1
Department of Cell Biology, The Scripps Research Institute, La Jolla, CA, USA
2
The Institute for Childhood and Neglected Diseases, The Scripps Research Institute, La Jolla, CA, USA
3
Department of Neuroscience, Tufts University, Boston, MA, USA
The use of molecular tools to study the neurobiology of complex behaviors has been hampered by an inability to target the desired changes to relevant groups of neurons. Specific memories and specific sensory representations are sparsely encoded by a small fraction of neurons embedded in a sea of morphologically and functionally similar cells. In this review we discuss genetics techniques that are being developed to address this difficulty. In several studies the use of promoter elements that are responsive to neural activity have been used to drive long-lasting genetic alterations into neural ensembles that are activated by natural environmental stimuli. This approach has been used to examine neural activity patterns during learning and retrieval of a memory, to examine the regulation of receptor trafficking following learning and to functionally manipulate a specific memory trace. We suggest that these techniques will provide a general approach to experimentally investigate the link between patterns of environmentally activated neural firing and cognitive processes such as perception and memory.
How does the brain represent the surrounding world in discrete percepts? How does the pattern of retinal activity produced by looking at the image of Halle Berry ultimately lead to the perception of the actress rather than a violin? How do the myriad different associations with Halle Berry (actress, specific roles) or violin (inanimate object, music) form through learning and memory? These are basic questions at the heart of many areas of neuroscience. They have been approached by a variety of techniques that fall into two broad categories, watching brain activity and disrupting brain function. In this review we will discuss the development of genetic techniques that bridge the divide between these two approaches and allow the targeting of molecular changes specifically to anatomically dispersed neural representations that are activated by discrete environmental stimuli. These new tools allow the establishment of causal relationships between the activation of sparsely distributed neural ensembles and changes at the behavioral level.
The use of single unit recordings in awake behaving animals provides an exquisitely precise measure of the temporal activity of neurons. This has been used to extract information about how the brain encodes information by studying the correlation between neuronal activity and the presentation of specific sensory stimuli. The best-studied example is probably in the primate visual system where a hierarchical pathway has been defined (Van Essen et al., 1992
). Neurons in the primary visual cortex (V1) fire in response to very general visual features such as orientation whereas following processing through the ventral visual pathway, neurons in inferior temporal cortex respond to complex object features. This visual information is then relayed to the medial temporal lobes, which integrate multimodal sensory information and play a critical role in memory.
Single unit recording studies in the medial temporal lobe in humans have detected neurons with responses to highly defined categories. In the limit, units were found that responded to the presentation of a single individual in a variety of contexts (Quiroga et al., 2005
). One neuron in the right anterior hippocampus responded to the actress Halle Berry, presented in a photograph, as a masked character (catwoman), as a drawing, or as the letter string “Halle Berry”. This level of response specificity shows that the cells are not tuned to general visual features common to images of Halle Berry but to the concept of the specific actress. The striking degree of responsive specificity of these units strongly suggests that they participate in the neural representation of specific individuals. However, these studies are still correlative in the sense that they allow us to watch neurons that fire in a manner suggesting a role in encoding specific information, but they do not allow us to disrupt these neurons specifically to test this hypothesis.
Historically much of what we know about the functional parceling of the brain has been obtained from lesion studies in experimental animals and in patients with damage to specific brain regions (Squire, 2004
). In learning and memory, studies of patient HM, who underwent a bilateral resection of the medial temporal lobe, have helped define this area, and in particular the hippocampal formation, as critical in the formation of long-lasting declarative memories. At a more molecular level both pharmacological and genetic manipulations have been used to test ideas about the cellular signaling mechanisms that underlie behavioral plasticity. However, each of these approaches is limited in that they act as sledgehammers, altering every neuron in a given brain region, when the electrophysiological studies suggest that it is really a very sparse group of neurons that is truly of interest in any given experimental context.
One approach to circuit analysis is to describe the precise pattern of wiring within specific processing units like the hippocampus or a cortical column. For example, in the hippocampus there is the classic tri-synaptic circuit where information from the entorhinal cortex enters through the dentate gyrus, is relayed to CA3 neurons via the mossy fiber pathway and then to CA1 neurons via the Schaffer collateral pathway and finally back out to the entorhinal cortex (Squire, 2004
). This connectivity diagram can be obtained to finer and finer levels of resolution and in principle an entire wiring diagram of a single brain at a single time point could be produced to the level of individual synaptic connections, similar to that obtained in C. elegans (White et al., 1986
). However, even if this were precisely defined down to the level of single synapses it is not likely that the mechanisms that give rise to a neural representation or memory trace would become apparent. As the example of the Halle Berry neuron indicates, these representations are likely to be quite sparse and embedded within a matrix of apparently identical neurons. The particular response patterns of an individual neuron are likely determined by the strength of specific synaptic connections that have been altered through experience. Moreover, even if one could explain how these specific firing patterns arise through circuit plasticity, it would be difficult to experimentally establish the contribution of a particular ensemble of neural activity to an actual representation of the environment.
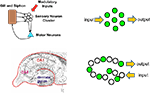
An alternate view is that what defines a circuit is the environmental contingencies that lead to its activation (Figure 1
). In the mammalian brain this is generally referred to as an ensemble code or neural representation of the particular environmental stimulus. In the case of simple systems or reflex pathways, the wiring diagram often predicts the location of the neural ensembles that encode specific environmental information. For example, in the Aplysia gill withdrawal reflex a group of sensory neurons are activated by tactile stimulation and synapse directly onto motor neurons to control withdrawal behavior (Kandel, 2001
). Here the primary sensory neurons are defined by their enervation of the gill and their activation by tactile stimulation of the gill. There is no further higher order processing prior to motor output. The behavioral plasticity of the withdrawal reflex is controlled by synaptic plasticity within these sensory neurons. Because of the uniformity and anatomical isolation of this group of cells, it has been possible to apply techniques for both watching and manipulating neurons within the context of a defined circuit (representation) in a behaving animal. The application of these convergent approaches has proven quite powerful in defining the cellular and molecular mechanisms that underlie behavioral plasticity in this system. The goal of this review is to discuss recent attempts to develop approaches that allow similar convergent molecular and physiological access to the more dispersed neural representations of the mammalian brain.
Figure 1. Clustered and distributed neural ensembles. Two examples of neural circuits are shown. The top panel shows a simplified version of the gill and siphon withdrawal circuit in Aplysia. The sensory neuron cell bodies are located adjacent to each other in a cluster and possess similar biochemistry and response properties. The bottom panel represents the hippocampal circuit of the mammalian brain. The green circles represent neurons that are activated by a specific pattern of sensory stimulation and that when activated contribute to a specific behavioral response. The hippocampal circuit, like many other circuits in the brain, responds to sensory stimulation with activation patterns that can not be predicted from their wiring diagram. Each of these neural ensembles involves a sparse subset of neurons that have an unpredictable spatial distribution. The Aplysia neurons are primary sensory neurons and their response properties can be predicted by their physical location in the ganglion.
One technique that has been used for many years to watch brain activity has taken advantage of a class of immediate early genes or IEGs that are expressed in response to high-level neural firing (Sagar et al., 1988
). The three most commonly used IEGs for this purpose are cfos, arc, and zif268. The expression of these genes is induced by action potential firing, and the �-life of the gene products are relatively short. Thus the expression pattern of IEGs in brain sections from an animal provides a record of the neural activity from several hours prior to sacrifice and has been used extensively to map brain activation from a wide variety of environmental stimulation and in learning and memory relevant paradigms (Guzowski et al., 2005
).
One of the limitations of this approach is that it provides only a single time point record of activity patterns, making it difficult to determine how plasticity modulates activity or even how stable this pattern of gene expression is in relation to an identical stimulus. This problem was addressed using the expression of the IEG arc (Guzowski et al., 1999
). By using fluorescent in situ hybridization to examine expression of arc mRNA they were able to detect the pattern of arc expression at two separate time points in the same animal. They took advantage of the fact that they could detect the expression of the arc precursor RNA while it was still in the nucleus as well as the mature mRNA which was present in the cytoplasm and dendrites. The nuclear arc signal represented very recent and ongoing expression reflecting neural activity several minutes prior to sacrifice of the animal, while the cytoplasmic signal reflected activity that had occurred 30 min or more prior. They used this approach to examine the consistency of activation of the hippocampus when an animal was repeatedly exposed to the same environment. They found that when animals were allowed to explore the same environment, they re-expressed arc in many of the same neurons that had also expressed it on the first exposure. This is a critical result in that it demonstrated for the first time that IEG expression could be used to consistently reflect patterns of activity associated with a discrete representation and provided results that were qualitatively and quantitatively similar to results obtained with electrophysiological recordings of the hippocampus.
The temporal information regarding neural activity that can be obtained using IEG expression is clearly limited relative to electrophysiological recordings. For example, while it is clear that high-level firing induces expression, it is not clear what the precise threshold is and how this might vary among different neuronal cell types. One advantage is that large brain regions can be surveyed and precise anatomical information can be obtained. A second advantage is that the promoter regulatory elements that confer neural activity dependence can, in theory, be used to drive expression of any linked heterologous transgene. This was first demonstrated in transgenic mice by (Smeyne et al., 1992
) using the cfos promoter to drive activity dependent expression of E. coli β-galactosidase. More recently, an axonally targeted β-galactosidase was expressed from the cfos promoter, providing the potential to trace the projections of specific active neuronal populations (Wilson et al., 2002
). The use of these promoter elements is general and provides the potential to introduce functional effector molecules directly into activated neural ensembles to allow their molecular manipulation.
The use of IEG promoters as tools for both watching and potentially manipulating functional neural circuits is limited in a number of ways. For example, the direct introduction of toxins or other molecular regulators via the cfos promoter could be complicated by developmental effects of their expression. In addition, it would be useful for many studies to allow the molecular change introduced into the activated neurons to be maintained for more prolonged periods than the short (minutes to hours) times afforded by the promoters themselves. We therefore set out in a recent study to develop a genetic system with the following features. (1) The expression of any transgene of interest should be linked to neural activity only during a specific experimenter controlled time window. (2) The transgene expressed in those active neurons should be maintained for a prolonged period, but no further labeling of active cells should occur following closure of the permissive time window.
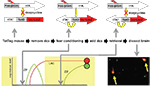
We achieved activity dependent regulation of transgene expression with these two features by combining elements of the tetracycline system for gene regulation with the cfos promoter as shown in Figure 2
(Reijmers et al., 2007
). The approach uses two separate transgenes in the same animal. The first uses the cfos promoter to drive expression of the tetracycline transactivator (tTA or TET-off). In mice carrying only this transgene high-level neural activity will result in the induction tTA, which is a transcription factor that can be blocked by the antibiotic Doxycycline (Dox). In the absence of Dox tTA drives expression of genes linked to a tetO-promoter sequence. The second transgene incorporates both a tetO-linked reporter (in this case the somato-axonal marker taulacZ) as well as a transcriptional feedback loop to maintain tetO-linked gene expression indefinitely once it is activated. The tTA (tTA*) in this construct was made Dox insensitive by introduction of a point mutation in the Tet binding domain. In the presence of Dox the tetO-linked reporter is not activated even in those neurons in which the cfos-linked tTA is expressed. However, if Dox is withdrawn then both taulacZ and tTA* are expressed, but only in those neurons that were active to a high enough level to induce the cfos-linked tTA. Once activated, the tTA* sets up a transcriptional feedback loop that can be maintained even in the presence of added Dox. In this manner discrete time windows for genetic tagging of active neurons can be opened and closed through the use of Dox. In the absence of Dox, any neuron that has sufficient induction of cfos-linked tTA to activate the feedback loop will persistently activate the taulacZ reporter, as well as any other tetO-linked transgene that is introduced into the mouse (Aiba and Nakao, 2007
). This expression will be maintained even when the time window for sampling active neurons is closed by the readministration of Dox. In this way a persistent record of neurons that were active during the off-Dox period can be maintained. We called this the TetTag mouse, which stands for TETracycline transactivator controlled genetic TAGging of active neural circuits.
Figure 2. The TetTag mouse. The figure summarizes an experiment from Reijmers et al. (2007)
. Mice carrying two transgenes were used. The first transgene uses the cfos promoter to drive expression of the tetracycline transactivator (tTA). tTA activates the tetO promoter in the absence but not presence of doxycycline (Dox). The second transgene uses the tetO promoter to drive expression of a Dox insensitive tTA (tTA*), which, once expressed, sets up a positive feedback loop that continuously drives expression of a β-galactosidase reporter coupled to the tau protein (taulacZ). Neurons activated during fear conditioning (while off Dox) were tagged with long-lasting expression of taulacZ (LAC; red circle). Mice were put back on food with doxycycline and a retrieval test was done 3 days later, followed by analysis of the brains 1 h after retrieval for expression of lacZ and zif268. Neurons activated during learning expressed lacZ and those active during retrieval expressed zif268 (ZIF; green circle). The number of neurons in the amygdala that expressed both LAC and ZIF, indicating that they were activate during both learning and retrieval, was positively correlated with the strength of the fear memory that the animal displayed.
We used the TetTag mouse to examine the neural circuit that mediates fear memory (Reijmers et al., 2007
). We asked whether neurons that are stimulated during learning in a Pavlovian fear-conditioning paradigm were reactivated during retrieval of the memory as shown in Figure 2
. We subjected TetTag mice to a learning trial consisting of paired presentations of a tone (CS) and a foot-shock (US). This results in a long-lasting fear memory for both the tone (cued conditioning) and the conditioning box in which the animals were shocked (context conditioning). The learning trials took place during a time window in which the animals were free from Dox, allowing activated neurons to be tagged with long-lasting expression of taulacZ. The animals were then returned to Dox to prevent further tagging of activate neurons, tested for retention of the memory in a retrieval trial, and sacrificed after 1 h for analysis using the endogenous IEG zif268 as a measure of recent neural activity. By comparing the expression of lacZ (activity during learning) and zif268 (activity during memory retrieval) we could determine the degree of circuit reactivation. We found that the number of reactivated neurons in the amygdala, a region critical in fear conditioning (LeDoux, 2007
), correlated with the retrieval of the fear memory.
We hypothesized that these reactivated neurons represent a component of the memory trace for conditioned fear. To test this idea we weakened the strength of the memory by extinction training; giving repeated CS presentations without the shock US. Animals were first fear conditioned while free from Dox to tag the learning activated neurons. The extinction training then took place following re-exposure to Dox (to prevent further labeling) and a memory retrieval trial was conducted 1 h prior to analysis. Extinction is never complete and varied significantly from animal to animal. We found that there was a significant correlation between the strength of the remaining fear response and the degree of circuit reactivation; animals with a high fear response during retrieval showed strong reactivation of the learning circuit while those with low fear responses showed a low degree of reactivation. In addition, we found some specificity in the anatomy of the responses so that reactivation in the basal amygdala was correlated with context fear while reactivation in the lateral amygdala was associated with the strength of the cued (tone) fear memory. These results are consistent with the known role of these subdivisions of the amygdala with the two different forms of fear memory (Quirk et al., 1995
; Herry et al., 2008
).
These results demonstrate that memory retrieval results in a reactivation of some of the same neurons that were active during the initial learning. We suggested that neurons activated by the US (shock) but also receiving weak CS inputs were altered during learning such that the CS alone could now activate them. In this way presentation of the CS alone after learning would recapitulate a portion of the aversive US leading to downstream fear responses. This approach represents a somewhat elaborate way of simply watching neural activity and the results are still purely correlative. However, by using a regulatable and binary genetic system, it should be relatively easy to introduce additional effector transgenes into the mice to control the activity or biochemistry of these neurons and directly test their role in memory. One recent study by Josselyn and coworkers has achieved this direct manipulation of neural ensembles associated with a specific memory trace using a somewhat different approach.
The use of neural activity to introduce genetic alterations into neurons offers the possibility of obtaining direct molecular control over the neurons that participate in a specific neural representation or memory trace. An alternate approach that realized this goal took advantage of the finding that certain molecular manipulations could recruit neurons to participate in control of a specific memory. In one recent study, (Han et al., 2007
), it was found that over expression of the transcription factor CREB in neurons resulted in their preferential recruitment into a fear memory trace. In this study, neurons in the amygdala were randomly infected with a viral vector that over expressed CREB and the animals were then trained in fear conditioning. They then performed a retrieval trial and examined the expression of the IEG arc in the amygdala. They found that the CREB over expressing neurons were more likely to be activated during the memory retrieval. This result suggests that these CREB over expressing neurons were predisposed to participate in the memory trace. The mechanism by which these neurons are preferentially recruited is unclear but it does demonstrate that, at least in the amygdala, there is a good deal of flexibility in which neurons can be used to encode a specific memory. The system is not hard wired at the level of individual neurons but there is a sort of competition, with CREB over expression favoring a neuron’s recruitment into the memory trace.
Josselyn and coworkers went on to take advantage of this CREB priming trick to directly manipulate a specific fear memory trace (Han et al., 2009
). In this study the viral vector that delivered CREB to amygdala neurons also carried a gene that allowed for the expression of the diphtheria toxin receptor (DTR) (Figure 3
). The CREB over expression recruited the neurons to the memory trace and expression of the DTR allowed for the selective ablation of these specific neurons with diphtheria toxin (DT). Ablation of the CREB over expressing neurons disrupted the fear memory while ablation of a similar number of random neurons in the amygdala did not. The memory effect was long lasting and specific (the same animals could learn a second fear association) demonstrating that this limited group of neurons played a critical role in the specific memory encoded during the CREB expression time window. This is the first example of the disruption of a specific memory within a distributed network.
Figure 3. Disrupting a specific memory in the mouse. The figure summarizes the experiments of Han et al. (2009)
. Transgenic mice were used that express an inducible diphtheria toxin receptor (iDTR). A viral vector expressing both CREB and CRE recombinase was injected into the amygdala of iDTR mice leading to the expression of both CRE and CREB in a random subset of neurons (circles with CREB/CRE). The CRE recombinase removed a transcriptional STOP sequence and allowed for expression of DTR in these CREB expressing neurons. The mice were then subjected to fear conditioning. An earlier study from the same authors (Han et al., 2007
) demonstrated that the CREB expressing neurons participate in the storage of the fear memory (green circles symbolize neurons that participate in the storage of the memory). After fear conditioning, mice were injected with diphtheria toxin (DT), which killed the CREB expressing neurons which participated in the encoding of that memory (red circles). This caused a significant reduction in the strength of the fear memory measured during retrieval.
While the ability of CREB over expression to recruit neurons to participate in a specific memory is interesting in its own right, it would be useful to have an approach to manipulate neurons that were naturally activated by any general environmental stimulus. This has recently been accomplished using a technique in which neurons expressing β-galactosidase can be specifically disrupted with a pharmacological agent (Koya et al., 2009
). The study used rats that carry a cfos-promoter driven β-galactosidase to label activated neurons. To manipulate the neurons they use a drug that is inactive in the absence of β-galactosidase (daun02) but can be hydrolyzed to a compound that can reduce Ca2+ dependent action potentials (Santone et al., 1986
). They examined context specific sensitization to cocaine, which is an associative paradigm where the response to a drug of abuse is potentiated when it is administered in the same environment in which it has been repeatedly taken. Animals were given repeated injections of cocaine over 1 week in context A to produce the context specific sensitization (measured as increased locomotor response to the drug). Following the training, a final sensitization trial to induce β-galactosidase was given and 90 min later daun02 was injected into the nucleus accumbens to disrupt the β-galactosidase expressing neurons. Previous studies had suggested that the nucleus accumbens was a critical site of plasticity mediating this behavior (Mattson et al., 2008
). The injected animals showed a reduction in the context specific component of the sensitization but retained normal responses to cocaine in a novel context B when tested several days later.
Like the results with the CREB over expression, this study suggests that a specific associative representation (context A + cocaine) is being interfered with selectively. While the behavioral results in this study are intriguing and the linkage to cfos based expression provides a potentially general approach for manipulating discrete neural representations, there are a number of important questions that remain to be addressed with this technique. First, the electrophysiological effect of the daun02 treatment was not examined directly in the neurons but inferred from studies in cell lines. Whether the effects are mediated by suppression Ca2+ dependent action potentials or some other effect of the treatment remains to be determined. The time course of any neural excitability or Ca2+ channel changes is also a critical parameter. The effect of the daun02 treatment was examined 3 days after the initial injection of the compound and it is unclear whether the observed effect was due to ongoing suppression of activity or to a persistent effect manifest during the initial treatment. Nevertheless, it demonstrates the general principles of this approach, which could be combined with the host of recently developed genetic regulators of neural activity.
While neural representations are encoded in the specific ensemble of neurons that are activated in response to a stimulus, the plasticity that molds these patterns of activation is thought to occur at the synapse. It has been known for some time that long-term memory lasting 24 h requires new gene expression initiated at the time of learning, while short-term memory lasting a few hours lacks this requirement (Davis and Squire, 1984
). Since the short and long-term memories for the same event presumably involve the same pattern of synaptic changes, it raises the question of how the required gene products exert their effects selectively on the appropriate synapses. A potential answer to this question was suggested by Frey and Morris in studies of long-term potentiation (LTP), a form of synaptic plasticity thought by many to underlie memory (Frey and Morris, 1997
, 1998
). They found that synaptic activity could produce a sort of molecular “tag” at a synapse that would allow it to utilize newly expressed gene products to maintain LTP for long periods. We recently used the cfos based genetic approach to demonstrate a similar mechanism in behavioral learning and memory.
A number of studies have implicated the regulated trafficking of the glutamate receptor GluR1 to synapses as an important mechanism in both LTP and fear learning (Kessels and Malinow, 2009
). In addition to allowing the genetic tagging of activated neurons, the IEG promoters like cfos show a very rapid onset and offset of expression, making them useful for cellular trafficking and turnover studies. We took advantage of this property to examine the trafficking of GluR1 following learning in the fear-conditioning paradigm (Matsuo et al., 2008
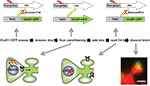
). Mice carrying both a cfos-tTA and tetO-GFPGluR1 fusion transgene were used in this study (Figure 4
). In the absence of Dox neural activity will induce a pulse of expression of the GFP tagged GluR1 and the distribution to synapses of this newly synthesized receptor can be followed histologically. Animals were fear conditioned in the absence of Dox to both induce a contextual fear memory and to induce synthesis of the GFP tagged receptor in activated neurons. The distribution of the receptor to dendritic spines (the site of most excitatory synapses) in the hippocampus was examined 24 h after the conditioning. We found that the receptor was not evenly distributed but present in only about 50% of spines, even in controls. In the fear conditioned animals we found a similar distribution except that there was an increase in trafficking to one morphological type of spine, the mushroom spines. This preferential trafficking only happened when the conditioned stimulus (CS: novel box) and the unconditioned stimulus (US: foot shock) were paired, but not when CS or US were presented separately. The newly synthesized GluR1 requires 2 h to begin to reach the dendritic spines, yet is somehow preferentially recruited to a specialized class of spine based on the associative conditioning that occurred 2 h prior to its arrival. This is indicative of a synaptic tagging event acting in behavioral memory similar to that described for LTP (Frey and Morris, 1997
).
Figure 4. Learning regulated targeting of glutamate receptors. The figure summarizes an experiment from Matsuo et al. (2008)
. Mice carrying two transgenes were used. The first transgene is identical to the one described in Figure 2 and uses the cfos promoter to drive expression of a tetracycline transactivator (tTA). The second transgene was a tetO-promoter GFP tagged glutamate receptor subunit (GluR1-GFP). Animals were fear conditioned in the absence of Dox to produce both a fear memory and a pulse of GFP-GluR1 expression in active neural ensembles in the hippocampus. The distribution of GFP-GluR1 in dendritic spines was analyzed 24 h following the conditioning using DiI to label all spines on a given neuron. Fear conditioning led to an increase in trafficking of the receptor specifically to mushroom type spines. This experiment demonstrates how genetic tools can be used to image a specific molecular event selectively within an activated neural circuit.
The ability to genetically manipulate activated neuronal ensembles or neurons participating in a sparsely encoded memory trace offers a number of advantages that are only beginning to be realized. A parallel line of technological development has focused on generating genetic tools for manipulating neuronal activity. The light regulated channelrhodopsin ChR, developed by Deisseroth and colleagues (Zhang et al., 2006
), allows for the very precise light regulated control of action potential firing in neurons expressing the channel. There is an expanding tool box of genetic effectors like ChR that are light or ligand controlled and can be used to either stimulate or suppress neural activity (Luo et al., 2008
). In addition, there are a number of similar effectors that can be used to regulate second messenger signaling pathways when expressed in heterologous cells (Isiegas et al., 2008
; Pei et al., 2008
; Airan et al., 2009
; Alexander et al., 2009
).
The combination of these new tools with activity based genetic delivery and multi time point brain activity mapping at cellular resolution will open up a variety of new questions to experimental analysis. To return to the initial discussion of neural representations, the fusion of these approaches should allow one to address the question of what neural firing patterns “mean” to the animal. To take the example of the Halle Berry neurons (actually the equivalent in genetically accessible animal models) one could ask what would be the consequences of silencing this specific group of neurons. Would the recognition of Halle Berry be inaccessible? What fraction of neurons in the representation need to be silenced in order to impair recognition? How does the silencing of a specific group of neurons in one brain region affect the activation patterns in downstream areas?
An alternate approach is to ask whether a representation can be built by experimenter driven stimulation of the appropriate neurons. In one recent study, Svoboda and colleagues delivered the ChR2 molecule to random populations of neurons in the somatosensory cortex. They found that stimulating as few as 300 neurons could be detected by the animals and used to alter behavior in a conditioning task (Huber et al., 2008
). However, the coordinated activation of those neurons presumably does not form any natural representation. Suppose that instead of a random group of neurons one could activate a subset of neurons that responded to a natural stimulus, say the tone CS in a fear-conditioning task. The artificial stimulation of those neurons paired with a foot shock would presumably lead to a conditioned fear of the extrinsic stimulation CS. But the more interesting question now is would the animal also fear a tone? If so would all the features of this sensory representation be maintained, for example frequency selectivity? With what fidelity does artificial stimulation of the tone representation in one brain region recapitulate the brain activity patterns produced by the natural tone itself? This type of approach should now be achievable and allow direct functional investigation of the structure of neural representations.
Finally, the ability to genetically alter activated neural ensembles provides an entre into a more specific biochemistry of the brain. In the GluR1 trafficking studies discussed above, the GFP tagged receptor provides not only a signal to watch molecular movements, but also a tag for specific biochemical analysis. For example, one could ask how the synapses from activated neurons that received new receptor differ biochemically from those that did not by using the antibodies to the GFP tag to affinity purify positive from negative material. A similar approach could be used to tag specific cellular compartments or molecular complexes so that biochemical studies can be limited to just the activated neurons, sparsely embedded in a matrix of inactive neurons and glia.
It should also be possible to improve the specificity of the genetic modifications using a variety of genetic tricks such that only neurons active in one brain region or active at time point 1 but not time point 2 would be tagged. The increased specificity along with the new genetic tools and biochemical tagging should provide a new level of circuit analysis in the brain and break down the barrier between watching neural firing and manipulating neural function.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alexander, G. M., Rogan, S. C., Abbas, A. I., Armbruster, B. N., Pei, Y., Allen, J. A., Nonneman, R. J., Hartmann, J., Moy, S. S., Nicolelis, M. A., McNamara, J. O., and Roth, B. L. (2009). Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron 63, 27–39.
Isiegas, C., McDonough, C., Huang, T., Havekes, R., Fabian, S., Wu, L. J., Xu, H., Zhao, M. G., Kim, J. I., Lee, Y. S., Lee, H. R., Ko, H. G., Lee, N., Choi, S. L., Lee, J. S., Son, H., Zhuo, M., Kaang, B. K., and Abel, T. (2008). A novel conditional genetic system reveals that increasing neuronal cAMP enhances memory and retrieval. J. Neurosci. 28, 6220–6230.
Koya, E., Golden, S. A., Harvey, B. K., Guez-Barber, D. H., Berkow, A., Simmons, D. E., Bossert, J. M., Nair, S. G., Uejima, J. L., Marin, M. T. Mitchell, T. B., Farquhar, D., Ghosh, S. C., Mattson, B. J., and Hope, B. T. (2009). Targeted disruption of cocaine-activated nucleus accumbens neurons prevents context-specific sensitization. Nat. Neurosci. 12, 1069–1073.