Bio-Swarm-Pipeline: a light-weight, extensible batch processing system for efficient biomedical data processing
1
Neuroimaging Core Facility, Genes, Cognition and Psychosis Program, National Institute of Mental Health/National Institutes of Health, Bethesda, MD, USA
2
Mood and Anxiety Disorders Program, National Institute of Mental Health/National Institutes of Health, Bethesda, MD, USA
A streamlined scientific workflow system that can track the details of the data processing history is critical for the efficient handling of fundamental routines used in scientific research. In the scientific workflow research community, the information that describes the details of data processing history is referred to as “provenance” which plays an important role in most of the existing workflow management systems. Despite its importance, however, provenance modeling and management is still a relatively new area in the scientific workflow research community. The proper scope, representation, granularity and implementation of a provenance model can vary from domain to domain and pose a number of challenges for an efficient pipeline design. This paper provides a case study on structured provenance modeling and management problems in the neuroimaging domain by introducing the Bio-Swarm-Pipeline. This new model, which is evaluated in the paper through real world scenarios, systematically addresses the provenance scope, representation, granularity, and implementation issues related to the neuroimaging domain. Although this model stems from applications in neuroimaging, the system can potentially be adapted to a wide range of bio-medical application scenarios.
Scientific workflow systems that are capable of tracking the details of data processing history can facilitate a number of fundamental requirements in everyday scientific research, such as scheduling batch processing on multiple computers, interpreting and comparing different results, sharing and reusing existing workflow, etc. In the scientific workflow research community, the information that describes the details of data processing history is referred to as “provenance” (also “lineage” or “pedigree”) (Simmhan et al., 2005
). Provenance management is a critical component of scientific workflow systems and most of the existing popular scientific workflow systems have a module for management of provenance information. For e.g., the Kepler workflow system is able to collect the provenance information (Ludäscher, 2006
), while the Taverna workflow system stores the provenance information for users to manage and reuse previous workflows (Oinn et al., 2004
). VisTrails is a provenance management system (PMS) that provides infrastructure for data exploration and visualization through workflows ( Callahan et al., 2006
; Silva et al., 2007
; Koop et al., 2008
). The Swift workflow system builds on and includes technology previously distributed as the GriPhyN Virtual Data System to capture the provenance (Zhao et al., 2007
). The Pegasus workflow system also uses the Virtual Data System to capture the provenance (Miles et al., 2008
). The VIEW workflow system manages the provenance data with a provenance management module (Lin et al., 2009
). The LONI workflow system has a provenance management framework to manage the provenance data (MacKenzie-Graham et al., 2008
).
Despite its importance, however, provenance modeling and management is still a relatively new area in the scientific workflow research community (Simmhan et al., 2005
) and the provenance model can vary from domain to domain (Freire et al., 2008
). In particular, although the VisTrails, Swift, VIEW and LONI workflow systems have been applied to neuroimaging, a number of provenance modeling and management issues that are specific to the neuroimaging domain have to be explored further:
(Q1) The provenance model varies from domain to domain and has to be identified and appropriately customized for the neuroimaging domain. First, as the neuroimaging databases, such as XNAT (Marcus et al., 2007
), HID (Keator et al., 2008
) and NDAR (Ndar 2009
), manage raw data provenance information, the neuroimaging workflow systems should be customized to work seamlessly with neuroimaging databases to minimize duplicated efforts and storage redundancy for provenance management. Second, as the neuroimaging domain involves domain specific user interaction and annotation, the provenance model should be extended to include this kind of information.
(Q2) The representation of the provenance is still not well addressed in the scientific workflow research community and needs to be adequately addressed in the neuroimaging domain. Improper representation of the provenance can result in huge redundancy. One way of minimizing the redundancy is to structure the provenance into layers of normalized components (Freire et al., 2008
). However, definition of the layers and components can still vary from domain to domain. In particular, this issue needs to be appropriately addressed in the neuroimaging domain.
(Q3) The provenance granularity can vary across domains, and has not been explicitly explored for the neuroimaging domain. The provenance can be recorded at different levels of granularity, i.e., varying levels of details. Improper selection of the granularity of provenance can produce inordinately large volume of provenance data bigger than the data it describes (Simmhan et al., 2005
), which may not be useful and may be hard to manage. In the neuroimaging workflow system, theoretically, the provenance granularity can be set at voxel-level, slice-level, volume-level, session/visit-level, subject-level, or group-level. Variability in granularity can result in a big difference in performance and storage overhead. The optimal provenance granularity for the neuroimaging workflow has hitherto not been explicitly explored in the existing literature.
(Q4) The provenance model can be implemented in many different ways which vary from application to application. The different approaches can vary in the way they capture, store and retrieve the provenance information. The capturing mechanism can be at various levels, i.e., at the OS-level, processing-level and workflow-level. The storage mechanism can be either file-system based or database based. The retrieval mechanism can be a special scripting language, like SQL or a visual user interface. When a new neuroimaging provenance model is created, the corresponding implementation issues have to be properly addressed as well.
In general, proper solutions for provenance modeling and management problems need to be explored for the neuroimaging domain. In this paper, we introduce the Bio-Swarm-Pipeline (BSP), a scientific workflow management system for bio-medical research developed at the Genes, Cognition and Psychosis Program (GCAP) of NIMH/NIH. It was designed to facilitate the fundamental requirements for everyday scientific research, such as scheduling batch processing on multiple computers, interpreting and comparing different results, sharing and reusing existing workflows, etc. This system is based on a new provenance model developed to meet the needs specific to a neuroimaging workflow management system. It systematically addresses the issues involved in the provenance modeling and management in the neuroimaging domain. First, by proper extension of the provenance model, the workflow management system can work seamlessly with existing neuroimaging databases and effectively reduce unnecessary storage and developing efforts. Second, by properly structuring provenance into two layers of six independent sub-provenance components, the BSP effectively minimizes the recording redundancy of provenance; Third, by proper determination of the provenance granularity, the BSP effectively eliminates unnecessary information, makes the system more light weighted and manageable; Fourth, by providing an optimal number of user interfaces, it makes provenance management and task scheduling an efficient and effective procedure. Finally, by taking swarm as analogy, an unsophisticated user with little or no knowledge in programming can easily capture the core concepts and understand how a task is processed by the system. Although this system stems from applications in the neuroimaging domain, the system can potentially be adapted to meet the requirements for a wide range of bio-medical application scenarios.
The remainder of this paper is organized as follows: in the methods section, we describe the BSP system architecture, highlight the structured provenance model, and demonstrate how it works with real examples; in the results section, we describe the current application status and impact of the system to the work at the GCAP of NIMH; and in the discussion section, we discuss how provenance modeling and management problems were addressed in the BSP. We also discuss some additional features and future extensions.
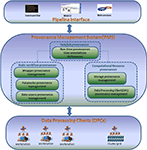
The BSP system architecture is made up of three layers – (I) pipeline interface layer, (II) PMS layer, and (III) data processing clients (DPCs) layer as shown in Figure 1
. The interface layer interacts with the PMS layer to submit data processing tasks and tracks the data processing provenance information. The DPC layer interacts with the PMS layer to perform data processing and updates provenance information. In this section, we will highlight layer II, i.e., PMS, and demonstrate how it works. We will also briefly introduce layer I and layer III.
Figure 1. Bio-Swarm-Pipeline system architecture.
Pipeline Interface
The interface enables the user to interact with the PMS to submit data processing tasks and tracks the data processing provenance information. Each interface is presented in the next section along with the provenance data that is managed.
Provenance Management System
The PMS manages a structured provenance model as shown in Figure 1
. The design and implementation is based on the MySQL relational database system. Conceptually, the model can be divided into two layers.
The first layer contains three sub-provenance components, i.e., task/job provenances, static workflow provenances and computational resource provenances. The task provenances record all the necessary information to reproduce a specific data processing result, including the run-time task provenances (such as result location, processing time, status) and user annotations as well as the references to static workflow provenances and the computational resource provenances.
In the second layer, the sub-provenance components are further decomposed. For example, the static workflow provenances are further divided into wrapper provenance, parameter provenance and data source provenance. The computational resource provenances are further divided into storage provenance and DPC provenance.
In this section, we will first introduce the static workflow and the computational resource provenances, and then introduce the task/job provenance. Later on in the discussing section, we will also discuss how this model systematically addresses the provenance modeling problems mentioned in Section “Introduction”.
Static workflow provenances
Static workflow provenances are specifications about workflows which can be shared across different tasks. This includes the specification of the wrappers, processing parameters and data sources.
Wrapper provenance management. The wrapper provenance management module manages the specification of the wrapper libraries for different data processing packages. Each data processing package (e.g. SPM (Friston et al., 1995
), AFNI (Cox, 1996
), VBM (Ashburner and Friston, 2000
), FreeSurfer (Dale et al., 1999
), etc.) is encapsulated by a wrapper so that they have a uniform calling interface like do + package_name + version + release. Each wrapper is uniquely identified by a wrapper ID so that the task/job provenances component can be simplified by referring to the wrapper ID.
Parameter provenance management. The parameter provenance management module manages the instantiated processing parameters for each given wrapper. It tracks everything related to the processing parameters that a user may be interested in. These include the parameter ID, the wrapper ID, the user login name who created the parameter set and the parameter body and some comments fields. Within the parameter ID, the user can easily interpret and compare the different results and check if they are generated from the same procedure.
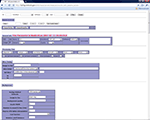
The parameter set is managed in the parameter management interface. As an illustration, in Figure 2
we use SPM-based first level data processing as an example to help the reader evaluate how the system works. In this illustration the processing parameters with parameter ID 11 are associated with wrapper SPM2. The parameter management interface allows the users to create new parameters or adapt from existing parameters. For the latter, users can first retrieve the parameters they want to duplicate from, and then click “borrow and create” button to make a new parameter. Then the user can modify the parameter as required. When a parameter set is first created, a unique parameter ID is automatically assigned to it. If the parameter set is derived from another parameter, users can add comments to indicate what the parent parameter ID is. This allows users to track the relationships among a family of related parameter sets.
Figure 2. Parameter management interface.
Data source provenance management. The data source provenance management module makes it easy for the workflow management system to inter-communicate with the neuroimaging database and other heterogeneous data sources and take input data from there. By default, BSP was designed to be work seamlessly with XNAT@GCAP neuroimaging database (Cheng et al., 2008
). It assumes that the raw data provenance information, such as the raw data locations, data acquisition parameters and subject demographics information are all managed by the neuroimaging database with XNAT like database schema (Marcus et al., 2007
). Therefore only references to the raw data provenance are kept in the system. The data source provenance component is able to retrieve raw data provenance information from the neuroimaging database when necessary.
Computational resource provenances
Computational resource provenances are specifications about computational resources that can be shared across different tasks. This includes specifications about the DPC and storage devices.
Data processing clients provenance management. The DPCs provenance management module manages the profiles of heterogeneous client workstations in the database. The profile describes the following information: hostname, system architecture, processor speed, memory capacity, operating system, network speed, available storage space, version of wrapper libraries and traffic lights, etc.
The storage provenance management. The storage provenance management module simplifies the dynamical management of the mappings between processing parameters and storage devices. The mappings are defined by the storage allocation rules in the format of (parameter_ID, output_dir, is_active), which specify a list of alternative output directories for each parameter IDs. When the available physical storage is below a certain threshold, the is_active flag will be automatically set to 0 by a daemon program so that the DPC will try to find the next available output directory with adequate space for ensuing tasks. If there is no output directory with adequate space, DPC will switch the task into “pending” status. When an output directory with adequate space is made available, DPC will automatically enable processing of the pending tasks.
Task/job provenances management
The task provenances record the most detailed information to reproduce an individual result. This includes the run-time task provenances (such as result location, processing time, status) and user annotations (such as data quality notes, etc.) as well as the references to static workflow provenances and the computational resource provenances defined above.
All the task provenance information is maintained in the task table of the database as shown in Figure 3
. Each task is identified by a unique ID in the task table. The task table specifies the following information for task processing: such as the priority of the task, the wrapper ID and the parameter ID required to process this task, and the name of the DPC (hostname) assigned to handle the task so that different tasks can be assigned to different hosts and processed in parallel, the directory where the results will be outputted, and the email addresses that should be notified upon completion of the tasks. Most of this information is collected before the task execution.
Figure 3. Structure of the task table in the database.
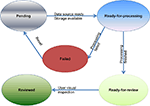
Moreover, the task status and quality annotations are customized to accommodate the requirements in neuroimaging domain. In our system, the possible status of a task can be “pending”, “ready-for-processing”, “failed”, “ready-for-review” or “reviewed” as shown in Figure 4
.
Figure 4. Flow chart of task status.
After a task is being created, if all the required data sources are ready and the storage space is available, the task will be set to the “ready-for-processing” status. Otherwise it will be set to a “pending” status. When processing has completed successfully, the pipeline is switched to “ready-for-review” status by the DPC. After the review is complete, it can be switched to the “reviewed” status by the user through the user interface, and the quality of the task will be marked as either + or − to indicate whether the results are usable. If the processing fails, the status will be set to “failed” by DPC, allowing the administrator to fix the error, and switch the status to “pending” or “ready-for-processing”.
To make the provenance model lightweight, the optimal granularity level of the provenance must be chosen. The available choices in the neuroimaging domain are voxel-level, slice-level, volume-level, session/visit-level, subject-level, and group-level. In most occasions, researchers may only be interested in the session/visit-level provenance information. The provenance information at this level is easily manageable, so the BSP provenance model is explicitly set at this level. However, the system can be easily extended to work on a different level of granularity when necessary.
A series of web user interfaces are provided to make it easier for the user to submit tasks, retrieve tasks and review the results on-line. In the following demonstration, we will again take SPM-based batch processing as an example to help the reader further evaluate the system.
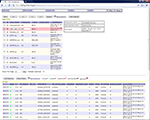
First, a user can submit tasks through the task management interface as shown in Figure 5
. Here, the user can create a new data processing project by click the “append” button. The user can then specify the project name, the parameter ID, the machine list to be used and a list of e-mail addresses where notifications can be sent upon the completion of the task. Afterwards, the user can click the “add session” button to add data and click the “place order” button to place data processing task (we also call them orders) for them. Multiple dataset (sessions) can be added to the project at the same time. Each dataset will be assigned to an individual task. If multiple machines are provided, the tasks will be split evenly among them. The processing status of each dataset is also available in the task management screen. As can be seen in the lower part of the Figure 5
, each processed session now has a green check mark on the left side of the row.
Figure 5. Task management interface.
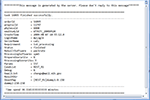
After the tasks have been submitted, the user can log off and wait for the process to finish. Upon the completion of each task, the user will get an email notification indicating the status of the task. An example of the email notification is shown in Figure 6
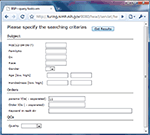
. If a task finishes without error, the user can log into the web interface, as shown in Figure 7
, to query the results by subject ID, task IDs or parameter ID.
Figure 6. Example of email notification.
Figure 7. Querying the processing results.
When the user provides the parameter ID and clicks the “get results” button (Figure 7
), the corresponding results of the tasks will be available for on-line review through the web user interface as shown in Figure 8
. In our illustration using SPM-based first level data processing the following results are made available for inspection: preprocessed results (lower left plot in Figure 8
), contrast map (middle plot in Figure 8
), quality control images and measures (right plot in Figure 8
). The user can add annotations concerning the quality of the results after visual inspection. The quality information can then be used in a query to filter the datasets.
Figure 8. Visual inspection interface.
There are also a number of command line tools used for simplifying routine administrative tasks such as task scheduling, traffic control and diagnosis.
Data Processing Clients
DPCs manage data processing in each client workstation by communicating with PMS. Each DPC is made up of two types of swarms, i.e., the manager-swarm (M-swarm) and the worker-swarm (W-swarm). The M-swarm, running as services on each local computer, manages (i.e., creates or kills) W-swarms in the local computer, retrieves tasks from the task table by local host name and task priority, and dispatches them to be processed by W-swarms. The W-swarm takes the task from M-swarm and processes them. The DPCs can be extended to incorporate computational resource(s) from a high-performance computing center like Beowulf cluster (Gropp et al., 2003
) by installing a customized DPC.
The BSP has been built, maintained and supported by GCAP since 2006. To date, around 130 workflows and above 32000 data processing tasks have been completed through this scientific workflow management system, with each task taking about 10–60 min. Currently the system supports SPM (for fMRI and VBM) and Freesurfer based data processing, but other popular packages or in-house packages can be easily integrated as well. The workflow system in its current status has been playing a critical role in the day-to-day neuroimaging research within GCAP, including but not limited to the following aspects:
• Improved productivity: due to its parallel data processing capabilities, the workflow system has refreshed data processing records in the past 2 years. The most recent accomplishment has been to successfully process almost 3000 high-resolution structural MRI preprocessing for VBM in <1 week, and nearly 5000 fMRI first level data processing for SPM in 2 weeks. This has been achieved using just one W-swarm per workstation because some of the packages were not multi-thread-able. The modern quad-core processor will be able to easily scale up to four W-swarms without compromising performance. Making the data processing packages multi-thread-able will make this a more efficient process. This kind of capability makes the BSP very efficient even when compared to the crowded Beowulf cluster.
• Improved efficiency of workflow within GCAP neuroimaging research groups. By enabling automated data processing of large datasets, it has made more time available for researchers to pursue more intellectually challenging tasks.
• Facilitates easy and efficient replication of results using identical parameters. This decreases the necessity to backup processed data and thereby decreases storage space requirements.
In this section first we discuss how the BSP provenance model addressed the provenance modeling and management problems mentioned in Section “Introduction” for the neuroimaging domain. Second we summarize the additional features of BSP along with the provenance model. Finally we list some possible future extensions.
The BSP Provenance Model Addresses the Provenance Modeling and Management Problems in the Neuroimaging Domain
The BSP provenance model has systematically addressed the provenance modeling and management problems for the neuroimaging domain (Q1–Q4) as outlined in Section “Introduction:”
(P1) the BSP provenance model is extended to cover the neuroimaging domain. First, the BSP is extended to work seamlessly with the XNAT@GCAP (Cheng et al., 2008
) neuroimaging data archiving system, i.e., the BSP data source component just keeps the references to the raw data provenance, such as the data acquisition parameters as well as the subject’s demographic information, and is able to retrieve the raw data provenance information from the XNAT@GCAP neuroimaging database as necessary. Therefore, the duplicated efforts and storage redundancy for the maintenance of the provenance information are minimized. Second, the BSP model is extended to include information specific to neuroimaging. Particularly, the system is customized to accommodate the domain specific user interactions for reviewing the quality of the images, for example the task status field is extended to include options like “ready-for-review”, “reviewed”, etc. After a task is reviewed, the annotation and comments related to the data quality can be stored. Special user interfaces (see Figures 2
, 5
, 7 and 8
) are also provided for the user to manage parameter sets, make queries, visually inspect the results, and manage the annotations. These extensions are different from existing workflow systems. For example, most workflows except LONI do not work with neuroimaging databases.
(P2) the BSP provenance model was structured into two layers of six independent sub-provenance components (i.e., wrapper provenance, parameter provenance, data source provenance, storage provenance, DPC provenance and task provenance) to minimize the recording of redundant information. Referring to Figure 3
, although the task provenance component tracks all the details necessary to reproduce the results, the storage overhead are very small, as most of the common information (such as the static workflow provenance and the computational resources provenance) is stored as references. In general, the BSP provenance model structure is quite different from that of other existing provenance models. For example, in VisTrails, the provenance model is structured into three layers: the workflow evolution, the workflow instance and the execution log (Freire et al., 2008
). In the LONI workflow system, the provenance model is divided into four components: the data provenance, the binary provenance, the executable provenance, the workflow provenance, the processing provenance (MacKenzie-Graham et al., 2008
). Although these systems have some features that are similar to the BSP provenance model, the overall structure is different.
(P3) in the BSP provenance model, the provenance granularity is explicitly selected to be at the session-level so that only information of interest to the user is tracked. However, there is no limitation if a user wants to extend the current model to include other levels of the provenance. Usually the provenance granularity for the neuroimaging domain is not explicitly specified in other workflows. As mentioned before, without explicitly specifying the level of granularity, the neuroimaging workflow system can potentially store too much detailed information – such as the provenance at the slice or voxel-level, which can result in a huge and unnecessary storage overhead. However, most users may not be interested in such fine-grained provenance.
(P4) the provenance model, implementation has been carefully chosen in the BSP to optimize the performance. First, most of the provenance information is collected prospectively (e.g., the wrapper provenance, parameter provenance, data source provenance, storage provenance, DPC provenance are all specified before task execution). Only little information in task provenance is collected retrospectively. Compared to the OS-level capturing mechanism, which needs to filter through all the system calls and files touched during the execution of a task’s, this approach is more efficient. Second, the BSP provenance model is based on a relational database system. In comparison to file-based provenance storage system, the data storage is optimized by the database system, and the query/retrieval stage is more flexible and efficient.
Although the BSP provenance model originates in the neuroimaging domain, it can be potentially adapted to cover many other bio-medical domains as well.
Additional Features of the BSP
Along with the BSP provenance model, here we would like to summarize some additional features of the BSP in general.
• BSP is parallel in nature
In contrast to workflow systems that are primarily designed to handle inter-package heterogeneities but do not facilitate parallel processing, the BSP allows optimal distribution of multiple data processing tasks across a number of computers to maximize the throughputs.
• BSP is light weighted
This is because: (A) The swarm is conceptually simple, an unsophisticated user can capture the core concepts and understand how a task is processed by the system fairly quickly without having to read the whole manual; (B) The system boundary is properly tailored, so that duplicated work is avoided; (C) The redundancy of provenance data is minimized as the provenance model is highly normalized; (D) The granularity of the provenance is set at session level, the unnecessary provenance information is effectively ignored.
• BSP is built on top of the relational database
This is a big advantage of the BSP over workflow systems that are not bundled with a database. With the powerful MySQL database and SQL language, routine management tasks such as wrapper management, DPCs management, task/job management, data source management and storage management can be very easy and flexible.
• BSP is reliable
The failure of one machine will not affect the data processing on another in the network, and is therefore easy to identify and recover from failure.
• BSP is scalable
– As there is no communication between the different processing tasks, the throughputs of the workflow system increases almost linearly with the number of workstations.
– A work station can join or leave the workflow system at any time without affecting the overall batch processing.
• BSP is extensible
– The workflow system can be extended to cover different data processing packages as long as the appropriate wrappers are provided.
– The DPCs are extensible. For example, a high performance computing center like Beowulf cluster can be treated as a DPC and managed by the workflow system
– Data sources can be extended to accommodate a wide range of different data sources as long as the appropriate data source adaptors are provided.
The BSP is flexible and has a number of other advantages. For example, when compared to the Beowulf cluster, it is: (1) capable of applying complicated and flexible data processing management; (2) free from limited license issue (e.g., the Beowulf cluster usually limits the number of Matlab licenses to 16 for each user); (3) no need to transfer data and results back and forth as is required between Beowulf and the local file systems; and (4) no waiting time (in comparison to the high performance computing center).
Possible Extensions
As some of the provenance management is currently conducted through command line, more user friendly interfaces will be provided in the new release. These include interfaces for: (1) wrapper management; (2) storage management; (3) task re-scheduling and traffic control; (4) data source management.
The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
This work was supported by GCAP, DIRP, NIMH, NIH. We would like to thank everyone in the GCAP neuroimaging core for their suggestions and feedbacks. We would also like to thank the reviewers for the informative comments and suggestions.
Currently we are creating extensions and developing a deployable release. A link to the downloadable version as well as the educational material will be made available at The Neuroimaging Informatics Tools and Resources Clearinghouse (http://www.nitrc.org/
).
Keator, D. B., Grethe, J. S., Marcus, D., Ozyurt, B., Gadde, S., Murphy, S., Pieper, S., Greve, D., Notestine, R., Bockholt, H. J., and Papadopoulos, P. (2008). A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN). IEEE Trans. Inf. Technol. Biomed. 12, 162–172.
NDAR (2009), National Database for Autism Research (NDAR). http://ndar.nih.gov/ndarpublicweb/
.