A Salmonella Regulator Modulates Intestinal Colonization and Use of Phosphonoacetic Acid
- 1Department of Microbial Pathogenesis and Immunology, College of Medicine, Texas A&M University Health Science Center, Bryan, TX, USA
- 2Department of Clinical Sciences, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, USA
- 3Paul G. Allen School for Global Animal Health, College of Veterinary Medicine, Washington State University, Pullman, WA, USA
- 4Department of Medial Microbiology and Immunology, School of Medicine, University of California Davis, Davis, CA, USA
Many microorganisms produce phosphonates, molecules characterized by stable carbon-phosphorus bonds that store phosphorus or act as antimicrobials. The role of phosphonates in the marine biosphere is well characterized but the role of these molecules in the intestine is poorly understood. Salmonella enterica uses its virulence factors to influence the host immune response to compete with the host and normal microflora for nutrients. Salmonella cannot produce phosphonates but encodes the enzymes to use them suggesting that it is exposed to phosphonates during its life cycle. The role of phosphonates during enteric salmonellosis is unexplored. We have previously shown that STM3602, encoding a putative regulator of phosphonate metabolism, is needed for colonization in calves. Here, we report that the necessity of STM3602 in colonization of the murine intestine results from multiple factors. STM3602 is needed for full activation of the type-3 secretion system-1 and for optimal invasion of epithelial cells. The ΔSTM3602 mutant grows poorly in phosphonoacetic acid (PA) as the sole phosphorus source, but can use 2-aminoethylphosphonate. PhnA, an enzyme required for PA breakdown, is not controlled by STM3602 suggesting an additional mechanism for utilization of PA in S. Typhimurium. Finally, the requirement of STM3602 for intestinal colonization differs depending on the composition of the microflora. Our data suggest that STM3602 has multiple regulatory targets that are necessary for survival within the microbial community in the intestine. Determination of the members of the STM3602 regulon may illuminate new pathways needed for colonization of the host.
Introduction
In the intestine the microflora and the host vie for nutrients that vary in availability along the length of the intestine. Salmonella Typhimurium (STm) uses multiple strategies to gain nutrients and survive in this niche. Non-typhoidal Salmonella, including STm, secrete effectors via the type 3-secretion system-1 (TTSS-1). The effectors SipA, SopB, SopD, SopA, and SopE2 promote invasion, an influx of inflammatory cells, predominantly neutrophils, and alter the composition of the microbial flora (Zhang et al., 2002; Raffatellu et al., 2005). STm uses the tetrathionate and nitrate, produced in the intestine as a result of the inflammatory response, as terminal electron acceptors (Winter et al., 2010; Lopez et al., 2012). In addition, STm uses host-derived sugars, such as ethanolamine released by microbial damage in the intestine, as energy sources during inflammation (Thiennimitr et al., 2011). Metal scavenging through salmochelin and high-affinity transporters allow STm to acquire essential nutrients during infection (Raffatellu et al., 2009; Liu et al., 2012). While these are some essential mechanisms salmonellae use to acquire nutrients and replicate in the intestine, these mechanisms may represent only a small fraction of the metabolic potential of STm in the intestine.
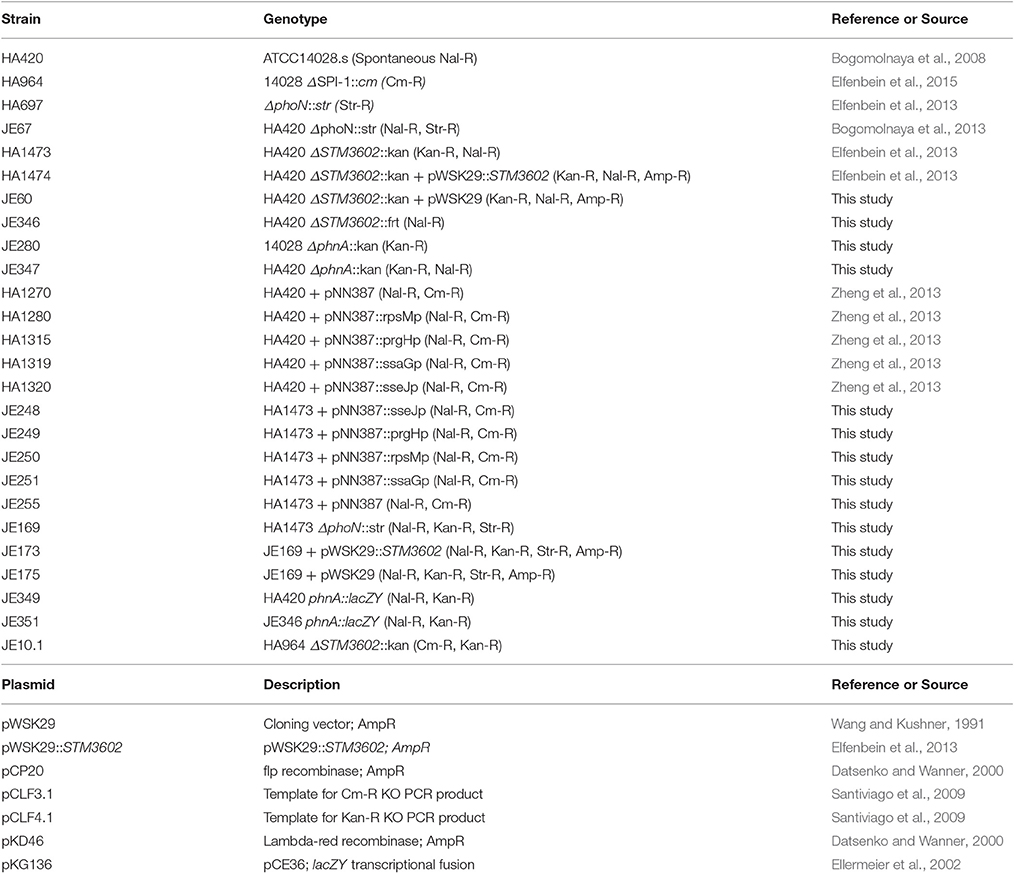
Phosphonates are molecules with stable carbon-phosphorus bonds. Microorganisms produce these molecules to store phosphorus during periods of phosphate limitation (Villarreal-Chiu et al., 2012). In addition to a metabolic utility of these molecules, some of them have potent antimicrobial activity, including fosfomycin and fosmidomycin, and may be produced by microbes as antibiotics (Seto and Kuzuyama, 1999; Metcalf and van der Donk, 2009). The carbon-phosphorus bond is produced by coupled enzymes phosphoenolpyruvate phosphomutase (Ppm) and phosphonopyruvate decarboxylase (Ppd) to generate phosphonoacetaldehyde that is then converted to 2-aminoethylphosphonate (2-AEP) by AEP transaminase (Metcalf and van der Donk, 2009; Villarreal-Chiu et al., 2012). These are the first steps in the production of compounds containing the stable carbon-phosphorus bond.
Although Salmonella lacks genes for the biosynthesis of phosphonates, it can utilize these compounds (Jiang et al., 1995; Errey and Blanchard, 2006). Salmonella has two loci annotated for degradation of phosphonates via the phosphonatase pathway, phnABO and phnVUTSRWX (Jiang et al., 1995). There are four genes annotated for phosphonate degradation, phnW (STM0431; 2-AEP-pyruvate aminotransferase), phnX (STM0432; phosphonatase), phnA (STM4289; phosphonoacetate hydrolase), and phnO (STM4287; aminoalkylphosphonic acid N-acetyltransferase) (Figure 1; Jiang et al., 1995; Errey and Blanchard, 2006). The phnVUTSRWX locus is under the control of the pho regulon and is activated during phosphate limitation (Jiang et al., 1995). Escherichia coli degrades phosphonates using the C-P lyase system encoded within a 14 gene operon (phnCDEFGHIJKLMNO; Metcalf and Wanner, 1993). Thus, the mechanism for phosphonate degradation seems to be different between Salmonella and its close relative.
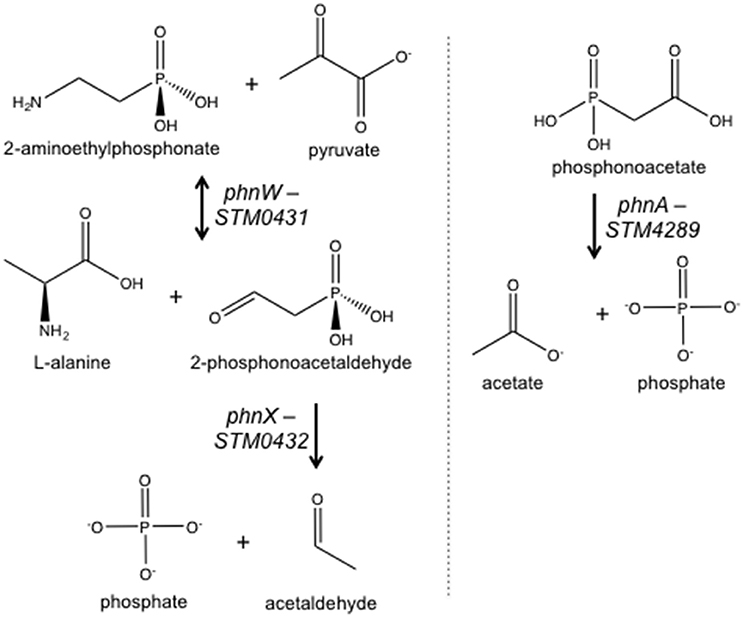
Figure 1. Salmonella enzymes with a putative role in phosphonate metabolism based on annotation. See text for description. Not included is PhnO (STM4287), aminoalkylphosphonic acid N-acetyltransferase.
We previously determined that a mutant lacking STM3602 colonized ligated ileal loops in calves poorly (Elfenbein et al., 2013). In the calf model, Salmonella Typhimurium is inoculated into intestinal segments with undisturbed microflora and promotes a profound neutrophilic inflammatory response (Frost et al., 1997; Santos et al., 2002). Salmonellae must compete with the intestinal microbiota and withstand the host inflammatory response to survive in this model. Here we report that STM3602 is necessary for colonization of the intestine of mice regardless of the composition of the microflora. A ΔSTM3602 mutant does not fully activate the TTSS-1 and has reduced epithelial cell invasion. However, this gene does not influence TTSS-2 expression and intracellular replication. The ΔSTM3602 mutant cannot grow if phosphonoacetic acid (PA) is provided as the sole source of phosphorus, but is capable of growth in 2-AEP. Finally, the gene encoding the known enzyme for PA breakdown, PhnA, does not appear to be under the control of STM3602, suggesting a different mechanism for utilization of PA. Our data suggest that STM3602 may contribute to the regulation of various processes needed for STm to thrive in the intestine.
Materials and Methods
Bacterial Strains and Plasmids
All bacterial strains are derivatives of ATCC 14028s and are listed in Table 1. Unless stated otherwise, bacteria were grown in Luria-Bertani (LB) broth or on LB agar supplemented with the following antibiotics as appropriate: kanamycin (50 mg/L), nalidixic acid (50 mg/L), carbenicillin (100 mg/L), streptomycin (100 mg/L), or chloramphenicol (20 mg/L). Mutants were constructed by a modification of the λ–red recombination technique and antibiotic cassettes were removed as described (Datsenko and Wanner, 2000; Santiviago et al., 2009). Chromosomal transcriptional fusions to lacZY were constructed as described previously (Ellermeier et al., 2002). All mutations were moved to a clean genetic background by bacteriophage P22-mediated transduction (Sternberg and Maurer, 1991).
Mouse Infections
The Texas A&M University and University of California Davis Institutional Animal Care and Use Committees approved all animal experiments (approval numbers TAMU 2012-084 and 2011-167; UCD 19001). Mouse experiments were performed using 10–12 week old female C57BL/6J or CBA/J mice as indicated (Jackson Laboratories) as previously described (Barthel et al., 2003).
In the acute murine colitis model, C57BL/6J mice were administered 20 mg streptomycin in 75 μL sterile water by gavage. Twenty-four hours after treatment, mice were infected with ~108 CFU of an equivalent mixture of WT and mutant bacteria in 100 μL volume by gavage. Feces were collected 24 h after infection. Mice were euthanized 96 h post-infection and organs harvested, homogenized, serially diluted, and plated on LB agar with appropriate antibiotics for enumeration of CFU.
In the chronic murine colitis model, CBA/J mice were administered 20 mg streptomycin in 75 μL water or 75 μL sterile water by gavage. Forty-eight hours after treatment, mice were infected as above. Feces were collected on the indicated days and mice euthanized 14 days post-infection. Competitive index was determined by comparing the ratio of WT to mutant bacteria in the tissue to that of the inoculum.
Germ-free Swiss Webster mice were bred and housed under germ-free conditions inside gnotobiotic isolators (Park Bioservices, LLC). Weekly cultures were grown to monitor the germ-free status of the mice. For experiments, male and female 6–8-week-old mice were transferred to a biosafety cabinet and maintained in sterile cages for the duration of the experiment. Mice were infected with ~108 CFU of an equivalent mixture of WT and mutant bacteria in 100 μL volume by gavage. Mice were euthanized 24 h post-infection, cecal and colon contents were homogenized, serially diluted, and plated on LB agar with appropriate antibiotics for enumeration of CFU. The competitive index was determined by normalizing the ratio of WT to mutant bacteria in intestinal contents to that of the inoculum.
Invasion Assays
Cell lines were purchased from American Type Culture Collection (ATCC) and used within 15 passages. HeLa cells (human cervical adenocarcinoma epithelial, ATCC CCL-2) were grown as recommended by ATCC. HeLa cells were seeded in 24-well plates at 5 × 104 cells/well ~24 h prior to infection.
Invasion assays were performed as previously described (Ibarra et al., 2010). Late-log phase cultures were prepared by inoculating 10 ml LB-Miller broth with 0.3 ml overnight culture. Flasks were grown at 37°C with aeration for 3 h. Bacteria were collected by centrifugation at 8,000 × g for 90 s, resuspended in an equal volume of Hanks' buffered saline solution (HBSS, Mediatech) and added directly to mammalian cells seeded in 24-well plates for 10 min. The multiplicity of infection was ~50:1 (bacteria:eukaryotic cell). Non-internalized bacteria were removed by aspiration, monolayers washed three times in HBSS and then incubated in growth media until 30 min post-infection. Gentamicin was added at 50 μg/ml from 30 to 90 min post-infection to kill extracellular bacteria and the media was replaced with media containing 10 μg/ml gentamicin from 90 min post infection. For enumeration of intracellular bacteria, monolayers were washed once in phosphate-buffered saline, solubilized in 0.2% (w/v) sodium deoxycholate and serial dilutions were plated on LB agar.
ß-Galactosidase Assays
For induction of SPI-1 expression, bacterial cells bearing plasmid constructs were grown overnight in LB with appropriate antibiotics. Overnight cultures were diluted 1:100 and incubated at 37°C with agitation for 3 h. SPI-2 inducing media was used as described (5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 8 μM MgCl2, 337 μM KH2PO4, 80 mM MES, 0.3% (v/v) glycerol, 0.1% (v/v) casamino acids, pH 5.8) (Coombes et al., 2004). Cells were grown overnight in SPI-2 inducing media, diluted 1:50, and incubated for an additional 24 h to evaluate SPI-2 expression. To evaluate the expression of phnA in the presence of PA, overnight cultures were diluted 1:100 in LB with 40 mM MOPS at pH 6.8 with the indicated phosphorus source. Cultures were incubated at 37°C with agitation for 3 h.
ß-galactosidase activity was determined using standard methodology (Miller, 1972). Briefly, bacterial cells were pelleted by centrifugation and resuspended in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM ß-mercaptoethanol). Culture density was determined by OD600. Bacteria were permeabilized with chloroform and 0.1% (w/v) SDS prior to addition of substrate (o-nitrophenyl-β-D-galactoside; ONPG 4 mg/mL). The reactions were performed at 28°C and were stopped with 1 M Na2CO3 for determination of OD420 and OD550. β-galactosidase activity (Miller units) was calculated using the following equation: 1000 × [OD420–(1.75 × OD550)] / [time × volume × OD600].
Phosphonate Growth
Modifications were made to a phosphorus-limited minimal medium (Neidhardt et al., 1974) to assess the ability of different bacterial strains to utilize different phosphorus sources. The final media composition (MMMM) was as follows: 20 mM NH4Cl, 2.5 mM Na2SO4, 80 mM NaCl, 0.35 mM CaCl2, 20 mM KCl, 40 mM MOPS, 1 mM MgSO4, 0.01 mM FeSO4, 0.2% (w/v) glucose, pH 6.8. Phosphorus sources were Na2HPO4 (Pi, Sigma), 2-aminoethylphosphonate (2-AEP, Sigma), and phosphonoacetic acid (PA, Sigma) added at the concentrations indicated. Bacterial strains were grown overnight at 37°C with agitation in MMMM with Na2HPO4 at the indicated concentration. Overnight cultures were pelleted and pellets were resuspended in an equal volume MMMM supplemented with the indicated phosphorous sources. After re-centrifugation and resuspension in the starting volume or the original culture, bacteria were diluted 1:100 into fresh MMMM with indicated phosphorus source. Aliquots were removed, serially diluted, and plated to determine CFU.
Data Analysis
All data were log transformed prior to analysis. Statistical significance was determined using Student's t-test or analysis of variance with significance set at P < 0.05.
Results
STM3602 in Intestinal Colonization
STM3602 is necessary for STm to colonize the intestine of calves in ligated ileal loops (Elfenbein et al., 2013). We used the murine colitis model to further dissect the function of this gene during infection (Barthel et al., 2003). In this model, mice are treated with high doses of streptomycin prior to infection promoting STm colonization and development of a neutrophilic inflammatory response. We found that the ΔSTM3602 mutant is shed in lower numbers than the wild type organism in the feces by 24 h post-infection, and colonizes Peyer's patches and mesenteric lymph nodes at lower levels than the wild type organism at 4 days post-infection (Figure 2). This colonization defect was reversed by complementation in trans. Colonization of the cecum by the ΔSTM3602 mutant was highly variable in different mice making it difficult to determine with statistical confidence whether or not mutants lacking STM3602 have a colonization defect in the cecum. Regardless, our results are consistent with previous data suggesting a requirement of STM3602 in colonization of the intestine in the presence of a profound neutrophilic inflammatory response.
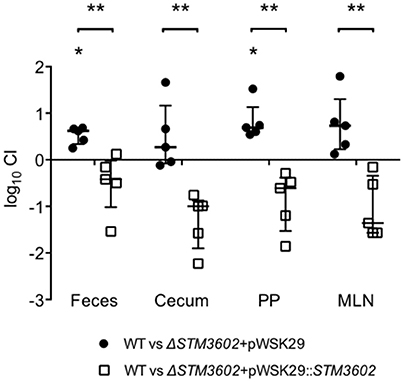
Figure 2. During acute colitis, the ΔSTM3602 mutant colonizes the murine Peyer's patches and mesenteric lymph nodes poorly. Ten C57BL/6 mice were treated with streptomycin (20 mg) then infected with ~108 CFU of an equivalent mixture of WT (JE67) and ΔSTM3602 mutant bearing an empty plasmid (JE175) or complementing plasmid (JE173) 24-h after antibiotic treatment. Feces (F) were collected 24 h after infection and mice were euthanized 96-h post-infection for collection of Peyer's patches (PP), mesenteric lymph nodes (MLN), and cecum (C). The competitive index (CI) was determined by comparing the ratio of WT to mutant in the tissue to that of the inoculum. Each data point represents a single mouse with median and interquartile range indicated by horizontal bars. Statistical significance was determined by Student's t-test, and is indicated by an asterisk (*), and statistically significant differences between groups is indicated by two asterisks (**) with P < 0.05.
STM3602 and SPI-1 Regulation
Salmonellae possess two type-3 secretion systems, encoded by Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2). The TTSS-1 and its associated effectors are essential for invasion of non-phagocytic epithelial cells and penetration of the intestinal epithelium (Zhang et al., 2002; Raffatellu et al., 2005). The TTSS-2 is needed for maintenance of the Salmonella-containing vacuole and intracellular replication (Figueira and Holden, 2012). We hypothesized that the intestinal colonization defect of ΔSTM3602 mutants could be due to reduced expression of TTSS-1.
We used plasmids containing lacZY under the control of promoters of genes on SPI-1 or SPI-2 to determine whether deletion of STM3602 resulted in an effect on activation of these promoters (Zheng et al., 2013). Using a plasmid containing lacZY under the control of the prgH promoter (a TTSS-1 structural gene), we found that the prgH promoter is activated less efficiently in SPI-1 inducing conditions in bacteria lacking STM3602 than in the WT (Figure 3A). Consistent with this result, deletion of STM3602 also results in a similar reduction in invasion in cultured epithelial cells (Figure 3B). Conversely, the ΔSTM3602 mutant is capable of activation of TTSS-2 apparatus (ssaG) and effector promoters (sseJ; Figure 3C) as well as survival within cultured epithelial cells to the same level as the wild type (Figure 3D). These results suggest that STM3602 plays a role in the complex regulatory network of the TTSS-1.
Figure 3. The ΔSTM3602 mutant has reduced invasion efficiency into cultured epithelial cells due to poor activation of SPI-1. (A) Activation of a terminal SPI-1 promoter (prgHp-lacZY) in SPI-1 inducing conditions as determined by ß-galactosidase activity. (B) Invasion efficiency of ΔSTM3602 (JE60) and complemented ΔSTM3602 (HA1474) mutants into HeLa epithelial cell monolayers normalized to the efficiency of the WT (HA420) at 1 h post-infection. (C) Activation of two SPI-2 terminal promoters (sseJp-lacZY and ssaGp-lacZY) in SPI-2-inducing conditions as determined by ß-galactosidase activity. (D) Fold-replication of ΔSTM3602 and complemented ΔSTM3602 mutants in HeLa cells; 7 h post-infection/1 h post-infection normalized to WT fold-replication. Bars represent the mean ± SD. Assays were performed on three separate occasions. Statistical significance (*) was determined by Student's t-test with P < 0.05.
Growth in Phosphonoacetate as Sole Phosphorus Source
STM3602 belongs to the GntR family of transcriptional regulators and is a part of the PhnR clade (Marchler-Bauer et al., 2015). This gene shares 29.2% sequence similarity and 45.1% amino acid identity with PhnR (STM0430; Sievers et al., 2011). We hypothesized that STM3602 might regulate one or both of the phosphonate utilization loci in STm. We characterized the survival and growth of the ΔSTM3602 mutant in media containing either 2-AEP or PA as the sole phosphorus source. When strains were grown in the presence of 5 mM PA, the ΔSTM3602 mutant replicated poorly (Figure 4A). However, this mutant grows normally in 5 mM 2-AEP or Pi (Figures 4B,C) as sole phosphorus sources. PA is degraded to acetate and inorganic phosphate (Figure 1). One possible explanation for the phenotype of the ΔSTM3602 mutant in PA-containing medium is that this mutant may be unable to utilize or excrete acetate. However, the ΔSTM3602 mutant exhibits similar growth kinetics to WT in the presence of both 5 mM acetate and 5 mM Pi (Figure 4D) suggesting a mechanism specific to use of PA. Overall, these data suggest a role for STM3602 in phosphonoacetate utilization.
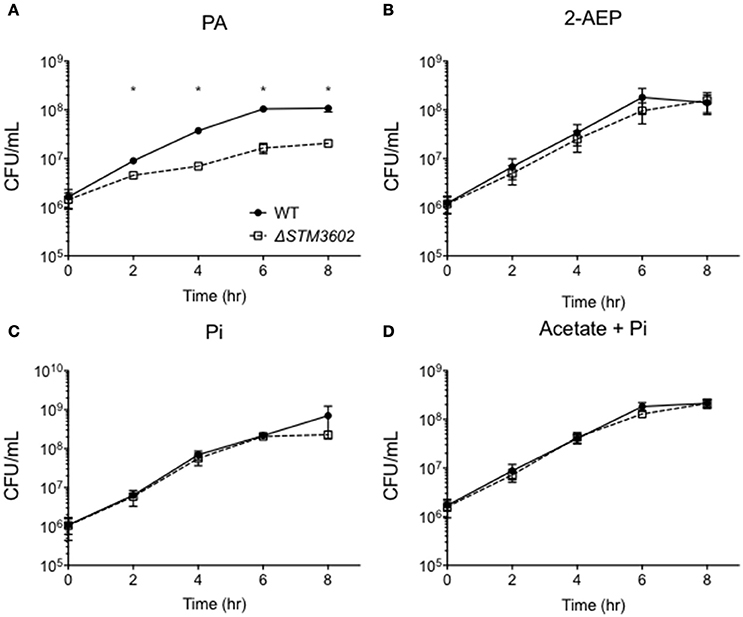
Figure 4. STM3602 is required for adequate growth with phosphonoacetic acid as a sole phosphorus source. (A–D) Growth curve of WT (HA420) and ΔSTM3602 (HA1473) in a minimal medium supplemented with 5 mM PA (A), 2-AEP (B), Pi (C) and 5 mM sodium acetate with 5 mM Pi (D). Bacteria were grown overnight in MMMM with 5 mM Pi then diluted 1:100 into media with the indicated phosphorus and/or carbon additions. CFU were determined every 2 h on three independent occasions. Data points represent mean ± SEM. CFU data were log transformed and statistical significance determined by ANOVA. Asterisk (*) indicates significant difference between ΔSTM3602 and WT with P < 0.05.
Next, we determined the growth kinetics of the ΔSTM3602 mutant in media with 5 mM PA after phosphorus deprivation. When bacteria were grown in phosphorus-limiting conditions (0.5 mM Pi) and transferred to media containing 10 times more phosphorus in the form of PA, the ΔSTM3602 mutant lost viability over the course of the 6-h experiment (Figure 5). A mutant deleted for phnA, a gene that encodes the phosphonoacetate hydrolase, had growth kinetics similar to the WT in these conditions (Figure 5). These results suggest that STM3602 is needed for activation of pathways necessary for growth in the presence of PA and that there is a pathway for metabolism of this compound that operates independently of phnA.
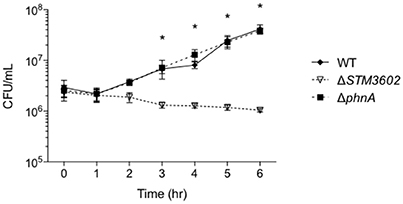
Figure 5. Growth in PA as a sole phosphorous source requires STM3602 but not phnA. Bacterial strains were grown overnight in 0.5 mM Pi and diluted 1:100 into media with 5 mM PA. CFU were determined hourly. CFU data were log transformed and statistical significance determined by ANOVA. (*) indicates significant difference between ΔSTM3602 and WT with P < 0.05.
Expression of phnA
The ΔSTM3602 mutant grows poorly in the presence of PA. We hypothesized that STM3602 regulates the phnABO operon because the annotation of phnA suggests that PhnA may degrade PA. In order to test this hypothesis, we generated a mutant strain bearing a lacZY fusion to the first 10 amino acids of phnA to monitor transcription from the native phnA promoter. Using this construct, we found that in rich media the expression of the phnA promoter was not affected by the addition of PA at varying concentrations (Figure 6). In addition, deletion of STM3602 did not affect the expression of phnA in these conditions during log phase growth (Figure 6), a condition where STM3602 is expressed (Kroger et al., 2013). These data suggest that STM3602 does not regulate phnA in rich medium in the presence or absence of PA.
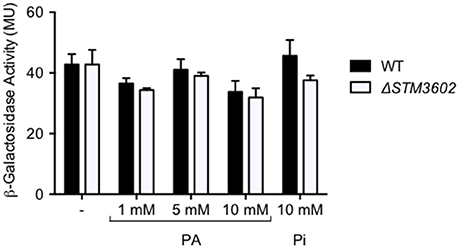
Figure 6. Deletion of STM3602 does not affect phnA expression. The expression of phnA::lacZY was determined in log phase cultures in rich medium by measuring ß-galactosidase activity in both the WT (black bars, JE349) and ΔSTM3602 mutant (white bars, JE351) backgrounds. Bars represent the mean ± SEM of 3 independent experiments.
Impact of Microbiota on ΔSTM3602 Intestinal Colonization
The first phosphonate to be discovered, 2-AEP is present in both ruminal and duodenal contents of sheep (Ankrah et al., 1989). This compound is associated with ruminal protozoal and bacterial populations and is less abundant in defaunated animals compared with those with a normal microbial composition. Phosphonoacetic acid is one product of 2-AEP metabolism mediated by the enzyme PhnY, phosphonoacetaldehyde oxidase (Agarwal et al., 2014). Although salmonellae lack a gene with this annotated function, phyla found in the murine intestinal microflora including Cyanobacteria, Proteobacteria and Firmicutes encode genes to produce PA from 2-AEP (Stecher et al., 2007; Martinez et al., 2010; Villarreal-Chiu et al., 2012).
We hypothesized that the colonization defect of the ΔSTM3602 mutant might correlate with microbial colonization of the host intestine. We used the chronic carriage mouse model to determine the longer-term kinetics of the colonization defect of this mutant. In streptomycin pre-treated mice (altered microbiota) in competitive infection with the WT, the ΔSTM3602 mutant failed to colonize the intestine beginning very early at 1-day post-infection. In animals with an intact microbiota (no streptomycin pretreatment) the colonization defect of the ΔSTM3602 mutant took longer to develop becoming apparent at 5 days post infection (Figure 7A). Thus, the colonization defect of the ΔSTM3602 mutant occurs much earlier in animals with disrupted microbiota than with intact microbiota.
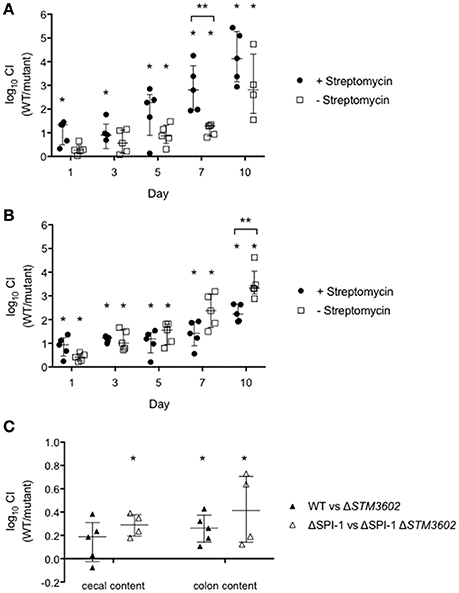
Figure 7. The colonization defect of the ΔSTM3602 mutant is independent of the host inflammatory response and microbial composition. (A,B) Ten CBA/J mice were treated with streptomycin (20 mg; closed circles) or an equivalent volume of sterile water (open squares) and infected 48 h later with ~108 CFU of an equivalent mixture of (A) WT (HA420) and ΔSTM3602 mutant (HA1473) or (B) ΔSPI-1 (HA964) and ΔSPI-1 ΔSTM3602 (JE10.1) mutant by gavage. Feces were collected on the indicated days. (C) Nine germ free Swiss Webster mice were infected with 108 CFU of an equivalent mixture of WT and ΔSTM3602 mutant (closed circles) or ΔSPI-1 and ΔSPI-1 ΔSTM3602 mutant (open squares) by gavage. Animals were sacrificed 1 day post-infection and bacterial numbers enumerated from cecal and colon contents. Significant difference in CI (WT/mutant) is indicated by an asterisk (*) and difference between groups is indicated by two asterisks (**) with P < 0.05. Analyses for statistical significance determined as in Figure 2.
One possible explanation for the reduced ability of the ΔSTM3602 mutant to colonize the murine intestine is due to reduced expression of the TTSS-1 (Figures 3A,B). To rule out this possibility, we performed a competitive infection experiment in a ΔSPI-1 genetic background (ΔSPI-1 mutant vs. ΔSPI-1ΔSTM3602 double mutant). In these experiments, the double ΔSPI-1ΔSTM3602 mutant colonizes the intestine poorly on the first day post-infection in animals regardless of whether the intestinal microflora were intact or disrupted (Figure 7B). The phenotype of the ΔSTM3602 mutant was significantly stronger 10 days post-infection in the animals that began with disrupted microflora. These data suggest that the observed colonization defects are unlikely to be due solely to poor activity of the TTSS-1 but more likely a result of inability to compete in the intestine.
Finally, we evaluated the fitness of the ΔSTM3602 mutant using competitive infections in germ-free mice to eliminate any effects of microbiota on colonization efficiency of the ΔSTM3602 mutant. In the germ free murine model, the ΔSTM3602 mutant exhibited a small but significant colonization defect in the cecal contents in the WT and ΔSPI-1 genetic backgrounds (Figure 7C) but only a significant colonization defect in the colon contents in the ΔSPI-1 genetic background. Taken together, these data suggest that STM3602 is required for colonization of the murine intestine regardless of the host inflammatory response or the microbial composition of the intestine.
Discussion
We report that the putative GntR family regulator STM3602 is necessary for colonization of the murine intestine, consistent with the reduced ability of this mutant to colonize the bovine intestine (Elfenbein et al., 2013). Two possible mechanisms for these colonization defects are: (1) a role for STM3602 in the activation of the TTSS-1 and/or (2) a role in use of the microbial-derived product, phosphonoacetic acid. Our data are consistent with a regulatory role of STM3602 in modulating STm virulence and use in the intestine of the infected animal.
2-AEP was the first phosphonate discovered (Horiguchi and Kandatsu, 1959). This compound is found in low amounts in feed and is associated with bacterial and protozoan populations within the intestine (Ankrah et al., 1989). 2-AEP is also found in mammalian tissues likely from assimilation from microbial and feed sources because mammals lack the enzymes to produce such molecules (Shimizu et al., 1965; Alhadeff and Daves, 1971; Metcalf and van der Donk, 2009). Numerous marine bacterial phyla contain genes for the biosynthesis and degradation of phosphonates (Villarreal-Chiu et al., 2012). However, the production of phosphonates by microbes is not restricted to the marine biosphere. The leading bacterial phyla containing phosphonate biosynthetic genes from mammalian and bird microbiome metagenomes are Firmicutes, Proteobacteria, and Bacteroidetes (Yu et al., 2013). These phyla are abundant in the murine cecum, although their relative abundance is substantially altered following streptomycin treatment (Stecher et al., 2007).
A mutant in ΔSTM3602 has both a colonization defect in the intestine of mammals and fails to utilize phosphonoacetic acid as a sole phosphorus source. The relationship between these phenotypes remains unclear. We and others have previously shown that a ΔSTM3602 mutant is defective for intestinal colonization of calves and pigs (Carnell et al., 2007; Elfenbein et al., 2013). Here we found that the ΔSTM3602 mutant also poorly colonizes the intestine of mice. This colonization defect persists regardless of the character of intestinal microbiota, and is also present both in the presence and absence of TTSS-1 induced inflammation. Thus, the host may act as a source of phosphonates, having absorbed them from the microflora or feed sources and incorporated them into phosphonolipids (Shimizu et al., 1965; Alhadeff and Daves, 1971). Further studies evaluating the phosphonate content of intestinal fluid and tissue in animals will be very informative.
The only gene in the STm genome with putative function to catalyze the conversion of phosphonoacetate to inorganic phosphate and acetate is phnA. No studies have evaluated the regulation of phnABO. Our data suggest that phnA is expressed at low levels in rich medium and that the expression of phnA is not induced in the presence of PA nor is it required for growth with PA as a sole phosphorus source. Our data also suggest that the expression of phnA is not affected by deletion of STM3602. We measured phnA expression during log phase growth, a time when STM3602 is expressed (Kroger et al., 2013). The phosphorus content of LB broth is undefined, and it is possible that the expression profile of phnA would differ in the defined media in the presence of different phosphorus sources. The ΔSTM3602 mutant is unable to utilize PA as a sole phosphorus source but the ΔphnA mutant can. One reason may be that the ΔSTM3602 mutant fails to activate a transporter for PA. While this would explain the lack of growth in the ΔSTM3602 mutant, it does not explain the finding that the ΔphnA mutant grows with WT efficiency with PA as a sole phosphorous source. This finding suggests that there may be an additional mechanism for catalysis of PA encoded elsewhere in the genome that is likely under the regulatory control of STM3602.
STM3602 is a member of the GntR family of regulators, characterized by N-terminal DNA-binding domains and variable C-terminal small ligand binding domains (Haydon and Guest, 1991; Gorelik et al., 2006). The crystal structure of PhnF of both E. coli and M. smegmatis, a regulator of the C-P lyase pathway for phosphonate catalysis and member of the GntR family of regulators has been solved and a small molecule ligand has been modeled into a binding site although the actual ligand remains unknown (Gorelik et al., 2006; Gebhard et al., 2014). STM3602 shares a conserved domain with PhnF, but has only 23.4% amino acid identity across the entire protein. Salmonella lacks the gene phnF but has a putative regulator of phosphonate utilization, phnR (STM0430). STM3602 has 45.1% amino acid identity with PhnR (Sievers et al. 2011). The GC content of STM3602 and phnR differ substantially (49 and 59%, respectively), consistent with recent acquisition of STM3602 likely with a mechanism independent of the function of phnR.
Non-typhoidal Salmonella use the TTSS-1 to invade normally non-phagocytic intestinal epithelial cells and induce a strong neutrophilic inflammatory response (Tsolis et al., 1999; Zhang et al., 2002; Raffatellu et al., 2005). The central importance of this pathogenicity island is illustrated by the fact that salmonellae lacking a functional TTSS-1 are avirulent in calf, pig, and mouse models of infection (Tsolis et al., 1999; Zhang et al., 2002; Barthel et al., 2003; Coombes et al., 2005). The regulatory network controlling the expression of the TTSS-1 is intricate. Transcriptional regulation is carefully controlled by regulatory proteins encoded both within SPI-1 and located elsewhere on the chromosome (Ong et al., 2010). The master regulator of the TTSS-1, hilA, is encoded within SPI-1 and integrates regulatory input from numerous sources to activate operons necessary for the production of protein components of the TTSS-1 and its associated effectors (reviewed in Ellermeier and Slauch, 2007). We have shown a minor role for STM3602 in the regulation of SPI-1 and invasion of tissue cultured epithelial cells. However, a ΔSTM3602 ΔSPI-1 double mutant was more severely compromised for colonization of the murine intestine than a mutant lacking only SPI-1 (Figure 7). This finding suggests that STM3602 plays a role in intestinal colonization that is not fully explained by inadequate activation of SPI-1. However, the role of STM3602 in regulation of SPI-1 remains an interesting area of further study to contribute to the breadth of knowledge on this essential virulence mechanism.
The genes neighboring STM3602 have recently been described as important for utilization of fructose-asparagine during colonization of the mouse intestine (Ali et al., 2014; Sabag-Daigle et al., 2016). These genes (STM3602-STM3598; fraRBDAE) share a similar GC content (47.7–51.3%) so were likely acquired together. However, it is unclear whether these genes are co-transcribed from the same promoter or whether they function in the same genetic pathway. The colonization defect of the ΔSTM3601 mutant compared with its isogenic WT is substantially greater than that of the ΔSTM3602 mutant after 1 day infection in germ-free animals (Figure 7C), suggesting that these genes may not operate in the same genetic pathway (Ali et al., 2014). Further studies are needed to determine whether these genes belong to an operon or have shared functions during infection.
We have confirmed that STM3602 is needed for colonization of the mammalian intestine. We report that a deletion mutant lacking STM3602 grows poorly with PA as a sole phosphorus source and that STM3602 has no effect on the expression of phnA, the enzyme annotated for utilization of this compound. Finally, we show that STM3602 plays a minor role in the regulation of the TTSS-1. Phosphonates are compounds that are produced by microorganisms and have been identified in the intestine of mammals as a result of microbial colonization. Further, studies evaluating the role of STM3602 in co-culture with microorganisms known to produce diverse phosphonates will elucidate the full complement of compounds to which this regulator responds. In addition, definition of the STM3602 regulon may elucidate novel mechanisms for phosphonate transport or utilization in the large complement of genes of unknown function scattered within the genome of STm.
Author Contributions
JE and HA designed the experiments, analyzed the data, and drafted the manuscript. JE, LK, AS, and FF performed the experiments. JE, LK, FF, AB, and HA edited the manuscript.
Funding
This work was supported by NIH grant R01 AI03646 and to HA. JE was supported by Office of Research Infrastructure Programs/OD NIH grant 8T32OD011083. LK was supported by start-up funds from the Paul G. Allen School for Global Animal Health. Work in the AB laboratory is supported by NIH AI096528.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agarwal, V., Peck, S. C., Chen, J.-H., Borisova, S. A., Chekan, J. R., van der Donk, W. A., et al. (2014). Structure and function of phosphonoacetaldehyde dehydrogenase: the missing link in phosphonoacetate formation. Chem. Biol. 21, 125–135. doi: 10.1016/j.chembiol.2013.11.006
Alhadeff, J. A., and Daves, G. D. Jr. (1971). 2-Aminoethylphosphonic acid: distribution in human tissues. Biochim. Biophys. Acta 244, 211–213. doi: 10.1016/0304-4165(71)90139-5
Ali, M. M., Newsom, D. L., González, J. F., Sabag-Daigle, A., Stahl, C., Steidley, B., et al. (2014). Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine. PLoS Pathog. 10:e1004209. doi: 10.1371/journal.ppat.1004209
Ankrah, P., Loerch, S. C., and Dehority, B. A. (1989). Occurrence of 2-aminoethylphosphonic acid in feeds, ruminal bacteria and duodenal digesta from defaunated sheep. J. Anim. Sci. 67, 1061–1069. doi: 10.2527/jas1989.6741061x
Barthel, M., Hapfelmeier, S., Quintanilla-Martinez, L., Kremer, M., Rohde, M., Hogardt, M., et al. (2003). Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71, 2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003
Bogomolnaya, L. M., Andrews, K. D., Talamantes, M., Maple, A., Ragoza, Y., and Vazquez-Torres, A. (2013). The ABC-type efflux pump MacAB protects Salmonella enterica serovar typhimurium from oxidative stress. MBio 4:e00630–13. doi: 10.1128/mbio.00630-13
Bogomolnaya, L. M., Santiviago, C. A., Yang, H. J., Baumler, A. J., and Andrews-Polymenis, H. L. (2008). ‘Form variation’ of the O12 antigen is critical for persistence of Salmonella typhimurium in the murine intestine. Mol. Microbiol. 70, 1105–1119. doi: 10.1111/j.1365-2958.2008.06461.x
Carnell, S. C., Bowen, A., Morgan, E., Maskell, D. J., Wallis, T. S., and Stevens, M. P. (2007). Role in virulence and protective efficacy in pigs of Salmonella enterica serovar Typhimurium secreted components identified by signature-tagged mutagenesis. Microbiology 153, 1940–1952. doi: 10.1099/mic.0.2006/006726-0
Coombes, B. K., Brown, N. F., Valdez, Y., Brumell, J. H., and Finlay, B. B. (2004). Expression and Secretion of Salmonella Pathogenicity Island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 279, 49804–49815. doi: 10.1074/jbc.M404299200
Coombes, B. K., Coburn, B. A., Potter, A. A., Gomis, S., Mirakhur, K., Li, Y., et al. (2005). Analysis of the contribution of Salmonella Pathogenicity Islands 1 and 2 to Enteric disease progression using a novel bovine Ileal loop model and a murine model of infectious enterocolitis. Infect. Immun. 73, 7161–7169. doi: 10.1128/IAI.73.11.7161-7169.2005
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Elfenbein, J. R., Endicott-Yazdani, T., Porwollik, S., Bogomolnaya, L. M., Cheng, P., Guo, J., et al. (2013). Novel determinants of intestinal colonization of Salmonella enterica serotype Typhimurium identified in bovine enteric infection. Infect. Immun. 81, 4311–4320. doi: 10.1128/IAI.00874-13
Elfenbein, J. R., Knodler, L. A., Nakayasu, E. S., Ansong, C., Brewer, H. M., Bogomolnaya, L., et al. (2015). Multicopy single-stranded DNA directs intestinal colonization of enteric pathogens. PLoS Genet. 11:e1005472. doi: 10.1371/journal.pgen.1005472
Ellermeier, C. D., Janakiraman, A., and Slauch, J. M. (2002). Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290, 153–161. doi: 10.1016/S0378-1119(02)00551-6
Ellermeier, J. R., and Slauch, J. M. (2007). Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10, 24–29. doi: 10.1016/j.mib.2006.12.002
Errey, J. C., and Blanchard, J. S. (2006). Functional annotation and kinetic characterization of PhnO from Salmonella enterica. Biochemistry 45, 3033–3039. doi: 10.1021/bi052297p
Figueira, R., and Holden, D. W. (2012). Functions of the Salmonella Pathogenicity Island 2 (SPI-2) type III secretion system effectors. Microbiology 158(Pt 5), 1147–1161. doi: 10.1099/mic.0.058115-0
Frost, A. J., Bland, A. P., and Wallis, T. S. (1997). The early dynamic response of the calf ileal epithelium to Salmonella typhimurium. Vet. Pathol. Online 34, 369–386. doi: 10.1177/030098589703400501
Gebhard, S., Busby, J. N., Fritz, G., Moreland, N. J., Cook, G. M., Lott, J. S., et al. (2014). Crystal Structure of PhnF, a GntR-Family transcriptional regulator of phosphate transport in Mycobacterium smegmatis. J. Bacteriol. 196, 3472–3481. doi: 10.1128/JB.01965-14
Gorelik, M., Lunin, V. V., Skarina, T., and Savchenko, A. (2006). Structural characterization of GntR/HutC family signaling domain. Protein Sci. 15, 1506–1511. doi: 10.1110/ps.062146906
Haydon, D. J., and Guest, J. R. (1991). A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 79, 291–295. doi: 10.1111/j.1574-6968.1991.tb04544.x
Horiguchi, M., and Kandatsu, M. (1959). Isolation of 2-aminoethane phosphonic acid from rumen protozoa. Nature 184(Suppl. 12), 901–902. doi: 10.1038/184901b0
Ibarra, J. A., Knodler, L. A., Sturdevant, D. E., Virtaneva, K., Carmody, A. B., Fischer, E. R., et al. (2010). Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella–host cell interactions in vitro. Microbiology 156, 1120–1133. doi: 10.1099/mic.0.032896-0
Jiang, W., Metcalf, W. W., Lee, K. S., and Wanner, B. L. (1995). Molecular cloning, mapping, and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J. Bacteriol. 177, 6411–6421. doi: 10.1128/jb.177.22.6411-6421.1995
Kröger, C., Colgan, A., Srikumar, S., Handler, K., Sivasankaran, S. K., Hammarlof, D. L., et al. (2013). An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 14, 683–695. doi: 10.1016/j.chom.2013.11.010
Liu, J. Z., Jellbauer, S., Poe, A. J., Ton, V., Pesciaroli, M., Kehl-Fie, T. E., et al. (2012). Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11, 227–239. doi: 10.1016/j.chom.2012.01.017
Lopez, C. A., Winter, S. E., Rivera-Chávez, F., Xavier, M. N., Poon, V., Nuccio, S.-P., et al. (2012). Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. MBio 3:e00143–12. doi: 10.1128/mBio.00143-12
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Martinez, A., Tyson, G. W., and DeLong, E. F. (2010). Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ. Microbiol. 12, 222–238. doi: 10.1111/j.1462-2920.2009.02062.x
Metcalf, W. W., and van der Donk, W. A. (2009). Biosynthesis of phosphonic and phosphinic acid natural products. Annu. Rev. Biochem. 78, 65–94. doi: 10.1146/annurev.biochem.78.091707.100215
Metcalf, W. W., and Wanner, B. L. (1993). Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene 129, 27–32. doi: 10.1016/0378-1119(93)90692-V
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratories.
Neidhardt, F. C., Bloch, P. L., and Smith, D. F. (1974). Culture medium for enterobacteria. J. Bacteriol. 119, 736–747.
Ong, S. Y., Ng, F. L., Badai, S. S., Yuryev, A., and Alam, M. (2010). Analysis and construction of pathogenicity island regulatory pathways in Salmonella enterica serovar Typhi. J. Integr. Bioinform. 7, 63–96. doi: 10.2390/biecoll-jib-2010-145
Raffatellu, M., George, M. D., Akiyama, Y., Hornsby, M. J., Nuccio, S. P., Paixao, T. A., et al. (2009). Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486. doi: 10.1016/j.chom.2009.03.011
Raffatellu, M., Wilson, R. P., Chessa, D., Andrews-Polymenis, H., Tran, Q. T., Lawhon, S., et al. (2005). SipA, SopA, SopB, SopD, and SopE2 Contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect. Immun. 73, 146–154. doi: 10.1128/IAI.73.1.146-154.2005
Sabag-Daigle, A., Blunk, H. M., Sengupta, A., Wu, J., Bogard, A. J., Ali, M. M., et al. (2016). A metabolic intermediate of the fructose-asparagine utilization pathway inhibits growth of a Salmonella fraB mutant. Sci. Rep. 6:28117. doi: 10.1038/srep28117
Santiviago, C. A., Reynolds, M. M., Porwollik, S., Choi, S. H., Long, F., Andrews-Polymenis, H. L., et al. (2009). Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. doi: 10.1371/journal.ppat.1000477
Santos, R. L., Zhang, S., Tsolis, R. M., Bäumler, A. J., and Adams, L. G. (2002). Morphologic and molecular characterization of Salmonella typhimurium infection in neonatal calves. Vet. Pathol. Online 39, 200–215. doi: 10.1354/vp.39-2-200
Seto, H., and Kuzuyama, T. (1999). Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat. Prod. Rep. 16, 589–596. doi: 10.1039/a809398i
Shimizu, H., Kakimoto, Y., Nakajima, T., Kanazawa, A., and Sano, I. (1965). Isolation and identification of 2-aminoethylphosphonic acid from bovine brain. Nature 207, 1197–1198. doi: 10.1038/2071197a0
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Stecher, B., Robbiani, R., Walker, A. W., Westendorf, A. M., Barthel, M., Kremer, M., et al. (2007). Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189. doi: 10.1371/journal.pbio.0050244
Sternberg, N. L., and Maurer, R. (1991). Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 204, 18–43.
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108, 17480–17485. doi: 10.1073/pnas.1107857108
Tsolis, R. M., Adams, L. G., Ficht, T. A., and Bäumler, A. J. (1999). Contribution of Salmonella typhimuriumvirulence factors to Diarrheal disease in Calves. Infect. Immun. 67, 4879–4885.
Villarreal-Chiu, J. F., Quinn, J. P., and McGrath, J. W. (2012). The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front. Microbiol. 3:19. doi: 10.3389/fmicb.2012.00019
Wang, R. F., and Kushner, S. R. (1991). Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100, 195–199. doi: 10.1016/0378-1119(91)90366-J
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Yu, X., Doroghazi, J. R., Janga, S. C., Zhang, J. K., Circello, B., Griffin, B. M., et al. (2013). Diversity and abundance of phosphonate biosynthetic genes in nature. Proc. Natl. Acad. Sci. U.S.A. 110, 20759–20764. doi: 10.1073/pnas.1315107110
Zhang, S., Santos, R. L., Tsolis, R. M., Stender, S., Hardt, W.-D., Bäumler, A. J., et al. (2002). The Salmonella enterica serotype typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce Diarrhea in Calves. Infect. Immun. 70, 3843–3855. doi: 10.1128/IAI.70.7.3843-3855.2002
Keywords: Salmonella, phosphonates, mice, infection, phosphonoacetic acid
Citation: Elfenbein JR, Knodler LA, Schaeffer AR, Faber F, Bäumler AJ and Andrews-Polymenis HL (2017) A Salmonella Regulator Modulates Intestinal Colonization and Use of Phosphonoacetic Acid. Front. Cell. Infect. Microbiol. 7:69. doi: 10.3389/fcimb.2017.00069
Received: 20 January 2017; Accepted: 23 February 2017;
Published: 15 March 2017.
Edited by:
Alfredo G. Torres, University of Texas Medical Branch, USAReviewed by:
Charles Martin Dozois, Institut National de la Recherche Scientifique, CanadaCarmen R. Beuzón, University of Málaga, Spain
Copyright © 2017 Elfenbein, Knodler, Schaeffer, Faber, Bäumler and Andrews-Polymenis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Helene L. Andrews-Polymenis, handrews@medicine.tamhsc.edu
 Johanna R. Elfenbein
Johanna R. Elfenbein Leigh A. Knodler
Leigh A. Knodler Allison R. Schaeffer1
Allison R. Schaeffer1  Franziska Faber
Franziska Faber Andreas J. Bäumler
Andreas J. Bäumler Helene L. Andrews-Polymenis
Helene L. Andrews-Polymenis