1
UMR 5170 CNRS, Centre des Sciences du Goût, Dijon, France
2
Departamento de Bioquímica, Instituto de Química, Universidade de São Paulo, São Paulo, Brazil
3
General Olfaction and Sensing Program on a European Level, Centre Européen des Sciences du Goût, Dijon, France
Taste receptors for sweet, bitter and umami tastants are G-protein-coupled receptors (GPCRs). While much effort has been devoted to understanding G-protein-receptor interactions and identifying the components of the signalling cascade downstream of these receptors, at the level of the G-protein the modulation of receptor signal transduction remains relatively unexplored. In this regard a taste-specific regulator of G-protein signaling (RGS), RGS21, has recently been identified. To study whether guanine nucleotide exchange factors (GEFs) are involved in the transduction of the signal downstream of the taste GPCRs we investigated the expression of Ric-8A and Ric-8B in mouse taste cells and their interaction with G-protein subunits found in taste buds. Mammalian Ric-8 proteins were initially identified as potent GEFs for a range of Gα subunits and Ric-8B has recently been shown to amplify olfactory signal transduction. We find that both Ric-8A and Ric-8B are expressed in a large portion of taste bud cells and that most of these cells contain IP3R-3 a marker for sweet, umami and bitter taste receptor cells. Ric-8A interacts with Gα-gustducin and Gαi2 through which it amplifies the signal transduction of hTas2R16, a receptor for bitter compounds. Overall, these findings are consistent with a role for Ric-8 in mammalian taste signal transduction.
Taste receptors are the sensors through which the tastants dissolved in the saliva are detected. Over the years evidence has accumulated supporting the importance of G-protein mediated signaling in taste detection. Initially, electrophysiological studies implicated cyclic nucleotides in taste transduction (Avenet et al., 1988
; Tonosaki and Funakoshi, 1988
). Subsequently, gustducin, a G-protein alpha subunit expressed in taste receptor cells was shown to be involved in bitter, sweet, and glutamate detection in mice (McLaughlin et al., 1992
; Wong et al., 1996
; He et al., 2004
). More recently and importantly, Tas1Rs and Tas2Rs, two families of G-protein-coupled-receptors (GPCRs) that are respectively involved in the detection of certain sweet, umami and bitter tasting compounds were discovered (Hoon et al., 1999
; Adler et al., 2000
; Chandrashekar et al., 2000
; Matsunami et al., 2000
; Nelson et al., 2001
, 2002
; Zhao et al., 2003
). GPCR signaling cascades involve the activation of G-proteins as well as subsequent stimulation of effector enzymes by the free alpha or beta-gamma subunits of the activated heterotrimeric G-protein (Oldham and Hamm, 2008
). Accessory proteins such as regulators of G-protein signaling (RGS) and GDP/GTP exchange factors (GEFs) which regulate the activation state of the G-protein, have been shown to modulate GPCR signaling in a variety of cell types (Klattenhoff et al., 2003
; Siderovski and Willard, 2005
; Neitzel and Hepler, 2006
). GEFs act as signal amplifiers by promoting the exchange of GDP to GTP on the receptor-activated Gα subunits whereas RGS promote the hydrolysis of GTP thus attenuating the activation of the effector (Wilkie and Kinch, 2005
; Jean-Baptiste et al., 2006
).
In vision and olfaction, two senses involving GPCRs as main detectors, RGS9-1 and RGS2 respectively have been reported to attenuate light or odorant responses (Sinnarajah et al., 2001
; Nishiguchi et al., 2004
) while Ric-8B a putative GEF for Gαolf that is abundant in olfactory sensory neurons amplifies odorant receptor signaling (Von Dannecker et al., 2005
, 2006
). Ric-8B has one other known mammalian homologue, Ric-8A, which was shown to enhance Gαq-mediated ERK activation by GPCRs (Nishimura et al., 2006
) as well as interact and display GEF activity for Gαi1, Gαq and Gαo but not Gαs in vitro (Tall et al., 2003
). The GEF activity of Ric-8A on the Gαι subunit has been extensively studied in receptor-independent G-protein-mediated events regulating microtubules pulling forces during cell division (Tall and Gilman, 2005
). In this system the action of Ric-8A is similar to that of a GPCR while the GDI activity of GPR/GoLoco motif-containing proteins resemble that of Gβγ subunits (Thomas et al., 2008
).
In gustation, RGS21 has been reported to be expressed specifically in taste cells and to play a role in gustducin signaling (von Buchholtz et al., 2004
); however, no reports of GEFs further controlling this system have been made. To study whether GEFs are involved in the transduction of the signal downstream of the taste GPCRs we investigated the expression of Ric-8A and Ric-8B in mouse taste cells and their interaction with G-protein subunits found in taste buds.
Animals
French guidelines for the use and the care of laboratory animals were followed, and experimental protocols were approved by the animal ethic committee of the University of Burgundy. Six-week-old C57BL/6J mice housed in a controlled environment (constant temperature and humidity, darkness from 8 pm to 8 am) were used for all experiments. They were fed a standard laboratory chow ad libitum (UAR A04, Usine d’Alimentation Rationnelle, France).
Yeast Two-Hybrid Interactions and Co-Immunoprecipitation
The entire open reading frame of mouse Ric-8A, RGS21, gustducin, Gαt2, Gαi2, Gαolf, or GPR domains 1–4 of AGS1, AGS2, AGS3 or GoLoco domain of AGS4, PBP, Pins, RGS2, RGS9, were PCR amplified from C57BL6/J mice heart, testis or circumvallate papillae cDNA using specific primers (Operon, Germany) containing a Sal I (forward primer) or Not I (reverse primer) restriction site. Ric-8B was from Von Dannecker et al. 2006
.
(5′ > 3′): Ric-8A(S) CGAGGTCGACTGAGCCCCGGGCAGTTGCG,
Ric-8A(AS) CTTAGCGGCCGCTCAGTCAGGATCTGAGTCAGG,
RGS21(S) TTGTCGACCTCGAGGCCAGTGAAATGCTGTTTC,
RGS21(AS) TTGTCGACGCGGCCGCTTACAGGAAAGGCAG,
Gαgus(S) AAAGCACGCGTGATGGGAAGTGGAATTAGTTCAG,
Gαgus(AS) CAAAGCGGCCGCTCAGAAGAGCCCACAGTCTTTGAGGTT,
Gαt2(S) CGAGGTCGACTGGGAGTGGCATCAGTGCT,
Gαt2(AS) GAATGCGGCCGCTTAAAAGAGCCCACAGTCCTTGA,
Gαi2(S) GGAATTCCCACCATGGGCTGGACCTGTAG,
Gαi2(AS) AGCGGCCGCGAAGAGGCCACAGTCCTTC,
Gαolf (S) CGAGGTCGACTGGGTGTTTGGGCAACAGC,
Gαolf (AS) GAATGCGGCCGCTCACAAGAGTTCGTACTGCTTG,
AGS1(S) CGAGGTCGACGAAACTGGCCGCGATGATC,
AGS1(AS) CTTAGCGGCCGCCTAACTGATGACACAGCG,
AGS2(S) CGAGGTCGACGGAAGACTTCCAGGCCTC,
AGS2(AS) CTTAGCGGCCGCTCAGATGGACAGTCCGAAG,
AGS3(S) CGAGGTCGACTATTCCCAGGGCCCCGTC,
AGS3(AS) CTTAGCGGCCGCTTAGCTGGCACCCGGTG,
AGS4(S) CGAGGTCGACGGAGGCTGAAAGACCCCAG,
AGS4(AS) CTTAGCGGCCGCTCAGCAGGTGTGTGTAGG,
PBP(S) CGAGGAATTCTGGCCGCCGACATCAGC,
PDB(AS) CTTAGCGGCCGCCTACTTCCCTGACAGCTG,
Pins(S) CGAGGTCGACAATCAGTTCAGACACGATTG,
Pins(AS) CTTAGCGGCCGCTTATTTTCCCGAATGCTTAAA,
RGS2(S) CGAGGTCGACTATGCAAAGTGCCATGTTCCTG,
RGS2(AS) CTTAGCGGCCGCTCATGTAGCATGGGGCTC,
RGS9(S) CGAGGTCGACGGTGGAGATCCCAACCAAGATG,
RGS9(AS) CTTAGCGGCCGCTCACTGGGTGATGTCCACGG.
After amplification with PFU (Stratagene, USA) according to the manufacturer’s specifications, the products of the expected size were subcloned into pSTBlue-1 according to the manufacturer’s specifications (Novagen, USA) and sequenced before subsequent subcloning as a fusion into the Sal I and Not I sites of either pDBLeu (bait vector) or pEXP (prey vector) of the Proquest two-hybrid system (Invitrogen, USA).
Competent Mav203 yeast cells (Invitrogen, USA) were co-transformed with 200 ng of each prey and bait vector and grown 48 h at 30°C on minimal media plates without leucine and tryptophan. Two colonies were then collected and each was dissolved separately into 500 μl of water before spotting 10 μl of each solution side by side onto plates lacking leucine histidine and tryptophan but containing either 10, 25 or 50 mM 3-AT to test the strength of the interaction. After 24 h at 30°C, the plates were replica cleaned using a velour cloth before an additional incubation of 48–72 h at 30°C prior to scoring growth.
For co-immunoprecipitations the open reading frame of Gαi2, Gαt2 and Gαgus were subcloned into pDisplay (Invitrogen, USA) in frame with the HA epitope. Prior to subcloning into pCDNA3 (Invitrogen, USA) a Flag tag was inserted at the N terminus of Ric-8A. All constructs were verified by sequencing. These constructs as well as a Flag-tagged-Ric-8B (Von Dannecker et al., 2006
) were transfected in various combinations into HEK 293T cells seeded in six well plates. Forty-eight hours later the cells were harvested and lysed following the manufacturer’s directions. The lysate was incubated overnight at 4°C with 2 μg of mouse monoclonal anti-Flag (Kodak, USA) or 12CA5 mouse monoclonal anti-HA antibody (Roche, Switzerland) and protein A Sepharose (Roche, Switzerland) following the manufacturer’s directions. After washing the immunoprecipitate four times with lysis buffer the samples were eluted by boiling for 5 min in sample buffer (Bio-Rad, USA) and subjected to SDS-PAGE and Western blot analysis. For Western blot hybridization the antibodies were used at a dilution of 1/1000. The membrane was subsequently processed using the components of the chemiluminescent detection kit according to the manufacturer’s instructions (GE Healthcare, USA).
RT-PCR and Quantitative RT-PCR
Total RNA was isolated from various tissues, using RNAeasy (Qiagen, Germany) according to the manufacturer’s instructions. First strand cDNA was synthesized using 500 ng of total RNA with Superscript II (Invitrogen, USA). With the exception of foliate, fungiform and palate papillae, for each tissue three pools of mRNA, each from three mice, were isolated to perform triplicate RT reactions and triplicate qPRC reactions. For non quantitative PCR, the three RT reactions were pooled for each tissue.
Quantitative RT-PCR was performed on a Mini Opticon (Biorad, USA). Reactions (50 μl) contained 1× iQTMSYBR green supermix (Biorad, USA), 100 nM of each primer, and 1 μl of a 1:5 dilution of the appropriate RT reaction.
Amplification efficiency (E) was determined for each primer pair on triplicate 5-fold serial dilutions of mouse brain cDNA. Relative quantification was performed in relation to cyclophilin expression using the Pfaffl algorithm with kinetic PCR efficiency correction (REST-MCS) (Pfaffl, 2001
; Pfaffl et al., 2002
). Results are expressed as expression ratio (R) derived from the equation:
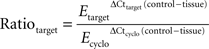
where the reference gene is cyclophilin (cyclo).
RT-PCR reactions (25 μl) contained 1× Taq mastermix (Qiagen, Germany), 0.4 μM of each primer, 1 μl of appropriate RT reaction (water for control). Cycling parameters: 95°C for 2 min then 35 cycles of 95°C for 30 s; 60 or 58°C for 40 s, 72°C for 40 s, and final elongation at 72°C for 2 min. Products (13 μl) were run onto 1.4% agarose Seakem TAE gels (Cambrex, USA). Primers used were (5′ > 3′): Ric-8A (S) CACGAAGGATCCTTAGAGTTCATG, Ric-8A (AS) GCACATTCTGTCAACACGTTCA, F78a: CAAAGAGAGAGTGGATAGCCTGC, R810b: GCAACTCCTCTTTTGGTTTTGC, cyclophilin (S): CAGACGCCACTGTCGCTTT, cyclophilin (AS): TGTCTTTGGAACTTTGTCTGCAA, F89: GCAAAACCAAACATTAATCTTATCACT, R910: AAGCAACTCCTCTCTGGAAAGTTTAT, F6: GGAAGCAGCTATAGAGAGGGTCTAA, R7: CCAGCTTATTTCTCACGGTTGA.
Immunohistochemistry and Western Blotting
Four- to six-week-old male C57BL6/J mice were perfused with 4% paraformaldehyde. Circumvallate papillae were excised and allowed to soak in 30% sucrose overnight before being snap-frozen in isopentane chilled with liquid nitrogen. The tissue was then embedded in OCT compound (Tissue-Tek, Japan) and processed using a cryostat into 14-μm sections. Sections were air-dried for 2 h at room temperature and stored at −80°C. On the day of the experiment sections were rehydrated in 0.1 M phosphate saline buffer (PBS, pH 7.4) for 10 min and blocked in 5% goat serum (Sigma, USA), 0.2% Triton X-100 in PBS for 15 min at room temperature then incubated overnight at 4°C with either a 1/100 dilution of the rabbit polyclonal anti-Ric-8B antibody (Kerr et al., 2008
) a 1/100 dilution of the rabbit polyclonal anti-Ric-8A antibody (Proteintech group, Inc., Chicago, IL, USA) or a 1/400 dilution of the rabbit polyclonal anti-gustducin antibody (SCBT, Santa-Cruz, USA). After washing, sections were next incubated for 2 h at room temperature with a 1/600 dilution of Alexa-488-conjugated anti-rabbit IgG secondary antibodies (Molecular probes, USA). After washing and counterstaining with Hoescht 33258 (Sigma, USA) slides were mounted in gel/mount (Biomeda Corp., USA) and analyzed under an Axioskop microscope equipped with an Axiocam MRc5 (Zeiss, Germany). For double-labelling, the sections were incubated simultaneously with a 1/200 dilution of the mouse monoclonal anti-IP3R-3 (BD Biosciences, USA) and either a 1/100 dilution of the rabbit polyclonal anti-Ric-8B antibody or a 1/100 dilution of the rabbit polyclonal anti-Ric-8A antibody (Proteintech group, Inc., Chicago, IL, USA) overnight at 4°C, washed prior to incubation with secondary antibodies for 2 h at room temperature (1/600 dilution of Alexa-488-conjugated goat anti-rabbit IgG and 1/600 dilution of Alexa-555-conjugated goat anti-mouse IgG (Molecular Probes, USA). Slides were analyzed under a TCS4D confocal microscope (Leica, Germany).
For Western blotting, circumvallate papillae from 4- to 6-week-old male C57BL6/J mice were collected and mechanically dissociated in standard lysis buffer using a dounce homogenizer on ice. 20 μg of the total protein extract was loaded on a 10% SDS-PAGE gel (Bio-Rad, USA), transferred onto a hybond-P, PVDF membrane (GE Healthcare, USA) and incubated overnight at 4°C with either a 1/100 dilution of the rabbit polyclonal anti-Ric-8B antibody, a 1/100 dilution of the rabbit polyclonal anti-Ric-8A antibody (Proteintech group, Inc., Chicago, IL, USA) or a 1/1000 dilution of a mouse monoclonal anti-β−actin antibody (Sigma, USA). The membrane was subsequently processed using the components of the ECL kit according to the manufacturer’s instructions (GE Healthcare, USA).
HEK 293T cells in DMEM supplemented with 10% fetal calf serum were grown in six wells plates until 50% confluence was reached. Cells were transfected with pCDNA3 alone or containing 5′Flag-Ric8A, or 5′Flag-Ric8B. Transfections were carried out using Lipofectamine LTX (Invitrogen, USA) according to the manufacturer’s protocol. 48 h after transfection the cells were harvested and lysate prepared. 15 μg of total protein extract was loaded on a denaturing 4–12% BisTris PAGE gel (Invitrogen, USA), transferred onto a hybond-P, PVDF membrane (GE Healthcare, USA) and incubated overnight with either a 1/100 dilution of the rabbit polyclonal anti-Ric-8B antibody, a 1/300 dilution of the rabbit polyclonal anti-Ric-8A antibody (Proteintech group, Inc., Chicago, IL, USA) or a 1/1000 dilution of mouse monoclonal anti-Flag (Kodak, USA). The membrane was subsequently processed using the components of the ECL kit according to the manufacturer’s instructions (GE Healthcare, USA). The next day, the membranes were stripped and reprobed with a 1/1000 dilution of a mouse monoclonal anti-β−actin antibody (Sigma, USA).
In Situ Hybridization
The procedure used for in situ hybridization of 16-μm sections of fresh frozen circumvallate papillae from C57BL6/J male mice was described previously (Matsunami et al., 2000
). Briefly, the sections were hybridized (58°C) overnight to hydrolysed digoxigenin-labeled cRNA probes prepared from cloned segments of cDNAs encoding Ric-8A (NCBI reference: NM_053194 nt 75-1667), Ric-8B (NCBI reference: AY940666 nt 99-1781) or gustducin (NCBI reference: BC147841 nt 125-1043). After washing, the sections were incubated with anti-Dig AP and subsequently developed using a mix of NBT-BCIP (Roche, Switzerland) according to the manufacturer’s instructions. Slides were then mounted with vectamount (Vector laboratories USA) and analyzed under an Axioskop microscope equipped with an Axiocam MRc5 (Zeiss, Germany).
cAMP Assay
Intracellular cAMP production was monitored using a cell-based reporter gene assay relying on transcriptional regulation of secreted human placental alkaline phosphatase (SEAP) by five 5′-cyclic AMP (cAMP) response elements (CREs). This assay is based on the premise that intracellular cAMP activates protein kinase A which in turn phosphorylates the transcription factor CREB (cAMP response element binding protein) ultimately leading to SEAP production. SEAP activity is then determined using 4-methylumbelliferyl phosphate (4-MUP) as substrate.
The coding sequence of hTas2R16, a receptor activated by salicin, was PCR amplified using specific primers and fused to the first 44 amino acids of Bovine rhodopsin before subcloning into pCDNA3 (Invitrogen, USA). The coding sequence of the mouse Gαi2 or Ric-8A were PCR amplified using specific primers and subcloned into pEF6 (Invitrogen, USA) and pcDNA 3.1 (Invitrogen) respectively. All constructs were verified by DNA sequencing. HEK 293T cells in DMEM supplemented with 10% fetal calf serum were grown in 96 wells plates until 80–95% confluence was reached. Cells were transfected with different combinations of plasmid constructs coding for Tas2R16, D1R (Von Dannecker et al., 2005
), Gαi2, Ric-8A and pCRE-SEAP (Durocher et al., 2000
). Transfections were carried out using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s protocol. 24 h after transfection the media was replaced by 200 μl of serum free DMEM containing the different agonists (dopamine, salicin) and incubated for 6 h at 37°C. 200 μl of the media from each well was transferred to a new plate, heated to 65°C for 30 min and spun to remove debris. 100 μl of each sample was mixed with 100 μl of the SEAP buffer (30 mM Tris pH 8.0, 0.06%BSA, 1 mM L-homoarginine, 10 mM MgCl2, 20% diethanolamine, 3.6 mg 4-methylumbelliferyl phosphate) and incubated for 1 h at 37°C. Fluorescence was measured at a wavelength of 449 nm using a Viktor 1420 microplate reader (PerkinElmer, USA). The deactivation rate was calculated by dividing each deactivation value (dopamine + salicin) by the corresponding activation value (dopamine alone). Statistical significance was evaluated by paired Student’s t-test.
Ric-8A and Ric-8B are Expressed in Taste Tissue
In the oral cavity most taste buds are found on the palate and on the tongue. The lingual structures harboring the taste buds are called taste papillae. In order to investigate Ric-8 expression in palate as well as in fungiform, foliate and circumvallate taste papillae we conducted RT-PCR using primers specific for Ric-8A and Ric-8B on RNA extracted from these tissues together with olfactory mucosa, vomeronasal organ, eye, brain and testis. As a control for taste bud rich tissue, the expression of gustducin and Gαi2, reportedly two of the most abundantly expressed G-protein alpha subunits in taste cells (Kusakabe et al., 2000
), was also analyzed.
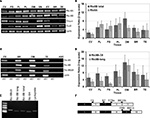
As can be seen in Figure 1
A, messenger RNAs for Ric-8A and Ric-8B are detected in fungiform, foliate and circumvallate papillae as well as in all other examined tissues. We also find as previously reported (Kusakabe et al., 2000
) that gustducin and Gαi2 are present in all three taste papillae including palatal taste buds.
Figure 1. Ric-8A and Ric-8B variants are expressed in tongue taste papillae. (A) RT-PCR on total RNA from various tissues using specific primers for Ric-8A, Ric-8B, gustducin, Gαi2 and cyclophilin. CV: circumvallate papillae, FG: fungiform papillae, FL: foliate papillae, PL: Palate, OM: olfactory mucosa, VN: vomeronasal organ, EY: Eye, BR: whole brain, TE: testis. (B) q-PCR showing the relative abundance of Ric-8B (both variants) and Ric-8A. Ric-8A is predominantly expressed over Ric-8B in palate, foliate and fungiform papillae but not in circumvallate papillae. (C) RT-PCR on a restricted panel of mRNAs using specific primers for Ric-8A, Ric-8B (long variant only), Ric-8BΔ9 only. For each tissue +RT: with Reverse transcriptase, -RT: without reverse transcriptase. CV: circumvallate papillae, BR: whole brain, OM: olfactory mucosa, TE: testis. (D) q-PCR showing the relative abundance of Ric-8B splicing variants. Ric-8BΔ9 is the major variant in palate, foliate and fungiform papillae but not in circumvallate papillae. (E) RT-PCR on brain RNA with the various primer pairs used for q-PCR. Note the presence of a single band of the expected size and the absence of primer-dimers for each primer pair tested, thus validating them for SYBR green q-PCR. (F) Scheme showing the strategy of amplification of the two Ric-8B isoforms. Black and white rectangles denote coding exons in the Ric-8B gene and black arrows the location of the primers used. Primer pair F6-R7 amplifies both Ric-8B isoforms (Ric-8B total). Primer pair F78a-R810b amplifies the Ric-8BΔ9 isoform only (Ric-8BΔ9) while primer pair F89-R910 amplifies the Ric-8B long form only (Ric-8B long). Primer F78a anneals to both isoforms but this is corrected by taking primer pair efficiency into account in the calculation of the ratio.
In order to quantify the relative abundance of Ric-8A and Ric-8B mRNAs in taste tissue compared to that in olfactory mucosa, total brain and testis, we conducted a quantitative PCR experiment using primers specific for each gene. As seen in Figure 1
B the relative levels of Ric-8 mRNA do not vary widely amongst the tissues examined, with the exception of the olfactory mucosa in which Ric-8B is significantly more abundant in line with a previous report focusing on olfactory neurons (Von Dannecker et al., 2005
).
In foliate, palate and fungiform papillae Ric-8A mRNA is slightly more abundant than that of Ric-8B while the circumvallate papillae displays a profile similar to that of brain, testis and olfactory mucosa in which Ric-8B is prevalent over Ric-8A. In the olfactory epithelium, an alternatively spliced variant of Ric-8B called Ric-8BΔ9 in which exon 9 is missing has also been described (Von Dannecker et al., 2005
). We looked for the presence of this isoform in tissues in which Ric-8B is predominant. When using primers for Ric-8B flanking exon 9 or primers specific for the short form only we observe that both isoforms can be detected (Figure 1
C). Interestingly, quantification of the relative abundance of both isoforms shows a predominance of the short Δ9 form of Ric-8B in all assayed tissues, but more markedly so in taste tissues, especially foliate and fungiform papillae (Figure 1
D).
Prior to performing qPCR the specificity of all quantitative primer pairs were tested by conventional PCR to confirm the amplification of a single product of the expected size and the absence of primer-dimers (Figure 1
E). Moreover each qPCR experiment was immediately followed by melt curve analysis for each sample to ensure amplification specificity. The primers used for differential amplification of the various Ric-8B splice variants were located at the exon-exon boundaries (Figure 1
F).
Expression of Ric-8A and Ric-8B in Taste Papillae is Confined to Taste Bud Cells
We next determined whether Ric-8A and Ric-8B gene expression in taste papillae is confined to the taste bud cells. To do this, we conducted in situ hybridization experiments on sections of circumvallate papillae using probes specific for Ric-8A or Ric-8B.
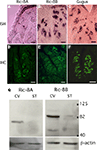
Figures 2
A,B show that Ric-8A and Ric-8B mRNAs are found in cells confined within the taste buds when compared with the expression pattern of gustducin, a taste bud specific G-protein (Figure 2
C). Although Ric-8B mRNA is clearly very abundant in taste buds we also noticed some staining in the basal layer of the tongue epidermis. This data is in line with an earlier study reporting the presence of Ric-8B messenger RNA in a human taste buds cDNA library (Rossier et al., 2004
).
Figure 2. The expression of Ric-8A and Ric-8B is confined to taste bud cells. (A–C) Sagittal sections of circumvallate papillae were hybridized with antisense probes to Ric-8A, Ric-8B or gustducin and visualized with an alkaline phosphatase colorimetric reaction. The purple signal is confined to taste buds. Section hybridized to the sense probes did not show any signal (not shown). (Scale bar = 50 μm). (D–F) Immunostaining of sagittal sections of circumvallate papillae using anti-Ric-8A, anti-Ric-8B or anti-gustducin rabbit polyclonal antibodies visualized with an anti-rabbit-Alexa488 secondary. The immunofluorescence is confined to the taste buds and labels 100% of taste buds. (Scale bar = 50 μm). (G) Western blotting analysis of circumvallate papillae (CV) or surrounding tissue (ST) extracts using anti-Ric-8A and Ric-8B antibodies. A major band around 70 kDa for Ric-8A and ∼88 kDa for Ric-8B is detected in CV indicating that these proteins are concentrated in taste buds. Bottom panels show β-actin loading controls. [Right margin: molecular weight marker bands (kDa)].
Using antibodies directed against Ric-8A (Figure 2
D) or Ric-8B (Figure 2
E) confirms that the labeling is more intense within the taste buds than in the surrounding tissue and very similar to what is observed for gustducin (Figure 2
F). To test the specificity of the antibodies against the Ric-8 proteins we performed a Western blotting analysis with protein extracts of taste bud-enriched tissue and surrounding tissue from mouse circumvallate papillae. As shown in Figure 2
G, the antibody against Ric-8A detected a major band at ∼70 kDa while the Ric-8B antibody detected a major band of ∼88 kDa. It is worth noting that the band is significantly stronger in taste bud-enriched tissue than in surrounding tongue tissue thus corroborating the staining pattern obtained in immunohistochemistry experiments. The size of the Ric-8A and Ric-8B bands are about 7 and 25 kDa bigger than the predicted molecular weight respectively indicating that these proteins likely undergo posttranslational modifications in this tissue. A similar observation has already been made in olfactory epithelium for Ric-8B (Kerr et al., 2008
).
Ric-8A and Ric-8B are Co-Localized with IP3R-3 in Type II Taste Bud Cells
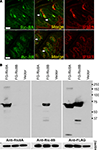
Taste bud cells are divided in at least four distinct cell types, which can be distinguished by the expression of specific markers. To analyze what type or subset of taste cells expresses Ric-8A and Ric-8B we performed double immunolabeling using an antibody specific to either Ric-8A or Ric-8B together with an antibody raised against IP3R-3, a marker of type II cells staining sweet, umami and bitter taste receptor cells. Our results (Figure 3
A) show that 100% of IP3R-3 positive cells are stained with Ric-8A or Ric-8B (Figure 3
A merge). The presence of Ric-8A and Ric-8B in taste-GPCR expressing cells is consistent with a role in taste transduction.
Additionally we find that a subpopulation (∼15%) of taste bud cells is immunopositive for Ric-8A or Ric-8B but negative for IP3R-3 (n = 100). The nature of these cells remains to be determined.
Figure 3. Ric-8 expression is not strictly confined to type II taste bud cells. (A) Double labeling immunohistochemistry on sagittal sections of circumvallate papillae using an antibody raised against IP3R-3 visualized with an Alexa555 coupled secondary antibody (red) and either an anti-Ric-8A (top row) or anti-Ric-8B (bottom row) antibody visualized with an Alexa488 secondary antibody (green). Middle panels are merged images showing co-localization. All IP3R-3 cells display Ric-8A and Ric-8B labeling but not all Ric-8A or Ric-8B cells are positive for IP3R-3 (white arrows) indicating that Ric-8A and Ric-8B expression extends to other cells beside IP3R-3 positive cells. (Scale bar top row = 20 μm; bottom row = 40 μm). (B) Western blotting analysis of whole cell lysates from cells transiently transfected with either Flag-Ric-8A (FG-Ric8A), Flag-Ric-8B (FG-Ric8B) or the expression vector alone (Vector) using anti-Ric-8A (left panel), anti-Ric-8B (middle panel) or anti-Flag (right panel) antibodies. A single band around 64 kDa for recombinant Flag-Ric-8A and ∼71 kDa for Flag-Ric-8B is detected in HEK 293T cells thus clearly showing that Ric-8 antibodies do not cross-react with the paralogous protein. Bottom panels show β-actin loading controls. [Right margin: molecular weight marker bands (kDa)].
The matching expression patterns of Ric-8A and Ric-8B which are both found in 100% of IP3R-3 positive cells suggest that they might be co-expressed in these cells, a prospect that we could not test directly given the fact that both antibodies against Ric-8A and Ric-8B are raised in rabbit. Nevertheless, to rule out the possibility that this pattern could be generated by cross-reactivity of the antibodies with the paralogous proteins a western blotting analysis testing the specificity of the antibodies against the two Flag-tagged recombinant Ric-8 proteins transiently expressed in HEK 293T cells was conducted. As seen in Figure 3
B no cross-reactivity was detected thus supporting the notion that these two proteins might be co-expressed.
Ric-8A Interacts with Alpha-Gustducin and Alpha-Transducin
Because a large percentage of IP3R-3 cells expresses gustducin (Clapp et al., 2001
), we used a directed two-hybrid interaction screen to investigate whether Ric-8A or Ric-8B can interact directly with gustducin.
We initially validated our assay by testing the previously reported interaction between Ric-8A and Gai2 as well as that between Ric-8B and Gαolf (Tall et al., 2003
; Von Dannecker et al., 2005
).
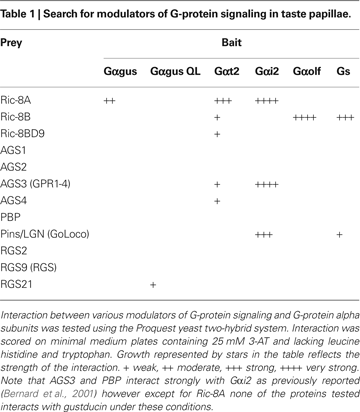
Figure 4
A shows that, as expected, Ric-8A strongly interacts with Gαi2 but not with Gαolf. More importantly we find that Gαt2 and to a lesser extent gustducin also interact with Ric-8A. Under the same conditions and as expected Ric-8B clearly interacts with Gαolf but not with Gαi2. We also observe a weak interaction with Gαt2 but none with gustducin. Ric-8BΔ9 was also included in the screen but no substantial interaction was detected (Table 1
).
Figure 4. Gustducin interacts with Ric-8A, but not Ric-8B. (A) Yeast two hybrid interaction assay. Mav203 yeasts cells were co-transformed with different combinations of bait (Gαolf, Gαt2, Gαgus or Gαi2) and prey (Ric-8A or Ric-8B) plasmids. Interaction was scored on minimal medium plates containing 3-AT and lacking histidine and tryptophan. Growth reflects the strength of the interaction. (B) Co-immunoprecipitation experiment showing the association between Flag-Ric-8A and HA-Gαi2 or HA-Gαgus. (*) denotes an artifact band (IgG) while (<) correspond to the Gα subunit band. IPFG: immunoprecipitation using an anti-Flag antibody; WBFG: western blotting membrane incubated with an anti-Flag antibody; WBHA: western blotting membrane incubated an anti-HA antibody.
As a control for the strength of the interaction several RGS (i.e. RGS2, RGS9 and RGS21) were also tested for their ability to interact with gustducin in the same assay. Our results confirm that RGS21 physically interacts with the constitutively activated form of gustducin (QL) as previously reported using a different test (von Buchholtz et al., 2004
), whereas Ric-8A is likely to interact with gustducin in its GDP-bound form as we find no interaction with the GTPase deficient Gαgus QL.
Next we validated these interactions in an heterologous expression system involving transient transfection of 5′-Flag-tagged-Ric-8 and 5′-HA-tagged-Gα-proteins expression constructs in HEK 293T cells followed by immunoprecipitation using an anti-Flag antibody. The results of this assay essentially corroborate the results obtained in yeast: Ric-8A interacts strongly with Gαi2 and Gαt2, and to a lesser extent with gustducin, while Ric-8B interacts weakly with Gαt2 but not with gustducin (Figure 4
B). Taken together these results strongly suggest that in taste buds Gαt2, and the abundantly represented gustducin and Gαi2, which have been reported to couple to taste receptors (Ueda et al., 2003
) are the most likely targets of Ric-8A in this tissue.
Ric-8A amplifies the Signal Downstream of hTas2R16
In a previous study it was shown that Ric-8B is particularly abundant in the olfactory sensory neurons where it acts as a signal amplifier for G-protein mediated signaling cascades (Von Dannecker et al., 2005
, 2006
). To test the possibility that Ric-8A’s expression in taste receptor cells plays a similar role in taste transduction we focused our study on hTas2R16, the human receptor for the bitter tastant salicin that was previously reported to be coupled to Gαi2 as well as Gαt2 (Ueda et al., 2003
).
To monitor the effect of hTas2R16 stimulation on intracellular cAMP levels we devised an heterologous expression system in HEK 293T cells involving transient co-transfection of the dopamine D1 receptor (D1R) which is known to be coupled to Gαs, together with hTas2R16 and Gαi2. Intracellular cAMP levels were monitored by a reporter enzyme (secreted alkaline phosphatase, SEAP) under the control of CRE.
In this system stimulation of the dopamine D1R receptor with dopamine triggers a rise of intracellular cAMP which is reduced in a dose depend way by stimulation of hTas2R16 with salicin (Figure 5
). Next we tested whether co-expression with Ric-8A affects the activity of hTas2R16. To do so HEK 293T cells were transiently transfected with D1R, hTas2R16, Gai2 and Ric-8A or a control expression construct. Upon stimulation of the cells with 10 μM dopamine and either 5 or 10 mM salicin, a dose dependent reduction in the cAMP levels was recorded (Figure 5
). The magnitude of this reduction (∼35%) was statistically significant between Ric-8A transfected versus transfected cells without Ric-8A vector suggesting that Ric-8A is able to promote hTas2R16 activity. Since this reduction in cAMP accumulation was stronger in cells transfected with Gαi2 than in cells which did not overexpress Gαi2 (data not shown) we conclude that Ric-8A is able to amplify hTas2R16 signal transduction through Gαi2.
Figure 5. Ric-8A enhances Gαi2 mediated decreases in cAMP concentration. Production of cAMP was measured in HEK 293T cells transfected with D1R, Tas2R16, Gαi2 with or without Ric-8A in the presence of 10 μM dopamine, or 10 μM dopamine plus 5 (white bars) or 10 mM (grey bars) salicin as indicated. The graph shows the mean and SD values of four independent experiments. The deactivation ratio is calculated by dividing values after a dopamine + salicin stimulation by the corresponding dopamine activation values. For 10 mM salicin Ric-8A enhancement of Gαi2 mediated deactivation was statistically significant. (*) p < 0.05, Paired Student’s t-test.
In taste cells two main families of GPCRs are involved in tastant detection and transduction. The Tas1Rs which recognize sweet and umami compounds and the Tas2Rs which are tuned to bitter compounds (Chandrashekar et al., 2006
). Upon stimulation with a tastant the receptor activates an heterotrimeric G-protein consisting of a Gα subunit and a Gβγ dimer. During that step the receptor bound Gα subunit undergoes an exchange of GDP for GTP which leads to the release of the GTP-Gα subunit from the Gβγ dimer. Both components are then capable of modulating effectors independently based on their affinity for that effector. This reaction can be further amplified by non GPCR GDP/GTP exchange factors which are specific for certain Gα subunits. The composition of the G-proteins coupled to the various GPCRs involved in taste detection is therefore germane to determining which signal transduction pathway is activated and how it might be regulated. Several studies have investigated the importance of G-proteins in taste transduction or the coupling of Tas1Rs and Tas2Rs to G-proteins. The report that knock-out mice for Gα-gustducin have diminished but not abolished sensitivity to sweet and bitter compounds (Wong et al., 1996
) which can be rescued by Gα-transducin expression together with heterologous expression studies showing that Gαgus is able to couple to the Tas1Rs and Tas2Rs are consistent with an important role of this G-protein in taste signal transduction (Li et al., 2002
; Ueda et al., 2003
; Sainz et al., 2007b
). Here we report for the first time that Gαgus and Gαt2 physically interact with Ric-8A a GDP/GTP exchange factor expressed in type II taste cells. In addition we show that Ric-8A is able to amplify Tas2R16 signaling through Gαi2. Interestingly, Tas2Rs have been reported to couple to all three Gα subunits interacting with Ric-8A namely Gαgus, Gαi2 and Gαt2 (Chandrashekar et al., 2000
; Ueda et al., 2003
; Sainz et al., 2007a
). Therefore we anticipate that Ric-8A might be a general modulator of bitter GPCR signaling (Figure 6
).
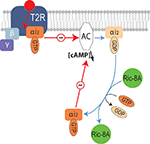
Figure 6. Molecular mechanisms underlying the modulation of bitter receptor signaling by Ric-8A. Diagram illustrating the signaling cascade downstream of bitter taste GPCR (T2R) modulated by Ric-8A. Upon activation of T2R the receptor coupled heterotrimeric G-protein separates into Gα-GTP and Gβγ, Gα-GTP will then inhibit the activity of adenylyl cyclase, subsequently the GTP bound to Gα will be hydrolyzed into GDP, Gα-GDP can then reassociate with Gγβ or interact with Ric-8A. By supporting the exchange of GDP by GTP, the GEF activity of Ric-8A will allow a new cycle of inhibition of the effector thus enhancing the signal.
Furthermore, we show that Ric-8A is expressed in all IP3R-3 positive taste bud cells, a good portion of which have been reported to be type II or taste receptor cells in mice (Clapp et al., 2001
). This finding and the fact that IP3R-3 links PLC-β2 signaling with bitter receptors support this argument. Note that some IP3R-3 positive cells have also been reported to express markers specific for type III cells such as SNAP-25 a synaptic protein found in mouse taste buds (DeFazio et al., 2006
); since we report that 100% of IP3R-3 positive cells are also positive for Ric-8A its function in these cell types may be linked to GPCRs present in these cells. Gαgus and Gαi have been shown to couple to Tas1Rs (Li et al., 2002
; Ozeck et al., 2004
; Sainz et al., 2007b
) and Gαt1 to play a role in behavioral and electrophysiological taste responses to umami (He et al., 2004
); it is therefore plausible that Ric-8A might amplify the signaling of these receptors as well.
Previous work reporting an interaction between Ric-8B and Gγ13 (Kerr et al., 2008
) or Gαq (Tall et al., 2003
), both found in taste bud cells (Kusakabe et al., 1998
; Huang et al., 1999
) together with our data showing that it is present in all IP3R-3 positive cells suggest that Ric-8B may effect G-protein signaling in these cells. One intriguing prospect involves a possible interaction with Gα14 a G-protein recently reported as co-expressed with the sweet taste receptor (Tas1R2–Tas1R3) (Shindo et al., 2008
; Tizzano et al., 2008
). These possibilities and its precise role in taste cells remain to be investigated.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Drs. V. Dionne (Boston University) and S. Simon (Duke University) for critical reading of the manuscript as well as C. Arnould and A. Gaudin for help with the confocal microscope. This work was supported by an Action Thematique Incitative sur Programme (CNRS) grant to Jean-Pierre Montmayeur, Région de Bourgogne post-doctoral fellowships to Lila Patrikainen and Zhenhui Liu as well as funds from GOSPEL to Fabienne Laugerette. Bettina Malnic and Daniel S. Kerr are supported by funds from FAPESP and CAPES.
Durocher, Y., Perret, S., Thibaudeau, E., Gaumond, M. H., Kamen, A., Stocco, R., and Abramovitz, M. (2000). A reporter gene assay for high-throughput screening of G-protein-coupled receptors stably or transiently expressed in HEK293 EBNA cells grown in suspension culture. Anal. Biochem. 284, 316–326.
Klattenhoff, C., Montecino, M., Soto, X., Guzman, L., Romo, X., Garcia, M. A., Mellstrom, B., Naranjo, J. R., Hinrichs, M. V., and Olate, J. (2003). Human brain synembryn interacts with Gsalpha and Gqalpha and is translocated to the plasma membrane in response to isoproterenol and carbachol. J. Cell Physiol. 195, 151–157.