Laboratory of Molecular Biology, National Institute of Mental Health, Bethesda, MD, USA
The availability of new tools for manipulating neuronal activity, coupled with the development of increasingly sophisticated techniques for targeting these tools to subsets of cells in living, behaving animals, is permitting neuroscientists to tease apart brain circuits by a method akin to classical mutagenesis. Just as mutagenesis can be used to introduce changes into an organism’s DNA to identify the genes required for a given biological process, changes in activity can be introduced into the nervous system to identify the cells required for a given behavior. If the changes are introduced randomly, the cells can be identifi ed without any prior knowledge of their properties. This strategy, which we refer to here as “neurotrapping,”has been implemented most effectively in Drosophila, where transgenes capable of either suppressing or stimulating neuronal activity can be reproducibly targeted to arbitrary subsets of neurons using so-called “enhancer-trap”techniques. By screening large numbers of enhancer-trap lines, experimenters have been able to identify groups of neurons which, when suppressed (or, in some cases, activated), alter a specifi c behavior. Parsing these groups of neurons to identify the minimal subset required for generating a behavior has proved difficult, but emerging tools that permit refined transgene targeting are increasing the resolution of the screening techniques. Some of the most recent neurotrapping screens have identified physiological substrates of behavior at the single neuron level.
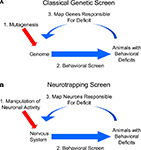
The fields of neuroscience and genetics face similar challenges: Both must explain how elementary components – neurons in the first case and genes in the second – interact to govern complex processes, such as behavior or development. Historically, the two fields have sought to meet their respective challenges in very different ways. Neuroscientists, having identified the basic unit of neural function early on, took a bottom-up approach, seeking to explain behavior and other integrative aspects of the brain by charting the connectivity of its component neurons and correlating their patterns of activity. Geneticists, lacking any knowledge of the basic unit of inheritance, were forced to take a different approach: They started at the top, working from the organismal phenotypes that resulted from unknown changes in the genetic material, and made their way down to the gene. The genetic approach was systematized by coupling methods for blindly inducing genetic changes with selective screens to identify mutations that affected particular biological processes of interest (Figure 1
A). The nature of the gene and the identity of the particular genes involved in diverse biological phenomena were elucidated by this powerful, unbiased approach.
Historically, neuroscience research has lacked a counterpart to the unbiased screen of genetics: While electrophysiological, pharmacological, anatomical, and genetic manipulations have each provided productive avenues to perturbing brain function, there has been no general method for systematically and randomly altering neuronal activity in freely behaving animals to identify the neurons underlying a given behavior. Such a method would usefully complement classic circuit-mapping techniques by supplying causal links to circuit maps derived from tracing synaptic connections and correlating activities. Fortunately, the steady advance of genetic technologies is beginning to provide neuroscientists with the basic elements of such a method. This method, to which we apply the previously-introduced term “neurotrapping” (White et al., 2001a
), is illustrated in Figure 1
B. By analogy to mutagenic screens, neurotrapping involves the perturbation of neuronal, rather than gene, function, and uses not chemical mutagens, but instead, transgenes whose products inhibit (or in some cases stimulate) the activity of neurons in which they are expressed. The expression of these transgenes in random subsets of cells in the brains of living animals constitutes a process analogous to mutagenesis. By identifying animals that exhibit deficits in a behavior of interest as a consequence of inactivating different subsets of neurons, neurotrapping can be used to interrogate the nervous system to identify those cells that are essential for a given behavior, just as mutagenesis is used to interrogate the genome to identify genes required for a given biological process.
Figure 1. Neurotrapping screens are analogous to genetic screens. (A) In a genetic screen, perturbations are introduced into the genome by mutagenesis, and animals bearing these mutations are analyzed for deficits in a behavior of interest. These heritable deficits are then traced back to the change(s) in the genetic material that are their source. (B) In a neurotrapping screen the activity of neurons, rather than genes, is perturbed. This type of screen requires genetic tools such as those listed in Table 1
for manipulating neuronal activity, and a means of targeting these tools to particular neurons. Unlike traditional methods of circuit-mapping, neurotrapping identifies the functional components of a neural network without knowledge of synaptic connectivity (which often represents only part of a neuron’s signaling capacity) or response properties (which may only coincidentally correlate with performance of a particular behavior).
It is worth pointing out that while neurotrapping and mutagenesis are analogous techniques, there are differences between them. Perhaps the most important relates to the relative scale of the perturbations made: In contrast to mutagenesis, which is usually performed so as to affect the function of single genes, the transgene expression systems used for neurotrapping typically lead to the manipulation of multiple neurons – often hundreds to thousands. While this broad targeting is probably often necessary to insure that enough neurons are functionally compromised to give rise to a behavioral deficit (either by inducing multiple changes that have small effects, or by compromising compensatory networks in addition to a primary circuit), it also means that identifying the essential neurons responsible for the deficit is often difficult. Indeed, neurotrapping has proved most feasible when the random targeting of gene expression can be performed reproducibly, allowing the same set of neurons to be repeatedly probed from animal-to-animal and generation-to-generation. This is possible when three conditions obtain: the targeting patterns are heritable, neuronal identities are relatively fixed, and behavioral circuits are more-or-less hard-wired. These conditions are largely met in Drosophila, where neurotrapping has been primarily developed. We review these developments here, first discussing the tools used for neuronal targeting and manipulation, and then, after reviewing early studies, focusing on recent innovations. These innovations have facilitated neurotrapping methodologies and have raised the prospects that neurotrapping will become as potent an engine of circuit discovery as random mutagenesis has been in elucidating transcriptional and signaling networks in genetics.
A profound advantage of Drosophila in screen-based studies of neural function is the availability of powerful gene-targeting technologies based on the Gal4-UAS system (Figure 2
). This expression system (Figure 2
A), consists of two types of transgenic fly line, one that expresses the yeast transcription factor Gal4 in specific groups of cells under the control of a particular enhancer, and another that bears a transgene of interest downstream of the Gal4 binding site, which is called the “upstream activating sequence” (i.e. UAS). When flies from two such lines are mated, Gal4 drives expression of the transgene in the same cells in which Gal4 itself is expressed. Diverse manipulations of the same set of neurons can therefore be made at will by matching a single Gal4 driver with different UAS-transgenes. By the same token, multiple Gal4 lines with different expression patterns can be matched with a single UAS-transgene to perform identical manipulations on many different neuronal groups. The targeting of arbitrary groups of neurons is accomplished using so-called “enhancer-trap” lines, which are made by allowing the Gal4 gene to randomly integrate into the genome. In transgenic flies made by this method, genomic enhancers that normally regulate endogenous genes near the integration site determine the expression pattern of Gal4. Libraries of Gal4 enhancer-trap lines that have diverse expression patterns typically serve as the starting point for neurotrapping studies. Once an enhancer-trap line of interest has been identified (for example, one that alters a specific behavior when driving a suppressor of neuronal activity), its pattern of expression can be further refined to identify the subset of neurons essential for the behavior using strategies such as those illustrated in Figures 2
B–D. These strategies are described in the context of specific studies in greater detail below.
Figure 2. Methods of transgene targeting. (A) The Gal4-UAS system described in the text. Left panel: the schematic depicts the transgene construct for Gal4 (blue outline) on one fly chromosome, and the transgene effector construct (green rectangle) on another fly chromosome. The Gal4 gene lies downstream of the promoter/enhancer element, P1, which dictates its pattern of expression. The effector transgene lies downstream of five Gal4 binding sites (i.e. “UAS,” black ovals). In flies bearing both constructs, neurons that express Gal4 protein (blue shapes) also express the effector protein (green circles). Right panel: schematic of the fly CNS depicting coincident expression of Gal4 (blue outline) and effector (green oval). (B) Subtractive restriction of effector gene expression using Gal80. Left panel: In cells that express the gene encoding Gal80 (brown outline) under the control of the promoter/enhancer P2, Gal80 protein (brown circles) will inhibit Gal4 by binding to its transcription activation domain and thus block effector gene expression (red X). Right panel: if P1 and P2 have overlapping expression patterns, Gal4 activity, and therefore effector expression, is eliminated in the region of overlap (i.e. in Gal80 positive cells, brown). (C) Random restriction using the “flp-out Gal80” system. Left panel: if the Gal80 transgene is placed downstream of the ubiquitously active tubulin promoter (tub) and is flanked by sites (triangles) that permit excision by heat-shock induced flp-recombinase activity, the Gal80 gene will be deleted in random subsets of cells in animals subjected to heat shock. Only cells that express Gal4, but not Gal80, will also express the effector gene. Right panel: Effector expression (green) is limited to cells within the Gal4 expression pattern (blue outline) that lack Gal80 expression. (D) Combinatorial restriction using Split Gal4. Left panel: If the DNA-binding (blue, DBD) and transcription activation (yellow, AD) domains of the Gal4 molecule are independently targeted to different neuronal groups using promoters P1 and P2, Gal4 activity will be reconstituted and the effector transgene expressed only in cells at the intersection of the P1 (blue) and P2 (yellow) expression patterns, as depicted in the right panel.
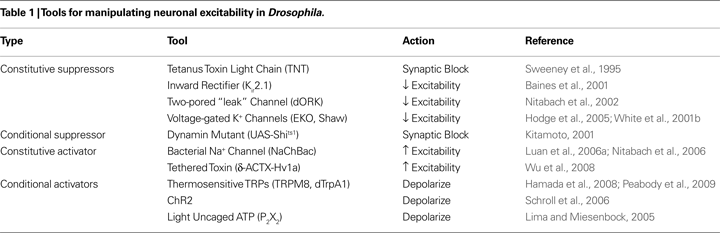
The types of UAS-transgenes that have been used to manipulate neural function in Drosophila are listed in Table 1
, which focuses on those whose products broadly affect neuronal function and are not cell-type specific. The products of most of these genes either suppress synaptic transmission or act to inhibit or enhance neuronal excitability. Some act constitutively, while others can be used to perform acute manipulations. The most relevant from the standpoint of the neurotrapping studies reviewed here are those that suppress either synaptic transmission or membrane excitability. These have received the greatest use for the simple reason that suppression of neuronal function allows one to determine whether a particular neuron or set of neurons is necessary for governing a particular behavior. It should be noted that many of the tools listed in Table 1
are discussed in detail in the Frontiers in Molecular Neuroscience Special Topic titled “Genetic techniques and circuit analysis” (see Han et al., 2009
; Hodge, 2009
; Holford et al., 2009
; Kasuya et al., 2009
). Those used specifically in Drosophila in the context of circuit mapping are also comprehensively treated in recent reviews by Holmes et al. (2007) and Simpson (2009)
. Interested readers are referred to these reviews for details.
The first cellular screen of neuronal function in Drosophila by Sweeney et al. (1995)
was facilitated by two developments: the coupling of the Gal4-UAS system to enhancer-trap technologies by Brand and Perrimon (1993) and the cloning of tetanus toxin light chain (TNT) by Mochida et al. (1990)
. TNT selectively cleaves the neuronal isoform of synaptobrevin in Drosophila and thus suppresses synaptic transmission in the neurons that express it. Using UAS-TNT, Sweeney and colleagues screened a collection of enhancer trap-Gal4 lines and identified several that produced deficits in an olfactory escape response. The expression pattern of one line that produced particularly strong deficits was investigated and found to include neurons that projected into the leg nerves, but no further identification of the locus of TNT action was undertaken.
Although this early use of UAS-TNT as a screening tool was occasionally emulated, as in the investigation of larval locomotion by Suster et al. (2003)
, UAS-TNT was most extensively applied to investigating the functions of candidate neurons or brain regions already suspected of playing a role in particular behaviors (for a review of these studies see Martin et al., 2002
). Gal4 enhancer-trap lines were typically screened anatomically to identify those that expressed in neurons of interest, and these lines were then used to suppress synaptic transmission so that the behavioral consequences could be observed. In this way, the neural components of various sensory and motor systems were investigated, as were neurons contributing to functionally interesting structures within the central nervous system, such as the mushroom bodies and the central complex.
Because of their focus on pre-selected groups of candidate neurons, these early UAS-TNT studies bear a closer relationship to reverse-genetic approaches than to the “forward” approaches that make use of random mutagenesis. In part, forward approaches to neuronal screens were handicapped by the broad expression patterns of most Gal4 enhancer-trap lines. Indeed, even in studies where the enhancer-trap lines were selected because their expression included a particular set of neurons, concomitant expression outside the region of interest often meant that the effects of suppression could not be attributed with complete confidence to the silencing of the target neurons. This problem could sometimes be ameliorated by comparing results obtained with several lines that had overlapping expression patterns (see for example Suster et al., 2003
; Broughton et al., 2004
), but, in general, when expression cannot be restricted to a single group of neurons – a condition often difficult to meet – some ambiguity will remain.
A second impediment to the implementation of unbiased screening strategies using UAS-TNT was the inability of investigators to temporally restrict UAS-TNT expression. The fact that synaptic transmission was constitutively compromised meant that synaptic suppression during development, and not acute suppression at the time of testing, might account for the observed behavioral deficits. There was also no guarantee that TNT would affect the same neurons, or neurons governing the same functions, during development and adulthood, since many enhancer-trap expression patterns are dynamic and change as the animal grows. The broad range of expression over development also meant that synaptic silencing was frequently lethal due to TNT expression in some subset of neurons essential for viability. An elegant solution to the problem of temporal control was developed by Kitamoto with his introduction of UAS-Shits1, a UAS-construct encoding a temperature-sensitive mutant of Shibiri, the Drosophila ortholog of dynamin (Kitamoto, 2001
). Shits1, as is described more extensively in the Frontiers in Molecular Neuroscience Special Topic titled “Genetic techniques and circuit analysis” (see Kasuya et al., 2009
), is a dominant inhibitor of synaptic vesicle recycling and thereby blocks synaptic transmission. Using flies bearing the UAS-Shits1 construct, Kitamoto demonstrated that synaptic suppression could, in many cases, be induced by a simple temperature shift within a matter of minutes.
Like UAS-TNT, UAS-Shits1 has been since used in many studies to inhibit synaptic transmission in candidate neurons for various processes, in some cases with only minimal pre-selection of enhancer-trap lines. In the latter cases, the resulting activity screens are essentially “forward” neurotrapping screens. For example, Pitman et al. (2006)
screened 92 enhancer-trap lines with UAS-Shits1 that were pre-selected for their relatively restricted expression within different brain regions. These lines were chosen, however, because their expression patterns collectively covered most of the fly central nervous system. By using UAS-Shits1 to inhibit neurons in each of these patterns, the authors were able to demonstrate a specific role of the mushroom bodies in sleep.
Similarly, in a study of larval locomotion, Hughes and Thomas (2007)
anatomically pre-screened approximately 1800 Gal4 enhancer-trap lines and selected only those that expressed in small subsets of CNS or PNS neurons. They then performed a secondary screen on the chosen lines using UAS-Shits1, which allowed them to identify two classes of sensory neurons that provide proprioceptive feedback critical for larval crawling. A similar screen that was carried out without anatomical pre-selection was conducted by Song et al. (2007) and arrived at similar conclusions. Song et al. used UAS-Shits1 to functionally screen approximately 1000 Gal4 enhancer-trap lines and identified 10 that drastically reduced peristaltic rhythm and locomotion. All 10 were found upon subsequent investigation to express strongly in peripheral sensory neurons. One of these lines had no CNS expression, and, as in the study of Hughes and Thomas, led to the identification of multidendritic neurons as essential components of the circuitry required for normal peristalsis during larval locomotion.
It is perhaps not surprising that the reported neurotrapping screens often involve the identification of sensory neurons. The accessibility of these neurons to manipulation and physiological recording has long made them a favorite object of study and the availability of specific drivers for subsets of them has facilitated their identification in functional screens for the circuitry underlying specific behaviors. Another example is a pilot screen reported by Suh et al. (2004)
, which sought to identify neurons involved in a chemosensory-mediated avoidance response. This screen of approximately 250 Gal4 enhancer-trap lines uncovered one line that helped confirm the participation of a subclass of odorant receptor neurons in this behavior.
Fewer neurotrapping screens have reported identifying components of central circuits involved in more complex behaviors. One notable exception is a study by Katsov and Clandinin (2008)
which used UAS-Shits1 to screen approximately 400 enhancer-trap lines for specific visual processing defects using a high-throughput behavioral assay. These authors identified one line in which expression was limited to two brain regions, one of which (the mushroom body), could be ignored because its ablation did not cause the visual processing defect. Remarkably, this left only a small set of three to four bilaterally represented neurons in each optic lobe as the presumptive circuit components responsible for the processing defect.
Although UAS-Shits1 affords excellent temporal control of synaptic transmission under many circumstances, it can also perturb basic cellular functions. These effects can lead to ambiguities in the interpretation of manipulations made with this tool (see Kitamoto, 2002b
). An alternate method for temporally regulating synaptic block is the flp-out TNT method introduced by Keller et al. (2002)
, which employs a construct with a transcription stop cassette interposed between the UAS and the TNT transgene. This stop cassette can be excised by the flp recombinase, which can be ubiquitously activated by heat shock in flies bearing the flp transgene expressed behind the heat-shock promoter.
More general temporal control can be achieved by techniques that permit conditional transgene expression such as the GeneSwitch system (Osterwalder et al., 2001
; Roman et al., 2001
) or the TARGET system (McGuire et al., 2004
). The latter uses a temperature-sensitive mutant of the Gal4 inhibitor Gal80 (i.e. Gal80ts), which is capable of repressing Gal4-mediated transcription at lower, restrictive temperatures, but not at higher, permissive temperatures. This technique has been particularly useful in suppressing the expression of TNT during development to prevent lethality, and then releasing it in the adult where it can suppress circuit function. This approach was used in two recent studies (Gordon and Scott, 2009
; Zhu et al., 2009
). The general applicability of the TARGET system in temporally regulating Gal4-mediated transgene transcription also means that it can be used with constitutive suppressors of neuronal activity besides TNT (see Table 1
).
…And Restriction of Expression Pattern
Gal80 (in its native, non-temperature-sensitive form) has also been useful for narrowing down sets of neurons to identify those that specifically function in a behavioral circuit. The general utility of Gal80 in restricting the range of Gal4 activity was first demonstrated by Lee and Luo with their introduction of the MARCM system (Lee and Luo, 2001
), which is routinely used to limit Gal4 activity to individual neurons. Due to its inherently mosaic expression, the MARCM system does not permit reproducible expression of effectors in the same cells from animal to animal, and has not typically been used in neurotrapping screens. However, Gal80 itself has proved useful in evaluating the lines generated by such screens (Figure 2
B) as was first demonstrated by Kitamoto, who used Gal80 to suppress the cholinergic subset of neurons within the expression pattern of a particular Gal4 enhancer-trap line (Kitamoto, ,b2002a
). This line was isolated from a screen designed to analyze the neural basis of courtship behavior. When subjected to transient suppression, males resulting from crosses of this line to UAS-Shits1 engaged in aberrant male–male courtship. By selectively relieving suppression in the cholinergic neurons of this pattern using a Gal80 construct expressed behind the choline acetyltransferase promoter, Kitamoto showed that male courtship patterns returned to normal. By this means he was able to demonstrate that cholinergic neurons within the expression pattern of the enhancer-trap line were responsible for the behavioral deficit seen in response to synaptic block.
While this example illustrates the utility of Gal80 in restricting an expression pattern to isolate a cell group of interest, it also illustrates one of the challenges of this approach. Because Gal80-mediated restriction represents a negative selection strategy that subtracts neurons from a pattern of interest (see Figure 2
B), these neurons are then no longer available for manipulation. Further narrowing of the pattern to identify the minimal set of behaviorally relevant neurons therefore becomes difficult. A positive selection strategy that does not share this drawback couples the use of Gal80 to the flp recombinase in such a way that the behavioral phenotype of interest is seen only in conjunction with restricted expression patterns. This “flp-out Gal80” system, illustrated in Figure 2
C relies on the stochastic activation of flp by heat shock and can lead to the restoration of Gal4 activity in small sets of cells within an expression pattern, in some cases as few as one. This method was recently used in conjunction with a neurotrapping screen by Gordon and Scott (2009)
to identify individual neurons involved in a sensorimotor taste circuit.
The latter study is perhaps the most elegant implementation to date of the neurotrapping strategy and provides a good example of what can be done using some of the latest tools. Starting with a suppressor screen of 534 Gal4 enhancer-trap lines, Gordon and Scott set out to identify neurons involved in taste-related behaviors. They used Gal80ts to prevent developmental mortality and identified 47 lines that gave rise to adult deficits in a sensorimotor taste reflex. These deficits fell into four different categories, including motor deficits. By analyzing one of the six motor defective lines using the flp-out Gal80 technique, they were able to identify and characterize a single pair of motor neurons that were necessary for the execution of a subprogram of the taste reflex. In addition to the synaptic blocker, TNT, the authors used a potent suppressor of excitability, Kir2.1, to conduct their screen and also made use of the light-sensitive channel, ChannelRhodopsin2 (i.e. ChR2). Indeed, by using ChR2 to selectively stimulate the pair of motor neurons identified using TNT and Kir2.1, they were able to demonstrate that activation of these neurons was sufficient to induce the same behavioral subprogram that was lacking in the flies in which these neurons were suppressed.
While the study of Gordon and Scott convincingly demonstrates the efficacy of existing neurotrapping techniques in Drosophila, emerging technologies promise to facilitate both the consistent targeting of small groups of neurons and the rational dissection of a large group of neurons into smaller subsets. In addition, an ever broadening palette of tools for manipulating excitability promises to increase the range of manipulations of neuronal activity that can be made for neurotrapping. Each of these is discussed in turn in the sections below.
Restriction of Transgene Expression
Unbiased transgene targeting is at the heart of neurotrapping. Given the large number of neurons in even the modestly-sized brain of the fruitfly, it is clear that generating Gal4 lines to target each individual neuron would be overwhelming and quite possibly also unproductive. Redundancy of function both within and between circuits will undoubtedly obscure the behavioral contributions of individual neurons in many cases. On the other hand, if one wants to target multiple neurons, it is unclear what number should be targeted: Targeting too many neurons risks hitting multiple circuits and confounding the behavioral effects of any given manipulation; on the other hand, targeting too few neurons risks missing circuits that are broadly distributed and rely on the participation of many weakly contributing components.
The data from neurotrapping screens conducted with Gal4 enhancer-trap lines indicates that 1–10% of lines can be expected to produce a phenotype of interest when used to drive suppressors, but only one to two per 1000 will have an expression pattern small enough to analyze easily using existing methods. This would suggest that having more Gal4 lines with small expression patterns would be beneficial. Pfeiffer et al. (2008)
recently described a method for generating such lines using random 3 kb fragments of putative regulatory DNA from the flanking and intronic regions of genes known to be expressed in the nervous system. They report that Gal4 lines made with 44 such enhancer fragments labeled only about a quarter as many central brain neurons on average (approximately 100) as conventional enhancer-trap lines. Happily, these authors are in the process of making several thousand such lines using putative enhancer regions from many different genes. They estimate that these lines will collectively provide coverage of most neurons in the Drosophila brain.
Even enhancer-trap lines with small expression patterns, however, will at best identify a set of candidate neurons in neurotrapping screens. This set will then have to be winnowed down to the minimal subset that gives rise to the behavioral deficit of interest when suppressed. In addition to the flp-out Gal80 method, there are several other combinatorial techniques that use the flp recombinase to restrict transgene expression. These include the UAS > STOP > TNT system described above, and UAS > STOP > Shits1, which was introduced by Stockinger et al. (2005)
. One drawback of these techniques as they are traditionally used (i.e. in combination with hs-flp) is that excision of the FRT-flanked cassette is stochastic, and the restricted expression patterns are therefore not reproducible from animal to animal.
An alternative methodology that provides reproducible expression patterns is the Split Gal4 system introduced by Luan et al. (2006b)
. In this system (Figure 2
D), the Gal4 molecule is split into its two component domains, the DNA-binding domain (DBD) and transcription activation domain (AD), which can then be independently targeted to different neuronal groups. Gal4 transcriptional activity, and therefore the expression of UAS transgenes, is thus restricted to the intersection of the expression patterns of the two domains. By making a fly line in which the expression of one domain (e.g. the DBD) is restricted to a cell group of interest, and crossing this line to enhancer-trap lines made with the other domain (i.e. the AD), one can produce many different patterns of expression in subsets of the original group (Luan et al., 2006b
; Luan and White, 2007
). These patterns are reproducible from animal-to-animal and can be generated at will.
The Split Gal4 system can be used in neurotrapping screens in two principal ways. In the first, a primary screen can be conducted using enhancer-trap (ET) lines made with one component (e.g. ET-DBD) while the other component is expressed throughout the nervous system (e.g. the AD driven by a pan-neuronal promoter). Each ET-DBD line that yields a behavioral deficit of interest in the primary screen can then be used in a secondary screen with AD enhancer-trap lines. In this way, the patterns of the individual ET-DBD lines from the primary screen can be subdivided in the secondary screen. Alternatively, if a Gal4 enhancer-trap line has been isolated in a neurotrapping screen, it can be converted into a Split Gal4 line by P-element swap (Sepp and Auld, 1999
) to subdivide its expression pattern. Gao et al. (2008)
have recently demonstrated the efficacy of converting an existing enhancer-trap Gal4 line into a DBD line which shares the expression pattern of the original. The process of swapping a Split Gal4 component for Gal4 could also be streamlined by using Gal4 constructs that permit recombinase-mediated cassette exchange (RMCE, Horn and Handler, 2005
). In any case, application of the Split Gal4 technique provides a strategy for systematically parsing the pattern of an identified enhancer-trap line into smaller groups so that their function can be analyzed.
Beyond Synaptic Silencing: Excitability Suppression and Neuronal Stimulation
Although most neurotrapping screens conducted thus far have used UAS-TNT or UAS-Shits1, these tools will not block communication through electrical synapses or necessarily impede signaling by hormones and neuromodulatory factors (Thum et al., 2006
). In principle, neuronal function can be more universally suppressed by inhibition of membrane excitability, which should block all signaling mechanisms used by neurons. As described in the Frontiers in Molecular Neuroscience Special Topic titled “Genetic techniques and circuit analysis” (see Hodge, 2009
) numerous tools have been developed to block neuronal excitability, from the potent inward rectifying Kir2.1 K+-channel to the relatively weak EKO channel. As noted above, UAS-Kir2.1 was used in conjunction with UAS-TNT to perform the neurotrapping study of Gordon and Scott (2009)
, though no differences in the efficacy of the two tools was reported.
In contrast, TNT and EKO display clear differences in efficacy in suppressing wing expansion behaviors (Luan et al., 2006a
). These behaviors are regulated by the hormone bursicon, which is expressed in a subset of neurons that express the neuropeptide CCAP. In contrast to EKO, TNT poorly blocks wing expansion when expressed in CCAP-expressing neurons and Kir2.1, because of its greater potency, causes developmental lethality at the pupal stage. Luan et al. (2006a)
therefore chose to use the EKO channel to conduct a screen of enhancer-trap lines to identify neurons involved in wing expansion. This screen identified 24 lines (from a total of 114) that produced wing expansion deficits when crossed to EKO. Using a CCAP-Gal80 construct, 23 of these lines were shown to act by suppressing CCAP-expressing neurons. Because of its selective expression in a subset of bursicon-secreting neurons, one of these lines permitted a functional dissection of the bursicon-expressing group.
Although there is, as yet, no conditional suppressor of excitability available for use in Drosophila, an increasing number of options are available for acutely stimulating neurons using either light or temperature (see Table 1
). As indicated by the use of ChR2 to stimulate motor neurons in the study of Gordon and Scott (2009)
, or the recent use of the temperature-sensitive TRP channels to selectively stimulate neurons promoting wakefulness (Parisky et al., 2008
) or wing expansion (Peabody et al., 2009
), these tools can be used to identify sets of central neurons that are capable of inducing behavioral programs. More generally, they have the potential of being used to identify neurons that act as “command systems” in generating behavioral outputs. Neurotrapping screens using activators rather than suppressors of neuronal activity thus offer an additional important tool for exploring behavioral control in Drosophila.
In addition to screens that involve suppressing and activating neurons, the general strategy described here can be applied to study any neuronal function of interest. Indeed, screens have been carried out using numerous other effectors, including UAS-transformer to feminize neurons in studies of courtship behavior (Ferveur et al., 1995
), UAS-PKAinh, a PKA inhibitor, to study brain regions sensitive to alcohol (Rodan et al., 2002
), and UAS-mC*, a constitutively active form of PKA, to study neural substrates of sleep (Joiner et al., 2006
). With the recent availability of UAS-RNAi constructs to virtually every gene in the Drosophila genome (Dietzl et al., 2007
), diverse neurotrapping strategies can now be envisaged that ask not only which neurons are necessary for executing different behaviors, but which genes in those neurons are necessary.
With recent advances, neurotrapping screens are beginning to realize their full potential. Although technical challenges to implementing them remain, the most recent advances provide proof that success is possible and suggest that whole circuits will soon yield to the neurotrapping approach. Whether all circuits can be “cracked” by this approach remains to be determined, but the answer is soon likely to depend less on the availability of techniques than it will on biology. Like all nervous systems, the Drosophila nervous system is densely interconnected and we have much to learn about how it is organized: How many behaviors are governed by “command” systems as opposed to “multifunctional” networks that broadly coordinate the animal’s responses to environmental inputs? How are conflicts between motor outputs resolved and what mechanisms govern behavioral choice? What compensatory mechanisms and circuits exist to allow for behavioral flexibility in the face of environmental challenge or developmental damage? How are complex behavioral programs assembled? All these questions remain, but given recent developments, neurotrapping in Drosophila seems poised to provide at least some of the answers.
The answers derived from Drosophila will undoubtedly inform our thinking about the functional architecture of other nervous systems, including our own. Will it be possible to also gain direct insights into the function of vertebrate nervous systems from neurotrapping studies? Despite the technical challenges of longer generation times, larger chromosome numbers, and increased costs encountered in working with vertebrate preparations and the fact that neuronal identities in such organisms are often epigenetically sculpted and less stereotyped between individuals than they are in Drosophila, the answer is almost certainly yes. Indeed, the feasibility of neurotrapping in fish has already been demonstrated by Wyart et al. (2009)
. Using multiple zebrafish Gal4 enhancer-trap lines that drive expression in spinal cord neurons, these authors expressed a light-activated cation channel to identify the stimulatory inputs to the swim central pattern generator in the spinal cord.
Demonstrations of neurotrapping’s feasibility in mammalian preparations may well follow. Sophisticated approaches for gene targeting and neuronal manipulation are available in the mouse (Deisseroth et al., 2006
; Luo et al., 2008
), and have already been extensively used in what might be called “reverse” neurotrapping studies in which targeted neuronal manipulations have been used to demonstrate the functional roles of defined neuronal populations (see for example Yu et al., 2004
; Adamantidis et al., 2007
; Tan et al., 2008
; Sohal et al., 2009
; Tsai et al., 2009
). A particularly elegant example is the study of Kim et al. (2009)
, who used an intersectional expression system to synaptically silence a subclass of serotonergic neurons and show that silencing correlated with a reduction in anxiety-related behaviors. Because the technical challenges mentioned above are a particular impediment to neurotrapping in the mouse, progress in this animal may have to exploit different strategies than those used in Drosophila. It would be particularly helpful if random neuronal manipulations could be correlated with behavioral changes in single animals. This would be facilitated if advances in gene expression profiling of mouse neurons (Lein et al., 2007
; Ng et al., 2009
) could be leveraged to identify the neurons randomly targeted by viral transduction. Whether such strategies will prove tractable will be decided by future developments. However, as the work reviewed here indicates, technical development is unlikely to limit progress for long, and neurotrapping, in Drosophila and in other organisms, is likely to play an increasing role in elucidating the circuitry that governs behavior.
The authors declare no commercial or financial relationships that could be construed as a potential conflict of interest related to this work.
We dedicate this review to the memory of Thomas Osterwalder who coined the term “neurotrapping” and who died of cancer in 2005. We thank Grace Gray and Dave Sandstrom for critically reading the manuscript. Benjamin White is supported by the Intramural Research Program of the National Institute of Mental Health.
Holmes, T. C., Sheeba, V., Mizrak, D., Rubovszky, B., and Dahdal, D. (2007). Circuit-breaking and behavioral analysis by molecular genetic manipulation of neural activity in Drosophila. In Invertebrate Neurobiology, G. North, and R. J. Greenspan, eds (Cold Spring Harbor, NY, Cold Spring Harbor Laboratory Press), pp. 19–52.
Kasuya, J., Ishimoto, H., and Kitamoto, T. (2009). Neuronal mechanisms of learning and memory revealed by spatial and temporal suppression of neurotransmission using shibirets1, a temperature-sensitive dynamin mutant gene in Drosophila melanogaster. Front. Mol. Neurosci. 2. doi: 10.3389/neuro.02.011.2009.
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., Boe, A. F., Boguski, M. S., Brockway, K. S., Byrnes, E. J., Chen, L., Chen, L., Chen, T. M., Chin, M. C., Chong, J., Crook, B. E., Czaplinska, A., Dang, C. N., Datta, S., Dee, N. R., Desaki, A. L., Desta, T., Diep, E., Dolbeare, T. A., Donelan, M. J., Dong, H. W., Dougherty, J. G., Duncan, B. J., Ebbert, A. J., Eichele, G., Estin, L. K., Faber, C., Facer, B. A., Fields, R., Fischer, S. R., Fliss, T. P., Frensley, C., Gates, S. N., Glattfelder, K. J., Halverson, K. R., Hart, M. R., Hohmann, J. G., Howell, M. P., Jeung, D. P., Johnson, R. A., Karr, P. T., Kawal, R., Kidney, J. M., Knapik, R. H., Kuan, C. L., Lake, J. H., Laramee, A. R., Larsen, K. D., Lau, C., Lemon, T. A., Liang, A. J., Liu, Y., Luong, L. T., Michaels, J., Morgan, J. J., Morgan, R. J., Mortrud, M. T., Mosqueda, N. F., Ng, L. L., Ng, R., Orta, G. J., Overly, C. C., Pak, T. H., Parry, S. E., Pathak, S. D., Pearson, O. C., Puchalski, R. B., Riley, Z. L., Rockett, H. R., Rowland, S. A., Royall, J. J., Ruiz, M. J., Sarno, N. R., Schaffnit, K., Shapovalova, N. V., Sivisay, T., Slaughterbeck, C. R., Smith, S. C., Smith, K. A., Smith, B. I., Sodt, A. J., Stewart, N. N., Stumpf, K. R., Sunkin, S. M., Sutram, M., Tam, A., Teemer, C. D., Thaller, C., Thompson, C. L., Varnam, L. R., Visel, A., Whitlock, R. M., Wohnoutka, P. E., Wolkey, C. K., Wong, V. Y., Wood, M., Yaylaoglu, M. B., Young, R. C., Youngstrom, B. L., Yuan, X. F., Zhang, B., Zwingman, T. A., and Jones, A. R. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176.
Ng, L., Bernard, A., Lau, C., Overly, C. C., Dong, H. W., Kuan, C., Pathak, S., Sunkin, S. M., Dang, C., Bohland, J. W., Bokil, H., Mitra, P. P., Puelles, L., Hohmann, J., Anderson, D. J., Lein, E. S., Jones, A. R., and Hawrylycz, M. (2009). An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 12, 356–362.
Nitabach, M. N., Wu, Y., Sheeba, V., Lemon, W. C., Strumbos, J., Zelensky, P. K., White, B. H., and Holmes, T. C. (2006). Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J. Neurosci. 26, 479–489.
Pfeiffer, B. D., Jenett, A., Hammonds, A. S., Ngo, T. T. B., Misra, S., Murphy, C., Scully, A., Carlson, J. W., Wan, K. H., Laverty, T. R., Mungall, C., Svirskas, R., Kadonaga, J. T., Doe, C. Q., Eisen, M. B., Celniker, S. E., and Rubin, G. M. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 9715–9720.