Virtual Nuclear Envelope Breakdown and Its Regulators in Fission Yeast Meiosis
- 1Graduate School of Frontier Biosciences, Osaka University, Suita, Japan
- 2Cell Biology Group, Advanced ICT Research Institute Kobe, National Institute of Information and Communications Technology, Kobe, Japan
- 3Graduate School of Science, Department of Biology, Osaka University, Toyonaka, Japan
Ran, a small GTPase, is required for the spindle formation and nuclear envelope (NE) formation. After NE breakdown (NEBD) during mitosis in metazoan cells, the Ran-GTP gradient across the NE is lost and Ran-GTP becomes concentrated around chromatin, thus affecting the stability of microtubules and promoting the assembly of spindle microtubules and segregation of chromosomes. Mitosis in which chromosomes are segregated subsequent to NEBD is called “open mitosis.” In contrast, many fungi undergo a process termed “closed mitosis” in which chromosome segregation and spindle formation occur without NEBD. Although the fission yeast Schizosaccharomyces pombe undergoes a closed mitosis, it exhibits a short period during meiosis (anaphase of the second meiosis; called “anaphase II”) when nuclear and cytoplasmic proteins are mixed in the presence of intact NE and nuclear pore complexes (NPC). This “virtual” nuclear envelope breakdown (vNEBD) involves changes in the localization of RanGAP1, an activator of Ran-GTP hydrolysis. Recently, Nup132, a component of the structural core Nup107-160 subcomplex of the NPC, has been shown to be involved in the maintenance of the nuclear cytoplasmic barrier in yeast meiosis. In this review, we highlight the possible roles of RanGAP1 and Nup132 in vNEBD and discuss the biological significance of vNEBD in S. pombe meiosis.
Introduction
In eukaryotic cells, the nucleus is enclosed by a nuclear envelope (NE) in interphase. The NE is a double membrane structure which separates the nucleoplasm from the cytoplasm. Macromolecules are transported between the nucleus and the cytoplasm across the NE through nuclear pores which are formed by large protein complexes called nuclear pore complexes (NPCs; Reichelt et al., 1990). The NPC has an eight-fold rotational symmetry structure (Hinshaw et al., 1992; Akey and Radermacher, 1993; Kiseleva et al., 1996) and is composed of ~30 kinds of proteins called nucleoporins (Yang et al., 1998). Nucleoporins can be classified into several groups according to their localization and function: membrane-integrated nucleoporins, cytoplasmic filaments, scaffold subcomplexes, adaptor subcomplexes, central channels, and nuclear baskets (Rout et al., 2000; Cronshaw et al., 2002; Mans et al., 2004; Osmani et al., 2006; Alber et al., 2007; DeGrasse et al., 2009; Iwamoto et al., 2009; Tamura et al., 2010; Asakawa et al., 2014). Distinct nucleoporin subclasses are involved in Ran-dependent nucleocytoplasmic transport as described below.
Nucleocytoplasmic transport depends on the activity of Ran, a small GTPase (Moore and Blobel, 1993; Moroianu and Blobel, 1995). Ran exists in two forms and is bound to either GTP or GDP (Scheffzek et al., 1995; Vetter et al., 1999). The GTP-bound form of Ran (Ran-GTP) is predominantly nuclear but also exists at low levels in the cytoplasm (Bischoff and Ponstingl, 1991b). This gradient of Ran-GTP between the nucleus and the cytoplasm establishes and maintains the direction of nucleocytoplasmic transport (Izaurralde et al., 1997; Nachury and Weis, 1999; Kaláb et al., 2006). The gradient of Ran-GTP is generated based on the localization of two important proteins called Ran GTPase activating protein 1 (RanGAP1) and Ran guanine nucleotide exchange factor (RanGEF/RCC1; Izaurralde et al., 1997; Nachury and Weis, 1999; Kaláb et al., 2006). RanGAP1, which converts Ran-GTP to Ran-GDP, is cytoplasmic (Bischoff et al., 1994; Becker et al., 1995; Seewald et al., 2002). In humans, RanGAP1 has been shown to require sumoylation within its C-terminal domain by Nup358/RanBP2 for localization to NPCs (Matunis et al., 1996, 1998; Mahajan et al., 1997; Saitoh et al., 1997). Nup358/RanBP2, a cytoplasmic filament nucleoporin, is a small ubiquitin-like modifier (SUMO) E3 ligase complex that catalyzes SUMO post-translational modification to target proteins (Pichler et al., 2002). In this way, RanGAP1 activity is concentrated at the cytoplasmic side of the NPC. On the other hand, RanGEF/RCC1, which converts Ran-GDP to Ran-GTP, is a chromatin-associated protein and its activity is concentrated in the nucleus (Ohtsubo et al., 1987, 1989; Bischoff and Ponstingl, 1991a,b; Figure 1A). This biased localization of RanGAP1 and RanGEF/RCC1 generates the gradient of Ran-GTP between the nucleus and the cytoplasm.
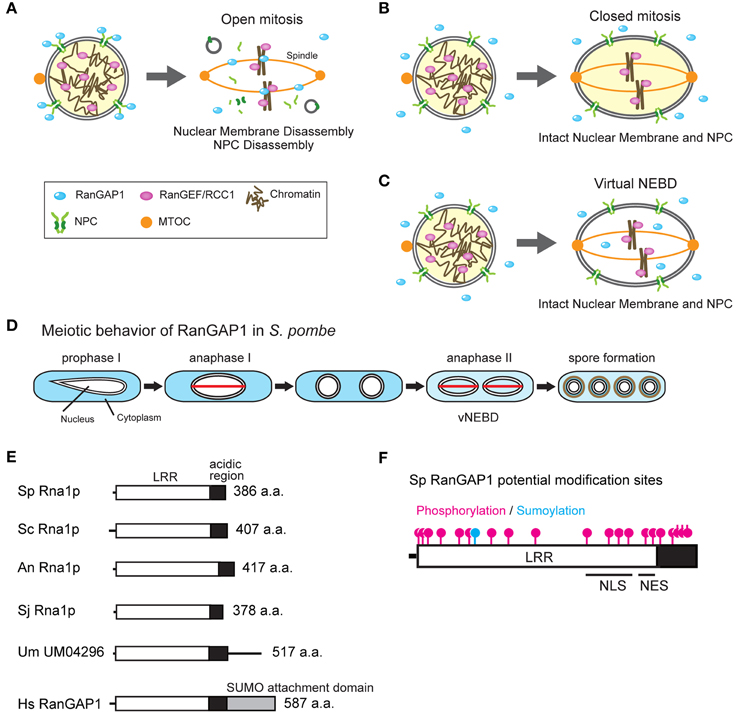
Figure 1. (A-C) Open and closed mitosis, and virtual nuclear envelope breakdown. (A) Interphase nucleus (left) and open mitosis (right) in metazoa. In interphase, the RanGAP1 is associated with the nuclear pore complex (NPC) whereas the RanGEF/RCC1 is associated with chromatin, maintaining the Ran-GTP gradient. In open mitosis, the nuclear membrane and NPCs are disassembled at the beginning of mitosis and nuclear and cytoplasmic proteins are mixed together. Mitotic spindle is elongated from both poles of the microtubule organizing center (MTOC) and capture kinetochores. SUMO-conjugated RanGAP1 associates with mitotic kinetochores and spindle microtubules and the remaining RanGAP1 diffuses throughout the cell. The disassembled nuclear membranes (shown as gray circles) are absorbed into the ER (Ellenberg et al., 1997; Yang et al., 1997). (B) Interphase nucleus (left) and closed mitosis (right) in fungi. In interphase, the Ran-GTP gradient is maintained by cytoplasmic RanGAP1 and chromatin-bound RanGEF/RCC1. In closed mitosis, the nuclear membrane and the NPCs remain intact, and the Ran-GTP gradient across the NE is maintained. Mitotic spindle is formed in the nucleus between the MTOCs which are penetrated into the NE. (C) Virtual nuclear envelope breakdown. During meiosis II in S. pombe, both the nuclear membrane and NPCs remain intact but RanGAP1 enters the nucleus, resulting in the abolishment of the Ran-GTP gradient. (D) Behavior of RanGAP1 during meiosis. RanGAP1 (blue) remains in the cytoplasm during the entire meiotic process, except anaphase II. During anaphase II, RanGAP1 diffuses throughout the cell. The red lines shown in anaphase I and II represent spindles. (E) Domain structure of RanGAP1. Molecular domains of RanGAP1 in eukaryotes are shown. Leucine-rich repeat (LRR; white box), acidic region (black box), and SUMO attachment domain (gray box) are indicated. Sp, S. pombe; Sc, S. cerevisiae; An, A. nidulans; Sj, S. japonicus; Um, U. maydis; Hs, Homo sapiens. U. maydis homolog UM04296 contains an additional amino acid sequence after the acidic region; however, the function of this sequence is unknown. (F) Potential post-translational modification sites, NLS, and NES in S. pombe RanGAP1. Magenta and blue indicate potential phosphorylation and sumoylation sites, respectively, that have been predicted using the sequence of S. pombe RanGAP1 (Asakawa et al., 2011). The putative NLS and NES have been predicted by Feng et al. (1999).
For nucleocytoplasmic transport, most proteins and RNA are carried by transport receptors of the importin β superfamily such as importin β or exportin. These proteins are structurally similar (reviewed in Conti et al., 2006), but are distinguished by their binding properties to cargos upon binding to Ran-GTP. Importin β releases its cargo when it binds to Ran-GTP in the nucleus and binds to its cargo in the cytoplasm where Ran-GTP is hydrolyzed to GDP. In contrast, exportin binds to its cargo in the nucleus when it binds to Ran-GTP and releases its cargo in the cytoplasm where Ran-GTP is hydrolyzed to GDP (reviewed in Cook et al., 2007). In mammals, there are 14–20 members of the importin β/exportin superfamily (reviewed in Kimura and Imamoto, 2014). Thus, the Ran-GTP gradient plays a critical role in determining the direction of nucleocytoplasmic transport.
Variation of Mitosis
During mitosis in metazoa, NE breakdown (NEBD) occurs (open mitosis) (reviewed in Güttinger et al., 2009; Figure 1A). NEBD causes diffusion of nuclear and cytoplasmic molecules within the entire cell, resulting in loss of the compartmentalization of guanine nucleotide-bound forms of Ran on either side of the NE. Simultaneously, SUMO-conjugated RanGAP1 associates with mitotic kinetochores and spindle microtubules and the remaining RanGAP1 diffuses throughout the cell (Joseph et al., 2002, 2004). In contrast, RanGEF/RCC1 remains associated with chromatin and thus the Ran-GTP gradient is shifted and remained only close regions around chromosomes (Kaláb et al., 2006). In metazoa, NEBD is essential for chromosome segregation because mitotic spindles are formed from cytosolic microtubule organizing centers (MTOCs) or centrosomes that exist at opposite ends of the cell and function to capture kinetochores after NEBD. In contrast, many fungi undergo mitosis without NEBD (closed mitosis) (Heath, 1980; Figure 1B). During closed mitosis, the spindle is formed in the nucleus between the spindle pole bodies (equivalent to centrosomes in metazoa) which are embedded in the NE, and chromosomes segregate without NEBD (Figure 1B). This manner of mitosis is observed in many fungi including yeasts.
In addition to closed mitosis, recent research has revealed unique types of mitoses in some fungal species (De Souza et al., 2004). In the filamentous fungus Aspergillus nidulans, “semi-open” mitosis has been reported (De Souza et al., 2004). During mitosis in A. nidulans, the NE remains largely intact but NPCs undergo partial disassembly. In this type of mitosis, peripheral nucleoporins that form cytoplasmic filaments, adaptor subcomplexes, central channels, and nuclear baskets are disassembled, leaving structural core nucleoporins such as the Nup107-160 subcomplex and membrane-integrated nucleoporins in the NE (De Souza et al., 2004; Osmani et al., 2006). Disassembly of these nucleoporins disrupts the compartmentalization of the nucleus and alters the localization of A. nidulans RanGAP1 during mitosis, resulting in the relocalization of RanGAP1, which is conventionally localized only to the cytoplasm during interphase, to both the cytoplasm and the nucleus during mitosis (De Souza et al., 2004). Nucleoporin disassembly in A. nidulans requires NIMA and CDK kinases. Particularly, the kinase activity of NIMA is associated with the phosphorylation of nucleoporin Nup98 in A. nidulans (De Souza et al., 2004). This is similar to NPC disassembly observed during open mitosis in metazoans where Nup98 is phosphorylated by NIMA and CDK kinases. The hypo-phosphorylated mutant of Nup98 delays NPC disassembly (Laurell et al., 2011).
The fission yeast Schizosaccharomyces japonicus exhibits another type of semi-open mitosis in which the nuclear envelope ruptures in anaphase (Aoki et al., 2011; Yam et al., 2011; Gu et al., 2012). In this organism, the mitotic spindle is formed in the nucleus and undergoes elongation in the limited nuclear space to form a bent spindle (Yam et al., 2011). The NE of S. japonicus is ruptured in the medial region when the nucleus elongates during anaphase (Aoki et al., 2011; Yam et al., 2011). The semi-open mitosis in S. japonicus involves APC/C activity that induces the degradation of Oar2, a 3-oxoacyl-[acyl-carrier-protein] reductase (Aoki et al., 2013). Oar2 is a conserved protein that elongates fatty acids and makes phospholipids, a source for cellular membranes (Schneider et al., 1997). The degradation of Oar2 likely decreases the sources required for membrane synthesis and leads to the breakage of the NE in S. japonicus during mitosis.
The corn smut basidiomycete fungus Ustilago maydis undergoes “open” mitosis, in which the nuclear envelope is disassembled upon entry into mitosis. This organism shows growth dimorphism of yeast and hyphae forms. In the yeast form of U. maydis, the nucleus elongates and moves from the mother cell to the daughter cell. When the nucleus reaches the daughter cell, the NE ruptures at the leading edge of the stretched nucleus and recedes into the mother cell (Straube et al., 2005). During this process, NPCs disassemble and disperse into the cytoplasm similar to that observed in organisms undergoing open mitosis (Theisen et al., 2008). These data indicate that these special modes of mitosis in A. nidulans, U. maydis, and S. japonicus require partial disruption of the NE integrity or alteration of NPC composition.
vNEBD and Its Regulators
RanGAP1
A special mechanism that reduces the Ran-GTP gradient during chromosome segregation in fission yeast S. pombe has been reported. S. pombe undergoes closed mitosis and meiosis I during which nuclear transport activity is maintained. However, during anaphase in meiosis II (i.e., anaphase II), nuclear and cytoplasmic molecules are mixed similar to open mitosis. Interestingly, both the NE and NPCs are maintained in this phase and thus this phenomenon is called “virtual” NEBD (vNEBD; Figure 1C; Arai et al., 2010; Asakawa et al., 2010).
vNEBD coincides with altered RanGAP1 localization from the cytoplasm to the nucleus (see “anaphase II” in Figure 1D). RCC1/RanGEF remains associated with chromatin throughout anaphase II (Arai et al., 2010; Asakawa et al., 2010). Ectopic expression of RanGAP1 fused with a nuclear localization signal (NLS) results in the mislocalization of GFP-NLS from the nucleus to the cytoplasm (Asakawa et al., 2010). These observations suggest that altered RanGAP1 localization abolishes the Ran-GTP gradient at the nucleus. Mobile characteristics of RanGAP1 in S. pombe may be due to its protein structure (Figure 1E). In humans, RanGAP1 is sumoylated at the C-terminal and is targeted to the cytoplasmic filaments of NPCs (Matunis et al., 1996; Mahajan et al., 1997; Saitoh et al., 1997). In organisms undergoing closed mitosis, including S. pombe, RanGAP1 does not have the C-terminal sumoylation domain (Figure 1E). In fact, S. pombe RanGAP1 is a cytoplasmic protein and is not associated with the NE (Figure 1C; Arai et al., 2010; Asakawa et al., 2010). On the other hand, S. pombe RanGAP1 contains several residues that may undergo post-translational modifications, including phosphorylation and N-terminal sumoylation (Figure 1F). Amino acid substitution of some of these residues in endogenous RanGAP1 results in cell death, thus these residues are important for the function of RanGAP1 (Asakawa et al., 2011). However, it is unknown whether these residues are required for vNEBD.
RanGAP1 of budding yeast Saccharomyces cerevisiae contains an NLS and a nuclear export signal (NES) that regulate its localization (Feng et al., 1999). These NLS and NES sequences are also conserved in S. pombe RanGAP1 (Figure 1F; Feng et al., 1999). Although it is unclear whether these sequences function as NLS and NES, it has been observed that S. pombe RanGAP1 binds to histone H3 and is required for heterochromatin assembly during vegetative growth (Nishijima et al., 2006), suggesting that RanGAP1, at least in part, is actively transported into the nucleus. This may explain how alteration of RanGAP1 localization actively occurs during vNEBD.
Nucleoporins
A recent study showed that there are eight non-essential nucleoporins, the mutation of which is associated with defective production of spores per ascus (Asakawa et al., 2014), suggesting their involvement in vNEBD. The phenotype of the nucleoporin gene nup132 disruption mutant has been reported recently (Yang et al., 2015). S. pombe Nup132 is a homolog of Nup133, which is a component of a conserved Nup107-160 subcomplex of NPCs (Lutzmann et al., 2002; Harel et al., 2003; Walther et al., 2003; Baï et al., 2004; Osmani et al., 2006; DeGrasse et al., 2009; Tamura et al., 2010; Figure 2A). The S. pombe nup132 mutant shows two major meiotic phenotypes (Figure 2B): (1) untimely kinetochore assembly during prophase of meiosis I (prophase I) and (2) loss of barrier function of the NE during anaphase of meiosis I (anaphase I; Yang et al., 2015). Notably, in this mutant, overall nuclear permeability during meiotic prophase I is not affected.
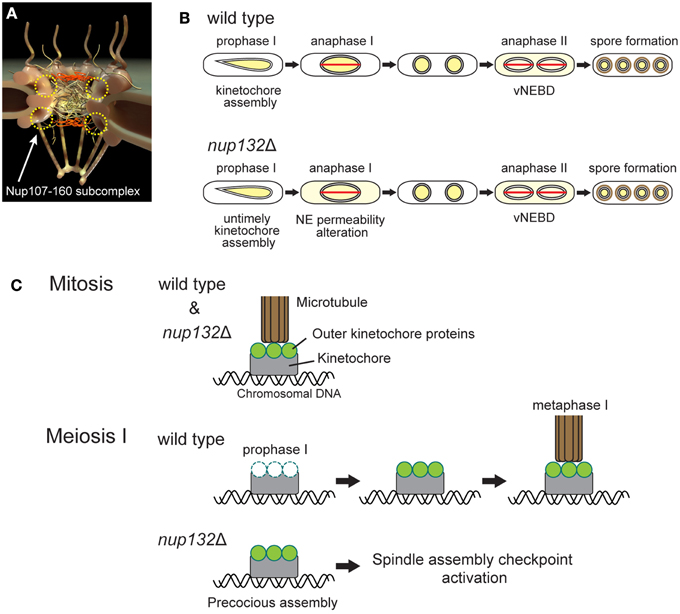
Figure 2. Meiotic phenotypes of nup132 deletion mutant cells. (A) NPC structure. The yellow circles indicate the putative positions of the Nup107-160 subcomplex. (B) Comparison of nuclear protein behavior between wild-type and nup132 mutant (nup132Δ) cells. Yellow indicates the localization site of nuclear proteins. The red lines shown in anaphase I and II represent spindles. (C) Attachment of kinetochores and microtubules during mitosis (upper panel) and meiosis I (lower two panels). During mitosis in wild-type and nup132Δ cells, kinetochores (gray) are connected to microtubules (brown) through outer kinetochore proteins (green). During meiotic prophase I in wild-type cells, the outer kinetochore proteins (green) disassemble and reassemble at the kinetochores to form normal kinetochores for meiosis I. In contrast, nup132Δ cells show a precocious assembly of the outer kinetochore proteins (green).
Our discovery raises the question of how Nup132 regulates both kinetochore assembly and NE permeability during meiosis I in S. pombe. Interestingly, the metazoan Nup107-160 subcomplex is known to re-localize to the kinetochores upon NEBD and functions in kinetochore-microtubule attachment (Loïodice et al., 2004; Orjalo et al., 2006; Zuccolo et al., 2007; Mishra et al., 2010), hinting at relocalization of Nup132 to the kinetochore during S. pombe meiosis I. In addition, Nup133, a human homolog of S. pombe Nup132, is required for proper localization of RanGAP1 at the kinetochore in human cells (Zuccolo et al., 2007). However, because of closed meiosis I in yeast, Nup132 remains at the NE (Asakawa et al., 2010), and there is no evidence of Nup132 relocalization to the kinetochores. The effect of Nup132 on kinetochores in the absence of NEBD should be investigated in future studies.
Inadequate kinetochore-microtubule attachment in the nup132 mutant activates the spindle assembly checkpoint and delays meiosis I progression (Figure 2C; Yang et al., 2015). The duration of meiosis II is also prolonged in this mutant, but this prolongation is independent of the spindle assembly checkpoint (Yang et al., 2015). Alternatively, loss of barrier function of the NE during anaphase I might induce defects in subsequent nuclear division during meiosis II. This is consistent with the overexpression of Sid1, a component of the septation initiation network (SIN) signaling pathway, that induces the precocious leakage of nuclear proteins through the NE during meiosis I, leading to defects in chromosome segregation (Arai et al., 2010). These results highlight the importance of temporal control of vNEBD during anaphase II.
As mentioned above, Nup132 depletion results in the loss of barrier function of the NE during anaphase I (Yang et al., 2015), suggesting that altered permeability of the NE depends on the function of only one nucleoporin during meiosis. This is supported by the fact that depletion of S. cerevisiae Nup133 results in Ran being uniformly localized in the cell rather than being enriched in the nucleus (Gao et al., 2003). How does Nup132 regulate the permeability or barrier function of the NE? One explanation is that in both S. pombe and S. cerevisiae, Nup132/Nup133 is required for the uniform distribution of NPCs along the NE (Doye et al., 1994; Li et al., 1995; Pemberton et al., 1995; Baï et al., 2004) and that unevenly distributed NPCs hamper nucleocytoplasmic transport (Steinberg et al., 2012). A second explanation is that Nup132 depletion may disrupt the structural integrity of NPCs because Nup132 is a component of the Nup107-160 subcomplex that forms the NPC scaffold structure. Thus, Nup132 depletion may lead to the partial disassembly of NPCs and induce “semi-open” meiosis I. A third explanation is that Nup132 depletion induces early vNEBD during anaphase I through yet unknown mechanisms involved in cell cycle progression. Identification of GFP-fused nucleoporins and NE proteins in the nup132 mutant will help determine whether changes in nuclear permeability during anaphase I involve NPC disassembly or NE rupture.
Functional alterations in Nup132 or other nucleoporins may change the barrier function of the NE without inducing NPC disassembly. Post-translational modifications in nucleoporins may alter the barrier function of NPCs. In mammals, nucleoporin Nup50 is phosphorylated by ERK kinase, which in turn changes the affinity of importin β to NPCs (Kosako et al., 2009). Thus, the barrier function of NPCs during meiosis in S. pombe may be regulated by phosphorylation or other post-translational modifications of nucleoporins. Further studies are required to elucidate the role of post-translational modifications of Nup132 or other nucleoporins in the regulation of barrier function of NPCs and vNEBD during meiosis in S. pombe.
Cdc2 Kinase
The timing of vNEBD corresponds to meiosis II during which S. pombe cells produce spores, and a correlation of vNEBD with spore formation has been established (Arai et al., 2010; Asakawa et al., 2010). Spore formation and meiotic nuclear division are coordinately regulated by Cdc2 kinase (Nakaseko et al., 1984; Grallert and Sipiczki, 1991). Similar to that observed during mitosis, Cdc2 kinase activity increases at the onset of meiosis I and dramatically decreases upon the completion of meiosis I. Transition from meiosis I to meiosis II also requires an increase in Cdc2 activity. The mes1 mutant, which is deficient in blocking the degradation of cyclin Cdc13 (Izawa et al., 2005), arrests before meiosis II due to insufficient re-activation of Cdc2 for starting meiosis II (Izawa et al., 2005; Kimata et al., 2008, 2011). Moreover, the mes1 mutant shows no vNEBD (Arai et al., 2010; Asakawa et al., 2010) and is defective in spore formation (Shimoda et al., 1985; Izawa et al., 2005), suggesting a correlation between them. In contrast, the tws1 mutant, a meiosis-specific allelic mutant of Cdc2 (MacNeill et al., 1991), does not undergo meiosis II and forms two diploid spores after meiosis I (Nakaseko et al., 1984). This mutant allele is thought to affect interactions between Cdc2 and its binding proteins (MacNeill et al., 1991). In the tws1 mutant, nuclear proteins diffuse to the cytoplasm and RanGAP1 localizes to the nucleus during meiosis I (Arai et al., 2010; Asakawa et al., 2010), suggesting that hypophosphorylation of a Cdc2 substrate during meiosis I may possibly drive vNEBD. It is unclear whether Cdc2 regulates vNEBD through nucleoporins and/or RanGAP1 or through lipid metabolism pathways similar to that observed during semi-open mitosis in S. japonicus (Aoki et al., 2013).
Significance of vNEBD
vNEBD occurs specifically during anaphase II and results in the diffusion of nuclear proteins into the cytoplasm and cytoplasmic proteins into the nucleus, which is similar to that observed during open mitosis. vNEBD may allow cytoplasmic proteins to function in the nucleus during or after anaphase II. Occurrence of vNEBD is correlated with spore formation (Arai et al., 2010; Asakawa et al., 2010). S. pombe produces spores under nutritionally starved conditions. These spores are metabolically inactive until they experience growth-favorable conditions (Shimoda and Nakamura, 2004). Because of their dormancy, it is tempting to think that the nuclei of these spores are transcriptionally silent. NPCs are suggested to be involved in gene regulation and chromatin organization (Therizols et al., 2006; Zuccolo et al., 2007; Van de Vosse et al., 2013; Pascual-Garcia and Capelson, 2014; Breuer and Ohkura, 2015; Yang et al., 2015). Based on these data and the fact that nuclear RanGAP1 triggers heterochromatin formation in S. pombe (Nishijima et al., 2006), we propose that functional alterations in NPCs that accompany vNEBD enable global chromatin reorganization for generating dormant nuclei.
Conclusion
The phenomenon of vNEBD suggests the importance of “opening” the gate between the nucleus and cytoplasm during meiosis. Nucleoporins and RanGAP1 may play key roles during open meiosis without physically breaking down the NE in S. pombe. This provides an example in which the regulated barrier function of the NE plays an important role for regulating meiotic processes in eukaryotes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. D.B. Alexander for critical reading of this paper. This work was supported by JSPS KAKENHI: Grant Number 26440098 to HA, 13F03384 to HY, 26251037 to YH, and 26291007 and 25116006 to TH. HY was a JSPS postdoctoral fellow.
References
Akey, C. W., and Radermacher, M. (1993). Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol. 122, 1–19. doi: 10.1083/jcb.122.1.1
Alber, F., Dokudovskaya, S., Veenhoff, L. M., Zhang, W., Kipper, J., Devos, D., et al. (2007). The molecular architecture of the nuclear pore complex. Nature 450, 695–701. doi: 10.1038/nature06405
Aoki, K., Hayashi, H., Furuya, K., Sato, M., Takagi, T., Osumi, M., et al. (2011). A Breakage of the nuclear envelope by an extending mitotic nucleus occurs during anaphase in Schizosaccharomyces japonicus. Genes Cells 16, 911–926. doi: 10.1111/j.1365-2443.2011.01540.x
Aoki, K., Shiwa, Y., Takada, H., Yoshikawa, H., and Niki, H. (2013). Regulation of nuclear envelope dynamics via APC/C is necessary for the progression of semi-open mitosis in Schizosaccharomyces japonicus. Genes Cells 18, 733–752. doi: 10.1111/gtc.12072
Arai, K., Sato, M., Tanaka, K., and Yamamoto, M. (2010). Nuclear compartmentalization is abolished during fission yeast meiosis. Curr. Biol. 20, 1913–1918. doi: 10.1016/j.cub.2010.09.004
Asakawa, H., Hiraoka, Y., and Haraguchi, T. (2011). Physical breakdown of the nuclear envelope is not necessary for breaking its barrier function. Nucleus 2, 523–526. doi: 10.4161/nucl.2.6.16117
Asakawa, H., Kojidani, T., Mori, C., Osakada, H., Sato, M., Ding, D.-Q., et al. (2010). Virtual breakdown of the nuclear envelope in fission yeast meiosis. Curr. Biol. 20, 1919–1925. doi: 10.1016/j.cub.2010.09.070
Asakawa, H., Yang, H.-J., Yamamoto, T. G., Ohtsuki, C., Chikashige, Y., Sakata-Sogawa, K., et al. (2014). Characterization of nuclear pore complex components in fission yeast Schizosaccharomyces pombe. Nucleus 5, 149–162. doi: 10.4161/nucl.28487
Baï, S. W., Rouquette, J., Umeda, M., Faigle, W., Loew, D., Sazer, S., et al. (2004). The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol. Cell Biol. 24, 6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004
Becker, J., Melchior, F., Gerke, V., Bischoff, F. R., Ponstingl, H., and Wittinghofer, A. (1995). RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J. Biol. Chem. 270, 11860–11865. doi: 10.1074/jbc.270.20.11860
Bischoff, F. R., Klebe, C., Kretschmer, J., Wittinghofer, A., and Ponstingl, H. (1994). RanGAP1induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. U.S.A. 91, 2587–2591. doi: 10.1073/pnas.91.7.2587
Bischoff, F. R., and Ponstingl, H. (1991a). Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354, 80–82. doi: 10.1038/354080a0
Bischoff, F. R., and Ponstingl, H. (1991b). Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc. Natl. Acad. Sci. U.S.A. 88, 10830–10834. doi: 10.1073/pnas.88.23.10830
Breuer, M., and Ohkura, H. (2015). A negative loop within the nuclear pore complex controls global chromatin organization. Genes Dev. 29, 1789–1794. doi: 10.1101/gad.264341.115
Conti, E., Müller, C. W., and Stewart, M. (2006). Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 16, 237–244. doi: 10.1016/j.sbi.2006.03.010
Cook, A., Bono, F., Jinek, M., and Conti, E. (2007). Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671. doi: 10.1146/annurev.biochem.76.052705.161529
Cronshaw, J. M., Krutchinsky, A. N., Zhang, W., Chait, B. T., and Matunis, M. J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915–927. doi: 10.1083/jcb.200206106
DeGrasse, J. A., DuBois, K. N., Devos, D., Siegel, T. N., Sali, A., Field, M. C., et al. (2009). Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics 8, 2119–2130. doi: 10.1074/mcp.M900038-MCP200
De Souza, C. P., Osmani, A. H., Hashmi, S. B., and Osmani, S. A. (2004). Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14, 1973–1984. doi: 10.1016/j.cub.2004.10.050
Doye, V., Wepf, R., and Hurt, E. C. (1994). A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 13, 6062–6075.
Ellenberg, J., Siggia, E. D., Moreira, J. E., Smith, C. L., Presley, J. F., Worman, H. J., et al. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193–1206. doi: 10.1083/jcb.138.6.1193
Feng, W., Benko, A. L., Lee, J.-H., Stanford, D. R., and Hopper, A. K. (1999). Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J. Cell Sci. 112, 339–347.
Gao, H., Sumanaweera, N., Bailer, S. M., and Stochaj, U. (2003). Nuclear accumulation of the small GTPase Gsp1p depends on nucleoporins Nup133p, Rat2p/Nup120p, Nup85p, Nic96p, and the acetyl-CoA carboxylase Acc1p. J. Biol. Chem. 278, 25331–25340. doi: 10.1074/jbc.M301607200
Grallert, B., and Sipiczki, M. (1991). Common genes and pathways in the regulation of the mitotic and meiotic cell cycles of Schizosaccharomyces pombe. Curr. Genet. 20, 199–204. doi: 10.1007/BF00326233
Gu, Y., Yam, C., and Oliferenko, S. (2012). Divergence of mitotic strategies in fission yeasts. Nucleus 3, 220–225. doi: 10.4161/nucl.19514
Güttinger, S., Laurell, E., and Kutay, U. (2009). Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell. Biol. 10, 178–191. doi: 10.1038/nrm2641
Harel, A., Orjalo, A. V., Vincent, T., Lachish-Zalait, A., Vasu, S., Shah, S., et al. (2003). Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11, 853–864. doi: 10.1016/S1097-2765(03)00116-3
Heath, B. (1980). Variant mitoses in lower eukaryotes: indicators of the evolution of mitosis? Int. Rev. Cytol. 64, 1–80. doi: 10.1016/S0074-7696(08)60235-1
Hinshaw, J. E., Carragher, B. O., and Milligan, R. A. (1992). Architecture and design of the nuclear pore complex. Cell 69, 1133–1141. doi: 10.1016/0092-8674(92)90635-P
Iwamoto, M., Mori, C., Kojidani, T., Bunai, F., Hori, T., Fukagawa, T., et al. (2009). Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate Tetrahymena. Curr. Biol. 19, 843–847. doi: 10.1016/j.cub.2009.03.055
Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I. W., and Görlich, D. (1997). The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 16, 6535–6547. doi: 10.1093/emboj/16.21.6535
Izawa, D., Goto, M., Yamashita, A., Yamano, H., and Yamamoto, M. (2005). Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature 434, 529–533. doi: 10.1038/nature03406
Joseph, J., Liu, S. T., Jablonski, S. A., Yen, T. J., and Dasso, M. (2004). The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14, 611–617. doi: 10.1016/j.cub.2004.03.031
Joseph, J., Tan, S. H., Karpova, T. S., McNally, J. G., and Dasso, M. (2002). SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156, 595–602. doi: 10.1083/jcb.200110109
Kaláb, P., Pralle, A., Isacoff, E. Y., Heald, R., and Weis, K. (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697–701. doi: 10.1038/nature04589
Kimata, Y., Kitamura, K., Fenner, N., and Yamano, H. (2011). Mes1 controls the meiosis I to meiosis II transition by distinctly regulating the anaphase-promoting complex/cyclosome coactivators Fzr1/Mfr1 and Slp1 in fission yeast. Mol. Biol. Cell 22, 1486–1494. doi: 10.1091/mbc.E10-09-0774
Kimata, Y., Trickey, M., Izawa, D., Gannon, J., Yamamoto, M., and Yamano, H. (2008). A mutual inhibition between APC/C and its substrate Mes1 required for meiotic progression in fission yeast. Dev. Cell 14, 446–454. doi: 10.1016/j.devcel.2007.12.010
Kimura, M., and Imamoto, N. (2014). Biological significance of the importin-β family-dependent nucleocytoplasmic transport pathways. Traffic 15, 727–748. doi: 10.1111/tra.12174
Kiseleva, E., Goldberg, M. W., Daneholt, B., and Allen, T. D. (1996). RNP export is mediated by structural reorganization of the nuclear pore basket. J. Mol. Biol. 260, 304–311. doi: 10.1006/jmbi.1996.0401
Kosako, H., Yamaguchi, N., Aranami, C., Ushiyama, M., Kose, S., Imamoto, N., et al. (2009). Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat. Struct. Mol. Biol. 16, 1026–1035. doi: 10.1038/nsmb.1656
Laurell, E., Beck, K., Krupina, K., Theerthagiri, G., Bodenmiller, B., Horvath, P., et al. (2011). Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144, 539–550. doi: 10.1016/j.cell.2011.01.012
Li, O., Heath, C. V., Amberg, D. C., Dockendorff, T. C., Copeland, C. S., Snyder, M., et al. (1995). Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol. Biol. Cell 6, 401–417. doi: 10.1091/mbc.6.4.401
Loïodice, I., Alves, A., Rabut, G., Van Overbeek, M., Ellenberg, J., Sibarita, J. B., et al. (2004). The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell 15, 3333–3344. doi: 10.1091/mbc.E03-12-0878
Lutzmann, M., Kunze, R., Buerer, A., Aebi, U., and Hurt, E. (2002). Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21, 387–397. doi: 10.1093/emboj/21.3.387
MacNeill, S. A., Creanor, J., and Nurse, P. (1991). Isolation, characterisation and molecular cloning of new mutant alleles of the fission yeast p34cdc2+ protein kinase gene: identification of temperature-sensitive G2-arresting alleles. Mol. Gen. Genet. 229, 109–118. doi: 10.1007/BF00264219
Mahajan, R., Delphin, C., Guan, T., Gerace, L., and Melchior, F. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97–107. doi: 10.1016/S0092-8674(00)81862-0
Mans, B. J., Anantharaman, V., Aravind, L., and Koonin, E. V. (2004). Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 3, 1612–1637. doi: 10.4161/cc.3.12.1316
Matunis, M. J., Coutavas, E., and Blobel, G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470. doi: 10.1083/jcb.135.6.1457
Matunis, M. J., Wu, J., and Blobel, G. (1998). SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 140, 499–509. doi: 10.1083/jcb.140.3.499
Mishra, R. K., Chakraborty, P., Arnaoutov, A., Fontoura, B. M., and Dasso, M. (2010). The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 12, 164–169. doi: 10.1038/ncb2016
Moore, M. S., and Blobel, G. (1993). The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 365, 661–663. doi: 10.1038/365661a0
Moroianu, J., and Blobel, G. (1995). Protein export from the nucleus requires the GTPase Ran and GTP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 92, 4318–4322. doi: 10.1073/pnas.92.10.4318
Nachury, M. V., and Weis, K. (1999). The direction of transport through the nuclear pore can be inverted. Proc. Natl. Acad. Sci. U.S.A. 96, 9622–9627. doi: 10.1073/pnas.96.17.9622
Nakaseko, Y., Niwa, O., and Yanagida, M. (1984). A meiotic mutant of the fission yeast Schizosaccharomyces pombe that produces mature asci containing two diploid spores. J. Bacteriol. 157, 334–336.
Nishijima, H., Nakayama, J., Yoshioka, T., Kusano, A., Nishitani, H., Shibahara, K., et al. (2006). Nuclear RanGAP is required for the heterochromatin assembly and is reciprocally regulated by histone H3 and Clr4 histone methyltransferase in Schizosaccharomyces pombe. Mol. Biol. Cell 17, 2524–2536. doi: 10.1091/mbc.E05-09-0893
Ohtsubo, M., Kai, R., Furuno, N., Sekiguchi, T., Sekiguchi, M., Hayashida, H., et al. (1987). Isolation and characterization of the active cDNA of the human cell cycle gene (RCC1) involved in the regulation of onset of chromosome condensation. Genes Dev. 1, 585–593. doi: 10.1101/gad.1.6.585
Ohtsubo, M., Okazaki, H., and Nishimoto, T. (1989). The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 109, 1389–1397. doi: 10.1083/jcb.109.4.1389
Orjalo, A. V., Arnaoutov, A., Shen, Z., Boyarchuk, Y., Zeitlin, S. G., Fontoura, B., et al. (2006). The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell 17, 3806–3818. doi: 10.1091/mbc.E05-11-1061
Osmani, A. H., Davies, J., Liu, H. L., Nile, A., and Osmani, S. A. (2006). Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17, 4946–4961. doi: 10.1091/mbc.E06-07-0657
Pascual-Garcia, P., and Capelson, M. (2014). Nuclear pores as versatile platforms for gene regulation. Curr. Opin. Genet. Dev. 25, 110–117. doi: 10.1016/j.gde.2013.12.009
Pemberton, L. F., Rout, M. P., and Blobel, G. (1995). Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc. Natl. Acad. Sci. U.S.A. 92, 1187–1191. doi: 10.1073/pnas.92.4.1187
Pichler, A., Gast, A., Seeler, J. S., Dejean, A., and Melchior, F. (2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120. doi: 10.1016/S0092-8674(01)00633-X
Reichelt, R., Holzenburg, A., Buhle, E. L. Jr., Jarnik, M., Engel, A., and Aebi, U. (1990). Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol. 110, 883–894. doi: 10.1083/jcb.110.4.883
Rout, M. P., Aitchison, J. D., Suprapto, A., Hjertaas, K., Zhao, Y., and Chait, B. T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–651. doi: 10.1083/jcb.148.4.635
Saitoh, H., Pu, R., Cavenagh, M., and Dasso, M. (1997). RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc. Natl. Acad. Sci. U.S.A. 94, 3736–3741. doi: 10.1073/pnas.94.8.3736
Scheffzek, K., Klebe, C., Fritz-Wolf, K., Kabsch, W., and Wittinghofer, A. (1995). Crystal structure of the nuclear Ras-related protein Ran in its GDP-bound form. Nature 374, 378–381. doi: 10.1038/374378a0
Schneider, R., Brors, B., Bürger, F., Camrath, S., and Weiss, H. (1997). Two genes of the putative mitochondrial fatty acid synthase in the genome of Saccharomyces cerevisiae. Curr. Genet. 32, 384–388. doi: 10.1007/s002940050292
Seewald, M. J., Körner, C., Wittinghofer, A., and Vetter, I. R. (2002). RanGAP mediates GTP hydrolysis without an arginine finger. Nature 415, 662–666. doi: 10.1038/415662a
Shimoda, C., Hirata, A., Kishida, M., Hashida, T., and Tanaka, K. (1985). Characterization of meiosis-deficient mutants by electron microscopy and mapping of four essential genes in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 200, 252–257. doi: 10.1007/BF00425432
Shimoda, C., and Nakamura, T. (2004). “Control of late meiosis and ascospore formation,” in The Molecular Biology of Schizosaccharomyces pombe, ed R. Egel (Berlin: Springer), 311–327.
Steinberg, G., Schuster, M., Theisen, U., Kilaru, S., Forge, A., and Martin-Urdiroz, M. (2012). Motor-driven motility of fungal nuclear pores organizes chromosomes and fosters nucleocytoplasmic transport. J. Cell Biol. 198, 343–355. doi: 10.1083/jcb.201201087
Straube, A., Weber, I., and Steinberg, G. (2005). A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J. 24, 1674–1685. doi: 10.1038/sj.emboj.7600644
Tamura, K., Fukao, Y., Iwamoto, M., Haraguchi, T., and Hara-Nishimura, I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22, 4084–4097. doi: 10.1105/tpc.110.079947
Theisen, U., Straube, A., and Steinberg, G. (2008). Dynamic rearrangement of nucleoporins during fungal “open” mitosis. Mol. Biol. Cell 19, 1230–1240. doi: 10.1091/mbc.E07-02-0130
Therizols, P., Fairhead, C., Cabal, G. G., Genovesio, A., Olivo-Marin, J. C., Dujon, B., et al. (2006). Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 172, 189–199. doi: 10.1083/jcb.200505159
Van de Vosse, D. W., Wan, Y., Lapetina, D. L., Chen, W. M., Chiang, J. H., Aitchison, J. D., et al. (2013). A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152, 969–983. doi: 10.1016/j.cell.2013.01.049
Vetter, I. R., Nowak, C., Nishimoto, T., Kuhlmann, J., and Wittinghofer, A. (1999). Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398, 39–46. doi: 10.1038/17969
Walther, T. C., Alves, A., Pickersgill, H., Loïodice, I., Hetzer, M., Galy, V., et al. (2003). The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113, 195–206. doi: 10.1016/S0092-8674(03)00235-6
Yam, C., He, Y., Zhang, D., Chiam, K. H., and Oliferenko, S. (2011). Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr. Biol. 21, 1314–1319. doi: 10.1016/j.cub.2011.06.052
Yang, H.-J., Asakawa, H., Haraguchi, T., and Hiraoka, Y. (2015). Nup132 modulates meiotic spindle attachment in fission yeast by regulating kinetochore reassembly. J. Cell Biol. 211, 295–308. doi: 10.1083/jcb.201501035
Yang, L., Guan, T., and Gerace, L. (1997). Integral membrane proteins of the nuclear envelope are dispersed throughout the endoplasmic reticulum during mitosis. J. Cell Biol. 137, 1199–1210. doi: 10.1083/jcb.137.6.1199
Yang, Q., Rout, M. P., and Akey, C. W. (1998). Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell 1, 223–234. doi: 10.1016/s1097-2765(00)80023-4
Keywords: closed mitosis, fission yeast, meiosis, nuclear envelope breakdown, RanGAP1, Nup132
Citation: Asakawa H, Yang H-J, Hiraoka Y and Haraguchi T (2016) Virtual Nuclear Envelope Breakdown and Its Regulators in Fission Yeast Meiosis. Front. Cell Dev. Biol. 4:5. doi: 10.3389/fcell.2016.00005
Received: 24 July 2015; Accepted: 15 January 2016;
Published: 02 February 2016.
Edited by:
Patrizia Lavia, National Research Council, ItalyCopyright © 2016 Asakawa, Yang, Hiraoka and Haraguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tokuko Haraguchi, tokuko@nict.go.jp
 Haruhiko Asakawa1
Haruhiko Asakawa1  Tokuko Haraguchi
Tokuko Haraguchi