Cellular reprogramming for understanding and treating human disease
- 1Epigenetics Laboratory, Department of Anatomy, Howard University, Washington, DC, USA
- 2InSilico Medicine, Emerging Technology Center, Johns Hopkins University Eastern, Baltimore, MD, USA
In the last two decades we have witnessed a paradigm shift in our understanding of cells so radical that it has rewritten the rules of biology. The study of cellular reprogramming has gone from little more than a hypothesis, to applied bioengineering, with the creation of a variety of important cell types. By way of metaphor, we can compare the discovery of reprogramming with the archeological discovery of the Rosetta stone. This stone slab made possible the initial decipherment of Egyptian hieroglyphics because it allowed us to see this language in a way that was previously impossible. We propose that cellular reprogramming will have an equally profound impact on understanding and curing human disease, because it allows us to perceive and study molecular biological processes such as differentiation, epigenetics, and chromatin in ways that were likewise previously impossible. Stem cells could be called “cellular Rosetta stones” because they allow also us to perceive the connections between development, disease, cancer, aging, and regeneration in novel ways. Here we present a comprehensive historical review of stem cells and cellular reprogramming, and illustrate the developing synergy between many previously unconnected fields. We show how stem cells can be used to create in vitro models of human disease and provide examples of how reprogramming is being used to study and treat such diverse diseases as cancer, aging, and accelerated aging syndromes, infectious diseases such as AIDS, and epigenetic diseases such as polycystic ovary syndrome. While the technology of reprogramming is being developed and refined there have also been significant ongoing developments in other complementary technologies such as gene editing, progenitor cell production, and tissue engineering. These technologies are the foundations of what is becoming a fully-functional field of regenerative medicine and are converging to a point that will allow us to treat almost any disease.
Introduction
Paradigm Shifts and Seeing Things in New Ways
Stem cell biology is sometimes thought of as a field that offers promises that can't be kept; for example the potential to one day treat heart attacks or repair spinal cord injuries. But focusing on regenerative medicine's so-far small impact on patient care, misses a shift in our understanding of cells so radical that it has rewritten the rules of biology in less than two decades.
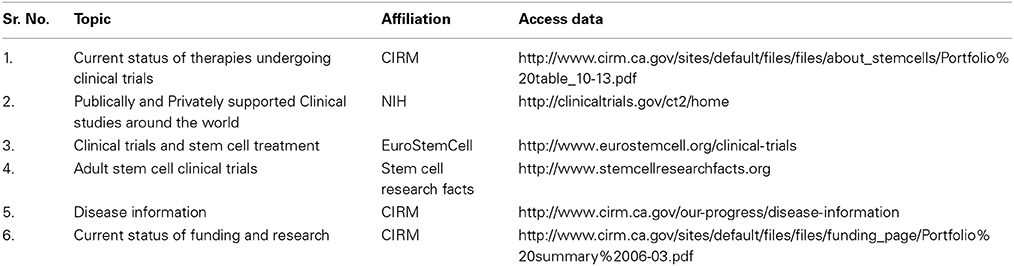
How much have things changed? By way of analogy, the Rosetta stone was a stone slab found in 1799 that bore parallel inscriptions in Greek, Demotic characters, and Egyptian hieroglyphics that made possible an unprecedented decipherment of the latter. We propose that the discovery of the mechanisms for producing and modulating stem cells, in particular induced pluripotent stem cells, will have an equally profound impact in understanding human health and disease (Figure 1). Stem cells can be conceptualized as “cellular Rosetta stones” because they are enabling us to create and compare cells with diseases or cellular phenomena that are poorly understood (non-deciphered Hieroglyphics), with isogenic cells that are completely normal (well-understood, Ancient Greek or Demotic). Because of this, reprogrammed stem cells will be the quintessential tool for studying epigenetics, aging, cancer, and regeneration. This new possibility will open up opportunities to study cells that go awry in disease and to one day use patients' own cells to heal them.
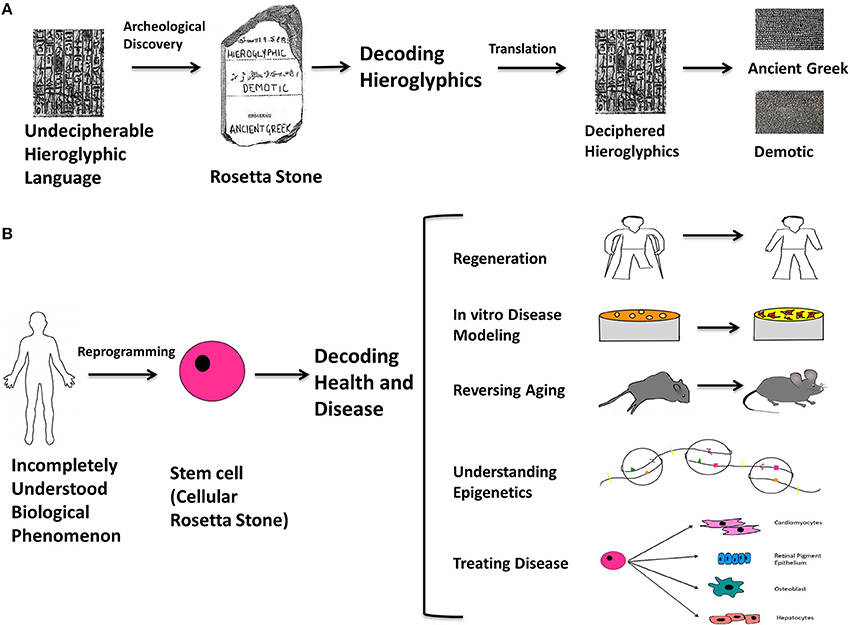
Figure 1. The Rosetta stone analogy. The Rosetta stone is an archeological slab with the same text in three different languages; Hieroglyphics, Demotic, and Ancient Greek, and its discovery proved to be a turning point in understanding the Hieroglyphic language. By direct comparison of the three languages, it was possible to decipher previously unintelligible Hieroglyphics from the other two known languages. An analogy can be made with respect to stem cells: they can be thought of as “cellular Rosetta stones” because they are key to understanding the multi-faceted mysteries underlying human health and disease. Stem cells can be conceptualized as “cellular Rosetta stones” because they are enabling us to create and compare cells with diseases or cellular phenomena that are poorly understood (non-deciphered Hieroglyphics), with isogenic cells that are completely normal (well-understood, Ancient Greek or Demotic). This is the contribution of stem cells toward modeling and treating diseases and generating functional cell types. In addition, they are becoming quintessential for studying epigenetics, aging, cancer and regeneration. It won't be long before they emerge as the ultimate practical tool and make their mark on the conceptualization of biological science in terms of understanding molecular and cellular events and treating debilitating diseases. By using reprogramming as the means to make cellular Rosetta stones, it will be possible to form a universal understanding of currently unknown biological phenomena and develop an accurate philosophy for cellular processes, disease and therapy. (A) Illustrates the rationale behind the way the Rosetta stone was used to decode hieroglyphics. (B) Illustrates the analogously unexplained biological phenomena to which stem cells can be made with various characteristics on an isogenic background and used to understand them; for example (1) limb-regeneration (2) disease modeling at the cellular level (3) treating organismal aging (4) understanding epigenetic mechanisms underlying diseases (5) generating lineage specific cell types to treat degenerative and chronic diseases, or acute injuries.
Stem cells are emerging as the ultimate tool for understanding biological processes at the molecular and cellular level, as well as treating debilitating diseases. By using reprogramming as the means to make “cellular Rosetta stones,” it will be possible to form an accurate understanding of currently unknown biological phenomena, and perhaps eventually develop a universal synthesis of cell biology (Figure 1).
In this review we present a comprehensive summary of the history of cellular reprogramming from its initial conception to the present day. We also describe how these techniques are being used to better understand basic biology and human disease, and lay the foundations for a fully functional discipline of regenerative medicine.
Therefore, to begin, what exactly are stem cells?
Stem Cells
Stem cells are undifferentiated cells that possess two unique characteristics:
(1) The ability to self-renew through numerous cycles of cell division while maintaining an undifferentiated state, and
(2) Pluripotent potential with the ability to generate progenitors of multiple lineages (International Stem Cell et al., 2007; Mitalipov and Wolf, 2009). These qualities confer unique regenerative abilities upon stem cells, and make them a desirable commodity in the endeavor to replenish, regenerate and repair human tissues (Gurtner et al., 2007; Wu et al., 2007). There are essentially two types of stem cells in mammals: somatic stem cells (SSCs, also sometimes called progenitor cells), and embryonic stem cells (ESCs) (Niwa et al., 2000).
SSCs are found in various adult tissues, such as bone marrow, adipose tissue and blood (including umbilical cord blood) (Jiang et al., 2002; Terai et al., 2006; Gimble et al., 2007; Luna-Zurita and Bruneau, 2013). They are partially differentiated cells found throughout the body that possess the capacity to divide in order to replenish damaged tissue. They are able to differentiate into more than one cell type, but unlike ESCs, they are restricted to a specific cellular lineage. The advantage of SSCs is that their production doesn't require the destruction of embryos, or reprogramming (see below) (Gardner, 2002).
ESCs are produced from eggs derived from the inner cell mass of fertilized embryos. Contrasted with SSCs, ESCs are pluripotent, and can differentiate into all of the three primary germ layers (ectoderm, endoderm, and mesoderm) and their derivatives. ESCs are characterized by long-term self-renewal, and can be grown in cell culture as an undifferentiated, pluripotent population. Regulation of pluripotency networks is important for maintaining the undifferentiated state of such cells in culture, or during differentiation to obtain desired cell types. The transcription factor (TF), Oct 3/4 is the master regulator of pluripotency, and its precise levels during development are responsible for the differentiation of ESCs into specific lineages, whereas repression of Oct 3/4 results in loss of pluripotency and formation of trophoectoderm (Niwa et al., 2000).
ESCs can be directed to differentiate into a particular cell type through alteration of culture conditions and/or the supplementation of differentiation signals. Understanding the differentiation process has provided insights into de-differentiation and trans-differentiation strategies as well. Dedifferentiation is the formation of pluripotent or multipotent stem cells from terminally differentiated somatic cells, i.e., reverting to a state of increased developmental plasticity, and becoming ready to accept a new identity (Halley-Stott et al., 2013). Transdifferentiation is the process in which a particular somatic cell is switched from one lineage-specific identity to a completely different identity (Graf, 2011; Vierbuchen and Wernig, 2012); in other words, the direct conversion of one type of somatic cell into another type, bypassing the intermediate step of dedifferentiation.
The discovery of ESCs (Evans and Kaufman, 1981; Martin, 1981) eventually prompted the search for discovering artificial dedifferentiation techniques to confer the properties of ESCs onto somatic cells by altering epigenomic activity, such that the derived cells are pluripotent and capable of giving rise to embryonic-like stem cells. These techniques are collectively referred to as cellular reprogramming. But before we describe these various techniques, we will provide some background on the history of how we arrived at today's reprogramming technology.
History and Development of Cellular Reprogramming
In 1909, Ethel Browne Harvey, who was known for her work on sea urchins, was the first to show that cell transplants could induce a secondary axis of polarity in the host. Harvey's experiments were the basis for the discovery of Spemann's organizer (Lenhoff, 1991). In 1928, Hans Spemann and Hilde Mangold, in a quest to discover the factors responsible for embryonic determination and cell differentiation, performed classical embryology experiments with salamanders and demonstrated cell-to-cell induction, in which a group of cells or organizing centers signal differentiation in neighboring cells and hence regulate their fate in the embryo (De Robertis, 2006). The cells responsible for this kind of phenomenon came to be known as the Spemann organizer, which over subsequent decades led to many experiments in molecular embryology aimed at finding inducing factors responsible for early embryonic determination and cell fate (Grunz, 2001). Further, Spemann had proposed an experiment to determine whether differentiated cells could be restored to an embryonic state, or if the cells continued to remain specialized (Subramanyam, 2013). Spemann reasoned that if a nucleus from a differentiated cell implanted in a previously enucleated egg developed into a normal embryo, this would prove that the transplanted nucleus retained a genome fully capable of directing all types of differentiation. In other words, a differentiated nucleus could still be totipotent.
Somatic Cell Nuclear Transfer
In 1938, Spemann published an account of his experiments with a prototypical nuclear transfer technique (Spemann, 1938). Using a piece of hair wrapped around a newly-fertilized salamander egg, he separated the egg's nucleus on one side, with the cytoplasm on the other. After the nucleated side divided four times, creating a 16-cell embryo, he removed the hair and allowed a nucleus to slide back into the separated cytoplasm. Cell division now began on this side as well, and by putting the hair loop back again tightly, he broke the two embryos apart. The result was a twin set of salamanders.
This important work showed that the nucleus remained totipotent after undergoing four divisions, but Spemann wondered whether nuclei from much older embryos, or even adult animals, had similar potential. He wrote that transplanting an older nucleus into an egg would be a “fantastical experiment.” What he was proposing is what eventually became known as somatic cell nuclear transfer (SCNT). However, for the next 14 years, scientists struggled with making it work.
But in 1952 Briggs and King performed the first successful SCNT experiment showing the reversal of cellular identity. They showed that the transfer of a frog nucleus from a blastula cell to an enucleated egg gave rise to a cleaved blastula (Briggs and King, 1952). This was followed by the pioneering experiments of John Gurdon in 1962, involving transfer of nuclei from terminally differentiated frog intestinal epithelium cells into enucleated eggs to produce normal tadpoles, which proved that the nucleus contains all the genetic information required to give rise to all the differentiated cells in the organism (Gurdon, 1962). After incremental improvements in SCNT techniques by various laboratories over the intervening three decades, the cloning of “Dolly” in 1996 was a major breakthrough, and the first ever successful mammalian cloning experiment, which involved transplanting quiescent nuclei from cultured adult sheep mammary gland cells into enucleated sheep eggs (Campbell et al., 1996). The creation of SCNT-derived human ESCs required further improvements in technology and almost another two decades, but in 2013, after a long and challenging pursuit, Mitalipov and colleagues finally succeeded in creating SCNT-derived human embryonic stem cell (hESC) lines (Tachibana et al., 2013).
SCNT has, over the decades, evolved as a technique, which can be used for reproductive cloning as well as to produce lines of hESCs. The attraction of using an egg for nuclear reprogramming is that, in nature, it has near 100% efficiency in reprogramming sperm nuclei (Gurdon and Melton, 2008; Teperek and Miyamoto, 2013). The egg uses molecular chaperones and enzymes to erase the epigenetic signatures during synkaryon formation and cell division, resulting in extremely efficient reprogramming (Kikyo and Wolffe, 2000). Variations in the technique used to produce offspring through SCNT such as improvement of processes like oocyte maturation, enucleation, donor nucleus transfer, activation and culture, along with prior epigenetic modification of the donor nucleus, can improve the efficiency of the process (Campbell et al., 2007).
Induced Pluripotency
Once SCNT in mammals was achieved, researchers wondered if the same result could be achieved without using eggs. Thanks to the pioneering experiments of Yamanaka, it is now possible to create close equivalents of ESCs without using embryos by engineering their creation in vitro. In 2006 and 2007, Yamanaka and colleagues identified conditions that allowed somatic cells to be genetically reprogrammed into induced pluripotent stem cells (iPSCs) that are almost as potent as ESCs. This was a significant breakthrough and was achieved by transfection of mouse and human fibroblasts with the transcription factors (TFs) Oct4, Sox2, Klf4, and c-Myc, (OSKM) using retroviral vectors to induce a “forced” expression of specific genes that results in iPSC formation (Takahashi and Yamanaka, 2006; Okita et al., 2007). Subsequent manipulation using different reprogramming factors such as Nanog rendered the reprogramming process more efficient (Wernig et al., 2007). There are now many different such pathways of inducing pluripotency, because different combinations of reprogramming factors can all achieve complete reprogramming (Takahashi, 2012). In addition to such reprogramming factors, introduction of improvements such as p53 knockdown and telomerase overexpression, along with small molecules that exert epigenetic effects (see Section Small Molecules below) are now being used routinely (Batista, 2014; Campos-Sanchez and Cobaleda, 2014). Besides using different combinations of TFs, iPSCs have also been induced using methods other than retroviral vectors, such as DNA and RNA (integrating plasmids, episomal plasmids and transposons), cell penetrating peptides, and small molecules (see Small Molecules below) (Li et al., 2014).
At present, a primary concern is the ability to achieve pluripotency in iPSCs while maintaining the functionality of ESC's—irrespective of the type of somatic cells used at the outset. For example, it is possible that less differentiated cells of the hematopoietic lineage reprogram more efficiently than differentiated cells, suggesting that the use of SSCs/progenitor cells as starting material could maximize the efficiency of reprogramming (Eminli et al., 2009). This may be because less differentiated progenitors possess less condensed chromatin in specific regions, which is more accessible to reprogramming factors. Also progenitors in general may be more susceptible to the disruption of their transcriptional networks, or possess a transcriptome that has higher resemblance to the transcriptome of ESCs (Papp and Plath, 2011).
A very recent refinement to induced pluripotency has been achieved which allows the in vitro reprogramming of cells back to a ground state of pluripotency that is even closer to ESCs. Novel human naïve stem cells have been derived by adding a unique combination of cytokines and small molecule inhibitors to the standard reprogramming cocktail (Gafni et al., 2013). These ground state stem cells closely resemble mouse iPSCs and ESCs (relatively more pluripotent than human ESCs or iPSCs), exhibiting hallmarks of naïve pluripotency that include driving Oct 4 transcription by its distal enhancer, retaining a pre-inactivation X chromosome state and a global reduction in DNA methylation and an H3K27me3 repressive chromatin mark deposition on developmental regulatory gene promoters (Gafni et al., 2013). The epigenetic changes induced by naïve human stem cell medium (NSHM) conditions indicated that naïve conditions are likely to resolve previously described technical phenotypes of epigenetic memory, lineage differentiation biases and aberrant reprogramming in human iPSCs and ESCs, thus providing a stable source for treating and modeling diseases (Gafni et al., 2013).
Also, recent studies in mice have shown that the in vivo transitory induction of OSKM generated teratomas, indicating that in situ reprogramming can give rise to iPSCs from a variety of cell types, and these in vivo iPSCs more closely resembled ESCs than in vitro generated iPSCs, with a more primitive and plastic state (Abad et al., 2013) (Figure 3). These in vivo iPSCs can further generate desired cell types in the presence of appropriate signals for re-differentiation (shown in Figure 3).
The most attractive application of induced pluripotency using the newest techniques is of course the production of patient-specific iPSCs for replacement of damaged, aged, or non-functional tissue, which will be discussed in later sections. While both ESCs and iPSCs are desirable for reasons of potency, the availability of ESCs is hindered by practical (egg availability) and ethical concerns, while iPSC production still remains relatively inefficient.
Cell Fusion
Another possible alternative to SCNT is cell fusion, in which two cells fuse together in the presence of agents like Sendai virus, polyethylene glycol (PEG), chimeric hemagglutinins, or an electric pulse to give rise to homokaryons from the same type of cells or heterokaryons from different types of cell (Soza-Ried and Fisher, 2012). In the case of heterokaryon formation, the dominant cell is the larger and actively dividing partner that imposes its own pattern of gene expression on the other. But cells fused in this way do not usually proliferate well, limiting their therapeutic value (Gurdon and Melton, 2008).
Cell-extract Treatment
In recent years, cell extracts (usually extracted from embryonic-like stem cells) have been used to derive pluripotent cells that can give rise to diverse cell lineages following differentiation (Patel and Yang, 2010). The use of extracts derived from pluripotent cells such as ESCs can also trigger the formation of ESC-like colonies with the upregulation of pluripotency genes and the downregulation of somatic genes such as Lamin A (LMNA) (Alberio et al., 2006). Notably, remodeling of the nuclear lamina has been used as a marker for reprogramming events in experiments involving the incubation of Xenopus laevis somatic cells in egg-extract in which reprogramming events could be tracked by the resulting nuclear configuration which acted as an indicator of increased plasticity (Alberio et al., 2006). LMNA is also a marker of differentiation of human ESC's into somatic cells (Constantinescu et al., 2006).
Small Molecules
Another recent addition to the different types of reprogramming techniques is the use of small molecules, with or without a combination of TFs. It is thought that small molecules might lead to a better clinical approach because they involve fewer genetic manipulations than TF-based reprogramming (Pandian et al., 2014). A recent breakthrough study showed that a combination of just seven small-molecules was enough to chemically reprogram mouse embryonic fibroblasts (MEFs) such that Oct4 was dispensable for reprogramming (Hou et al., 2013). Such chemically induced pluripotent stem cells (CiPSCs) were similar to ESCs in terms of gene expression profile and epigenetic state, and did not require any exogenous expression of master pluripotency genes at all (Hou et al., 2013).
Transdifferentiation/direct Reprogramming
Transdifferentiation techniques might make it possible to produce patient-specific cells in a faster and more efficient manner by avoiding the intermediate stage of iPSC formation, and thus preventing the risk of tumorigenesis and immunogenicity associated with iPSCs (Ma et al., 2013) (Figure 3). For example, transdifferentiation studies in vivo have already demonstrated the conversion of adult murine pancreatic exocrine cells to β-cell-like populations using the TFs Ngn3, Pdx1, and Mafa (Zhou et al., 2008).
iPSC TF-based transdifferentiation is a new method of transdifferentiation that uses the transient overexpression of iPSC related TFs and cell-type-specific signals (growth factors, cytokines, and small molecules) to reprogram somatic cells to diverse lineage-specific cells or multipotent progenitors without transitioning through the iPSC stage (Ma et al., 2013). An example of such inter-lineage transdifferentiation is the generation of expandable neural progenitors from fibroblasts by the transient induction of OSKM along with appropriate signaling inputs (Kim et al., 2011). Another example is the conversion of human dermal fibroblasts to multipotent hematopoetic progenitors of myeloid, erythroid and megakaryocytic lineages via induction of OCT4 and its specific binding to regulatory regions of hematopoetic-specific genes along with cytokine supplementation (Szabo et al., 2010).
Regeneration
Finally, the process of regeneration involves local dedifferentiation of somatic cells into a blastema composed of SSCs, which are multipotent and capable of proliferating, and re-differentiating into different lineages, and which can repopulate the damaged or degenerated tissue with functional cells. In human adults somatic cells cannot normally dedifferentiate and the process of wound healing involves scar tissue formation caused by the deposition of collagen, which is the most abundant part of the extracellular matrix secreted during wound healing. Thus, as compared to lower vertebrates like salamanders that are capable of complete regeneration with very little or no scar tissue, human adults have limited regenerative capacity. Regenerative capacity in humans seems to have diminished during the course of evolution compared to our lower vertebrate cousins as a mechanism to fight cancer.
However, regeneration in humans is not completely non-existent. The developing fetus is capable of scarless wound healing and this capacity is lost in gestation. For example, in the case of cardiac regeneration in neonates, the same extracellular matrix that contributes to scar formation might be involved in signaling an increase in proliferative capacity (Porrello and Olson, 2014). Thus, the much-coveted mammalian fetal regeneration capacity including wound healing without scar-tissue formation is a result of the extracellular matrix modulators, growth factors as well as cytokines (Colwell et al., 2005).
In spite of having wide-ranging information about the regenerative capacity of invertebrates, absolute knowledge of the limits of mammalian regenerative capacity is obscure because our understanding of mammalian stem cell biology usually comes from isolated stem cells, rather than from actually studying them in vivo. Given the fact that cells are dynamic entities and their properties change according to the environment they are in, the actual information on the number and functionality of stem cells in vivo is not accurately known (Sanchez Alvarado and Yamanaka, 2014).
Apart from harvesting the intrinsic regenerative potential of stem cells, mammalian regeneration can be induced through the administration of TFs known to locally dedifferentiate cells (transient) followed by re-differentiation. In a landmark study, it was shown that the upregulation of a single transcription factor, FOXN1, resulted in regeneration of the thymus in aged mice to an extent that it restored thymopoesis, a ground breaking example of in vivo mammalian regeneration (Bredenkamp et al., 2014). Such recent efforts in regeneration research have helped to advance our knowledge to the point where restoration of regenerative potential in mammals is perhaps within reach.
All of the above cellular reprogramming methods remodel chromatin and cause epigenetic changes in the target cells. But before we describe the application of the above methods to understanding and treating disease, we will first provide an explanation of what exactly is meant by “epigenetics,” with a brief summary of this rapidly developing field.
Epigenetics
Overview of Epigenetics
Epigenetics is the study of heritable changes in gene activity exclusive of direct modification of the DNA sequence, and includes DNA methylation and post-translational histone modification (Berger et al., 2009). The epigenome is a collection of the DNA methylation states and covalent modifications of histone proteins along the genome, and it differs in each cell type. Epigenetic mechanisms play an important role in the control of gene expression by organizing the nuclear architecture of chromosomes, restricting or facilitating TF access to DNA, and preserving a memory of past transcriptional activities (Rivera and Ren, 2013).
Epigenetic theory explains how the genome and environment work in tandem, involving mechanisms that affect DNA by regulating gene expression (Weinhold, 2006), and this interaction can operate across the entire human lifespan (Kanherkar et al., 2014).
Epigenetic modifications are of course crucially important for driving a cell toward an appropriate function during differentiation. The complexity of signaling during differentiation causes a cell's DNA to acquire specific epigenetic marks that restrict the expression of specific genes, and this inactivation survives cell division. DNA methylation is a signaling tool that occurs naturally on cytosine bases at CpG island promoter sequences and inactivates genes (Phillips, 2008). This type of epigenetic modification is associated with regulation of gene transcription, X-chromosome inactivation, and regulation of cellular development and differentiation (Bird, 2007). Another method of gene regulation is through the remodeling of chromatin. Remodeling occurs by post-translational modification of the amino acids that make up histone proteins via acetylation, methylation, phosphorylation and ubiquitination (Lunyak and Rosenfeld, 2008). Such epigenetic signatures are maintained by histone modifying enzymes such as histone acetyltransferases and histone methyltransferases (known as the “writers”) and histone demethylases and histone deacetylases (known as the “erasers”) which act as co-activators or co-repressors of OSKM respectively at different stages of reprogramming, and thereby influence iPSC formation (Apostolou and Hochedlinger, 2013) (Figure 2).
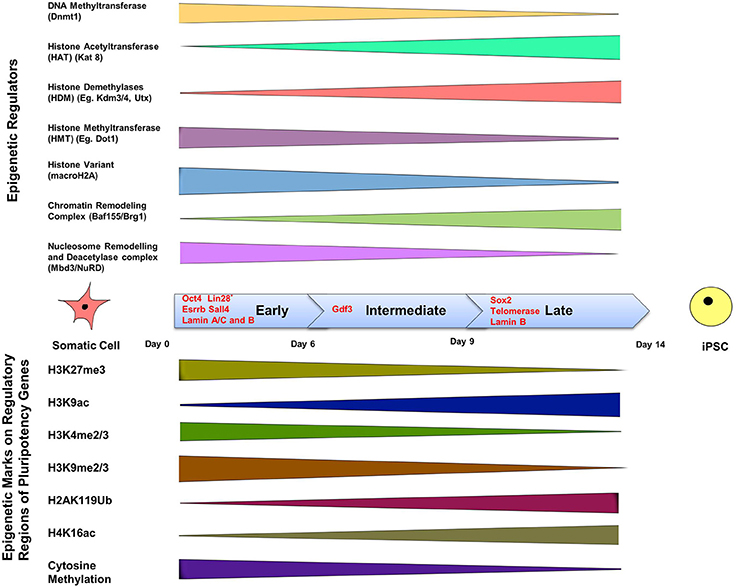
Figure 2. The epigenetics of induced pluripotency. The process of induced pluripotency using OSKM involves turning on pluripotency genes and turning off genes responsible for the maintenance of a differentiated somatic cell state. The timeline of formation of an iPSC can be divided into three stages; early, intermediate, and late, and involves the presence of different transcription factors during each of these phases. Transcription factors Oct4, Esrrb, and Sall4, are expressed during the early stage. Lin 28, suggested to be expressed in the early phase, is a controversial marker of early stages of reprogramming. Gdf3 is expressed during the intermediate stage, and Sox2 during the late stage. Lamins A/C and B are expressed in somatic cells but not in iPSCs, whereas telomerase expression is upregulated in iPSCs, but not in somatic cells. The color bars tapering toward either side of the timeline indicate a decline in expression or activity of the epigenetic regulator or epigenetic mark. The top panel shows the range of activity of some well-defined epigenetic regulators of chromatin in a somatic cell during the three phases. Methyl group-adding enzymes are shown on the very top, next is histone acetyltranferase that adds acetyl groups, followed by demethylases that remove methylation marks; the presence of histone variant: macroH2A, and finally chromatin remodeling complexes are depicted in the last two rows. The bottom panel includes the list of known epigenetic tags present on regulatory regions of pluripotency genes and their presence during the early, late and intermediate phases of induction in a somatic cell to pluripotency using reprogramming factors. DNA methylation on pluripotency genes decreases in the course of reprogramming because of a decline in the activity of Dnmt (DNA Methyltransferase), which is responsible for methylation. Histone acetylation increases during reprogramming due to the increased activity of histone acetyltransferase. Insufficient histone acetylation and hypermethylated DNA are the “epigenetic barriers,” which need to be overcome during reprogramming. Reprogramming leads to acquisition of active histone marks (e.g., H3K9ac) and loss of repressive histone marks (H3K4me2) on pluripotency genes, which facilitates the opening up of a compact chromatin structure and thereby allowing exposure of pluripotency gene promoters and binding of pluripotency factors like Oct4. (Buganim et al., 2012; Hansson et al., 2012; Loh and Lim, 2012; Apostolou and Hochedlinger, 2013; Liang and Zhang, 2013; Luna-Zurita and Bruneau, 2013; Papp and Plath, 2013).
Epigenetics during Cellular Reprogramming
Cellular reprogramming affects epigenetics at two different levels: firstly, through histone modification at the level of chromatin, and secondly through regulation of epigenetic marks on pluripotency genes and lineage-specifying genes that promote differentiation of various progenitors cells (Festuccia et al., 2013; Van Oevelen et al., 2013; Watanabe et al., 2013) (Figure 2). A major mechanism affecting epigenetic plasticity is genome-wide methylation and its regulation (Watanabe et al., 2013); nevertheless, it has to be accompanied by other epigenetic modulators that affect the state of pluripotency, totipotency, and their erasure (Loh and Lim, 2012). These modulators include histone acetylases and deacetylases such as Brg1 and Baf155 (Figure 2). While initial efforts to improve the reprogramming efficiency targeted the roles of transcriptional regulators, current studies suggest the involvement of signaling pathways in this process, and that the upregulation and downregulation of major signaling pathways can help in improving pluripotency reprogramming, lineage reprogramming and/or cell differentiation (Fritz et al., 2014).
The role of epigenetics during cellular reprogramming can be shown figuratively as an “epigenetic landscape.” An epigenetic landscape graphically represents the process of cell fate decision during development. Such a landscape is in reality a product of complex gene networks that are epigenetically regulated but can be represented figuratively in the form of a “mountain” with numerous “valleys.” Cells (represented by balls) reside in the valleys (epigenetically stable networks) and a cell can go down any valley depending upon which stimuli it receives to attain a differentiated state (Figure 3). Such epigenetic landscapes and the related complex gene regulatory networks that regulate cell fate can be used to predict the TFs associated with particular cell fates (Lang et al., 2014).
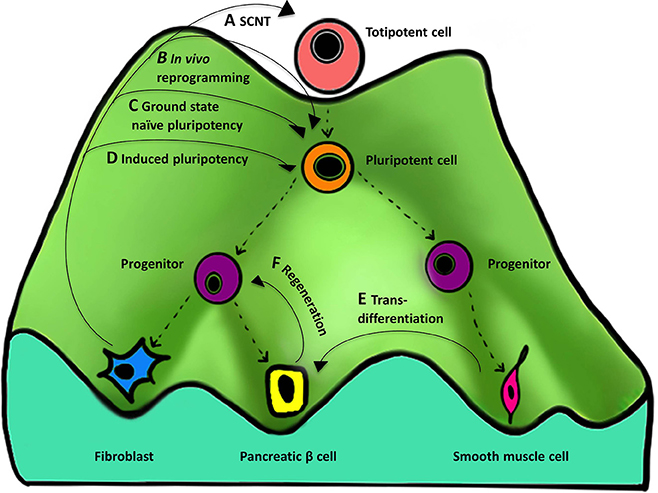
Figure 3. Importance of the “epigenetic landscape” in cellular reprogramming. An epigenetic landscape represents the process of cell fate decisions during development and is a graphical rendering of complex regulatory networks. The figure represents such an epigenetic landscape denoted by a “mountain” with its various “valleys.” The top of the mountain displays a totipotent cell that is completely undifferentiated and represents ultimate “stemness,” whereas at the bottom of the mountain are differentiated cells resulting from differentiation of their predecessors. Proceeding from the top of the landscape to the bottom, the cell changes from undifferentiated (totipotent and pluripotent), to partially differentiated (progenitor), to terminally differentiated (eg., fibroblast). (A–F) represent the different methods and their course on an epigenetic landscape, showing reversal of cellular identity to a more primitive state. There are four types of methods that are currently known for reprogramming to (A) totipotency, (B,C) near totipotency, and (D) pluripotency. (For simplicity, we have excluded reprogramming methods using only small molecules, cell-extract treatment, and cell fusion that can also result in ESCs-like cell formation). (A) SCNT gives rise to a totipotent cell capable of forming a complete organism. (B) In vivo reprogramming using OSKM generates iPSCs that are additionally capable of contributing to the trophoectoderm lineage and express embryonic and extra-embryonic markers. (C) In vitro reprogramming to ground state naïve pluripotency using NHSM media generates iPSCs more similar to mouse naïve ESCs. (D) Induced pluripotency generates iPSCs similar to but less pluripotent than (A–C). Other processes that also involve manipulation of the epigenetic regulatory networks include (E) transdifferentiation—one differentiated cell type (smooth muscle cell) is converted to another (pancreatic beta cell) and (F) regeneration—derivation of a progenitor-like cell from a differentiated cell during wound healing.
Reprogramming cell fate works through the manipulation of networks governing an epigenetic state. Different types of reprogramming can result in producing undifferentiated cells with varying degree of “stemness” by moving the cell back to the top of the mountain. Four cellular reprogramming methods, namely SCNT, and induced pluripotency (in vivo, in vitro, and in vitro using a special media) are noteworthy and are described below.
The totipotent cell resides at the very top of the landscape, and currently SCNT is the only method capable of reprogramming a somatic cell to a totipotent cell capable of forming a complete organism (Figure 3A). Below this would be a near-totipotent stem cell that has been generated through one of the following two methods: in vivo reprogramming using OSKM that generates iPSCs that are additionally capable of contributing to the trophoectoderm lineage and express embryonic and extra-embryonic markers (Abad et al., 2013) (Figure 3B), or in vitro reprogramming to ground state naïve pluripotency using NHSM media (see Section Induced Pluripotency above) that generates iPSCs more similar to mouse naïve ESCs and demonstrates the potential to overcome problems related to epigenetic memory and lineage differentiation biases (Gafni et al., 2013) (Figure 3C). The conventional in vitro method of TF-based reprogramming generates iPSCs that are similar to ESCs but often have problems related to epigenetic memory and lack totipotent cell-like features (Papp and Plath, 2011; De Los Angeles and Daley, 2013) (Figure 3D).
Other processes that also involve manipulation of the epigenetic regulatory networks include transdifferentiation (Figure 3E) and regeneration (Figure 3F). In transdifferentiation one differentiated cell type is converted to another through direct reprogramming (Takahashi, 2012), by avoiding going all the way back up the mountain to an intermediate pluripotent state. Regeneration is similar in that it results in derivation of SSCs or progenitor-like cells from a differentiated cell during wound healing, for example the formation of a blastema (an undifferentiated cell mass) in amphibians (Gurtner et al., 2008) that involves the cell going half-way back up the mountain.
A new framework has been developed in order to elucidate the role of epigenetic landscapes in reprogramming that combines the techniques of whole genome expression profiling and spin glass physics (Lang et al., 2014). This model has verified that partially reprogrammed cells co-express TFs associated with multiple cell fates, has reproduced known reprogramming protocols, and has the potential to generate new reprogramming protocols to create novel cell types (Lang et al., 2014). Defining epigenetic landscapes can not only help to gain better insights into reprogramming processes, but also to design highly efficient protocols. This can be done by targeting epigenetic modifiers involved in the differentiation process at specific positions in the landscape, as well as identifying easy access points in the hierarchy at which reprogramming can be induced for high efficiency.
Now that reprogramming technology has been described in detail we will move on to practical applications.
Modeling Human Disease In vitro
We can group human diseases into three broad categories: genetic, epigenetic and acute environmental (Cherry and Daley, 2012). Modeling of all three types is possible in vitro using stem cells, and an excellent way to study the intricate mechanisms and pathways underlying the etiology and pathophysiology of disease (Figure 4). Stem cells in general are ideal for creating “disease-in-a-dish” models because of their capacity for self-renewal and differentiation, their potential for recapitulating disease pathogenesis, and also their amenability for developing and testing therapeutics (Sterneckert et al., 2014).
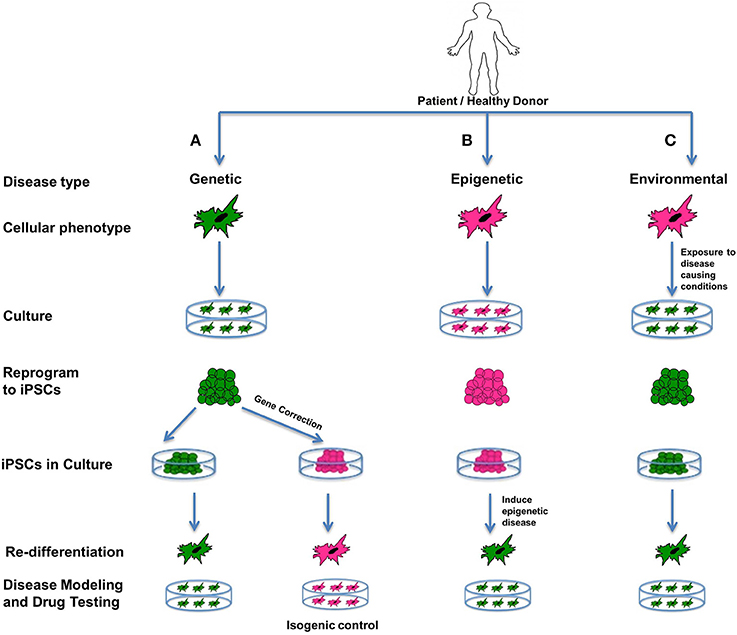
Figure 4. Disease modeling. iPSCs are an excellent source for modeling genetic, epigenetic, and environmental diseases. Such cellular models representing diseased phenotypes can be used for understanding the interplay between the genetics, epigenetics and environment involved in the disease, and can expose unknown details about disease pathophysiology, and can be used for screening drugs. In the figure all green cells represent diseased cells, and all pink cells represent healthy cells. (A) Genetic diseases can be modeled by reprogramming diseased cells to iPSCs and then re-differentiating them to produce a diseased phenotype. Additionally, these iPSCs can be corrected for the genetic mutation involved in the disease using gene-editing technology. On re-differentiation, corrected iPSCs produce healthy cells that can be used as isogenic controls. (B) Epigenetic diseases can be modeled using healthy cells that are reprogrammed to iPSCs and then induced toward an epigenetic disease state by recapitulating an environment containing the epigenetic factor(s) contributing to the disease. If iPSCs retain an epigenetic mark when in culture, or after being redifferentiated to the desired cell type, it indicates that the epigenetic mark is permanent and is likely to be passed on to offspring or carried by germ-line cells. It can also mean that the particular cell type is predisposed to retaining that epigenetic mark. Patient-specific models can be used as special models, as they can involve known epigenetic factors contributing to the disease. (C) Acute environmental diseases can be modeled using healthy cells by exposing them to a disease-causing environment that results in genetic damage or instability in the cells. For disease modeling, such cells can be reprogrammed to iPSCs and redifferentiated to diseased phenotypes. All of the above models can help us gain better insight into the diverse factors affecting a complex disease in terms of susceptibility, prognosis as well outcomes.
The first and simplest types of diseases are monogenic disorders. For example Fanconi anemia (FA) has been studied using patient-derived, integration-free iPSCs with and without in situ gene correction (Liu et al., 2014). These FA-iPSCs are also useful for drug screening and shortlisting compounds that can trigger their hematopoietic differentiation potential (Liu et al., 2014) (Figure 4A). Likewise, iPSCs have been derived from cells obtained from patients suffering from a wide and rapidly-growing number of diseases such as Hutchinson-Gilford Progeria Syndrome (HGPS, see below), cystic fibrosis, Parkinson's disease, familial amyotrophic lateral sclerosis (ALS), dyskeratosis congenita, epilepsy, autism, spinal muscular atrophy, Huntington's disease, adenosine deaminase severe combined immunodeficiency, sickle cell anemia, long QT syndrome type I, glycogen storage disease type 1a, Alzheimer's disease, diabetes, and Down's syndrome and can be differentiated toward disease-specific lineages in order to study the genotype-phenotype interactions of the disease, as well as to screen new drugs that could be capable of reversing cellular pathology (Liu et al., 2011a; Unternaehrer and Daley, 2011; Cherry and Daley, 2012; Ess, 2013; Miller et al., 2013; Abdelalim et al., 2014; Acimovic et al., 2014; Hibaoui et al., 2014; Mohamet et al., 2014) (Figure 4A).
Monogenic diseases are easy to model in culture, but it is becoming increasingly clear that most diseases are a manifestation of altered genetics and/or epigenetics, with factors like age and environment complicating things further. It is necessary to consider such interlinked effects on different cell types in the body, especially in the case of late-onset disease, and for such purposes iPSCs can serve as an excellent model (Csobonyeiova et al., 2014). Thus, in addition to modeling monogenic diseases, cellular diseased phenotypes derived from iPSCs can be useful in studying complex diseases having genetic and epigenetic components, or having unknown sporadic genetic or acute environmental etiologies (Grskovic et al., 2011; Zhu et al., 2011). Such cellular phenotypes are also useful for screening drugs and understanding the pathophysiology of complex diseases (Grskovic et al., 2011).
Epigenetic diseases can be subtly contrasted with environmental diseases based on the type and extent of damage caused by the epigenetic or environmental factor. Specifically, when iPSCs are exposed to epigenetic factors, the iPSCs and redifferentiated cells may gain temporary or permanent epigenetic marks and manifest the diseased phenotype. Modeling epigenetic diseases in such a way can elucidate the trans-generational potential of the epigenetic factors i.e., whether the epigenetic mark is strong enough to pass on through the germ line to the next generation. Compared to epigenetic diseases, acute environmental insults do not cause heritable genetic or epigenetic effects, but rather the effect is seen at the somatic DNA sequence or protein level (Cherry and Daley, 2012). For example, an acute environmental disease like melanoma that is frequently caused by excessive sunburn (ultraviolet ray damage) can lead to direct DNA damage in the form of pyrimidine dimers (Anna et al., 2007). Such sporadic environmental insults, including spontaneous deaminations, change the DNA sequence as well as alter the cellular biochemistry through upregulation of DNA repair processes, alter expression of cell cycle regulators, or possibly cause apoptosis (Fulda et al., 2010).
Using patient-specific iPSCs we can recapitulate the suspected effects of the environmental or epigenetic component known to have contributed to the patient's disease in order to understand its severity or mechanism of action before, during, or after reprogramming (Figures 4B,C). Cells from any source can be epigenetically manipulated to mimic epigenetic (Figure 4B) or acute environmental (Figure 4C) alterations contributing to disease. Such models can help us gain better insight into the environmental or epigenetic factors affecting a complex disease in terms of susceptibility, prognosis as well as outcomes. Patient-specific models can also be used as special models, as they can involve known epigenetic changes contributing to the disease.
iPSCs can also be used for modeling disease at the organ level as well as understanding systemic diseases. For example human iPSCs can be used for in vitro development of complex structures like the retina by spatiotemporally recapitulating the in vivo steps of retina formation and making it possible to test future therapies in vitro (Zhong et al., 2014).
Treating Human Disease
Beyond modeling disease, reprogramming can also be used to treat disease. While the technology of cellular reprogramming has been developed and refined there have also been significant ongoing developments in other complementary technologies such as gene editing, progenitor cell production, and tissue engineering that we will describe below. These are converging to a point that will allow us to treat almost any disease. Personalized medicine may soon become a reality, with cellular harvest and reprogramming, genetic engineering and tissue engineering becoming a routine procedure. These technologies are the foundations of what is becoming a fully-functional field of regenerative medicine.
In vitro Reprogramming and Complementary Technologies
Induced pluripotent stem cells
The greatest advantage of using in vitro reprogramming is that we can generate patient-specific iPSCs (autologous) and eliminate the threat of rejection, or have well-characterized iPSC cell-lines (allogenic). Reprogramming in vitro is often more useful when used with complementary technologies like gene editing to treat genetic diseases by fixing the causative mutation and then repopulating damaged or non-functional tissue with the desired healthy cell type (Figure 5A).
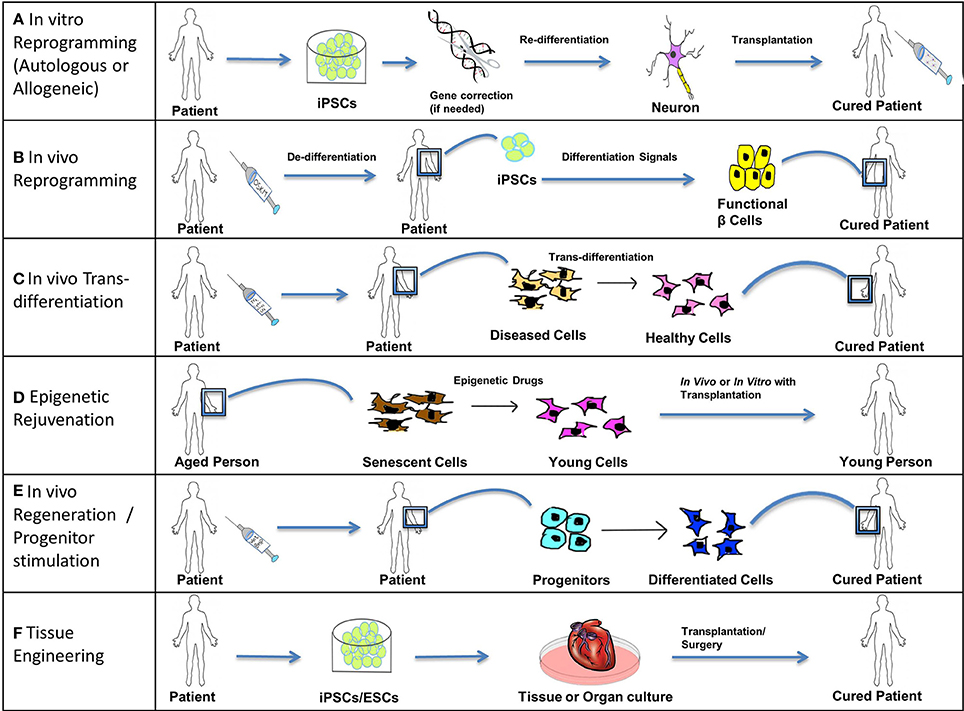
Figure 5. Treating disease. Stem cells (iPSCs, ESCs, and SSCs/Progenitors) can be used in the following ways to treat acute, chronic, and degenerative diseases. (A) In vitro reprogramming. iPSCs reprogrammed in vitro using different combinations of transcription factors can be further differentiated to specific lineages and these cells can be transplanted into the patient suffering from a degenerative disease or injury. For example, autologous iPSCs generated from fibroblasts of a patient suffering from Parkinson's disease can be differentiated into functional dopamine secreting neurons and can be transplanted into the patient's brain through surgery. In cases where solely transplanting functional cells is inadequate to completely cure a genetic disease, a permanent fix of the genetic mutation involved is required. Thus, an additional step of gene correction can be performed on the patient-specific iPSCs using gene-editing technologies like CRISPR/Cas. Allogeneic iPSCs might be useful to treat traumatic acute injuries like spinal cord injury, as they would be available immediately when required. (B) In vivo reprogramming. iPSCs can be generated in vivo at the site of non-functional or damaged tissue (by administration of reprogramming factors like OSKM) and can then be differentiated toward a specific lineage (by the administration of differentiation signals). For example, iPSCs generated in vivo from pancreatic cells of a patient suffering from diabetes can be differentiated into functional β cells secreting insulin and thus restore the lost function by repopulating the pancreatic tissue with functional β cells. (C) In vivo trans-differentiation. A somatic cell can be trans-differentiated in vivo into a different type of somatic cell using transcription factors, growth factors, etc. such that the new cell type helps to restore the lost function of the diseased or damaged tissue. This process is noteworthy as it avoids the intermediate step of reprogramming somatic cells to pluripotency; however this process is not yet well-established. (D) Epigenetic rejuvenation. The idea behind epigenetic rejuvenation is that epigenetic drugs can be used to convert senescent cells to young cells such that aging is reversed without affecting the differentiation or specialized function of the cell. This can be done in vivo directly or in vitro followed by transplantation of the rejuvenated cells into the patient. (E) In vivo regeneration/progenitor cell stimulation. Progenitors can be induced to differentiate into their successors by injecting differentiation signals like Wnt and Bmp proteins in order to achieve regeneration in vivo, which will restore lost function of damaged or diseased tissue. This process avoids procedures involving induction of pluripotency as well as transplantation into the patient. (F) Tissue Engineering. Patient-specific iPSCs (with optional gene correction step) or ESCs can be used to culture tissues and organs in vitro using specialized scaffolds and organ molds or using bioprinting. Such cultured organs can then be transplanted into a patient through advanced surgical procedures to restore the function of an entire organ or organs, in the case of chronic or systemic diseases, as well as to treat acute illnesses like myocardial infarction with cardiac tissue transplants.
Generation of iPSCs enables in vitro coordination of differentiation into cells of all three germ layers (endoderm, mesoderm, and ectoderm). Where the aim is simply to repopulate the non-functional or damaged tissue with functional cells, transplantation with iPSC-derived cells could cure the patient. For example, mouse embryonic fibroblasts or mouse pancreas-derived epithelial cells reprogrammed to iPSCs and then differentiated to functional pancreatic beta cells were capable of treating diabetes in mice (Jeon et al., 2012). These iPSC-derived beta-like cells released insulin and on transplantation into non-obese mice, normalized the blood-sugar level (Jeon et al., 2012). If treating a disease involved fixing a genetic mutation permanently, then gene-editing technologies (which are explained in detail in section Gene editing using CRISPR/Cas9, ZFN, TALENs etc) can be used as an additional step before differentiating the iPSCs into the desired cell type. For example in mice, the correction of a mutation in iPSCs by gene therapy using homologous recombination, followed by autologous transplantation back into the mouse, has led to the replacement of diseased cells and rejuvenation of functional tissue in single-gene defects like sickle cell anemia. Such procedures have overcome problems associated with immune rejection and validated the role of iPSCs in regenerative medicine (Hanna et al., 2007; Takahashi and Yamanaka, 2013). However, autologous iPSCs will incur a comparatively higher medical cost compared to allogenic ones, and their creation could take a longer time compared to allogenic iPSCs. Also they might not be available immediately when required for the treatment of acute injuries like spinal cord injury, implying the need for HLA-matched donor allogenic iPSCs (Takahashi and Yamanaka, 2013), unless they can be prepared beforehand.
One of the most promising cell types derived from iPSCs are neural cells (Svendsen, 2013). Neurons derived from iPSCs demonstrate great promise in revitalizing and repopulating the central nervous system, and have promise for use in the development of neurologically based disease models such as Huntington's and Parkinson's diseases (Kriks et al., 2011; Liu et al., 2013b).
Potential risks associated with clinical iPSC applications include the possibility that viral or other agents used for reprogramming could trigger harmful immune or inflammatory responses (Zhao et al., 2011; Guha et al., 2013). The prevention of vector or transgene integration into the host genome or residual transgene expression once reprogramming has been achieved is important in order to avoid such unwanted responses. This problem has now been resolved with the use of non-integrating episomal vectors to create integration-free iPSCs (Hu et al., 2011; Su et al., 2013). The use of non-viral vectors such as the transposon-based systems like Sleeping Beauty (SB) and piggyBac (PB) offer a cost-effective and safe alternative (Talluri et al., 2014). Several experiments to eliminate the use of more than one or two reprogramming factors during induced pluripotency have also been conducted (Radzisheuskaya and Silva, 2014). For example easily accessible dermal papilla cells (specialized mesenchymal cells endogenously expressing Sox2, Klf4, Myc) from the hair follicles of skin have been reprogrammed with Oct4 alone and without any other chemical compounds or small molecules (Tsai et al., 2011).
Gene editing using CRISPR/Cas9, ZFN, TALENs etc
As stated, the use of the combined techniques of reprogramming and genetic engineering followed by cellular therapy can potentially correct a genetic defect and repopulate a degenerated or damaged tissue with the required cell type and cure a genetic disease. Such technology can also be used to manipulate and study model organisms as well (Gaj et al., 2013).
Gene editing technologies are methods used to fix an endogenous mutation, or to make a gene inactive for the purpose of repairing or deleting a mutation. Recent progress in gene editing technology has made it possible to induce a site-specific DNA cleavage followed by repair. This results in high precision genome editing in cultured cells and whole organisms; useful in designing therapeutic treatments and creating models in vitro and in vivo (Kim and Kim, 2014).
Gene editing technology is based on the use of RNA sequences or proteins that bind to specific DNA sequences and recruit nucleases to introduce double strand breaks in the target sequence. Once DNA damage occurs, homologous or non-homologous end joining processes repair the DNA sequence. With homologous recombination, the wrong nucleotides/s can be replaced with the correct ones to repair a genetic mutation. Current precise gene editing methods include the following three types depending on the type of repair and nucleases involved: 1. Zinc finger nucleases (ZFN), 2. Transcription activator-like effector nuclease (TALEN), and 3. Clustered regularly interspaced short palindromic repeats (CRISPR).
1. ZFN- This is the most commonly used type at present, and is comprised of a eukaryotic zinc finger binding protein domain coupled with a nuclease.
2. TALEN- This type similarly uses a transcription activator-like effector (TALE) domain coupled with a nuclease.
ZNFs and TALENs include programmable, sequence specific DNA binding modules linked to a non-specific DNA cleavage domain and are capable of introducing a variety of genetic modifications (Gaj et al., 2013).
3. CRISPR- This is the newest type of gene editing technology based on RNA interference and uses CRISPR and CRISPR associated genes (Cas) from bacterial and archeal immune systems. The CRISPR loci have short palindromic repeats (24–37 base pairs) separated by unique spacer sequences of equal length that can integrate foreign DNA, [which in the source bacteria/archea can be used as a type of memory in the case of a future attack by the same foreign DNA (Richter et al., 2013)]. When a CRISPR sequence is transcribed, the foreign DNA is transcribed along with it. The processed RNA in the form of crRNA and tracrRNA is responsible for sequence specific silencing by recognizing the target foreign DNA sequence and degrading it with Cas proteins that have nuclease activity (Gaj et al., 2013).
This RNA-guided genome editing tool could be used to correct disease- associated genes in zygotes and human cell lines (High et al., 2014). The CRISPR/Cas system can effectively replace the traditional method of producing mutant mice by sequential recombination since this technology allows a highly efficient single-step approach for production of ES cells with mutations in multiple genes, as well as generation of mutant mice by direct embryo manipulation (Wang et al., 2013).
CRISPR- based genome editing is also useful in disease modeling because it is very efficient at generating allelic isogenic models to study diseases in vitro. For example the combination of CRISPR technology and an antibody-based method to screen recombination events has been used to generate genetically modified human cell lines for the study of Huntington's disease (An et al., 2014). The Type II CRISPR/Cas9 system is gaining special recognition for its ability to accelerate the production of cell lines, especially in combination with reprogramming, and the production of animal models with a desired genetic makeup for drug screening and pre-clinical evaluation. Also it seems to be a desirable candidate for gene therapy alone (Yingze Zhao, 2014).
In vivo Reprogramming
Other potential ways of using iPSCs for treating diseases would be by generating them in vivo. These in vivo iPSCs can further generate desired cell types in the presence of appropriate signals for re-differentiation (Figure 5B). As described in section Induced Pluripotency and Epigenetics during cellular reprogramming, in vivo transitory induction of OSKM in mice led to in situ reprogramming and gave rise to iPSCs from a variety of cell types (Abad et al., 2013). These iPSCs represented totipotent-like features and they resembled ESCs more closely compared to in vitro generated iPSCs (Abad et al., 2013). Another in vivo study showed the generation of induced adult neuroblasts (iANBs) from resident astrocytes using a single TF Sox2 and these iANBs formed mature neurons when targeted for differentiation using noggin, BDNF or histone deacetylases inhibitor (Niu et al., 2013).
In vivo Trans-differentiation
Transdifferentiation is the process in which a particular somatic cell is switched from one lineage-specific identity to a completely different identity (Graf, 2011; Vierbuchen and Wernig, 2012); in other words, the direct conversion of one type of somatic cell into another type, bypassing the intermediate step of dedifferentiation (Figure 5C). As described in Section Transdifferentiation/direct reprogramming transdifferentiation in vivo converted adult murine pancreatic exocrine cells to β-cell like population using the transcription factors Ngn3, Pdx1, and Mafa (Zhou et al., 2008). Another example is the in vivo transdifferentiation of human peripheral blood CD34+ cells into non-hematopoietic lineages such as cardiomyocytes (Yeh et al., 2003). However, it is important to note that such processes occur only under the conditions of severe tissue damage (Yeh et al., 2003).
Epigenetic Rejuvenation
Epigenetic rejuvenation is the concept of erasing an “aged” epigenome to obtain an epigenome that corresponds to the one seen in a “young” healthy cell along with maintaining its differentiated state and rejuvenating its function (Manukyan and Singh, 2012). The idea behind epigenetic rejuvenation is to treat aged cells from patients in vitro and transplant them back into the tissue and replace aged or unhealthy, damaged cells (Manukyan and Singh, 2012) (Figure 5D). (Please refer to Section Hutchinson-Gilford Progeria Syndrome for additional details.)
In vivo Regeneration/progenitor Cell Stimulation
In vivo regeneration involves the repair of tissue damage by stimulation of endogenous stem cells (SSCs/progenitors) by growth factors, cytokines, transcription factors, and second messengers (Figure 5E). A promising strategy to treat degenerative and chronic disorders resulting from loss of function of a particular type of cell, is through harnessing the intrinsic regenerative potential of the progenitor cells which are responsible for creating the specific type of damaged cell and restoring their lost function. Studies involving the use of regenerative factors like developmental signals that play a role in the regeneration of a damaged tissue, suggest that the latent/dormant intrinsic potential can be triggered to harness the in vivo regeneration capacity in a disease or trauma-struck individual (Figure 5E). Such changes during regeneration are accompanied by epigenetic modifications. A few examples of signaling pathways and differentiation factors, which are being studied and could potentially activate regeneration (thus should be considered as suitable targets) are described below.
1. Wnt, a ligand working in conjunction with a receptor, involved in the transcription of β- catenin promotes morphogenesis and has the capacity to initiate the regeneration of broken bone to treat bone deformities or injuries, as well as cause Muller progenitors to differentiate into retinal progenitors, thereby ameliorating retinal trauma, and possibly blindness (Minear et al., 2010; Liu et al., 2013a).
2. Platelet-derived growth factor (PDGF) and insulin-like growth factor (IGF) have been shown to stimulate the regenerative capacity of mammalian skeletal and neural tissue (Rutherford et al., 1992; Mason et al., 2000).
3. Bone morphogenic protein (BMP) is essential for ESC renewal whereas basic Fibroblast growth factor (bFGF) increases the differentiation potential of MSCs into chondrogenic, osteogenic and adipogenic lineages and epidermal growth factor receptor (EGFR) increases the proliferative potential and survival capacity of MSCs (Bhattacharyya et al., 2012).
4. Lin28 is expressed during embryogenesis as it plays an important role in pluripotency and development, also its overexpression promotes epidermal hair regrowth, and digit repair in mice by altering the bioenergetic state during tissue repair following enhanced translation of metabolic enzymes that increase glycolysis and oxidative phosphorylation (Shyh-Chang et al., 2013).
5. The transient inactivation of two tumor suppressors ARF (alternative reading frame protein) and RB (retinoblastoma protein) in mammalian cells resulted in dedifferentiation restoring regenerative capacity and produced differentiated cell types when cultured in the presence of proper signaling factors suggesting that terminally differentiated mammalian cells can be induced for regeneration without neoplastic transformation (Pajcini et al., 2010).
6. Hox genes play an important role in embryonic pattern formation and can be programmed such that their previous epigenetic memory is erased resulting in plasticity, as observed in iPSCs, that can in turn be used to trigger regenerative capability of injured tissue (Wang et al., 2009).
Tissue Engineering
Tissue engineering is the field of engineering tissues, or organs grown from in vitro cultures of stem cell-derived differentiated cells, followed by their transplantation (Figure 5F).
Tissue engineering is the de novo production of tissues or organs using starting material like cells, extracellular matrix and scaffolds for supporting the three dimensional process of tissue formation (Fisher and Mauck, 2013). This technology has been translated into actual therapies for skin replacement and cartilage repair (Berthiaume et al., 2011). The first step in tissue engineering is to procure the desired cell types required for constructing the tissue or organ. iPSCs as well ESCs serve as a good source for producing the cells through directed differentiation. For example, iPSC-derived multipotent neural crest stem cells (NCSCs) have been used to engineer nerve conduits by seeding NCSCs into nanofibrous tubular scaffolds and such nerve conduits were used for regenerating sciatic nerve (Wang et al., 2011). Tissue engineering uses conventional methods for scaffold fabrication that are probably inadequate (Seol et al., 2014), but the latest bioprinting technology can construct 2-D and 3-D tissues and organs using additive manufacturing technology that allows precise placement of cells, biomaterial and biomolecules in an adequately fabricated scaffold (Seol et al., 2014).
Clinical Trials for Stem Cell Therapy in Humans
The U.S. Food and Drug Administration defines somatic cell therapy as the administration to humans of autologous, allogeneic or xenogeneic living non-germ line cells, other than transfusion blood products, which have been manipulated, processed, propagated or expanded ex vivo, or are drug-treated (Health et al., 2001; Parson, 2006; Bieback et al., 2011; George, 2011).
With the FDA approving the first ESCs Phase I clinical trials for acute spinal cord injury in 2011, the science behind stem cell biology and regenerative medicine made its way to practical applications in the real world (Table 1). Extensive efforts from the California Institute of Regenerative Medicine (CIRM), National institute of Health (NIH) and other organizations around the world has allowed stem cell therapies to progress into clinical trials for a variety of diseases, many of which have made significant progress. Clinical trials using mesenchymal stem cells (MSCs) for treating bone and cartilage diseases, neurodegenerative diseases, heart diseases, gastrointestinal diseases, autoimmune/immune rejection diseases, and cancer have shown a lot of progress with massive funding from CIRM, their collaborating partners and others (Trounson et al., 2011) (Table 1).
The most advanced clinical trials listed by EuroStemCell include testing of therapies for bone, skin, corneal disease and injury, the majority of which use mesenchymal stem cells. The NIH lists two active clinical trials for the eyes diseases Age related Macular Degeneration (AMD) and Stargardt's muscular dystrophy using cells derived from human ESCs, being produced by Advanced Cell Technology. Other sources like bone marrow stem cells for treating cancer and blood disorders, human central nervous system stem cells for AMD and human spinal cord stem cells for ALS are also in clinical trials. Human MSCs are being used in clinical trials by Osiris therapeutics for Type 1 diabetes, cardiomyopathies, Graft vs. host disease and Crohn's disease. More details about these ongoing clinical trials can be found through the links for online resources provided in Table 1.
Candidate Diseases for Treatment using Reprogramming
In this section, we provide specific examples of how reprogramming and complementary technologies can be used to treat and eventually cure human disease.
Aging and Age-related Diseases
A few decades ago, Richard Cutler proposed that aging was a result of cells drifting away from their proper state of differentiation due to changes in gene expression, through a process he termed “dysdifferentiation” (Kator et al., 1985). This process could be reinterpreted today as epigenetic drift. These changes are so predictable that recently an “epigenetic clock” has been formulated that can very accurately predict the age of an individual based solely on the methylation of specific genes in genomic DNA (Horvath, 2013). Indeed, epigenetic changes represent a major aspect of aging, and epigenetic manipulation may allow reversal of these damaging effects (Campisi and Vijg, 2009; Munoz-Najar and Sedivy, 2011).
It has been proposed that if we can reverse the epigenetic changes associated with aging, we could actually reverse aging itself. This epigenetic approach to reversing aging has been termed “epigenetic rejuvenation” (see Section Epigenetic Rejuvenation and Figure 5D). Although the very existence of animals cloned by SCNT proves that epigenetic rejuvenation is possible in principle, only recently have in vitro experiments on human cells shown that this is possible. Indeed, even centenarian fibroblasts can be rejuvenated, and young fibroblasts cultured to senescence can be reprogrammed to a pluripotent state (Lapasset et al., 2011; Villeda et al., 2011; Loffredo et al., 2013). Recently, very detailed experiments have shown that age-related and tissue-specific DNA methylation patterns remain erased in mesenchymal stem cells produced by induced pluripotency (Frobel et al., 2014).
Epigenetic reversal also crucially includes reactivation of telomerase expression, which resets the chromosome “aging clock” back to a young state. Reprogramming “old” cells to “young” cells will eventually enable the repopulation of an aged and/or degenerated tissue with cells capable of extended proliferation. Perhaps this strategy could eventually be applied to the whole body.
An example of a type of epigenetic rejuvenation already practically applied is heterochronic parabiosis: experiments with two animals of a different age joined together surgically, showed that tissue-specific stem cells from an aged animal could be rejuvenated by exposing them to a “young” environment (Rando and Chang, 2012; Conboy et al., 2013). This “young” environment involves factors such as chemokine's, cytokines and rejuvenating Wnt and TGFβ signaling pathways (Conboy et al., 2013). On the other hand “young” stem cells adopt a more aged structure and function (Conboy et al., 2005). Unlike reprogramming, heterochronic parabiosis maintains the differentiated states of the cells and rejuvenates the regenerative potential of old cells in the milieu of a young environment, thus uncoupling dedifferentiation and rejuvenation (Conboy et al., 2013). This type of rejuvenation might have an epigenetic basis regulated by a switch that could possibly convert a cell from “old” to “young” or vice versa.
Aging also involves mitochondrial dysfunction as well as defective mitochondrial DNA. SCNT can be used to correct diseases caused by defective mitochondrial DNA (mtDNA) because in NT-ESCs the mtDNA exclusively comes from the oocyte, and only the nucleus from the donor/patient, thus making it possible to avoid defective mtDNA being carried over from the donor/patient (Tachibana et al., 2013). Surprisingly, induced pluripotency rejuvenates mitochondira, but in a different way to SCNT (Suhr et al., 2010).
Hutchinson-Gilford Progeria Syndrome
Hutchinson-Gilford Progeria Syndrome (HGPS) is a segmental premature aging disease characterized by symptoms of premature aging, with a life expectancy in the early teens or twenties. The single point mutation responsible for causing this disease occurs in position 1824 (cytosine is replaced with thymine) of the Lamin A (LMNA) gene on chromosome 1. Lamin A is an intermediate filament protein that stabilizes the inner membrane of the nuclear envelope. The mutation results in a defective Lamin A that produces an abnormal protein called Progerin. Ultimately, this results in abnormally shaped, dysfunctional nuclei that in turn disrupt cell division and other cellular functions (Eriksson et al., 2003; McClintock et al., 2007; Korf, 2008).
Somatic cells from Progeria patients can be reprogrammed to iPSCs and the LMNA mutation can be corrected using gene-editing technology (see above). The fact that ESCs and iPSCs do not express LMNA (Constantinescu et al., 2006), allows the efficient correction of the mutation in iPSCs, and any disease-related pathology can be avoided in iPSC-derived differentiated cells. Notably, HGPS iPSCs exhibit downregulation of lamin A/C and Progerin, causing a lack of progeroid features in the iPSC population, rendering these iPSCs normal with respect to nuclear morphology, pluripotency, and epigenetic profiles (Liu et al., 2011a). HGPS iPSCs corrected for the LMNA mutation can then be differentiated into normal somatic cells (Liu et al., 2011b). These iPSC-derived differentiated cells could eventually be transplanted into patients using cellular therapy to repopulate the aged tissue with healthy proliferative cells free of the original Progeria mutation.
Age-related macular degeneration
Age-related macular degeneration (AMD) is a complex multifactorial diseases caused by the degeneration of the photoreceptors and retinal pigment epithelium (RPE) of the eye (Yonekawa and Kim, 2014). It is the most common cause of visual impairment in the elderly (+60 years) and it is broadly classified into two clinical categories, namely, the wet form (neovascular or exudative) and dry form (Cook et al., 2008). AMD, which is primarily known to be an age-related disease, is also affected by other risk factors like genetics, patient history, smoking, trauma, etc (Chakravarthy et al., 2010). Data from pre-clinical studies suggests that transplantation with ESCs-derived RPE or patient-specific iPSC-derived RPE (with correction of the gene mutation) may prevent photoreceptor degeneration in animal models of RPE degeneration (Schwartz et al., 2012; Heller and Martin, 2014). With the advent of techniques in in vivo reprogramming, another promising approach to cure AMD is to target the intrinsic potential of neural progenitors in a patient and drive their differentiation into RPE through the administration of appropriate differentiation signals. It has already been suggested that retinal stem cells (RSCs) that are multipotent, rare cells in the pigmented ciliary epithelium of peripheral retina can be enriched an directed to differentiate into photoreceptors using factors influencing neural retinal development (Ballios et al., 2012). Specialized knowledge of such differentiation and proliferation signals with the development of advanced techniques toward delivery and transplantation of stem cells or RPE lead us one step closer to the development of clinical therapies for AMD.
Epigenetic Diseases
Epigenetic diseases are caused by altered expression of genes caused by abnormal changes in the epigenetic profile of a cell. Reprogramming can be used to reset the epigenetic profile of any cell so that the cell becomes pluripotent. It can then be differentiated to obtain the cell type with a desired epigenetic profile. Such procedures followed by cellular therapy might be a potential treatment for epigenetic diseases caused by known epigenetic factors. Let us consider the example of polycystic ovary syndrome (PCOS) as an epigenetic disease.
PCOS is a heterogeneous endocrine disorder responsible for a large proportion of female subfertility (Azziz et al., 2004). The hallmarks of the disease are anovulation, excessive androgenic hormones, and insulin resistance. The PCOS phenotype is characterized by hypersecretion of both luteinizing hormone as a result of compromised hypothalamic sensitivity, and insulin in response to increased abdominal adiposity (Escobar-Morreale et al., 2005). Familial clustering of cases and the heritability of endocrine and metabolic aspects of PCOS indicate a dominant regulatory gene with incomplete penetrance and strongly indicate a genetic etiology (Diamanti-Kandarakis et al., 2006).
As described in Section Epigenetics above, environmental factors can induce epigenetic alterations, which potentially involve trans-generational inheritance of methylation patterns. The exposure of female rats to a mixture of pesticides showed that the first and third generation offspring were afflicted with PCOS (Manikkam et al., 2012). This demonstrated that PCOS might have an epigenetic basis caused by a differential methylation pattern in the diseased offspring. In humans aberrant gene methylation has been reported in patients with PCOS, suggesting a role for a bZIP TF, CEBPB, in insulin resistance and regulation of the interleukin-1 response element in the IL-6 gene and other genes important for the immune and inflammatory components of this disease (Natsuka et al., 1992; Harries et al., 2012; Shen et al., 2013). Inappropriate epigenetic reprogramming is a contributing factor in PCOS. Epigenetic alterations of peroxisome proliferator-activated receptor gamma 1 (PPARG1), nuclear co-repressor 1 (NCOR1), and histone deacetylase 3 (HDAC3) genes in granulosa cells are induced by hyperandrogenism (Qu et al., 2012).
Recently, PCOS-derived hESCs have been successfully isolated from the inner cell masses of blastocysts. These hESCs retain pluripotency, which allows the dissection of the in vitro pathogenesis of the disease. Eventually cell therapies will be developed for this disease.
Combined Genetic and Epigenetic Diseases
Some diseases long known to have genetic causes are now thought to also have epigenetic bases. Just as with epigenetic diseases, such diseases can be treated using reprogramming to re-establish the normal epigenetic profile but with the addition of gene correction. One of the most well-known such diseases is cancer.
Cancer
Cancer remains the leading cause of death worldwide with over 7.6 million deaths (13%) in 2008 (Jemal et al., 2011). Cancer does of course have a partly genetic basis: most human cancers are caused by gene mutations in one or more components of the p53 and/or the pRB pathway (Campisi, 2001). And most tumor suppressor genes can be classified into two types; Caretaker tumor suppressors which prevent DNA damage and Gatekeeper tumor suppressors which eliminate potential neoplastic cells via apoptosis (Campisi, 2005).
But besides the genetic mutations, cancer is also an epigenetic disease. The dysdifferentiation hypothesis of aging and cancer emphasizes the importance of instability of the differentiated state of cells due to improper gene regulation in the development of cancer (Zs-Nagy et al., 1988). Cancer is comprised of the following eight hallmarks, some of which may have a wholly epigenetic etiology: sustained proliferative signaling, evading growth suppressors, enabling replicative immortality, inducing angiogenesis, resisting cell death, activating invasion and metastasis, reprogramming of energy metabolism and evading immune destruction. (Hanahan and Weinberg, 2011). Reprogramming of metabolic pathways contributes to the development of the disease, which is evidently seen in cancer with upregulation of oxidative phosphorylation and glycolysis-associated tarnscriptomes (Lisanti et al., 2014). For example, cancer-associated fibroblasts found in tumor microenvironments have a distinctive energy metabolism reprogramming phenotype responsible for proliferation, migration, invasion and epithelial-mesenchymal transition (Tang et al., 2014).
Cancer cells have altered DNA methylation profiles such that the overall methylated DNA is less than that seen in normal cells. A study testing epigenetics as a molecular marker system for cancer noted 12 genes to be associated with promoter hypermethylation causing silencing of their respective genes. These were functional in cell cycle regulation, tumor suppression, DNA repair, metastatic potential and apoptosis and have been linked to 15 major tumor types, making epigenetics a significant tool in cancer detection and typing (Esteller et al., 2001). Epigenetic changes in cancer are becoming well-noted and epigenetic drugs like DNA methylation inhibitors and histone deacetylase inhibitors have shown promising results in clinical trials and are less toxic than the conventional chemotherapy drugs (Chen et al., 2014). Epigenetic modulators may be used to inhibit self-renewal and survival of quiescent cancer stem cells (CSCs) by making them sensitive to targeted or conventional chemotherapy (Li and Bhatia, 2011).
Other diseases
Genetic factors contributing to epigenetic variation like DNA sequence variants, differential expressions of genes that regulate chromatin remodeling and DNA methylation, can contribute toward an epigenetic etiology of a disease (Bjornsson et al., 2004). Familial forms of neurodegenerative diseases like Alzheimer's disease, Huntington's disease, Parkinson's disease, and ALS are known to have a genetic basis but increasing evidence points toward epigenetic alterations like DNA methylation, histone tail modifications, and RNA mediated mechanisms in sporadic forms (Kleivi, 2006).
Some autoimmune diseases such as systemic lupus erythematous, rheumatoid arthritis, multiple sclerosis, and type 1 diabetes mellitus suggest the contribution of environment to a genetically pre-disposed trait (Hewagama and Richardson, 2009). For example, Immunodeficiency chromosomal-instability facial anomalies syndrome (ICF) is one such disease, which is caused by mutation in the gene for DNA methyl transferase (DNMT3B) that further leads to abnormal methylation causing varied gene expression (Ehrlich, 2003; Bjornsson et al., 2004; Rodenhiser and Mann, 2006).
Other examples include the downregulation of the Reelin gene (RELN) in schizophrenia which leads to the reduction of RELN protein by 50% compared to a normal brain and this has been linked to an epigenetic modulation of the promoter of the RELN gene through increased expression of DNMT1 (Costa et al., 2002; Sharma, 2005; Rodenhiser and Mann, 2006), and Rett syndrome, mostly caused by a mutation in the gene MeCP2 that produces methyl-CpG- binding protein, is an X linked dominant neurodevelopmental disorder (Bienvenu et al., 2000). The loss of the MeCP2 protein results in loss of transcriptional repression and hence inappropriate transcription of downstream genes during brain development (Van Den Veyver and Zoghbi, 2001; Rodenhiser and Mann, 2006).
Infectious Diseases
One of the most promising strategies to treat an infectious disease is to replace a receptor on a host target cell, which is used by a pathogen to gain entry into the cell, with a mutated one. This makes the receptors on the target cell non-functional and blocks the entry of the pathogen into the cell, preventing its multiplication and the progression of the disease. One important disease that may eventually be universally treated and perhaps eradicated this way is acquired immunodeficiency syndrome (AIDS). AIDS is a disease of the human immune system caused by infection with human immunodeficiency virus (HIV). It is a major public health concern worldwide, with more than 30 million people infected (Kallings, 2008; Unaids, 2013). There is no cure or vaccine at present, but antiretroviral treatment (ART) can slow the course of the disease and result in near-normal life expectancy. ART reduces the risk of death and complications from the disease, but is expensive and associated with side-effects. We need to find better treatments.
Of note, one of the molecular mechanisms of HIV infection and resulting AIDS is depletion of CD4+ T cells. All cells susceptible to HIV infection (CD4 T cells, macrophages, and dendritic cells) are derived from hematopoietic stem cells. Following infection, most CD4+ T cell loss occurs in the intestinal mucosa which harbors the majority of the lymphocytes found in the body (Mehandru et al., 2004).
C-C chemokine receptor type 5, also known as CCR5, is a protein on the surface of white blood cells. It functions as a receptor for chemokines allowing T cells to hone in on target tissues and organs. HIV often uses CCR5 to enter and infect host cells. The majority of mucosal CD4+ T cells express the CCR5 protein, which HIV uses as a co-receptor to gain access to the cells (Brenchley et al., 2004). HIV seeks out and destroys CCR5-expressing CD4+ T cells during acute infection. However, certain individuals carry a mutation known as CCR5-Δ32, which renders the receptor non-functional and prevents entry of HIV into the cells. In humans the CCR5 gene is located on chromosome 3. A Δ32 mutation results in the genetic deletion of a portion of the CCR5 gene. Homozygous carriers of this mutation are resistant to certain strains of HIV-1 infection (Samson et al., 1996). This mutation, although having negative effects on T-cell function, can protect these individuals against HIV.
Future reprogramming technology may allow generation of CCR5-deficient HSCs derived from hESCs/iPSCs, perhaps by creating iPSCs from a CCR5-deficient individual (Ledran et al., 2008; Hutter et al., 2009), or even better from an existing patient using gene editing technology. Diseases such as HIV could potentially be cured by means of hematopoietic cell transplantation whereby HSCs or even fibroblasts are first dedifferentiated into continuously growing iPSC lines using induced reprogramming, followed by gene editing to introduce the Δ32 mutation and then directed differentiation toward the desired hematopoietic lineage followed by bone marrow transplantation (Kambal et al., 2011). The use of engineered zinc finger nucleases (ZFNs) to disrupt the CCR5 gene in human HSCs in vivo suggested that ZFN mediated autologous HSC modification could provide a permanent supply of HIV-resistant CD34+ HSC progeny which as well would be capable of immune reconstitution and long-term protection against viral replication (Holt et al., 2010).
In principle this basic idea could be applied to other infectious diseases that rely on a single or even multiple receptors to gain entry into human cells.
Conclusion
The last decade has seen a boom in the field of stem cell biology and regenerative medicine coupled with extensive studies on the epigenetics involved in cellular reprogramming and disease. Nuclear reprogramming experiments beginning in the early 1950's introduced the concept of generation of pluripotent cells from somatic cells through nuclear transfer. The discovery of hESCs in the late 1990's acquainted us with a source for designing therapies to treat a broad range of diseases. Following that came the invention of iPSC technology, bringing stem cell production and its use in medicine out of an ethical quandary. Today, stem cell therapies have advanced into clinical trials and we predict that it will not be long before regenerative medicine units become established at health care centers.
Similarly, epigenetic studies have revolutionized the classical genetic approach toward studying and treating hereditary diseases. As we embark on this journey to understand every aspect of the interlocking pathways and processes underlying aging, cancer, regeneration, and repair, we still need to completely decipher the epigenetic code in order to design and implement effective and safe stem-cell based therapies to treat age-related diseases, acute injuries and perhaps chronic disorders.
A full understanding of epigenetics, reprogramming, senescence, cancer, and regeneration will drive remarkable progress in achieving cellular and rejuvenation therapies as well as designing drugs for diseases with an epigenetic basis. The best approach is to embrace all of these fields in a unifying synthesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Authors would like to thank Dr. Donald Orlic for critical review of the manuscript.
References
Abad, M., Mosteiro, L., Pantoja, C., Canamero, M., Rayon, T., Ors, I., et al. (2013). Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature 502, 340–345. doi: 10.1038/nature12586
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Abdelalim, E. M., Bonnefond, A., Bennaceur-Griscelli, A., and Froguel, P. (2014). Pluripotent stem cells as a potential tool for disease modeling and cell therapy in diabetes. Stem Cell Rev. 10, 327–337. doi: 10.1007/s12015-014-9503-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Acimovic, I., Vilotic, A., Pesl, M., Lacampagne, A., Dvorak, P., Rotrekl, V., et al. (2014). Human pluripotent stem cell-derived cardiomyocytes as research and therapeutic tools. Biomed. Res. Int. 2014:512831. doi: 10.1155/2014/512831
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Alberio, R., Campbell, K. H., and Johnson, A. D. (2006). Reprogramming somatic cells into stem cells. Reproduction 132, 709–720. doi: 10.1530/rep.1.01077
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
An, M. C., O'brien, R. N., Zhang, N., Patra, B. N., De La Cruz, M., Ray, A., et al. (2014). Polyglutamine disease modeling: epitope based screen for homologous recombination using CRISPR/Cas9 system. PLoS Curr. 6:ecurrents.hd.0242d2e7ad72225efa72f6964589369a. doi: 10.1371/currents.hd.0242d2e7ad72225efa72f6964589369a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anna, B., Blazej, Z., Jacqueline, G., Andrew, C. J., Jeffrey, R., and Andrzej, S. (2007). Mechanism of UV-related carcinogenesis and its contribution to nevi/melanoma. Expert Rev. Dermatol. 2, 451–469. doi: 10.1586/17469872.2.4.451
Apostolou, E., and Hochedlinger, K. (2013). Chromatin dynamics during cellular reprogramming. Nature 502, 462–471. doi: 10.1038/nature12749
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Azziz, R., Woods, K. S., Reyna, R., Key, T. J., Knochenhauer, E. S., and Yildiz, B. O. (2004). The prevalence and features of the polycystic ovary syndrome in an unselected population. J. Clin. Endocrinol. Metab. 89, 2745–2749. doi: 10.1210/jc.2003-032046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ballios, B. G., Clarke, L., Coles, B. L., Shoichet, M. S., and Van Der Kooy, D. (2012). The adult retinal stem cell is a rare cell in the ciliary epithelium whose progeny can differentiate into photoreceptors. Biol. Open 1, 237–246. doi: 10.1242/bio.2012027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Batista, L. F. (2014). Telomere biology in stem cells and reprogramming. Prog. Mol. Biol. Transl. Sci. 125, 67–88. doi: 10.1016/B978-0-12-397898-1.00003-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berger, S. L., Kouzarides, T., Shiekhattar, R., and Shilatifard, A. (2009). An operational definition of epigenetics. Genes Dev. 23, 781–783. doi: 10.1101/gad.1787609
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Berthiaume, F., Maguire, T. J., and Yarmush, M. L. (2011). Tissue engineering and regenerative medicine: history, progress, and challenges. Annu. Rev. Chem. Biomol. Eng. 2, 403–430. doi: 10.1146/annurev-chembioeng-061010-114257
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhattacharyya, S., Kumar, A., and Lal Khanduja, K. (2012). The voyage of stem cell toward terminal differentiation: a brief overview. Acta Biochim. Biophys. Sin. (Shanghai). 44, 463–475. doi: 10.1093/abbs/gms027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bieback, K., Kinzebach, S., and Karagianni, M. (2011). Translating research into clinical scale manufacturing of mesenchymal stromal cells. Stem Cells Int. 2010:193519. doi: 10.4061/2010/193519
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bienvenu, T., Carrie, A., De Roux, N., Vinet, M. C., Jonveaux, P., Couvert, P., et al. (2000). MECP2 mutations account for most cases of typical forms of Rett syndrome. Hum. Mol. Genet. 9, 1377–1384. doi: 10.1093/hmg/9.9.1377
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bird, A. (2007). Perceptions of epigenetics. Nature 447, 396–398. doi: 10.1038/nature05913
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bjornsson, H. T., Fallin, M. D., and Feinberg, A. P. (2004). An integrated epigenetic and genetic approach to common human disease. Trends Genet. 20, 350–358. doi: 10.1016/j.tig.2004.06.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bredenkamp, N., Nowell, C. S., and Blackburn, C. C. (2014). Regeneration of the aged thymus by a single transcription factor. Development 141, 1627–1637. doi: 10.1242/dev.103614
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenchley, J. M., Schacker, T. W., Ruff, L. E., Price, D. A., Taylor, J. H., Beilman, G. J., et al. (2004). CD4+T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200, 749–759. doi: 10.1084/jem.20040874
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Briggs, R., and King, T. J. (1952). Transplantation of living nuclei from blastula cells into enucleated Frogs' eggs. Proc. Natl. Acad. Sci. U.S.A. 38, 455–463.
Buganim, Y., Faddah, D. A., Cheng, A. W., Itskovich, E., Markoulaki, S., Ganz, K., et al. (2012). Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150, 1209–1222. doi: 10.1016/j.cell.2012.08.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campbell, K. H., Fisher, P., Chen, W. C., Choi, I., Kelly, R. D., Lee, J. H., et al. (2007). Somatic cell nuclear transfer: past, present and future perspectives. Theriogenology 68(Suppl. 1), S214–S231. doi: 10.1016/j.theriogenology.2007.05.059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campbell, K. H., McWhir, J., Ritchie, W. A., and Wilmut, I. (1996). Sheep cloned by nuclear transfer from a cultured cell line. Nature 380, 64–66. doi: 10.1038/380064a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campisi, J. (2001). Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 11, S27–S31. doi: 10.1016/S0962-8924(01)02151-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campisi, J. (2005). Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120, 513–522. doi: 10.1016/j.cell.2005.02.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campisi, J., and Vijg, J. (2009). Dos damage to DNA and other macromolecules play a role in aging? If so, how? J. Gerentol. A. Biol. Sci. 64, 4. doi: 10.1093/gerona/gln065
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campos-Sanchez, E., and Cobaleda, C. (2014). Tumoral reprogramming: plasticity takes a walk on the wild side. Biochim. Biophys. Acta. doi: 10.1016/j.bbagrm.2014.07.003. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chakravarthy, U., Wong, T. Y., Fletcher, A., Piault, E., Evans, C., Zlateva, G., et al. (2010). Clinical risk factors for age-related macular degeneration: a systematic review and meta-analysis. BMC Ophthalmol. 10:31. doi: 10.1186/1471-2415-10-31
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, Q. W., Zhu, X. Y., Li, Y. Y., and Meng, Z. Q. (2014). Epigenetic regulation and cancer (Review). Oncol. Rep. 31, 523–532. doi: 10.3892/or.2013.2913
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cherry, A. B., and Daley, G. Q. (2012). Reprogramming cellular identity for regenerative medicine. Cell 148, 1110–1122. doi: 10.1016/j.cell.2012.02.031
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Colwell, A. S., Longaker, M. T., and Lorenz, H. P. (2005). Mammalian fetal organ regeneration. Adv. Biochem. Eng. Biotechnol. 93, 83–100. doi: 10.1007/b99972
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conboy, I. M., Conboy, M. J., Wagers, A. J., Girma, E. R., Weissman, I. L., and Rando, T. A. (2005). Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433, 760–764. doi: 10.1038/nature03260
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conboy, M. J., Conboy, I. M., and Rando, T. A. (2013). Heterochronic parabiosis: historical perspective and methodological considerations for studies of aging and longevity. Aging Cell 12, 525–530. doi: 10.1111/acel.12065
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Constantinescu, D., Gray, H. L., Sammak, P. J., Schatten, G. P., and Csoka, A. B. (2006). Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells 24, 177–185. doi: 10.1634/stemcells.2004-0159
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cook, H. L., Patel, P. J., and Tufail, A. (2008). Age-related macular degeneration: diagnosis and management. Br. Med. Bull. 85, 127–149. doi: 10.1093/bmb/ldn012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Costa, E., Chen, Y., Davis, J., Dong, E., Noh, J. S., Tremolizzo, L., et al. (2002). REELIN and schizophrenia: a disease at the interface of the genome and the epigenome. Mol. Interv. 2, 47–57. doi: 10.1124/mi.2.1.47
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Csobonyeiova, M., Polak, S., Koller, J., and Danisovic, L. (2014). Induced pluripotent stem cells and their implication for regenerative medicine. Cell Tissue Bank. doi: 10.1007/s10561-014-9462-9. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Los Angeles, A., and Daley, G. Q. (2013). Stem cells: reprogramming in situ. Nature 502, 309–310. doi: 10.1038/nature12559
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Robertis, E. M. (2006). Spemann's organizer and self-regulation in amphibian embryos. Nat. Rev. Mol. Cell Biol. 7, 296–302. doi: 10.1038/nrm1855
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Diamanti-Kandarakis, E., Kandarakis, H., and Legro, R. S. (2006). The role of genes and environment in the etiology of PCOS. Endocrine 30, 19–26. doi: 10.1385/ENDO:30:1:19
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ehrlich, M. (2003). The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin. Immunol. 109, 17–28. doi: 10.1016/S1521-6616(03)00201-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eminli, S., Foudi, A., Stadtfeld, M., Maherali, N., Ahfeldt, T., Mostoslavsky, G., et al. (2009). Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat. Genet. 41, 968–976. doi: 10.1038/ng.428
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eriksson, M., Brown, W. T., Gordon, L. B., Glynn, M. W., Singer, J., Scott, L., et al. (2003). Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423, 293–298. doi: 10.1038/nature01629
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Escobar-Morreale, H. F., Luque-Ramirez, M., and San Millan, J. L. (2005). The molecular-genetic basis of functional hyperandrogenism and the polycystic ovary syndrome. Endocr. Rev. 26, 251–282. doi: 10.1210/er.2004-0004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ess, K. C. (2013). Patient heal thyself: modeling and treating neurological disorders using patient-derived stem cells. Exp. Biol. Med. (Maywood). 238, 308–314. doi: 10.1177/1535370213480713
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Esteller, M., Corn, P. G., Baylin, S. B., and Herman, J. G. (2001). A gene hypermethylation profile of human cancer. Cancer Res. 61, 3225–3229.
Evans, M. J., and Kaufman, M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156.
Festuccia, N., Osorno, R., Wilson, V., and Chambers, I. (2013). The role of pluripotency gene regulatory network components in mediating transitions between pluripotent cell states. Curr. Opin. Genet. Dev. 23, 504–511. doi: 10.1016/j.gde.2013.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fisher, M. B., and Mauck, R. L. (2013). Tissue engineering and regenerative medicine: recent innovations and the transition to translation. Tissue Eng. Part B Rev. 19, 1–13. doi: 10.1089/ten.TEB.2012.0723
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fritz, A. L., Mao, S. R., West, M. G., and Schaffer, D. V. (2014). A medium-throughput analysis of signaling pathways involved in early stages of stem cell reprogramming. Biotechnol. Bioeng. doi: 10.1002/bit.25336. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frobel, J., Hemeda, H., Lenz, M., Abagnale, G., Joussen, S., Denecke, B., et al. (2014). Epigenetic rejuvenation of mesenchymal stromal cells derived from induced pluripotent stem cells. Stem Cell Reports 3, 414–422. doi: 10.1016/j.stemcr.2014.07.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fulda, S., Gorman, A. M., Hori, O., and Samali, A. (2010). Cellular stress responses: cell survival and cell death. Int. J. Cell Biol. 2010:214074. doi: 10.1155/2010/214074
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gafni, O., Weinberger, L., Mansour, A. A., Manor, Y. S., Chomsky, E., Ben-Yosef, D., et al. (2013). Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286. doi: 10.1038/nature12745
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gaj, T., Gersbach, C. A., and Barbas, C. F. 3rd. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gardner, R. L. (2002). Stem cells: potency, plasticity and public perception. J. Anat. 200, 277–282. doi: 10.1046/j.1469-7580.2002.00029.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
George, B. (2011). Regulations and guidelines governing stem cell based products: clinical considerations. Perspect. Clin. Res. 2, 94–99. doi: 10.4103/2229-3485.83228
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gimble, J. M., Katz, A. J., and Bunnell, B. A. (2007). Adipose-derived stem cells for regenerative medicine. Circ. Res. 100, 1249–1260. doi: 10.1161/01.RES.0000265074.83288.09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Graf, T. (2011). Historical origins of transdifferentiation and reprogramming. Cell Stem Cell 9, 504–516. doi: 10.1016/j.stem.2011.11.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grskovic, M., Javaherian, A., Strulovici, B., and Daley, G. Q. (2011). Induced pluripotent stem cells–opportunities for disease modeling and drug discovery. Nat. Rev. Drug Discov. 10, 915–929. doi: 10.1038/nrd3577
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grunz, H. (2001). Developmental biology of amphibians after Hans Spemann in Germany. Int. J. Dev. Biol. 45, 39–50.
Guha, P., Morgan, J. W., Mostoslavsky, G., Rodrigues, N. P., and Boyd, A. S. (2013). Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell 12, 407–412. doi: 10.1016/j.stem.2013.01.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gurdon, J. B. (1962). The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J. Embryol. Exp. Morphol. 10, 622–640.
Gurdon, J. B., and Melton, D. A. (2008). Nuclear reprogramming in cells. Science 322, 1811–1815. doi: 10.1126/science.1160810
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gurtner, G. C., Callaghan, M. J., and Longaker, M. T. (2007). Progress and potential for regenerative medicine. Annu. Rev. Med. 58, 299–312. doi: 10.1146/annurev.med.58.082405.095329
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gurtner, G. C., Werner, S., Barrandon, Y., and Longaker, M. T. (2008). Wound repair and regeneration. Nature 453, 314–321. doi: 10.1038/nature07039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Halley-Stott, R. P., Pasque, V., and Gurdon, J. B. (2013). Nuclear reprogramming. Development 140, 2468–2471. doi: 10.1242/dev.092049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hanna, J., Wernig, M., Markoulaki, S., Sun, C. W., Meissner, A., Cassady, J. P., et al. (2007). Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 318, 1920–1923. doi: 10.1126/science.1152092
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hansson, J., Rafiee, M. R., Reiland, S., Polo, J. M., Gehring, J., Okawa, S., et al. (2012). Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2, 1579–1592. doi: 10.1016/j.celrep.2012.10.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Harries, L. W., Pilling, L. C., Hernandez, L. D., Bradley-Smith, R., Henley, W., Singleton, A. B., et al. (2012). CCAAT-enhancer-binding protein-beta expression in vivo is associated with muscle strength. Aging Cell 11, 262–268. doi: 10.1111/j.1474-9726.2011.00782.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Health, U. S. D. O., Human Services, F., and Drug Administration, C. F. B. E. (2001). Guidance for human somatic cell therapy and gene therapy. Hum. Gene Ther. 12, 303–314. doi: 10.1089/10430340150218431
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heller, J. P., and Martin, K. R. (2014). Enhancing RPE cell-based therapy outcomes for AMD: the role of Bruch's membrane. Transl. Vis. Sci. Technol. 3:11. doi: 10.1167/tvst.3.3.11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hewagama, A., and Richardson, B. (2009). The genetics and epigenetics of autoimmune diseases. J. Autoimmun. 33, 3–11. doi: 10.1016/j.jaut.2009.03.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hibaoui, Y., Grad, I., Letourneau, A., Sailani, M. R., Dahoun, S., Santoni, F. A., et al. (2014). Modeling and rescuing neurodevelopmental defect of down syndrome using induced pluripotent stem cells from monozygotic twins discordant for trisomy 21. EMBO Mol. Med. 6, 259–277. doi: 10.1002/emmm.201302848
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
High, K., Gregory, P. D., and Gersbach, C. (2014). CRISPR technology for gene therapy. Nat. Med. 20, 476–477. doi: 10.1038/nm.3566
Holt, N., Wang, J., Kim, K., Friedman, G., Wang, X., Taupin, V., et al. (2010). Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 28, 839–847. doi: 10.1038/nbt.1663
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biol. 14:R115. doi: 10.1186/gb-2013-14-10-r115
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hou, P., Li, Y., Zhang, X., Liu, C., Guan, J., Li, H., et al. (2013). Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341, 651–654. doi: 10.1126/science.1239278
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, K., Yu, J., Suknuntha, K., Tian, S., Montgomery, K., Choi, K. D., et al. (2011). Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood 117, e109–e119. doi: 10.1182/blood-2010-07-298331
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hutter, G., Nowak, D., Mossner, M., Ganepola, S., Mussig, A., Allers, K., et al. (2009). Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 360, 692–698. doi: 10.1056/NEJMoa0802905
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
International Stem Cell, I., Adewumi, O., Aflatoonian, B., Ahrlund-Richter, L., Amit, M., Andrews, P. W., et al. (2007). Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25, 803–816. doi: 10.1038/nbt1318
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jemal, A., Bray, F., Center, M. M., Ferlay, J., Ward, E., and Forman, D. (2011). Global cancer statistics. CA Cancer J. Clin. 61, 69–90. doi: 10.3322/caac.20107
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jeon, K., Lim, H., Kim, J. H., Thuan, N. V., Park, S. H., Lim, Y. M., et al. (2012). Differentiation and transplantation of functional pancreatic beta cells generated from induced pluripotent stem cells derived from a type 1 diabetes mouse model. Stem Cells Dev. 21, 2642–2655. doi: 10.1089/scd.2011.0665
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jiang, Y., Jahagirdar, B. N., Reinhardt, R. L., Schwartz, R. E., Keene, C. D., Ortiz-Gonzalez, X. R., et al. (2002). Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418, 41–49. doi: 10.1038/nature00870
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kallings, L. O. (2008). The first postmodern pandemic: 25 years of HIV/AIDS. J. Intern. Med. 263, 218–243. doi: 10.1111/j.1365-2796.2007.01910.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kambal, A., Mitchell, G., Cary, W., Gruenloh, W., Jung, Y., Kalomoiris, S., et al. (2011). Generation of HIV-1 resistant and functional macrophages from hematopoietic stem cell-derived induced pluripotent stem cells. Mol. Ther. 19, 584–593. doi: 10.1038/mt.2010.269
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kanherkar, R. R., Bhatia-Dey, N., and Csoka, A. B. (2014). Epigenetics across the human lifespan. Front. Cell Dev. Biol. 2:49. doi: 10.3389/fcell.2014.00049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kator, K., Cristofalo, V., Charpentier, R., and Cutler, R. G. (1985). Dysdifferentiative nature of aging: passage number dependency of globin gene expression in normal human diploid cells grown in tissue culture. Gerontology 31, 355–361.
Kikyo, N., and Wolffe, A. P. (2000). Reprogramming nuclei: insights from cloning, nuclear transfer and heterokaryons. J. Cell Sci. 113(Pt 1), 11–20.
Kim, H., and Kim, J. S. (2014). A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334. doi: 10.1038/nrg3686
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J., Efe, J. A., Zhu, S., Talantova, M., Yuan, X., Wang, S., et al. (2011). Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 108, 7838–7843. doi: 10.1073/pnas.1103113108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kleivi, K. (2006). Advances in the genetics and epigenetics of gene regulation and human disease. Genome Biol. 7:325. doi: 10.1186/gb-2006-7-8-325
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Korf, B. (2008). Hutchinson-Gilford progeria syndrome, aging, and the nuclear lamina. N. Engl. J. Med. 358, 552–555. doi: 10.1056/NEJMp0800071
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kriks, S., Shim, J. W., Piao, J., Ganat, Y. M., Wakeman, D. R., Xie, Z., et al. (2011). Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature 480, 547–551. doi: 10.1038/nature10648
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lang, A. H., Li, H., Collins, J. J., and Mehta, P. (2014). Epigenetic landscapes explain partially reprogrammed cells and identify key reprogramming genes. PLoS Comput. Biol. 10:e1003734. doi: 10.1371/journal.pcbi.1003734
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lapasset, L., Milhavet, O., Prieur, A., Besnard, E., Babled, A., Ait-Hamou, N., et al. (2011). Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 25, 2248–2253. doi: 10.1101/gad.173922.111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ledran, M. H., Krassowska, A., Armstrong, L., Dimmick, I., Renstrom, J., Lang, R., et al. (2008). Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell 3, 85–98. doi: 10.1016/j.stem.2008.06.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lenhoff, H. M. (1991). Ethel Browne, Hans Spemann, and the discovery of the discovery of the oganiser phenomenon. Biol. Bull. 181, 72–80.
Li, J., Song, W., Pan, G., and Zhou, J. (2014). Advances in understanding the cell types and approaches used for generating induced pluripotent stem cells. J. Hematol. Oncol. 7, 50. doi: 10.1186/s13045-014-0050-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, L., and Bhatia, R. (2011). Stem cell quiescence. Clin. Cancer Res. 17, 4936–4941. doi: 10.1158/1078-0432.CCR-10-1499
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liang, G., and Zhang, Y. (2013). Embryonic stem cell and induced pluripotent stem cell: an epigenetic perspective. Cell Res. 23, 49–69. doi: 10.1038/cr.2012.175
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lisanti, S., Tavecchio, M., Chae, Y. C., Liu, Q., Brice, A. K., Thakur, M. L., et al. (2014). Deletion of the mitochondrial chaperone TRAP-1 uncovers global reprogramming of metabolic networks. Cell Rep. 8, 671–677. doi: 10.1016/j.celrep.2014.06.061
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, B., Hunter, D. J., Rooker, S., Chan, A., Paulus, Y. M., Leucht, P., et al. (2013a). Wnt signaling promotes muller cell proliferation and survival after injury. Invest. Ophthalmol. Vis. Sci. 54, 444–453. doi: 10.1167/iovs.12-10774
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, G. H., Barkho, B. Z., Ruiz, S., Diep, D., Qu, J., Yang, S. L., et al. (2011a). Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature 472, 221–225. doi: 10.1038/nature09879
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, G. H., Suzuki, K., Li, M., Qu, J., Montserrat, N., Tarantino, C., et al. (2014). Modeling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat. Commun. 5, 4330. doi: 10.1038/ncomms5330
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, G. H., Suzuki, K., Qu, J., Sancho-Martinez, I., Yi, F., Li, M., et al. (2011b). Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell 8, 688–694. doi: 10.1016/j.stem.2011.04.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, Y., Weick, J. P., Liu, H., Krencik, R., Zhang, X., Ma, L., et al. (2013b). Medial ganglionic eminence-like cells derived from human embryonic stem cells correct learning and memory deficits. Nat. Biotechnol. 31, 440–447. doi: 10.1038/nbt.2565
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Loffredo, F. S., Steinhauser, M. L., Jay, S. M., Gannon, J., Pancoast, J. R., Yalamanchi, P., et al. (2013). Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 153, 828–839. doi: 10.1016/j.cell.2013.04.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Loh, K. M., and Lim, B. (2012). Epigenetics: actors in the cell reprogramming drama. Nature 488, 599–600. doi: 10.1038/488599a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Luna-Zurita, L., and Bruneau, B. G. (2013). Chromatin modulators as facilitating factors in cellular reprogramming. Curr. Opin. Genet. Dev. 23, 556–561. doi: 10.1016/j.gde.2013.07.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lunyak, V. V., and Rosenfeld, M. G. (2008). Epigenetic regulation of stem cell fate. Hum. Mol. Genet. 17, R28–R36. doi: 10.1093/hmg/ddn149
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, T., Xie, M., Laurent, T., and Ding, S. (2013). Progress in the reprogramming of somatic cells. Circ. Res. 112, 562–574. doi: 10.1161/CIRCRESAHA.111.249235
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Manikkam, M., Tracey, R., Guerrero-Bosagna, C., and Skinner, M. K. (2012). Pesticide and insect repellent mixture (permethrin and DEET) induces epigenetic transgenerational inheritance of disease and sperm epimutations. Reprod. Toxicol. 34, 708–719. doi: 10.1016/j.reprotox.2012.08.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Manukyan, M., and Singh, P. B. (2012). Epigenetic rejuvenation. Genes Cells 17, 337–343. doi: 10.1111/j.1365-2443.2012.01595.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Martin, G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638.
Mason, J. L., Jones, J. J., Taniike, M., Morell, P., Suzuki, K., and Matsushima, G. K. (2000). Mature oligodendrocyte apoptosis precedes IGF-1 production and oligodendrocyte progenitor accumulation and differentiation during demyelination/remyelination. J. Neurosci. Res. 61, 251–262. doi: 10.1002/1097-4547(20000801)61:3%3C251::AID-JNR3%3E3.0.CO;2-W
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McClintock, D., Ratner, D., Lokuge, M., Owens, D. M., Gordon, L. B., Collins, F. S., et al. (2007). The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS ONE 2:e1269. doi: 10.1371/journal.pone.0001269
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mehandru, S., Poles, M. A., Tenner-Racz, K., Horowitz, A., Hurley, A., Hogan, C., et al. (2004). Primary HIV-1 infection is associated with preferential depletion of CD4+T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200, 761–770. doi: 10.1084/jem.20041196
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, J. D., Ganat, Y. M., Kishinevsky, S., Bowman, R. L., Liu, B., Tu, E. Y., et al. (2013). Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 13, 691–705. doi: 10.1016/j.stem.2013.11.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Minear, S., Leucht, P., Jiang, J., Liu, B., Zeng, A., Fuerer, C., et al. (2010). Wnt proteins promote bone regeneration. Sci. Transl. Med. 2, 29ra30. doi: 10.1126/scitranslmed.3000231
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mitalipov, S., and Wolf, D. (2009). Totipotency, pluripotency and nuclear reprogramming. Adv. Biochem. Eng. Biotechnol. 114, 185–199. doi: 10.1007/10_2008_45
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mohamet, L., Miazga, N. J., and Ward, C. M. (2014). Familial Alzheimer's disease modeling using induced pluripotent stem cell technology. World J. Stem Cells 6, 239–247. doi: 10.4252/wjsc.v6.i2.239
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Munoz-Najar, U., and Sedivy, J. M. (2011). Epigenetic control of aging. Antioxid. Redox Signal. 14, 241–259. doi: 10.1089/ars.2010.3250
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Natsuka, S., Akira, S., Nishio, Y., Hashimoto, S., Sugita, T., Isshiki, H., et al. (1992). Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood 79, 460–466.
Niu, W., Zang, T., Zou, Y., Fang, S., Smith, D. K., Bachoo, R., et al. (2013). In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 15, 1164–1175. doi: 10.1038/ncb2843
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Niwa, H., Miyazaki, J., and Smith, A. G. (2000). Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376. doi: 10.1038/74199
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Okita, K., Ichisaka, T., and Yamanaka, S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317. doi: 10.1038/nature05934
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pajcini, K. V., Corbel, S. Y., Sage, J., Pomerantz, J. H., and Blau, H. M. (2010). Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell 7, 198–213. doi: 10.1016/j.stem.2010.05.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pandian, G. N., Taniguchi, J., and Sugiyama, H. (2014). Cellular reprogramming for pancreatic beta-cell regeneration: clinical potential of small molecule control. Clin. Transl. Med. 3:6. doi: 10.1186/2001-1326-3-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Papp, B., and Plath, K. (2011). Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Res. 21, 486–501. doi: 10.1038/cr.2011.28
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Papp, B., and Plath, K. (2013). Epigenetics of reprogramming to induced pluripotency. Cell 152, 1324–1343. doi: 10.1016/j.cell.2013.02.043
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parson, A. (2006). The long journey from stem cells to medical product. Cell 125, 9–11. doi: 10.1016/j.cell.2006.03.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Patel, M., and Yang, S. (2010). Advances in reprogramming somatic cells to induced pluripotent stem cells. Stem Cell Rev. 6, 367–380. doi: 10.1007/s12015-010-9123-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Porrello, E. R., and Olson, E. N. (2014). A neonatal blueprint for cardiac regeneration. Stem Cell Res. doi: 10.1016/j.scr.2014.06.003. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qu, F., Wang, F. F., Yin, R., Ding, G. L., El-Prince, M., Gao, Q., et al. (2012). A molecular mechanism underlying ovarian dysfunction of polycystic ovary syndrome: hyperandrogenism induces epigenetic alterations in the granulosa cells. J. Mol. Med. (Berl.) 90, 911–923. doi: 10.1007/s00109-012-0881-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Radzisheuskaya, A., and Silva, J. C. (2014). Do all roads lead to Oct4? the emerging concepts of induced pluripotency. Trends Cell Biol. 24, 275–284. doi: 10.1016/j.tcb.2013.11.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rando, T. A., and Chang, H. Y. (2012). Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell 148, 46–57. doi: 10.1016/j.cell.2012.01.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Richter, H., Randau, L., and Plagens, A. (2013). Exploiting CRISPR/Cas: interference mechanisms and applications. Int. J. Mol. Sci. 14, 14518–14531. doi: 10.3390/ijms140714518
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rivera, C. M., and Ren, B. (2013). Mapping human epigenomes. Cell 155, 39–55. doi: 10.1016/j.cell.2013.09.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rodenhiser, D., and Mann, M. (2006). Epigenetics and human disease: translating basic biology into clinical applications. CMAJ 174, 341–348. doi: 10.1503/cmaj.050774
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rutherford, R. B., Niekrash, C. E., Kennedy, J. E., and Charette, M. F. (1992). Platelet-derived and insulin-like growth factors stimulate regeneration of periodontal attachment in monkeys. J. Periodont. Res. 27, 285–290.
Samson, M., Libert, F., Doranz, B. J., Rucker, J., Liesnard, C., Farber, C. M., et al. (1996). Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725. doi: 10.1038/382722a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sanchez Alvarado, A., and Yamanaka, S. (2014). Rethinking differentiation: stem cells, regeneration, and plasticity. Cell 157, 110–119. doi: 10.1016/j.cell.2014.02.041
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schwartz, S. D., Hubschman, J. P., Heilwell, G., Franco-Cardenas, V., Pan, C. K., Ostrick, R. M., et al. (2012). Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet 379, 713–720. doi: 10.1016/S0140-6736(12)60028-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Seol, Y. J., Kang, H. W., Lee, S. J., Atala, A., and Yoo, J. J. (2014). Bioprinting technology and its applications. Eur. J. Cardiothorac. Surg. 46, 342–348. doi: 10.1093/ejcts/ezu148
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, R. P. (2005). Schizophrenia, epigenetics and ligand-activated nuclear receptors: a framework for chromatin therapeutics. Schizophr. Res. 72, 79–90. doi: 10.1016/j.schres.2004.03.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shen, H. R., Qiu, L. H., Zhang, Z. Q., Qin, Y. Y., Cao, C., and Di, W. (2013). Genome-wide methylated DNA immunoprecipitation analysis of patients with polycystic ovary syndrome. PLoS ONE 8:e64801. doi: 10.1371/journal.pone.0064801
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shyh-Chang, N., Zhu, H., Yvanka De Soysa, T., Shinoda, G., Seligson, M. T., Tsanov, K. M., et al. (2013). Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 155, 778–792. doi: 10.1016/j.cell.2013.09.059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Soza-Ried, J., and Fisher, A. G. (2012). Reprogramming somatic cells towards pluripotency by cellular fusion. Curr. Opin. Genet. Dev. 22, 459–465. doi: 10.1016/j.gde.2012.07.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sterneckert, J. L., Reinhardt, P., and Scholer, H. R. (2014). Investigating human disease using stem cell models. Nat. Rev. Genet. 15, 625–639. doi: 10.1038/nrg3764
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Su, R. J., Baylink, D. J., Neises, A., Kiroyan, J. B., Meng, X., Payne, K. J., et al. (2013). Efficient generation of integration-free ips cells from human adult peripheral blood using BCL-XL together with Yamanaka factors. PLoS ONE 8:e64496. doi: 10.1371/journal.pone.0064496
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Subramanyam, D. (2013). Cellular reprogramming—Turning the clock back. Reson. 18, 514–521. doi: 10.1007/s12045-013-0069-4
Suhr, S. T., Chang, E. A., Tjong, J., Alcasid, N., Perkins, G. A., Goissis, M. D., et al. (2010). Mitochondrial rejuvenation after induced pluripotency. PLoS ONE 5:e14095. doi: 10.1371/journal.pone.0014095
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Svendsen, C. N. (2013). Back to the future: how human induced pluripotent stem cells will transform regenerative medicine. Hum. Mol. Genet. 22, R32–R38. doi: 10.1093/hmg/ddt379
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Szabo, E., Rampalli, S., Risueno, R. M., Schnerch, A., Mitchell, R., Fiebig-Comyn, A., et al. (2010). Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468, 521–526. doi: 10.1038/nature09591
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tachibana, M., Amato, P., Sparman, M., Gutierrez, N. M., Tippner-Hedges, R., Ma, H., et al. (2013). Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 153, 1228–1238. doi: 10.1016/j.cell.2013.05.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takahashi, K. (2012). Cellular reprogramming–lowering gravity on Waddington's epigenetic landscape. J. Cell Sci. 125, 2553–2560. doi: 10.1242/jcs.084822
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takahashi, K., and Yamanaka, S. (2013). Induced pluripotent stem cells in medicine and biology. Development 140, 2457–2461. doi: 10.1242/dev.092551
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Talluri, T. R., Kumar, D., Glage, S., Garrels, W., Ivics, Z., Debowski, K., et al. (2014). Non-viral reprogramming of fibroblasts into induced pluripotent stem cells by sleeping beauty and piggybac transposons. Biochem. Biophys. Res. Commun. 450, 581–587. doi: 10.1016/j.bbrc.2014.06.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tang, S., Yang, L., Tang, X., and Liu, M. (2014). The role of oxidized ATM in the regulation of oxidative stress-induced energy metabolism reprogramming of CAFs. Cancer Lett. 353, 133–144. doi: 10.1016/j.canlet.2014.07.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Teperek, M., and Miyamoto, K. (2013). Nuclear reprogramming of sperm and somatic nuclei in eggs and oocytes. Reprod. Med. Biol. 12, 133–149. doi: 10.1007/s12522-013-0155-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Terai, S., Ishikawa, T., Omori, K., Aoyama, K., Marumoto, Y., Urata, Y., et al. (2006). Improved liver function in patients with liver cirrhosis after autologous bone marrow cell infusion therapy. Stem Cells 24, 2292–2298. doi: 10.1634/stemcells.2005-0542
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Trounson, A., Thakar, R. G., Lomax, G., and Gibbons, D. (2011). Clinical trials for stem cell therapies. BMC Med. 9:52. doi: 10.1186/1741-7015-9-52
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsai, S. Y., Bouwman, B. A., Ang, Y. S., Kim, S. J., Lee, D. F., Lemischka, I. R., et al. (2011). Single transcription factor reprogramming of hair follicle dermal papilla cells to induced pluripotent stem cells. Stem Cells 29, 964–971. doi: 10.1002/stem.649
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Unternaehrer, J. J., and Daley, G. Q. (2011). Induced pluripotent stem cells for modeling human diseases. Philos Trans. R. Soc. Lond. B Biol. Sci. 366, 2274–2285. doi: 10.1098/rstb.2011.0017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Den Veyver, I. B., and Zoghbi, H. Y. (2001). Mutations in the gene encoding methyl-CpG-binding protein 2 cause Rett syndrome. Brain Dev. 23(Suppl. 1), S147–S151. doi: 10.1016/S0387-7604(01)00376-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Van Oevelen, C., Kallin, E. M., and Graf, T. (2013). Transcription factor-induced enhancer modulations during cell fate conversions. Curr. Opin. Genet. Dev. 23, 562–567. doi: 10.1016/j.gde.2013.07.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vierbuchen, T., and Wernig, M. (2012). Molecular roadblocks for cellular reprogramming. Mol. Cell 47, 827–838. doi: 10.1016/j.molcel.2012.09.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Villeda, S. A., Luo, J., Mosher, K. I., Zou, B., Britschgi, M., Bieri, G., et al. (2011). The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477, 90–94. doi: 10.1038/nature10357
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, A., Tang, Z., Park, I. H., Zhu, Y., Patel, S., Daley, G. Q., et al. (2011). Induced pluripotent stem cells for neural tissue engineering. Biomaterials 32, 5023–5032. doi: 10.1016/j.biomaterials.2011.03.070
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, H., Yang, H., Shivalila, C. S., Dawlaty, M. M., Cheng, A. W., Zhang, F., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. doi: 10.1016/j.cell.2013.04.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, K. C., Helms, J. A., and Chang, H. Y. (2009). Regeneration, repair and remembering identity: the three Rs of hox gene expression. Trends Cell Biol. 19, 268–275. doi: 10.1016/j.tcb.2009.03.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Watanabe, A., Yamada, Y., and Yamanaka, S. (2013). Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20120292. doi: 10.1098/rstb.2012.0292
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weinhold, B. (2006). Epigenetics: the science of change. Environ. Health Perspect. 114, A160–A167. doi: 10.1289/ehp.114-a160
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wernig, M., Meissner, A., Foreman, R., Brambrink, T., Ku, M., Hochedlinger, K., et al. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324. doi: 10.1038/nature05944
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, D. C., Boyd, A. S., and Wood, K. J. (2007). Embryonic stem cell transplantation: potential applicability in cell replacement therapy and regenerative medicine. Front. Biosci. 12, 4525–4535. doi: 10.2741/2407
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yeh, E. T., Zhang, S., Wu, H. D., Korbling, M., Willerson, J. T., and Estrov, Z. (2003). Transdifferentiation of human peripheral blood CD34+-enriched cell population into cardiomyocytes, endothelial cells, and smooth muscle cells in vivo. Circulation 108, 2070–2073. doi: 10.1161/01.CIR.0000099501.52718.70
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yingze Zhao, Y. Y. A. Y. W. (2014). Developing CRISPR/Cas9 technologies for research and medicine. MOJ Cell Sci. Report 1, 7.
Yonekawa, Y., and Kim, I. K. (2014). Clinical characteristics and current treatment of age-related macular degeneration. Cold Spring Harb. Perspect. Med. doi: 10.1101/cshperspect.a017178. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, T., Zhang, Z. N., Rong, Z., and Xu, Y. (2011). Immunogenicity of induced pluripotent stem cells. Nature 474, 212–215. doi: 10.1038/nature10135
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhong, X., Gutierrez, C., Xue, T., Hampton, C., Vergara, M. N., Cao, L. H., et al. (2014). Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 5, 4047. doi: 10.1038/ncomms5047
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, Q., Brown, J., Kanarek, A., Rajagopal, J., and Melton, D. A. (2008). In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455, 627–632. doi: 10.1038/nature07314
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, H., Lensch, M. W., Cahan, P., and Daley, G. Q. (2011). Investigating monogenic and complex diseases with pluripotent stem cells. Nat. Rev. Genet. 12, 266–275. doi: 10.1038/nrg2951
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: reprogramming, stem cells, aging, disease, epigenetics
Citation: Kanherkar RR, Bhatia-Dey N, Makarev E and Csoka AB (2014) Cellular reprogramming for understanding and treating human disease. Front. Cell Dev. Biol. 2:67. doi: 10.3389/fcell.2014.00067
Received: 18 June 2014; Accepted: 27 October 2014;
Published online: 12 November 2014.
Edited by:
Dan S. Kaufman, University of Minnesota, USAReviewed by:
In-Hyun Park, Yale University, USAShree Ram Singh, National Cancer Institute, USA
George Scaria, University of Minnesota, USA
Copyright © 2014 Kanherkar, Bhatia-Dey, Makarev and Csoka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonei B. Csoka, Epigenetics Laboratory, Department of Anatomy, Howard University, 520 W St. NW, Mudd 431, Washington, DC 20059, USA e-mail: antonei.csoka@howard.edu
 Riya R. Kanherkar
Riya R. Kanherkar Naina Bhatia-Dey
Naina Bhatia-Dey Evgeny Makarev2
Evgeny Makarev2  Antonei B. Csoka
Antonei B. Csoka