Cloning and phylogenetic analysis of NMDA receptor subunits NR1, NR2A and NR2B in Xenopus laevis tadpoles
Watson School of Biological Sciences, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA
N-methyl-D-aspartate receptors (NMDARs) play an important role in many aspects of nervous system function such as synaptic plasticity and neuronal development. NMDARs are heteromers consisting of an obligate NR1 and most commonly one or two kinds of NR2 subunits. While the receptors have been well characterized in some vertebrate and invertebrate systems, information about NMDARs in Xenopus laevis brain is incomplete. Here we provide biochemical evidence that the NR1, NR2A and NR2B subunits of NMDARs are expressed in the central nervous system of X. laevis tadpoles. The NR1-4a/b splice variants appear to be the predominant isoforms while the NR1-3a/b variants appear to be expressed at low levels. We cloned the X. laevis NR2A and NR2B subunits and provide a detailed annotation of their functional domains in comparison with NR2A and NR2B proteins from 10 and 13 other species, respectively. Both NR2A and NR2B proteins are remarkably well conserved between species, consistent with the importance of NMDARs in nervous system function.
N-methyl-D-aspartate receptors (NMDARs) are ligand-gated ionotropic glutamate receptors that are important mediators for neuronal events such as synaptic plasticity, learning and memory, neuronal development and circuit formation, and have been implicated in various neuronal disorders (Cull-Candy et al., 2001
; Dingledine et al., 1999
; Hua and Smith, 2004
; Riedel et al., 2003
; Waxman and Lynch, 2005
). NMDARs are heteromers consisting of two obligate NR1 subunits and two NR2 (NR2A-D) or NR3 subunits (NR3A-B) (Cull-Candy and Leszkiewicz, 2004
). In addition to the subunit diversity, the NR1 subunit is alternatively spliced yielding eight possible isoforms that are determined by the inclusion or deletion of exons 5, 21 and 22, called N1, C1 and C2 respectively (Zukin and Bennett, 1995
). The spliced exons not only modulate the biophysical properties of the receptor (Traynelis et al., 1995
, 1998
; Zhang et al., 1994
; Zheng et al., 1994
) but also differentially affect the trafficking behavior of NR1 (Ehlers et al., 1995
; Holmes et al., 2002
; Mu et al., 2003
; Standley et al., 2000
), while the NR2 subunits determine the kinetics of the receptor as well as its trafficking behavior (Barria and Malinow, 2002
; Monyer et al., 1994
; Vicini et al., 1998
). The examination of sequence conservation across species provides an indication of critical amino acids and domains required for NMDAR function in nervous system plasticity.
Expression of the NR1 isoforms and the NR2 subunits is developmentally and spatially regulated (Laurie and Seeburg, 1994
; Monyer et al., 1994
), indicating a functional significance for specific NMDAR subunit compositions during nervous system development. While NMDARs have been cloned and very well studied in other vertebrate (Cox et al., 2005
; Dingledine et al., 1999
; Laurie et al., 1997
; Monyer et al., 1994
; Moriyoshi et al., 1991
; Zarain-Herzberg et al., 2005
) and invertebrate systems (Brockie et al., 2001
; Xia et al., 2005
), NMDARs in Xenopus laevis have only been partially characterized (Schmidt et al., 2006
; Soloviev et al., 1996
). The NR1 splice variants NR1-4a and -4b were the only splice variants cloned from an adult frog cDNA library (Soloviev et al., 1996
). It is unknown, however, if this is also applicable for developing X. laevis tadpoles. Electrophysiological studies in X. laevis optic tectum suggest the presence of NR2A- and NR2B-containing NMDARs (Aizenman and Cline, 2007
; Cline et al., 1996
), and the cloning of X. laevis NR2B has recently been reported (Schmidt and Hollmann, 2008
), however, biochemical evidence of the presence of the NR2A and NR2B subunits in X. laevis brain or the cloning of the complete NR2A sequence has not been described yet.
Here, we provide biochemical evidence for the presence of NR1, NR2A and NR2B in the central nervous system of X. laevis tadpoles. Furthermore, we characterized the NR1 splice variants in the developing tadpole, and confirmed the predominance of the NR1-4a/b isoforms but also found low-level expression of the NR1-3a/b isoforms. We cloned the X. laevis NR2A and NR2B subunits and analyzed their phylogenetic relationships with NR2A and NR2B proteins from other species. A detailed annotation of the functional residues between species revealed a remarkably high degree of sequence conservation of X. laevis NR2A and NR2B, suggesting that analysis of NMDAR function in the nervous system of X. laevis is likely to provide important insights into aspects of NMDAR function that span multiple phyla.
All chemicals were from Sigma, unless otherwise noted. PCR was done in a Mastercycler Gradient PCR machine (Eppendorf). Tadpoles were anaesthetized in 0.02% MS-222.
Western Blot
All experimental procedures were approved by Cold Spring Harbor Laboratory IACUC. Whole brains of stage 47/48 tadpoles were dissected on dry ice and homogenized in lysis buffer (in mM: 10 Tris pH7.4, 60 octyl glucoside, 1 EGTA pH8, 0.5 DTT, 0.5 PMSF, 5 µg/ml leupeptin, 20 µg/ml soybean trypsin inhibitor, 0.1% SDS, 1% Triton X100). An equal amount of protein per lane was separated by 8% SDS-PAGE and transferred onto nitrocellulose membrane that were probed with primary antibodies at 1:100–500: α-NR1, α-NR2B (both BD PharMingen), α-NR2A (crude rabbit serum JH 1817, gift from the Huganir Lab), α-NR1-C1 (crude rabbit serum JH 2079, gift from the Huganir Lab), α-NR1-N1, α-NR1-C2 and α-NR1-C2’ (all three Novus Biologicals) and incubated with the HRP-conjugated α-goat or α-mouse secondary antibody (Bio-Rad, 1:3000).
RT-PCR-Analysis
Total RNA was extracted from whole brains of stage 47/48 tadpoles (TRIZOL, Invitrogen), the mRNA was prepared with the Poly(A) Quik mRNA Isolation Kit (Stratagene) and reverse transcribed with the C. therm. Polymerase (Roche). The C-terminal ends of NR1 cDNA were amplified with TaqBead Hot Start Polymerase (Promega) according to the manufacturer’s protocol with primers 5′ and 3′ to the C-terminal splice sites based on the published X. laevis NR1 sequence (GenBank accession no.: X94156), 5′ primer [CAAGGATGCCCGTAGGAAG], 3′ primer [CAAACGATCACGTGAGACTG] for 94°C 2 min, 31 cycles of [94°C 30 s, 60°C 30 s, 72°C 3 min 15 s], 72°C 5 min 50 s. In addition, the same PCR reaction was performed on three different cDNA libraries present in the lab. The PCR product included varying fragment sizes depending on the exon sizes: C1 + C2 561 bp, C1 + C2′ 516 bp, C2 447 bp, C2′ 402 bp. The fragments were inserted with a TA-Cloning Kit (Promega) into the pGEM-T vector and sequenced.
Cloning of Xenopus NR2A and NR2B – cDNA Library Screen, 5′ and 3′ Race
The probes for the library screen were a cocktail of the full-length rat NR2A and NR2B cDNAs and PCR-amplified 450–600 bp fragments that cover the entire length of rat NR2A and rat NR2B cDNAs. Eight primer pairs were chosen for NR2A, and nine for NR2B. The PCR fragments were amplified with TaqBead Polymerase (Promega) at 94°C 2 min, 30 cycles [94°C 30 s, 68°C 30 s, 72°C 3 min], 72°C 5 min. The NR2A full-length probe was excised from pCIGFPNR2A with NheI/NotI/SpeI after GFP, to yield a 5136-bp fragment. The NR2B full-length probe was excised from pCIGFPNR2B with EcoRI/NheI/SpeI after GFP, to yield a 4816-bp fragment. We screened 500,000 pfu containing a cDNA Lambda Zap I library made from stage 50–56 Xenopus tadpole brains (Lambda ZAP Premade Library manual; Stratagene). The probes were labeled with [32P] dCTP with the RediPrimeII random prime labeling system (Amersham) and used at 0.5–2 × 106 cpm/ml. Lifts were hybridized, and cDNA excision from positive plaques yielded a 1.7- and 2.3-kb fragment, identified by BLAST to be Xenopus NR2B and NR2A, respectively. 5′ and 3′ RACE was performed using the SMART RACE cDNA Amplification kit (Clontech) to assemble the full-length sequences in silico. The GenBank accession nos. are EU275164 (NR2A) and EU275164 (NR2B).
Bioinformatic Analysis
The amino acid sequences of NR2A and NR2B from different species were obtained from NCBI’s EntrezProtein, with the links obtained from a BLAST search or assembled by hand using BLAT on UCSC’s Genome Bioinformatics page. If the entire sequence could not be fully reconstructed, then the partial sequence was used. The GenBank accession nos. for NR2A were: Chimp - AAV74321, Dog - XM_547132, Human - NP_000824, Mouse - BAA01069, Rat - NP_036705 and Squirrel Monkey - AAV74276. Chicken, Cow, Opossum and X. tropicalis NR2A protein sequences were assembled by hand. For NR2B, the GenBank accession nos. were: Cow - XP_617989, Dog - NP_001008719, Electric Fish - AAN65280, Human - NP_000825, Mouse - NP_032197 and Rat - NP_036706. Chicken, Chimp, Fugu, Opossum, Tetraodon, Rhesus Macaque and X. tropicalis NR2B protein sequences were assembled by hand. The accession nos. for Caenorhabditis elegans and Drosophila melanogaster NR2 were NP_506694, and AAT40461, respectively. Phylogenetic relationships were analyzed using the Neighbor Joining algorithm on MacVector (Accelrys). Distances were calculated using the “Uncorrected (“p”)” option. Confidence values of the nodes were determined by bootstrapping with 1000 repetitions.
Detection of NR1, NR2A and NR2B in X. laevis Tadpole Brain
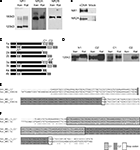
In order to provide biochemical evidence for the presence of NR1, NR2A and NR2B subunits in X. laevis brain, we performed western blots on stage 47/48 tadpoles. Since antibodies generated against X. laevis-specific NMDAR subunits are not available, we probed with antibodies generated against the rat homologs and found cross-reactive bands for X. laevis NR1 (∼120 kD), NR2A (∼180 kD) and NR2B (∼180 kD) that run approximately at the same size as the bands found in rat whole brain lysate (Figure 1
A). The X. laevis NR1 protein is slightly smaller than rat NR1. This could be due to the predominance of the smallest NR1 splice variant, NR1-4b, in X. laevis (Soloviev et al., 1996
; this paper), which is less abundant in rat (Durand et al., 1992
, 1993
; Hollmann et al., 1993
; Sugihara et al., 1992
). We confirmed the cross-reactivity of the NR1 and NR2A antibodies by expressing the X. laevis NR1 (Soloviev et al., 1996
) and X. laevis NR2A cDNAs in heterologous cells (Figure 1
B). This biochemical analysis together with previous electrophysiological data (Aizenman and Cline, 2007
; Cline et al., 1996
) indicates that NR1, NR2A and NR2B are abundantly expressed X. laevisbrain.
Figure 1. Detection of NMDAR subunits and splice variants in X. laevis tadpole brain. (A) Immunoblots for NR1, NR2A and NR2B were done on whole brain lysates of stage 47/48 tadpoles (Xen) with antibodies generated against homologous rat proteins. Immunoreactive bands of about the same size as in rat whole brain lysate (Rat) were found for all three subunits. (B) Western blots of whole cell lysates of HEK293 cells transfected with X. laevis the NR1-4a/b splice variant or NR2A cDNA, and their mock-transfected controls. (C) Cartoon of NR1 splice variants and the alternatively spliced exons (N1, C1, C2) and C-terminal ends (C2′) that they contain. (D) Cross-reactive bands for exons N1 and C1, as well as alternative C-terminal end C2′ in X. laevis were detected with antibodies to rat homologs by Western blot on whole brain extracts from stage 47/48 tadpoles and rat. No cross-reactive band was detected for C2. (E,F) C-terminal ends of NR1 mRNAs were amplified by RT-PCR and PCR with primer pairs 5′ to the spliced region and in the 3′UTR. (E) Twenty-six out of 29 nucleotide sequences contained only C2′ (Xen_NR1_C2′). They aligned perfectly to the published X. laevis NR1 sequence (Xen_NR1_X94156). (F) Two out of 29 nucleotide sequences contained C1 and C2′ (Xen_NR1_C1_C2′), indicating that NR1-3a/b also exists in X. laevis. The alignment to rat NR1-3a (Rat_NR1-3a) shows a very high degree of sequence conservation. Identical residues are marked with (*).
Characterization of NR1 Splice-Variants in X. laevis
We further characterized which NR1 splice isoforms are expressed in X. laevis tadpoles and performed Western blots on whole brain extracts of stage 47 tadpoles using antibodies generated against rat NR1 isoforms containing the alternatively spliced exons N1, C1, C2 and the alternative C-terminal end C2′ (Figure 1
C). We found cross-reactive bands of approximately 120 kD in size for NR1 proteins containing the N-terminal exon N1, as well as the C-terminal exons C1 and C2′, but not for the C2 cassette (Figure 1
D). These results suggest that the splice variants NR1-3a, -3b, -4a and -4b (Figure 1
C) are expressed in X. laevis brain.
In addition we analyzed the NR1 splice variant component in X. laevis by RT-PCR with mRNA from stage 47 tadpoles, as well as PCR from three different X. laevis cDNA libraries. To amplify all possible C-terminal variants, we used X. laevis-specific primers (Soloviev et al., 1996
) targeted to the common regions 5′ and 3′ to the C-terminal splice sites. Sequencing of the subcloned PCR fragments showed that 26 out of 29 sequenced fragments contained only the C2′ cassette. The sequences aligned perfectly to the published X. laevis NR1 sequence (Figure 1
E). Two sequence fragments contained full-length C1 and C2′ cassettes, which were 85% identical at the nucleotide level to rat NR1-3a (up to the stop codon; Figure 1
F). Another sequence contained only a fragment of C1 and C2′ and was thus not classified. Therefore, the majority of mRNAs in X. laevis is either the NR1-4a or -4b splice variant.
Together, the Western blot and the RT-PCR experiments suggest that NR1-4a/b are the most abundantly expressed splice variants of NR1 in X. laevis, while NR1-3a/b are expressed at low levels. We were unable to detect NR1-1a/b and NR1-2a/b by western blot or RT-PCR, but we cannot rule out that they could be expressed at very low levels or at a different developmental stage. These results extend the previous findings (Soloviev et al., 1996
) by demonstrating that NR1-4a/b is the most abundant splice isoform in X. laevis tadpoles.
Cloning of X. laevis NR2A and NR2B and Phylogenetic Analysis
Sequence information on the NR2 subunits has recently been reported for the NR2B (Schmidt and Hollmann, 2008
), but is still lacking for the NR2A subunit. In addition, their degree of sequence conservation between X. laevis and other species has not been described. To determine the sequence of X. laevis NR2A and NR2B we performed a cDNA library screen, using a cDNA library made from X. laevis tadpoles and a cocktail of probes that was specific to the entire rat cDNA sequence of each NR2A and NR2B. We obtained a 2.3- and 1.7-kb fragment for NR2A and NR2B, respectively, and completed the sequence information making use of 5′ and 3′ RACE. We obtained 5041 nucleotides for NR2A, 4353 of which comprise the coding sequence. This translates into 1451 amino acids, which is slightly smaller than rat NR2A, which is 1464 amino acids long. For NR2B 4952 nucleotides in total were obtained, of which 4347 are the coding sequence. The protein is therefore made up of 1449 amino acids, which is smaller than the 1482 amino acids of rat NR2B.
A phylogenetic tree was constructed with the Neighbor Joining algorithm to analyze the evolutionary relationships of 11 NR2A and 14 NR2B protein sequences from different species, and of the NR2 proteins from C. elegans and D. melanogaster (Figure 2
). The invertebrates C. elegans and D. melanogaster are most distantly related to all other species and have only one type of NR2 subunit (Xia et al., 2005
; wormbase.org). The NR2A and NR2B subunits, in contrast, are present in all vertebrates and diverge evolutionarily as expected (Figure 2
). Sequence information for fish was only available for NR2B and this group of species contains the most ancestral forms of NR2B. The fish are followed by the amphibians, X. laevis and X. tropicalis, which cluster together in a branch for both NR2A and NR2B. Chicken, as a representative of birds, splits off next followed by the marsupial opossum, and then cow and dog. The rodents, rat and mouse, are clustered together, as well as the primate group, where human and chimp are most closely related (Figure 2
).
Figure 2. Phylogenetic trees of NR2A and NR2B. The phylogenetic relationships of 11 NR2A and 14 NR2B proteins from different species and the NR2 proteins from C. elegans and D. melanogaster were analyzed using the Neighbor Joining algorithm. The confidence values of the nodes were determined after bootstrapping with 1000 repetitions and are shown at the nodes. The trees were rooted by aligning two unrelated sequences, mouse GABA-A-a1 and GABA-A-a2 (gray italics). The cloned X. laevis NR2A and NR2B are bold, sequences that were assembled by hand are in italics and the remaining sequences have GenBank accession numbers.
Comparison of Sequence Conservation of X. laevis NR2A and NR2B
Xenopus laevis NR2A and NR2B are remarkably well conserved between species at the protein sequence level. We aligned the amino acid sequences of X. laevis NR2A with 10 other species (Figure 4
) and X. laevis NR2B with 13 other species (Figure 5
). C. elegans and D. melanogaster were omitted from the sequence alignment and analysis because they only have one kind of NR2 subunit.
Even though some sequences are only available as fragments for NR2A (squirrel monkey and X. tropicalis) and NR2B (tetraodon, fugu and X. tropicalis), the degree of sequence identity is striking (Figures 3
–5
). The number and percentage of conserved residues and domains were determined in comparison to the sequence of the species where they were characterized and are listed in Figure 3
. Taking the rat and X. tropicalis sequences as examples for species that are, respectively, quite distantly and closely related to X. laevis, we find that for X. laevis and rat, NR2A is 73% identical and NR2B is 85% identical at the amino acid level (Figure 3
). Sequence similarities are even higher at 91% for NR2A and 93% for NR2B. From the sequence that is available for X. tropicalis, the X. laevis and X. tropicalis protein sequences are 95% identical for NR2A and 96% identical for NR2B (Figure 3
). The sequence similarity between the two frog species for both NR2A and NR2B protein is 99%.
Figure 3. Conserved residues in X. laevis NR2A and NR2B. (A) Cartoon of the annotated functional domains and residues of NR2A. (B) Table of the number and percentage of conserved residues and domains of NR2A from 11 different species. The overall amino acid identity was determined in comparison to the X. laevis NR2A protein. (C) Cartoon of the annotated functional domains and residues of NR2B. (D) Table of the number and percentage of conserved residues and domains of NR2B from 14 different species. The overall amino acid identity was determined in comparison to the X. laevis NR2B protein. The number or percentage of conserved residues was determined in comparison to the sequence of the species where they were first characterized (mostly rat). Functional sites that are not conserved between all species are highlighted in gray.
Figure 4. Protein sequence alignment of NR2A from 11 different species. The protein sequences of X. laevis NR2A and 10 other species were aligned with the ClustalW algorithm. When full sequences were not available or could not be assembled by hand, incomplete sequences were used. Amino acids identical to the reference sequence are depicted as (.). Lack of sequence is symbolized by (−). A (*) below the alignment signifies amino acid identity, while (:) and (.) indicate the degree of amino acid similarity. NR2A is highly conserved between species. The membrane-associated segments M1–M4, other structurally and functionally important domains and amino acids, as well as known protein–protein interaction domains and phosphorylation sites are highlighted. Descriptions in italics indicate sites that are not conserved between species.
Figure 5. Protein sequence alignment of NR2B from 14 different species. The protein sequences of X. laevis NR2B and 13 other species were aligned with the ClustalW algorithm. When full sequences were not available or could not be assembled by hand, incomplete sequences were used. Amino acids identical to the reference sequence are depicted as (.). Lack of sequence is symbolized by (−). A (*) below the alignment signifies amino acid identity, while (:) and (.) indicate the degree of amino acid similarity. NR2B is very highly conserved between species. The membrane-associated segments M1–M4, other structurally and functionally important domains and amino acids, as well as known protein–protein interaction domains and phosphorylation sites are highlighted. Descriptions in italics indicate sites that are not conserved between species.
In our detailed analysis of the NR2A sequence (Figure 4
) we find that almost all residues that have been identified as important for high affinity Zn2+ binding (Fayyazuddin et al., 2000
; Paoletti et al., 2000
) are conserved. The exceptions are the first arginine, R37, changed from histidine and the last aspartate, D258, changed from glutamate. However, both changes are relatively conservative, since arginine and histidine are both basic amino acids, while aspartate and glutamate are acidic amino acids. The functional consequences of these modifications may therefore be small or negligible, and would need to be tested experimentally.
All of the residues and domains that are known to be important for the biophysical function of the receptor are conserved. The residues that are important for glutamate binding (Anson et al., 1998
; Chen et al., 2005
) are identical in all species. The pre-M1 segment, suggested to dynamically link ligand binding and channel gating (Krupp et al., 1998
) and the four membrane-associated segments, M1–M4, are almost identical in all species. Crucial residues for the proper function of NR2A such as the asparagine and serine residues, N608 and S609 that constitute the selectivity filter, as well as the WGP signature of the pore loop, and the SYTANLAAF signature of the M3 segment are conserved (Kuner et al., 1996
, 2003
; Wollmuth et al., 1996
, 1998
). In the C-terminal tail, the proximal endocytosis motif YXXΦ (Scott et al., 2004
; Vissel et al., 2001
) is present in all species. Interestingly, the distal endocytosis LL motif (Lavezzari et al., 2004
) is changed to a tyrosine and isoleucine both in X. laevis and X. tropicalis. Critical phosphorylation sites for Src kinase (Zheng et al., 1998
) and CaMKII (Omkumar et al., 1996
) are present in all species, whereas only some PKC phosphorylation sites are conserved (Gardoni et al., 2001
; Jones and Leonard, 2005
). The isoleucine and asparagine residues, 5′ to the CaMKII phosphorylation site, have been identified in rat to interfere with the strength of CaMKII binding to NR2A (Mayadevi et al., 2002
). Interestingly, this motif is conserved in the Xenopus species, as well as in rodents but not in primates, dog, cow, opossum or chicken. This suggests that this motif may not play a critical role in the interaction of CaMKII with NR2A or that CaMKII co-evolved to compensate for the sequence changes. Finally, the PDZ class I binding motif, E(T/S)XV, has been conserved in all species. This motif has been found to be crucial for receptor trafficking, downstream signaling and interaction with the postsynaptic density (Barria and Malinow, 2002
; Sheng and Pak, 2000
).
A detailed analysis of the NR2B sequence (Figure 5
) shows that almost all residues that have been identified as important for the binding of the antagonist ifenprodil (Perin-Dureau et al., 2002
) have been conserved. The only exception is a phenylalanine, F171, which in fish is changed to tyrosine. All of the regions that are important for the function of NR2B as a glutamate receptor have been conserved. The residues important for glutamate binding (Laube et al., 1997
) are identical in all species, as is a lysine residue, K486 that has been found to facilitate strong binding of the antagonists APV and CPP (Laube et al., 1997
). Furthermore, the pre-M1 segment and the four membrane-associated segments are conserved. Almost all residues are identical and there are only few variations in individual species in M1–M4. The critical asparagine and serine residues of the selectivity filter, N613 and S614, as well as the WGP signature of the pore loop, and the SYTANLAAF signature of the M3 segment are present in all species (Kuner et al., 1996
, 2003
). Three endocytosis motifs have been identified in NR2B (Roche et al., 2001
; Scott et al., 2004
). In contrast to NR2A, the proximal endocytosis YXXΦ motif (Scott et al., 2004
) is not conserved in either Xenopus species and only partially conserved in fish. However, a second endocytosis YXXΦ motif located distally has been characterized to be important for AP2-binding and clathrin-mediated endocytosis (Lavezzari et al., 2003
; Roche et al., 2001
) and is identical in all species. Unlike NR2A, the distal endocytosis LL motif (Roche et al., 2001
) is present in most species including Xenopus. The CaMKII (Omkumar et al., 1996
), PKC (Liao et al., 2001
) and Fyn kinase (Nakazawa et al., 2001
) phosphorylation sites are also identical across species. The lysine, K1270, and LRRQ residues that have been identified as important for interaction with CaMKII (Strack et al., 2000
) are only partially conserved as the latter is changed to LHRQ in both Xenopus species. Particularly, the changed arginine, R1277, was identified as an important residue for CaMKII interaction (Strack et al., 2000
). Potentially the remaining residues compensate for this missing residue or Xenopus does not critically rely on this motif for interaction with CaMKII. Finally, the PDZ class I binding domain of NR2B, E(T/S)XV, is present in all species. As for NR2A, the PDZ binding domain is critical for receptor trafficking, interaction with the postsynaptic density and downstream signaling (Barria and Malinow, 2002
; Prybylowski et al., 2005
; Sheng and Pak, 2000
).
NR1 Splice-Variants
We found that the NR1-4a/b splice-variants are the most abundantly expressed NR1 isoforms in developing X. laevis brain, as previously reported for adult frog brain (Soloviev et al., 1996
), and that the NR1-3a/b isoforms are expressed at lower levels. The developmental difference of the animals from which the cDNA libraries were generated might explain why Soloviev et al. detected only NR1-4a/b in their study of adult Xenopus. Cloning and characterization of NR1 isoforms in rat, human and chick have detected the spatial and temporal coexistence of multiple isoforms in the nervous system (Foldes et al., 1993
, 1994
; Laurie and Seeburg, 1994
; Lee-Rivera et al., 2003
; Zarain-Herzberg et al., 2005
). Similar to our findings in X. laevis, studies in zebrafish and electric fish detected NR1-3a/b and NR1-4a/b splice variants but not NR1-1a/b and NR1-2a/b isoforms (Bottai et al., 1998
; Cox et al., 2005
). In contrast to mammals, this selective expression of a subset of NR1 splice variants seems to be conserved among amphibians and fish and might well distinguish them from other species.
Conservation of NR2A and NR2B Subunits
We provided biochemical evidence that NR2A and NR2B are highly abundant in X. laevis brain in accordance with previous observations (Aizenman and Cline, 2007
; Cline et al., 1996
). The X. laevis NR2A and NR2B cDNA sequences demonstrate that both are remarkably conserved across species, including all major functional domains. This high degree of sequence conservation is found for some neuronal proteins in X. laevis such as CPG15, a GPI-linked protein that can regulate dendritic growth (Nedivi et al., 1998
), which is 73% identical to rat (Nedivi et al., 2001
), but not for others, for example X. laevis NOGO-A/RTN-4, a myelin-associated axon growth inhibitor, which is only 34% identical to its rat counterpart (Klinger et al., 2004
).
Conserved domains include the glutamate and antagonist binding sites, the membrane-associated segments and the residues that are important for ion selectivity, consistent with functional studies showing that the biophysical properties of NMDA receptors in Xenopus are comparable to other species (Wollmuth et al., 1996
, 1998
; Anson et al., 1998
; Chen et al., 2005
; Fayyazuddin et al., 2000
; Kuner et al., 1996
; Laube et al., 1997
; Paoletti et al., 2000
; Perin-Dureau et al., 2002
; Schmidt and Hollmann, 2008
; ). Moreover, residues that determine NMDAR function as a signaling partner to the postsynaptic density are also preserved throughout evolution. However, there are some interesting differences in a few regulatory domains. Remarkably, for NR2A, the isoleucine and asparagine residues, I1274 and N1275 that have been determined in rat to weaken the interaction with CaMKII (Mayadevi et al., 2002
) are conserved only in frogs and rodents, but not in other species. This implies that either the residues are not functionally relevant in vivo as biochemical experiments would suggest or regulation of the interaction with CaMKII differs between species. Alternately CaMKII itself could be changed in a compensatory manner. In addition, there are differences in the distal LL endocytosis motif of NR2A, as the two leucines are conserved in most species but not in chicken and Xenopus. In both frog species, LL is replaced by tyrosine and isoleucine, Y1306 and I1307. The proximal YXXΦ endocytosis motif, in contrast, is conserved. Conversely in NR2B, the proximal YXXΦ endocytosis motif (Scott et al., 2004
) is changed in both frog species from YWQF to FWQL, while the distal YXXΦ endocytosis motif (Roche et al., 2001
) is conserved. These observations in both NR2A and NR2B would suggest that the Xenopus genus potentially utilizes one of the endocytic sites depending on the NR2 subunit or that these changes are otherwise compensated for. It remains to be investigated if there is some differential regulatory function behind this.
Functional Analysis of NR2A and NR2B Expression in Xenopus Neurons
A logical extension of the analysis presented here is to conduct a structure function study of the effect of expressing full-length or mutant NR2 subunits in neurons. In a previous study we overexpressed in Xenopus neurons the same full-length Xenopus NR2A or rat NR2B constructs shown in Figure 1 and found that they lengthened and shortened, respectively, the decay time of evoked synaptic NMDAR-mediated currents. Morpholino-mediated knockdown of the NR2 subunits had the opposite effects on kinetics of synaptic responses, consistent with the predicted function of the NR2A and 2B subunits on synaptic function (Ewald et al., 2008
). We then carried out a functional analysis of the effect of overexpression and knockdown of NR2A and NR2B subunits on structural plasticity of Xenopus optic tectal neurons in vivo in an effort to identify subunit-specific effects on arbor development. Whereas the dendritic arbors of control tectal neurons have local clusters of dendritic branches, expression of either full-length NR2A or NR2B significantly decreased local branch clusters and resulted in a dispersed branching pattern. Detailed analysis of time-lapse image data indicates that neurons overexpressing NR2B are more dynamic than those overexpressing NR2A or control neurons. Our previous work has shown that a 4-h period of exposure to a simulated motion stimulus increases dendritic arbor growth rate, and that the enhanced growth rate requires glutamatergic synaptic transmission (Sin et al., 2002
). Surprisingly, overexpression of either NR2A or NR2B prevented the experience-dependent structural plasticity. These data, together with a recent report in rodent barrel cortex (Espinosa et al., 2009
) suggest that a highly regulated ratio of NR2A to NR2B levels underlies dendritic arbor development and experience-dependent plasticity in the CNS of the intact animal. The data further suggest that a structure function analysis of NMDAR subunits in neurons requires an appreciation of the complex nature of cellular effects of manipulating subunit expression.
In conclusion, we found that from a phylogenetic perspective both NR2A and NR2B show a high degree of evolutionary conservation, which underscores the potential importance of the NMDAR subunits in nervous system function. This suggests that the study of NMDARs in the nervous system of X. laevis is likely to provide important insights into the universal aspects of NMDAR function.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr Richard Huganir for generously providing the α-NR1-C1 and α-NR2A antibodies, Dr Andrew Neuwald for comments on the manuscript and members of the Cline laboratory for helpful discussions. This work was funded by NEI: EY011261.
Chen, P. E., Geballe, M. T., Stansfeld, P. J., Johnston, A. R., Yuan, H., Jacob, A. L., Snyder, J. P., Traynelis, S. F., and Wyllie, D. J. (2005). Structural features of the glutamate binding site in recombinant NR1/NR2A N-methyl-D-aspartate receptors determined by site-directed mutagenesis and molecular modeling. Mol. Pharmacol. 67, 1470–1484.
Soloviev, M. M., Brierley, M. J., Shao, Z. Y., Mellor, I. R., Volkova, T. M., Kamboj, R., Ishimaru, H., Sudan, H., Harris, J., Foldes, R. L., Grishin, E. V., Usherwood, P. N., and Barnard, E. A. (1996). Functional expression of a recombinant unitary glutamate receptor from Xenopus, which contains N-methyl-D-aspartate (NMDA) and non-NMDA receptor subunits. J. Biol. Chem. 271, 32572–32579.