Statistical learning analysis in neuroscience: aiming for transparency
- 1 Center for Cognitive Neuroscience, Dartmouth College, Hanover, NH, USA
- 2 Department of Psychological and Brain Sciences, Dartmouth College, Hanover, NH, USA
- 3 Department of Experimental Psychology, University of Magdeburg, Magdeburg, Germany
- 4 Center for Behavioral Brain Sciences, Magdeburg, Germany
Encouraged by a rise of reciprocal interest between the machine learning and neuroscience communities, several recent studies have demonstrated the explanatory power of statistical learning techniques for the analysis of neural data. In order to facilitate a wider adoption of these methods, neuroscientific research needs to ensure a maximum of transparency to allow for comprehensive evaluation of the employed procedures. We argue that such transparency requires “neuroscience-aware” technology for the performance of multivariate pattern analyses of neural data that can be documented in a comprehensive, yet comprehensible way. Recently, we introduced PyMVPA, a specialized Python framework for machine learning based data analysis that addresses this demand. Here, we review its features and applicability to various neural data modalities.
Introduction
In recent years, there has been an increasing interest in applying multivariate pattern (MVP) analysis to neural data, especially from functional magnetic resonance imaging (fMRI; Haynes and Rees, 2006 ; Norman et al., 2006 ; O’Toole et al., 2007 ), more than a decade after the first application in studies employing positron emission tomography (PET; Moeller and Strother, 1991 ; Kippenhahn et al., 1992 ) and fMRI (e.g., McIntosh et al., 1996 ). Employing multivariate statistical learning methods allows refocusing research on how information is encoded, instead of exclusively looking at where it could be detected in the brain (O’Toole et al., 2007 ). Pioneering work on this topic has been done by Haxby et al. (2001) , and subsequently by Hanson et al. (2004) who revealed a combinatorial encoding of object category information in human ventral temporal cortex. Shortly after these initial studies, Kamitani and Tong (2005) showed that the already known fine-grained columnar representation of visual orientation in primary visual cortex is represented in fMRI data, despite a coarser spatial resolution. Along this line, Miyawaki et al. (2008) demonstrated that even more information is contained in the fMRI signal, by being able to reconstruct actual visual stimuli from blood oxygenation level-dependent (BOLD) response patterns in early visual cortex. Unlike the aforementioned studies, their model was able to generalize from unstructured basic visual feature s to geometric shapes and letters.
In addition to the most popular MVP analysis approach (i.e., classification), analysis of the full covariance structure of a dataset can also be used to investigate similarity structures of brain response patterns. This transformation of neural responses into the domain of stimulus space (O’Toole et al., 2007 ) represents a powerful tool for exploratory analysis that can, for example, help to deduce the purpose of a functional processing unit of the brain. Their flexibility even allows for comparative analyses across different species. By employing such a technique, Kriegeskorte et al. (2008) showed striking correlations between the similarity structure of object category representations in the ventral processing stream of humans and monkeys.
Finally, MVP analyses are suitable and have been used to validate computational models of processing streams in the human brain. Kay et al. (2008) , for example, constructed a receptive-field model of early visual processing, and trained it on BOLD-response patterns in early visual cortex, associated with the presentation of over a thousand natural images. They then used this model to identify more than a hundred novel images using a relatively simple nearest-neighbor classification algorithm, hence providing evidence for the plausibility of the model architecture.
This recent progress in neuroscience research was made possible by the work on statistical learning methods that has been done mostly in the NIPS 1 community over the past decades. Despite significant differences in terminology and methods, many cognitive neuroscientists are nevertheless familiar with the concept of MVP classification, because of common roots in connectionism in psychological research in the 1980s and early 1990s. From a conceptual point of view, studying classifier performance when predicting category labels of brain response patterns is very similar to the analysis of behavioral data of humans performing a categorization task (e.g., in a typical two-alternative-forced-choice paradigm). Procedures, such as those originating in signal detection theory (Green and Swets, 1966 ), are well understood and provide familiar measures (e.g., d′ and receiver operating characteristics curves, ROC) to assess the quality of human or algorithmic classifier model performances. More recent research on kernel-based methods in machine learning (ML) shows sometimes striking similarity with categorization models in cognitive psychology, or neuron tuning curves in theoretical neuroscience (Jäkel et al., 2009 ).
However, there are certain problems associated with the adoption of statistical learning methods to answer neuroscientific questions. This article will point out some critical aspects that significantly impact this emerging field.
The Need for Transparency
The examples of studies employing MVP analyses listed in the previous section offer a glimpse at the explanatory power and flexibility of these techniques. However, as with all other methods, one has to be careful to obey limitations and requirements of a particular method, since inappropriate use (e.g., double-dipping; Kriegeskorte et al., 2009 ) or interpretation might completely invalidate conclusions drawn from such analysis. Therefore, a proper assessment of the value of a scientific study requires knowledge about whether employed methods were used appropriately. Obviously, this judgment becomes more difficult with increasing complexity of the analysis procedure.
The use of terms like “mind-reading” or “decoding” may be taken as an indication that MVP analyses are automatically deciphering the encoding of information in the brain. However, the generalization accuracy of a model alone (even if it is perfect) does not justify concluding that it identified the true neural signal of an underlying cognitive process. For example, a classifier is simply distinguishing between abstract patterns. Without an appropriate experimental design and further investigation, it remains unknown what stimulus property (if any) is reflected in the data. In the context of psychological experiments, confounds always limit the interpretability of experimental results. However, the enormous flexibility of statistical learning techniques theoretically allows them to pick up any signal in a dataset (e.g., differences in stimulation frequency). Consequently, studies employing these methods have to be carefully planned to ensure their validity, but (even given an appropriate experimental design) the suitability assessment of methods remains difficult if they are not a part of the standard toolbox of scientists and reviewers in a certain field, since in this case there is no common ground to base an evaluation on.
Unfortunately, this is the current situation of MVP analysis techniques of fMRI data. For the conventional univariate statistical parametric mapping (SPM) based approach, there is a huge amount of literature that allows one to derive at least a reasonable guess of many parameters that have to be considered in each analysis. Based on this literature, any scientist evaluating a particular study should be able to decide whether a reasonable spatial filtering kernel was used during processing, or whether the data was modeled with an appropriate hemodynamic response function. For MVP analysis of fMRI, there is only a handful of articles concerned with the evaluation of different methods. This is, of course, not very surprising, since the total number of studies using this approach is negligible in comparison to, at least, 15 years of very productive SPM-based fMRI data analysis.
There is an increasing number of studies that employ MVP analysis aiming to answer actual scientific questions despite the absence of a tested set of good practices. Ideally, the validity of such studies is established by replication. If an effect is found using similar or even different methods, by different research groups using different data acquisition equipment, its existence will generally be accepted. However, in the context of MVP analyses of fMRI data, replicating a study is hindered by at least two main factors. First, in contrast to the ML community, datasets are typically not available to the general public. The second factor is that published studies generally only contain a verbal description of the applied analysis.
This second factor is much less important for studies employing conventional SPM-based fMRI data analysis, since the majority is performed by using one of the popular fMRI toolkits, like AFNI 2 (Cox, 1996 ), BrainVoyager 3 (Goebel et al., 2006 ), FSL 4 (Smith et al., 2004 ), or SPM 5 (Friston et al., 1994 ). The behavior of these toolkits is known, they are available to the whole research community, and the algorithms they implement are published in peer-reviewed publications. All these aspects allow one to summarize an analysis by listing relatively few parameters.
For an MVP analysis the situation is different. In the absence of an established toolkit for neuroscientific research, an analysis generally involves the combination of many different tools, combined with custom, usually unpublished code. For a researcher intending to replicate a study, translating a verbal – potentially incomplete or too superficial – description into running analysis code is a lengthy and error-prone task that turns a replication attempt into a costly project.
To make a replication effort worthwhile for the majority of MVP analysis-based studies, analysis descriptions should be provided in a form that captures every detail of a procedure and is nevertheless comprehensible by scientists in the field. Ideally, a publication should be accompanied by the actual source code that was used to run an analysis, in addition to a verbal description (Hanke et al., 2009b ). The source code, by definition, provides the highest possible descriptive level. Access to the source code can immediately facilitate validation and replication efforts, enabling the potential for timely feedback with respect to newly developed analysis strategies, while simultaneously fostering the use of successful methods.
Attracting Researchers to Fathom the Full Potential of MVP Analysis
Up to now, only a small fraction of the available algorithms and procedures originating from research on statistical learning has been tested regarding their applicability to neuroimaging data, and cognitive science research questions and paradigms. Potential implications of a particular classification algorithm, or feature selection procedure on the interpretation of the results, are yet to be fully explored. A systematic evaluation and formulation of guidelines for MVP analyses of neural data represents a herculean task that requires the joint effort of the neuroscience community. Fortunately, an initial set of articles providing an overview of various important aspects of MVP analysis for different target audiences has appeared (e.g., Jäkel et al., 2009 ; Mur et al., 2009 ; Pereira et al., 2009 ). However, while there are versatile ML libraries and statistical learning frameworks, such as Weka 6 (Hall et al., 2009 ) and Shogun 7 (Sonnenburg et al., 2006 ), none of them is specifically geared toward the analysis of neuroimaging data. In addition, other neuroimaging-aware MVPA software and pipeline packages described in the literature are closed-source, covered by a restrictive license, or intentionally focused on a specific analysis technique (Rex et al., 2003 ; McIntosh et al., 2004 ; Strother et al., 2004 ).
PyMVPA
To facilitate the exploration, application, and evaluation of MVP analysis in the context of neuroimaging research, we recently introduced a free and open-source, cross-platform analysis framework, called PyMVPA 8 (Hanke et al., 2009a ,b ). It aims to bridge the gap between the rich set of algorithms and procedures originating from research on statistical learning, and the specific properties and requirements of neuroimaging data.
PyMVPA uses the Python 9 programming language to achieve this goal. The ability of this generic scripting environment to access a huge code base in various languages, combined with its syntactical simplicity, makes it an ideal tool for implementing and sharing ideas among scientists from numerous fields and with heterogeneous methodological backgrounds. The recent Python in Neuroscience 10 special issue of the journal Frontiers in Neuroinformatics listed a number of versatile neuroscience projects and software libraries that make use of Python, and extend it as a rich high-performance computing environment.
The approach of PyMVPA is to combine as many of the available building blocks as possible into a consistent framework that allows one to build processing pipelines of arbitrary complexity. Whenever feasible, we used existing software implementations in various programming languages, instead of rewriting algorithms in Python itself. This strategy strives to focus users on a single implementation of an algorithm and by that, increase the likelihood to detect and fix errors.
A key feature of PyMVPA is that it abstracts the peculiarities of neuroimaging datasets and exposes them as a generic data array that is compatible with most ML software packages. However, information relevant to the neuroscience context (e.g., spatial metrics of fMRI voxels) is retained and accessible within the framework. This allows for transparent conversion of datasets or results – from a generic representation (e.g., a numerical vector) into the native data space (e.g., a brain volume).
This aspect of PyMVPA offers the possibility to write and test “neuroscience-aware” algorithms that are suitable to address the underlying goal of cognitive neuroscience: to understand how the brain performs the information processing that results in complex behavior. That goal has important implications for almost every single step of an analysis procedure. Consider, for example, a feature selection algorithm. In ML research, feature selection is usually performed to remove unimportant information from a dataset that does not improve, or even has a negative impact on, the generalization of a particular classifier. In the neuroscience context, however, the primary focus is not on the accuracy level, but on the structure and origin of the information that allows for correct predictions to be made – the accuracy simply has to be reasonably high to allow for an interpretation of the model at all. Careless removal of features that provide redundant information can have significant side-effects on the conclusions that can be drawn from an analysis. PyMVPA is explicitly designed to address the demand for methodologies that acknowledge the specifics of cognitive neuroscience research. The mentioned exposure of spatial data metrics inside the framework, for example, allows to easily develop “neuroscience-aware” methods that aggregate information from generic feature-based algorithms to derive decisions regarding the selection of whole functional areas.
All processing units in the PyMVPA framework, such as classifiers or other dataset measures, are designed to have compatible interfaces that allow for modular adjustments to an analysis pipeline. For example, a particular statistical learning algorithm can be replaced for another, without having to adjust any other part of the intended analysis pipeline (e.g., preprocessing, feature selection, or cross-validation procedures). This property makes it very easy to benchmark the performance of various algorithms on a particular dataset – which represents a key task when evaluating generic algorithms in the neuroscience domain.
Additionally, PyMVPA provides extensive debugging and logging functionality, aggregation, and intermediate storage of results; plus convenient ways to interface with generic, as well as domain-specific visualization software. Altogether, the availability of these basic housekeeping features allow researchers to focus on aspects important for neuroscience research, instead of low-level software engineering tasks.
Despite its high-level programming interface (Hanke et al., 2009a ), PyMVPA nevertheless allows one to modify its behavior in great detail. Its modular architecture is easily extensible with novel algorithms and support for additional data formats. Like PyMVPA itself, virtually all software that it interfaces with is free and open-source. This makes it an ideal environment for in-depth verification, validation, and comparison of existing and newly developed algorithms.
Uniform Analysis of Various Data Modalities
Statistical learning methods are generic and applicable to a wide range of datasets. PyMVPA acknowledges this fact and offers an abstraction layer that can represent data regardless of its original dimensionality or dataspace metrics, while nevertheless exposing these properties inside the framework. Using this functionality, we have shown in Hanke et al. (2009b) that PyMVPA can be used to implement a unified analysis pipeline for a whole spectrum of neural data modalities . Despite modality-specific data properties, PyMVPA allowed for easy and straightforward initial preprocessing (e.g., input, detrending, normalization), and analysis with statistical learning methods (Figure 1 ). To address the original research questions, the results could be conveniently visualized and interpreted in the original domain-specific data space. Using this uniform approach, we were not only able to replicate previous findings, but also to obtain additional, sometimes thought-provoking results. To encourage other researchers to verify and extend the suggested methodology, the complete source code and a sample fMRI dataset were made available alongside the publication.
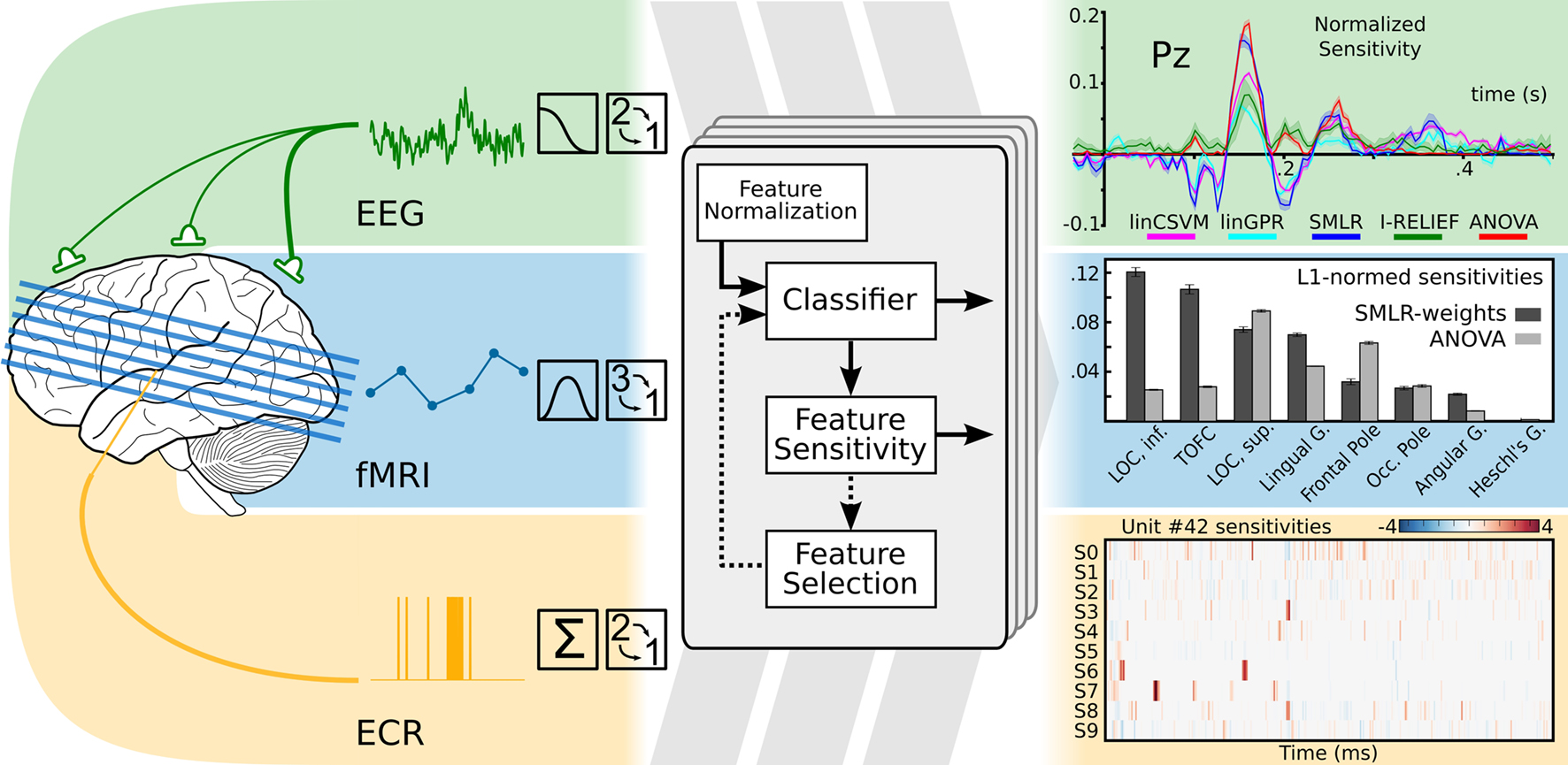
Figure 1. Modality-independent data analysis with PyMVPA. On the left side: typical preprocessing steps for data from electroencephalography (EEG), functional magnetic resonance imaging (fMRI), and extra-cellular recordings (ECR) are shown. After initial modality-specific preprocessing, PyMVPA transforms data into a simple array representation that is compatible with generic machine learning software implementations. At the final stage of an analysis, PyMVPA allows for easy back-projection of the results into the original modality-specific data space. Examples are modified from Hanke et al. (2009b) .
Data modality-independent analysis opens the door to combine the power of the plethora of available invasive and non-invasive data recording techniques. While they are all measuring reflections of the same underlying neural signals, each of them is offering a unique set of properties in terms of spatio-temporal resolution, signal to noise, data acquisition cost, applicability to humans, and the corresponding neural correlates that result from the measurement process. Neuroscientists often focus on only one or a smaller subset of these neural modalities, partly due to the kinds of questions investigated, and partly due to the cost of learning to analyze data from these different modalities. The availability of a generic framework, such as PyMVPA, that provides a uniform processing pipeline, can encourage the exchange of available methodologies among research communities specialized in the analysis of a particular data modality.
Conclusions
The emerging field of MVP analysis of neural data is beginning to complement the established analysis techniques, and has great potential for novel insights into the functional architecture of the brain. Even if an MVP analysis is not a mind-reader (Logothetis, 2008 ), it undoubtedly represents a major step forward in the analysis of the brain function. Its flexibility acknowledges the complexity of neural signals, and hence can help to advance the understanding of brain function. However, there are many open questions on how the wealth of statistical learning algorithms can be applied optimally in this domain. Although evaluating use cases and identifying potential pitfalls in the neuroscience context is an ambitious and demanding task, it urgently needs to be done to ensure valid analysis and unbiased conclusions.
PyMVPA is a generic framework that is explicitly tailored toward MVP analyses of neural data. Its versatility on data from various modalities has already been shown by us and others (Hanke et al., 2009b ; Sun et al., 2009 ). The offered transparency in expressing complex processing pipelines hopefully will facilitate the systematic evaluation of statistical learning methods in the neuroscience context, and will serve as a solid foundation for collaborative and derivative research efforts.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Michael Hanke was supported by the German Academic Exchange Service (grant: PPP-USA D/05/504/7). Yaroslav O. Halchenko was supported by the National Science Foundation (grant: SBE 0751008) and the James McDonnell Foundation (grant: 220020127). Stefan Pollmann was supported by Deutsche Forschungsgemeinschaft (PO 548/8-1).
Footnotes
- ^ Neural Information Processing Systems http://nips.cc/
- ^ http://afni.nimh.nih.gov/afni
- ^ http://www.brainvoyager.com
- ^ http://www.fmrib.ox.ac.uk/fsl
- ^ http://www.fil.ion.ucl.ac.uk/spm
- ^ http://www.cs.waikato.ac.nz/ml/weka
- ^ http://www.shogun-toolbox.org
- ^ http://www.pymvpa.org
- ^ http://www.python.org
- ^ http://www.frontiersin.org/neuroinformatics/specialtopics/8/
Key Concepts
Statistical learning: A field of science related to ML that aims at exploiting statistical properties of data to construct robust models, and to assess their convergence and generalization performances.
Feature: A variable that represents a dimension in a dataset. This might be the output of a single sensor, such as a voxel, or a refined measure reflecting a specific aspect of data, such as a specific spectral component.
Classifier: A model that maps an arbitrary feature space into a discrete set of labels.
Machine learning: A field of computer science that aims at constructing methods, such as classifiers, to integrate available knowledge extracted from existing data.
Generalization: An ability of a model to perform reliably well on any novel data in the given domain.
Feature selection: A technique that targets detection of features relevant to a given problem, so that their selection improves generalization of the constructed model.
Neural data modality: A reflection of neural activity collected using some available instrumental method (e.g., EEG, fMRI, etc.).
Cross-validation: A technique to assess the generalization of the constructed model by the analysis of accuracy of the model predictions on a presumably independent dataset.
References
Cox, R. W. (1996). AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–173.
Friston, K., Jezzard, P., and Turner, R. (1994). Analysis of functional MRI time-series. Hum. Brain Mapp. 1, 153–171.
Goebel, R., Esposito, F., and Formisano, E. (2006). Analysis of functional image analysis contest (FIAC) data with Brainvoyager QX: from single-subject to cortically aligned group general linear model analysis and self-organizing group independent component analysis. Hum. Brain Mapp. 27, 392–401.
Green, D. M., and Swets, J. A. (1966). Signal Detection and Recognition by Human Observers. New York, Wiley.
Hall, M., Frank, E., Holmes, G., Pfahringer, B., Reutemann, P., and Witten, I. H. (2009). The WEKA data mining software: an update. SIGKDD Explor. 11, 10–18.
Hanke, M., Halchenko, Y. O., Sederberg, P. B., Hanson, S. J., Haxby, J. V., and Pollmann, S. (2009a). PyMVPA: a Python toolbox for multivariate pattern analysis of fMRI data. Neuroinformatics 7, 37–53.
Hanke, M., Halchenko, Y. O., Sederberg, P. B., Olivetti, E., Fründ, I., Rieger, J. W., Herrmann, C. S., Haxby, J. V., Hanson, S. J., and Pollmann, S. (2009b). PyMVPA: a unifying approach to the analysis of neuroscientific data. Front. Neuroinformatics 3:3. doi: 10.3389/neuro.11.003.2009.
Hanson, S., Matsuka, T., and Haxby, J. (2004). Combinatorial codes in ventral temporal lobe for object recognition: Haxby (2001) revisited: is there a “face” area? Neuroimage 23, 156–166.
Haxby, J. V., Gobbini, M. I., Furey, M. L., Ishai, A., Schouten, J. L., and Pietrini, P. (2001). Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science 293, 2425–2430.
Haynes, J.-D., and Rees, G. (2006). Decoding mental states from brain activity in humans. Nat. Rev. Neurosci. 7, 523–534.
Jäkel, F., Schölkopf, B., and Wichmann, F. A. (2009). Does cognitive science need kernels? Trends Cogn. Sci. 13, 381–388.
Kamitani, Y., and Tong, F. (2005). Decoding the visual and subjective contents of the human brain. Nat. Neurosci. 8, 679–685.
Kay, K. N., Naselaris, T., Prenger, R. J., and Gallant, J. L. (2008). Identifying natural images from human brain activity. Nature 452, 352–355.
Kippenhahn, J. S., Barker, W. W., Pascal, S., Nagel, J., and Duara, R. (1992). Evaluation of a neural-network classifier for pet scans of normal and Alzheimer’s disease subjects. J. Nucl. Med. 33, 1459–1467.
Kriegeskorte, N., Mur, M., Ruff, D. A., Kiani, R., Bodurka, J., Esteky, H., Tanaka, K., and Bandettini, P. A. (2008). Matching categorical object representations in inferior temporal cortex of man and monkey. Neuron 60, 1126–1141.
Kriegeskorte, N., Simmons, W. K., Bellgowan, P. S. F., and Baker, C. I. (2009). Circular analysis in systems neuroscience: the dangers of double dipping. Nat. Neurosci. 12, 535–40.
McIntosh, A. R., Bookstein, F. L., Haxby, J. V., and Grady, C. L. (1996). Spatial pattern analysis of functional brain images using partial least squares. Neuroimage 3, 143–57.
McIntosh, A. R., Chau, W. K., and Protzner, A. B. (2004). Spatiotemporal analysis of event-related fmri data using partial least squares. Neuroimage 23, 764–75.
Miyawaki, Y., Uchida, H., Yamashita, O., Sato, M., Morito, Y., Tanabe, H. C., Sadato, N., and Kamitani, Y. (2008). Visual image reconstruction from human brain activity using a combination of multiscale local image decoders. Neuron 60, 915–29.
Moeller, J. R., and Strother, S. (1991). A regional covariance approach to the analysis of functional patterns in positron emission tomographic data. J. Cereb. Blood Flow Metab. 11, 121–135.
Mur, M., Bandettini, P. A., and Kriegeskorte, N. (2009). Revealing representational content with pattern-information fMRI – an introductory guide. Soc. Cogn. Affect. Neurosci. 4, 101–109.
Norman, K. A., Polyn, S. M., Detre, G. J., and Haxby, J. V. (2006). Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends Cogn. Sci. 10, 424–430.
O’Toole, A. J., Jiang, F., Abdi, H., Penard, N., Dunlop, J. P., and Parent, M. A. (2007). Theoretical, statistical, and practical perspectives on pattern-based classification approaches to the analysis of functional neuroimaging data. J. Cogn. Neurosci. 19, 1735–1752.
Pereira, F., Mitchell, T., and Botvinick, M. (2009). Machine learning classifiers and fMRI: a tutorial overview. Neuroimage 45, 199–209.
Rex, D. E., Ma, J. Q., and Toga, A. W. (2003). The LONI pipeline processing environment. Neuroimage 19, 1033–48.
Smith, S. M., Jenkinson, M., Woolrich, M. W., Beckmann, C. F., Behrens, T. E. J., Johansen-Berg, H., Bannister, P. R., De Luca, M., Drobnjak, I., Flitney, D. E., Niazy, R. K., Saunders, J., Vickers, J., Zhang, Y., De Stefano, N., Brady, J. M., and Matthews, P. M. (2004). Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23, 208–219.
Sonnenburg, S., Raetsch, G., Schaefer, C., and Schoelkopf, B. (2006). Large scale multiple kernel learning. J. Mach. Learn. Res. 7, 1531–1565.
Strother, S., La Conte, S., Kai Hansen, L., Anderson, J., Zhang, J., Pulapura, S., and Rottenberg, D. (2004). Optimizing the fmri data-processing pipeline using prediction and reproducibility performance metrics: I. A preliminary group analysis. Neuroimage 23, 196–207.
Sun, D., van Erp, T. G., Thompson, P. M., Bearden, C. E., Daley, M., Kushan, L., Hardt, M. E., Nuechterlein, K. H., Toga, A. W., and Cannon, T. D. (2009). Elucidating an mri-based neuroanatomic biomarker for psychosis: classification analysis using probabilistic brain atlas and machine learning algorithms. Biol. Psychiatry 66, 1055–1060.
Keywords: machine learning, Python, PyMVPA, MVPA
Citation: Hanke M, Halchenko YO, Haxby JV and Pollmann S (2010) Statistical learning analysis in neuroscience: aiming for transparency. Front. Neurosci. 4,1: 38-43 doi:10.3389/neuro.01.007.2010
Received: 25 September 2009;
Paper pending published: 14 November 2009;
Accepted: 24 November 2009;
Published online: 15 May 2010
Edited by:
Rolf Kötter, Radboud University Nijmegen, NetherlandsReviewed by:
Stephen C. Strother, Baycrest, Canada; University of Toronto, CanadaRolf Kötter, Radboud University Nijmegen, Netherlands
Copyright: © 2010 Hanke, Halchenko, Haxby and Pollmann. This is an open-access publication subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Michael Hanke & Dr. Yaroslav O. Halchenko, Dartmouth College, Department of Psychological and Brain Sciences, 419 Moore Hall, Hanover, NH, 03755, USA, michael.hanke@gmail.com, yaroslav.o.halchenko@onerussian.com