Shiga Toxin Subtypes of Non-O157 Escherichia coli Serogroups Isolated from Cattle Feces
- 1Department of Diagnostic Medicine and Pathobiology, Kansas State University, Manhattan, KS, USA
- 2Veterinary Diagnostic Laboratory, Kansas State University, Manhattan, KS, USA
Shiga toxin producing Escherichia coli (STEC) are important foodborne pathogens responsible for human illnesses. Cattle are a major reservoir that harbor the organism in the hindgut and shed in the feces. Shiga toxins (Stx) are the primary virulence factors associated with STEC illnesses. The two antigenically distinct Stx types, Stx1 and Stx2, encoded by stx1 and stx2 genes, share approximately 56% amino acid sequence identity. Genetic variants exist within Stx1 and Stx2 based on differences in amino acid composition and in cytotoxicity. The objective of our study was to identify the stx subtypes in strains of STEC serogroups, other than O157, isolated from cattle feces. Shiga toxin gene carrying E. coli strains (n = 192), spanning 27 serogroups originating from cattle (n = 170) and human (n = 22) sources, were utilized in the study. Shiga toxin genes were amplified by PCR, sequenced, and nucleotide sequences were translated into amino acid sequences using CLC main workbench software. Shiga toxin subtypes were identified based on the amino acid motifs that define each subtype. Shiga toxin genotypes were also identified at the nucleotide level by in silico restriction fragment length polymorphism (RFLP). Of the total 192 STEC strains, 93 (48.4%) were positive for stx1 only, 43 (22.4%) for stx2 only, and 56 (29.2%) for both stx1 and stx2. Among the 149 strains positive for stx1, 132 (88.6%) were stx1a and 17 (11.4%) were stx1c. Shiga toxin 1a was the most common subtype of stx1 among cattle (87.9%; 123/140) and human strains (100%; 9/9) of non-O157 serogroups. Of the total 99 strains positive for stx2, 79 were stx2a (79.8%), 11 (11.1%) were stx2c, 12 (12.1%) were stx2d. Of the 170 strains originating from cattle feces, 58 (34.1%) were stx2a subtype, 11 (6.5%) were stx2c subtype, and 11 were of subtype stx2d (6.5%). All but one of the human strains were positive for stx2a. Three strains of cattle origin were positive for both stx2a and stx2d. In conclusion, a number of non-O157 STEC serogroups harbored by cattle possess a wide variety of Shiga toxin subtypes, with stx1a and stx2a being the most predominant stx subtypes occurring individually or in combination. Cattle are a reservoir of a number of non-O157 STEC serogroups and information on the Shiga toxin subtypes is useful in assessing the potential risk as human pathogens.
Introduction
Shiga toxin producing Escherichia coli (STEC) are major foodborne pathogens responsible for human illnesses, characterized by non-bloody to bloody diarrhea, sometimes leading to complications of hemolytic uremic syndrome (HUS), particularly in children (Gyles, 2007). Escherichia coli O157:H7 is the major serotype responsible for many of the STEC illness outbreaks in humans. However, there is increasing incidence of outbreaks associated with non-O157 STEC in recent years, particularly O26, O45, O103, O111, O121, and O145, referred to as top six non-O157 STEC. According to FoodNet sites, incidence of top six non-O157 STEC infections increased from 0.12 per 100,000 population in 2,000 to 0.95 per 100,000 population in 2010 (Gould et al., 2013). Non-O157 STEC associated illnesses range from cases of sporadic to major outbreaks, and clinically, from mild watery diarrhea to life threatening complications of HUS, similar to STEC O157 infections (Johnson et al., 2006). Cattle are a major reservoir of O157 and non-O157 STEC, which harbor the organisms in the hindgut and shed in the feces. Consumption of water, beef and fresh produce contaminated with cattle feces leads to human illnesses. In addition to O157 and the six top non-O157, cattle do harbor and shed in the feces a number of other serogroups of STEC (Bettelheim, 2007; Hussein, 2007).
Shiga toxins (Stx) are the major virulence factors of STEC. Shiga toxins (Stx) belong to the AB5 family of protein toxins, with an enzymatically active A moiety and a B moiety involved in binding to the host cell receptor. The A subunit is responsible for the cleavage of N-glycosidic bond in the 28 s rRNA of 60 s ribosomal subunit, which leads to cytotoxicity (Endo et al., 1988; Fraser et al., 1994). The two antigenically distinct Stx types, Stx1 and Stx2, encoded by stx1 and stx2 genes, share approximately 56% amino acid sequence similarity (Strockbine et al., 1986; Weinstein et al., 1988). Although Stx1 and Stx2 are structurally similar, they differ in cellular distribution and cytotoxicity. Shiga toxin 1 is located in the periplasmic space of the bacterial cell, whereas Stx2 is in the extracellular fraction (Shimizu et al., 2009). Basu et al. (2016) have shown that A1 subunit of Stx2 has a higher affinity for ribosomes and a higher catalytic activity compared to A1 subunit of Stx1 (Basu et al., 2016), which makes Stx2 more cytotoxic than Stx1. Shiga toxin 2 is reported to be 400 times more toxic than Stx1 in a murine infection model (Tesh et al., 1993). Shiga toxin 2 is more commonly associated with complications of HUS than Stx1 (Ethelberg et al., 2004; Brooks et al., 2005). Variants exist within stx1 (stx1a, stx1c, and stx1d) and stx2 (stx2a, stx2b, stx2c, stx2d, stx2e, stx2f, and stx2g) families based on differences in amino acid compositions and in the degree of cytotoxicity. The outcome of human illness associated with STEC strains has been shown to be influenced by Stx subtypes (Friedrich et al., 2002). Shiga toxin 2a and Stx2c are the major subtypes produced by E. coli O157:H7 strains associated with HUS in humans (Persson et al., 2007). Therefore, identifying the subtypes of (Stx) is important to assess the potential risk for human illnesses associated with STEC infections. Subtyping method based on restriction fragment length polymorphism of PCR products (PCR-RFLP) has been developed to identify stx1 (stx1a, stx1c, and stx1d) and stx2 (stx2, stx2v-ha, stx2v-hb, stx2g, stx2-NV206, and stx2-EC1586) genotypes at the nucleotide level based on their unique restriction patterns (Gobius et al., 2003; Beutin et al., 2007). However, the demerits of this method are the lack of consistency in the nomenclature, and misinterpretation of stx subtypes due to single nucleotide changes (Scheutz et al., 2012). Scheutz et al. (2012) standardized the Stx nomenclature by designating stx1 subtypes as stx1a, stx1c and stx1d, and stx2 subtypes as stx2a, stx2b, stx2c, stx2d, stx2e, stx2f, and stx2g based on amino acid sequence similarity (Scheutz et al., 2012). There is a paucity of data on Shiga toxin subtypes carried by non-O157 E. coli serogroups isolated from cattle feces in the United States. The objective of our study was to determine the subtypes of stx1 and stx2 in non-O157 E. coli serogroups isolated from cattle feces.
Materials and Methods
Strains
Shiga toxin gene-positive E. coli strains (n = 192) spanning 27 non-O157 E. coli serogroups isolated from cattle feces (n = 170), and human clinical cases (n = 22), available in our culture collection, were used in the study. A majority of strains belonged to the top six non-O157 E. coli serogroups: O26 (n = 16), O45 (n = 4), O103 (n = 54), O111 (n = 21), O121 (n = 4), and O145 (n = 27). The other non-O157 E. coli serogroups included O6 (n = 2), O8 (n = 3), O15 (n = 1), O22 (n = 1), O38 (n = 2), O39 (n = 3), O74 (n = 3), O88 (n = 3), O91 (n = 2), O96 (n = 3), O104 (n = 18), O113 (n = 3), O116 (n = 3), O117 (n = 3), O130 (n = 4), O141 (n = 3), O146 (n = 1), O153 (n = 1), O163 (n = 2), O171 (n = 3), and O172 (n = 2). Cattle strains were isolated from feces, primarily from commercial feedlots (Renter et al., 2005; Noll et al., 2015; Shridhar et al., 2016; Cull et al., 2017). A few human clinical strains, obtained from Michigan State University (MSU) and the Kansas Department of Health and Environment (KDHE), were also included in the study. The strains, stored at −80° C on cryo beads (CryoCare™, Key Scientific Products, Round Rock, TX), were streaked onto blood agar plates (Remel, Lenexa, KS), colonies were suspended in 50 μl distilled water, boiled for 10 min, centrifuged at 9,300 × g for 5 min, and supernatant was used for PCR amplification and subsequent sequencing of the amplicons.
Primers Design
Nucleotide sequences of stx1 and stx2 of E. coli O157 and non-O157 E. coli were downloaded from NCBI Genbank. The sequences were aligned using CLC Main Workbench (CLC Bio, Cambridge, MA), and the conserved regions flanking the Shiga toxin genes were selected to design the oligonucleotide primers for amplification of stx1 and stx2 genes (Table 1). Sequences of stx2f subtype appeared to be more divergent from the other subtypes, therefore, a separate set of primers were designed for amplification (Table 1). Primers were obtained from Integrated DNA technologies (Coralville, Iowa).
PCR Assay Conditions
The stx genes were amplified by touchdown PCR method, where the annealing temperature of each cycle was lowered gradually to avoid amplification of non-specific sequences (Don et al., 1991). PCR was performed using Eppendorf Mastercycler (Eppendorf, Hamburg, Germany).
PCR amplification protocol for stx1 included an initial denaturation at 94°C for 5 min, 10 cycles of touch-down PCR (denature: 94°C for 30 s, annealing: 56–51°C (Δ-0.5°C) for 30 s; and extension: 72°C for 1 min 45 s) followed by 30 cycles of regular PCR (denature: 94°C for 30 s, annealing: 51°C for 30 s; and extension: 72°C for 1 min 45 s). PCR amplification protocol for stx2 included an initial denaturation at 94°C for 5 min, 10 cycles of touch-down PCR (denature: 94°C for 30 s, annealing: 47–44°C (Δ-0.3°C) for 30 s; and extension: 72°C for 1 min 45 s) followed by 30 cycles of regular PCR (denature: 94°C for 30 s, annealing: 44°C for 30 s; and extension: 72°C for 1 min 45 s). All PCR reagents were obtained from TaKaRa Bio USA, Inc. (CA).
Sequencing and Data Analyses
Amplified PCR products were visualized using a Qiaxcel capillary electrophoresis system (Qiagen, Valencia, CA) and purified using a QIAquick PCR purification kit (Qiagen). The purified PCR products were measured by a spectrophotometer (NanoDrop-Thermo Scientific, Wilmington, DE) to assess the DNA concentration and purity. PCR products and primers were shipped to Genewiz, Inc., (South Plainfield, NJ) for nucleotide sequencing. The chromatogram data were visualized using the CLC Main Workbench software for further analysis. All sequences were individually analyzed for conflicts, and secondary peaks, and consensus sequences were produced. Nucleotide sequences were translated to amino acid sequences after removing intergenic sequences. Shiga toxin subtypes were determined based on the amino acid motifs that define each stx subtype (Scheutz et al., 2012).
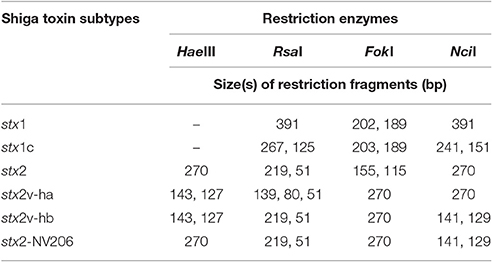
In silico Restriction Fragment Length Polymorphism (RFLP)
A subset of strains (n = 68) were subjected to in silico RFLP for identification of stx1 (stx1, stx1c, and stx1d) and stx2 (stx2, stx2v-ha, stx2v-hb, stx2-NV206, stx2g, and stx2-EC1586) genotypes. For stx1, a 391–392 bp fragment starting with GAYTATCATGGACAAGACTCYGTT and ending with TGACGATACYTTTACAGTTAAAGTGG was digested with RsaI, FokI, and NciI enzymes using CLC Main Workbench software to identify the unique restriction sites for each stx1 genotype. For stx2, a 270 bp fragment starting with ATGAAGAAGATGTTTATG and ending with CAGTTTAATAATGACTGA was digested with HaeIII, RsaI, FokI, and NciI enzymes (Beutin et al., 2007) using CLC Main Workbench software. Restriction patterns of stx1 and stx2 sequences of the non-O157 STEC strains were compared to that of the reference sequences from the NCBI database to identify the stx1 and stx2 genotypes.
Cloning and Sequencing of PCR Products
Three strains belonging to serogroups O96, O113, and O130 that revealed double peaks in the chromatograms of stx2 sequences were subjected to cloning. PCR products were cloned using TOPO® TA Cloning kits and protocols by Invitrogen-Life Technologies (Grand Island, NY). Up to 12 transformants were selected and grown in Luria Bertani broth (LB; Becton, Dickinson Co., Sparks, MD) containing carbenicillin for 2 h. Clones obtained from the LB broth were subjected to sequencing using flanking M13 forward and reverse primers. The sequences were analyzed as mentioned above to identify the Shiga toxin subtypes.
Results
Of the total 192 non-O157 STEC strains (170 cattle and 22 human strains) belonging to 27 serogroups tested in the study, 93 (48.4%) were positive for stx1 only, 43 (22.4%) for stx2 only, and 56 (29.2%) for both stx1 and stx2. A total of 149 strains belonging to 23 serogroups were positive for stx1, and of those 132 (88.6%) were stx1a and 17 (11.4%) were stx1c (Table 2). Of the 140 stx1 positive strains originating from cattle feces, 123 (87.9%) were stx1a subtype, and 17 (12.1%) were stx1c. Shiga toxin 1a was the only stx1 subtype found in human STEC strains (9/9; 100%). The stx1a was also the most common subtype of stx1 identified in top six non-O157 E. coli serogroups (Table 2). None of the strains were positive for more than one stx1 subtype.
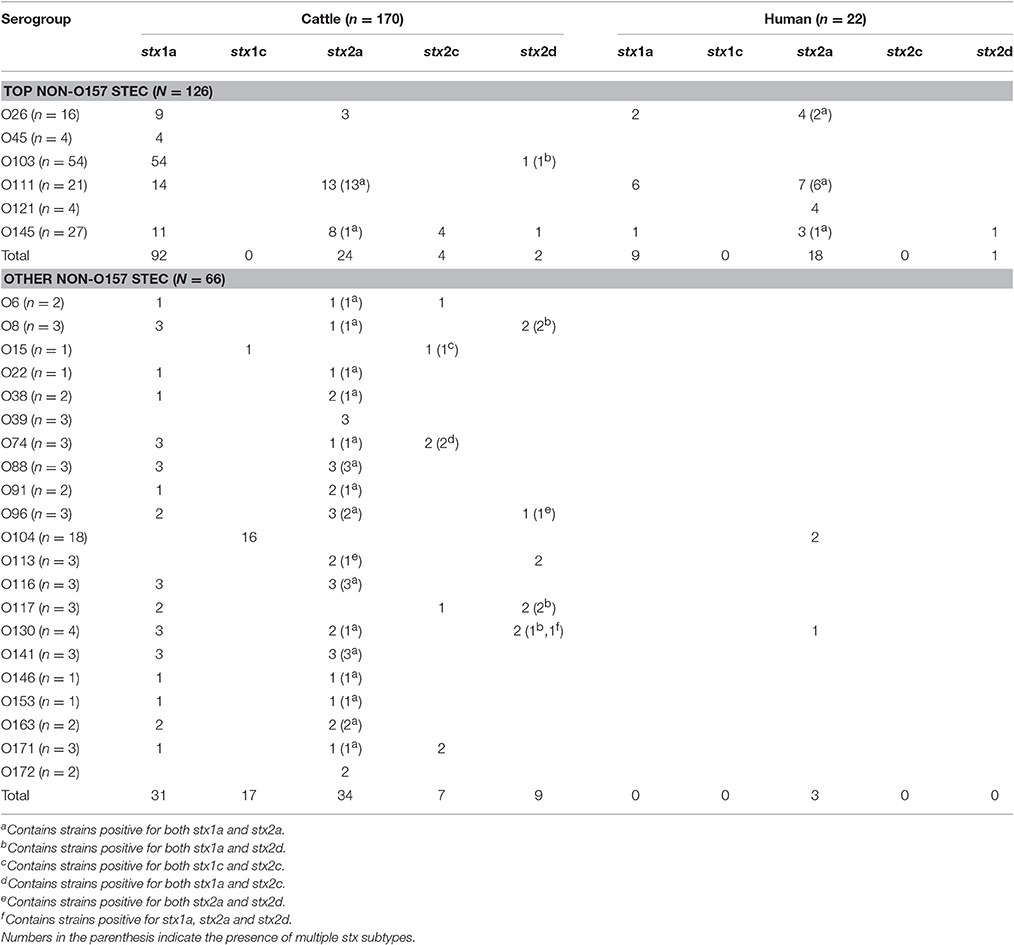
Table 2. Shiga toxin subtype distribution in non-O157 Shiga toxin-producing E. coli (STEC) serogroups of cattle and human origin (n = 192).
A total of 99 strains belonging to 25 serogroups were positive for stx2, and of those 79 were stx2a (79.8%), 11 (11.1%) were stx2c, 12 (12.1%) were stx2d. Three stx2 positive-strains belonging to O96, O113 and O130 serogroups of cattle origin revealed double peaks in the chromatogram of stx2 sequences, suggesting the presence of more than one stx2 subtype in the same strain. Subsequent cloning and sequencing revealed that all three strains carried a combination of stx2a and stx2d. All human clinical strains (n = 22), except one, were of subtype stx2a. Only one strain belonging to serogroup O145 carried stx2d. All strains positive for stx2c were of bovine origin (Table 2, Figure 1). Shiga toxin subtypes 2e, 2f, and 2g were not detected in any of the strains tested. A majority of cattle strains (n = 170) possessed stx1a subtype (123/170; 72.4%) followed by stx2a (58/170; 34.1%; Figure 1). However, a majority of human strains carried stx2a (21/22; 95.5%) followed by stx1a (9/22; 40.9%). Shiga toxin 1a was most commonly found in combination with stx2a in strains (n = 46) belonging to 18 serogroups. Six strains belonging to O8, O103, O117, and O130 carried a combination of stx1a and stx2d. Two strains of serogroup O74 carried a combination of stx1a and stx2c and a strain of serogroup O15 was positive for a combination of stx1c and stx2c. A strain of serogroup O130 was positive for three subtypes, stx1a, stx2a, and stx2d (Table 2).
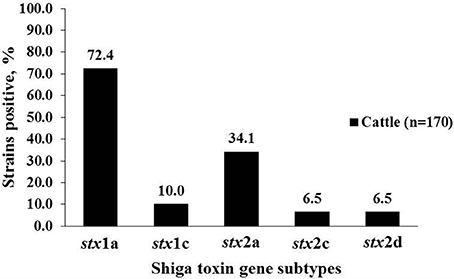
Figure 1. Percentage of stx1 and stx2 subtypes in non-O157 Shiga toxin-producing Escherichia coli (STEC) strains isolated from cattle (n = 170).
The sequences from a subset of strains (n = 68) were also analyzed by in silico RFLP. The sizes of fragments generated by restriction enzyme digestion are shown in Table 3. The two stx1 genotypes (stx1 and stx1c) determined by in silico RFLP corresponded to the two subtypes (stx1a and stx1c) determined based on the amino acid motifs. For stx2, genotypes, such as stx2, stx2-NV206, and stx2v-ha determined based on the restriction patterns corresponded to stx2a, stx2d, and stx2c subtypes, respectively, determined based on amino acid sequence. However, some strains positive for stx2v-hb gene (based on restriction patterns) corresponded to stx2c and some to stx2d subtype (based on amino acid sequence motifs). Restriction patterns of stx1 and stx2 genotypes determined by in silico RFLP are shown in Figure 2.
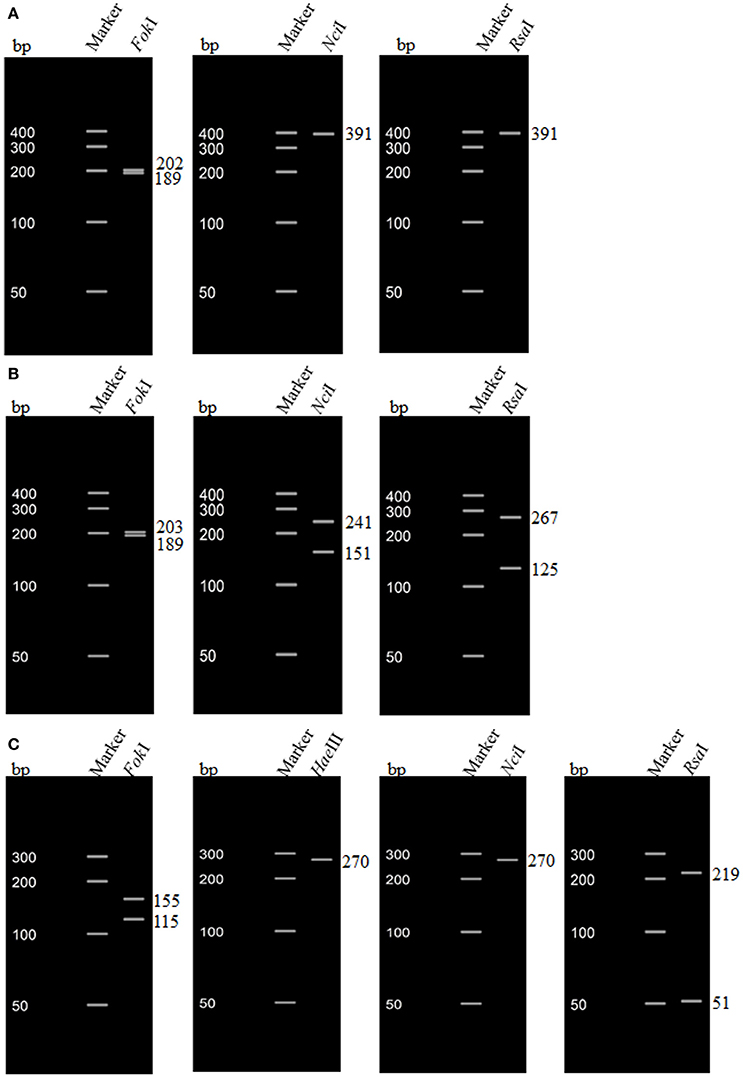
Figure 2. In silico restriction fragment length polymorphism (RFLP) of stx1 and stx2 genes of non-O157 Shiga toxin-producing Escherichia coli (STEC) strains. (A) RFLP pattern of stx1a of a non-O157 STEC strain (O103 serogroup) isolated from cattle feces; (B) RFLP pattern of stx1c of a non-O157 STEC strain (O15 serogroup) isolated from cattle feces; (C) RFLP pattern of stx2 of a non-O157 STEC strain (O145 serogroup) isolated from cattle feces.
Discussion
Shiga toxins are the major virulence factors of STEC, which are responsible for foodborne illnesses, including life threatening complications of HUS in humans. Shiga toxins exist as several subtypes, which vary in their cytotoxicity, and therefore in the extent of their involvement in human illness (Friedrich et al., 2002; Persson et al., 2007; Fuller et al., 2011). Fuller et al. (2011) demonstrated that stx2a and stx2d subtypes were more potent than stx2b, stx2c, and stx1 based on in vitro (using primary human renal proximal tubule epithelial cells and Vero cells) and in vivo (using mice) experiments (Fuller et al., 2011). Karve and Weiss (2014) demonstrated stronger binding of stx2a, stx2c, and stx2d to a mixture of Gb3 and glycolipids when compared to stx1 and stx2b. Determining the Shiga toxin subtype carried by the STEC strains is important to estimate the risk of human illness associated with specific serotype or source of transmission. Studies have shown that STEC strains carrying stx2a and stx2c are most commonly associated with HUS in humans (Friedrich et al., 2002; Persson et al., 2007; Iyoda et al., 2014). Production of elastase activatable Stx2d subtype in STEC strains has been reported to be a predictor of severity of clinical illness (Bielaszewska et al., 2006). The cleavage of C-terminal amino acids of A2 peptide of Stx2d by elastase has been reported to increase the cytotoxicity of this Shiga toxin subtype (Kokai-Kun et al., 2000; Melton-Celsa et al., 2002). The Stx2e subtype is most commonly associated with porcine STEC, however, it is also associated with STEC from asymptomatic humans and fresh produce (Friedrich et al., 2002; Beutin et al., 2008; Feng and Reddy, 2013). Although pigeons are a primary reservoir of stx2f carrying E. coli strains, this subtype has also been isolated from human diarrheic patients (Schmidt et al., 2000; Prager et al., 2009). There are studies reporting the distribution of Shiga toxin subtypes in STEC strains isolated from humans and fresh produce (Friedrich et al., 2002; Feng and Reddy, 2013). There are very limited studies on the distribution of Shiga toxin subtypes in cattle, particularly of non-O157 serogroups, in the United States.
In this study, we identified Shiga toxin subtypes associated with 27 serogroups of non-O157 STEC strains of cattle (n = 170) and human (n = 22) origin based on the amino acid sequences deduced from nucleotide sequences. Additionally, we also identified stx genotypes by in silico RFLP, based on the restriction patterns obtained after in silico digestion of nucleotide sequences with specific restriction enzymes. The most common subtype of stx1 carried by non-O157 STEC strains of cattle and human origin was stx1a. A similar finding was also reported in a study on cattle and human STEC strains in Canada (Chui et al., 2015). Tostes et al. (2017) reported that stx1a and stx2a are the most common subtypes in both E. coli O157 and non-O157 E. coli strains of human and bovine origin in Alberta (Tostes et al., 2017). In the present study, a majority of the cattle STEC strains (21.8%; 37/170) and human STEC strains (40.9%; 9/22) carried a combination of stx1a and stx2a. A majority of E. coli O157:H7 strains isolated from outbreaks and sporadic cases in Canada carried a combination of stx1a and stx2a (Chui et al., 2015). In our study, 17 (10%) cattle STEC strains carried stx1c, while none of the human STEC strains carried this subtype. STEC strains carrying stx1c have been isolated from asymptomatic humans and patients with uncomplicated diarrhea (Friedrich et al., 2003). However, stx1c carrying E. coli O78 was isolated from a 2-week old boy suffering from bacteremia and HUS in Finland (Lienemann et al., 2012).
Shiga toxin subtype 2a was the most common stx2 subtype (41.1%; 79/192) found in cattle and human sources. It was the most common subtype detected in STEC strains isolated from patients suffering from HUS (Persson et al., 2007) and those isolated from fresh produce (Feng and Reddy, 2013). It was also the most common subtype of stx2 in STEC strains isolated from cattle in Australia (Brett et al., 2003) and France (Bertin et al., 2001). The second most frequent subtype of stx2 associated with STEC strains isolated from cattle was stx2c, however, none of the human STEC strains included in this study contained stx2c. The stx2c subtype has been reported in STEC strains isolated from human patients suffering from HUS (Persson et al., 2007). It was the third most common subtype (next to stx2d) associated with STEC strains isolated from fresh produce (Feng and Reddy, 2013). Tostes et al. (2017) have reported the distribution of stx1 and stx2 subtypes associated with E. coli O157 and non-O157 E. coli serogroups of bovine and human origin in Alberta, and found that the stx2c subtype was found only in E. coli O157 strains (Tostes et al., 2017). Shiga toxin 2c was the most common subtype associated with STEC O157 strains of cattle origin in Italy, however, none of the STEC O157 strains harbored stx2a (Bonardi et al., 2015). However, in our study, 6.5% of the non-O157 STEC strains isolated from cattle carried stx2c subtype. In the present study, only a small proportion (6.5%) of the bovine strains carried stx2d subtype, and only one human STEC strain carried stx2d. Tasara et al. (2008) have reported stx2d carrying STEC strains isolated from cattle and sheep (Tasara et al., 2008). Presence of stx2d carrying STEC strains in human patients was reported to be significantly associated with HUS (Bielaszewska et al., 2006). In our study, three cattle strains carried two different stx2 subtypes (stx2a and stx2d). STEC strains carrying more than one stx2 subtypes have been reported in previous studies (Persson et al., 2007; Scheutz et al., 2012). The significance of multiple stx2 subtypes with regard to virulence of STEC has not been studied yet. The presence of multiple stx subtypes in STEC is assumed to be the result of recombination of stx phages (Ashton, 2015).
We also identified Shiga toxin genotypes by in silico RFLP. Some strains positive for stx2v-hb gene (based on restriction patterns; Beutin et al., 2007) corresponded to stx2c and some to stx2d subtype (based on amino acid sequence motifs). Single nucleotide changes within the restriction sites could lead to misinterpretation of stx subtypes based on the restriction pattern (Scheutz et al., 2012). In conclusion, a number of non-O157 STEC serogroups isolated from cattle possessed a wide variety of Shiga toxin subtypes, with stx1a and stx2a being the most predominant stx subtypes occurring individually or in combination. Cattle harbor a number of non-O157 STEC serogroups and identification of the Shiga toxin subtypes is useful in assessing the potential risk to cause human illnesses.
Author Contributions
Conceived and designed the experiments: JB, TN; Performed the experiments: PS, CS, XS, LN; Contributed reagents/materials/analysis tools: JB, TN; Wrote the paper: PS, TN, JB.
Funding
Research was supported by the Agriculture and Food Research Initiative Grant No. 2012-68003-30155 from the USDA National Institute of Food and Agriculture.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Neil Wallace and Emily A. Slone for their assistance with this project. This is contribution no. 17-259-J of the Kansas Agricultural Experiment Station.
References
Ashton, P. (2015). Insight into Shiga toxin genes encoded by Escherichia coli O157 from whole genome sequencing. Peer J. 3:e739. doi: 10.7717/peerj.739
Basu, D., Li, X. P., Kahn, J. N., May, K. L., Kahn, P. C., and Tumer, N. E. (2016). The A1 subunit of Shiga toxin 2 has higher affinity for ribosomes and higher catalytic activity than the A1 subunit Shiga toxin 1. Infect. Immun. 84, 149–161. doi: 10.1128/IAI.00994-15
Bertin, Y., Boukhors, K., Pradel, N., Livrelli, V., and Martin, C. (2001). Stx2 subtyping of shiga toxin-producing Escherichia coli isolated from cattle in france: detection of a new Stx2 subtype and correlation with additional virulence factors. J. Clin. Microbiol. 39, 3060–3065. doi: 10.1128/JCM.39.9.3060-3065.2001
Bettelheim, K. A. (2007). The non-O157 shiga-toxigenic (Verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit. Rev. Microbiol. 33, 67–87. doi: 10.1080/10408410601172172
Beutin, L., Krüger, U., Krause, G., Miko, A., Martin, A., and Strauch, E. (2008). Evaluation of major types of shiga toxin 2e-producing Escherichia coli bacteria present in food, pigs, and the environment as potential pathogens for humans. Appl. Environ. Microbiol. 74, 4806–4816. doi: 10.1128/AEM.00623-08
Beutin, L., Miko, A., Krause, G., Pries, K., Haby, S., Steege, K., et al. (2007). Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by a combination of serotyping and molecular typing of Shiga toxin genes. Appl. Environ. Microbiol. 73, 4769–4775. doi: 10.1128/AEM.00873-07
Bielaszewska, M., Friedrich, A. W., Aldick, T., Schürk-Bulgrin, R., and Karch, H. (2006). Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43, 1160–1167. doi: 10.1086/508195
Bonardi, S., Alpigiani, I., Tozzoli, R., Vismarra, A., Zecca, V., Greppi, C., et al. (2015). Shiga toxin-producing Escherichia coli O157, O26 and O111 in cattle faeces and hides in Italy. Vet. Rec. Open 2:e000061. doi: 10.1136/vetreco-2014-000061
Brett, K. N., Hornitzky, M. A., Bettelheim, K. A., Walker, M. J., and Djordjevic, S. P. (2003). Bovine non-O157 shiga toxin 2-containing Escherichia coli isolates commonly possess stx(2-EDL933) and/or stx(2vhb) subtypes. J. Clin. Microbiol. 41, 2716–2722. doi: 10.1128/JCM.41.6.2716-2722.2003
Brooks, J. T., Sowers, E. G., Wells, J. G., Greene, K. D., Griffin, P. M., Hoekstra, R. M., et al. (2005). Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192, 1422–1429. doi: 10.1086/466536
Chui, L., Li, V., Fach, P., Delannoy, S., Malejczyk, K., Patterson-Fortin, L., et al. (2015). Molecular profiling of Escherichia coli O157:H7 and non-O157 strains isolated from humans and cattle in Alberta, Canada. J. Clin. Microbiol. 53, 986–990. doi: 10.1128/JCM.03321-14
Cull, C. A., Renter, D. G., Dewsbury, D. M., Noll, L. W., Shridhar, P. B., Ives, S. E., et al. (2017). Feedlot- and pen-level prevalence of Enterohemorrhagic Escherichia coli in feces of commercial feedlot cattle in two major U.S. cattle feeding areas. Foodborne Pathog. Dis. doi: 10.1089/fpd.2016.2227. [Epub ahead of print].
Don, R. H., Cox, P. T., Wainwright, B. J., Baker, K., and Mattick, J. S. (1991). 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. doi: 10.1093/nar/19.14.4008
Endo, Y., Tsurugi, K., Yutsudo, T., Takeda, Y., Ogasawara, T., and Igarashi, K. (1988). Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 171, 45–50.
Ethelberg, S., Olsen, K. E., Scheutz, F., Jensen, C., Schiellerup, P., Enberg, J., et al. (2004). Virulence factors for hemolytic uremic syndrome, Denmark. Emerg. Infect. Dis. 10, 842–847. doi: 10.3201/eid1005.030576
Feng, P. C., and Reddy, S. (2013). Prevalences of shiga toxin subtypes and selected other virulence factors among shiga-toxigenic Escherichia coli strains isolated from fresh produce. Appl. Environ. Microbiol. 79, 6917–6923. doi: 10.1128/AEM.02455-13
Fraser, M. E., Chernaia, M. M., Kozlov, Y. V., and James, M. N. (1994). Crystal structure of the holotoxino from Shigella dysenteriae at 2.5 A resolution. Nat. Struct. Mol. Biol. 1, 59–64. doi: 10.1038/nsb0194-59
Friedrich, A. W., Bielaszewska, M., Zhang, W. L., Pulz, M., Kuczius, T., Ammon, A., et al. (2002). Escherichia coli harboring shiga toxin 2 gene variants: frequency and association with clinical symptoms. Am. J. Infect. Dis. 185, 74–84. doi: 10.1086/338115
Friedrich, A. W., Borell, J., Bielaszewska, M., Fruth, A., Tschape, H., and Karch, H. (2003). Shiga Toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41, 2448–2453. doi: 10.1128/JCM.41.6.2448-2453.2003
Fuller, C. A., Pellino, C. A., Flagler, M. J., Strasser, J. E., and Weiss, A. A. (2011). Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 79, 1329–1337. doi: 10.1128/IAI.01182-10
Gobius, K. S., Higgs, G. M., and Desmarchelier, P. M. (2003). Presence of Activatable shiga toxin genotype (stx2d) in shiga toxigenic Escherichia coli from livestock sources. J. Clin. Microbiol. 41, 3777–3783. doi: 10.1128/JCM.41.8.3777-3783.2003
Gould, L. H., Mody, R. K., Ong, K. L., Clogher, P., Cronquist, A. B., Garman, K. N., et al. (2013). Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000-2010: epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog. Dis. 10, 453–460. doi: 10.1089/fpd.2012.1401
Gyles, C. L. (2007). Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85(Suppl. 13), E45–E62. doi: 10.2527/jas.2006-508
Hussein, H. S. (2007). Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 85(Suppl. 13), E63–E72. doi: 10.2527/jas.2006-421
Iyoda, S., Manning, S. D., Seto, K., Kimata, K., Isobe, J., Etoh, Y., et al. (2014). Phylogenetic clades 6 and 8 of enterohemorrhagic Escherichia coli O157:H7 with particular stx subtypes are more frequently found in isolates from hemolytic uremic syndrome patients than from asymptomatic carriers. Open Forum. Infect. Dis. 1:ofu061. doi: 10.1093/ofid/ofu061
Johnson, K. E., Thorpe, C. M., and Sears, C. L. (2006). The emerging clinical importance of non-O157 shiga toxin—producing Escherichia coli. Clin. Infect. Dis. 43, 1587–1595. doi: 10.1086/509573
Karve, S. S., and Weiss, A. A. (2014). Glycolipid binding preferences of shiga toxin variants. PLoS ONE 9:e101173. doi: 10.1371/journal.pone.0101173
Kokai-Kun, J. F., Melton-Celsa, A. R., and O'Brien, A. D. (2000). Elastase in intestinal mucus enhances the cytotoxicity of Shiga toxin type 2d. J. Biol. Chem. 275, 3713–3721. doi: 10.1074/jbc.275.5.3713
Lienemann, T., Salo, E., Rimhanen-Finne, R., Rönnholm, K., Taimisto, M., Hirvonen, J. J., et al. (2012). Shiga toxin–producing Escherichia coli serotype O78:H(–) in family, finland, 2009. Emerg. Infect. Dis. 18, 577–581. doi: 10.3201/eid1804.111310
Melton-Celsa, A. R., Kokai-Kun, J. F., and O'Brien, A. D. (2002). Activation of Shiga toxin type 2d (Stx2d) by elastase involves cleavage of the C-terminal two amino acids of the A2 peptide in the context of the appropriate B pentamer. Mol. Microbiol. 43, 207–215. doi: 10.1046/j.1365-2958.2002.02733.x
Noll, L. W., Shridhar, P. B., Dewsbury, D. M., Shi, X., Cernicchiaro, N., Renter, D. G., et al. (2015). A comparison of culture- and PCR-based methods to detect six major non-O157 serogroups of shiga toxin-producing Escherichia coli in cattle feces. PLoS ONE 10:e0135446. doi: 10.1371/journal.pone.0135446
Persson, S., Olsen, K. E., Ethelberg, S., and Scheutz, F. (2007). Subtyping Method for Escherichia coli Shiga Toxin (Verocytotoxin) 2 Variants and correlations to clinical manifestations. J. Clin. Microbiol. 45, 2020–2024. doi: 10.1128/JCM.02591-06
Prager, R., Fruth, A., Siewert, U., Strutz, U., and Tschäpe, H. (2009). Escherichia coli encoding Shiga toxin 2f as an emerging human pathogen. Int. J. Med. Microbiol. 299, 343–353. doi: 10.1016/j.ijmm.2008.10.008
Renter, D. G., Morris, J. G. Jr., Sargeant, J. M., Hungerford, L. L., Berezowski, J., Ngo, T., et al. (2005). Prevalence, risk factors, O serogroups, and virulence profiles of Shiga toxin-producing bacteria from cattle production environments. J. Food Prot. 68, 1556–1565. doi: 10.4315/0362-028X-68.8.1556
Scheutz, F., Teel, L. D., Beutin, L., Pierard, D., Buvens, G., Karch, H., et al. (2012). Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 50, 2951–2963. doi: 10.1128/JCM.00860-12
Schmidt, H., Scheef, J., Morabito, S., Caprioli, A., Wieler, L. H., and Karch, H. (2000). A new shiga toxin 2 Variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66, 1205–1208. doi: 10.1128/AEM.66.3.1205-1208.2000
Shimizu, T., Ohta, Y., and Noda, M. (2009). Shiga toxin 2 is specifically released from bacterial cells by two different mechanisms. Infect. Immun. 77, 2813–2823. doi: 10.1128/IAI.00060-09
Shridhar, P. B., Noll, L. W., Shi, X., Cernicchiaro, N., Renter, D. G., Bai, J., et al. (2016). Escherichia coli O104 in feedlot cattle feces: prevalence, isolation and characterization. PLoS ONE 11:e0152101. doi: 10.1371/journal.pone.0152101
Strockbine, N. A., Marques, L. R., Newland, J. W., Smith, H. W., Holmes, R. K., and O'Brien, A. D. (1986). Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect. Immun. 53, 135–140.
Tasara, T., Bielaszewska, M., Nitzsche, S., Karch, H., Zweifel, C., and Stephan, R. (2008). Activatable Shiga toxin 2d (Stx2d) in STEC strains isolated from cattle and sheep at slaughter. Vet. Microbiol. 131, 199–204. doi: 10.1016/j.vetmic.2008.03.001
Tesh, V. L., Burris, J. A., Owens, J. W., Gordon, V. M., Wadolkowski, E. A., O'Brien, A. D., et al. (1993). Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect. Immun. 61, 3392–3402.
Tostes, R., Goji, N., Amoako, K., Chui, L., Kastelic, J., DeVinney, R., et al. (2017). Subtyping Escherichia coli virulence genes isolated from feces of beef cattle and clinical cases in alberta. Foodborne Pathog. Dis. 14, 35–42. doi: 10.1089/fpd.2016.2199
Keywords: shiga toxin, subtypes, non-O157 E. coli, cattle, human
Citation: Shridhar PB, Siepker C, Noll LW, Shi X, Nagaraja TG and Bai J (2017) Shiga Toxin Subtypes of Non-O157 Escherichia coli Serogroups Isolated from Cattle Feces. Front. Cell. Infect. Microbiol. 7:121. doi: 10.3389/fcimb.2017.00121
Received: 09 February 2017; Accepted: 24 March 2017;
Published: 11 April 2017.
Edited by:
Alfredo G. Torres, University of Texas Medical Branch, USAReviewed by:
Analía Inés Etcheverría, National University of Central Buenos Aires, ArgentinaTim Reuter, Alberta Ministry of Agriculture and Forestry, Canada
Peter Feng, United States Food and Drug Administration, USA
Copyright © 2017 Shridhar, Siepker, Noll, Shi, Nagaraja and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianfa Bai, jbai@vet.k-state.edu
T. G. Nagaraja, tnagaraj@vet.k-state.edu
 Pragathi B. Shridhar1
Pragathi B. Shridhar1  T. G. Nagaraja
T. G. Nagaraja