. It could be possible that additional variants in the same gene have similar effect but are unidentified. Such is the case of double muscling in cattle (12, 13). This technology has been successfully used to identify genetic variants that cause differences in productivity of milk (14, 15), muscling (15, 16), and fertility (17). Combining GWAS with high-throughput sequencing procedures will enable the identification of unknown causative genetic variants that could improve productivity of cattle.
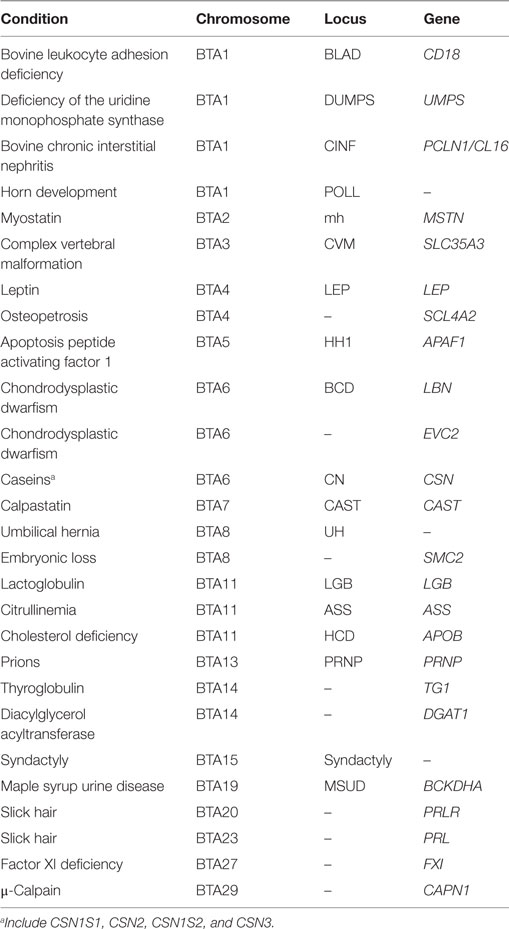
Several genes have been identified as having an association with productivity and health-related traits in cattle. This review is not meant to be a comprehensive list of genes, given that science continually discovers the association of genes with economically important traits. This review is meant to provide information on relevant conditions that could potentially impact the productivity and health of the cattle industry, and not to detail modifications in the genome that results in each condition. This is because several conditions are produced by different variants in the DNA sequence, as it is the case for double muscling (MSTN) in beef cattle where different SNP and insertion/deletions are responsible for the condition (18, 19). For simplicity, this review was organized by bovine chromosomes, rather than by the influence of the gene in productivity or health of cattle. Relevant conditions will be discussed. Table 1 summarizes the genetic conditions discussed in this review.
Bovine Leukocyte Adhesion Deficiency (BLAD)
Although this autosomal recessive, eventually lethal condition was recognized in Holstein cattle, it is reported to be segregating in other cattle breeds where Holstein genetics were introduced (20). It is characterized by reduced expression of functional β2-integrins on all leukocytes (21). β2-integrins are adhesion proteins that are the primary effectors of neutrophil adhesion to receptors on endothelial cells of postcapillary venules and the subsequent egress of neutrophils through intercellular cell junctions into extravascular tissues to defend the host against normal flora and pathogens. Animals affected with this condition have abnormally low levels of β2-integrins on all leukocytes; however, the reduced expression on neutrophils produces inadequate innate immunity against microbes in all tissues. Animals affected with this condition show severe pneumonia, ulcerative gingivitis, periodontitis, papillomatosis, dermatophytosis, tooth loss, poor wound healing, and slow growth (1, 21–28). A single point mutation at position 383 in the transcribed RNA in the CD18 gene was identified as the causative mutation. The substitution results in the replacement of an aspartic acid with a glycine at position 128 of the protein (D128G). CD18 resides on chromosome 1 (29, 30). This mutation has been identified worldwide as responsible for the condition (31). A genetic test is available to identify carriers of the condition (29) and has been used to virtually eradicate the clinical condition from the Holstein breed within a period of not more than 5 years (32) without negatively impacting genetic merit for performance traits (33).
Deficiency of the Uridine Monophosphate Synthase
This is an autosomal recessive lethal condition resulting from a deficiency of an enzyme that catalyzes the conversion of orotic acid to uridine-5′-monophosphate, which is the precursor of cytosine and thymine (components of DNA). Animals with this condition lack growth during embryonic development. Embryo mortality occurs approximately at 40 days of gestation (34). Carriers of this condition have a high incidence of return to estrus and long open day periods. The uridine monophosphatase synthase gene resides on chromosome 1 (35, 36). The substitution of a cytosine to a thymine at codon 405 of the gene has been identified as responsible for the condition (37), so it is possible to identify carriers of the condition.
Bovine Chronic Interstitial Nephritis (BCIN)
This is a kidney condition characterized by interstitial fibrosis with inflammatory cell infiltration. This condition has been identified in the Wagyu, or Japanese Black Cattle. Animals with this condition have delayed growth and high levels of ureic nitrogen in blood (38). The gene resides in chromosome 1, and the causal mutation has been identified (39). Producers interested in producing export beef genetics using this breed, especially to the Japanese market, will be required to establish that breeding animals are not carriers of the condition.
Horn Development
Horns may be a problem when handling cattle. It has been estimated that de-horning cattle, as well as loss of meat due to bruising from horns, represents a cost of approximately $25 million annually to the United States beef industry (40). Losses could be eliminated if carriers of alleles responsible for horn development are identified.
The locus of the gene responsible for horn development was detected on chromosome 1 and was termed POLL (40, 41). A locus has been recently identified on cattle chromosome 1 and is known in cattle and buffalo only. Wunderlich et al. (42) narrowed the region to a 2.5-Mb region on chromosome 1. Cargill et al. (43) identified 13 SNPs associated with horn development in this region, proposing several genes as responsible for the condition by sequencing a 1.6-kb region. Seichter et al. (44) identified nine additional SNPs in this region for the horn/polled condition. The gene from this locus is highly expressed in fetal tissue of horned animals, as compared with tissue from polled animals (45). It may be possible to select toward polled animals by using genomics information of horn development.
Myostatin (MSTN)
Double muscling or muscle hypertrophy was recognized and documented in the nineteenth century (46). The locus that causes double muscling (mh locus) in cattle was localized on the telomeric end of chromosome 2 (12). At that time, the mh locus was being studied to identify the gene responsible for double muscling in cattle, a deletion in the transforming growth factor β in mice showed similar effects in mice (47). This protein was later named myostatin, and the symbol of the gene in the double muscling locus on chromosome 2 is MSTN. Later, it was demonstrated that this was the same gene that caused double muscling in cattle (48). After the identification of the gene responsible for double muscling, several studies were directed to identify SNPs associated with double muscling in cattle (13, 49, 50).
Growth and carcass traits were evaluated in double-muscled cattle before myostatin was identified. True double-muscled animals, or animals with two copies of the allele that produces double muscling, are heavier at birth (46). Casas et al. (51) found that double-muscled animals have up to 20% more calving difficulty than non-double-muscled breeds, but animals with only one copy of the MSTN allele increase muscling without having calving difficulty. From this, it can be concluded that use of true double-muscled animals can be problematic under rangeland conditions due to the need to assist the cow during calving. However, if this gene was to be used under rangeland conditions, it is necessary to correctly manage the herd to produce animals with one copy of the gene, which would increase muscle mass without calving problems, avoiding the production of double-muscled calves. Regarding carcass traits, Arthur (46) indicates that double-muscled animals have up to 30% more muscle mass than non-double-muscled animals. However, further studies detected an increase of 17% in muscle yield and 66% less fat in double-muscled animals (52). Meat of animals with double muscling has also been associated with tender meat (46, 53). Myostatin, as the gene that produces double muscling, can be considered in animal production to increase muscle mass, without increasing calving difficulty, only if managed in terminal crosses.
Complex Vertebral Malformation (CVM)
Complex vertebral malformation is an autosomal recessive lethal condition of Holsteins observed in premature and mature calves (54). Congenital growth retardation, malformed vertebrae, and symmetric arthrogryposis typically characterize the condition although some morphological variations including various cardiac abnormalities occur (55, 56). Moreover, significant effects on reproductive performance and herd life have been reported (57). The causal point mutation (a G to T transversion) has been found to reside in an allele of a Golgi-resident transporter of UDP-N-acetylglucosamine encoded by SLC35A3 on BTA3 that results in a valine substitution at position 180 with a phenylalanine (V180F) (58). This allele was carried by many of the same animals that carried BLAD, and the allelic frequency of the mutant allele had reached as high as 20–30% in many countries before a rapid genetic diagnostic test was available to prevent further transmission of this mutant allele (58).
Leptin (LEP)
Leptin is the hormone produced by the obesity gene (ob). It is secreted by adipocytes, and it has been associated with feed consumption and energy balance in mice and humans. The leptin gene is located on bovine chromosome 4. A genetic marker was identified in the sequence (59) and in the promoter region of the bovine gene (60). Buchanan et al. (59) proposed that the missense mutation in the gene sequence could be considered the causative mutation in differences of fat deposits in cattle. Barendse et al. (61) indicated that there is no association between genetic markers in the leptin gene with fat in cattle; however, additional studies have supported the theory that different alleles of the gene are associated with differences in fat in cattle (60, 62, 63). The use of genetic markers in the leptin gene could be of use in beef production.
Osteopetrosis
The condition osteopetrosis was identified in cattle, which is also known as marble bone disease. Animals are characterized by forming exaggeratedly dense bones. This is the result of a deficiency in the number, or lack of function, of osteoclasts (64). Animals with this condition are usually stillborn, slightly premature with small body size. They often display distinctive features in the skull like flat skull, impacted molars, shortened mandibles, and protruding tongue (65, 66). A deletion in the SCL4A2 gene has been associated with the condition in Red Angus. The gene resides on chromosome 4 (64).
Apoptosis Peptide Activating Factor 1 (APAF1) Truncation
A haplotype named HH1 was initially identified on BTA5 using high-density SNP genotyping and associated with a decrease in conception rates and an increase in stillbirths in Holstein cattle (67). The causal mutation has now been identified to be a result of a truncation of the APAF1 protein and has been traced to the bull Pawnee Farm Arlinda Chief (Chief), a bull born in 1962 that sired several other prominent bulls used for artificial insemination, some of which have since had their genomes sequenced (68, 69). Through the use of artificial insemination, Chief also produced over 16,000 daughters, 500,000 granddaughters, and 2 million great-granddaughters (69). Owing to the widespread use of his genetics, the estimated cumulative number of spontaneous abortions caused by APAF1 truncation over the three decades that Chief’s alleles became highly frequent to be more than 100,000 in the United States and nearly 500,000 worldwide.
Chondrodysplastic Dwarfism
Dwarfism has been studied in cattle and other species. The principal characteristic of this condition is the abnormal bone ossification of extremities. It has also been associated with other conditions in the animal. Different genes produce dwarfism in different species. Chondrodysplastic dwarfism has been identified in the Japanese Brown Cattle. Economic losses can be attributed to this condition if the breed is used in breeding schemes. The locus responsible for this condition was identified on chromosome 6 (70). Two different DNA changes were identified in the LIMBIN gene associated with dwarfism. The first was a mutation that produces an alternative splicing site in the gene, and the other was a deletion in the gene (71). Takeda et al. (71) proposed these changes in the gene as responsible for chondrodysplastic dwarfism in Japanese Brown cattle.
Genetic variants in the Ellis van Creveld Syndrome 2 gene have also been proposed as responsible for chondrodysplastic dwarfism in Tyrolean Grey cattle (72). This gene also resides on chromosome 6. A 2-bp deletion was identified as responsible for the condition. The deletion produces a premature stop codon and thus a loss of function in the protein (72). Identification of mutations for the chondrodysplastic dwarfism allows genetic testing with the objective to eliminate this condition in cattle.
Caseins (CSN)
Milk proteins in ruminants have been comprehensively studied due to their importance in milk composition and cheese-making properties (73, 74). Dalgleish and Corredig (74) have described in detail the structure of the casein micelles and its changes during the processing of milk. Farrell et al. (73) reported the current nomenclature of the proteins of the cow in milk, with emphasis on caseins. Polymorphisms identified in the genes of caseins in bovine have also been summarized (75). Caseins comprise 80% of total proteins in milk.
There are four casein molecules produced (AlphaS1, AlphaS2, Beta, and Kappa). These proteins are coded by four genes on chromosome 6. The order of these genes on the chromosome is CSN1S1, CSN2, CSN1S2, and CSN3. The genomic region is referred to as the CN locus because these four genes are tightly linked (74, 75). Of particular interest in cheese production has been the kappa-casein genetic variants. Farrell et al. (73) have described in detail the genomic differences among variants in each casein gene. The B variant or allele is of particular interest because it has been associated with increased cheese production (76). Differences between the A and the B alleles from the kappa-casein protein is the substitution of an isoleucine for a threonine at position 136 and the substitution of an alanine for an aspartic acid in position 148 of the protein (73). SNPs associated with variants in the casein genes have been developed (77).
Using high-throughput genotyping technology, additional associations for casein production have been identified. A GWAS for bovine milk caseins and lactalbumin was done in Dutch Holstein Friesian cows (78). Several genomic regions were associated with the proportion of different caseins in milk. Schopen et al. (78) concluded that the proportion of genetic variance explained by the SNP on chromosomes 6, 11, and 14 could be explained by the casein locus on chromosome 6, beta-lactoglobulin on chromosome 11, and diglyceride acyltransferase-1 (DGAT1) on chromosome 14. Gambra et al. (79), performing a GWAS in a Holstein X Jersey population, identified six additional SNP, on chromosome 6, associated with caseins in milk. Given the proximity of the casein genes in a single locus, it has been suggested that SNP in the casein genes can be used in haplotypes instead of selecting each variant independently (75, 77). Different alleles may be suitable for management to increase milk production or to increase cheese production in dairy production systems.
Calpastatin (CAST)
Meat tenderness is one of the most important factors for consumer satisfaction. The calpastatin proteolytic axis has been identified as an important process to established meat tenderness. Calpastatin is the regulator of m-Calpain and μ-Calpain. m-Calpain and μ-Calpain are proteolytic enzymes responsible for the breakage of muscle fibers, producing postmortem tenderization of meat (80). Calpastatin is the natural inhibitor of calpains in this proteolytic system.
The gene that produces calpastatin (CAST) is located in bovine chromosome 7, and a genetic marker was identified within this gene. An association between meat tenderness and this genetic marker in the calpastatin gene has been observed in several studies (81, 82). This makes calpastatin a suitable gene to develop genetic markers associated with meat tenderness in other breeds, especially for breeds with known tough meat.
Umbilical Hernia
Umbilical hernia is a bovine defect observed after birth. It consists of the protrusion of the intestine or other organs through the abdominal wall at the umbilicus. In Israeli Holstein populations, frequency of retained placenta increased 7% in first-parity cows without umbilical hernia, compared with 18% in first-parity cows with umbilical hernia (83).
The locus responsible for umbilical hernia resides on chromosome 8. The gene is still unidentified, but the gene resides within an 8-Mb region (83). Genomics can be of assistance in identifying the gene and the causative mutation for the condition. Marker-assisted selection could be used to select against umbilical hernia to reduce costs associated with placenta retention.
Embryonic Loss
Fertility is an important factor that affects the dairy industry. The ability to produce offspring is a major component of milk production. Embryo loss hampers milk production for the cow, and it is an important economic factor in the dairy industry. A locus on chromosome 8 has been identified as responsible for death loss in Holsteins. Fritz et al. (84) identified that the Holstein Haplotype 3 (HH3) was responsible for embryo losses in French Holstein. Daetwyler et al. (85) had the same conclusion when evaluated HH3 using information from the 1,000 Bull Genomes Project.1 The condition was identified by the lack of homozygous individuals for a particular haplotype in the United States Holstein population. The mutation causing embryonic loss was estimated to reside in the Structural Maintenance of Chromosomes Protein-2 (SMC2) gene on chromosome 8. McClure et al. (17) confirmed the effect of the haplotype, within the SMC2 gene, as responsible for embryonic loss in Holstein.
Lactoglobulin (LGB)
The beta-lactoglobulin is the major protein in whey. Although LGB is not implicated in the coagulation process of milk, different variants of this gene affect renneting properties of raw milk. The LGB gene resides on chromosome 11, and 11 different variants have been described. The most common variants are alleles A and B in most dairy breeds; however, allele C is common in Jersey, and alleles D and E are common in other breeds (73, 75). The sequence of the variant B for this protein is considered the standard. Differences between variants B and A are the change of a glycine for an aspartic acid in position 64 and the substitution of an alanine for a valine in position 118 of the protein (73). Variants A and B have different properties affecting milk. The B variant of this protein denatures faster than variant A; therefore, heat stability is higher for the latter variant (86). Jakob and Puhan (86) indicate that kappa-casein reacts faster with the B variant, when compared to the A variant of LGB. SNP in the gene that produces the protein can be used to improve cheese production.
Citrullinemia
This autosomal recessive lethal genetic condition is characterized by high levels of ammonia in blood (87). This is due to a deficiency in activity of the enzyme argininosuccinate synthase that is produced by the gene ASS1. The enzyme is a key component of the urea cycle (1, 34). Animals with this condition are unable to excrete ammonia and have neurologic symptoms, producing perinatal mortality. The gene resides on chromosome 11, and the gene has been identified. The initial report demonstrated the first use in animal production of the polymerase chain reaction (PCR) combined with an endonuclease enzyme (AvaII) digestion to identify a restriction fragment length polymorphism of a fragment of the gene to identify carriers of the condition (88). Current technology would be capable to identify the SNP responsible for the condition.
Cholesterol Deficiency in Cattle
A recessive condition has been recently identified in Holstein cattle. Calves show severe hypocholesterolemia and die soon after birth due to diarrhea (89). Kipp et al. (89) identified a region of chromosome 11 associated with this condition on chromosome 11. The analysis of the pedigree of calves, known to have died from this condition, where traced to a predominant Canadian Holstein bull (Maughlin Storm). It was concluded that he was a carrier of the condition. Further analysis of this condition was pursued by Menzi et al. (90). An insertion of 1.3 kb of a transposable long terminal repeat (LTR) element within exon 5 of the APOB gene was identified (90). The LTR element identified is a vestige of viral DNA inserted in the host. These elements have also been identified in human (91). The insertion results in a frameshift that starts at amino acid 135 and produces a 97% truncation of the 4,567 amino acid long apolipoprotein B (90). Results from Menzi et al. (90) can be used as a genetic test to identify carriers of the condition in Holstein.
Prion (PRNP) Diseases
Bovine spongiform encephalopathy (BSE) is a neurodegenerative fatal condition affecting cattle. This condition is a transmissible spongiform encephalopathy that is similar to scrapie in sheep, chronic wasting disease in deer, and Creutzfeld–Jacob disease (CJD) in humans. BSE is caused by an accumulation of an abnormally folded isoform of the prion protein in central nervous system tissues. While the vast majority of cases of BSE were the result of ingesting feed derived from animal origin protein products contaminated with infected central nervous system tissues, today there are recognized atypical BSE cases that are thought to arise spontaneously and are not attributed to a transmissible origin in feedstuffs (92). Defined by their atypical molecular profiles on western blots when compared with classical BSE, atypical BSE cases tend to occur in older cattle than what is seen with classical BSE. One study of PRNP haplotypes from six atypical BSE cases has suggested a genetic determinant in or near PRNP influencing susceptibility of cattle to atypical BSE (93). Although believed to be extremely rare (94), one H-type BSE case (95) was associated with a heritable E211K mutation in the prion protein gene (PRNP) and is now thought to represent the bovine ortholog of the most common form of genetic CJD (E200K) in humans (96). Intracranial inoculation of infected brain material from the E211K BSE case into cattle possessing the 211K allele has demonstrated a very rapid onset of clinical BSE, consistent with a genetic form of BSE (97). A comprehensive review is available (98). The locus of the prion protein resides on chromosome 13 and consists of three exons. Initial studies identified three gene variants, differing in the number of repeats for eight peptides. Five, six, and seven repeats of the peptides were identified (99, 100). Seabury et al. (101), besides evaluating the repeats, studied the promoter region without identifying an association. However, there were differences in allelic frequencies in the intronic region of the gene. Additional studies have identified two indels in the bovine PRNP promoter region that have been studied for their association with BSE (102–104). Others examined the prevalence of the indel polymorphisms among selected cattle populations and found no association with the development of experimentally transmitted TSEs in cattle (105–107); this provided evidence that genetic factors associated with resistance to classical BSE in cattle do not provide resistance to cattle naturally infected with atypical BSE, thus suggesting that atypical BSE progresses via an alternative pathogenesis route compared to classical BSE and therefore is most likely a spontaneous prion disease in cattle.
Thyroglobulin (TG1)
The thyroglobulin gene (TG1) resides on chromosome 14. The first association of genetic markers in this gene was reported by Thaller et al. (108). Since then, several studies have established the association of this gene with intra- and extra-muscular fat in Bos taurus (109, 110). Although fat could be considered a secondary trait in beef production, the use of genetic markers for this trait could be of value in evaluating marbling in carcasses of B. taurus or Bos indicus origin.
Diacylglycerol Acyltransferase (DGAT1)
The gene that produces this protein is known as DGAT1, which is localized on chromosome 14, neighboring the TG1 gene (108, 111). This marker was originally associated with fat yield in milk (111), and additional studies have validated this association (112). This genetic marker has also been associated with marbling and fat thickness in beef cattle (108, 113, 114). This genetic marker residing in this gene is associated with fat production in dairy and beef cattle.
Syndactyly
This genetic condition is also known as “Mule Foot.” It is characterized by the fusion or stenosis of the functional phalanges in the bovine. The condition precludes natural mount in sires. This genetic condition presents susceptibility to hyperthermia due to high environmental temperatures. The locus resides on chromosome 15; however, the causative gene or change in the DNA has not been identified (115). Microsatellite markers in the neighboring region have been used to identify carriers of the condition (116). The LRP4 gene has been proposed as the candidate for the condition in this chromosomal region, and SNPs in the gene have been associated with the condition (117).
Maple Syrup Urine Disease
Animals with this condition have sweet-smelling urine. It is a progressive neurological condition, resulting in the inability of the animal to walk and ultimately results in death. The condition is the result of a mutation in the BCKADH gene. The enzyme has four subunits (Ea-alpha, E1-beta, E2, and E3). A mutation in the E1-alpha subunit, from a cytosine to a thymine, produces an incomplete enzyme (changes from a glutamine to a stop codon) in Hereford cattle. Animals with this condition have elevated levels of isocaproic acid which is the substrate of the enzyme (118, 119).
The Pre-E1-alpha subunit of the branched chain alpha-ketoacid dehydrogenase gene resides in chromosome 19. The mutation in Hereford was first identified, and further studies established that a different mutation was responsible for the condition in Shorthorn (118–120). Additional studies are needed to determine the causative mutation in Shorthorn.
Slick Hair
This condition should be of utmost importance for milk production under tropical and subtropical conditions. Slick hair coat has been observed in tropical breeds of B. taurus and has been studied in the Senepol and Carora breeds. These animals have very short, slick hair coats (121). Although it is unknown where the condition originated, it is known that Criollo cattle in the Americas possess this condition. The capability to preserve normal body temperature during heat stress conditions is an important trait in tropical and subtropical cattle. Heat stress is a problem for animals not adapted to tropical conditions, negatively impacting milk production. Senepol cattle with slick hair are capable of maintaining a lower body temperature when compared with Senepol without this condition. Senepol cattle are known to be as heat-tolerant as Brahman cattle (121, 122). Mariasegaram et al. (123) reported that the locus responsible for slick hair resided on chromosome 20. Further studies confirmed that the slick hair locus resides on chromosome 20 (124). Dikmen et al. (124) indicate that three potential candidate genes are in the region where the slick hair locus resides (SKP2, SPEF2, and PRLR). Littlejohn et al. (125) propose mutations in the prolactin receptor (PRLR) on chromosome 20 and the prolactin gene (PRL) on chromosome 23, as responsible for the slick hair condition. Littlejohn et al. (125) indicate that animals with the slick hair condition produced more milk than animals without the condition. Furthermore, Dikmen et al. (124) show that Holstein cattle with the slick hair condition produce more milk than Holstein cattle without the condition. This condition in dairy cattle under tropical conditions should be of economic importance if introduced.
Factor XI Deficiency
First reported in 1975 (126), Factor XI deficiency in Holstein cattle was later shown by planned matings to be inherited as an autosomal recessive trait (127). Factor XI is a plasma serine protease critical for activation of the intrinsic blood coagulation cascade. Factor XI-deficient cattle can be asymptomatic or exhibit symptoms including prolonged bleeding times following injections or insect bites, production of bloody milk and anemia. Although first reported in an 8-year-old steer, there are reports of lower calving and survival rates, and increased susceptibility to infectious diseases (128). The initial estimate of heterozygote frequency based upon activated partial thromboplastin times ranged between 8 and 17% (127). The mutation causing Factor XI deficiency is a 76-bp insertion within exon 12 that introduces a premature stop codon, thus resulting in a truncated Factor XI protein that is missing the functional serine protease domain responsible for proteolytic activation of Factor XI (129). Limited conscientious testing of bulls entering artificial insemination programs through the use of the activated partial thromboplastin time test reduced the allelic frequency of the mutant allele to 1.2% among 419 animals genotyped from the Dairy Bull DNA Repository at the time the DNA mutation was identified and a DNA PCR diagnostic test was available (129).
μ-Calpain (CAPN1)
This protein is a component of the calpastatin proteolytic axis. Its gene is located on chromosome 29, and the gene symbol is CAPN1. The first SNP identified in this gene was associated with meat tenderness in B. taurus breeds (130). Additional SNP were developed and found to be of better use than the original in B. indicus cattle (81, 131–133). Currently, there are several efforts to identify SNP associated with meat tenderness in beef cattle from B. indicus origin (131, 134, 135) and other native breeds (136). There are also attempts to associate SNP of CAPN1 with meat quality traits (137). Genetic markers in CAPN1 are suitable to be used by producers in Latin America to increase meat quality.
Final Remarks
Relevant information regarding genetic conditions affecting productivity and health of cattle is available. Such is the case of the Quantitative Trait Loci Data Base, or QTLdb,2 which maintains current information of genomic regions that have been associated with traits of economic importance in cattle. The database contains information for 81,652 quantitative trait loci detected in the cattle genome. Information includes information for health, meat and carcass, milk production, growth, reproduction, and exterior traits of cattle (138). Similarly, the Online Mendelian Inheritance in Animals3 currently contains a list of 494 cattle genetic traits or disorders, of which 229 are known to be inherited in a Mendelian fashion. There are 130 traits or disorders of which the point mutation, or quantitative trait nucleotide, is known. In the current database, there is information for 181 traits in which cattle can be used as a model for humans (139). These sources are readily available to cattle producers, students, and researchers in the field of cattle genomics.
Recognition of genetic conditions is an important component in animal production. Screening for genetic conditions needs to be assessed by the producer, according to the production system and breeds used. Several genetic conditions would improve productivity, while others would be deleterious. For each production system, it is necessary to screen the herd for known genetic conditions.
There are genetic conditions that would improve productivity (i.e., slick hair). For these genetic conditions, it would be possible to select for favorable alleles within the herd. This would increase productivity and make the production system more efficient. This is important where productivity is limited by hazardous environmental conditions.
The producer may want to introduce genetic conditions in the herd, or completely avoid them (i.e., myostatin). These genetic conditions need to be carefully assessed if they were to be introduced in the herd. It would increase productivity by increasing amount of muscle mass and salable meat. However, adequate management of the herd needs to take place to avoid detrimental effects that would result in economic losses to the producer. These losses would be in the form of expenses due to calving difficulty, or losses of the calf due to poor management.
If the goal is to sell to international markets, there are genetic conditions that producers need to be aware (i.e., BCIN). These conditions have only been identified in specific breeds, and they would not represent a problem in other breeds. However, if the producer decides to introduce these breeds for international trade, screening for genetic conditions needs to be undertaken before productivity is hindered.
Deleterious genetic conditions need to be recognized (i.e., syndactyly). These genetic conditions will limit productivity of animals expressing the trait. Animals should be eliminated from the herd to benefit the production system. A point of caution is that in spite of current genetic testing being highly effective at accurately identifying carrier animals of particular traits, leukochimerism can result in erroneous test results if blood is used as a convenient DNA source; test results must always be confirmed with an independent tissue source not subject to mixed genotypes (140).
As advances in molecular biology continue, the impact on our ability to more rapidly detect traits of economic importance will only improve. Use of haplotype reconstruction and haplotracking with a database of >1 million cattle4 will enable discovery of various traits (many of which may be unrecognized) that impact livestock performance and production.
Author Note
Mention of trade name, proprietary product, or specified equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable.
Author Contributions
EC and MK wrote and reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This document is an intramural project of the USDA/ARS National Animal Disease Center. The funding was provided by USDA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
References
1. Healy PJ. Testing for undesirable traits in cattle: an Australian perspective. J Anim Sci (1996) 74:917–22. doi:10.2527/1996.744917x
2. Mai MD, Rychtarova J, Zink V, Lassen J, Guldbrandtsen B. Quantitative trait loci for milk production and functional traits in two Danish cattle breeds. J Anim Breed Genet (2010) 127:469–73. doi:10.1111/j.1439-0388.2010.00869.x
3. Mai MD, Sahana G, Christiansen FB, Guldbrandtsen B. A genome-wide association study for milk production traits in Danish Jersey cattle using a 50K single nucleotide polymorphism chip. J Anim Sci (2010) 88:3522–8. doi:10.2527/jas.2009-2713
4. Cole JB, Wiggans GR, Ma L, Sonstegard TS, Lawlor TJ Jr, Crooker BA, et al. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genomics (2011) 12:408. doi:10.1186/1471-2164-12-408
5. Casas E, Shackelford SD, Keele JW, Stone RT, Kappes SM, Koohmaraie M. Quantitative trait loci affecting growth and carcass composition of cattle segregating alternate forms of myostatin. J Anim Sci (2000) 78:560–9. doi:10.2527/2000.783560x
6. Casas E, Stone RT, Keele JW, Shackelford SD, Kappes SM, Koohmaraie M. A comprehensive search for quantitative trait loci affecting growth and carcass composition of cattle segregating alternative forms of the myostatin gene. J Anim Sci (2001) 79:854–60. doi:10.2527/2001.794854x
7. Snelling WM, Allan MF, Keele JW, Kuehn LA, McDaneld T, Smith TP, et al. Genome-wide association study of growth in crossbred beef cattle. J Anim Sci (2010) 88:837–48. doi:10.2527/jas.2009-2257
8. Snelling WM, Allan MF, Keele JW, Kuehn LA, Thallman RM, Bennett GL, et al. Partial-genome evaluation of postweaning feed intake and efficiency of crossbred beef cattle. J Anim Sci (2011) 89:1731–41. doi:10.2527/jas.2010-3526
9. Neibergs HL, Seabury CM, Wojtowicz AJ, Wang Z, Scraggs E, Kiser JN, et al. Susceptibility loci revealed for bovine respiratory disease complex in pre-weaned Holstein calves. BMC Genomics (2014) 15:1164. doi:10.1186/1471-2164-15-1164
10. Casas E, Hessman BE, Keele JW, Ridpath JF. A genome-wide association study for the incidence of persistent bovine viral diarrhea virus infection in cattle. Anim Genet (2015) 46:8–15. doi:10.1111/age.12239
11. Djari A, Esquerré D, Weiss B, Martins F, Meersseman C, Boussaha M, et al. Gene-based single nucleotide polymorphism discovery in bovine muscle using next-generation transcriptomic sequencing. BMC Genomics (2013) 14:307. doi:10.1186/1471-2164-14-307
12. Charlier C, Coppieters W, Farnir F, Grobet L, Leroy PL, Michaux C, et al. The mh gene causing double-muscling in cattle maps to bovine chromosome 2. Mamm Genome (1995) 6:788–92. doi:10.1007/BF00539005
13. Grobet L, Poncelet D, Royo LJ, Brouwers B, Pirottin D, Michaux C, et al. Molecular definition of an allelic series of mutations disrupting the myostatin function and causing double-muscling in cattle. Mamm Genome (1998) 9:210–3. doi:10.1007/s003359900727
14. Jiang L, Liu X, Yang J, Wang H, Jiang J, Liu L, et al. Targeted resequencing of GWAS loci reveals novel genetic variants for milk production traits. BMC Genomics (2014) 15:1105. doi:10.1186/1471-2164-15-1105
15. Shin DH, Lee HJ, Cho S, Kim HJ, Hwang JY, Lee CK, et al. Deleted copy number variation of Hanwoo and Holstein using next generation sequencing at the population level. BMC Genomics (2014) 15:240. doi:10.1186/1471-2164-15-240
16. Tsuda K, Kawahara-Miki R, Sano S, Imai M, Noguchi T, Inayoshi Y, et al. Abundant sequence divergence in the native Japanese cattle Mishima-Ushi (Bos taurus) detected using whole-genome sequencing. Genomics (2013) 102:372–8. doi:10.1016/j.ygeno.2013.08.002
17. McClure MC, Bickhart D, Null D, Vanraden P, Xu L, Wiggans G, et al. Bovine exome sequence analysis and targeted SNP genotyping of recessive fertility defects BH1, HH2, and HH3 reveal a putative causative mutation in SMC2 for HH3. PLoS One (2014) 9:e92769. doi:10.1371/journal.pone.0092769
18. Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet (1997) 17:71–4. doi:10.1038/ng0997-71
19. Dunner S, Miranda ME, Amigues Y, Cañón J, Georges M, Hanset R, et al. Haplotype diversity of the myostatin gene among beef cattle breeds. Genet Sel Evol (2003) 35:103–18. doi:10.1186/1297-9686-35-1-103
20. Paiva DS, Fonseca I, Pinto IS, Ianella P, Campos TA, Caetano AR, et al. Incidence of bovine leukocyte adhesion deficiency, complex vertebral malformation, and deficiency of uridine-5-monophosphate synthase carriers in Brazilian Girolando cattle. Genet Mol Res (2013) 12:3186–92. doi:10.4238/2013.August.29.2
21. Kehrli ME Jr, Schmalstieg FC, Anderson DC, Van der Maaten MJ, Hughes BJ, Ackermann MR, et al. Molecular definition of the bovine granulocytopathy syndrome: identification of deficiency of the Mac-1 (CD11b/CD18) glycoprotein. Am J Vet Res (1990) 51:1826–36.
22. Kehrli ME Jr, Ackermann MR, Shuster DE, van der Maaten MJ, Schmalstieg FC, Anderson DC, et al. Animal model of human disease: bovine leukocyte adhesion deficiency: β2 integrin deficiency in Young Holstein cattle. Am J Path (1992) 140:1489–92.
23. Ackermann MR, Kehrli ME Jr, Morfitt DC. Ventral dermatitis and vasculitis in a calf with bovine leukocyte adhesion deficiency. J Am Vet Med Assoc (1993) 202:413–5.
24. Gilbert RO, Rebhun WC, Kim CA, Kehrli ME Jr, Shuster DE, Ackermann MR. Clinical manifestations of leukocyte adhesion deficiency in cattle: 14 cases (1977-1991). J Am Vet Med Assoc (1993) 202:445–9.
25. Kehrli ME Jr, Shuster DE, Ackermann MR, Smith CW, Anderson DC, Dore M, et al. Clinical and immunological features associated with bovine leukocyte adhesion deficiency. In: Lipsky PE, Rothlein R, Kishimoto TK, Faanes RB, Smith CW, editors. Structure, Function, and Regulation of Molecules Involved in Leukocyte Adhesion. New York: Springer-Verlag (1993). p. 314–27.
26. Lienau A, Stöber M, Kehrli ME, Tammen I, Schwenger B, Kuczka A, et al. Bovine leukocyte adhesion deficiency: clinical picture and differential diagnosis. Dtsch Tierärztl Wochenschr (1994) 101:405–6.
27. Nagahata H, Nochi H, Tamoto K, Taniyama H, Noda H, Morita M, et al. Bovine leukocyte adhesion deficiency: neutrophil function and pathological analysis. Am J Vet Res (1994) 55:40–8.
28. Ackermann MR, Kehrli ME Jr, Laufer JA, Nusz LT. Alimentary and respiratory tract lesions in eight medically fragile Holstein cattle with bovine leukocyte adhesion deficiency (BLAD). Vet Pathol (1996) 33:273–81. doi:10.1177/030098589603300303
29. Shuster DE, Kehrli ME Jr, Ackermann MR, Gilbert RO. Identification and prevalence of a genetic defect that causes leukocyte adhesion deficiency in Holstein cattle. Proc Natl Acad Sci U S A (1992) 89:9225–9. doi:10.1073/pnas.89.19.9225
30. Rexroad CE, Schlapfer JS, Yang Y, Harlizius B, Womack JE. A radiation hybrid map of bovine chromosome one. Anim Genet (1999) 30:325–32. doi:10.1046/j.1365-2052.1999.00504.x
31. Vatasescu-Balcan RA, Manea MA, Georgescu SE, Dinischiotu A, Tesio CD, Costache M. Evidence of single point mutation inducing BLAD disease in Romanian Holstein-derived cattle breed. Biotechnol Anim Husb (2007) 23:375–81. doi:10.2298/BAH0701375V
32. Schutz E, Scharfenstein M, Brenig B. Implication of complex vertebral malformation and bovine leukocyte adhesion deficiency DNA-based testing on disease frequency in the Holstein population. J Dairy Sci (2008) 91:4854–9. doi:10.3168/jds.2008-1154
33. Powell RL, Norman HD, Cowan CM. Relationship of bovine leukocyte adhesion deficiency with genetic merit for performance traits. J Dairy Sci (1996) 79:895–9. doi:10.3168/jds.S0022-0302(96)76438-X
34. Citek J, Blahova B. Recessive disorders – a serious health hazard? J Appl Biomed (2004) 2:187–94.
35. Ryan AM, Gallagher DS Jr, Schober S, Schwenger B, Womack JE. Somatic cell mapping and in situ localization of the bovine uridine monophosphate synthase gene (UMPS). Mamm Genome (1994) 5:46–7. doi:10.1007/BF00360568
36. Harlizius B, Schober S, Tammen I, Simon D. Isolation of the bovine uridine monophosphate synthase gene to identify the molecular basis of DUMPS in cattle. J Anim Breed Genet (1996) 113:303–9. doi:10.1111/j.1439-0388.1996.tb00620.x
37. Schwenger B, Schober S, Simon D. DUMPS cattle carry a point mutation in the uridine monophosphate synthase gene. Genomics (1993) 16:241–4. doi:10.1006/geno.1993.1165
38. Kobayashi N, Hirano T, Maruyama S, Matsuno H, Mukoujima K, Morimoto H, et al. Genetic mapping of a locus associated with bovine chronic interstitial nephritis to chromosome 1. Anim Genet (2000) 31:91–5. doi:10.1046/j.1365-2052.2000.00589.x
39. Hirano T, Hirotsune S, Sasaki S, Kikuchi T, Sugimoto Y. A new deletion mutation in bovine claudin-16 (CL-16) deficiency and diagnosis. Anim Genet (2002) 33:118–22. doi:10.1046/j.1365-2052.2002.00844.x
40. Brenneman RA, Davis SK, Sanders JO, Burns BM, Wheeler TC, Turner JW, et al. The polled locus maps to BTA1 in a Bos indicus x Bos taurus cross. J Hered (1996) 87:156–61. doi:10.1093/oxfordjournals.jhered.a022975
41. Georges M, Drinkwater R, King T, Mishra A, Moore SS, Nielsen D, et al. Microsatellite mapping of a gene affecting horn development in Bos taurus. Nat Genet (1993) 4:206–10. doi:10.1038/ng0693-206
42. Wunderlich KR, Abbey CA, Clayton DR, Song Y, Schein JE, Georges M, et al. A 2.5-Mb contig constructed from Angus, Longhorn and horned Hereford DNA spanning the polled interval on bovine chromosome 1. Anim Genet (2006) 37:592–4. doi:10.1111/j.1365-2052.2006.01538.x
43. Cargill EJ, Nissing NJ, Grosz MD. Single nucleotide polymorphisms concordant with the horned/polled trait in Holsteins. BMC Res Notes (2008) 1:128. doi:10.1186/1756-0500-1-128
44. Seichter D, Russ I, Rothammer S, Eder J, Förster M, Medugorac I. SNP-based association mapping of the polled gene in divergent cattle breeds. Anim Genet (2012) 43:595–8. doi:10.1111/j.1365-2052.2011.02302.x
45. Wiedemar N, Tetens J, Jagannathan V, Menoud A, Neuenschwander S, Bruggmann R, et al. Independent polled mutations leading to complex gene expression differences in cattle. PLoS One (2014) 9:e93435. doi:10.1371/journal.pone.0093435
46. Arthur PF. Double muscling in cattle: a review. Aust J Agric Res (1995) 46:1493–515. doi:10.1071/AR9951493
47. McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature (1997) 387:83–90. doi:10.1038/387083a0
48. McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci U S A (1997) 94:12457–61. doi:10.1073/pnas.94.23.12457
49. Dunner S, Charlier C, Farnir F, Brouwers B, Canon J, Georges M. Towards interbreed IBD fine mapping of the mh locus: double-muscling in the Asturiana de los Valles breed involves the same locus as in the Belgian Blue cattle breed. Mamm Genome (1997) 8:430–5. doi:10.1007/s003359900462
50. Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res (1997) 7:910–6.
51. Casas E, Keele JW, Fahrenkrug SC, Smith TP, Cundiff LV, Stone RT. Quantitative analysis of birth, weaning, and yearling weights and calving difficulty in Piedmontese crossbreds segregating an inactive myostatin allele. J Anim Sci (1999) 77:1686–92. doi:10.2527/1999.7771686x
52. Casas E, Bennett GL, Smith TP, Cundiff LV. Association of myostatin on early calf mortality, growth, and carcass composition traits in crossbred cattle. J Anim Sci (2004) 82:2913–8. doi:10.2527/2004.82102913x
53. Wheeler TL, Shackelford SD, Casas E, Cundiff LV, Koohmaraie M. The effects of Piedmontese inheritance and myostatin genotype on the palatability of longissimus thoracis, gluteus medius, semimembranosus, and biceps femoris. J Anim Sci (2001) 79:3069–74. doi:10.2527/2001.79123069x
54. Agerholm JS, Bendixen C, Andersen O, Arnbjerg J. Complex vertebral malformation in Holstein calves. J Vet Diagn Invest (2001) 13:283–9. doi:10.1177/104063870101300401
55. Agerholm JS, Andersen O, Almskou MB, Bendixen C, Arnbjerg J, Aamand GP, et al. Evaluation of the inheritance of the complex vertebral malformation syndrome by breeding studies. Acta Vet Scand (2004) 45:133–7. doi:10.1186/1751-0147-45-133
56. Agerholm JS, Bendixen C, Arnbjerg J, Andersen O. Morphological variation of “complex vertebral malformation” in Holstein calves. J Vet Diagn Invest (2004) 16:548–53. doi:10.1177/104063870401600609
57. Nielsen US, Aamand GP, Andersen O, Bendixen C, Nielsen VH, Agerholm JS. Effects of complex vertebral malformation on fertility traits in Holstein cattle. Livest Prod Sci (2003) 79:233–8. doi:10.1016/S0301-6226(02)00170-7
58. Thomsen B, Horn P, Panitz F, Bendixen E, Petersen AH, Holm LE, et al. A missense mutation in the bovine SLC35A3 gene, encoding a UDP-N-acetylglucosamine transporter, causes complex vertebral malformation. Genome Res (2006) 16:97–105. doi:10.1101/gr.3690506
59. Buchanan FC, Fitzsimmons CJ, Van Kessel AG, Thue TD, Winkelman-Sim DC, Schmutz SM. Association of a missense mutation in the bovine leptin gene with carcass fat content and leptin mRNA levels. Genet Sel Evol (2002) 34:105–16. doi:10.1186/1297-9686-34-1-105
60. Nkrumah JD, Li C, Yu J, Hansen C, Keisler DH, Moore SS. Polymorphisms in the bovine leptin promoter associated with serum leptin concentration, growth, feed intake, feeding behavior, and measures of carcass merit. J Anim Sci (2005) 83:20–8. doi:10.2527/2005.83120x
61. Barendse W, Bunch RJ, Harrison BE. The leptin C73T missense mutation is not associated with marbling and fatness traits in a large gene mapping experiment in Australian cattle. Anim Genet (2005) 36:86–8. doi:10.1111/j.1365-2052.2004.01224.x
62. Kononoff PJ, Deobald HM, Stewart EL, Laycock AD, Marquess FL. The effect of a leptin single nucleotide polymorphism on quality grade, yield grade, and carcass weight of beef cattle. J Anim Sci (2005) 83:927–32. doi:10.2527/2005.834927x
63. Schenkel FS, Miller SP, Ye X, Moore SS, Nkrumah JD, Li C, et al. Association of single nucleotide polymorphisms in the leptin gene with carcass and meat quality traits of beef cattle. J Anim Sci (2005) 83:2009–20. doi:10.2527/2005.8392009x
64. Meyers SN, McDaneld TG, Swist SL, Marron BM, Steffen DJ, O’Toole D, et al. A deletion mutation in bovine SLC4A2 is associated with osteopetrosis in Red Angus cattle. BMC Genomics (2010) 11:337. doi:10.1186/1471-2164-11-337
65. Leipold HW, Doige CE, Kaye MM, Cribb PH. Congenital osteopetrosis in Aberdeen Angus calve. Can Vet J (1970) 11:181–5.
66. Huston K, Leipold H. Hereditary osteopetrosis in Aberdeen-Angus calves. II. – genetical aspects. Ann Genet Sel Anim (1971) 3:419–23. doi:10.1186/1297-9686-3-4-419
67. VanRaden PM, Olson KM, Null DJ, Hutchison JL. Harmful recessive effects on fertility detected by absence of homozygous haplotypes. J Dairy Sci (2011) 94:6153–61. doi:10.3168/jds.2011-4624
68. Larkin DM, Daetwyler HD, Hernandez AG, Wright CL, Hetrick LA, Boucek L, et al. Whole-genome resequencing of two elite sires for the detection of haplotypes under selection in dairy cattle. Proc Natl Acad Sci U S A (2012) 109:7693–8. doi:10.1073/pnas.1114546109
69. Adams HA, Sonstegard TS, VanRaden PM, Null DJ, Van Tassell CP, Larkin DM, et al. Identification of a nonsense mutation in APAF1 that is likely causal for a decrease in reproductive efficiency in Holstein dairy cattle. J Dairy Sci (2016) 99:6693–701. doi:10.3168/jds.2015-10517
70. Yoneda K, Moritomo Y, Takami M, Hirata S, Kikukawa Y, Kunieda T. Localization of a locus responsible for the bovine chondrodysplastic dwarfism (bcd) on chromosome 6. Mamm Genome (1999) 10:597–600. doi:10.1007/s003359901052
71. Takeda H, Takami M, Oguni T, Tsuji T, Yoneda K, Sato H, et al. Positional cloning of the gene LIMBIN responsible for bovine chondrodysplastic dwarfism. Proc Natl Acad Sci U S A (2002) 99:10549–54. doi:10.1073/pnas.152337899
72. Murgiano L, Jagannathan V, Benazzi C, Bolcato M, Brunetti B, Muscatello LV, et al. Deletion in the EVC2 gene causes chondrodysplastic dwarfism in Tyrolean Grey cattle. PLoS One (2014) 9:e94861. doi:10.1371/journal.pone.0094861
73. Farrell HM Jr, Jimenez-Flores R, Bleck GT, Brown EM, Butler JE, Creamer LK, et al. Nomenclature of the proteins of cows’ milk – sixth revision. J Dairy Sci (2004) 87:1641–74. doi:10.3168/jds.S0022-0302(04)73319-6
74. Dalgleish DG, Corredig M. The structure of the casein micelle of milk and its changes during processing. Annu Rev Food Sci Technol (2012) 3:449–67. doi:10.1146/annurev-food-022811-101214
75. Caroli AM, Chessa S, Erhardt GJ. Invited review: milk protein polymorphisms in cattle: effect on animal breeding and human nutrition. J Dairy Sci (2009) 92:5335–52. doi:10.3168/jds.2009-2461
76. Lucey J, Kelley J. Cheese yield. J Soc Dairy Tech (1994) 47:1–14. doi:10.1111/j.1471-0307.1994.tb01264.x
77. Lien S, Rogne S. Bovine casein haplotypes: number, frequencies and applicability as genetic markers. Anim Genet (1993) 24:373–6. doi:10.1111/j.1365-2052.1993.tb00343.x
78. Schopen GC, Visker MH, Koks PD, Mullaart E, van Arendonk JA, Bovenhuis H. Whole-genome association study for milk protein composition in dairy cattle. J Dairy Sci (2011) 94:3148–58. doi:10.3168/jds.2010-4030
79. Gambra R, Peñagaricano F, Kropp J, Khateeb K, Weigel KA, Lucey J, et al. Genomic architecture of bovine kappa-casein and beta-lactoglobulin. J Dairy Sci (2013) 96:5333–43. doi:10.3168/jds.2012-6324
80. Koohmaraie M. Biochemical factors regulating the toughening and tenderization processes of meat. Meat Sci (1996) 43S1:193–201. doi:10.1016/0309-1740(96)00065-4
81. Casas E, White SN, Wheeler TL, Shackelford SD, Koohmaraie M, Riley DG, et al. Effects of calpastatin and micro-calpain markers in beef cattle on tenderness traits. J Anim Sci (2006) 84:520–5. doi:10.2527/2006.843520x
82. Barendse W, Harrison BE, Hawken RJ, Ferguson DM, Thompson JM, Thomas MB, et al. Epistasis between calpain 1 and its inhibitor calpastatin within breeds of cattle. Genetics (2007) 176:2601–10. doi:10.1534/genetics.107.074328
83. Ron M, Tager-Cohen I, Feldmesser E, Ezra E, Kalay D, Roe B, et al. Bovine umbilical hernia maps to the centromeric end of Bos taurus autosome 8. Anim Genet (2004) 35:431–7. doi:10.1111/j.1365-2052.2004.01196.x
84. Fritz S, Capitan A, Djari A, Rodriguez SC, Barbat A, Baur A, et al. Detection of haplotypes associated with prenatal death in dairy cattle and identification of deleterious mutations in GART, SHBG and SLC37A2. PLoS One (2013) 8:e65550. doi:10.1371/journal.pone.0065550
85. Daetwyler HD, Capitan A, Pausch H, Stothard P, van Binsbergen R, Brøndum RF, et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet (2014) 46:858–65. doi:10.1038/ng.3034
86. Jakob E, Puhan Z. Technological properties of milk as influenced by genetic polymorphism of milk proteins – a review. Int Dairy J (1992) 2:157–78. doi:10.1016/0958-6946(92)90014-D
87. Healy PJ, Harper PA, Dennis JA. Bovine citrullinaemia: a clinical, pathological, biochemical and genetic study. Aust Vet J (1990) 67:255–8. doi:10.1111/j.1751-0813.1990.tb07780.x
88. Dennis JA, Healy PJ, Beaudet AL, O’Brien WE. Molecular definition of bovine argininosuccinate synthetase deficiency. Proc Natl Acad Sci U S A (1989) 86:7947–51. doi:10.1073/pnas.86.20.7947
89. Kipp S, Segelke D, Schierenbeck S, Reinhardt F, Reents R, Wurmser C, et al. A New Holstein Haplotype Affecting Calf Survival. Interbull Bulletin. Orlando, FL: Interbull (2015). p. 49–53.
90. Menzi F, Besuchet-Schmutz N, Fragnière M, Hofstetter S, Jagannathan V, Mock T, et al. A transposable element insertion in APOB causes cholesterol deficiency in Holstein cattle. Anim Genet (2016) 47(2):253–7. doi:10.1111/age.12410
91. Marchi E, Kanapin A, Magiorkinis G, Belshaw R. Unfixed endogenous retroviral insertions in the human population. J Virol (2014) 88:9529–37. doi:10.1128/JVI.00919-14
92. Guldimann C, Gsponer M, Drogemuller C, Oevermann A, Seuberlich T. Atypical H-type bovine spongiform encephalopathy in a cow born after the reinforced feed ban on meat-and-bone meal in Europe. J Clin Microbiol (2012) 50:4171–4. doi:10.1128/JCM.02178-12
93. Clawson ML, Richt JA, Baron T, Biacabe AG, Czub S, Heaton MP, et al. Association of a bovine prion gene haplotype with atypical BSE. PLoS One (2008) 3:e1830. doi:10.1371/journal.pone.0001830
94. Heaton MP, Keele JW, Harhay GP, Richt JA, Koohmaraie M, Wheeler TL, et al. Prevalence of the prion protein gene E211K variant in U.S. cattle. BMC Vet Res (2008) 4:25. doi:10.1186/1746-6148-4-25
95. Richt JA, Hall SM. BSE case associated with prion protein gene mutation. PLoS Pathog (2008) 4:e1000156. doi:10.1371/journal.ppat.1000156
96. Nicholson EM, Brunelle BW, Richt JA, Kehrli ME Jr, Greenlee JJ. Identification of a heritable polymorphism in bovine PRNP associated with genetic transmissible spongiform encephalopathy: evidence of heritable BSE. PLoS One (2008) 3:e2912. doi:10.1371/journal.pone.0002912
97. Greenlee JJ, Smith JD, West Greenlee MH, Nicholson EM. Clinical and pathologic features of H-type bovine spongiform encephalopathy associated with E211K prion protein polymorphism. PLoS One (2012) 7:e38678. doi:10.1371/journal.pone.0038678
98. Greenlee JJ, Greenlee MH. The transmissible spongiform encephalopathies of livestock. ILAR J (2015) 56:7–25. doi:10.1093/ilar/ilv008
99. Schlapfer I, Saitbekova N, Gaillard C, Dolf G. A new allelic variant in the bovine prion protein gene (PRNP) coding region. Anim Genet (1999) 30:386–7. doi:10.1046/j.1365-2052.1999.00526-5.x
100. Seabury CM, Honeycutt RL, Rooney AP, Halbert ND, Derr JN. Prion protein gene (PRNP) variants and evidence for strong purifying selection in functionally important regions of bovine exon 3. Proc Natl Acad Sci U S A (2004) 101:15142–7. doi:10.1073/pnas.0406403101
101. Seabury CM, Womack JE, Piedrahita J, Derr JN. Comparative PRNP genotyping of U.S. cattle sires for potential association with BSE. Mamm Genome (2004) 15:828–33. doi:10.1007/s00335-004-2400-6
102. Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, et al. Analysis of sequence variability of the bovine prion protein gene (PRNP) in German cattle breeds. Neurogenetics (2004) 5:19–25. doi:10.1007/s10048-003-0171-y
103. Sander P, Hamann H, Drögemüller C, Kashkevich K, Schiebel K, Leeb T. Bovine prion protein gene (PRNP) promoter polymorphisms modulate PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem (2005) 280:37408–14. doi:10.1074/jbc.M506361200
104. Juling K, Schwarzenbacher H, Williams JL, Fries R. A major genetic component of BSE susceptibility. BMC Biol (2006) 4:33. doi:10.1186/1741-7007-4-33
105. Brunelle BW, Hamir AN, Baron T, Biacabe AG, Richt JA, Kunkle RA, et al. Polymorphisms of the prion gene promoter region that influence classical bovine spongiform encephalopathy susceptibility are not applicable to other transmissible spongiform encephalopathies in cattle. J Anim Sci (2007) 85:3142–7. doi:10.2527/jas.2007-0208
106. Brunelle BW, Greenlee JJ, Seabury CM, Brown CE II, Nicholson EM. Frequencies of polymorphisms associated with BSE resistance differ significantly between Bos taurus, Bos indicus, and composite cattle. BMC Vet Res (2008) 4:36. doi:10.1186/1746-6148-4-36
107. Brunelle BW, Kehrli ME Jr, Stabel JR, Spurlock DM, Hansen LB, Nicholson EM. Short communication: allele, genotype, and haplotype data for bovine spongiform encephalopathy-resistance polymorphisms from healthy US Holstein cattle. J Dairy Sci (2008) 91:338–42. doi:10.3168/jds.2007-0423
108. Thaller G, Kühn C, Winter A, Ewald G, Bellmann O, Wegner J, et al. DGAT1, a new positional and functional candidate gene for intramuscular fat deposition in cattle. Anim Genet (2003) 34:354–7. doi:10.1046/j.1365-2052.2003.01011.x
109. Hou GY, Yuan ZR, Zhou HL, Zhang LP, Li JY, Gao X, et al. Association of thyroglobulin gene variants with carcass and meat quality traits in beef cattle. Mol Biol Rep (2011) 38:4705–8. doi:10.1007/s11033-010-0605-1
110. Bennett GL, Shackelford SD, Wheeler TL, King DA, Casas E, Smith TP. Selection for genetic markers in beef cattle reveals complex associations of thyroglobulin and casein1-s1 with carcass and meat traits. J Anim Sci (2013) 91:565–71. doi:10.2527/jas.2012-5454
111. Thaller G, Krämer W, Winter A, Kaupe B, Erhardt G, Fries R. Effects of DGAT1 variants on milk production traits in German cattle breeds. J Anim Sci (2003) 81:1911–8. doi:10.2527/2003.8181911x
112. Beribe MJ, Carignano HA, Lopez-Villalobos N, Poli MA, Roldan DL. QTL detection for fat yield on BTA14 using linkage disequilibrium based methods. 10th World Cong Genet Appl Livest Prod. Vancouver, BC (2014). Communication # 617.
113. Wu XX, Yang ZP, Shi XK, Li JY, Ji DJ, Mao YJ, et al. Association of SCD1 and DGAT1 SNPs with the intramuscular fat traits in Chinese Simmental cattle and their distribution in eight Chinese cattle breeds. Mol Biol Rep (2012) 39:1065–71. doi:10.1007/s11033-011-0832-0
114. Tait RG Jr, Shackelford SD, Wheeler TL, King DA, Keele JW, Casas E, et al. CAPN1, CAST, and DGAT1 genetic effects on preweaning performance, carcass quality traits, and residual variance of tenderness in a beef cattle population selected for haplotype and allele equalization. J Anim Sci (2014) 92:5382–93. doi:10.2527/jas.2013-7075
115. Charlier C, Farnir F, Berzi P, Vanmanshoven P, Brouwers B, Vromans H, et al. Identity-by-descent mapping of recessive traits in livestock: application to map the bovine syndactyly locus to chromosome 15. Genome Res (1996) 6:580–9. doi:10.1101/gr.6.7.580
116. Drogemuller C, Distl O. Genetic analysis of syndactyly in German Holstein cattle. Vet J (2006) 171:120–5. doi:10.1016/j.tvjl.2004.09.009
117. Drögemüller C, Leeb T, Harlizius B, Tammen I, Distl O, Höltershinken M, et al. Congenital syndactyly in cattle: four novel mutations in the low density lipoprotein receptor-related protein 4 gene (LRP4). BMC Genet (2007) 8:5. doi:10.1186/1471-2156-8-5
118. Healy PJ, Dennis JA. Molecular heterogeneity for bovine maple syrup urine disease. Anim Genet (1994) 25:329–32. doi:10.1111/j.1365-2052.1994.tb00366.x
119. Dennis JA, Healy PJ. Definition of the mutation responsible for maple syrup urine disease in Poll Shorthorns and genotyping Poll Shorthorns and Poll Herefords for maple syrup urine disease alleles. Res Vet Sci (1999) 67:1–6. doi:10.1053/rvsc.1998.0296
120. Zhang B, Healy PJ, Zhao Y, Crabb DW, Harris RA. Premature translation termination of the pre-E1 alpha subunit of the branched chain alpha-ketoacid dehydrogenase as a cause of maple syrup urine disease in Polled Hereford calves. J Biol Chem (1990) 265:2425–7.
121. Olson TA, Lucena C, Chase CC Jr, Hammond AC. Evidence of a major gene influencing hair length and heat tolerance in Bos taurus cattle. J Anim Sci (2003) 81:80–90. doi:10.2527/2003.81180x
122. Hammond AC, Chase CC Jr, Bowers EJ, Olson TA, Randel RD. Heat tolerance in Tuli-, Senepol-, and Brahman-sired F1 Angus heifers in Florida. J Anim Sci (1998) 76:1568–77. doi:10.2527/1998.7661568x
123. Mariasegaram M, Chase CC Jr, Chaparro JX, Olson TA, Brenneman RA, Niedz RP. The slick hair coat locus maps to chromosome 20 in Senepol-derived cattle. Anim Genet (2007) 38:54–9. doi:10.1111/j.1365-2052.2007.01560.x
124. Dikmen S, Khan FA, Huson HJ, Sonstegard TS, Moss JI, Dahl GE, et al. The SLICK hair locus derived from Senepol cattle confers thermotolerance to intensively managed lactating Holstein cows. J Dairy Sci (2014) 97:5508–20. doi:10.3168/jds.2014-8087
125. Littlejohn MD, Henty KM, Tiplady K, Johnson T, Harland C, Lopdell T, et al. Functionally reciprocal mutations of the prolactin signalling pathway define hairy and slick cattle. Nat Commun (2014) 5:5861. doi:10.1038/ncomms6861
126. Gentry PA, Crane S, Lotz F. Factor XI (plasma thromboplastin antecedent) deficiency in cattle. Can Vet J (1975) 16:160–3.
127. Gentry PA, Black WD. Prevalence and inheritance of factor XI (plasma thromboplastin antecedent) deficiency in cattle. J Dairy Sci (1980) 63:616–20. doi:10.3168/jds.S0022-0302(80)82980-8
128. Liptrap RM, Gentry PA, Ross ML, Cummings E. Preliminary findings of altered follicular activity in Holstein cows with coagulation factor XI deficiency. Vet Res Commun (1995) 19:463–71. doi:10.1007/BF01839334
129. Marron BM, Robinson JL, Gentry PA, Beever JE. Identification of a mutation associated with factor XI deficiency in Holstein cattle. Anim Genet (2004) 35:454–6. doi:10.1111/j.1365-2052.2004.01202.x
130. Page BT, Casas E, Heaton MP, Cullen NG, Hyndman DL, Morris CA, et al. Evaluation of single-nucleotide polymorphisms in CAPN1 for association with meat tenderness in cattle. J Anim Sci (2002) 80:3077–85. doi:10.2527/2002.80123077x
131. Riley DG, Chase CC Jr, Pringle TD, West RL, Johnson DD, Olson TA, et al. Effect of sire on mu- and m-calpain activity and rate of tenderization as indicated by myofibril fragmentation indices of steaks from Brahman cattle. J Anim Sci (2003) 81:2440–7. doi:10.2527/2003.81102440x
132. White SN, Casas E, Wheeler TL, Shackelford SD, Koohmaraie M, Riley DG, et al. A new single nucleotide polymorphism in CAPN1 extends the current tenderness marker test to include cattle of Bos indicus, Bos taurus, and crossbred descent. J Anim Sci (2005) 83:2001–8. doi:10.2527/2005.8392001x
133. Morris CA, Cullen NG, Hickey SM, Dobbie PM, Veenvliet BA, Manley TR, et al. Genotypic effects of calpain 1 and calpastatin on the tenderness of cooked M. longissimus dorsi steaks from Jersey x Limousin, Angus and Hereford-cross cattle. Anim Genet (2006) 37:411–4. doi:10.1111/j.1365-2052.2006.01483.x
134. Cafe LM, McIntyre BL, Robinson DL, Geesink GH, Barendse W, Greenwood PL. Production and processing studies on calpain-system gene markers for tenderness in Brahman cattle: 1. Growth, efficiency, temperament, and carcass characteristics. J Anim Sci (2010) 88:3047–58. doi:10.2527/jas.2009-2679
135. Cafe LM, McIntyre BL, Robinson DL, Geesink GH, Barendse W, Pethick DW, et al. Production and processing studies on calpain-system gene markers for tenderness in Brahman cattle: 2. Objective meat quality. J Anim Sci (2010) 88:3059–69. doi:10.2527/jas.2009-2679
136. Lee SH, Kim SC, Chai HH, Cho SH, Kim HC, Lim D, et al. Mutations in calpastatin and mu-calpain are associated with meat tenderness, flavor and juiciness in Hanwoo (Korean cattle): molecular modeling of the effects of substitutions in the calpastatin/mu-calpain complex. Meat Sci (2014) 96:1501–8. doi:10.1016/j.meatsci.2013.11.026
137. Casas E, Duan Q, Schneider MJ, Shackelford SD, Wheeler TL, Cundiff LV, et al. Polymorphisms in calpastatin and mu-calpain genes are associated with beef iron content. Anim Genet (2014) 45:283–4. doi:10.1111/age.12108
138. Hu ZL, Park CA, Reecy JM. Developmental progress and current status of the animal QTLdb. Nucleic Acids Res (2016) 44:D827–33. doi:10.1093/nar/gkv1233
139. Faculty of Veterinary Science, U. S. Online Mendelian Inheritance in Animals. (2011). Available from: http://omia.angis.org.au/
Keywords: genes, cattle, genetic conditions, beef, dairy, genomics
Citation: Casas E and Kehrli ME Jr. (2016) A Review of Selected Genes with Known Effects on Performance and Health of Cattle. Front. Vet. Sci. 3:113. doi: 10.3389/fvets.2016.00113
Received: 28 June 2016; Accepted: 28 November 2016;
Published: 15 December 2016
Edited by:
Ino Curik, University of Zagreb, CroatiaReviewed by:
James Reecy, Iowa State University, USAHans D. Daetwyler, La Trobe University, Australia
Copyright: © 2016 Casas and Kehrli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eduardo Casas, eduardo.casas@ars.usda.gov
 Eduardo Casas
Eduardo Casas Marcus E. Kehrli Jr.
Marcus E. Kehrli Jr.