Non-tuberculous Mycobacteria in South African Wildlife: Neglected Pathogens and Potential Impediments for Bovine Tuberculosis Diagnosis
- Tuberculosis Laboratory, Onderstepoort Veterinary Institute, Zoonotic Diseases, Agricultural Research Council, Onderstepoort, South Africa
Non-tuberculous mycobacteria (NTM) are not only emerging and opportunistic pathogens of both humans and animals, but from a veterinary point of view some species induce cross-reactive immune responses that hamper the diagnosis of bovine tuberculosis (bTB) in both livestock and wildlife. Little information is available about NTM species circulating in wildlife species of South Africa. In this study, we determined the diversity of NTM isolated from wildlife species from South Africa as well as Botswana. Thirty known NTM species and subspecies, as well as unidentified NTM, and NTM closely related to Mycobacterium goodii/Mycobacterium smegmatis were identified from 102 isolates cultured between the years 1998 and 2010, using a combination of molecular assays viz PCR and sequencing of different Mycobacterial house-keeping genes as well as single nucleotide polymorphism (SNP) analysis. The NTM identified in this study include the following species which were isolated from tissue with tuberculosis- like lesions in the absence of Mycobacterium tuberculosis complex (MTBC) implying their potential role as pathogens of animals: Mycobacterium abscessus subsp. bolletii, Mycobacterium gastri, Mycobacterium species closely related to Mycobacterium goodii/Mycobacterium smegmatis, Mycobacterium brasiliensis, Mycobacterium sinense JMD 601, Mycobacterium avium subsp. avium, Mycobacterium sp. GR-2007, Mycobacterium bouchedurhonense, and Mycobacterium septicum/M. peregrinum. Mycobaterium brasiliensis, Mycobacterium gastri, Mycobacterium sp. GR-2007, and a potential novel Mycobacterium species closely related to Mycobacterium goodii were found for the first time in this study to be potential pathogens of animals. Mycobacterium simiae was isolated from a sample originating from a tuberculin skin test positive reactor, demonstrating its potential to elicit inappropriate immune responses in animals that may interfere with diagnosis of tuberculosis by immunology. Mycobacterium abscessus subsp. bolletti was the most frequently detected NTM identified in 37 of the 102 isolates. Other NTM species were also isolated from animals not showing any pathological changes. Knowledge gained in this study contribute to the understanding of NTM species circulating in wild animals in South Africa and the pathogenic potential of certain species, whose role in disease causation need to be examined, as well as to a certain extent the potential of M. simiae to hamper the diagnosis of bTB.
Introduction
Non-tuberculous mycobacteria (NTM), otherwise known as “mycobacteria other than tuberculosis” (MOTT) or environmental mycobacteria (EM) are believed to be natural inhabitants of the environment, found as saprophytes, commensals, and symbionts in the ecosystem. Since their clinical relevance was unknown, these bacteria have been neglected for many years as they have always been recognized as just environmental contaminants or colonizers (Covert et al., 1999). It was only after the acquired immune deficiency syndrome (AIDS) epidemic that NTM attracted abrupt attention in certain countries (Gopinath and Singh, 2010). Finding immune compromised hosts, especially with the emergence of the AIDS pandemic, some NTM species are now recognized as potential opportunistic pathogens of humans (Mirsaeidi et al., 2014). However, other than a few known NTM pathogens of animals, including the well-known Mycobacterium avium subsp. paratuberculosis, the etiological agent of Johne's disease and other members of the Mycobacterium avium complex (MAC), reports of animal diseases caused by NTM are few.
Currently there are more than 150 non-tuberculous mycobacterial species listed in public databases and about a third of them have been implicated in diseases of humans. (http://www.bacterio.net; Botha et al., 2013). Because of the ubiquity of NTM, human infections have been reported from most geographic areas in the world and the geographical distribution is thought to be regional (Martin-Casabona et al., 2004; van Ingen et al., 2009). Mycobacterial diseases caused by NTM have been classified as skin lesions, localized lymphadenitis, TB-like pulmonary lesions and disseminated diseases (Primm et al., 2004). By far the most notable opportunistic NTM pathogens of animals and humans are the members of the Mycobacterium avium complex (MAC) consisting of closely related species and subspecies including Mycobacterium avium subsp. avium, Mycobacterium avium subsp. paratuberculosis, Mycobacterium avium subsp. hominissuis, Mycobacterium intracellulare, Mycobacterium sylvaticum, Mycobacterium colombiense, Mycobacterium bouchedurhonense, Mycobacterium timonense, Mycobacterium chimaera, Mycobacterium arosiense, Mycobacterium yongonense, and Mycobacterium marseillense (Cayrou et al., 2010). Other common NTM species reported to cause opportunistic mycobacterial diseases include Mycobacterium kansasii, Mycobacterium marinum, and Mycobacterium ulcerans. Animal infections by M. kansasii are very rare (Botha et al., 2013). Infections by Mycobacterium haemophilum, Mycobacterium szulgai, Mycobacterium fortuitum, Mycobacterium chelonae, Mycobacterium wolinskyi, Mycobacterium scrofulaceum, Mycobacterium abscessus, Mycobacterium xenopi, Mycobacterium genavense, Mycobacterium fortuitum, Mycobacterium porcicum, Mycobacterium farcinogens, Mycobacterium senegalense, and Mycobacterium genavense have also been reported in humans or animals. These NTM species have been reported to be etiology of mycobacterial diseases in humans and a wide range of animal species including livestock, wildlife amphibians, fish, reptiles, seals, birds, and rodents (Tortoli et al., 1998; Bercovier and Vincent, 2001; Martin-Casabona et al., 2004; van Ingen et al., 2008; Malama et al., 2014).
There are even fewer reports of NTM isolated from wildlife in Africa. Studies on isolation of NTM from animal sources have mainly focused on either livestock from slaughter houses or on NTM that were coincidentally isolated from animal tissue, including livestock and wildlife, while looking for Mycobacterium bovis (Kazwala et al., 1998; Tschopp et al., 2010). The extent of NTM infection and distribution in African wildlife is therefore largely unknown. In a study in Ethiopian cattle, Mycobacterium nonchromogenicum was isolated as a predominant species (Berg et al., 2009) while Mycobacterium terrae was isolated as a frequently occurring species in Ethiopian wildlife (Tschopp et al., 2010). A study from Chad found M. nonchromogenicum together with MAC and M. fortuitum to be common in humans and cattle (Diguimbaye-Djaïbe et al., 2006). Mycobacterium terrae was isolated from cattle in Tanzania (Kazwala et al., 1998; Cleaveland et al., 2007). M. nonchromogenicum has also been detected in small mammals and cattle in Tanzania (Durnez et al., 2011). A recent study in Tanzania identified Mycobacterium intracellulare, Mycobacterium lentiflavum, Mycobacterium fortuitum, and Mycobacterium chelonae-abscessus group as the most frequently isolated NTM from humans and wildlife (Katale et al., 2014). It should be noted that, isolation of NTM from an animal source does not necessarily imply an active disease status. Mycobacterium goodii was reported to have caused infection in a hyena in South Africa as well as in African rodents in Tanzania and M. kansasii has been isolated from wild animal (Bercovier and Vincent, 2001; van Helden et al., 2009; Durnez et al., 2011). M. farcinogenes and M. senegalense were reported to be responsible for bovine farcy, pathology found in Africa (Bercovier and Vincent, 2001). The environment is also of interest as the source of NTM infections (van Ingen et al., 2009). In Uganda, M. nonchromogenicum, M. fortuitum complex, M. avium complex, and M. gordonae were identified as most frequently detected species from environmental samples (Kankya et al., 2011). A study in Zaire also identified M. nonchromogenicum and M. terrae to be among the most prevalent NTM species in the environment (Portaels, 1995). In South Africa the following NTM have been identified as most occurring in different studies: Gcebe et al. (2013) have found the following NTM to be frequently occurring in African buffaloes, cattle and their environments: Mycobacterium terrae, M. nonchromogenicum, M. vaccae/M. vanbaalenii, and unidentified species closely related to Mycobacterium moriokense. They also noted an overlap in terms of the species isolated from the animal sources as well as the environments indicating that NTM are readily exchanged between the environment and animals (Gcebe et al., 2013). Botha et al. (2013) have reported isolation of the following NTM from different wildlife species including lions, rhinos, banded mongooses, cattle, baboons, elephants, and monkeys: M. abcessus, M. asiaticum, M. avium, M. brasilienses, M. chelonae, M. elephantis, M. engbackii, M. farcinogenes, M. fortuitum, M. gilven, M. gordonae, M. heraklionense, M. hiberniae, M. intracellulare, M. interjectum, M. lentiflavum, M. marseillense, M. moriokaense, M. nonchromogenicum, M. palustre, M. pulveris, M. paraffinicum, M. phlei, M. senegalense, M. simiae, M. sherrisii, M. sphagni, M. terrae, and M. vulneris. Mycobacterium gordonae and Mycobacterium gilvum were identified as the most frequently occurring NTM in biofilms, in a study by September et al. (2004).
In Africa, high interaction between wild animals especially captive and semi-captive, the environment, and humans seem to be very common posing a risk of NTM contamination of natural waters and transmission of NTM between animals and humans (Katale et al., 2014).
From a veterinary point of view, besides being opportunistic pathogens, some NTM species may colonize the host without development of any disease, but inappropriately prime the host's immune system hampering the immuno-diagnosis of bovine tuberculosis by tuberculin based assays (Vordermeier et al., 2007; Schiller et al., 2010). Furthermore, NTM may have a negative effect on vaccination of animals by Mycobacterium bovis Bacillus Calmette-Guérin (BCG; Poyntz et al., 2014). Incidents of bovine tuberculosis in different South African wildlife have been reported, and efforts for eradication or reduction of the diseases to minimal levels include the test and slaughter strategies (Hlokwe et al., 2014). Vaccination is also a possibility. Yet very little information is available on NTM circulating in wild animals in South Africa. Since, bovine tuberculosis (bTB) is endemic in South African wildlife conservation areas, specifically, in the Kruger National Park, incidences of NTM infection of wildlife may have been neglected probably due to the fact that focus has been on bTB, and NTM infection may be last on the agenda or perceived to be less important. The irony is that the interference of NTM in the diagnosis of bTB as well is in vaccination by BCG may hinder the bTB control/eradication efforts in wildlife of South Africa.
In this study, we determined the diversity of NTM species isolated from different wildlife species of South Africa and Botswana.
This study forms the basis for understanding the diversity of NTM species which are circulating in the South African wildlife as well as potential pathogens of different animal species as well as NTM that may have potentially induced in-appropriate immune responses which interfered with the diagnosis of tuberculosis. This information may hopefully aid in the efforts for control of bovine tuberculosis in wildlife through development of diagnostic assays and vaccination strategies.
Materials and Methods
Ethics Statement
This work was carried out from samples submitted for routine diagnostic purposes in the Tuberculosis laboratory: a Veterinary Laboratory of South Africa approved by the Department of Agriculture, Forestry, and Fisheries (DAFF) in compliance with the requirements of the Animal Diseases act no. 35 of 1984 as well as DAFF 001, 002, and 012 procedures.
Study Site
Isolates originating from samples collected for bovine tuberculosis or mycobacterial routine diagnosis from the Greater Kruger National Park (GKNP) complex, the National Zoological Gardens (NZG) of South Africa, and four private game reserves, were used in this study. The Greater Kruger National Park complex consists of several game parks/reserves and covers the area in the north east of South Africa, covering 19.485 km2. National Zoological Gardens of South Africa is located in Pretoria, north of the Gauteng Province north east of South Africa, covering 0.85 km2. Three of the private game reserves are located in the Mpumalanga Province, east of South Africa, and one is located in the Northern Cape Province, north west of South Africa. Isolates from mongoose and wildcat from Botswana were also included in the study.
Sample Collection from Wildlife
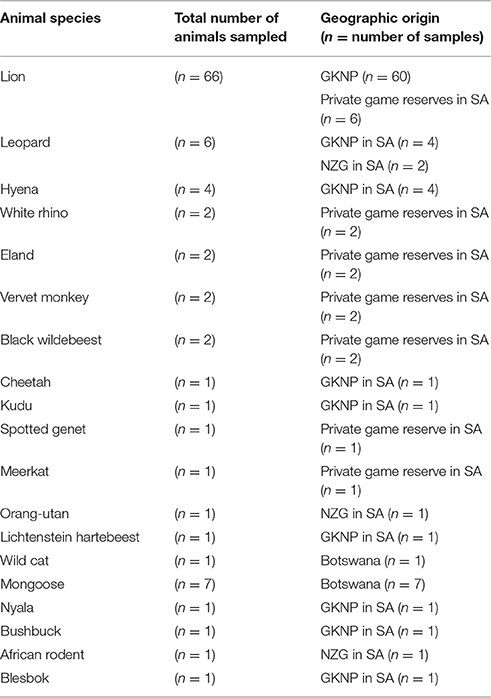
Samples were collected opportunistically from dead as well as live animals. Dead animals included road kills, animals that died from disease or old age and animals that were culled for bTB confirmation after they tested positive on tuberculin skin test (TST). One hundred and two (102) tissue samples including bronchial, mesenteric, thoracic, peripheral, abdominal, mammary, axial, popliteal, cranial, and head lymph nodes as well as lungs, liver, testicles, heart, tonsils, bone marrow, ulna, nose, bronchial washes, and elbow hygroma were collected between the years 1998 and 2010. Information regarding the different animal species sampled and their geographic origin is presented in Table 1.
Pathological Examination and Culture
Tissue samples to be processed for mycobacterial isolation were carefully examined macroscopically for formation of suspected tuberculous-like lesions (visible). The presence or absence of suspect bTB-like lesion was recorded for each sample.
All tissue samples were processed and cultured according to standard laboratory procedures in a Class II biosafety cabinet (Esco Technologies, South Africa) following biosafety guidelines as described in the OIE Terrestrial manual (OIE, 2015). Briefly, ~5 g of tissue samples were cut into small pieces and covered with 100 ml of sterile distilled water. The samples were homogenized at 4500 rpm using the Ultra-Turrax® homogenizer (Separation Scientific, SA). Seven milliliters of the homogenates were poured into two separate 15 ml falcon tubes, and decontaminated with 7 ml of 2% HCl and 7 ml of 4% NaOH, respectively. The suspensions were incubated at room temperature for 10 min and centrifuged again. Following centrifugation, sterile distilled water was added to the pellet. The pellet was inoculated onto four Löwenstein-Jensen (LJ) media slopes supplemented with pyruvate, as well as two slopes supplemented with glycerol and an antibiotic cocktail of polymyxin B, ampho-tericin B, carbenicillin, and trimethoprim (PACT) (National Health Laboratories, South Africa, and Becton Dickinson, South Africa). The slopes were incubated at 37°C and monitored weekly for mycobacterial growth. When growth of bacteria was observed, based on morphology of mycobacterial colonies, individual colonies were selected for Ziehl–Neelsen staining. The acid fast cultures were stored at −21°C for further analysis.
DNA Extraction
DNA was extracted using the heating method. Individual acid fast colonies were suspended in 100 μl of sterile distilled water and heated at 95°C in a heating block/or in boiling water for 25 min. The culture suspensions were stored at −70 or −20°C until further analysis.
In vitro Amplification and Sequencing of the Mycobacterial 16S rDNA and hsp65 Genes for NTM Speciation
PCR and sequencing of the 577 bp fragment of the mycobacterial 16S rDNA was used for mycobacterial speciation (Harmsen et al., 2003; Gcebe et al., 2013). Sequencing of the 439 bp hsp65 gene fragment was done for differentiation of some NTM species with similar, or identical 16S rRNA which could not be delineated using the 16S rDNA PCR-sequencing assay (Telenti et al., 1993).
16S rDNA PCR
PCR targeting a 577 bp fragment of mycobacterial 16S rDNA was performed using the primers: 16S-F (5′-AGA GTT TGA TCM TGG CTCAG-3′) and 16S-R (5′-GCG ACA AAC CAC CTA AGA G-3′). Culture suspensions were used as DNA template in a 25 μl PCR mixture containing 12.9 μl deionized water, 2.5 μl of 10X PCR buffer [160 mM; Tris Cl, KCl, (NH4)2SO4], 2 μl MgCl2 (25 mM), 1 μl dNTPs (10 mM), 0.1 μl Taq polymerase (Qiagen Hotstar Taq, Whitehead Scientific, South Africa), 5 μl of 5x Q-solution, 1 μl of each forward and reverse primers (50 pmol) and 1–2 μl DNA template. The PCR cycling parameters were as follows: initial denaturation at 95°C for 15 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 30 s and a final extension at 72°C for 10 min.
hsp65 PCR
Tb11 (5′-ACCAACGATGGTGTGTCCAT-3′) and Tb12 (5′-CTTGTCGAACCGCATACCCT-3′) primers were used for the amplification of the hsp65 gene fragment. A 50 μl PCR mixture containing 28.5 μl de-ionized water, 3 μl MgCl2 (25 mM), 1 μl dNTP mix (10 mM), 4.75 μl of 10x PCR buffer (160 mM) [Tris-HCl, MgCl2, Tween 20, (NH4)2,SO4], 0.75 μl Taq DNA Polymerase (5 U/μl; Supertherm™), 1 μl of each forward and reverse primers (50 pmol) and 10 μl of DNA template was assembled. The PCR cycling parameters were as follows: Forty-five cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and elongation at 72°C for 1 min and final extension at 72°C for 10 min.
The PCR products were sent to Inqaba Biotechnologies, South Africa for sequencing of the forward 16S rDNA strands and both hsp65 gene strands using an ABI sequencer. Sequences were edited manually and pairwise alignments undertaken using the BioEdit Sequence alignment editor (version 7.1.9) and Molecular Evolutionary Genetics Analysis (MEGA) platform (www.megasoftware.net; version 7; Kumar et al., submitted). The sequences were analyzed on the NCBI BLAST platform for species identification (https://blast.ncbi.nlm.nih.gov/Blast.cgi) by megablast.
PCR for Detection of Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis
To delineate between two members of MAC: i.e., M. avium subspecies paratuberculosis and M. avium subsp. avium which could not be differentiated by the genus specific 16S rDNA-sequencing assay, the MYCGEN-MYCAV PCR as described by Wilton and Cousins (1992) as well as amplification of the IS900 of the M. avium subsp. paratuberculosis (Cousins et al., 1999) was conducted in two separate PCR reactions using primers described below:
MYCGEN-F (5′-AGAGTTTGATCCTGGCTCAG-3′), MYCGEN-R (5′-TGCACACAGGCCACAAGGGA-3′) and MYCAV (5′-ACCAGAAGACATGCGTCTTG- 3′) primers were used for screening of the Mycobacterium avium subspecies avium isolates as described by Wilton and Cousins (1992). Briefly a 25 μl PCR mixture was prepared, containing 14 μl de-ionized water, 2 μl MgCl2 (25 mM), 1 μl dNTP mix (10 mM), 1 μl of 10x PCR buffer [160 mM; Tris-HCl, MgCl2, Tween 20, (NH4)2, SO4], 0.125 μl Taq DNA Polymerase (5 U/μl; Supertherm™), 0.5 μl of each primer (50 pmol), and 5 μl of DNA template. The PCR cycling parameters were as follows: Initial denaturation at 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 62°C for 3 min, and elongation at 75°C for 3 min.
IS900 (P90) forward: 5′-GAAGGGTGTTCGGGGCCGTC-3′ and IS900 (P91) reverse: 5′-GAGGTCGATCGCCCACGTGAC-3′ primers were used for screening of the isolates for M. avium subsp. paratuberculosis in 50 μl PCR reaction mixture containing 28 μl de-ionized water, 3 μl MgCl2 (25 mM), 2 μl dNTP mix (10 mM), 5 μl of 10x PCR buffer [160 mM; Tris -HCl, MgCl2, Tween 20, (NH4)2,SO4], 0.125 μl Taq DNA Polymerase (5 U/μl; Hotstar), 1 μl of each primer (50 pmol), and 10 μl of DNA template. The PCR cycling parameters were as follows: Initial denaturation at 95°C for 10 min, followed by fifty cycles of denaturation at 95°C for 1 min, annealing at 65°C for 75 s, and elongation at 72°C for 3 min. This was followed by a final elongation at 72°C for 10 min. Amplicons were analyzed by electrophoresis on a 1.5% agarose gel.
Alignment of the 16S rDNA Sequences and Phylogenetic Analysis
Multiple alignment of the part of the 16S rDNA sequences from representative isolate of each species identified in the study as well other NTM sequences (both slow and rapidly growing) extracted from NCBI Nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore) was performed using MEGA version 7.0 platform (Kumar et al., submitted). For phylogenetic analysis the 16S rDNA sequences were first trimmed at both the 5′ and the 3′ ends to encompass the most corresponding gene fragment sequences of mycobacteria deposited in NCBI Nucleotide database. Phylogenetic trees were constructed using the neighbor-joining method (Satou and Nei, 1987) and validated using the maximum composite likelihood method. One thousand bootstrap replicates were run and Nocardia farcinica was used as an outgroup.
Since M. smegmatis and M. goodii have similar 16S rDNA sequences, isolates belonging or closely related to these two species were differentiated by single nucleotide polymorphism (SNP) analysis of their 16S rDNA sequences (Figure 1; van Helden et al., 2008). Alignment of part of the isolate's 16S rDNA as well as that of M. goodii and M. smegmatis reference sequences extracted from NCBI Nucleotide (https://www.ncbi.nlm.nih.gov/nuccore) database was also performed using MEGA version 7. SNP identification was done manually.
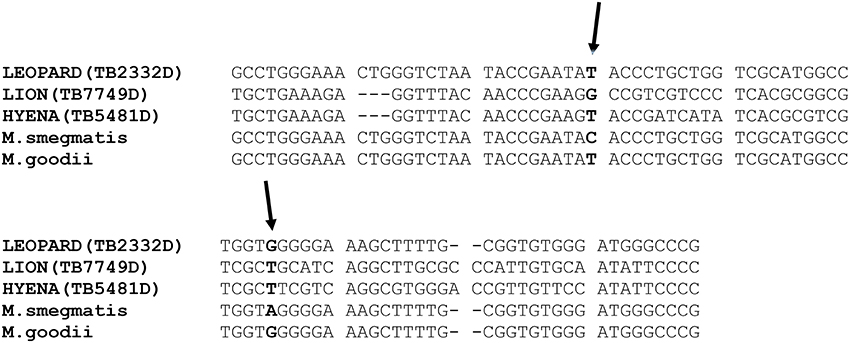
Figure 1. Alignment of part of the 16S rDNA sequences of three isolates and those of the reference M. goodii and M. smegmatis retrieved from NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide/) demonstrating similarity between the isolates from leopard to M. goodii as well as sequence differences between the isolates from hyena and lion to M. goodii and M. smegmatis. The 16S rDNA sequence from the lion was submitted to Genbank database and the accession number is KX670576. The two arrows point to single nucleotide differences that exist between M. smegmatis and M. goodii are in this part of the 16S rDNA.
Results
The diversity of NTM species isolated from different wild animals in South Africa is presented in Table 2. The 16S rDNA and hsp65 gene sequence analysis revealed that, of 102 isolates analyzed, 96 belong to 30 known Mycobacterium species, two belonged to a species closely related to Mycobacterium goodii and M. smegmatis and six represented species that could not be identified from the NCBI database (<95% identical to species in NCBI databse). SNP analysis of part of the 16S rDNA of two isolates closely related to M. goodii and M. smegmatis and M. smegmatis ATCC 19420 and M. goodii ATCC 700504 reference strains revealed differences between these two isolates and M. goodii and M. smegmatis, suggesting that these are potentially novel species. The 16S rDNA fragment sequence of one of the isolate closely related M. goodii/M. smegmatis originating from a lion sample with tuberculosis-like lesions was submitted to the Genbank database (https://www.ncbi.nlm.nih.gov/genbank) and the accession number is KX670576.
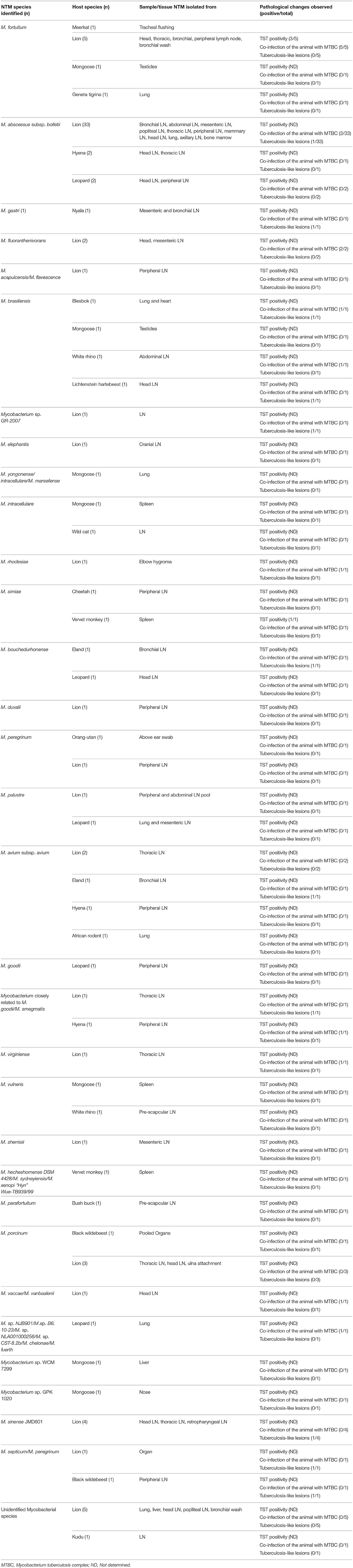
Of the 102 isolates analyzed, 37 (36.3%) were identified as Mycobacterium abscessus subsp. bolletii by sequencing of both 16S rDNA and the hsp65 gene fragments. Thirty-three of the 37 isolates originated from lion samples, two from hyenas and another two from leopards. The second most frequently isolated species was M. fortuitum (n = 8) isolated from lions (n = 5), meerkat (n = 1), geneta tigrina (n = 1), and mongoose (n = 1); followed by M. sinense JMD601 (n = 4) with all isolates originating from lion samples; M. brasiliensis (n = 4) isolated from blesbok (n = 1), mongoose (n = 1), white rhino (n = 1) and Lichtenstein hartebeest (n = 1); and finally M. porcinum (n = 4) isolated from lions (n = 3) and a black wildebeest (n = 1). The other NTM species were identified in low numbers of <4 and these include: M. goodii, M. gastri, M. fluoranthenivorans, M. acapulcensis/M. flavescence, Mycobacterium sp. GR-2007, M. elephantis, M. intracellulare, M. yongonense/intracellulare/M. marseilense, M. rhodesiae, M. simiae, M. bouchedurhonense, M. duvalii, M. peregrinum, M. palustre, M. avium subsp. avium, M. virginiense, M. sherrisiii, M. vulneris, M. hecheshornense DSM 4428/M. sydneyiensis/M. xenopi “Hyn” Wue-TB939/99, M. parafortuitum, M. sp. NJB901/M.sp. B6. 10-23/M. sp. NLA001000256/M. sp. CST-8.2b/M. chelonae/M. fuerth, M. vaccae/M. vanbaalenii, Mycobacterium sp. WCM 7299, Mycobacterium sp. GPK 1020, M. septicum/M. peregrinum, and Mycobacterium species closely related to M. goodii/M. smegmatis.
Phylogenetic relatedness of the isolates representative of each species identified in this study, and other Mycobacterium species, based on the 16S rDNA sequences is illustrated by the phylogenetic tree in Figure 2.

Figure 2. Phylogenetic tree constructed using neighbor joining method, illustrating the genetic position of the isolates. The species that the isolates were found to belong to are shown in parenthesis next to each isolate. Genbank accession numbers for the reference sequences are also shown in parenthesis. The tree is based on the partial 16S rDNA gene sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. Nocardia farcinica was used as an out group sequence. Evolutionary analyses were conducted in MEGA7.
To investigate if the NTM species isolated could have potentially caused tuberculous-like or cross-reactive immune responses, records relating to pathological changes including the formation of tuberculosis-like lesions and the tuberculin skin test positivity of the animals were analyzed. Information about whether more than one Mycobacterium species including Mycobacterium tuberculosis complex (MTBC) or another NTM were isolated from a single sample showing tuberculosis-like lesion was analyzed in-order to eliminate the possibility or any doubt or confusion about which mycobacterial species could have potentially caused lesions. However, we did not investigate other pathogens other than Mycobacteria as potential causes of lesions, a move that could have strengthened findings of certain NTM species identified in this study, as opportunistic pathogens of animals. A similar analysis was done at animal level (Investigating if isolation of more than one Mycobacterium species from the same animal). Likewise, information about isolation of more than one Mycobacterium species from the TST positive reactors was also investigated for elimination of any doubt about which mycobacteria could have elicited an immune response. The summary of the data is presented in Table 2.
Tuberculosis-like lesions were observed in the mesenteric and bronchial lymph nodes of nyala; lung, heart and head lymph node of blesbok, and haartebeest; bronchial lymph nodes of an eland and a lion; retropharyngeal lymph nodes and lung of lion. In all these cases, pure cultures of the following NTM species or subspecies were isolated and no any other mycobacterial species was isolated from the same animal or sample: M. abscessus subsp. bolletii isolated from lion, Mycobacterium gastri was isolated from nyala; Mycobacterium brasiliensis from blesbok and hartebeest; Mycobacterium avium subsp. avium from eland; Mycobacterium sp. GR-2007, Mycobacterium sinense JMD601, Mycobacterium species closely related Mycobacterium goodii/Mycobacterium smegmatis, and Mycobacterium septicum/M. peregrinum were all isolated from lions. Co-infection of the host animal species with M. bovis/M. tuberculosis and the following NTM species was observed: M. abscessus subspecies bolletii, M. fluoranthenivorans, M. brasiliensis, M. rhodesiae, Mycobacterium species closely related to M. goodii/M. smegmatis, M. virginiense, M. vaccae/M. vanbaalenii and finally, M. sp. NJB901/M.sp. B6. 10-23/M. sp. NLA001000256/M. sp. CST-8.2b/M. chelonae/M. fuerth in different animal species as shown in Table 2.
A pure culture of Mycobacterium simiae was isolated from TST positive reactor vervet monkey. No other mycobacterial species was isolated from this animal. In other cases, where NTM were isolated, there were either no visible lesions or no information regarding TST (Table 2).
Discussion
In this study 30 known NTM species, a species closely related to Mycobacterium goodii and M. smegmatis (comprising of two isolates) as well-unidentified species (comprising of six isolates) were detected from 102 isolates originating from wild animals in South Africa and Botswana, collected between the years 1998 and 2010. These findings demonstrate a wide diversity of NTM species circulating in the wildlife ecosystems of Southern Africa. Furthermore, isolation of strains belonging to unidentified species suggests the occurrence of a number of uncharacterized potentially novel NTM species in wildlife. In this study except for a species found to be closely related to M. goodii and M. smegmatis, they were not associated with any pathological changes in their hosts and as such, their roles in causing diseases and in eliciting an immune response are not fully known. With the use of a combination of molecular assays viz sequencing of the 16S rDNA and the hsp65 genes as well as SNP analysis of the 16S rDNA, screening of the isolates for IS900 and Mycobacterium avium specific 16S rDNA gene fragments, we were able to delineate closely related species as well as identification of some NTM to subspecies level. The NTM identified in this study comprise species of which data for their clinical and veterinary significance have been reported previously (known pathogens), which have been previously described as opportunistic pathogens; as well as species that have not been previously reported to cause any diseases. The isolation of these NTM groups (known pathogens as well as those whose pathogenicity has not been reported in animals) from tissue samples with tuberculosis-like lesions in this study suggests their potential role as sources of infection in those particular animals. These include Mycobacterium abscessus subsp. bolletii, Mycobacterium gastri, Mycobacterium closely related to Mycobacterium goodii, Mycobacterium brasiliensis, Mycobacterium sinense JMD 601, Mycobacterium avium subsp. avium, Mycobacterium sp. GR-2007, Mycobacterium septicum/M. peregrinum, and Mycobacterium bouchedurhonense. Mycobacterium abscessus subspecies bolletii together with Mycobacterium abscessus subspecies abscessus and Mycobacterium abscessus subspecies massiliense constitute the Mycobacterium abscessus group. These rapidly growing Mycobacteria (RGM) have been identified not only as sources of pulmonary infections but as sources of nosocomial, skin and soft tissue infections, osteoarticular infections as well as disseminated diseases in humans (Viana-Niero et al., 2008; Piersimoni and Scarparo, 2009; Zelazyn et al., 2009; Colombo et al., 2012). Another significance of these NTM is their high resistance to antibiotics (Nessar et al., 2012). The different subspecies however differ from each other in their antibiotic resistance profile (Nessar et al., 2012). Information on the pathogenic role of Mycobacterium abscessus subspecies bolletii in animals is still scarce. This is the first study to report M. abscessus subspecies bolletii as a probable cause of lesions in a peripheral lymph node of a lion in South Africa. Mycobacterium abcessus has been isolated in various fish species and has been described to cause diseases in humans (Bercovier and Vincent, 2001). In a study in Asia, M. abscessus was found to be the second most clinically relevant NTM, after Mycobacterium avium subsp. avium, responsible for pulmonary diseases (Simons et al., 2011). M. abscessus has also been previously reported in animals of South Africa, including wildlife, but not necessarily as a cause of diseases (Botha et al., 2013; Gcebe et al., 2013). In three other cases in this study, co-infection with M. bovis and this NTM was observed in lions. Isolation of M. abscessus subspecies bolletii together with M. bovis from sample presenting tuberculosis-like lesions could indicate the role of this NTM as an opportunistic pathogen. Contrary, to what has been reported in wildlife NTM diversity studies in Ethiopia, Tanzania, and buffaloes in South Africa, M. abscessus subsp. bolletii was isolated most frequently in this study mainly from lions (Tschopp et al., 2010; Gcebe et al., 2013; Katale et al., 2014).
Only a few reports of Mycobacterium gastri as a cause of disseminated and pulmonary mycobacteriosis diseases in humans have been published (Velayati et al., 2005; Guerra et al., 2008). Our study presents a first report of Mycobacterium gastri isolation in South Africa from an animal species (nyala) which presented tuberculosis -like lesions. Investigation of the pathogenic role of this NTM in animals is therefore necessary.
Mycobacterium avium subspecies avium is a well-known and mostly studied animal pathogen (Rastogi et al., 2001). We isolated this species/subspecies from a bronchial lymph node of an eland with lesions, confirming its status as an animal pathogen. It was also isolated from four other animal samples: hyena, African rodent, and two lions without any visible lesions.
Mycobacterium sinense JMD601 is a recently described pathogenic NTM, first isolated from a human patient with tuberculosis- like disease (Zhang et al., 2013). Rónai et al. (2016) have reported isolation of this species from wild boar, fallow deer, swine, and cattle from Hungary. We isolated this pathogenic NTM from a lion which also showed tuberculosis-like lesions, as well as three other lion samples with no apparent lesions.
Mycobacterium brasiliensis has been isolated from humans before, but its clinical relevance is still largely unknown (Chin'ombe et al., 2016). Reports of isolation of this species from animals are scarce. In this study we isolated this species from a Lichtenstein hartebeest head lymph node without MTBC, presenting tuberculosis like-lesions, suggesting its pathogenic potential in this animal species. This NTM was isolated in this study from samples from three other animals namely, blesbok, mongoose, and white rhino, showing no visible lesions. Botha et al. (2013) have reported isolation of this NTM from wildlife of South Africa. It is important to examination this NTM's role in disease causation in animals.
Mycobacterium bouchedurhonense was isolated in this study from an eland bronchial lymph node, representing tuberculosis-like lesions. This recently described member of MAC is being reported for the first time in South African wildlife.
We have also isolated Mycobacterium sp. GR-2007 from a lymph node of a lion with lesions. This is not a validly published Mycobacterium species, and there are no reports of its occurrence. However, its isolation from a lymph node with a tuberculosis-like lesion may imply its pathogenic nature and the need for further characterization of this NTM.
Mycobacterium goodii is a RGM species closely related to M. smegmatis. Mycobacterium goodii was reported to have caused infection in a hyena in South Africa as well as in African rodents in Tanzania (van Helden et al., 2009; Durnez et al., 2011). We identified a Mycobacterium species closely related to M. goodii and M. smegmatis from a lion lymph node with tuberculosis-like lesions. This NTM is not in the public databases and as such it could be a potential novel NTM species and therefore needs to be further characterized with a particular focus on its pathogenicity.
It has also been hypothesized that mycobacterial infections do not necessarily need to be established and maintained by animal species, colonization without casing disease may be enough for development of immune responses. Some NTM species, notably Mycobacterium kansasii, Mycobacterium marinum, and M. fortuitum have been reported to induce anti-mycobacterial immune responses that interfere with the diagnosis of bovine tuberculosis either by TST or gamma interferon tests (Vordermeier et al., 2007; Michel et al., 2011). Mycobacterium simiae was isolated in this study from as sample originating from a tuberculin skin test positive monkey, demonstrating its potential to elicit inappropriate immune responses in monkeys that may interfere with diagnosis of tuberculosis by immunological tests (Schiller et al., 2010). It may therefore be important to investigate the role of this NTM in eliciting cross-reactive immune responses against Mycobacterium bovis antigens. M. simiae has been reported to cause diseases in monkeys not presenting any specific pathology and has also been isolated from fish (Bercovier and Vincent, 2001). This species has previously been reported in cattle, African buffaloes, and the environments in South Africa (Gcebe et al., 2013). It has also been isolated from cattle and humans in Tanzania and from the environment in Uganda (Kankya et al., 2011; Katale et al., 2014).
We also identified other species that have been shown to be of clinical significance in human mycobacterial diseases in several studies, which in this study were isolated from specimen without lesions. These notably include members of the Mycobacterium avium complex: M. yongonense/intracellulare/M. marseilense, M. intracellulare as well as M. sherrisii, Mycobacterium porcinum, and M. virginiense. Mycobacterium sherrisii is a slow growing emerging human Mycobacterial pathogen, particularly isolated from HIV positive patients who may develop invasive and disseminated mycobacteriosis diseases (van Ingen et al., 2011). Botha et al. (2013) had reported isolation of this Mycobacterium species from wild animals in South Africa (Botha et al., 2013). Mycobacterium virginiense is a recently described member of the M. terrae complex reported to cause major tenosynovitis/osteomyelitis in humans (Vasireddy et al., 2016). Here, this species was isolated from a lion co-infected with M. bovis and its possible role as an opportunistic pathogen of animals need to be investigated. We report isolation of this Mycobacterium species for the first time in animals in South Africa. Mycobacterium porcinum was first isolated from porcine lymph nodes presenting tuberculosis-like lymphadenitis (Tsukamura et al., 1983). In this study, this species was isolated from lions and black wildebeest. The relevance of the MAC species, viz M. yongonense/M. marseilense/M. intracellulare in disease causation or possible cross-reactivity with M. bovis antigens was not established in our case. Mycobacterium yongonense is a recently described member of MAC isolated from patients with pulmonary symptoms (Kim et al., 2013) while M. intracellulare is a known pathogen of humans (van Ingen et al., 2009). This MAC species has been isolated from cattle in Tanzania, and Uganda as well as from wildlife in South Africa (Kankya et al., 2011; Botha et al., 2013; Katale et al., 2014). Other NTM species isolated in this study from specimen not presenting tuberculosis-like lesions include M. fortuitum, M. fluoranthenivorans, M. elephantis, M. vulneris, M. rhodesiae, M. duvalii, M. peregrinum, M. palustre, M. vaccae/M. vanbaalenii, M. parafortuitum, Mycobacterium sp. WCM 7299, and Mycobacterium sp. GPK 1020. These NTM have been isolated from animals as well the environment in several studies (Bercovier and Vincent, 2001; Botha et al., 2013; Gcebe et al., 2013, 2016; Katale et al., 2014). Mycobacterium fortuitum and M. vulneris have both been found to be associated with pulmonary infections in humans (Simons et al., 2011; Hoefsloot et al., 2013). The role of these mycobacterial species as causative agents of disease as well as their possible influence in bovine tuberculosis diagnostics has not been established in this study.
In conclusion, we have elucidated the diversity of NTM species circulating in different wild animals in South Africa. Thirty known NTM species/subspecies as well as unidentified NTM, were detected in this study, of which nine were isolated from animal specimen showing tuberculosis-like lesions, highlighting the importance of these NTM as potential pathogens of animals. In addition, isolation of unidentified NTM species, suggests the occurrence of a number of uncharacterized potentially novel NTM species in wildlife, whose roles are unknown. Isolation of Mycobacterium simiae from a positive TST reactor vervet monkey free of MTBC demonstrate this NTM species' ability to induce cross-reactive immune responses against MTBC antigens. Mycobacterium abscesses subsp. bolletti was found to be the most frequently occurring NTM, isolated mainly from lions. Knowledge gained in this study contribute to the understanding of NTM species circulating in wild animals in South Africa and the pathogenic potential of certain species as well as to a certain extent their potential to hamper bTB immunological diagnostic assays.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Mr. I-Chang Lee and the Tuberculosis laboratory personnel for technical support as well as the Agricultural Research Council for financial support.
References
Bercovier, H., and Vincent, V. (2001). Mycobacterium infections in domestic and wild animals due to Mycobacterium marinum, M. fortuitum, M. chelonae, M. porcicum, M. farcinogenes, M. smegmatis, M. scrofulaceum, M. xenopi, M. kansasii, M. simiae and M. genavense. Rev. Sciet. Tech. Off. Int. Epiz. 20, 265–290. PMID:11288516. doi: 10.20506/rst.20.1.1269
Berg, S., Firdessa, R., Habtamu, M., Gadisa, E., Mengistu, A., Yamuah, L., et al. (2009). The burden of mycobacterial disease in Ethiopian cattle: implications for public health. PLoS ONE 4:e5068. doi: 10.1371/journal.pone.0005068
Botha, L., Gey van Pittius, N. C., and Helden, P. D. (2013). Mycobacteria and disease in southern Africa. Transbound. Emerg. Dis. 60, 147–156. doi: 10.1111/tbed.12159
Cayrou, C., Turenne, C., Behr, M. A., and Drancourt, M. (2010). Genotyping of Mycobacterium avium complex organisms using multispacer sequence typing. Microbiology 156, 687–694. doi: 10.1099/mic.0.033522-0
Chin'ombe, N., Muzividzi, B., Munemo, E., and Nziramasanga, P. (2016). Molecular identification of nontuberculosis-like mycobacteria in humans in Zimbabwe using 16S ribosequencing. Open Microbiol. J. 10, 113–123. doi: 10.2174/1874285801610010113
Cleaveland, S., Shaw, D. J., Mfinanga, S. G., Shirima, G., Kazwala, R. R., Eblate, E., et al. (2007). Mycobacterium bovis in rural Tanzania: risk factors for infection in human and cattle populations. Tuberculosis 87, 30–43. doi: 10.1016/j.tube.2006.03.001
Colombo, C., Rubino, R., Di Carlo, P., Mammina, C., Bonura, C., Siracusa, L., et al. (2012). Probable disseminated Mycobacterium abscessus subspecies bolletii infection in a patient with idiopathic CD4+ T lymphocytopenia: a case report. J. Med. Case Rep. 6:277. doi: 10.1186/1752-1947-6-277
Cousins, D. V., Whittington, R., Marsh, I., Masters, A., Evans, R. J., and Kluver, P. (1999). Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS 900-like sequences detectable by IS 900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13, 431–442. doi: 10.1006/mcpr.1999.0275
Covert, T. C., Rodgers, M. R., Reyes, A. L., and Stelma, G. N. Jr. (1999). Occurrence of non-tuberculosis-like mycobacteria in environmental samples. Appl. Environ. Microbiol. 65, 2492–2496.
Diguimbaye-Djaïbe, C., Vincent, V., Schelling, E., Hilty, M., Ngandolo, R., Mahamat, H. H., et al. (2006). Species identification of non-tuberculous mycobacteria from humans and cattle of Chad. Schweiz. Arch. Tierheilkd. 148, 251–256. doi: 10.1024/0036-7281.148.5.251
Durnez, L., Katakweba, A., Sadiki, H., Katholi, C. R., Kazwala, R. R., Machang'u, R. R., et al. (2011). Mycobacteria in terrestrial small mammals on cattle farms in Tanzania. Vet. Med. Int. 2011:495074. doi: 10.4061/2011/495074
Gcebe, N., Michel, A., Gey van Pittius, N., and Rutten, V. (2016). Comparative genomics and proteomic analysis of four non-tuberculosis-like Mycobacterium species and Mycobacterium tuberculosis complex: occurrence of shared immunogenic proteins. Front. Microbiol. 7:795. doi: 10.3389/fmicb.2016.00795
Gcebe, N., Rutten, V., Gey van Pittius, N. C., and Michel, A. (2013). Prevalence and distribution of non-tuberculosis-like mycobacteria (NTM) in cattle, African Buffaloes (Syncerus caffer) and their environments in South Africa. Transbound. Emerg. Dis. 60, 74–84. doi: 10.1111/tbed.12133
Gopinath, K., and Singh, S. (2010). Non-tuberculosis-like mycobacteria in TB-endemic countries: are we neglecting the danger? PLoS Negl. Trop. Dis. 4:e615. doi: 10.1371/journal.pntd.0000615
Guerra, L. J., Morán, B. A., Lorenzana, D. L. V. R., and Fernández, N. I. (2008). [Cavited pneumonia with Mycobacterium gastri positive culture smear]. Rev. Clin. Esp. 208, 237–238. doi: 10.1016/S0014-2565(08)71729-6
Harmsen, D., Dostal, S., Roth, A., Niemann, S., Rothgänger, J., Sammeth, M., et al. (2003). RIDOM: comprehensive and public sequence database for identification of Mycobacterium species. BMC Infect. Dis. 3:26. doi: 10.1186/1471-2334-3-26
Hlokwe, T. M., Van Helden, P., and Michel, A. L. (2014). Evidence of increasing intra and inter-species transmission of Mycobacterium bovis in South Africa: are we losing the battle? Prev. Vet. Med. 115, 10–17. doi: 10.1016/j.prevetmed.2014.03.011
Hoefsloot, W., Van Ingen, J., Andrejak, C., Ängeby, K., Bauriaud, R., Bemer, P., et al. (2013). The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: an NTM-NET collaborative study. Eur. Respir. J. 42, 1604–1613. doi: 10.1183/09031936.00149212
Kankya, C., Muwonge, A., Djønne, B., Munyeme, M., Opuda-Asibo, J., Skjerve, E., et al. (2011). Isolation of non-tuberculous mycobacteria from pastoral ecosystems of Uganda: public health significance. BMC Public Health 11:320. doi: 10.1186/1471-2458-11-320
Katale, B. Z., Mbugi, E. V., Botha, L., Keyyu, J. D., Kendall, S., Dockrell, H. M., et al. (2014). Species diversity of non-tuberculosis-like mycobacteria isolated from humans, livestock and wildlife in the Serengeti ecosystem, Tanzania. BMC Infect. Dis. 14:616. doi: 10.1186/s12879-014-0616-y
Kazwala, R. R., Daborn, C. J., Kusiluka, L. J. M., Jiwa, S. F. H., Sharp, J. M., and Kambarage, D. M. (1998). Isolation of Mycobacterium species from raw milk of pastoral cattle of the Southern Highlands of Tanzania. Trop. Anim. Health Prod. 30, 233–239. doi: 10.1023/A:1005075112393
Kim, B. J., Math, R. K., Jeon, C. O., Yu, H. K., Park, Y. G., and Kook, Y. H. (2013). Mycobacterium yongonense sp. nov., a slow-growing non-chromogenic species closely related to Mycobacterium intracellulare. Int. J. Syst. Evol. Microbiol. 63(Pt 1), 192–199. doi: 10.1099/ijs.0.037465-0
Malama, S., Munyeme, M., Mwanza, S., and Muma, J. B. (2014). Isolation and characterization of non-tuberculous mycobacteria from humans and animals in Namwala District of Zambia. BMC Res. Notes 7:622. doi: 10.1186/1756-0500-7-622
Martin-Casabona, N., Bahrmand, A. R., Bennedsen, J., Thomsen, V. Ø., Curcio, M., Fauville-Dufaux, M., et al. (2004). Non-tuberculous mycobacteria: patterns of isolation. A multi-country retrospective survey. Int. J. Tuberc. Lung. Dis. 8, 1186–1193.
Michel, A. L., Cooper, D., Jooste, J., De Klerk, L. M., and Jolles, A. (2011). Approaches towards optimising the gamma interferon assay for diagnosing Mycobacterium bovis infection in African buffalo (Syncerus caffer). Prev. Vet. Med. 98, 142–151. doi: 10.1016/j.prevetmed.2010.10.016
Mirsaeidi, M., Farshidpour, M., Allen, M. B., Ebrahimi, G., and Falkinham, J. O. (2014). Highlights on advances in non-tuberculosis-like mycobacterial disease in North America. Biomed Res. Int. 2014:919474. doi: 10.1155/2014/919474
Nessar, R., Cambau, E., Reyrat, J. M., Murray, A., and Gicquel, B. (2012). Mycobacterium abscessus: a new antibiotic nightmare. J. Antimicrob. Chemother. 67, 810–818. doi: 10.1093/jac/dkr578
Piersimoni, C., and Scarparo, C. (2009). Extrapulmonary infections associated with nontuberculous mycobacteria in immunocompetent persons. Emerg. Infect. Dis. 15, 1351–1358. doi: 10.3201/eid1509.081259
Portaels, F. (1995). Epidemiology of mycobacterial diseases. Clin. Dermatol. 13, 207–222. doi: 10.1016/0738-081X(95)00004-Y
Poyntz, H. C., Stylianou, E., Griffiths, K. L., Marsay, L., Checkley, A. M., and McShane, H. (2014). Non-tuberculous mycobacteria have diverse effects on BCG efficacy against Mycobacterium tuberculosis. Tuberculosis 94, 226–237. doi: 10.1016/j.tube.2013.12.006
Primm, T. P., Lucero, C. A., and Falkinham, J. O. III. (2004). Health impacts of environmental Mycobacteria. Clin. Microbiol. Rev. 17, 98–106. doi: 10.1128/CMR.17.1.98-106.2004
Rastogi, N., Legrand, E., and Sola, C. (2001). The mycobacteria: an introduction to nomenclature and pathogenesis [cell envelope]. Rev. Sci. Tech. 20, 21–54. doi: 10.20506/rst.20.1.1265
Rónai, Z., Eszterbauer, E., Csivincsik, Á., Guti, C. F., Dencső, L., Jánosi, S., et al. (2016). Detection of wide genetic diversity and several novel strains among non-avium nontuberculosis-like mycobacteria isolated from farmed and wild animals in Hungary. J. Appl. Microbiol. 121, 41–54. doi: 10.1111/jam.13152
Satou, N., and Nei, M. (1987). The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Schiller, I., Oesch, B., Vordermeier, H. M., Palmer, M. V., Harris, B. N., Orloski, K. A., et al. (2010). Bovine tuberculosis: a review of current and emerging diagnostic techniques in view of their relevance for disease control and eradication. Transbound. Emerg. Dis. 57, 205–220. doi: 10.1111/j.1865-1682.2010.01148.x
September, S. M., Brözel, V. S., and Venter, S. N. (2004). Diversity of nontuberculoid Mycobacterium species in biofilms of urban and semiurban drinking water distribution systems. Appl. Environ. Microbiol. 70, 7571–7573. doi: 10.1128/AEM.70.12.7571-7573.2004
Simons, S., van Ingen, J., Hsueh, P.-R., Van Hung, N., Dekhuijzen, P. N. R., Boeree, M. J., et al. (2011). Nontuberculous mycobacteria in respiratory tract infections, Eastern Asia. Emer. Infect. Dis. 17, 343–349. doi: 10.3201/eid170310060
Telenti, A., Marchesi, F., Balz, M., Bally, F., Böttger, E. C., and Bodmer, T. (1993). Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31, 175–178.
Tortoli, E., Besozzi, G., Lacchini, C., Penati, V., Simonetti, M. T., and Emler, S. (1998). Pulmonary infection due to Mycobacterium szulgai, case report and review of the literature. Eur. Respir. J. 11, 975–977. doi: 10.1183/09031936.98.11040975
Tschopp, R., Berg, S., Argaw, K., Gadisa, E., Habtamu, M., Schelling, E., et al. (2010). Bovine tuberculosis in Ethiopian wildlife. J. Wildlife Dis. 46, 753–762. doi: 10.7589/0090-3558-46.3.753
Tsukamura, M., Nemoto, H., and Yugi, H. (1983). Mycobacterium porcinum sp. nov. a porcine pathogen. IJSB 33, 162–165. doi: 10.1099/00207713-33-2-162
van Helden, P. D., and Parsons, S. D. Gey van Pittius, N. C. (2009). “Emerging” mycobacteria in South Africa. J. S. Afr. Vet. Assoc. 80, 210–214. doi: 10.4102/jsava.v80i4.209
van Helden, P. D., van Pittius, N. C. G., Warren, R. M., Michel, A., Hlokwe, T., Morar, D., et al. (2008). Pulmonary infection due to Mycobacterium goodii in a spotted hyena (Crocuta crocuta) from South Africa. J. Wildl. Dis. 44, 151–154. doi: 10.7589/0090-3558-44.1.151
van Ingen, J., Boeree, M. J., de Lange, W. C., de Haas, P. E., Dekhuijzen, P. R., and van Soolingen, D. (2008). Clinical relevance of Mycobacterium szulgai in the Netherlands. Clin. Infect. Dis. 46, 1200–1205. doi: 10.1086/529443
van Ingen, J., Boeree, M. J., Kösters, K., Wieland, A., Tortoli, E., Dekhuijzen, P. R., et al. (2009). Proposal to elevate Mycobacterium avium complex ITS sequevar MAC-Q to Mycobacterium vulneris sp. nov. Int. J. Syst. Evol. Microbiol. 59, 2277–2282. doi: 10.1099/ijs.0.008854-0
van Ingen, J., Tortoli, E., Selvarangan, R., Coyle, M. B., Crump, J. A., Morrissey, A. B., et al. (2011). Mycobacterium sherrisii sp. nov., a slow-growing non-chromogenic species. Int. J. Syst. Evol. Microbiol. 61, 1293–1298. doi: 10.1099/ijs.0.024752-0
Vasireddy, R., Vasireddy, S., Brown-Elliott, B. A., Wengenack, N. L., Eke, U. A., Benwill, J. L., et al. (2016). Mycobacterium arupense, Mycobacterium heraklionense, and a newly proposed species,“Mycobacterium virginiense” sp. nov., but not Mycobacterium nonchromogenicum, as species of the Mycobacterium terrae complex causing tenosynovitis and osteomyelitis. J. Clin. Microbiol. 54, 1340–1351. doi: 10.1128/JCM.00198-16
Velayati, A. A., Boloorsaze, M. R., Farnia, P., Mohammadi, F., Karam, M. B., Soheyla-Zahirifard, M. B., et al. (2005). Mycobacterium gastri causing disseminated infection in children of same family. Pedetr Pulmonol. 39, 284. doi: 10.1002/ppul.20186
Viana-Niero, C., Lima, K. V. B., Lopes, M. L., da Silva Rabello, M. C., Marsola, L. R., Brilhante, V. C. R., et al. (2008). Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J. Clin. Microbiol. 46, 850–855. doi: 10.1128/JCM.02052-07
Vordermeier, H. M., Brown, J., Cockle, P. J., Franken, W. P., Arend, S. M., Ottenhoff, T. H., et al. (2007). Assessment of cross-reactivity between Mycobacterium bovis and M. kansasii ESAT-6 and CFP-10 at the T-cell epitope level. Clin. Vaccine Immunol. 14, 1203–1209. doi: 10.1128/CVI.00116-07
Wilton, S., and Cousins, D. (1992). Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Appl. 1, 269–273. doi: 10.1101/gr.1.4.269
Zelazyn, A. M., Root, J. M., Shea, Y. R., Colombo, R. E., Shamputa, I. C., Stock, F., et al. (2009). Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J. Clin. Microbiol. 47, 1985–1995. doi: 10.1128/JCM.01688-08
Keywords: non-tuberculous mycobacteria, South African wildlife, bovine tuberculosis, 16S rDNA sequencing, immune responses, potential pathogens
Citation: Gcebe N and Hlokwe TM (2017) Non-tuberculous Mycobacteria in South African Wildlife: Neglected Pathogens and Potential Impediments for Bovine Tuberculosis Diagnosis. Front. Cell. Infect. Microbiol. 7:15. doi: 10.3389/fcimb.2017.00015
Received: 29 August 2016; Accepted: 11 January 2017;
Published: 30 January 2017.
Edited by:
Adel M. Talaat, University of Wisconsin-Madison, USAReviewed by:
Lei Wang, Nankai University, ChinaTyler C. Thacker, National Animal Disease Center (USDA-ARS), USA
Copyright © 2017 Gcebe and Hlokwe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nomakorinte Gcebe, gceben@arc.agric.za
 Nomakorinte Gcebe
Nomakorinte Gcebe Tiny M. Hlokwe
Tiny M. Hlokwe