1
Department of Neurobiology, The Alexander Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem, Israel
2
Institute for Life Sciences and Interdisciplinary Center for Neural Computation, The Hebrew University of Jerusalem, Jerusalem, Israel
Advances in neuroanatomy and computational power are leading to the construction of new digital brain atlases. Atlases are rising as indispensable tools for comparing anatomical data as well as being stimulators of new hypotheses and experimental designs. Brain atlases describe nervous systems which are inherently plastic and variable. Thus, the levels of brain plasticity and stereotypy would be important to evaluate as limiting factors in the context of static brain atlases. In this review, we discuss the extent of structural changes which neurons undergo over time, and how these changes would impact the static nature of atlases. We describe the anatomical stereotypy between neurons of the same type, highlighting the differences between invertebrates and vertebrates. We review some recent experimental advances in our understanding of anatomical dynamics in adult neural circuits, and how these are modulated by the organism’s experience. In this respect, we discuss some analogies between brain atlases and the sequenced genome and the emerging epigenome. We argue that variability and plasticity of neurons are substantially high, and should thus be considered as integral features of high-resolution digital brain atlases.
Since the days of Ramón y Cajal, it was clearly demonstrated that neurons come in different “flavors”. Each neuron within a local circuit may look and function differently even when the neurons are closely packed together. Today we know that differences are evident at almost any scale and any level, ranging from gene expression profiles to the structure of dendritic and axonal arbors. Because it is still not technically possible to study all neurons within a tissue simultaneously, we are subjected to study one or a few neurons at a time in different animals. Since neurons are different, the issue of variability arises. This problem becomes even greater when comparisons are made across studies and across laboratories. To date, there is no common reference frame, and detailed experimental data are rarely shared for purposes of comprehensive comparisons. Therefore, it is desirable to have a shared atlas based on experimental data. When such atlases become available, they could provide a road map for comparative studies across brain regions and species.
In anatomical terms, a new atlas means reviving Cajal’s efforts with modern tools, but now taking into consideration not only the two-dimensional dendritic and axonal structures but possibly molecular signatures (Lein et al., 2007
), and even complete synaptic connectivity patterns (Lichtman et al., 2008
). Furthermore, atlases containing shared data from many research groups would allow in-depth morphological analysis, which could shed light on the relation between arbor structure and function (see e.g., Wen et al., 2009
). Indeed, in the past few years there have been a number of efforts to construct high-resolution brain atlases of standardized brains of invertebrates (see e.g., Rein et al., 2002
; Brandt et al., 2005
; Kurylas et al., 2008
). A similar effort has been extremely successful in genetics where the “master atlas” of our era, the genome, has been comprehensively mapped and sequenced. In analogy to the genome, it would then be possible to search the atlas for any combination of axonal projection patterns and/or dendritic arborization of one’s favorite neuron. Atlases would not only prompt higher quality data but would be a valuable tool for stimulating new hypotheses and experimental designs.
What should a brain atlas contain? What are the main challenges that arise from the numerous degrees of freedom making up brain circuits? Here, we discuss some advantages and limitations of such futuristic atlases. Specifically, we focus on the limitations imposed by stereotypy and structural plasticity in mature brains and discuss to what extent these would affect future brain atlases of invertebrates and vertebrates alike. We limit our discussion to mature nervous systems. A discussion of atlases in the context of ontogenetic development is equally beneficial (see e.g., Huetteroth et al., also in this issue), however, given space considerations it is out of the scope of this review.
Several sources of variability call for consideration in future atlases; namely, those arising from inter-neuronal variability within an animal and those arising from inter-animal variability between neurons of the same type. Inter-animal variability has been quantified most powerfully in invertebrates where neurons can be identified individually. One of the first quantitative anatomical reports was carried out on visual neurons of the small crustacean Daphnia magna (Macagno et al., 1973
). These authors reconstructed axonal projections of identified neurons and revealed that their gross anatomical features are reproducible but that finer details are not. Since then, numerous studies have reported similar observations in a variety of species, using better staining methods and more powerful analytical tools (Mizrahi et al., 2000
; Marin et al., 2002
; Wong et al., 2002
). In numerous invertebrate systems (e.g. cockroaches, crickets, flies, leeches and honeybees) single neurons can be identified anatomically with relative ease (Coggeshall and Fawcett, 1964
; Daley et al., 1981
; Johansen et al., 1989
; Bodnar et al., 1991
; Hammer and Menzel, 1995
). Notably, however, anatomical identity may not be generally applicable to all invertebrate species. For example, in the lobster Panulirus Interruptus some electrophysiologically identified neurons could not be identified morphologically (Thuma et al., 2009
).
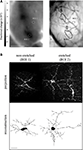
What exactly is variable between the morphology of different identified neurons? Quantitative analysis of identified neurons has revealed substantial variations in high-order dendritic and axonal branching patterns, whereas a neuron’s morphological identity is based primarily on its low-order or primary branching pattern (Mizrahi et al., 2000
). In line with those observations, a rigorous analysis of insect neurons recently showed that anatomical identity is determined by the neuropile area covered by the neurites, and not by the complete branching topology (Figures 1
A,B; Cuntz et al., 2008
). A similar principle might also apply to vertebrate neurons of the same type, but this has not been quantified (see e.g., hippocampal CA1 pyramidal neurons; Figure 1
C). In the context of atlases, the gross area of neuronal arborization may be used to infer (or reject) potential synaptic partners and possible functions. However, based on projection patterns alone, one cannot deduce either the function or the precise synaptic connectivity patterns of a network with any certainty. To study synaptic connectivity or function, higher resolution methods, such as electron microscopy and/or electrophysiology should be used. This is a major limitation of anatomical atlases which will be hard to overcome.
Figure 1. Structural stereotypy in invertebrate and vertebrate neurons. (A,B) Identified neurons in the fly visual system. (A) Reconstructions of different types of lobula plate tangential cells from the blowfly (HSE, HSN, VS2 and VS4) within the lobula plate (gray). (B) Left: Neuronal reconstructions of identified neurons superimposed and sorted according to their cell type. Cells were aligned along their axonal axis (red lines). Right: the corresponding dendrite spanning fields of the reconstructed neurons. Neuronal anatomical identity is determined by the neuropile area that is covered by the neurites, and not by the complete branching topology. (C) Qualitative anatomical stereotypy in vertebrate neurons. Reconstructions of eight rat hippocampal CA1 pyramidal cells. Note that the gross field covered by the neurites exhibits a similar trend in all eight neurons. Stereotypy seems to be less robust as compared to invertebrate identified neurons. Scale bar: 200 μm. Panels (A,B) are adapted from Cuntz et al. (2008)
, with permission. Panel (C) is adapted from Scorcioni et al. (2004)
, with permission.
In vertebrates, and specifically in mammals, the precise nature of stereotypy, variability and synaptic specificity is not known. In principle, stereotypy and variability could have been empirically measured using a similar logic to that used in invertebrate identified neurons. However, unlike invertebrates, there are no known “identified neurons” in the vertebrate CNS as each neuronal subtype may have at least dozens to hundreds of replicas in the circuit (with a known exception – the “Mauthner neurons” in fish; Zottoli, 1978
). Moreover, in mammals there is still no consensus on basic anatomical features such as cell numbers, borders between brain regions, cell types and cell subtypes (Bota et al., 2003
). Thus, it would be hard to provide compelling statistical evidence for stereotypy and variability in mammalian neurons based on sparse sampling and datasets that might not be composed of homogenous populations of cells. Accordingly, welcome efforts are now underway to establish widely approved classification criteria for defining neuronal subtypes based on anatomical, physiological and molecular characteristics (e.g., Ascoli et al., 2008
). These efforts are paving the way for community-approved definitions of vertebrate neuronal subtypes, which would be highly beneficial for future vertebrate brain atlases.
An even more challenging issue is to determine synaptic connectivity patterns between neurons, and include these in atlases. Due to lack of resolution, it is currently impossible to determine the existence of synapses between sets of neurons based on their neurite morphology. The most adequate anatomical method that can show synaptic connectivity is electron microscopy (EM). Recently developed methods, such as “array tomography”, are now also providing new tools for exploring the interface between molecular signatures and synaptic connectivity (Micheva and Smith, 2007
).
Current efforts, which focus on using EM, are now underway to study synaptic connectivity in dense, highly interconnected, neural tissue. Specifically, EM sampling techniques are being designed to reconstruct “everything” (but primarily synapses) within a volume of tissue (Denk and Horstmann, 2004
; see also the early work of Brenner and colleagues on c.elegans, by Ward et al., 1975
, and the more recent work on c.elegans by Chklovskii and colleagues, Chen et al., 2006
). This effort has been termed “connectomics” and dares to the challenges imposed by the complexity of the nervous system. There is no doubt that even optimistic connectomists still face serious technical and computational challenges for reconstructing the mammalian connectome. But once these are overcome, the connectome is expected to provide direct empirical data and finite numbers on issues such as inter-animal variability and the level of synaptic specificity in large neural circuits (Lichtman and Sanes, 2008
).
Recently, the first connectome in mammals has been solved (the intermuscularis muscle connectome in mice; Lu et al., 2009
). Notably, this peripheral nervous system connectome was solved by light microscopy rather than by EM (which was possible due to the large size of neuromuscular synapses). Motor units from the same or different animals were compared and an unexpectedly large variability was revealed. Motor unit connectomes from the left and right sides of the same individual were not less different than connectomes of different individuals. This result points to what might be a fundamental difference in organization principles between vertebrate and invertebrate identified neurons (e.g., compare to Goodman, 1978
; Mizrahi et al., 2000
). In addition, if we assume that the peripheral nervous system represents a more rigid system than the CNS, then connectome variability in the CNS (which is still not known) is expected to be even higher. These results imply that the nervous system in mammals is not as genetically “hard-wired” as it might seem in invertebrates (Bentley, 1975
) and may rely on different organization principles during development, and/or adulthood. The mechanisms underlying higher variability in mammalian systems are not clear. While the basic cellular principles of experience-dependent plasticity are not necessarily different in vertebrates and invertebrates, speculatively, experience-dependent plasticity may play a more dominant role in synapse formation in adult mammals, as compared to invertebrates. In this respect, there is still no compelling evidence for mammalian “identified neurons”. Thus, anatomical atlases of mammalian species should include multiple examples of each individual neuron they attempt to describe; certainly more than would be needed in invertebrate atlases. Otherwise, the atlas would not be much more useful than any isolated anatomical reference.
An atlas is a highly useful tool for describing a stereotypical, static system but might not be as useful for highly variable, dynamic systems. The more variable and dynamic the system is, the less likely an atlas will recapitulate it faithfully. Therefore, the extent to which the brain is structurally stereotyped and static determines the constraints and general validity of brain atlases. In biology, neurons stand out in their significant capacity to change and adapt; a capacity usually referred to as “plasticity”. There is ample evidence for physiological plasticity. Synapses can potentiate within minutes (Madison et al., 1991
; Zucker and Regehr, 2002
), receptive fields can rapidly change (Dorris et al., 2000
; Gandolfo et al., 2000
; Fritz et al., 2003
; Geffen et al., 2007
) and firing patterns of the same neuronal subtype are rarely the same. On longer time scales (hours to days), neuronal structure is also dynamic (Bhatt et al., 2009
). To what extent can we predict one brain from the next in such an ever-changing system? Is structural plasticity an Achilles Heel for brain atlases?
Direct evidence for the degree of structural dynamics is continuously emerging in the last decade, mainly due to advances in genetic labeling and imaging techniques (Young and Feng, 2004
). The first studies became possible with the development of the “thy-1 XFP” transgenic mice (Feng et al., 2000
). These publicly available mice have fluorescently labeled neurons that are sparsely scattered throughout the brain and are bright enough so that they could be readily imaged in vivo. In vivo time-lapse measurements are the experiments of choice for studying structural plasticity since they minimize variability. In principal, digital brain atlases, would be based on averaging across several specimens and across time. Thus, it is possible that many discoveries regarding experience dependant plasticity would still rely on revealing deviations from this average. In this context, the advantage of in vivo time-lapse experiments is that they eliminate some of the inter-individual variation discussed above. This allows the detection of smaller, yet significant, changes. Actually, before the era of in vivo time-lapse imaging, experimentally explored structural plasticity was limited to large-scale changes (see e.g., Woolley, 1999
), missing some of the more subtle differences that are currently being unveiled.
Two papers, using in vivo time-lapse imaging, by Trachtenberg et al. (2002) and Grutzendler et al. (2002)
prompted a series of experiments that started to reveal the extent of structural plasticity in mature brains in more detail. To date, almost all the available data regarding structural dynamics (at least in mature brains) have been obtained from mammals and particularly from mice (Holtmaat and Svoboda, 2009
).
One consistent observation is that neurites of adult projection neurons are largely stable. In both neocortical pyramidal neurons and olfactory bulb mitral cells only negligible changes were found in dendritic structure over periods of weeks (Grutzendler et al., 2002
; Trachtenberg et al., 2002
; Mizrahi and Katz, 2003
). But unlike projection neurons, neocortical interneurons exhibit structural changes at rates of ∼15% over several weeks (Lee et al., 2006
). This level of structural plasticity is evident only at specific laminae, suggesting that both the brain region and the cell-type may determine the levels of structural plasticity (Lee et al., 2008
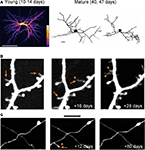
). One extreme example of structural plasticity of dendrites comes from our work on adult-born periglomerular interneurons in the olfactory bulb. Periglomerular interneurons show high levels of dendritic structural plasticity reaching up to 40% per week (Figure 2
A; Mizrahi, 2007
). Notably however, periglomerular interneurons are unique neurons as they are born during adulthood, and are thus relatively immature compared to most other neurons in the brain.
Figure 2. Structural plasticity in the adult mammalian brain. (A) In vivo dendritic structural plasticity of adult-born periglomerular interneurons. Left panel: average intensity projection image of 9 imaging sessions of the same developing periglomerular neuron (12 h apart). The color scale bar indicates how many times a dendritic branch was located in a given region (from 1 to 9). Bright colors represent stable dendrites, and dark colors represent dynamic dendrites. Right panel: reconstructions of a mature periglomerular neuron, imaged 7 days apart. Both developing and mature periglomerular neurons exhibit extensive dendritic dynamics. Scale bars: 50 and 10 μm (left and right panel, respectively). (B,C) Images from in vivo time lapse imaging of dendritic spines (B) and axons (C) from the neocortex. The images are examples of the same dendritic and axonal segments that were imaged up to 28 or 80 days apart. Note both the addition (arrowhead) and loss (arrows) of dendritic spines (B) and axonal segments (C). Scale bars: 5 μm. Panel (A) is adapted from Mizrahi (2007)
, with permission. Panels (B,C) are adapted from Holtmaat et al. (2009)
, with permission.
In mammals, axons are thinner than dendrites and more difficult to follow in vivo. The few studies that followed axonal morphology over time found that axons are also largely stable. Specifically, time-lapse experiments revealed only ∼4% of de novo branch formation or elimination per month, with ∼25–60% of the branches that exhibited short distance (few micrometers) elongation and/or retraction per month (De Paola et al., 2006
). Interestingly, like dendritic dynamics, axonal dynamics vary as a function of cell-type (e.g., thalamocortical axons are less dynamic than intracortical axons; De Paola et al., 2006
; Stettler et al., 2006
). Together, these results suggest that the extent of dendritic and axonal plasticity varies across cell-types, and should thus be tested for each neuronal population separately.
In contrast to the relative stability of dendrites and axons, dendritic spines are far more plastic structures (Figure 2
B). Since new spines are a good approximation of synapses (Knott et al., 2006
), this suggests that synaptic connectivity within the mammalian CNS is in an ongoing state of change (Trachtenberg et al., 2002
; Zuo et al., 2005
; Hofer et al., 2009
). Structural plasticity of spines under normal conditions can reach up to 50% per week (Trachtenberg et al., 2002
). Interestingly, dendritic spine dynamics also differ between different cell-types and brain regions. For example, in the barrel cortex, the spines of L2/3 pyramidal neurons are less dynamic than those of L5 neurons. L5 pyramidal neurons in the barrel cortex are also more dynamic than L5 pyramidal neurons in the visual cortex (Holtmaat et al., 2005
). This idea has received additional support from experiments, in which rewiring visual input to the auditory cortex at birth failed to alter dendritic spine dynamics (Majewska et al., 2006
). Presynaptic boutons, which are also used as an approximation of synapses, were also found to exhibit cell-type specific dynamics (up to 50% turnover per month; Figure 2
C; De Paola et al., 2006
), further supporting the existence of ongoing changes in CNS synaptic connectivity. Notably, structural dynamics are homeostatically regulated such that the overall morphology is kept constant (e.g., total dendritic branch length, spine density; Trachtenberg et al., 2002
; Mizrahi, 2007
). This homeostasis might bear functional significance (Samsonovich and Ascoli, 2006
).
Structural dynamics may have direct implications on the attempts to reconstruct connectomes (and atlases; see also below). If structural dynamics reflect changes in neuronal connectivity, rather than just strengthening/weakening of synapses between previously connected neurons, then connectomes are also expected to be dynamic. While this issue remains unclear, it might well be that several connectomes would need to be constructed before synaptic connectivity patterns could be inferred with “connectomic precision”.
The studies discussed above were made with mice housed under normal housing conditions. This prompts the question of whether and how experience might affect structural plasticity. Indeed, several studies have explored this relationship, mainly in primary sensory areas such as the mouse barrel cortex, visual cortex and olfactory bulb (Trachtenberg et al., 2002
; Mizrahi and Katz, 2003
; Zuo et al., 2005
; Mizrahi, 2007
; Livneh et al., 2009
). In one extreme case, Keck et al. (2008)
recently showed that a focal retinal lesion leads to a dramatic increase in spine turnover rate in the visual cortex. Remarkably, within 2 months following the lesion, an almost complete replacement of spines was observed. In a similar study from the same group, Hofer et al. (2009)
explored the structural correlates of ocular dominance plasticity (Hubel et al., 1977
). In the adult, after the closure of all known developmental critical periods, binocular neurons remain plastic in response to a second monocular deprivation. This occurs only if the animal had previously underwent a first monocular deprivation when it was younger, during the relevant developmental critical period (Hofer et al., 2006
). To continue this work, Hofer et al. (2009)
correlated these functional changes with the addition of new persistent spines, which may provide the structural changes required for subsequent functional shifts (Hofer et al., 2009
). Similarly, functional changes in the barrel cortex due to whisker trimming have also been shown to be correlated with modified structural dynamics (Trachtenberg et al., 2002
).
Consistent with structural dynamics under normal housing conditions, different types of neurons seem to undergo different degrees of experience-dependent structural plasticity (Mizrahi and Katz, 2003
; Holtmaat et al., 2006
; Hofer et al., 2009
; Livneh et al., 2009
). For example, while L5 visual neocortical neurons undergo experience-dependent structural plasticity, L2/3 neurons do not (Hofer et al., 2009
). Recently, we explored experience-dependent structural plasticity of adult-born periglomerular neurons in response to sensory enrichment with a low concentration odor mixture of two simple odors. Enrichment increased the size of these neurons’ dendritic tree, as well as their number of putative synapses. Remarkably, this effect was specific only to neurons, which were located in sensory-active regions of the olfactory bulb, even within the same tissue (Figure 3
; Livneh et al., 2009
). These data further support the view that the levels of structural plasticity in different neurons can vary dramatically, even within the same local network.
Figure 3. Specificity of experience-dependent plasticity during adult-born neuron development. (A) Intrinsic signal odor map in response to a two-odor mixture (left panel), and the corresponding blood vessel map on the surface of the olfactory bulb (right panel). Sensory-active domains are marked by a black contour in the blood vessel map. Two regions of interest (ROIs), containing the neurons shown in B, are indicated by dotted circles. ROI 1 is in a sensory-non-active domain, while ROI 2 is within a sensory-active domain. Scale bar: 1 mm. (B) Two adult-born periglomerular neurons expressing PSD95-GFP as a proxy for synapses from the same experiment shown in A. Left: a periglomerular neuron from a non-enriched domain (non-active, ROI 1), Right: a periglomerular neuron from an enriched domain (active, ROI 2). Top, maximum projection images of the original Z stacks. Bottom, two-dimensional view of the reconstructed neurons at the top. Asterisks mark putative synapses. Scale bar: 20 μm. Sensory enrichment increased both the size of the neurons’ dendritic tree and the number of their putative synapses. Adapted from Livneh et al. (2009)
, with permission.
How can an atlas possibly reflect the continuous structural changes which neurons undergo? One possibility is that there is actually no need to describe structural dynamics in atlases. In most adult animals, structural dynamics, over a time scale of one to several weeks, are usually on the order of 5–20% of the total population. This means that any atlas will be a good approximation (80–95% correct) of a neuron’s morphology during its lifetime. A 5–20% error is a figure which might in any case be confounded with inter-individual variability and as such, is satisfactory. However, even a 10% change in synaptic connectivity (morphologically reflected as dendritic spine dynamics) can have dramatic effects on neuronal function. Consequently, small changes in many neurons can lead to substantial changes in the entire network’s function (Hofer et al., 2009
). Thus, we propose that, as more information becomes available, digital integrated shared atlases could provide not only information regarding the molecular subtype of neurons, their laminar position, and their connectivity; but also their expected structural dynamics.
As discussed above, structural dynamics are also a reflection of (or at least are correlated with) experience-dependent plasticity. Notably, however, experience-dependent plasticity was consistently demonstrated following rather extreme “experience” manipulations (e.g., whisker trimming, retinal lesion, prolonged eye closure, sensory enrichment and nostril occlusion). In line with these extreme manipulations, altered neuronal structural dynamics have been described in pathological conditions, such as ischemia, Alzheimer’s disease, and prion disease (Tsai et al., 2004
; Zhang et al., 2005
; Brown et al., 2007
; Fuhrmann et al., 2007
). All these experimental manipulations can be viewed as composing a spectrum that ranges from severe brain pathology (in which complete turnover may occur), to learning tasks which supposedly underlie everyday learning (in which only subtle anatomical changes are expected). While experience-dependent structural plasticity has mostly been correlated with extreme and pathological manipulations, it still remains to be determined whether more subtle (i.e., relatively natural) manipulations are also correlated with detectable structural changes (Polley et al., 2004
; Megevand et al., 2009
). Recent work by Gan and colleagues has correlated motor learning with the formation and selective stabilization of a small fraction of new dendritic spines in the motor cortex. This work suggests that structural plasticity indeed has the potential to underlie a wider array of everyday learning and memory tasks (Yang et al., 2009
).
The incorporation of the wide range of structural dynamics into brain atlases is not trivial. This is primarily due to an insufficient amount of information regarding most cell-types. Due to technical limitations, structural plasticity in vivo has largely been studied in superficial layers of the mammalian brain like the neocortex and the olfactory bulb. As a result, we actually know very little about most other parts of the CNS with respect to their structural dynamics. One might expect that some regions like the hippocampus or the amygdala, which are associated with numerous processes of learning and memory might be far more structurally plastic than others, but this remains to be seen. Given that the primary sensory regions in the neocortex and the olfactory bulb already show fair levels of structural plasticity and diversity, we are probably (and also literally) only scratching the surface.
As a relatively young field, many questions still remain unresolved regarding structural plasticity in the adult brain. Will deeper brain regions also exhibit ongoing and experience-dependent structural plasticity? Do mature neurons in adult invertebrates or non-mammalian vertebrates also share this capacity for experience- dependent structural plasticity? Is the experience-dependent structural plasticity observed under rather extreme experimental manipulations similar to the plasticity that underlies real life experiences, such as learning? Are most forms of functional plasticity correlated with structural plasticity? Time will tell.
Another recent and well-known atlas of sorts is the sequenced genome. The genomes of more than 180 organisms have now been sequenced (Genome News Network
1
)and thus a tremendous amount of data has accumulated in easily-accessible databases. Sequenced genomes allow easy and comprehensive analyses of inter- and intra-species variability, and its relation to pathological conditions. Can high-resolution brain atlases conceptually benefit from the currently more advanced “genome atlas”?
One major lesson to learn from genomes is that it was well worth the effort. The sequencing of the genome and the establishment of shared online genomic databases have greatly advanced our understanding of genome function and evolution. This has allowed for a quantitative assessment of the actual extent of inter-individual genetic variability. Additionally, comparative genomics has enabled the evaluation of the extent and sources of inter-species genetic variability, which has led to a multitude of insights in the field of evolution, and has also advanced our basic understanding of the areas in the genome, which are important for function (protein coding sequences, regulatory elements, etc.; Miller et al., 2004
). Furthermore, comparative genomics have facilitated the exploration of the genetic sources of numerous pathological conditions (e.g., Down syndrome; Antonarakis et al., 2004
). From a researcher’s perspective, the sequenced genome, combined with freely-available online databases, has also expedited scientific progress as it has made many everyday scientific practices considerably easier. In analogy to brain atlases, how variable is the genome? Is genomic variability a constraint?
The most common inter-individual genomic variation is single nucleotide polymorphism (SNP), which has been associated with various traits such as disease susceptibility, and even with perception (Keller et al., 2007
; Shastry, 2007
). In addition to SNPs, other forms of DNA sequence variation exist, arising for example from inter-individual copy number variation (Beckmann et al., 2007
) and from the transposition of transposable DNA elements, which comprise ∼20% of mammalian genomes (Lander et al., 2001
; Waterston et al., 2002
; Gibbs et al., 2004
). Inter-individual copy number variation may partially explain inherited and sporadic traits (Beckmann et al., 2007
), and the transposition of transposable DNA elements has been shown, for example, to affect the differentiation fate of neuronal precursors (Muotri et al., 2005
). Despite these sources of genomic variation, the vast majority of the genome sequence is considered to be relatively stable between and within individuals (at least compared to the variability of the nervous system). Indeed, it is commonly thought that the genomes of two randomly selected individuals are ∼0.1% different (Shastry, 2007
; The Human Genome Project
2
). Notably, in absolute numbers, it is estimated that any two individuals will have several million differences in their genome, a number which emphasizes that this relatively small variability (0.1%) is not negligible at all.
On top of sequence variability, another striking layer of complexity comes from epigenetics. Despite a relatively stable genomic sequence, epigenetic modifications are highly dynamic. At the cellular level, there is accumulating evidence for the impact of epigenetic modifications such as DNA methylation, chromatin remodeling, nucleosome positioning, and histone modifications (most notably methylation and acetylation). These epigenetic modifications affect gene expression, and consequently various cellular processes from stem cell differentiation and cancer to synaptic plasticity (Levenson and Sweatt, 2005
; Meshorer and Misteli, 2006
; Smith et al., 2007
). At the level of the entire organism, epigenetic modifications are gaining increasing experimental attention as well. For example, although monozygotic twins are epigenetically indistinguishable during the early years of life, older monozygotic twins exhibit considerable differences in their overall epigenetic profile (Fraga et al., 2005
). Furthermore, epigenetic modifications have been implicated as a possible molecular mechanism for various types of experience-dependent plasticity in both the developing and mature nervous system (Fagiolini et al., 2009
; Sweatt, 2009
). For example, maternal care has been shown to affect the hypothalamus-pituitary-adrenal stress axis through epigenetic modifications of stress-related genes such as the glucocorticoid receptor (Meaney and Szyf, 2005
).
Thus, given the importance of epigenetics and using a similar approach to the Human Genome Project, efforts are now underway to map the human epigenome, starting with DNA methylation (The Human Epigenome Project
3
). This project’s official rationale is that epigenetic modifications “…constitute(s) the main and so far missing link between genetics, disease and the environment that is widely thought to play a decisive role in the etiology of virtually all human pathologies
4
.” Thus, sequencing the genome has also led to the realization that large-scale mapping of further layers of complexity (e.g., epigenetics) will most likely be equally beneficial. Accordingly, we suggest that high-resolution brain atlases should be analogous to a combination of the genome and the epigenome. But in what way are sequenced genomes and brain atlases analogous?
Despite the notable differences in the extent of inter-and intra-individual variability of these two systems, they are analogous in the sense that the basic components of the system (nucleotides/neurons) are relatively stable. Hence, much will be gained from high-resolution mapping of the system’s components and their respective position. However, on top of this relative stability, both systems have developed a remarkable capacity for change (epigenetics/neuronal plasticity). This analogy is further strengthened by the fact that the capacity to change, on top of a relatively constant background, seems to be a general mechanism by which biological systems (from single cells to organisms) maintain normal function, while constantly adapting to their ever-changing environment.
Efforts to construct high-resolution brain atlases face numerous challenges including inter- and intra-animal variability. Overcoming these challenges will substantially advance our understanding of the organization of neural circuits, and their relation to brain function. In this review, we described structural dynamics (primarily in mammals) as a further layer of complexity, which poses yet another challenge to take into consideration. Digital brain atlases could be a convenient platform for the integration of structural dynamics into pre-existing neuroanatomical (and molecular) data. In this way, seemingly static brain atlases will also be able to capture the dynamic nature of the brain.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank E. Meshorer, A. Darvasi, S. Druckmann and members of the Mizrahi lab for useful discussions and for critically commenting on the manuscript.
Ascoli, G. A., Alonso-Nanclares, L., Anderson, S. A., Barrionuevo, G., Benavides-Piccione, R., Burkhalter, A., Buzsaki, G., Cauli, B., Defelipe, J., Fairen, A., Feldmeyer, D., Fishell, G., Fregnac, Y., Freund, T. F., Gardner, D., Gardner, E. P., Goldberg, J. H., Helmstaedter, M., Hestrin, S., Karube, F., Kisvarday, Z. F., Lambolez, B., Lewis, D. A., Marin, O., Markram, H., Munoz, A., Packer, A., Petersen, C. C., Rockland, K. S., Rossier, J., Rudy, B., Somogyi, P., Staiger, J. F., Tamas, G., Thomson, A. M., Toledo-Rodriguez, M., Wang, Y., West, D. C., and Yuste, R. (2008). Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 9, 557–568.
Fraga, M. F., Ballestar, E., Paz, M. F., Ropero, S., Setien, F., Ballestar, M. L., Heine-Suner, D., Cigudosa, J. C., Urioste, M., Benitez, J., Boix-Chornet, M., Sanchez-Aguilera, A., Ling, C., Carlsson, E., Poulsen, P., Vaag, A., Stephan, Z., Spector, T. D., Wu, Y. Z., Plass, C., and Esteller, M. (2005). Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. U.S.A. 102, 10604–10609.
Gibbs, R. A., Weinstock, G. M., Metzker, M. L., Muzny, D. M., Sodergren, E. J., Scherer, S., Scott, G., Steffen, D., Worley, K. C., Burch, P. E., Okwuonu, G., Hines, S., Lewis, L., DeRamo, C., Delgado, O., Dugan-Rocha, S., Miner, G., Morgan, M., Hawes, A., Gill, R., Celera, Holt, R. A., Adams, M. D., Amanatides, P. G., Baden-Tillson, H., Barnstead, M., Chin, S., Evans, C. A., Ferriera, S., Fosler, C., Glodek, A., Gu, Z., Jennings, D., Kraft, C. L., Nguyen, T., Pfannkoch, C. M., Sitter, C., Sutton, G. G., Venter, J. C., Woodage, T., Smith, D., Lee, H. M., Gustafson, E., Cahill, P., Kana, A., Doucette-Stamm, L., Weinstock, K., Fechtel, K., Weiss, R. B., Dunn, D. M., Green, E. D., Blakesley, R. W., Bouffard, G. G., De Jong, P. J., Osoegawa, K., Zhu, B., Marra, M., Schein, J., Bosdet, I., Fjell, C., Jones, S., Krzywinski, M., Mathewson, C., Siddiqui, A., Wye, N., McPherson, J., Zhao, S., Fraser, C. M., Shetty, J., Shatsman, S., Geer, K., Chen, Y., Abramzon, S., Nierman, W. C., Havlak, P. H., Chen, R., Durbin, K. J., Egan, A., Ren, Y., Song, X. Z., Li, B., Liu, Y., Qin, X., Cawley, S., Cooney, A. J., D’Souza, L. M., Martin, K., Wu, J. Q., Gonzalez-Garay, M. L., Jackson, A. R., Kalafus, K. J., McLeod, M. P., Milosavljevic, A., Virk, D., Volkov, A., Wheeler, D. A., Zhang, Z., Bailey, J. A., Eichler, E. E., Tuzun, E., Birney, E., Mongin, E., Ureta-Vidal, A., Woodwark, C., Zdobnov, E., Bork, P., Suyama, M., Torrents, D., Alexandersson, M., Trask, B. J., Young, J. M., Huang, H., Wang, H., Xing, H., Daniels, S., Gietzen, D., Schmidt, J., Stevens, K., Vitt, U., Wingrove, J., Camara, F., Mar Alba, M., Abril, J. F., Guigo, R., Smit, A., Dubchak, I., Rubin, E. M., Couronne, O., Poliakov, A., Hubner, N., Ganten, D., Goesele, C., Hummel, O., Kreitler, T., Lee, Y. A., Monti, J., Schulz, H., Zimdahl, H., Himmelbauer, H., Lehrach, H., Jacob, H. J., Bromberg, S., Gullings-Handley, J., Jensen-Seaman, M. I., Kwitek, A. E., Lazar, J., Pasko, D., Tonellato, P. J., Twigger, S., Ponting, C. P., Duarte, J. M., Rice, S., Goodstadt, L., Beatson, S. A., Emes, R. D., Winter, E. E., Webber, C., Brandt, P., Nyakatura, G., Adetobi, M., Chiaromonte, F., Elnitski, L., Eswara, P., Hardison, R. C., Hou, M., Kolbe, D., Makova, K., Miller, W., Nekrutenko, A., Riemer, C., Schwartz, S., Taylor, J., Yang, S., Zhang, Y., Lindpaintner, K., Andrews, T. D., Caccamo, M., Clamp, M., Clarke, L., Curwen, V., Durbin, R., Eyras, E., Searle, S. M., Cooper, G. M., Batzoglou, S., Brudno, M., Sidow, A., Stone, E. A., Payseur, B. A., Bourque, G., Lopez-Otin, C., Puente, X. S., Chakrabarti, K., Chatterji, S., Dewey, C., Pachter, L., Bray, N., Yap, V. B., Caspi, A., Tesler, G., Pevzner, P. A., Haussler, D., Roskin, K. M., Baertsch, R., Clawson, H., Furey, T. S., Hinrichs, A. S., Karolchik, D., Kent, W. J., Rosenbloom, K. R., Trumbower, H., Weirauch, M., Cooper, D. N., Stenson, P. D., Ma, B., Brent, M., Arumugam, M., Shteynberg, D., Copley, R. R., Taylor, M. S., Riethman, H., Mudunuri, U., Peterson, J., Guyer, M., Felsenfeld, A., Old, S., Mockrin, S., and Collins, F. (2004). Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521.
Holtmaat, A., Bonhoeffer, T., Chow, D. K., Chuckowree, J., De Paola, V., Hofer, S. B., Hubener, M., Keck, T., Knott, G., Lee, W. C., Mostany, R., Mrsic-Flogel, T. D., Nedivi, E., Portera-Cailliau, C., Svoboda, K., Trachtenberg, J. T., and Wilbrecht, L. (2009). Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc 4, 1128–1144.
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., Funke, R., Gage, D., Harris, K., Heaford, A., Howland, J., Kann, L., Lehoczky, J., LeVine, R., McEwan, P., McKernan, K., Meldrim, J., Mesirov, J. P., Miranda, C., Morris, W., Naylor, J., Raymond, C., Rosetti, M., Santos, R., Sheridan, A., Sougnez, C., Stange-Thomann, N., Stojanovic, N., Subramanian, A., Wyman, D., Rogers, J., Sulston, J., Ainscough, R., Beck, S., Bentley, D., Burton, J., Clee, C., Carter, N., Coulson, A., Deadman, R., Deloukas, P., Dunham, A., Dunham, I., Durbin, R., French, L., Grafham, D., Gregory, S., Hubbard, T., Humphray, S., Hunt, A., Jones, M., Lloyd, C., McMurray, A., Matthews, L., Mercer, S., Milne, S., Mullikin, J. C., Mungall, A., Plumb, R., Ross, M., Shownkeen, R., Sims, S., Waterston, R. H., Wilson, R. K., Hillier, L. W., McPherson, J. D., Marra, M. A., Mardis, E. R., Fulton, L. A., Chinwalla, A. T., Pepin, K. H., Gish, W. R., Chissoe, S. L., Wendl, M. C., Delehaunty, K. D., Miner, T. L., Delehaunty, A., Kramer, J. B., Cook, L. L., Fulton, R. S., Johnson, D. L., Minx, P. J., Clifton, S. W., Hawkins, T., Branscomb, E., Predki, P., Richardson, P., Wenning, S., Slezak, T., Doggett, N., Cheng, J. F., Olsen, A., Lucas, S., Elkin, C., Uberbacher, E., Frazier, M., Gibbs, R. A., Muzny, D. M., Scherer, S. E., Bouck, J. B., Sodergren, E. J., Worley, K. C., Rives, C. M., Gorrell, J. H., Metzker, M. L., Naylor, S. L., Kucherlapati, R. S., Nelson, D. L., Weinstock, G. M., Sakaki, Y., Fujiyama, A., Hattori, M., Yada, T., Toyoda, A., Itoh, T., Kawagoe, C., Watanabe, H., Totoki, Y., Taylor, T., Weissenbach, J., Heilig, R., Saurin, W., Artiguenave, F., Brottier, P., Bruls, T., Pelletier, E., Robert, C., Wincker, P., Smith, D. R., Doucette-Stamm, L., Rubenfield, M., Weinstock, K., Lee, H. M., Dubois, J., Rosenthal, A., Platzer, M., Nyakatura, G., Taudien, S., Rump, A., Yang, H., Yu, J., Wang, J., Huang, G., Gu, J., Hood, L., Rowen, L., Madan, A., Qin, S., Davis, R. W., Federspiel, N. A., Abola, A. P., Proctor, M. J., Myers, R. M., Schmutz, J., Dickson, M., Grimwood, J., Cox, D. R., Olson, M. V., Kaul, R., Shimizu, N., Kawasaki, K., Minoshima, S., Evans, G. A., Athanasiou, M., Schultz, R., Roe, B. A., Chen, F., Pan, H., Ramser, J., Lehrach, H., Reinhardt, R., McCombie, W. R., de la Bastide, M., Dedhia, N., Blocker, H., Hornischer, K., Nordsiek, G., Agarwala, R., Aravind, L., Bailey, J. A., Bateman, A., Batzoglou, S., Birney, E., Bork, P., Brown, D. G., Burge, C. B., Cerutti, L., Chen, H. C., Church, D., Clamp, M., Copley, R. R., Doerks, T., Eddy, S. R., Eichler, E. E., Furey, T. S., Galagan, J., Gilbert, J. G., Harmon, C., Hayashizaki, Y., Haussler, D., Hermjakob, H., Hokamp, K., Jang, W., Johnson, L. S., Jones, T. A., Kasif, S., Kaspryzk, A., Kennedy, S., Kent, W. J., Kitts, P., Koonin, E. V., Korf, I., Kulp, D., Lancet, D., Lowe, T. M., McLysaght, A., Mikkelsen, T., Moran, J. V., Mulder, N., Pollara, V. J., Ponting, C. P., Schuler, G., Schultz, J., Slater, G., Smit, A. F., Stupka, E., Szustakowski, J., Thierry-Mieg, D., Thierry-Mieg, J., Wagner, L., Wallis, J., Wheeler, R., Williams, A., Wolf, Y. I., Wolfe, K. H., Yang, S. P., Yeh, R. F., Collins, F., Guyer, M. S., Peterson, J., Felsenfeld, A., Wetterstrand, K. A., Patrinos, A., Morgan, M. J., de Jong, P., Catanese, J. J., Osoegawa, K., Shizuya, H., Choi, S., and Chen, Y. J. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921.
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., Boe, A. F., Boguski, M. S., Brockway, K. S., Byrnes, E. J., Chen, L., Chen, T. M., Chin, M. C., Chong, J., Crook, B. E., Czaplinska, A., Dang, C. N., Datta, S., Dee, N. R., Desaki, A. L., Desta, T., Diep, E., Dolbeare, T. A., Donelan, M. J., Dong, H. W., Dougherty, J. G., Duncan, B. J., Ebbert, A. J., Eichele, G., Estin, L. K., Faber, C., Facer, B. A., Fields, R., Fischer, S. R., Fliss, T. P., Frensley, C., Gates, S. N., Glattfelder, K. J., Halverson, K. R., Hart, M. R., Hohmann, J. G., Howell, M. P., Jeung, D. P., Johnson, R. A., Karr, P. T., Kawal, R., Kidney, J. M., Knapik, R. H., Kuan, C. L., Lake, J. H., Laramee, A. R., Larsen, K. D., Lau, C., Lemon, T. A., Liang, A. J., Liu, Y., Luong, L. T., Michaels, J., Morgan, J. J., Morgan, R. J., Mortrud, M. T., Mosqueda, N. F., Ng, L. L., Ng, R., Orta, G. J., Overly, C. C., Pak, T. H., Parry, S. E., Pathak, S. D., Pearson, O. C., Puchalski, R. B., Riley, Z. L., Rockett, H. R., Rowland, S. A., Royall, J. J., Ruiz, M. J., Sarno, N. R., Schaffnit, K., Shapovalova, N. V., Sivisay, T., Slaughterbeck, C. R., Smith, S. C., Smith, K. A., Smith, B. I., Sodt, A. J., Stewart, N. N., Stumpf, K. R., Sunkin, S. M., Sutram, M., Tam, A., Teemer, C. D., Thaller, C., Thompson, C. L., Varnam, L. R., Visel, A., Whitlock, R. M., Wohnoutka, P. E., Wolkey, C. K., Wong, V. Y., Wood, M., Yaylaoglu, M. B., Young, R. C., Youngstrom, B. L., Yuan, X. F., Zhang, B., Zwingman, T. A., and Jones, A. R. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176.
Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., Antonarakis, S. E., Attwood, J., Baertsch, R., Bailey, J., Barlow, K., Beck, S., Berry, E., Birren, B., Bloom, T., Bork, P., Botcherby, M., Bray, N., Brent, M. R., Brown, D. G., Brown, S. D., Bult, C., Burton, J., Butler, J., Campbell, R. D., Carninci, P., Cawley, S., Chiaromonte, F., Chinwalla, A. T., Church, D. M., Clamp, M., Clee, C., Collins, F. S., Cook, L. L., Copley, R. R., Coulson, A., Couronne, O., Cuff, J., Curwen, V., Cutts, T., Daly, M., David, R., Davies, J., Delehaunty, K. D., Deri, J., Dermitzakis, E. T., Dewey, C., Dickens, N. J., Diekhans, M., Dodge, S., Dubchak, I., Dunn, D. M., Eddy, S. R., Elnitski, L., Emes, R. D., Eswara, P., Eyras, E., Felsenfeld, A., Fewell, G. A., Flicek, P., Foley, K., Frankel, W. N., Fulton, L. A., Fulton, R. S., Furey, T. S., Gage, D., Gibbs, R. A., Glusman, G., Gnerre, S., Goldman, N., Goodstadt, L., Grafham, D., Graves, T. A., Green, E. D., Gregory, S., Guigo, R., Guyer, M., Hardison, R. C., Haussler, D., Hayashizaki, Y., Hillier, L. W., Hinrichs, A., Hlavina, W., Holzer, T., Hsu, F., Hua, A., Hubbard, T., Hunt, A., Jackson, I., Jaffe, D. B., Johnson, L. S., Jones, M., Jones, T. A., Joy, A., Kamal, M., Karlsson, E. K., Karolchik, D., Kasprzyk, A., Kawai, J., Keibler, E., Kells, C., Kent, W. J., Kirby, A., Kolbe, D. L., Korf, I., Kucherlapati, R. S., Kulbokas, E. J., Kulp, D., Landers, T., Leger, J. P., Leonard, S., Letunic, I., Levine, R., Li, J., Li, M., Lloyd, C., Lucas, S., Ma, B., Maglott, D. R., Mardis, E. R., Matthews, L., Mauceli, E., Mayer, J. H., McCarthy, M., McCombie, W. R., McLaren, S., McLay, K., McPherson, J. D., Meldrim, J., Meredith, B., Mesirov, J. P., Miller, W., Miner, T. L., Mongin, E., Montgomery, K. T., Morgan, M., Mott, R., Mullikin, J. C., Muzny, D. M., Nash, W. E., Nelson, J. O., Nhan, M. N., Nicol, R., Ning, Z., Nusbaum, C., O’Connor, M. J., Okazaki, Y., Oliver, K., Overton-Larty, E., Pachter, L., Parra, G., Pepin, K. H., Peterson, J., Pevzner, P., Plumb, R., Pohl, C. S., Poliakov, A., Ponce, T. C., Ponting, C. P., Potter, S., Quail, M., Reymond, A., Roe, B. A., Roskin, K. M., Rubin, E. M., Rust, A. G., Santos, R., Sapojnikov, V., Schultz, B., Schultz, J., Schwartz, M. S., Schwartz, S., Scott, C., Seaman, S., Searle, S., Sharpe, T., Sheridan, A., Shownkeen, R., Sims, S., Singer, J. B., Slater, G., Smit, A., Smith, D. R., Spencer, B., Stabenau, A., Stange-Thomann, N., Sugnet, C., Suyama, M., Tesler, G., Thompson, J., Torrents, D., Trevaskis, E., Tromp, J., Ucla, C., Ureta-Vidal, A., Vinson, J. P., Von Niederhausern, A. C., Wade, C. M., Wall, M., Weber, R. J., Weiss, R. B., Wendl, M. C., West, A. P., Wetterstrand, K., Wheeler, R., Whelan, S., Wierzbowski, J., Willey, D., Williams, S., Wilson, R. K., Winter, E., Worley, K. C., Wyman, D., Yang, S., Yang, S. P., Zdobnov, E. M., Zody, M. C., and Lander, E. S. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562.