Whole genome sequencing of extended-spectrum β-lactamase producing Klebsiella pneumoniae isolated from a patient in Lebanon
- 1Department of Natural Sciences, School of Arts and Sciences, Lebanese American University, Byblos, Lebanon
- 2University of California Davis Genome Center, Davis, CA, USA
- 3School of Medicine, Lebanese American University, Byblos, Lebanon
Objective: The emergence of extended-spectrum β-lactamase (ESBL)-producing bacteria is now a critical concern. The ESBL-producing Klebsiella pneumoniae constitutes one of the most common multidrug-resistant (MDR) groups of gram-negative bacteria involved in nosocomial infections worldwide. In this study we report on the molecular characterization through whole genome sequencing of an ESBL-producing K. pneumoniae strain, LAU-KP1, isolated from a stool sample from a patient admitted for a gastrointestinal procedure/surgery at the Lebanese Amrican University Medical Center-Rizk Hospital (LAUMCRH) in Lebanon.
Methods: Illumina paired-end libraries were prepared and sequenced, which resulted in 4,220,969 high-quality reads. All sequence processing and assembly were performed using the A5 assembly pipeline.
Results: The initial assembly produced 86 contigs, for which no scaffolding was obtained. The final collection of contigs was submitted to GenBank. The final draft genome sequence consists of a combined 5,632,663 bases with 57% G+C content. Automated annotation was performed using the RAST annotation server. Sequencing analysis revealed that the isolate harbored different β-lactamase genes, including blaoxa−1, blaCTX−M−15, blaSHV−11, and blaTEM−1b. The isolate was also characterized by the concomitant presence of other resistance determinants most notably acc(6′)-lb-cr and qnrb1. The entire plasmid content was also investigated and revealed homology with four major plasmids pKPN-IT, pBS512_2, pRSF1010_SL1344, and pKPN3.
Conclusions: The potential role of K. pneumonia as a reservoir for ESBL genes and other resistance determinants is along with the presence of key factors that favor the spread of antimicrobial resistance a clear cause of concern and the problem that Carbapenem-non-susceptible ESBL isolates are posing in hospitals should be reconsidered through systematic exploration and molecular characterization.
Introduction
Extended-spectrum β-lactamases (ESBL) are enzymes produced by many bacterial species as a means for defense against β-lactam drugs with the genes encoding for those enzymes being mainly located on mobile genetic elements (Pfeifer et al., 2010). ESBL enzymes confer antimicrobial resistance to a wide spectrum of β-lactams such as penicillins, aztreonam, as well as first, second, and third generation cephalosporins (Jacoby and Medeiros, 1991; Bradford, 2001). β-lactamases are widespread and were detected in a wide range of Enterobacteriaceae (Bradford, 2001) predominantly in Escherichia coli, and Klebsiella pneumoniae, as well as in other non-enteric microorganisms, such as Pseudomonas aeruginosa, Acinetobacter baumanni (Naas et al., 1999; Spanu et al., 2002; Goossens and Grabein, 2005; Paterson and Bonomo, 2005), and Capnocytophaga ochracea (Rosenau et al., 2000). In addition, Shigella species have been increasingly reported to be resistant to β-lactams including third-generation cephalosporins (Levesque et al., 1995).
K. pneumoniae is an important human pathogen causing nosocomial infections (Podschun and Ullmann, 1998). ESBL-producing K. pneumoniae is one of the most common multidrug-resistant (MDR) groups of gram-negative bacteria worldwide (Breurec et al., 2013). Infections caused by ESBL-producing K. pneumoniae are often associated with the urinary and respiratory tracts along with epidemic clones causing outbreaks in intensive care units (Peirana et al., 2012).
There are four molecular classes of β-lactamases. Classes A, C, and D. β-lactamases possess an active-site serine, while class B β-lactamases are metalloenzymes requiring Zn molecule for their activity (Ambler et al., 1991; Bush et al., 1995). CTX-M is a dominant ESBL family in K. pneumoniae strains, but TEM and SHV enzymes in addition to members of the three classes of β-lactamases (B, C, D) are also common (Breurec et al., 2013). Carbapenems are usually used for the treatment of infections caused by ESBL producing Enterobacteriaceae. Increasing rates of ESBL-producing isolates has led to the overuse of carbapenems creating a pressure and triggering resistance. Resistance to carbapenems may be associated with the production of carbapenemases including class A KPCs, class B metallo-β-lactamases (VIM, IMP, or NDM-1) or the class D OXA-type enzymes (Queenan and Bush, 2007; Matar et al., 2008; Nordmann et al., 2009).
The aim of this study is to characterize an ESBL-producing K. pneumoniae isolated from a stool sample from a patient admitted for a gastrointestinal procedure/surgery in a Lebanese hospital using whole genome sequencing (WGS).
Materials and Methods
Ethical Approval
The University's Institutional Review Board approved the study (UMCRH.AF.12/Dec/2012).
Study Design and Bacterial Isolate
Samples were collected from the University Medical Center-Rizk Hospital (UMC-RH). All patients included in the study were screened for the presence of ESBL-producing Enterobacteriaceae by means of three consecutive rectal swab or stool cultures, as per regular screening for ESBL detection. The first fecal sample was collected at the same time as the other preoperative tests. The second and third samples were collected later, latest day 2-post surgery. K. pneumoniae LAU-KP1 sequenced in this study was isolated from a 62-year-old woman working as practitioner nurse or nurse's assistant. She was admitted for radical resection of an abdominal tumor and repair of abdominal hernia. The patient had other than her cancer, recurrent urinary tract infections. She had been using amoxicillin-clavulanate, metronidazol, and cefriaxon in the 3 months prior to presentation. She had been admitted to hospital in the year prior to surgery, but was not known to be a carrier of multidrug resistant organism and no one in her household was known to be a carrier of multidrug resistant organisms.
Screening for ESBL
The standard disk diffusion method with combined discs (ceftazidime/clavulanate and cefotaxime/clavulanate) was used for screening. The standard E-test method using double sided strips containing on one side ceftazidime or cefotaxime and on the other the same antibiotic combined with clavulanate was used to assess for screening and confirming ESBL production in all strains according to the recommendations of the CLSI. E. coli ATCC25922 and K. pneumoniae ATCC700603 were used as the quality control strains.
Antimicrobial Testing
Antimicrobial susceptibility test by the disk diffusion method was performed to determine the resistance patterns of the isolates to 17 antibiotics: cefotaxime, ceftriaxone, ceftazidime, aztreonam, ceftazidime/clavulanate, cefotaxime/clavulanate, cephalothin, cefuroxime sodium, cefepime, gentamicin, amikacin, ciprofloxacin, tetracycline, ampicillin, amoxicillin/clavulanate, piperacillin, piperacillin/tazobactam, imipenem, meropenem, ertapenem, and sulfamethoxazole/trimethoprim [Oxoid, England; disc contents according to Clinical and Laboratory Standards Institute (CLSI) guidelines]. All antimicrobial testing was performed on Mueller-Hinton agar by the flooding technique and data interpreted according to the CLSI guidelines.
DNA Isolation and Sequencing
Bacterial DNA was extracted using the Nucleospin kit (Macherey-Nagel) and following the manufacturer's instructions.
Genome Sequencing
Genomic DNA (gDNA) was used as input for library preparation using the Illumina TruSeq DNA library preparation kit (Illumina). Bioruptor® NGS was used to sonicate 1 μg of sample DNA (100 μl final sonication volume in TE buffer) for 8 min of 30 s on/off cycles at 4°C. Sheared DNA was used as input for library preparation using the Illumina TruSeq DNA library preparation kit (Illumina). The gDNA was subjected to end-repair, A-tailing, ligation of adaptors including sample-specific barcodes as recommended in the manufacturer's protocol. After ligation of the adaptors, and to reduce the number of fragments smaller than 500 bp, fragments between 500 and 1000 bp were selected using a Pippin Prep™ DNA size selection system (Sage Science). The resulting library was quantified by quantitative PCR in triplicate at 1:1000 and using the Kapa library quantification kit (Kapa Biosystems, Woburn, MA) and as recommended by the manufacturer's protocol. The resultant library size was assessed using an Agilent Bioanalyzer with the High Sensitivity DNA Kit. The library was multiplexed, clustered, and sequenced on an Illumina MiSeq with paired-end 500 cycles protocol to read a length of 250 bp.
Genome Assembly
Genome assembly was performed de novo using A5 with default parameters (Tritt et al., 2012). This pipeline automates the processes of data cleaning, error correction, contig assembly, scaffolding and quality control.
Genome Analysis
The assembly was uploaded and annotated using RAST server (http://rast.nmpdr.org). The service identified protein-encoding, rRNA and tRNA genes, assigned functions to the genes, and predicted which subsystems are represented in the genome (Larsen et al., 2012).
The multi-locus sequence type for the isolate was determined from the WGS data. The presence of known acquired resistance genes was determined by mapping the data from the isolate to an online database. The ResFinder web server (www.genomicepidemiology.org) was used to identify acquired antimicrobial resistance genes in the WGS data, using a threshold of 98.00% identity (ID) (Zankari et al., 2012). ResFinder detects the presence of whole resistance genes, but not functional integrity and expression or resistance due to acquired variation in housekeeping genes. Based on the ResFinder results, a predicted phenotype was determined using phenotypes from original published studies of the genes found.
A K. pneumoniae concatenated marker gene maximum-likelihood tree was constructed using all available assembled K. pneumoniae genomes on the NCBI ftp server. The genomes were first processed with PhyloSift (Darling et al., 2014) using –besthit and –isolate options to get alignments to concatenated conserved housekeeping genes. The tree was made using FastTree (Price et al., 2010). The tree was edited and visualized using Dendroscope (Huson and Scornavacca, 2012). Clades with zero branch lengths were collapsed to a single representative. For visualization of LAU-KP1, the assembly was aligned against the reference strain K. pneumoniae Ecl8's (GCF_000315385) using progressiveMauve (Darling et al., 2010). Ring representation was obtained using BRIG 0.95 (Alikhan et al., 2011). Ecl8 is the K. pneumoniae isolate which was closest to LAU-KP1 in the concatenated marker gene tree that had a complete genome.
Nucleotide Sequence Accession Number
The draft sequence of the K. pneumoniae LAU-KP1 have been deposited at DDBJ/EMBL/GenBank under the accession no. AYQE00000000.1.
Results and Discussion
To the best of our knowledge we report for the first time the isolation of a CTX-M-15, SHV-11, KPC-producing K. pneumoniae belonging to ST336 in Lebanon. This clone belongs to CG17 and was isolated from a 62-year-old woman working as practitioner nurse or nurse's assistant. She was admitted for radical resection of an abdominal tumor and repair of abdominal hernia. The patient had other than her cancer, recurrent urinary tract infections.
In K. pneumoniae MLST is based on genetic variation in seven housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB, and tonB) (Diancourt et al., 2005). ST allele profile of ST336 is: 2-1-1-1-72-4-4, while ST258, being the most common KPC-producing isolate (Woodford et al., 2011), is: 3-3-1-1-1-1-79 (http://www.pasteur.fr/mlst). Screening patients for rectal carriage by Baraniak et al. (2013) revealed the prevalence of CG17 in France, highlighting the role of human mobility in spreading MDR K. pneumoniae clones.
LAU-KP1 was found to be resistant to the following drugs: ciprofloxacin, cefotaxime, aztreonam, and ceftazidime. It was intermediate to amoxicillin clavulanic acid, and sensitive to imipenem and gentamycin. Analysis of the data extracted from the draft genome sequence however, revealed that LAU-KP1 was characterized by the presence of multiple resistance determinants, most notably was the concomitant presence of acc(6′)lb-cr, qnr, SHV-11, and CTX-M-15. CTX-M-15 detection was consistent with the recent predominance of CTX-M and CTX-M-15 in particular (Rossolini et al., 2008), which additionally was found to be more common in K. pneumoniae than in E. coli in rehabilitation units (Baraniak et al., 2013). Rodrigues et al. (2014) reported recently that the increase in the incidence of CTX-M-15 and diverse SHV ESBL types observed in a Portuguese hospital was associated with the increase of MDR K. pneumoniae epidemic clones (ST15, ST147, ST336). Harbouring the qnrB1 (quinolone resistance) was another important finding that additionally distinguished LAU-KP1. Previously Breurec et al. (2013) revealed that qnrB was predominant among strains from Africa while qnrS was mainly detected in strains from Vietnam.
Resistance to carbapenems involves multiple mechanisms including alterations in outer membrane permeability mediated by the loss of porins, upregulation of efflux systems along with hyper production of AmpC β-lactamases or more commonly the production of carbapenemases (Chen et al., 2014). Class A carbapenemases include the K. pneumoniae carbapenemase (KPC). These enzymes hydrolyze all β-lactams including monobactams (El-Herte et al., 2012). Carbapenem resistant Enterobacteriaceae have spread in Northeastern USA and to other countries including Argentina, Greece, Italy, and China (Nordmann and Poirel, 2014). Transmission of the KPC gene can be mediated through horizontal gene transfer on mobile elements including transposons and plasmids (Munoz-Price and Quinn, 2009). Spread of the KPC-producing K. pneumonia has been linked to a major multi locus sequence type (MLST-ST) ST258 and its variants (Cuzon et al., 2010; Chen et al., 2014). LAU-KP1 harbored INCFIIK plasmids and DNA comparison showed the presence of sequences with high homology to plasmid pKPN-IT and pKPN3. This was consistent with the finding that the dissemination of CTX-M-15 was primarily associated with IncFIIK plasmid in K. pneumoniae isolates (Coelhoa et al., 2010). Earlier Garcia-Fernandez et al. (2012) detected in K. pneumoniae ST258 IncFIIK-FIB like plasmid and was named as pKPN-IT. This plasmid was highly related to plasmid pKPN3. The plasmid pKPN-IT was shown to also acquire Fec-like iron (III) dicitrate transport system and a class I integron carrying trimethoprim resistance, both of which were also detected in LAU-KP1.
β-lactamase class C, the presence of which renders bacteria to become resistant to most β-lactams including cephamycins and β-lactam/β-lactamase combinations (Doi and Paterson, 2007) was and along with macrolide-specific efflux pump MacAB-TolC important factors increasing the resistance of LAU-KP1. Periplasmic membrane fusion proteins (MFPs) are essential components of the type I protein secretion systems and drug efflux pumps in Gram-negative bacteria (Tikhonova et al., 2007). MacA is a peripheral membrane protein of the MFP family and a component of the MacAB macrolide efflux transporter complex. MacB on the other hand, is an integral membrane protein. Similar to other MFP-dependent transporters from E. coli, MacAB requires the outer membrane channel TolC for its function (Kobayashi et al., 2001; Tikhonova et al., 2007).
Additionally, a key feature of these MDR efflux systems is their ability to extrude a broad spectrum of substrates, including various antimicrobial agents (Bambeke et al., 2000; Putman et al., 2000; Poole, 2001). One important family of drug transporters that contribute to multidrug resistance (MDR) in gram-negative bacteria are the resistance-nodulation-cell division (RND) efflux systems, which consist of an inner membrane transporter, a periplasmic fusion protein, and an outer membrane protein (Zgurskaya and Nikaido, 2000). Detecting CmeABC operon in LAU-KP1 was an important feature. CmeABC is an energy-dependent efflux system contributing to the intrinsic resistance of Campylobacter to diverse antimicrobial agents. Disruption of the CmeABC resulted in MICs that were 8-fold lower for ciprofloxacin, 2-fold for norfloxacin and nalidixic acid, 256-fold for cefotaxime, and 32-fold for ampicillin (Lin et al., 2002).
In K. pneumoniae MLST is based on genetic variation in seven housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB, and tonB) (Diancourt et al., 2005). ST allele profile of ST336 is: 2-1-1-1-72-4-4, while ST258, being the most common KPC-producing isolate (Woodford et al., 2011), is: 3-3-1-1-1-1-79 (http://www.pasteur.fr/mlst). Screening patients for rectal carriage by Baraniak et al. (2013) revealed the prevalence of CG17 in France, highlighting the role of human mobility in spreading MDR K. pneumoniae clones.
Furthermore, the phylogenetic analysis of LAU-KP1 showed that it falls within a clade of other known multidrug resistant K. pneumoniae strains and demonstrated that there is considerable genomic diversity among the included isolates (Figure 1). Within the closely related isolates was K. pneumonia 7699 previously cultured from a perianal swab of a patient admitted to the intensive care unit of the University of Maryland Medical Center (UMMC) in Baltimore, MD (Hazen et al., 2014). The isolate had a 167-kb IncA/C plasmid encoding the FOX-5 β-lactamase, a CARB-2 β-lactamase, other antimicrobial resistance genes, and heavy metal resistance genes. On the other hand, a considerable phylogenetic difference was detected between LAU-Kp1 and K. pneumoniae ATCC BAA-2146, which is the first U.S. isolate found encoding the gene for the broad-spectrum and carbapenem-active metallo-β-lactamase NDM-1 (Hudson et al., 2014). Additionally, we mapped our contigs to the phylogenetically closest K. pneumoniae isolate with a complete genome, Ecl8, in order to visualize the genomic organization (Figure 2).

Figure 1. Phylogenetic tree of all assembled K. pneumoniae genomes available on the NCBI ftp server as of 2-11-15, based on a concatenated alignment of 37 conserved genes obtained using PhyloSift (Darling et al., 2014). The tree was constructed using default settings in FastTree (Price et al., 2010) and edited and visualized with Dendroscope v.3 (Huson and Scornavacca, 2012). Clades with zero branch lengths were collapsed to a single representative. K. pneumoniae LAU-KP1 is indicated by a square, and the reference genome used in Figure 2 is indicated by a diamond.
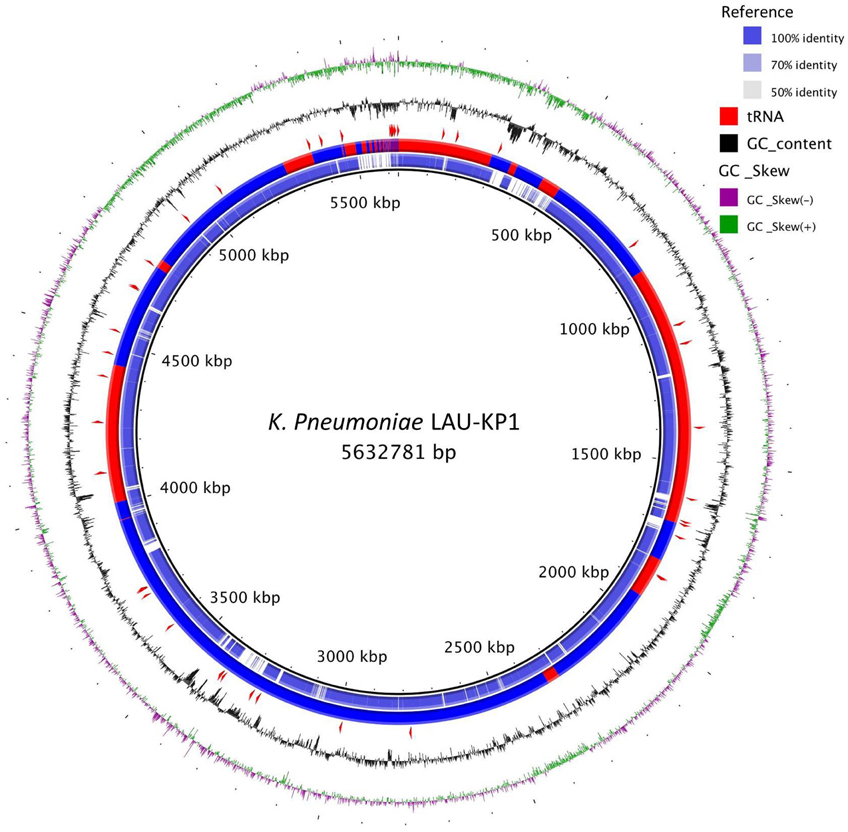
Figure 2. K. pneumoniae LAU-KP1 ring representation using BRIG 0.95 (Alikhan et al., 2011). The contigs from LAU-KP1 were ordered to match Ecl8's (GCF_000315385) genomic arrangement using progressiveMauve (Darling et al., 2010). Ecl8 is the K. pneumoniae isolate which was closest to LAU-KP1 in the tree shown in Figure 1 for which there was a finished genome available. The inner ring shows the percent identity comparing Ecl8 and LAU-KP1. The next ring alternating blue and red sections shows the contig delimitations for LAU-KP1. The next ring shows the location of tRNAs according to Prokka v1.7 (Seemann, 2014). The last two (outer) rings show the GC content and skew.
The detection of such a multidrug resistant strain of K. pneumoniae emphasizes the potential for transmission of highly resistant gram-negative pathogens that are difficult to treat. This merits more detailed future studies directed toward characterization of molecular mechanisms, environmental factors, and selection pressures promoting the spread of such strains. Detection of IncFIIK a virulence plasmid (Dolejska et al., 2013) known to evolve quickly by replicon diversification and acquisition of antibiotic resistance traits (Coelhoa et al., 2010; Carattoli, 2013), increases the potential role of K. pneumoniae as a reservoir for ESBL genes and other resistance determinants. The spread of antimicrobial resistance in this region could be attributed to different factors including: uncontrolled consumption of antimicrobial agents through self-medication, inappropriate antibiotic prescription, the substandard quality of some drugs, and a lack of effective measures to prevent nosocomial infections. The virtual lack of physical barriers between the community and hospital settings in the countries in this region and the increasing pressure from war immigrants along with poor living conditions may be also important contributory factors. High-resolution genotyping studies of isolates collected over larger spatial and temporal scales will be necessary to decipher the dynamics of the emergence of MDR K. pneumoniae clonal groups.
Therefore, we urgently need well-designed epidemiological and molecular studies to understand the dynamics of transmission, risk factors, and reservoirs for K. pneumoniae. This will provide a better insight into the emergence and spread of these multidrug resistant strains and hopefully lead to information essential in preventing infections and limiting the spread of such organisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Jacques Mokhbat, Mrs. Sanaa Zoghbi and the bacteriology team at the LAUMCRH for all their help and support.
References
Alikhan, N., Petty, K., Ben Zakour, N., and Beatson, S. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ambler, R., Coulson, A., Frere, J., Ghuysen, J., Joris, B., Forsman, M., et al. (1991). A standard numbering scheme for the class A β-lactamases. Biochem. J. 276, 269–270.
Bambeke, V., Balzi, F., and Tulkens, P. (2000). Antibiotic efflux pumps. Biochem. Pharmacol. 60, 457–470. doi: 10.1016/S0006-2952(00)00291-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Baraniak, R., Izdebski, J., Fiett, E., Sadowy, A., Adler, M., Kazma, J., et al. (2013). Comparative population analysis of Klebsiella pneumoniae with extended-spectrum β-lactamases colonizing patients in rehabilitation centers in four countries. Antimicrob. Agents Chemother. 57, 1992–1997. doi: 10.1128/AAC.02571-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bradford, P. A. (2001). Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–935. doi: 10.1128/CMR.14.4.933-951.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Breurec, S., Guessennd, N., Timinouni, M., Le, T., Cao, V., Ngandjio, A., et al. (2013). Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin. Microbiol. Infect. 19, 349–355. doi: 10.1111/j.1469-0691.2012.03805.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bush, K., Jacoby, G., and Medeiros, A. (1995). A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233. doi: 10.1128/AAC.39.6.1211
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chen, L., Mathema, B., Chavada, K., DeLeo, F., Bonomo, R., and Kreswirth, N. (2014). Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 22, 686–696. doi: 10.1016/j.tim.2014.09.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Coelhoa, A., González-Lópeza, J., Miróc, E., Alonso-Tarrésd, C., Mirelisb, B., Larrosaa, M., et al. (2010). Characterization of the CTX-M-15-encoding gene in Klebsiella pneumoniae strains from the Barcelona metropolitan area: plasmid diversity and chromosomal integration. Int. J. Antimicrob. Agents 36, 73–78. doi: 10.1016/j.ijantimicag.2010.03.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cuzon, G., Naas, T., Truong, H., Villegas, M., Wisell, K., Carmeli, Y., et al. (2010). Worldwide diversity of Klebsiella pneumoniae that produce β-lactamase blaKPC2 gene. Emerg. Infect. Dis. 16, 1349–1356. doi: 10.3201/eid1609.091389
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Darling, A., Jospin, G., Lowe, E., Matsen, A. I. V., Bik, H., and Eisen, J. (2014). PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 2:e243. doi: 10.7717/peerj.243
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Darling, A., Mau, B., and Perna, N. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. doi: 10.1371/journal.pone.0011147
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Diancourt, L., Passet, V., Verhoef, J., Grimont, A., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Doi, Y., and Paterson, D. (2007). Detection of plasmid-mediated class C β-lactamases. Int. J. Infect. Dis. 11, 191–197. doi: 10.1016/j.ijid.2006.07.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dolejska, M., Villa, L., Dobiasova, H., Fortini, D., Feudi, C., and Carattoli, A. (2013). Plasmid content of a clinically relevant Klebsiella pneumoniae clone from the Czech Republic producing CTX-M-15 and QnrB1. Antimicrob. Agents Chemother. 57, 1073–1076 doi: 10.1128/AAC.01886-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
El-Herte, R., Araj, G. F., Matar, G. M., Baroud, M., Kanafani, Z., and Kanj, S. (2012). Detection of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae producing NDM-1 in Lebanon. J. Infect. Dev. Ctries. 6, 457–461. doi: 10.3855/jidc.2340
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Garcia-Fernandez, A., Villa, L., Carta, C., Venditti, C., Giordano, A., Venditti, M., et al. (2012). Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56, 2143–2145. doi: 10.1128/AAC.05308-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Goossens, H., and Grabein, B. (2005). Prevalence and antimicrobial susceptibility data for extended-spectrum β-lactamases- and AmpC-producing Enterobacteriaceae from the MYSTIC program in Europe and the United States (1997-2004). Diagn. Microbiol. Infect. Dis. 53, 257–264. doi: 10.1016/j.diagmicrobio.2005.10.001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hazen, T., Zhao, L., Boutin, M., Stancil, A., Robinson, G., Harris, A., et al. (2014). Comparative genomics of an IncA/C multidrug resistance plasmid from Escherichia coli and Klebsiella isolates from intensive care unit patients and the utility of whole-genome sequencing in health care settings. Antimicrob. Agents Chemother. 58, 4814–4825. doi: 10.1128/AAC.02573-14
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hudson, C. M., Bent, Z., Meagher, R., and Williams, K. (2014). Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS ONE 9:e99209. doi: 10.1371/journal.pone.0099209
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Huson, D., and Scornavacca, C. (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 6, 1061–1067. doi: 10.1093/sysbio/sys062
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jacoby, G. A., and Medeiros, A. A. (1991). More extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 35, 1697–1704. doi: 10.1128/AAC.35.9.1697
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kobayashi, N., Nishino, K., and Yamaguchi, A. (2001). Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183, 5639–5644. doi: 10.1128/JB.183.19.5639-5644.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Levesque, C., Pich, L., Larose, C., and Roy, P. (1995). PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39, 185–191. doi: 10.1128/AAC.39.1.185
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lin, J., Michel, L., and Zhang, Q. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46, 2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Matar, G., Cuzon, G., Araj, G., Naas, T., Corkill, J., Kattar, M., et al. (2008). Oxacillinase-mediated resistance to carbapenems in Klebsiella pneumoniae from Lebanon. Clin. Microbiol. Infect. 14, 887–888. doi: 10.1111/j.1469-0691.2008.02059.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Munoz-Price, S., and Quinn, P. (2009). The spread of Klebsiella pneumoniae carbapenemases: a tale of strains, plasmids, and transposons. Clin. Infect. Dis. 49, 1739–1741. doi: 10.1086/648078
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Naas, T., Philippon, L., Poirel, L., Ronco, E., and Nordmann, P. (1999). An SHV-derived extended-spectrum β-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 1281–1284.
Nordmann, P., Cuzon, G., and Naas, T. (2009). The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 9, 228–236. doi: 10.1016/S1473-3099(09)70054-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Nordmann, P., and Poirel, L. (2014). The difficult-to-control spread of carbapenemase producers in Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 20, 821–830. doi: 10.1111/1469-0691.12719
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum, β-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/CMR.18.4.657-686.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Peirana, G., Sang, J., Pitondo-Silva, A., Laupland, K., and Pitout, J. (2012). Molecular epidemiology of extended-spectrum-β-lactamase-producing Klebsiella pneumoniae over a 10 year period in Calgary, Canada. J. Antimicrob. Chemother. 67, 1114–1120. doi: 10.1093/jac/dks026
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pfeifer, Y., Cullik, A., and Witte, W. (2010). Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 300, 371–379. doi: 10.1016/j.ijmm.2010.04.005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603.
Poole, K. (2001). Multidrug resistance in gram-negative bacteria. Curr. Opin. Microbiol. 4, 500–508. doi: 10.1016/S1369-5274(00)00242-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Price, M., Dehal, P., and Arkin, A. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Putman, M., Veen, H., and Konings, W. (2000). Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64, 672–693. doi: 10.1128/MMBR.64.4.672-693.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Queenan, A., and Bush, K. (2007). Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/CMR.00001-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rodrigues, C., Machadoa, E., Ramos, H., Peixea, L., and Novais, A. (2014). Expansion of ESBL-producing Klebsiella pneumoniae in hospitalized patients: a successful story of international clones (ST15, ST147,ST336) and epidemic plasmids (IncR, IncFIIK). Int. J. Med. Microbiol. 304, 1100–1108. doi: 10.1016/j.ijmm.2014.08.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rosenau, A., Cattier, B., Gousset, N., Harriau, P., and Quentin, R. (2000). Capnocytophaga ochracea: characterization of a plasmid encoded extended-spectrum TEM-17 β-lactamase in the phylum Flavobacter-Bacteroides. Antimicrob. Agents Chemother. 44, 760–762. doi: 10.1128/AAC.44.3.760-762.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rossolini, G., D'Andrea, M., and Mugnaioli, C. (2008). The spread of CTX-M-type extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14, 33–41. doi: 10.1111/j.1469-0691.2007.01867.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Spanu, T., Luzzaro, F., Perilli, M., Amicosante, G., Toniolo, A., and Fadda, G. (2002). Occurrence of extended-spectrum β-lactamases in members of the family Enterobacteriaceae in Italy: implications for resistance to β-lactams and other antimicrobial drugs. Antimicrob. Agents Chemother. 46, 196–202. doi: 10.1128/AAC.46.1.196-202.2002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tikhonova, E., Devroy, V., Lau, S., and Zgurskaya, H. (2007). Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol. Microbiol. 63, 895–910. doi: 10.1111/j.1365-2958.2006.05549.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tritt, A., Eisen, J., Facciotti, M., and Darling, A. (2012). An integrated pipeline for de novo assembly of microbial genomes. PLOS ONE 7:e42304. doi: 10.1371/journal.pone.0042304
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Woodford, N., Turton, J., and Livermore, D. (2011). Multi-resistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35, 736–755. doi: 10.1111/j.1574-6976.2011.00268.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zgurskaya, H., and Nikaido, H. (2000). Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37, 219–225. doi: 10.1046/j.1365-2958.2000.01926.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: ESBL, Klebsiella pneumoniae, whole genome sequencing, CTX-M-15, SHV-11
Citation: Tokajian S, Eisen JA, Jospin G, Farra A and Coil DA (2015) Whole genome sequencing of extended-spectrum β-lactamase producing Klebsiella pneumoniae isolated from a patient in Lebanon. Front. Cell. Infect. Microbiol. 5:32. doi: 10.3389/fcimb.2015.00032
Received: 28 November 2014; Accepted: 19 March 2015;
Published: 08 April 2015.
Edited by:
Ghassan M. Matar, American University of Beirut, LebanonReviewed by:
Valerio Iebba, ‘Sapienza’ University of Rome, ItalyNeta Sal-Man, Ben-Gurion University of the Negev, Israel
Copyright © 2015 Tokajian, Eisen, Jospin, Farra and Coil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sima Tokajian, Department of Natural Sciences, School of Arts and Sciences, Lebanese American University, P.O. Box 36, Block A, Byblos, Lebanon stokajian@gmail.com
 Sima Tokajian
Sima Tokajian Jonathan A. Eisen
Jonathan A. Eisen Guillaume Jospin
Guillaume Jospin Anna Farra
Anna Farra David A. Coil2
David A. Coil2