Understanding the pneumococcus: transmission and evolution
- Department of Microbiology, University of Ghana Medical School, Accra, Ghana
Streptococcus pneumoniae is part of the normal bacterial flora of the narsopharynx, but is also associated with several invasive and non-invasive diseases. Recently, there has been a plethora of research information on the pneumococcus, however, there are few comprehensive review papers discussing the research information. This paper provides a review of the pneumococcus in two vital areas related to its biology including transmission and evolution. Transmission of the pneumococcus is a highly efficient process that usually occurs through respiratory droplets from asymptomatic carriers. Following acquisition, the pneumococcus may only establish in the nasopharynx of the new host, or further progress to sites such as the lungs and cause disease. Pneumococcus transmission risk factors, as well as factors involved in its translocation from the nasophyarnx to diseases sites are still not fully understood. Pneumococcal evolution is dominated by recombination. The recombinational events usually involve genetic exchange with streptococci of the mitis group and some pneumococci are thought to exhibit hyper-recombination.
Introduction
The pneumococcus is one of the most virulent human pathogens and causes a wide range of infections, including invasive and non-invasive diseases. There are about one million new pneumococcal infections every year, majority of which occur among children <5 years, and the organism is responsible for 10–20% of all deaths in this age group (O'Brien et al., 2009). Until about two decade ago, little was known about Streptococcus pneumoniae. However, since this time there has been a plethora of research information contributing significantly to our understanding about this very important human pathogen. Unfortunately, there are few recent comprehensive review papers discussing the plethora of research information about the pneumococcus. This paper provides a review of the pneumococcus in two vital areas related to its biology including transmission and evolution.
S. pneumoniae Carriage and Diseases
S. pneumoniae is part of the normal bacterial flora of the upper respiratory tract of humans, and is mainly found in the nasopharynx. Carriage of the organism is more prevalent in children than adults; the colonization rate rises from birth until it peaks around the age of 1–2 years, and thereafter an age related decline is observed (Lloyd-Evans et al., 1996; Hussain et al., 2005). S. pneumoniae carriage normally lasts for a couple of weeks, and duration periods of more than 30 weeks are observed (Sleeman et al., 2008). A seasonal carriage trend has also been described, with peak rates occurring during January–March (Gray et al., 1980). Children acquire several different strains over time, and less immunogenic serotypes tend to be carried in the narsopharynx for a much longer period of time than the more immunogenic serotypes (Rosen et al., 1984; Obaro and Adegbola, 2002).
In developing countries, carriage rates are relatively higher especially, in children. In the Gambia, Lloyd-Evans et al. (1996) reported a carriage rate of 80% among children under five years of age, and a lower rate of 20% in adults. In a study by Gratten et al. (1986), it was found that 60% of infants in Papua New Guinea acquired S. pneumoniae already during the neonatal period, and all infants were colonized within the first 3 months of life (Gratten et al., 1986). Similar high S. pneumoniae carriage rates in the developing world have been reported in several other countries including Zambia (Frederiksen and Henrichsen, 1988), Pakistan (Mastro et al., 1993), The Philippines (Lankinen et al., 1994), Papua New Guinea (Gratten et al., 1986), and Bangladesh (Granat et al., 2007). In the developed world, S. pneumoniae carriage appears to be lower than rates found in the developing world. Aniasson et al. (1992) reported that in Sweden, only 12% of infants were colonized with S. pneumoniae at 3 months, 30% at 7 months, and 32% at 12–18 months. In the UK, a longitudinal study by Goldblatt et al. (2005) showed an overall prevalence rate of 25%, with carriage rates of 52 and 8% in children under 2 years and adults over 18 years of age, respectively. Labout et al. (2008) studied S. pneumoniae carriage in infants in the Netherlands and observed carriage rates of 8.3% at age 1.5 months, 31.3% at 6 months, and 44.5% at 14 months. The high pneumococcal carriage rates in the developing world appear to provide more opportunities for multiple carriage, as relatively higher multiple carriage rates have also been reported in the developing countries compared to the developing world (Gratten et al., 1994; Obaro et al., 1996; Brugger et al., 2010).
The major diseases caused by S. pneumoniae include pneumonia, meningitis, septicaemia, and otitis media. There are two types of pneumonia, namely, bronchial pneumonia and lobar pneumonia, and S. pneumoniae is a major cause of both types. In an infection of pneumonia, S. pneumoniae stimulates the immune system and causes migration of white blood cells to the lungs. The interaction of white blood cells, proliferating bacteria and excessive fluid define the presence of pneumonia (Tuomanen et al., 1995), which can be detected by a chest X-ray. Bacteraemia and septicaemia can occur in 20–30% of cases with pneumococcal pneumonia (Musher, 2004). S. pneumoniae meningitis perhaps has the highest case fatality rate (~40%) among the various pneumococcal infections and up to 50% of survivors suffer from debilitating sequalae such as mental retardation and motor deficiency (Bohr et al., 1984; Leimkugel et al., 2005). The clinical presentations of meningitis caused by S. pneumoniae are similar to other bacterial causes of acute meningitis and include severe headache, photophobia, neck stiffness, and fever. Otitis media is the most common of the pneumococcal diseases. The condition is an inflammation of the middle ear and up to 50% of the cases are caused by S. pneumoniae (Musher, 2004). The major clinical signs of the infection include limited mobility and enlarged tympanic membrane. It is reported that successful treatment with antibiotics can still lead to recurrent otitis media due to proximity of the middle ear to the nasopharynx where S. pneumoniae resides (Libson et al., 2005). Other S. pneumoniae diseases include conjunctivitis, acute tracheobronchitis, endometritis, peritonitis, endocarditis, arthritis, and osteomyelitis. However, these infections are relatively uncommon.
Transmission of S. pneumoniae
As humans are the main host for S. pneumoniae, successful transmission of S. pneumoniae among humans is crucial for survival of the organism, and without this the pneumococcus is likely to be eliminated. Transmission of S. pneumoniae occurs through respiratory droplets from people with pneumococcal disease or more commonly healthy individuals who carry the organism in the nasopharynx (Bogaert et al., 2004; Sleeman et al., 2005). Little is known about the risk factors of pneumococcal transmission, though certain risk factors including number of siblings and visits to general practitioners for mild upper respiratory disease have been identified (Sleeman et al., 2005). Additionally, higher rates of pneumococcal transmission are known to occur at certain sites including day care centers, military camps, and prisons (Givon-Lavi et al., 2002; Bogaert et al., 2004). Givon-Lavi et al. (2002) compared pneumococcal isolates recovered from children in day care centers with isolates recovered from younger siblings not attending day care by pulsed-field gel electrophoresis. This showed a high level of genetic similarity among isolates from the specific day care center the older sibling was attending and those isolated from younger siblings, which indicate that pneumococcal transmission may be a highly efficient process.
Following acquisition, the pneumococcus may establish in the nasopharynx of the new host, and in most cases this leads to asymptomatic colonization (Sleeman et al., 2008). However, occasionally, the newly acquired pneumococcus moves from the nasopharynx to other parts of the human host such as the lungs where it evades the host defence mechanisms and causes disease (Bogaert et al., 2004). Because asymptomatic carriers far exceed symptomatic individuals, most of the links in the transmission chain of person-to-person are not visible. In contrast, a respiratory disease such as measles is also transmitted person-to-person through the same route as pneumococcus but asymptomatic colonization does not happen, and each link in the transmission chain is evident as disease (Mrozek-Budzyn, 2010). There is evidence that the risk for progression of pneumococcus from asymptomatic colonization to disease seems to be greatest soon after acquisition and a complex interplay of factors are involved (Sleeman et al., 2005). Colonizing pneumococci strains may elicit an immune response that may eliminate them (Obaro and Adegbola, 2002). Additionally, the composition of the microflora of the nasopharynx, which is thought to contain more than 700 diverse species, may support or hinder colonization and invasion by symbiosis and/or competition (Aniasson et al., 1992; Harputluoglu et al., 2005). There are significant differences in the attack rate of different serotypes, where attack rates refer to the incidence of invasive pneumococcal disease per the incidence of pneumococcal acquisition. Pneumococcal serotypes such as 1, 4, 5, and 9A have high attack rates, while serotypes such as 9N, 16F, 20, and 38 have low attack rates; generally, attack rates are higher for serotypes which are carried for short time periods (Sleeman et al., 2005). Phase variation where pneumococcal variants have the same serotype but vary from opaque to transparent colonies, is thought to be important in the progression of S. pneumoniae from carriage to invasive disease (Weiser et al., 1994; Arai et al., 2011). This is because the opaque form has been commonly isolated from patient samples, while the transparent form is adapted to colonization of the nasopharynx. According to Ring et al. (1998), phase variation of the transparent type increases pneumococcal invasion into human brain microvascular endothelial cells as much as six-fold.
Evolution of S. pneumoniae
Evolution of the pneumococcal population is known to be dominated by recombination. Using MLST data, Feil et al. (2000) demonstrated that the rate of recombination in S. pneumoniae was 10 times higher than the rate of mutation, while for Neisseria meningitidis, the rate of mutation was five times higher than the rate of mutation. The high rate of pneumococcal recombination has recently been illustrated by the whole genome sequencing of 240 strains of a single lineage, ST 81 (Croucher et al., 2011a). This showed that over 700 recombinational events had occurred in this pneumococcal lineage and 74% of the genome length had undergone recombination in at least one isolate. Currently, it is unknown whether this observation holds for all pneumococcal lineages or whether certain geographical settings could lead to yet more recombination. In contrast to the frequent pneumococcal recombination, mutation is known to be more important than recombination in evolution of clonal bacteria such as Staphylococcus aureus (Feil et al., 2003). The high rates of pneumococcal recombination may be attributed to the relatively high density of repeat elements in the genome that may facilitate incorporation of foreign DNA into the S. pneumoniae chromosome and contribute to rearranging its structure. Aras et al. (2003) analysed the density of repeats for 51 prokaryotic genomes, and observed that S. pneumoniae had the greatest density of 1 every 500 bp. Until recently, two types of such repeat elements including BOX and RUPS elements were known to occur in the S. pneumoniae genome. However, a study by Croucher et al. (2011b) identified a third type of repeat element called Streptococcus pneumoniae Rho-independent Terminator-like Element (SPITE). Like the previously known BOX and RUPS repeat elements, SPRITE is thought to contribute to genome evolution of the pneumococcus and is important in termination of transcription (Croucher et al., 2011b). Another factor that contributes to the high S. pneumoniae recombination is the fact that the pan-genome of the organism is open which means that the pan-genome has an infinite size and thus provides a rapid response to diverse environments (Donati et al., 2010). Phylogenetic studies on S. pneumoniae have shown that, the high rates of recombination can result in the elimination of any deep-rooted phylogenetic signal (Feil and Spratt, 2001). However, this does not prevent the formation of distinct pneumococcal lineages or clones but makes such clones relatively unstable compared to several other bacteria (Feil et al., 2003; Donati et al., 2010).
S. pneumoniae interspecies recombination is usually related to genetic exchange with streptococci of the mitis group (Hanage et al., 2009). The mitis group of streptococci currently includes S. pneumoniae and 10 other members: S. oralis, S. mitis, S. infantis, S. sanguis, S. gordonii, S. pseudopneumoniae, S. cristatus, S. oligofermentans, S. parasanguinis, and S. peroris. S. pneumoniae which is the main pathogen in the group is closely related to S. oralis, and both species are believed to have evolved from a common ancestor (Kilian et al., 2008). Genetic exchange between S. pneumoniae and other mitis streptococci is facilitated by the co-habitation of these organisms in the nasopharynx, as well as the natural transformability of S. pneumoniae (Kilian et al., 2008). Based on sequence analysis of pbp genes, S. mitis and S. oralis have been found to donors of chromosomal DNA to S. pneumoniae in the evolution of mosaic penicillin-binding protein genes (Zerfass et al., 2009). Such mosaic pneumococcal genes from homologous recombination with the mitis group streptococci have also been observed in virulence genes such as lytA, nanA, pspA, and pspC (Johnston et al., 2010). Using MLST data, Hanage et al. (2009) investigated recombination among S. pneumoniae and other mitis group streptococci, and observed a distinct population of S. pneumoniae that exhibit hyper-recombination (Figure 1). This pneumococcal sub-population which was the main recipient of genetic material from other mitis group streptococci showed significantly higher levels of resistance for various antibiotics (penicillin, erythromycin, tetracycline, chloramphenicol, and cefotaxime) compared to other pneumococcal sub-populations which did not show evidence of recombination. While the basis of such a pneumococcal population exhibiting hyper-recombination is poorly understood, in terms of genome evolution, it is possible that the population may possess extraordinarily high density of repeat elements (Hoskins et al., 2001; Tettelin et al., 2001). Defects in the DNA mismatch repair system may also contribute to the hyper-recombination of this pneumococcal sub-population (Denamur and Matic, 2006; Hall and Henderson-Begg, 2006; Henderson-Begg et al., 2010). Very recently, Croucher et al. (2012) have demonstrated that the location and selective advantage of accessory genome loci may have the greatest mechanistic impact on homologous recombination occurring between lineages of the pneumococcal species. While there is a distinct pneumococcal population that exhibit hyper-recombination, there is no evidence of a pneumococcal population that exhibit hyper-mutation (Henderson-Begg et al., 2010).
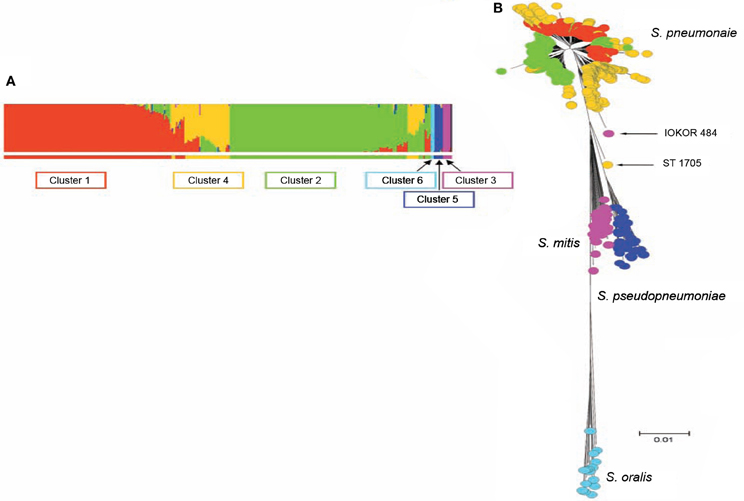
Figure 1. (A) Admixture analysis of 2024 distinct streptococcal genotypes of S. pneumoniae, S. oralis, S. mitis, and S. pseudopneumoniae based on Bayesian Analysis of Population Structure. (B) Minimum evolution tree constructed from concatenates of the different streptococci species. The streptococci fell into six clusters shown by the different colors (coloring of taxa in B corresponds to colors of BAPS clusters in A). Three BAPS clusters (including Clusters 1, 2, and 4) were associated with S. pneumoniae subpopulations, while the other three BAPS clusters namely, clusters 3, 5, and 6 were associated with S. mitis, S. pseudopneumoniae, and S. oralis, respectively. Relatively, S. pneumoniae cluster 4 appears heterogeneous and the isolates of this BAPS cluster are composed of a high proportion of mosaics due to genetic material from other clusters of S. pneumoniae and other streptococci. Adapted from Hanage et al. (2009).
Pneumococcal evolutionary events frequently occur at the capsular locus (cps), which encodes the pneumococcal capsule. The pneumococcal cps genes are flanked by the conserved dexB and aliA genes and synthesize capsule polysaccharide via the Wzx/Wzy-dependent pathway using the regulatory and processing genes wzg, wzh, wzd, and wze (Kolkman et al., 1998; Garcia et al., 2000). The only exceptions are serotypes 3 and 37 which utilize a synthase pathway for capsular biosynthesis (Cartee et al., 2001; Llull et al., 2001). Homologous recombination occurs at the flanking regions common among different serotypes. Each pneumococcal cps has a serotype specific region, where recombinational events lead to capsular switching and the possible formation of vaccine escape variants (Golubchik et al., 2012). Additionally, capsular recombinational events resulting in some pneumococcal capsular types may be associated with increase in virulence (Hu et al., 2012). It appears that some capsular genes undergo recombinational exchange among pneumococcal lineages more commonly than others. Several reasons may account for this observation, including differences in the genetic organization within the capsular loci (Bentley et al., 2006), the capabilities of the pneumococcal isolates to be transformed (Chen and Dubnau, 2004), and co-existence of isolates of different pneumococcal serotypes in the nasophyarnx. Within the pneumococcal cps loci, several mutational events have also been documented (Bentley et al., 2006), with some recent data suggesting that mutational events are also important in capsular switching (Sheppard et al., 2010).
Conclusions
Transmission of the pneumococcus is a highly efficient process that usually occurs through respiratory droplets from asymptomatic carriers. Following acquisition, the pneumococcus may only establish in the nasopharynx of the new host, or further progress to sites such as the lungs and cause disease. Pneumococcus transmission risk factors, as well as factors involved in its translocation from the nasophyarnx to diseases sites are still not fully understood. Pneumococcal evolution is dominated by recombination which may be attributed to the relatively high density of repeat elements in the genome and also the fact that the pan-genome of the organism is open. The recombinational events usually involve genetic exchange with streptococci of the mitis group and some pneumococci are thought to exhibit hyper-recombination.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aniasson, G., Alm, B., Andersson, B., Larsson, P., Nylén, O., Peterson, H., et al. (1992). Nasopharyngeal colonization during the first years of life. J. Infect. Dis. 165, 38–42.
Arai, J., Hotomi, M., Hollingshead, S. K., Ueno, Y., Briles, D. E., and Yamanaka, N. (2011). Streptococcus pneumoniae isolates from middle ear fluid and nasopharynx of children with acute otitis media exhibit phase variation. J. Clin. Microbiol. 49, 1646–1649.
Aras, R., Kang, J., Tschumi, A. I., Harasaki, Y., and Blaser, M. J. (2003). Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc. Natl. Acad. Sci. U.S.A. 100, 13579–13584.
Bentley, S. D., Aanensen, D. M., Mavroidi, A., Saunders, D., Rabbinowitsch, E., Collins, M., et al. (2006). Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. doi: 10.1371/journal.pgen.0020031
Bogaert, D., De Groot, R., and Hermans, P. W. (2004). Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4, 144–154.
Bohr, V., Paulson, O. B., and Rasmussen, N. (1984). Pneumococcal meningitis. Late neurologic sequelae and features of prognostic impact. Arch. Neurol. 41, 1045–1049.
Brugger, S. D., Frey, P., Aebi, S., Hinds, J., and Muhlemann, K. (2010). Multiple colonization with S. pneumoniae before and after introduction of the seven-valent conjugated pneumococcal polysaccharide vaccine. PLoS ONE 5:e11638. doi: 10.1371/journal.pone.0011638
Cartee, R. T., Forsee, W. T., Jensen, J. W., and Yother, J. (2001). Expression of the Streptococcus pneumoniae type 3 synthase in Escherichia coli. Assembly of type 3 polysaccharide on a lipid primer. J. Biol. Chem. 276, 48831–48839.
Chen, I., and Dubnau, D. (2004). DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249.
Croucher, N. J., Harris, S. R., Barquist, L., Parkhill, J., and Bentley, S. D. (2012). A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 8:e1002745. doi: 10.1371/journal.ppat.1002745
Croucher, N. J., Harris, S. R., Fraser, C., Quail, M. A., Burton, J., Van Der Linden, M., et al. (2011a). Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434.
Croucher, N. J., Vernikos, G. S., Parkhill, J., and Bentley, S. D. (2011b). Identification, variation and transcription of pneumococcal repeat sequences. BMC Genome 12:120. doi: 10.1186/1471-2164-12-120
Denamur, E., and Matic, I. (2006). Evolution of mutation rates in bacteria. Mol. Microbiol. 60, 820–827.
Donati, C., Hiller, N. L., Tettelin, H., Muzzi, A., Croucher, N. J., Angiuoli, S. V., et al. (2010). Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 11:R107. doi: 10.1186/gb-2010-11-10-r107
Feil, E. J., Cooper, J. E., Grundmann, H., Robinson, D. A., Enright, M. C., Berendt, A., et al. (2003). How clonal is Staphylococcus aureus? J. Bacteriol. 185, 3307–3316.
Feil, E. J., Smith, J. M., Enright, M. C., and Spratt, B. G. (2000). Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154, 1439–1450.
Feil, E. J., and Spratt, B. G. (2001). Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55, 561–590.
Frederiksen, B., and Henrichsen, J. (1988). Throat carriage of Streptococcus pneumoniae and Streptococcus pyogenes among infants and children in Zambia. J. Trop. Pediatr. 34, 114–117.
Garcia, E., Llull, D., Munoz, R., Mollerach, M., and Lopez, R. (2000). Current trends in capsular polysaccharide biosynthesis of Streptococcus pneumoniae. Res. Microbiol. 151, 429–435.
Givon-Lavi, N., Fraser, D., Porat, N., and Dagan, R. (2002). Spread of Streptococcus pneumoniae and antibiotic-resistant S. pneumoniae from day-care center attendees to their younger siblings. J. Infect. Dis. 186, 1608–1614.
Goldblatt, D., Hussain, M., Andrews, N., Ashton, L., Virta, C., Melegaro, A., et al. (2005). Antibody responses to nasopharyngeal carriage of Streptococcus pneumoniae in adults: a longitudinal household study. J. Infect. Dis. 192, 387–393.
Golubchik, T., Brueggemann, A. B., Street, T., Gertz, R. E. Jr., Spencer, C. C., Giannoulatou, E., et al. (2012). Pneumococcal genome sequencing tracks a vaccine escape variant formed through a multi-fragment recombination event. Nat. Genet. 44, 352–355.
Granat, S. M., Mia, Z., Ollgren, J., Herva, E., Das, M., Piirainen, L., et al. (2007). Longitudinal study on pneumococcal carriage during the first year of life in Bangladesh. Pediatr. Infect. Dis. J. 26, 319–324.
Gratten, M., Gratten, H., Poli, A., Carrad, E., Raymer, M., and Koki, G. (1986). Colonisation of Haemophilus influenzae and Streptococcus pneumoniae in the upper respiratory tract of neonates in Papua New Guinea: primary acquisition, duration of carriage, and relationship to carriage in mothers. Biol. Neonate 50, 114–120.
Gratten, M., Manning, K., Dixon, J., Morey, F., Torzillo, P., Hanna, J., et al. (1994). Upper airway carriage by Haemophilus influenzae and Streptococcus pneumoniae in Australian aboriginal children hospitalised with acute lower respiratory infection. Southeast Asian J. Trop. Med. Public Health 25, 123–131.
Gray, B. M., Converse, G. M., and Dillon, H. C. (1980). Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 142, 923–933.
Hall, L. M. C., and Henderson-Begg, S. K. (2006). Hypermutable bacteria isolated from humans – a critical analysis. Microbiology 152, 2505–2514.
Hanage, W. P., Fraser, C., Tang, J., Connor, T. R., and Corander, J. (2009). Hyper-recombination, diversity, and antibiotic resistance in pneumococcus. Science 324, 1454–1457.
Harputluoglu, U., Egeli, E., Sahin, I., Oghan, F., and Ozturk, O. (2005). Nasopharyngeal aerobic bacterial flora and Staphylococcus aureus nasal carriage in deaf children. Int. J. Pediatr. Otorhinolaryngol. 69, 69–74.
Henderson-Begg, S. K., Sheppard, C. L., George, R. C., Livermore, D. M., and Hall, L. M. (2010). Mutation frequency in antibiotic-resistant and -susceptible isolates of Streptococcus pneumoniae. Int. J. Antimicrob. Agents 35, 342–346.
Hoskins, J., Alborn, W. E., Arnold, J., Blaszczak, L. C., Burgett, S., Dehoff, B. S., et al. (2001). Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183, 5709–5717.
Hu, F. Z., Eutsey, R., Ahmed, A., Frazao, N., Powell, E., Hiller, N. L., et al. (2012). In vivo capsular switch in Streptococcus pneumoniae- analysis by whole genome sequencing. PLoS ONE 7:e47983. doi: 10.1371/journal.pone.0047983
Hussain, M., Melegaro, A., Pebody, R. G., George, R., Edmunds, W. J., Talukdar, R., et al. (2005). A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol. Infect. 133, 891–898.
Johnston, C., Hinds, J., Smith, A., Van Der Linden, M., Van Eldere, J., and Mitchell, T. J. (2010). Detection of large numbers of pneumococcal virulence genes in streptococci of the mitis group. J. Clin. Microbiol. 48, 2762–2769.
Kilian, M., Poulsen, K., Blomqvist, T., Håvarstein, L. S., Bek-Thomsen, M., Tettelin, H., et al. (2008). Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS ONE 3:e2683. doi: 10.1371/journal.pone.0002683
Kolkman, M. A., Van Der Zeijst, B. A., and Nuijten, P. J. (1998). Diversity of capsular polysaccharide synthesis gene clusters in Streptococcus pneumoniae. J. Biochem. (Tokyo) 123, 937–945.
Labout, J. A. M., Duijts, L., Arends, L. R., Jaddoe, V. W. V., Hofman, A., De Groot, R., et al. (2008). Factors associated with pneumococcal carriage in healthy Dutch infants: the generation R study. J. Pediatr. 153, 771–776.
Lankinen, K. S., Leinonen, M., Tupasi, T. E., Haikala, R., and Ruutu, P. (1994). Pneumococci in nasopharyngeal samples from Filipino children with acute respiratory infections. J. Clin. Microbiol. 32, 2948–2952.
Leimkugel, J., Adams Forgor, A., Gagneux, S., Pfluger, V., Flierl, C., Awine, E., et al. (2005). An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J. Infect. Dis. 192, 192–199.
Libson, S., Dagan, R., Greenberg, D., Porat, N., Trepler, R., Leiberman, A., et al. (2005). Nasopharyngeal carriage of Streptococcus pneumoniae at the completion of successful antibiotic treatment of acute otitis media predisposes to early clinical recurrence. J. Infect. Dis. 191, 1869–1875.
Lloyd-Evans, N., O'Dempsey, T. J., Baldeh, I., Secka, O., Demba, E., Todd, J. E., et al. (1996). Nasopharyngeal carriage of pneumococci in Gambian children and in their families. Pediatr. Infect. Dis. J. 15, 866–871.
Llull, D., Garcia, E., and Lopez, R. (2001). Tts, a processive β-glucosyltransferase of Streptococcus pneumoniae directs the synthesis of the branched type 37 capsular polysaccharide in pneumococcus and other gram-positive species. J. Biol. Chem. 276, 21053–21061.
Mastro, T. D., Nomani, N. K., Ishaq, Z., Ghafoor, A., Shaukat, N. F., Esko, E., et al. (1993). Use of nasopharyngeal isolates of Streptococcus pneumoniae and Haemophilus influenzae from children in Pakistan for surveillance for antimicrobial resistance. Pediatr. Infect. Dis. J. 12, 824–830.
Mrozek-Budzyn, D. (2010). The significance of epidemiological studies for progress of measles elimination. Przegl. Epidemiol. 64, 361–366.
Musher, D. M. (2004). “A pathogenic categorization of clinical syndromes caused by Streptococcus pneumoniae,” in The Pneumococcus, ed E. I. Tuomanen (Washington, DC: ASM Press), 211–220.
Obaro, S., and Adegbola, R. (2002). The pneumococcus: carriage, disease and conjugate vaccines. J. Med. Microbiol. 51, 98–104.
Obaro, S. K., Adegbola, R. A., Banya, W. A., and Greenwood, B. M. (1996). Carriage of pneumococci after pneumococcal vaccination. Lancet 348, 271–272.
O'Brien, K. L., Wolfson, L. J., Watt, J. P., Henkle, E., and Deloria-Knoll, M. (2009). Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374, 893–902.
Ring, A., Weiser, J. N., and Tuomanen, E. I. (1998). Pneumococcal trafficking across the blood-brain barrier. Molecular analysis of a novel bidirectional pathway. J. Clin. Invest. 102, 347–360.
Rosen, C., Christensen, P., Hovelius, B., and Prellner, K. (1984). A longitudinal study of the nasopharyngeal carriage of pneumococci as related to pneumococcal vaccination in children attending day-care centres. Acta Otolaryngol 98, 524–532.
Sheppard, C. L., Pichon, B., George, R. C., and Hall, L. M. (2010). Streptococcus pneumoniae isolates expressing a capsule with epitopes of both serotypes 6A and 6B. Clin. Vaccine Immunol. 17, 1820–1822.
Sleeman, K. L., Daniels, L., Gardiner, M., Griffiths, D., Deeks, J. J., Dagan, R., et al. (2005). Acquisition of Streptococcus pneumoniae and nonspecific morbidity in infants and their families: a cohort study. Pediatr. Infect. Dis. J. 24, 121–127.
Sleeman, K. L., Griffiths, D., Shackley, F., Diggle, L., Gupta, S., Maiden, M. C., et al. (2008). Capsular serotype-specific attack rates and duration of carriage of Streptococcus pneumoniae among adlosescents. J. Med. Microbiol. 57, 185–189.
Tettelin, H., Nelson, K. E., Paulsen, I. T., Eisen, J. A., Read, T. D., Peterson, S., et al. (2001). Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506.
Tuomanen, E. I., Austrian, R., and Masure, H. R. (1995). Pathogenesis of pneumococcal infection. N. Engl. J. Med. 332, 1280–1284.
Weiser, J. N., Austrian, R., Sreenivasan, P. K., and Masure, H. R. (1994). Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62, 2582–2589.
Keywords: pneumococcus, evolution, transmission, carriage, recombination
Citation: Donkor ES (2013) Understanding the pneumococcus: transmission and evolution. Front. Cell. Infect. Microbiol. 3:7. doi: 10.3389/fcimb.2013.00007
Received: 22 November 2012; Accepted: 12 February 2013;
Published online: 07 March 2013.
Edited by:
Didier Raoult, Université de la Méditerranée, FranceReviewed by:
Ross Fitzgerald, Roslin Institute, UKYongqun “Oliver” He, University of Michigan School of Medicine, USA
Didier Raoult, Université de la Méditerranée, France
Copyright © 2013 Donkor. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Eric S. Donkor, Department of Microbiology, University of Ghana Medical School, PO Box 4236, Accra, Ghana. e-mail: ericsdon@hotmail.com; eric.sampane-donkor@lshtm.ac.uk