Characterization of serine hydroxymethyltransferase GlyA as a potential source of D-alanine in Chlamydia pneumoniae
- Pharmaceutical Microbiology Section, Institute for Medical Microbiology, Immunology and Parasitology, University of Bonn, Bonn, Germany
For intracellular Chlamydiaceae, there is no need to withstand osmotic challenges, and a functional cell wall has not been detected in these pathogens so far. Nevertheless, penicillin inhibits cell division in Chlamydiaceae resulting in enlarged aberrant bodies, a phenomenon known as chlamydial anomaly. D-alanine is a unique and essential component in the biosynthesis of bacterial cell walls. In free-living bacteria like Escherichia coli, penicillin-binding proteins such as monofunctional transpeptidases PBP2 and PBP3, the putative targets of penicillin in Chlamydiaceae, cross-link adjacent peptidoglycan strands via meso-diaminopimelic acid and D-Ala-D-Ala moieties of pentapeptide side chains. In the absence of genes coding for alanine racemase Alr and DadX homologs, the source of D-Ala and thus the presence of substrates for PBP2 and PBP3 activity in Chlamydiaceae has puzzled researchers for years. Interestingly, Chlamydiaceae genomes encode GlyA, a serine hydroxymethyltransferase that has been shown to exhibit slow racemization of D- and L-alanine as a side reaction in E. coli. We show that GlyA from Chlamydia pneumoniae can serve as a source of D-Ala. GlyA partially reversed the D-Ala auxotrophic phenotype of an E. coli racemase double mutant. Moreover, purified chlamydial GlyA had racemase activity on L-Ala in vitro and was inhibited by D-cycloserine, identifying GlyA, besides D-Ala ligase MurC/Ddl, as an additional target of this competitive inhibitor in Chlamydiaceae. Proof of D-Ala biosynthesis in Chlamydiaceae helps to clarify the structure of cell wall precursor lipid II and the role of chlamydial penicillin-binding proteins in the development of non-dividing aberrant chlamydial bodies and persistence in the presence of penicillin.
Introduction
Acute and chronic diseases caused by Chlamydiaceae are a global health problem. The Gram-negative obligate intracellular pathogens depend on eukaryotic host cells to maintain their unique biphasic developmental cycle. One elusive phenomenon of the chlamydial biology has fascinated researches for two decades: for endobacteria, such as Chlamydiaceae, there is no need to resist osmotic challenges and a functional cell wall has not been detected in these pathogens so far (McCoy and Maurelli, 2006). Nevertheless, in the evolutionary process of adaptation to the host environment, Chlamydiaceae species conserved in their reduced genomes a nearly complete cell wall precursor biosynthesis pathway (Figure 1) and antibiotics that target cell wall biosynthesis are active (McCoy and Maurelli, 2006). Penicillin has no bactericidal effect, as seen in free-living bacteria, but induces a reversible state of persistence in Chlamydiaceae that is characterized by the formation of viable, enlarged, reticulate bodies. These persisting cells are called aberrant bodies (AB) and show resistance to azithromycin (Wyrick and Knight, 2004), the first-line treatment for chlamydial infections (CDC, 2010). Beta-lactam induced formation of non-dividing ABs has been observed in cell culture (Skilton et al., 2009) as well as in vivo (Phillips Campbell et al., 2012).
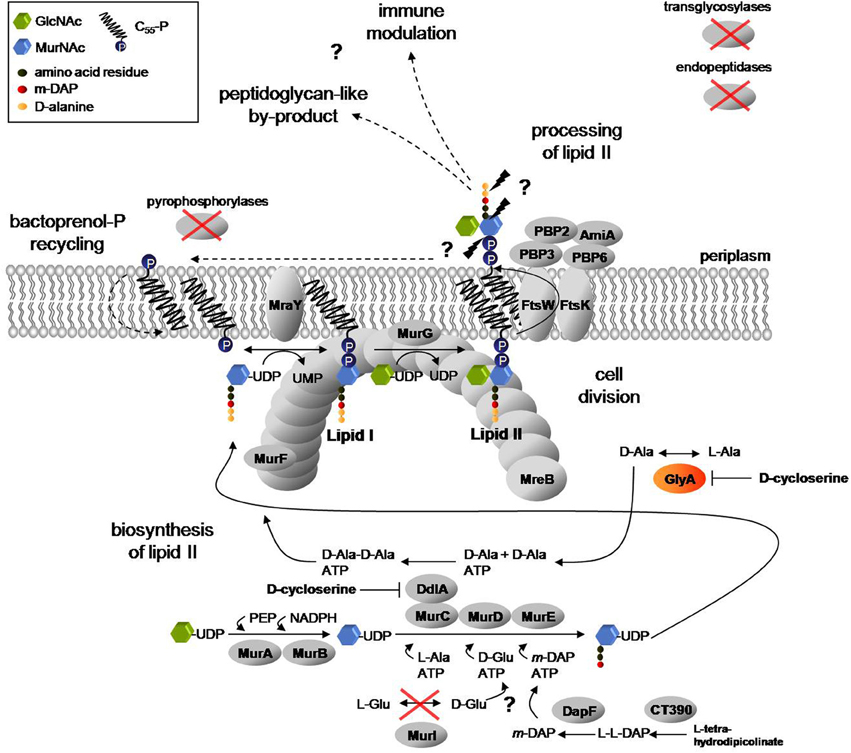
Figure 1. Proposed lipid II pathway in Chlamydiaceae. A complete cycle of lipid II biosynthesis, including translocation to the periplasm, processing and bactoprenol carrier recycling is required for coordinated function of the divisome machinery and modulation of Nod1 and Nod2 mediated host immune response to chlamydial muropeptides. Biosynthesis of lipid II takes place in the cytoplasm and at the inner leaflet of the cytoplasmic membrane. In two consecutive biosynthesis steps of L-Ala racemization and D-Ala ligation, catalyzed by GlyA and the MurC/Ddl fusion protein, the D-Ala-D-Ala dipeptide is produced. MurF adds the dipeptide to the nascent peptide chain to complete synthesis of the soluble precursor UDP-MurNAc-pentapeptide and to provide D-Ala-D-Ala moieties in the cell wall precursors for transpeptidation activity of the penicillin-binding proteins PBP2 and PBP3. Actin-ortholog MreB functionally organizes MurF, MraY, and MurG (Gaballah et al., 2011), the last three enzymes in lipid II biosynthesis, at the septum. The synthesized precursor is translocated to the periplasm and processed by the concerted activity of the PBP enzymes and amidase AmiA to allow for bactoprenol-P recycling. In the process, the rudimentary by-product found by Liechti et al. (2013), in which the peptide side chains are cross-linked by peptide bonds, might result (Ghuysen and Goffin, 1999) and released muropeptides might contribute to modulation of the host immune response (McCoy and Maurelli, 2006). Chlamydiaceae lack transglycosylases as well as endopeptidases and pyrophosphorylases described so far to link lipid II sugar units to form glycan chains, to cleave peptide bridges between cross-linked glycan chains and to dephosphorylate bactoprenol-PP, respectively. Moreover, the L-Glu racemase MurI is absent. Question marks and dashed arrows highlight steps of the pathway that remain to be clarified. (GlcNAc: N-acetylglucosamine; MurNAc: N-acetylmuramic acid).
In free-living bacteria, cell division must be highly coordinated with cell wall biosynthesis to maintain cell integrity. The need for tightly interconnecting both cell biological processes may be reflected by the partial overlap of components from both multi-protein machineries; e.g., the transpeptidase PBP3 (FtsI) is essential for the incorporation of cell wall building blocks at the septal cell wall and for cell division.
The bacterial cell wall consists of peptidoglycan, a polymer of long chains with alternating sugar units of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), which are cross-linked via flexible peptide bridges. Peptidoglycan is found in all eubacteria with the exception of some obligate intracellular species. Biosynthesis of peptidoglycan takes place in three stages (Figure 1). In the cytoplasm, six enzymes (MurA to MurF) catalyze the formation of the soluble precursor UDP-MurNAc-pentapeptide. Notably, the precursor contains D-Ala in positions 4 and 5 of the pentapeptide moiety. D-alanine is a unique and essential component in the biosynthesis of bacterial cell walls. The non-proteinogenic amino acid is synthesized by alanine racemases Alr and DadX and ligated by Ddl to form D-Ala-D-Ala. The dipeptide is attached to the amino acid in position 3 by the action of MurF to complete the pentapeptide side chain. In the first membrane-linked step, MraY catalyzes the synthesis of lipid I by transferring UDP-MurNAc-pentapeptide to the lipid carrier bactoprenol-phosphate (undecaprenyl-P). With the addition of UDP-GlcNAc, MurG synthesizes lipid II, the completed peptidoglycan cell wall building block. Lipid II is then translocated by the flippase FtsW to the outside of the cell and incorporated into the peptidoglycan network by the action of penicillin-binding proteins (PBPs) which exhibit transglycosylase and DD-transpeptidase activities.
The human cytosolic Pattern Recognition Receptors, Nod1 and Nod2, sensing bacterial cell wall fragments, recognize intracellular C. pneumoniae and subsequently mediate activation of the transcription factor NFkB which plays a key role in regulating the immune response to infection (McCoy and Maurelli, 2006). Nod1 and Nod2 receptor mediated recognition, together with the susceptibilty to penicillin, suggests that cell wall precursors/peptidoglycan fragments are synthesized by Chlamydiaceae during infection.
A nearly complete lipid II biosynthesis pathway has been found in genomes of Chlamydiaceae (Figure 1) (McCoy and Maurelli, 2006), and functional conservation of enzymes catalyzing cytoplasmic steps (MurA, MurC/Ddl, CT390, DapF, MurE, MurF) and the two membrane-linked steps (MraY and MurG) of cell wall precursor biosynthesis has been demonstrated (McCoy and Maurelli, 2006; McCoy et al., 2006; Henrichfreise et al., 2009; Patin et al., 2009, 2012). Chlamydiaceae genomes code for only two PBPs that serve as DD-transpeptidases in free-living bacteria. PBP2 and PBP3 are the putative targets of penicillin in Chlamydiaceae and cross-link adjacent peptidoglycan strands via meso-diaminopimelic acid and D-Ala-D-Ala moieties in E. coli. AmiA is the only ortholog of septal peptidoglycan hydrolyzing amidases found in chlamydial genomes. Moreover, Chlamydiaceae harbor a rudimentary set of cell division proteins, lacking the central organizer FtsZ, but comprising FtsW, FtsI (PBP3) and FtsK, and possess, despite their spherical shape, the cytoskeletal protein MreB (Gaballah et al., 2011). Chlamydial MreB was shown to interact with key components in lipid II biosynthesis and FtsK (Gaballah et al., 2011; Ouellette et al., 2012).
We proposed that retaining biosynthesis of lipid II in cell wall-lacking “minimal bacteria,” like Chlamydiaceae, may reflect a vital role of the lipid II pathway in prokaryotic cell division (Henrichfreise et al., 2009). Moreover, we discussed a model for the role of the conserved lipid II pathway in maintaining a functional divisome and contributing to modulation of host response in Chlamydiaceae (Figure 1). A recent study revealed the presence of cell wall sacculi in Protochlamydia, a genus of evolutionary older amoeba symbionts with less reduced genomes as compared to Chlamydiaceae (Pilhofer et al., 2013). In the pathogenic Chlamydiaceae, however, no functional cell wall but ring-like shaped structures were found and supposed to contain peptidoglycan-like material and localize to the division plane (Liechti et al., 2013). These findings are consistent with our model described above which implicates a crucial function of the PBP2 and PBP3 DD-transpeptidase activity in lipid II processing and sustaining a complete cycle of lipid II biosynthesis and recycling. PBP catalyzed DD-transpeptidation depends on the presence of the D-Ala-D-Ala terminus in the pentapeptide side chain of cell wall building blocks. Genomes of Chlamydiaceae do not encode homologs of the pyridoxal-5′-phosphate (PLP) cofactor requiring alanine racemases Alr and DadX. Therefore, the source of D-Ala and thus the presence of substrates for PBP2 and PBP3 activity in Chlamydiaceae have remained unclear for years.
We searched chlamydial genomes for genes encoding other PLP dependent proteins and found serine hydroxymethyltransferase GlyA to be conserved in all chlamydial genera. Serine hydroxymethyltransferases are found in eu- and prokaryotes and are well known for their function in reversible interconversion of serine and glycine using tetrahydrofolate as the one-carbon carrier. In addition, the enzymes show a particularly broad reaction specificity and catalyze other side reactions typical for PLP dependent enzymes, such as decarboxylation, transamination and retroaldol cleavage (Contestabile et al., 2001). Moreover, an alanine racemase co-activity was proven in vitro for GlyA from E. coli (Shostak and Schirch, 1988).
The aim of this study was to analyze GlyA as a potential source of D-Ala in Chlamydiaceae.
Here, we demonstrate that GlyA from C. pneumoniae is capable of the racemization of alanine in vivo and in vitro implicating that the enzyme can substitute for the absent alanine racemases and that D-Ala is self-synthesized in Chlamydiaceae.
Results
Racemization of Alanine in Chlamydiae
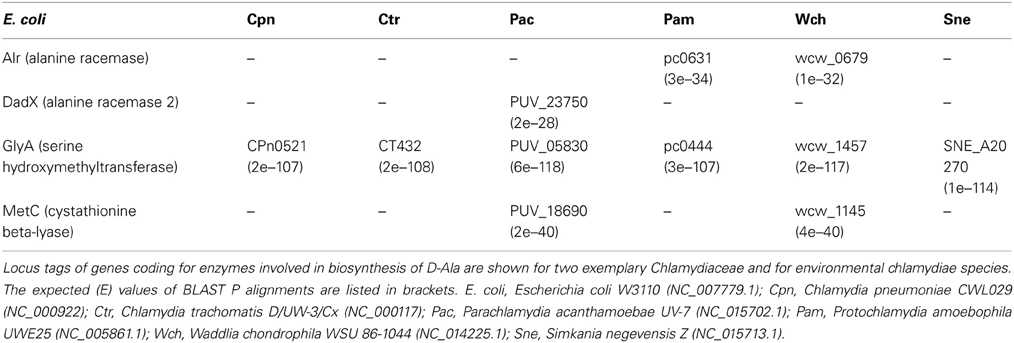
Using BLAST alignments, we searched Chlamydiaceae and enviromental chlamydiae genomes to identify genes coding for orthologs of E. coli PLP cofactor-requiring enzymes known to confer alanine racemization activity (Table 1). In contrast to the Chlamydiaceae and Simkania, the three environmental chlamydiae genera Parachlamydia, Protochlamydia, and Waddlia harbored one ortholog of the Alr or DadX alanine racemases. GlyA was the only enzyme to be encoded in Chlamydiaceae and in all enviromental chlamydiae.
In vivo Activity of GlyA from C. pneumoniae
In the absence of a tractable system to genetically manipulate C. pneumoniae we tested whether heterologous expressed C. pneumoniae GlyA (GlyACp) shows an effect on the D-Ala auxotrophic phenotype of an E. coli Δ alrΔdadX racemase double mutant strain. Our experiments in liquid and solid culture revealed that chlamydial GlyA did not completely reverse the need of exogenous D-Ala of the racemase mutant strain but favored its growth under D-Ala limited conditions (Figure 2). These findings suggest that GlyACp is a functional alanine racemase and capable of generating D-Ala in E. coli.
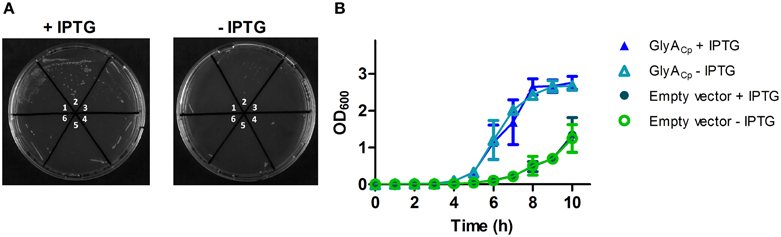
Figure 2. GlyACp exhibits in vivo activity in an E. coli racemase double mutant. A temperature sensitive Δ alrΔ dadX E. coli double mutant was transformed with pET21b-glyACp to allow for the expression of GlyACp in the cytoplasm. Independently generated transformants (1–3 containing pET21b-glyACp and 4–6 containing the empty vector) were grown on solid (A) or in liquid (B) LB medium under limited D-Ala growth conditions at 42°C. LB medium was supplemented with 5 mg/L D-Ala, 50 μM of cofactor PLP, 25 μg/ml thymine and 50 μg/ml ampicillin. Expression of GlyACp was induced by the addition of 0.1 mM IPTG.
Recombinant GlyACp has L-alanine Racemase Activity
To investigate the potential alanine racemase activity of the serine hydroxymethyltransferase GlyA from C. pneumoniae, we overexpressed recombinant GlyACp in E. coli and purified the Strep-tagged protein. In vitro activity of GlyACp was tested in a D-amino acid oxidase coupled enzymatic assay containing L-Ala and cofactor PLP. Alanine racemase from Bacillus stearothermophilus served as positive control. D-Ala that was produced by GlyA was converted to pyruvate by the activity of D-amino acid oxidase (DAAO) and colorimetrically quantified. The chlamydial GlyA converted L-Ala to D-Ala in vitro exhibiting weak racemase activity in comparision to the enzyme from B. stearothermophilus (Figure 3).
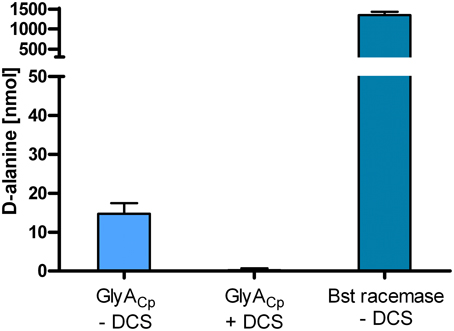
Figure 3. The purified serine hydroxymethyltransferase GlyACp has alanine racemase activity. In vitro activity of recombinant GlyACp was tested in a coupled enzymatic assay containing L-Ala and cofactor PLP. D-Ala that was produced by 1 μg GlyA or Bst racemase was converted to pyruvate by the activity of DAAO and colorimetrically quantified. (DCS: D-cycloserine; Bst racemase: alanine racemase from B. stearothermophilus).
GlyACp Racemase Activity is Sensitive to D-cycloserine
D-cycloserine is a structural analog of D-Ala and competitively inhibits activity of alanine racemases and D-Ala ligases from free-living bacteria (Strominger et al., 1960; Lambert and Neuhaus, 1972). The inhibitor has anti-chlamydial activity in chick embryo yolk sac infection (Moulder et al., 1963) which can be reversed by the addition of D-Ala. For the C. trachomatis D-Ala ligase Ddl which is encoded as a fusion with MurC, as typical for Chlamydiaceae, sensitivity to D-cycloserine has been proven before (McCoy and Maurelli, 2005). We performed activity assays for GlyACp in the presence of D-cycloserine and identified the enzyme to be a second target of D-cycloserine in Chlamydiaceae (Figure 3).
Discussion
The source of D-Ala in Chlamydiaceae and thus the presence of the transpeptidation substrates for PBPs is a crucial aspect of the chlamydial anomaly that will help to gain understanding of the penicillin induced persistence in these human pathogens. Penicillin and other beta-lactams structurally mimic the D-Ala-D-Ala terminus of the pentapeptide side chain of the lipid II cell wall building blocks and are recognized by the active sites of DD-transpeptidase PBPs. Beta-lactams are active against Chlamydiaceae in cell culture (McCoy and Maurelli, 2006; Skilton et al., 2009) and in vivo (Phillips Campbell et al., 2012) and the chlamydial monofunctional DD-transpeptidases PBP2 and PBP3, recovered in detergent-soluble fractions from whole cell preparations, bind [H3]-penicillin (Barbour et al., 1982). Moreover, the recombinant MurC/Ddl protein from C. trachomatis has been shown to specifically ligate D-Ala but not the L-Ala enantiomer to form alanine dipeptides and MurF was demonstrated to add these D-Ala-D-Ala dipeptides to the lipid II peptide side chain (McCoy and Maurelli, 2006; Patin et al., 2012). Moulder et al. (1963) proved anti-chlamydial activity of D-cycloserine in chick embryo yolk sac infection models and demonstrated the specific reversal of this effect by the addition of D-alanine (Moulder et al., 1963). D-cycloserine is a structural analog of D-Ala and well known as competitive inhibitor of alanine racemases and the D-Ala ligase Ddl in other bacteria (Strominger et al., 1960; Lambert and Neuhaus, 1972). Finally, feeding of replicating Chlamydia trachomatis with D-Ala-D-Ala probes revealed evidence for the incorporation of D-Ala into ring-like shaped peptidoglycan-like structures (Liechti et al., 2013). All in all, these data strongly indicate that D-Ala is present in the cells of Chlamydiaceae and plays an essential role in chlamydial cell biology. In the past, the mammalian host was discussed as a potential source of D-Ala (McCoy and Maurelli, 2006) and in Chlamydiaceae genomes a D-alanine permease (DagA_2) was annotated (Stephens et al., 1998) that could allow for the passive transport of D-Ala over the chlamydial cytoplasmic membrane. Though D-Ala is found almost exclusively in the microbial world and is not synthesized in mammalian cells, D-Ala was detected in trace quantities in mammalian urine and tissues, apparently due to breakdown products from intestinal and food bacteria (Guoyao, 2013). Nevertheless, experiments with Listeria monocytogenes and Shigella flexneri indicated that mammalian host cells cannot serve as source of D-Ala. Alanine racemase knockout mutants of both facultative intracellular species failed to survive within mammalian host cells unless exogenous D-Ala was added to the cell culture medium (McCoy and Maurelli, 2006). Based on these data, self-biosynthesis of D-Ala by alternative racemase activity is more likely the source of D-Ala in Chlamydiaceae.
For E. coli, besides the constitutive expressed Alr racemase and the catabolic DadX racemase, two other PLP dependent enzymes involved in the methionine pathway have been shown to confer L-Ala racemization as a side reaction. Overexpressed cystathionine beta-lyase MetC completely reversed the D-Ala auxotrophic phenotype of an E. coli racemase double mutant whereas racemase co-activity of serine hydroxymethyltransferase GlyA was not sufficient to allow for growth on D-Ala lacking medium (Kang et al., 2011). Recently, in vitro activity of MetC from Wolbachia indicated that the enzyme substitute for the absent alanine racemases in these endosymbionts of many arthropods and filarial nematodes. Wolbachia are excellent targets for anti-filarial therapy. Similar to Chlamydiaceae, these obligate intracellular bacteria lack a cell wall but treatment with lipid II biosynthesis blocking fosfomycin, results in enlarged Wolbachia cells (Vollmer et al., 2013). Among the Chlamydiales only the environmental Parachlamydia and Waddlia possess a MetC ortholog but all chlamydial genera harbor GlyA. The serine hydroxymethyltransferase is the only component of the methionine pathway that is encoded by Chlamydiaceae genomes and phylogenetic analysis indicated lateral transfer of the glyA gene from Actinobacteria to the common ancestor of Chlamydiales (Griffiths and Gupta, 2006). Moreover, transcription profiles revealed overlapping expression of the genes encoding GlyA, enzymes for lipid II biosynthesis (MurA to MurF, MraY, and MurG) as well as processing (PBP2, PBP3, and AmiA), the structural protein MreB, and the cell division proteins FtsW and FtsK (Belland et al., 2003). These data suggest an essential function of GlyA in chlamydial biology and correlation with the cellular processes of lipid II biosynthesis and cytokinesis.
We demonstrated L-Ala racemase activity for the GlyA enzyme from C. pneumoniae in vivo in an E. coli racemase double mutant and characterized the purified protein in vitro. Moreover, we identified GlyA as a second target of D-cycloserine besides MurC/Ddl in Chlamydiaceae.
Our results implicate that the enzyme can substitute for the absent alanine racemases and that D-Ala is present and self-synthesized in Chlamydiaceae. The observed weak alanine racemase activity of GlyACp cannot completely compensate D-Ala requirements of the E. coli racemase mutant to build a functional cell wall but might be sufficient to produce D-Ala in amounts that maintain the lipid II biosynthesis pathway in the cell wall-lacking Chlamydiaceae for the proposed functions of the cell wall precursor in co-ordination of cell division and modulation of the host immune response. Future research toward the elucidation of the chlamydial anomaly will include the isolation and structural characterization of lipid II building blocks and biochemical analysis of the penicillin binding proteins PBP2 and PBP3, the putative targets of penicillin whose activity depends on the presence of D-Ala-D-Ala in the pentapeptide side chain of lipid II.
Like other effectors, such as interferon-γ and tumor necrosis factor-α, penicillin can be used to induce persistence as an experimental tool to study chlamydiae/host interactions. Knowledge of the underlying mechanisms of penicillin induced formation of ABs will help to assess these results on the pathogenicity of Chlamydiaceae.
Moreover, analysis of the molecular biology of penicillin induced persistence is important to improve understanding of long-term infection in patients in particular as to the role of chlamydial cell wall precursors in immune modulation and refractory of ABs to anti-chlamydial agents.
Materials and Methods
Bacterial Strains and Growth Conditions
E. coli JM83 harboring the GlyACp expression vector was grown on Luria Bertani (LB) agar plates containing 30 μg/ml chloramphenicol and 100 μg/ml ampicillin, respectively. The temperature sensitive E. coli Δ alrΔ dadX racemase double mutant TKL-10 was maintained on LB agar plates containing 25 μg/ml thymine.
In vivo Complementation
E. coli TKL-10 Δ alrΔ dadX was transformed with pET21b-glyACp and grown in liquid or on solid LB medium supplemented with 5 mg/L D-Ala, 50 μM PLP, 25 μg/ml thymine, 50 μg/ml ampicillin and 0.1 mM IPTG at 42°C. For each experiment three independently generated transformants were used and controls with E. coli TKL-10 Δ alrΔ dadX harboring the empty pET21b vector were carried out.
Cloning of glyA
The glyA gene from C. pneumoniae GiD was amplified by PCR using primer glyACp_f (5′-ATGGTAGGTCTCAGGCCTTGCTAAAAGTTTTTGAGAAATTTAAGA-3′) and glyACp_r (5′-ATGGTAGGTCTCAGCGCTAACTAAAGCTTCTAAATCAATTTCAGG-3′) and cloned into pASK-IBA2c (IBA, Germany) using the BsaI restriction site to generate an N-terminal OmpA-leader peptide fused, C-terminal Strep-tagged protein for periplasmic overproduction. For cytoplasmic expression in complementation assays, glyA was amplified with primers glyACp_pET21_f (5′-CGTCTTTAGAAGCATATGCTAAAAG-3′) and glyACp_pET21_r (5′-GTCTCTGCGGCCGCAACTAAAGCTTC-3′) and cloned into pET21b (Novagen, VWR, Germany) using NdeI and NotI restriction sites.
Overproduction and Purification of GlyACp
E. coli JM83 cells, transformed with pASK-IBA2c-glyACp, were grown in no salt LB in presence of 30 μg/ml of chloramphenicol, 250 mM sucrose and 50 mM L-serine at 30°C. After induction at an OD600 of 1.2 with 200 ng/ml anhydrotetracyline (AHT), 50 μM PLP and 200 μM of folinic acid were added and the cells were incubated for 4 h at 25°C. The purification of GlyACp was performed using the protocol for cleared lysates recommended by the manufacturer (IBA, Germany) with small modifications: the buffers contained 2% N-lauroylsarcosine (or 0.1% N-lauroylsarcosine in the washing and elution buffer), 2 mM 1,4-dithiothreitol (DTT) and 50 μM PLP. Purity of the protein was controlled using SDS-PAGE.
In vitro GlyACp Activity Assay
Racemase activity of GlyACp was determined in a DAAO coupled enzymatic assay system as described previously with slight modifications (Francois and Kappock, 2007). Briefly, 1 μg GlyACp or DAAO were incubated with 50 mM L-alanine in a final volume of 60 μl for 16 h at 37°C in 50 mM KH2PO4, pH 8, 100 mM KCl, 80 μM PLP and 2 mM DTT. D-Ala that was derived from GlyACp racemization was deaminated into pyruvate by the activity of DAAO and indirectly quantified by determining the amount of produced pyruvate with a colorimetric assay using 2,4-dinitrophenylhydrazine (DNPH) as described before (Milner and Wood, 1976). Alanine racemase from B. stearothermophilus (Sigma-Aldrich, Germany) was used as positive control. PLP containing enzymes have been described to show weak transamination activity converting alanine to pyruvate. As a control for potential L-Ala transamination activity of GlyACp, we ran experiments in the absence of DAAO. No GlyACp catalyzed production of pyruvate was detected. For GlyACp inhibition assays, 10 mM D-cycloserine was added and both enzymatic steps were carried out consecutively with a step of heat deactivation in between as DAAO is sensitive to D-cycloserine inhibition.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Katja Mölleken for providing us with C. pneumoniae DNA and thank Fabian Grein and Jennifer Vollmer for fruitful discussions. Stefania De Benedetti holds a fellowship from the NRW International Graduate Research School Biotech-Pharma. Henrike Bühl and Christian Otten received a PhD fellowship from the Jürgen Manchot foundation. Ahmed Gaballah was associated member of the NRW International Graduate Research School Biotech-Pharma and received a PhD fellowship from the DAAD. Beate Henrichfreise was supported by the European Union (“New Antimicrobials” project, people programme, FP7). Support was received by the intramural funding scheme of the Medical Faculty of Bonn, BONFOR, and the Fonds der Chemischen Industrie. Hans-Georg Sahl and Beate Henrichfreise are members of the DFG Cluster of Excellence ImmunoSensation.
References
Barbour, A. G., Amano, K., Hackstadt, T., Perry, L., and Caldwell, H. D. (1982). Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J. Bacteriol. 151, 420–428.
Belland, R. J., Zhong, G., Crane, D. D., Hogan, D., Sturdevant, D., Sharma, J., et al. (2003). Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U.S.A. 100, 8478–8483. doi: 10.1073/pnas.1331135100
Centers for Disease Control and Prevention (CDC). (2010). Sexually transmitted diseases treatment guidelines. MMWR Recomm. Rep. 59, 1–110.
Contestabile, R., Paiardini, A., Pascarella, S., Di Salvo, M. L., D'aguanno, S., and Bossa, F. (2001). l-Threonine aldolase, serine hydroxymethyltransferase and fungal alanine racemase. A subgroup of strictly related enzymes specialized for different functions. Eur. J. Biochem. 268, 6508–6525. doi: 10.1046/j.0014-2956.2001.02606.x
Francois, J. A., and Kappock, T. J. (2007). Alanine racemase from the acidophile Acetobacter aceti. Protein Expr. Purif. 51, 39–48. doi: 10.1016/j.pep.2006.05.016
Gaballah, A., Kloeckner, A., Otten, C., Sahl, H. G., and Henrichfreise, B. (2011). Functional analysis of the cytoskeleton protein MreB from Chlamydophila pneumoniae. PLoS ONE 6:e25129. doi: 10.1371/journal.pone.0025129
Ghuysen, J. M., and Goffin, C. (1999). Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob. Agents Chemother. 43, 2339–2344.
Griffiths, E., and Gupta, R. S. (2006). Lateral transfers of serine hydroxymethyltransferase (glyA) and UDP-N-acetylglucosamine enolpyruvyl transferase (murA) genes from free-living Actinobacteria to the parasitic chlamydiae. J. Mol. Evol. 63, 283–296. doi: 10.1007/s00239-005-0286-x
Guoyao, W. (2013). Amino Acids: Biochemistry and Nutrition. Cleveland, OH: CRC Press; Taylor and Francis Group.
Henrichfreise, B., Schiefer, A., Schneider, T., Nzukou, E., Poellinger, C., Hoffmann, T. J., et al. (2009). Functional conservation of the lipid II biosynthesis pathway in the cell wall-less bacteria Chlamydia and Wolbachia: why is lipid II needed? Mol. Microbiol. 73, 913–923. doi: 10.1371/journal.pone.0025129
Kang, L., Shaw, A. C., Xu, D., Xia, W., Zhang, J., Deng, J., et al. (2011). Upregulation of MetC is essential for D-alanine-independent growth of an alr/dadX-deficient Escherichia coli strain. J. Bacteriol. 193, 1098–1106. doi: 10.1128/JB.01027-10
Lambert, M. P., and Neuhaus, F. C. (1972). Mechanism of D-cycloserine action: alanine racemase from Escherichia coli. W. J. Bacteriol. 110, 978–987.
Liechti, G. W., Kuru, E., Hall, E., Kalinda, A., Brun, Y. V., Vannieuwenhze, M., et al. (2013). A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature. doi: 10.1038/nature12892. [Epub ahead of print].
McCoy, A. J., Adams, N. E., Hudson, A. O., Gilvarg, C., Leustek, T., and Maurelli, A. T. (2006). L,L-diaminopimelate aminotransferase, a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc. Natl. Acad. Sci. U.S.A. 103, 17909–17914. doi: 10.1073/pnas.0608643103
McCoy, A. J., and Maurelli, A. T. (2005). Characterization of Chlamydia MurC-Ddl, a fusion protein exhibiting D-alanyl-D-alanine ligase activity involved in peptidoglycan synthesis and D-cycloserine sensitivity. Mol. Microbiol. 57, 41–52. doi: 10.1111/j.1365-2958.2005.04661.x
McCoy, A. J., and Maurelli, A. T. (2006). Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol. 14, 70–77. doi: 10.1016/j.tim.2005.12.004
Milner, Y., and Wood, H. G. (1976). Steady state and exchange kinetics of pyruvate, phosphate dikinase from Propionibacterium shermanii. J. Biol. Chem. 251, 7920–7928.
Moulder, J. W., Novosel, D. L., and Officer, J. E. (1963). Inhibition of the growth of agents of the psittacosis group by d-cycloserine and its specific reversal by d-alanine. J. Bacteriol. 85, 707–711.
Ouellette, S. P., Karimova, G., Subtil, A., and Ladant, D. (2012). Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol. Microbiol. 85, 164–178. doi: 10.1111/j.1365-2958.2012.08100.x
Patin, D., Bostock, J., Blanot, D., Mengin-Lecreulx, D., and Chopra, I. (2009). Functional and biochemical analysis of the Chlamydia trachomatis ligase MurE. J. Bacteriol. 191, 7430–7435. doi: 10.1128/JB.01029-09
Patin, D., Bostock, J., Chopra, I., Mengin-Lecreulx, D., and Blanot, D. (2012). Biochemical characterisation of the chlamydial MurF ligase, and possible sequence of the chlamydial peptidoglycan pentapeptide stem. Arch. Microbiol. 194, 505–512. doi: 10.1007/s00203-011-0784-8
Phillips Campbell, R., Kintner, J., Whittimore, J., and Schoborg, R. V. (2012). Chlamydia muridarum enters a viable but non-infectious state in amoxicillin-treated BALB/c mice. Microbes Infect. 14, 1177–1185. doi: 10.1016/j.micinf.2012.07.017
Pilhofer, M., Aistleitner, K., Biboy, J., Gray, J., Kuru, E., Hall, E., et al. (2013). Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat. Commun. 4:2856. doi: 10.1038/ncomms3856
Shostak, K., and Schirch, V. (1988). Serine hydroxymethyltransferase: mechanism of the racemization and transamination of D- and L-alanine. Biochemistry 27, 8007–8014. doi: 10.1021/bi00421a006
Skilton, R. J., Cutcliffen, L. T., Barlow, D., Wang, Y., Salim, O., Lambden, P. R., et al. (2009). Penicillin induced persistence in Chlamydia trachomatis: high quality time lapse video analysis of the developmental cycle. PLoS ONE 4:e7723. doi: 10.1371/journal.pone.0007723
Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., et al. (1998). Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759. doi: 10.1126/science.282.5389.754
Strominger, J. L., Ito, E., and Threnn, R. H. (1960). Competitive inhibition of enzymatic reactions by oxamycin. J. Am. Chem. Soc. 82, 998–999. doi: 10.1021/ja01489a058
Vollmer, J., Schiefer, A., Schneider, T., Jülicher, K., Johnston, K. L., Taylor, M. J., et al. (2013). Requirement of lipid II biosynthesis for cell division in cell wall-less Wolbachia, endobacteria of arthropods and filarial nematodes. Int. J. Med. Microbiol. 303, 140–149. doi: 10.1016/j.ijmm.2013.01.002
Keywords: chlamydial anomaly, persistence, aberrant bodies, D-alanine, alanine racemase, GlyA, penicillin, D-cycloserine
Citation: De Benedetti S, Bühl H, Gaballah A, Klöckner A, Otten C, Schneider T, Sahl H-G and Henrichfreise B (2014) Characterization of serine hydroxymethyltransferase GlyA as a potential source of D-alanine in Chlamydia pneumoniae. Front. Cell. Infect. Microbiol. 4:19. doi: 10.3389/fcimb.2014.00019
Received: 30 October 2013; Paper pending published: 10 January 2014;
Accepted: 03 February 2014; Published online: 26 February 2014.
Edited by:
Jan Rupp, University of Lübeck, GermanyReviewed by:
Guido Hansen, University of Lübeck, GermanyPatrick Viollier, University of Geneva, Switzerland
Copyright © 2014 De Benedetti, Bühl, Gaballah, Klöckner, Otten, Schneider, Sahl and Henrichfreise. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beate Henrichfreise, Pharmaceutical Microbiology Section, Institute for Medical Microbiology, Immunology and Parasitology, University of Bonn, Meckenheimer Allee 168, 53115 Bonn, Germany e-mail: bhenrich@uni-bonn.de
†Present address: Ahmed Gaballah, Medical Research Institute, Alexandria University, Alexandria, Egypt.
‡These authors have contributed equally to this work.
 Stefania De Benedetti‡
Stefania De Benedetti‡  Hans-Georg Sahl
Hans-Georg Sahl Beate Henrichfreise
Beate Henrichfreise