1
Psychiatry and Human Behavior, College of Medicine, University of California, Irvine, CA, USA
2
Brain Imaging and Analysis Center, Duke University, Durham, NC, USA
3
MIND Research Network, Albuquerque, NM, USA
4
Center for Research on Biological Systems, University of California San Diego, San Diego, CA, USA
5
Neuroinformatics Research Group, Washington University, Saint Louis, MO, USA
6
Brigham and Women’s Hospital, Harvard University, Boston, MA, USA
7
Department of Psychiatry, Duke University, Durham, NC, USA
Organizing and annotating biomedical data in structured ways has gained much interest and focus in the last 30 years. Driven by decreases in digital storage costs and advances in genetics sequencing, imaging, electronic data collection, and microarray technologies, data is being collected at an ever increasing rate. The need to store and exchange data in meaningful ways in support of data analysis, hypothesis testing and future collaborative use is pervasive. Because trans-disciplinary projects rely on effective use of data from many domains, there is a genuine interest in informatics community on how best to store and combine this data while maintaining a high level of data quality and documentation. The difficulties in sharing and combining raw data become amplified after post-processing and/or data analysis in which the new dataset of interest is a function of the original data and may have been collected by multiple collaborating sites. Simple meta-data, documenting which subject and version of data were used for a particular analysis, becomes complicated by the heterogeneity of the collecting sites yet is critically important to the interpretation and reuse of derived results. This manuscript will present a case study of using the XML-Based Clinical Experiment Data Exchange (XCEDE) schema and the Human Imaging Database (HID) in the Biomedical Informatics Research Network’s (BIRN) distributed environment to document and exchange derived data. The discussion includes an overview of the data structures used in both the XML and the database representations, insight into the design considerations, and the extensibility of the design to support additional analysis streams.
The biomedical science community has seen increased numbers of multi-site consortia driven in part by advances in speed and robustness of internet technologies, the demand for cross-scale data to understand fundamental disease processes, the need for experts from diverse domains to integrate and interpret the data, and the movement of science in general toward freely available information (Arzberger and Finholt, 2002
). The Science of Collaboratories website
1
lists 213 collaboratories since 1993. These consortia face increased challenges in managing, interpreting, and sharing data without informatics methods to clearly document necessary metadata at both the time of data collection and subsequent data processing and analysis.(Olsen et al., 2008
; Paton, 2008
) The difficulties in sharing and combining raw data become amplified after post-processing and/or data analysis in which the new dataset of interest is a function of the original data and may have been collected by multiple collaborating sites. Simple metadata, documenting which subject and version of data were used for a particular analysis, becomes complicated by the heterogeneity of the collecting sites yet is critically important to the interpretation and reuse of derived results. Numerous recent publications have discussed the benefits of documenting the origin and steps by which data were collected and derived (Foster et al., 2003
; Simmhan et al., 2005
; Zhao, et al., 2006
; MacKenzie-Graham et al., 2008
; Moreau et al. 2008
). Provenance, as defined by the Oxford English Dictionary, is “the source or origin of an object; its history and pedigree; a record of the ultimate derivation and passage of an item through its various owners” (Freire et al., 2008
). MacKenzie-Graham et al. (2008)
make a distinction between data provenance and processing provenance where the former refers to metadata describing how the original data was collected and the later referring to the processing original data undergoes after the initial collection. Both types of metadata are crucially important for subsequent use of the data by a single laboratory and the scientific community. In multi-site, distributed, collaboratories where information is dynamic in nature and not centrally managed, robust, scalable metadata management tools are essential (Moreau et al., 2008
).
The Biomedical Informatics Research Network (BIRN)
2
is a large multi-site consortia of individual test beds coalesced around a shared set of resources, developing standards, methods, and processing tools in a distributed, grid-enabled environment (Grethe et al., 2005
; Keator et al., 2006
). The BIRN enables scientists across disparate domains to securely and transparently share data and tools. The Function BIRN test bed (Keator et al., 2006
) brings together investigators developing data sharing standards, instrument calibration methods in the context of functional MRI (fMRI), novel statistical models, and advanced clinical/cognitive paradigms necessary to study the neural substrates of schizophrenia in a collaborative setting. Since its inception in 2002, FBIRN has prospectively collected over 400 fMRI human datasets collected during the protocol design and execution of four separate studies and thousands of agar phantom calibration datasets across the 11 participating sites. The datasets generally consisted of a minimum of five functional acquisitions and at least a T1-weighted structural acquisition. Details about the publically available data can be found at http://nbirn.org/bdr
.
Beyond prospective data collection, the FBIRN neuroinformatics working group, in collaboration with other BIRN test bed informatics groups, has developed data structures and software to dynamically track and document data acquired and analyzed as part of human imaging studies. The suite of tools forms a cooperative system for managing and documenting acquired and derived data entitled the FBIRN Federated Informatics Research Environment (FIRE)
3
. Data management in the federated environment of both the original and derived data is supported through three core components: the Human Imaging Database (HID)
4
for distributed/federated relational database support and web-enabled graphical user interface, the XML-Based Clinical Experiment Data Exchange (XCEDE2)
5
schema used to define valid XML documents for structured data/metadata storage and exchange, and data publication scripts to organize and transfer data to the distributed file system and send appropriate uniform resource locator (URL) links to the HID database. In this manuscript we introduce tools from the BIRN software suite used for documenting multi-site functional and structural neuroimaging analyses in a federated database and distributed data handling environment. The discussion centers around two data processing pipelines, one designed for multi-site preprocessing of fMRI data and the other, a structural analysis of schizophrenia in humans. Our intention is to provide the informatics community with insights into the data structures used and our view of the extensibility of this system.
Fbirn Neuroimaging Data Management and Workflows Overview
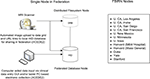
Scientific data management systems generally consist of at least a few core components: a back-end database for permanent, structured, data storage and efficient query, a front-end graphical user interface for client interaction, and an import/export mechanism to get data into and out of the database and share with collaborators (Keator et al., 2008
). These systems can exist entirely at a single site or be distributed geographically. FBIRN operates in a completely distributed environment. The suite of tools developed by FBIRN form the FIRE, providing management support of clinical, behavioral, and imaging data in a decentralized way using federated databases and a distributed file system (Figure 1
). Each site maintains its own HID database back-end and graphical user interface (Ozyurt et al., 2004a
,b
, 2006
; Keator et al., 2006
; Keator, 2009
). The HID is an open-source extensible database schema designed to support multi-site, federated, installations and inclusion of new data types without changing the core table space. The graphical user interface is a three-tier J2EE application supporting data input, single-site and multi-site query, data export, and core system administration tasks. More detailed information about HID can be found in the references and the software is available through the NITRC website
6
. Currently, within FBIRN, there are 11 federated installations managing 790 imaging visits and 4239 clinical assessments as collected across four prospective FBIRN studies and retrospective data contributed by the Brainscape repository of Washington University, St. Louis
7
. Clinical assessments collected are those common in studies of Schizophrenia such as SCID, Beckman Depression Inventory (BDI), North American Adult Reading Test (NAART), InterSePT scale, and many others. Details of publically available data can be found at http://nbirn.org/bdr
. Data files that are part of an imaging study are published to the Storage Resource Broker (SRB) distributed file system and cross-linked in the database using a URL string (Rajasekar et al., 2003
). The data publication process involves data reorganization into a standardized directory hierarchy, format conversions, and the creation of XCEDE2 XML (eXtensible Markup Language)
8
files containing minimal metadata about the experiment stored with the imaging files on the SRB. This process is facilitated by data publication scripts. The scripts use an XML formatted template which a site can configure using an XML editor or a provided GUI. The upload template consists of metadata describing the imaging series, visit, and project information. When available the information is automatically extracted from DICOM image headers. Information that is not available in the DICOM headers is input manually. The data publication scripts include schematron validation definitions which are prepared during study design to validate the data publication XML templates. Once the templates are created they can be reused with minor modifications to visit dates and subject IDs using the GUI provided with the publication scripts. The bulk of metadata describing the subject visit is stored in the database. Additional details about data provenance and management of the original collected data can be found in publications by Ozyurt et al. (2006) and Keator et al. (2006)
.
Figure 1. Schematic of original (raw) data entry and file registration process at a node in the federation and FBIRN node locations.
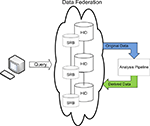
Once the data has been published into the federated system, it is available for processing. The FBIRN has developed quality assurance and image processing utilities optimized to work with data from the federated system. Data analysis and/or post-processing workflows currently instantiated in FBIRN share a few common steps. First, the datasets are located in the federation, either by browsing the low-level distributed file system or interacting with the HID graphical user interfaces to query and filter data collected in the federation. Once datasets of interest are identified, they are downloaded to the local system for computation (Figure 2
). The downloaded datasets contain both imaging data files and the XML metadata files stored with the dataset. Additional metadata exports from the HID database are also available during the downloading process if one is using the graphical user interface. Once data is downloaded, any number of analysis algorithms could be run and a new derived dataset created. If an investigator feels the derived dataset is of sufficient technical quality and scientific interest to others in the collaboratory, it should be published to the federation with sufficient processing provenance and searchable metadata such that others can effectively interpret and reuse the derived data. This overall process of documenting steps in an analysis pipeline, representing the provenance in a consistent and well documented way, and providing a means of querying derived data which references original subjects collected at geographically distributed sites in a robust and extensible manner were the motivations driving the informatics components presented here.
Figure 2. FBIRN data management discovery and analysis workflow.
Case Studies
Two analysis workflows will be referred to throughout the following sections, giving substantive context to the abstract informatics structures discussed. Each workflow has slightly different requirements for processing provenance and metadata storage. Together the case studies illustrate the robustness of the informatics structures.
Structural MRI analysis workflow
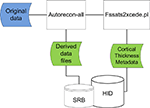
This workflow consists of a multi-site structural MRI analysis of schizophrenia. The imaging data consisted of 3D T1-weighted MRI images collected across consortium sites. The original images were shared using the data management components described in Section “FBIRN Neuroimaging Data Management and Workflows Overview.” The structural morphometric (StructMorph) analysis was performed across two participating sites. Data were analyzed with the FreeSurfer software
9
using a single program “autorecon-all”. The “autorecon-all” script calculates cortical and sub-cortical thickness statistics in two stages: a volumetric processing stage which includes noise correction, volumetric registration, and white matter segmentation, and a surface processing stage for cortical parcellation and thickness measurements. The “autorecon-all” is a black box processing script. Provenance documentation about which FreeSurfer binaries are called by “autorecon-all” were not provided with the analysis. It has a version number and compilation date that uniquely identifies the script but the details about what other modules it calls during the course of execution is hidden from the user. Cortical and sub-cortical thickness estimates from the structural processing pipeline were chosen by study investigators as metadata to make available for query in the database federation. All other images, intermediate files, and program specific outputs were made available on the distributed file system. Cortical thickness measurements are extracted from output files using the script “fsstats2xcede.pl”. The overall workflow is shown in Figure 3
. This case study is used to illustrate the process of extracting relevant analysis specific metadata, encapsulating it in XML, loading it into database tables, and making it available for query in the federated data management system in a generic way.
Figure 3. Original data files (blue) downloaded from data federation are processed using autorecon-all and cortical thickness data extracted (green). Resulting data files are loaded back into data federation.
fMRI data preprocessing workflow
The fMRI data preprocessing (PreProc) workflow consists of a multi-level pipeline with numerous intermediate derived results combined with original data inputs at various points in the workflow (Figure 4
). The complex nature of the workflow makes it an ideal test case for the informatics structures. This workflow was designed to provide an automated and consistent pre-processing pipeline for FBIRN studies. Preprocessing in fMRI is a general term describing any processing done after image reconstruction prior to statistical analysis of brain activation (Strother, 2006
). The PreProc pipeline consists of motion correction, slice timing correction, magnetic field inhomogeneity correction (B0), and spatial smoothing. For additional information on the FBIRN imaging processing pipeline used for the PreProc analysis, please visit www.nitrc.org/projects/fips/
. For this workflow, investigators were most interested in documenting the processing provenance. Unlike the StructMorph analysis discussed in Section “Structural MRI Analysis Workflow” in which the processing is treated as a single black box script, this workflow has many separate programs put together in a specific order. Changing the order and/or any of the parameter settings potentially alter the derived results. Investigators were most interested in carefully documenting the ordering of steps and the parameters used. Proper documentation of the PreProc workflow enables its use in higher order analyses without duplicating work. As the data federation grows, original data may be processed numerous times with slightly different steps or with different parameter settings and made available through the data management systems. It is therefore critically important to document the workflow as completely as possible given limited time and resources of investigators to enable maximum derived data reusability.
Figure 4. fMRI PreProc data preprocessing workflow. Original data inputs colored blue, intermediate derived data colored yellow, final derived data output colored green. Workflow transform modules Fips-mc-fsl, Fips-b0c-fsl, Fips-stc-fsl, and Fips-sm2-fsl contains variable numbers of sub-transform steps to produced intermediate and final derived results.
Derived Data Exchange Schema
The XML-Based Clinical Experiment Data Exchange (XCEDE2)
10
schema was designed for documenting research and clinical studies (Keator et al., 2006
). The schema defines components and constraints on those components required to form a valid XCEDE2 compliant XML document. Initially the focus of XCEDE2 was on human imaging studies but the schema contains many generic and extensible structures useful for a wider range of scientific domains. Development of the schema was a joint effort within BIRN and is the exchange medium for many database web services currently in use. The schema is flexible, providing mechanisms for linking to external output files and for storing analysis data directly in the XML document. XCEDE2 documents can be split into sub-documents and linked together using constructs of the schema. The data analysis portion of the XCEDE2 schema is the most relevant to the case studies and will be presented in more detail. For complete documentation of the schema readers are encouraged to visit the website. The analysis component of the XCEDE2 schema was designed as a generic container used for documenting results of analyses. An analysis in this context is composed of the “inputs” (i.e., the files and parameters used in an analysis or processing of data), a list of the application(s) or method(s) used in the analysis (provenance), and the resultant data (i.e., values and output files) (Figure 5
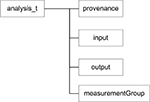
).
Figure 5. Base <analysis> component.
The format of the <input> and <output> components (Figure 6
A) are essentially identical. Using the ID attributes <dataID> and <analysisID>, they serve as pointers to other portions of the XCEDE dataset (in the same XML document or another XCEDE2-compliant XML file) that more fully describe the analysis or data consumed or written by this processing step.
Figure 6. <analysis_t> components of the XCEDE2 schema. The input/output components (panel A) used to reference input data and output derived data files and/or metadata. The measurement group component (panel B) used to store derived data values directly in XML formatted file. The provenance and processStep components (panel C) used for documenting processing pipeline specific metadata.
The <measurementGroup> component is used to store information and data related to the outcome of analyses (Figure 6
B). Each measurement group contains observations on an entity. Entities are used to give meaning to the measurements being stored. The entity element can reference any number of terminology sources and is composed of multiple nomenclature/termID pairs. The observation element of a measurementGroup contains the actual measurement values for the particular entity along with attributes defining the data type and units of the measurement. An example of the <measurementGroup>entry for the StructMorph analysis is shown in Figure 7
. The measurements for this analysis are related to curvature and thickness of particular anatomical parcellations of the cortex. The <measurementGroup> component is extensible in that any number of self-describing observations can be grouped together to record a derived data output complete with entity information. The nomenclature used in this example is the FreeSurfer native terminology thus giving meaning to an otherwise arbitrary anatomical location identifier. In the StructMorph analysis, there are many <measurementGroup> entries, one for each anatomical region analyzed. Hemispheric analyses are physically separated into different XCEDE2 files but could alternatively be contained within one file. The decision to separate results into multiple XCEDE2 files was to facilitate granularity of analysis summary downloads.
Figure 7. XCEDE2 XML entry for thickness and curvature derived data. Entity tags document terminology source “rh.aparc.annot” and term “caudalmiddlefrontal” which is the native term and source within FreeSurfer analysis software.
In thinking about how users would interact with the derived results, there were two methods that were most desirable to support in FBIRN. The first method is a direct query of the database, filtering on cortical thickness and/or curvature measurements by anatomical region for the StructMorph analysis shown in Figure 7
. To facilitate this use case, the parcellation results need to be loaded into the data management system. Web services for the HID database were developed in support of derived data loading using the XCEDE2 format. Effectively any derived result that can be represented using XCEDE2’s <analysis> component can be directly imported into the HID database without table space changes (see Section “Derived Data Database Schema” for database design). The intermediate representation of derived results in the form of an XCEDE2 file is important for downstream processing tools, data management systems, and structured data exchange. Tools that might otherwise not have access to a processing pipeline’s native output file formats can be written to parse XCEDE2 documents and obtain an agnostic view of derived results. For those pipeline stages that don’t directly export XCEDE2 data, it is a simple matter to create wrapper scripts that extract relevant summary data into XCEDE2 documents. The second method of derived data use in the FBIRN federation is downloading the entire analysis output and exploring the output within the analysis tool or pipeline itself. For this method of interaction, a user may just need to filter on some aspect of the processing provenance. For example, a user might query on all analyses performed using named pipeline PreProc, version 1.0. Additionally, the user might want to find all analyses that used a particular dataset as input. To support these use cases, structured documentation of original data and processing provenance is needed.
The <provenance> element of the <measurementGroup> component provides a mechanism for documenting processing provenance in an XCEDE2 compliant XML document (Figure 6
C). A typical <provenance> entry consists of many <processStep> blocks used to store metadata about the analysis pipeline itself. The schema provides elements for documenting program arguments, compiler and library information, platform and architecture, time stamping, and user identification. Typically in standalone analysis packages and in arbitrary processing pipelines constructed from multiple standalone applications, rich metadata is difficult to capture. Unless there has been concerted effort by software developers to provide provenance with analysis execution, it is up to the user to maintain accurate records. Workflow environments such as the LONI pipeline
11
and Fiswidgets
12
augment information provided by tool developers with enhanced pipeline metadata easing the burden of provenance documentation (Fissell et al., 2003
; MacKenzie-Graham et al., 2008
). The XCEDE2 schema provides flexibility in storing pipeline provenance alongside derived data. Figure 8
shows examples of processing provenance collected for the StructMorph and PreProc use cases. The complete provenance records for the analyses are quite long so selected segments have been extracted. To provide the ability to reconstruct arbitrarily complex pipelines, the data provenance schema in XCEDE2 supports multiple forks, merges, and/or parallel analysis streams. Currently, the XCEDE2 provenance <processStep> components have attributes “id” and “parent” that together are used to document complex tree structured processing pipelines. The schema does not put any restrictions on “parent” attributes allowing maximum flexibility at some expense of clarity. In the StructMorph use case, provenance wrapping scripts were written by FBIRN developers working directly with FreeSurfer software developers. In the PreProc use case, provenance was compiled by FBIRN developers using information available from only the standalone tools and linked together using XCEDE2 constructs consistent with the defined the pipeline. The <provenance> components in an XCEDE2 compliant export of an analysis are used directly by the HID database web services to store the processing pipeline description. The <measurementGroup> data is also parsed by the web service layer and loaded into the data management system (see Section “Derived Data Database Schema”).
Figure 8. Example XCEDE2 provenance blocks from PreProc (top) analysis and StructMorph (bottom) analyses.
Derived Data Database Schema
Cataloging derived data and metadata in the HID data management system is a vital step in making the analytic results available to BIRN collaborators and ultimately the wider scientific community. Because the BIRN infrastructure is inherently distributed and federated in nature, simple changes to database schema at one site becomes difficult and time consuming in the federation. Therefore, an important requirement for the database schema is a stable set of generic tables capable of storing processing pipeline provenance, interesting analytic results, and metadata about analyses complete with ontology and terminology source references. The table space should not change when presented with new derived data types and/or pipeline definitions. The StructMorph and PreProc analyses are interesting cases to test the stability of the data management schema. The StructMorph use case tests the capability of storing derived data values directly in the database and the automated query interface creation by the web application. The PreProc use case tests the table space for documenting multi-layered processing pipeline provenance. As shown in Figure 4
, the processing pipeline is complex with transforms composed of sub-transforms hierarchically, with inputs and outputs interleaved along with multiple intermediate states.
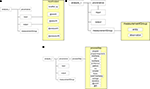
The database schema for documenting processing pipeline definitions consists of four core tables: nc_analysis, nc_analysisFlow, nc_analysisComponent, and nc_transformation (Figure 9
). Defining a processing pipeline is differentiated from any particular instantiation of that processing pipeline on actual data. The nc_transformation table serves as a generic bag of processes where each entry contains a reference name, reference version, package name, package version, and ontological information. The reference name and version are user-defined identifiers for the process whereas the package name and version corresponds to the name given by the process developers. The idea is to select processes from the nc_transformation table and put them together into pipelines. By adding the processes to the nc_transformation table, one can reuse tools in subsequent analytic pipelines. With respect to the use cases, the nc_transformation table contains entries for “autorecon-all” and “fsstats2xcede.pl” for the StructMorph analysis and “avwmerge”, “avwmaths++”, “mcflirt”, “nifti_tool”, “bet”, “fugue”, “flirt”, “slicetimer”, “mri_fwhm”, and “ip_32R” for the PreProc analysis. By comparing the list with Figure 4
there are four occurrences of the “nifti_tool” process in the pipeline but only a single entry in the nc_transformation bag of tools table. Next, the processing pipeline is assembled from the tools available in the nc_transformation table and the processing flow defined. The nc_analysisFlow table defines the flow through the processing tree defined in the nc_analysisComponent table.
Figure 9. Core HID tables for defining processing pipelines.
For the PreProc pipeline, the nc_analysisFlow table contains two entries, one for “autorecon-all” and one for “fsstats2xcede.pl”. The analysisid entry uniquely identifies the processing pipeline as described in the nc_analysis table’s name, version and ontology source fields. The componentid field in the nc_analysisFlow table references the component ID stored in the nc_analysisComponent table for a process (autorecon-all for example). The priorcomponentid field in the nc_analysisFlow table defines a component executing immediately prior to the current step of the pipeline. Any number of entries for prior components can be added to the nc_analysisFlow table for a given componentid providing flexibility in defining complex pipelines. The nc_analysisComponent table defines the hierarchical relationship between steps in the pipeline. The analysisid and transformationid fields reference the pipeline and processing steps. Fields parentcomponentid and nodelevel reference the parent processing step and the depth within the processing pipeline tree. The nodelevel field is used to both identify the first step in the processing pipeline tree (nodelevel = 1) and to group processing tasks into distinct levels (or depths). The parentcomponentid identifies the parent node in the pipeline. Cyclic operations in a graph representation of a processing pipeline where there are multiple executions of a particular step are duplicated in the current implementation. Database queries through the HID web interface can be constructed either as simple queries filtering on particular components of the pipeline (nc_analysisComponent table), on sequences of tools (nc_analysisComponent and nc_analysisFlow tables), and by overall pipeline named identifiers (nc_analysis table). More advanced concept and ontology based queries are also supported if the ontology fields are populated for the processing pipeline.
Pipeline metadata related to output formats from an analysis are described in a generic way similar to those used in HID for storing new data types (Ozyurt et al., 2004a
,b
, 2006
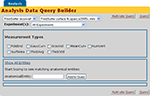
). The nc_extendedTuple table along with a number of accessory tables enables new classes of data to be described in a similar way as one constructs classes in programming languages such as C++ and Java. In the StructMorph use case, the extended tuples functionality is used to describe the anatomical thickness measurements that are loaded into the database from the XCEDE2 document discussed above. The database graphical user interface uses the extended tuples class definition to construct a query interface in the web application that is appropriate for basic logical queries over the results from an instantiation of the pipeline on actual data (Figure 10
). The mechanisms used by HID to automatically construct web based query forms are in active development and beyond the scope of this manuscript. Interested readers are encouraged to visit the NITRC HID website for further details and documentation.
Figure 10. HID web interface derived data query form for StructMorph analysis.
The instantiation of a processing pipeline and the resulting derived data is stored among a variety of HID tables linking the analysis files deposited in the data grid (SRB) with the pipeline metadata stored using the data class description discussed above. Because the databases are federated, it may not be the case that the original data used to produce a derived result are registered in the database where the pipeline outputs are to be stored. The provenance information stored in the XCEDE2 formatted output files includes information about which original data were used in the processing pipeline. This information is used by the HID import web service to determine whether the original data exists in the particular HID the derived result is being deposited or not. If the original data does not exist in the database, an entry is put into the nc_externalData table with information about which HID to contact for more detail about the original input data such as demographics, behavioral assessments, visit dates, etc. The HID federated query mechanism used to find information across the data federation is used in this context to provide additional information about the data included in an analysis pipeline. Interesting queries can be executed to locate all data processed with a particular pipeline and find which pipelines a particular dataset were used in, for example.
The StructMorph analysis was performed on 146 subjects collected across the FBIRN sites. The analysis was performed at two sites and the resulting derived data loaded into the HID systems at those two sites. Beyond the database and schema structures, code was written to convert the FreeSurfer cortical parcellation and volumetric segmentation output measures to XCEDE2 XML files. Instantiated pipeline provenance for each of the 146 runs was more difficult to obtain. Log files extracted from the processing tools were parsed for provenance and in some cases were unsatisfactory depending on the amount of information stored by the applications. The “fsstats2xcede.pl” tool was written within the FBIRN consortium and contained rich provenance information highlighting the need for either provenance wrappers around tools developed elsewhere or advocating the use of workflow environments such as LONI and Fiswidgets. Preliminary testing of metadata queries was successful, identifying the derived data consistently. The design of the derived data query pages required programmer input for clearer organization of form components.
The PreProc analysis was performed at one site after downloading the distributed data sets from the data federation and the resulting derived data loaded into the HID at the site performing the analysis. The database tables and XCEDE schema structures were sufficient in describing the more complicated processing pipeline. Investigators were initially interested in querying the PreProc data by filtering on pipeline provenance therefore the pipeline definition itself was used in test queries. For instance, a query to find all data derived using the “fugue” tool in the pipeline could be executed or a query to find the pipeline called “FIPS_MBTS_preprocs” (name stored in nc_analysis table for the PreProc pipeline, Figure 4
).
There were many analyses done using the data collected prospectively by the FBIRN consortium. Details about the data processing pipelines and results can be found in publications Friedman and Glover (2006)
, Magnotta and Friedman (2006)
, Friedman et al. (2008)
, Ford et al. (2009)
, Potkin and Ford (2009)
, Potkin et al. (2009a
,b)
and Wible et al. (2009)
. The StructMorph and PreProc workflows were chosen to illustrate two different use cases for the derived data constructs presented here. Design of the derived data system was focused on the capability to represent the derived data generated as part of the publications listed above. Currently the derived datasets are being loaded into the data management system using the components discussed in this manuscript.
The most challenging aspect has been obtaining sufficient provenance from the applications used for processing. Convincing the tool developers to output detailed provenance records is time consuming and difficult even when there is a good relationship between the developer and the users. Extracting provenance information from software log files is very demanding, error-prone, incomplete, and brittle. What has worked best for the FBIRN test bed, but far from satisfying, is a combination of working with developers (where possible) and scripting/automating analysis pipelines such that provenance is automatically documented during script execution. Processing pipelines are effectively wrapped with code to populate XCEDE formatted XML files with proper provenance detailing the analysis. There are no guarantees of provenance accuracy when wrapping pipelines. One could easily change a parameter and it not be reflected in the provenance output. FBIRN has found that regardless of the provenance capturing system and analysis automation method used, a human curator is invaluable for maintaining high quality data within the federation.
Storing and documenting derived results in data management systems along with important provenance information about the original input data and the pipeline itself in the context of a federated system is a challenging yet critically important endeavor. In large multi-site consortia where many geographically distributed investigators process the original data in different ways, providing a mechanism for them to contribute their work back to the federation and inform collaborators is desirable.
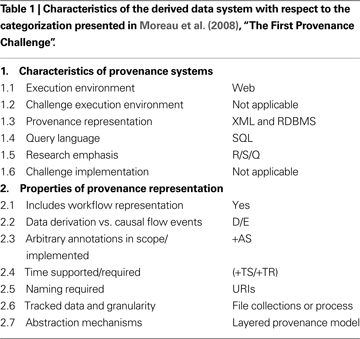
In the “The First Provenance Challenge” by Moreau et al. (2008)
, a challenge pipeline is presented along with a set of criteria to categorize and compare provenance systems (Moreau et al., 2008
). Table 1
describes the derived data system presented here in terms of the Moreau et al. (2008)
categorization criteria. The derived data system is capable of storing the provenance challenge workflow described in Moreau et al. (2008) and addressing all of the core provenance queries. Core queries Q5, Q8, and Q9 in Moreau et al. (2008)
filter on specific key-value pairs extracted from derived intermediate outputs or command line parameters of processing stage execution. Our system provides a very flexible method of allowing the researchers to specify which metadata and/or key-value pairs from the pipeline execution should be made query-able in the database graphical user interface (through the XCEDE XML representation, Section “Derived Data Exchange Schema”).
The derived data management system introduced here is a joint effort by many collaborators across the BIRN consortium and the authors believe have promise in facilitating knowledge discovery through collaborative, distributed, data collection and analysis. The design and implementation is still being tested on many derived datasets produced and published by consortium members. Further testing of the query capabilities and automatic creation of derived data query forms is needed. Ultimately the goal is to create a dynamic federated system where collaborators can download original data (or derived data), perform novel analyses, and contribute that information back to the federation in a consistent and well documented way with minimal programmer input. The generic structures presented here are a good start and have been useful to the FBIRN consortium.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported by U24-RR021992 to the Function Biomedical Informatics Research Network, U24-RR021382 to the Morphometry Biomedical Informatics Research Network, and U24-RR019701 to the Biomedical Informatics Research Network Coordinating Center (BIRN, http://www.nbirn.net
), that is funded by the National Center for Research Resources (NCRR) at the National Institutes of Health (NIH). The authors thank Randy Notestine at the University of California, San Diego for contributing the fMRI PreProc use case, Steven Potkin at the University of California, Irvine for his support of the work presented here, and the FBIRN consortium members for the data collection and support of the scientists and engineers involved in the project.
- ^ www.scienceofcollaboratories.org
- ^ www.nbirn.net
- ^ www.nitrc.org/projects/fbirn/
- ^ www.nitrc.org/projects/hid
- ^ www.xcede.org
- ^ http://www.nitrc.org/projects/hid/
- ^ www.brainscape.org
- ^ www.w3.org/XML
- ^ http://surfer.nmr.mgh.harvard.edu
- ^ www.xcede.org
- ^ http://pipeline.loni.ucla.edu
- ^ http://grommit.lrdc.pitt.edu/fiswidgets/
Ford, J. M., Roach, B. J., Jorgensen, K. W., Turner, J. A., Brown, G. G., Notestine, R., Bischoff-Grethe, A., Greve, D., Wible, C., Lauriello, J., Belger, A., Mueller, B. A., Calhoun, V., Preda, A., Keator, D., O’Leary, D. S., Lim, K. O., Glover, G., Potkin, S. G., and Mathalon, D. H. (2009). Tuning in to the voices: a multisite FMRI study of auditory hallucinations. Schizophr. Bull. 35, 58–66.
Friedman, L., Stern, H., Brown, G. G., Mathalon, D. H., Turner, J., Glover, G. H., Gollub, R. L., Lauriello, J., Lim, K. O., Cannon, T., Greve, D. N., Bockholt, H. J., Belger, A., Mueller, B., Doty, M. J., He, J., Wells, W., Smyth, P., Pieper, S., Kim, S., Kubicki, M., Vangel, M., and Potkin, S. G. (2008). Test–retest and between-site reliability in a multicenter fMRI study. Hum. Brain Mapp. 29, 958–972.
Grethe, J. S., Baru, C., Gupta, A., James, M., Ludascher, B., Martone, M., Papadopoulos, P. M., Peltier, S. T., Rajasekar, A., Santini, S., Zaslavsky, I. N., and Ellisman, M. H. (2005). Biomedical informatics research network: building a national collaboratory to hasten the derivation of new understanding and treatment of disease. From grid to healthgrid: proceedings of healthgrid 2005. Amsterdam, IOS Press.
Potkin, S. G., Turner, J. A., Brown, G. G., McCarthy, G., Greve, D. N., Glover, G. H., Manoach, D. S., Belger, A., Diaz, M., Wible, C. G., Ford, J. M., Mathalon, D. H., Gollub, R., Lauriello, J., O’Leary, D., van Erp, T. G., Toga, A. W., Preda, A., and Lim, K. O. (2009a). Working memory and DLPFC inefficiency in schizophrenia: the FBIRN study. Schizophr. Bull. 35, 19–31.
Wible, C. G., Lee, K., Molina, I., Hashimoto, R., Preus, A. P., Roach, B. J., Ford, J. M., Mathalon, D. H., McCarthey, G., Turner, J. A., Potkin, S. G., O’Leary, D., Belger, A., Diaz, M., Voyvodic, J., Brown, G. G., Notestine, R., Greve, D., and Lauriello, J. (2009). fMRI activity correlated with auditory hallucinations during performance of a working memory task: data from the FBIRN consortium study. Schizophr. Bull. 35, 47–57.