Peptide Nucleic Acid Knockdown and Intra-host Cell Complementation of Ehrlichia Type IV Secretion System Effector
- Department of Veterinary Biosciences, Ohio State University, Columbus, OH, United States
Survival of Ehrlichia chaffeensis depends on obligatory intracellular infection. One of the barriers to E. chaffeensis research progress has been the inability, using conventional techniques, to generate knock-out mutants for genes essential for intracellular infection. This study examined the use of Peptide Nucleic Acids (PNAs) technology to interrupt type IV secretion system (T4SS) effector protein expression in E. chaffeensis followed by intracellular complementation of the effector to determine its requirement for infection. Successful E. chaffeensis infection depends on the E. chaffeensis-specific T4SS protein effector, ehrlichial translocated factor-1 (Etf-1), which induces Rab5-regulated autophagy to provide host cytosolic nutrients required for E. chaffeensis proliferation. Etf-1 is also imported by host cell mitochondria where it inhibits host cell apoptosis to prolong its infection. We designed a PNA specific to Etf-1 and showed that the PNA bound to the target region of single-stranded Etf-1 RNA using a competitive binding assay. Electroporation of E. chaffeensis with this PNA significantly reduced Etf-1 mRNA and protein, and the bacteria's ability to induce host cell autophagy and infect host cells. Etf-1 PNA-mediated inhibition of ehrlichial Etf-1 expression and E. chaffeensis infection could be intracellularly trans-complemented by ectopic expression of Etf-1-GFP in host cells. These data affirmed the critical role of bacterial T4SS effector in host cell autophagy and E. chaffeensis infection, and demonstrated the use of PNA to analyze the gene functions of obligate intracellular bacteria.
Introduction
Discovered in 1986 and designated as a nationally notifiable disease in 1998 by the CDC, human monocytic ehrlichiosis (HME) is one of the most prevalent, life-threatening, emerging tick-borne diseases in the US (Paddock and Childs, 2003; CDC, 2015). HME is caused by infection with Ehrlichia chaffeensis, an obligatory intracellular bacterium in the order Rickettsiales (Paddock and Childs, 2003; Rikihisa, 2015). E. chaffeensis replicates within human monocytes-macrophages, and causes severe flu-like symptoms accompanied by hematologic abnormalities and signs of hepatitis. Currently, the only choice of treatment is the broad-spectrum antibiotic doxycycline, which is only effective if initiated early because delayed therapy initiation, for example due to misdiagnosis, can lead to severe complications or death. The presence of underlying illness or injury, stress, immunosuppression, and/or coinfection with other tick-borne pathogens can lead to severe complications or death in 2–5% of infected individuals (Paddock and Childs, 2003). No vaccines exist for HME. Tick-borne diseases have risen dramatically in the past two decades, and continue to rise (Paddock and Yabsley, 2007), underscoring the importance of developing a novel therapeutic approach for infections with tick-borne intracellular bacteria.
Because E. chaffeensis has a small genome of 1.176 Mb and relatively few genes encoding proteins needed for biosynthesis and metabolism, it cannot survive outside of eukaryotic host cells and relies heavily on host-derived nutrients for its replication (Rikihisa, 2015). As E. chaffeensis cannot survive anywhere else, with adequate knowledge and tools, targeting specific E. chaffeensis genes that enable its intracellular survival and proliferation may be ideal for new anti-Ehrlichia therapeutic strategies. However, one of the major barriers to E. chaffeensis research progress has been the inability to generate E. chaffeensis knockout mutants for genes essential for obligatory intracellular infection, using conventional techniques. Thus, it has not been feasible to fulfill molecular Koch's postulates by studying phenotypes of knockout mutants and restoring lost functions by complementation.
E. chaffeensis encodes a type IV (type IVa, VirB/D) secretion system (T4SS) that mediates the transport of bacterial DNA and/or proteins, referred to as effectors/substrates, across the bacterial membrane into the eukaryotic cell to deregulate or modulate target cell functions for the benefit of the bacteria (Alvarez-Martinez and Christie, 2009; Rikihisa, 2015). T4SS machinery and effectors are major proteins produced by E. chaffeensis, suggesting they merit such extensive energy expenditures by the pathogen (Bao et al., 2009; Lin et al., 2011). E. chaffeensis translocated factor 1 (Etf-1) is the first experimentally proven T4SS effector in the genus Ehrlichia (Liu et al., 2012). Etf-1 is highly produced and secreted by E. chaffeensis in human monocytes (Lin et al., 2011; Liu et al., 2012); neutralization of secreted Etf-1 by delivering anti-Etf-1 IgG into the cytoplasm of host cells inhibited cellular infection by E. chaffeensis (Liu et al., 2012), whereas ectopic Etf-1 expression enhanced E. chaffeensis infection (Lin et al., 2016). Secreted Etf-1 has two important functions: (1) it localizes to mitochondria and blocks host cell apoptosis, and thereby allows sufficient time for E. chaffeensis replication (Liu et al., 2012); and (2) it is required, via a novel protein-protein interaction, for E. chaffeensis-induced Rab5-regulated autophagy to make host cytosolic nutrients available for E. chaffeensis (Lin et al., 2016). Because Etf-1 regulates host cell functions, Etf-1 influences the entire intracellular bacterial population.
Unlike some facultative intracellular bacteria including the well-studied Legionella pneumophila, to date, no T4SS system knockout mutants have been obtained in any member of the order Rickettsiales, and stable targeted mutagenesis has not been achieved in E. chaffeensis (Cheng et al., 2013). Despite the recent use of Himar1 transposon random insertion mutagenesis in E. chaffeensis (Cheng et al., 2013), genetic manipulation of E. chaffeensis must overcome many difficulties, e.g., (1) the lack of resistance markers except for a single antibiotic (spectinomycin/streptomycin; tetracycline cannot be used as it is the only clinically effective antibiotic) limits screening for E. chaffeensis transformants; (2) the very slow process for selecting mutants, i.e., can take weeks due to poor transformation efficiencies, viability, and slow mutant bacterial growth (Cheng et al., 2013); and (3) the lack of plasmids or phages capable of replicating in this group of bacteria (family Anaplasmataceae).
Peptide Nucleic Acids (PNAs) are single-stranded synthetic nucleic acids with a pseudopeptide backbone in place of the phosphodiester-linked sugar and phosphate found in traditional oligos. PNAs are chemically stable, resistant to enzymatic degradation, with low tissue toxicity (Good and Nielsen, 1998). PNA-DNA binding is stronger and the specificity is higher than DNA-DNA binding (Nielsen and Egholm, 1999). Anti-sense PNAs that disrupt transcription and translation have been developed for gene therapy, genetic disorder diagnostics, infection prophylaxis, and molecular tools for nucleic acid manipulations (Nielsen et al., 1991; Good and Nielsen, 1998; Janowski et al., 2005; Abushahba et al., 2016). The PNA approach is advantageous for working with E. chaffeensis because PNAs do not require mutation/selection of bacteria. Furthermore, partial reduction of expression can be achieved allowing study of essential genes, where investigations of a phenotype by a true knockout are not possible. This approach, however, has not been used for obligatory intracellular bacteria, except for Rickettsia montanensis and Rickettsia typhi (Pelc et al., 2015), and complementation assays have never been accomplished after PNA knock down. In this study, we designed anti-sense PNAs to disrupt the T4SS effector Etf-1, and determined the effectiveness of target gene knockdown, blockage of host cell autophagy induction, and E. chaffeensis infection in human cells, and whether Etf-1 inhibition could be trans-complemented within host cells.
Materials and Methods
Cultivation of E. chaffeensis and Host Cells
E. chaffeensis Arkansas (Dawson et al., 1991) was cultured in the human monocytic leukemia cell line THP-1 (ATCC, Manassas, VA) in RPMI 1640 medium (Corning Cellgro, Manassas, VA) supplemented with 5% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) and 2 mM L-glutamine (Invitrogen, Grand Island, NY) at 37°C in 5% CO2 and 95% air in a humidified atmosphere as previously described (Barnewall and Rikihisa, 1994). Cells were monitored for 2–3 days for infection using HEMA 3 stain (Thermo Fisher, Waltham, MA) on cytocentrifuged specimens (Cytospin™4 Cytocentrifuge, Thermo Fisher), and passaged/harvested when the percentage of infected cells reached >95% as previously described (Barnewall et al., 1997). The human kidney epithelial cell line HEK293 (ATCC) was cultured in Dulbecco's minimal essential medium (DMEM; Corning Cellgro) supplemented with 10% FBS and 2 mM L-glutamine as previously described (Miura et al., 2011).
PNA Synthesis, Biotin Labeling, and RNA-PNA Hybridization
Custom PNA oligomers were synthesized, and PNA quality and quantity were verified by HPLC and MS (PNABio, Newbury Park, CA). An antisense oligomer was designed to the start codon of Etf-1 (Figure 1). A scrambled PNA control (CTL PNA; 3′-CACATATCTCGG-5′) also was designed. To ensure the specificity of the PNA sequence, each sequence was blasted against E. chaffeensis Arkansas whole genome (Dunning Hotopp et al., 2006). PNA oligomers were biotinylated using EZ-Link Sulfo-NHS-Biotin according to the manufacturer's instructions (Thermo Fisher). Labeled PNA was purified using Pierce C18 Spin Columns (Thermo Fisher). Dot blots were used to confirm the successful biotinylation of oligomers as previously described (Pelc et al., 2015). The membrane was developed using Pierce Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher) according to the manufacturer's instructions.
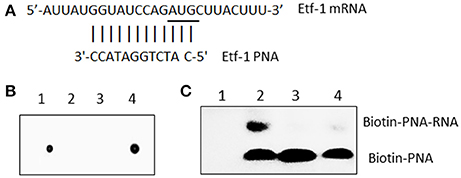
Figure 1. E. chaffeensis Etf-1 PNA bound to the target ssRNA. (A) Etf-1 PNA targeted the putative Shine-Dalgarno region and translational start site (underline). (B) Dot blot analysis of biotin-labeled Etf-1 and streptavidin-HRP-labeled CTL PNA. Dot 1–4: Biotin-CTL PNA, Unlabeled CTL PNA, Unlabeled Etf-1 PNA, and Biotin-Etf-1 PNA. (C) Electrophoresis migration shift assay of biotinylated Etf-1 PNA bound to ssRNA. Lane 1, no labeled PNA, lanes 2–4 labeled PNA incubated with 0, 10 ×, and 1 × non-labeled PNA, respectively.
To verify that Etf-1 PNA binds to the targeted region, the Etf-1 ssRNA molecule 5′-AUUAUGGUAUCCAGAUGCUUACUUU-3′ (bold nucleotides represent the RNA sequence corresponding to the designed PNA sequence, Figure 1) was synthesized by Sigma-Aldrich (St. Louis, MO). RNA-PNA hybridization was performed in a 15 μl reaction mixture with 2.0 μl (200 μM) ssRNA and 10 μM biotin-labeled Etf-1 PNA or 0, 10, and 100 μM of unlabeled PNA with 1 μl of RNaseOUT (Thermo Fisher) in 250 mM Tris buffer (pH 7.2) at room temperature for 30 min. The samples were run on a 2% agarose gel in TBE buffer (40 mM Tris, pH 8.3, 45 mM boric acid, and 1 mM EDTA) and transferred to an Amersham HyBond N+ membrane (GE Healthcare Life Sciences, Marlborough, MA) using a semi-dry blotting apparatus (WEP, Seattle, WA). Membranes were UV cross-linked in a Stratalinker® UV Crosslinker 1800 (Stratagene, San Diego, CA) and developed as above.
Transfection of Host Cell-Free E. chaffeensis
E. chaffeensis-infected THP-1 cells (~4 × 107 cells, >90% infected cells from two T75 flasks, ~100 bacteria/cell based on 16S rRNA qPCR) were harvested by centrifugation at 400 × g for 5 min. The pellet was resuspended in 20 ml of 300 mM sucrose and sonicated on ice twice at setting 2 for 5 s using a W-380 Sonicator (Heat Systems-Ultrasonics, Farmingdale, NY). Unbroken cells were removed by centrifugation at 1,000 × g for 5 min. The supernatant was collected after centrifugation at 1,700 × g for 5 min and passed through 2.7- and 5.0-μm GD/X nylon filters (Whatman, Florham Park, NJ) sequentially to remove cell debris, and finally centrifuged at 9,700 × g for 10 min as previously described (Liu et al., 2012). The bacterial pellet was resuspended in 200 μl of ice-cold 300 mM sucrose.
To deliver PNA, 3 μg of Etf-1 or CTL PNA in 10 μl nuclease-free water was mixed with 100 μl of host-cell free bacteria in 300 mM sucrose in a sterile 0.2 cm gap electroporation cuvette (Bio-Rad, Hercules, CA) on ice. E. chaffeensis bacteria were electroporated using a Gene PulserXcell Microbial System (Bio-Rad) by applying a ~9 ms pulse (2,000 V, 25 μF, 400 Ω). Immediately after electroporation, 400 μl of prewarmed RPMI with 5% FBS medium was added to the electroporated Ehrlichia. The PNA-Ehrlichia suspension was transferred to a T25 flask containing 5 × 105 THP-1 cells for internalization at 37°C for 90 min. The cells were washed to remove uninternalized bacteria, and continuously cultured in fresh medium. Ehrlichial and host mRNA and protein expression were measured respectively at 36 and 48 h post-transfection (pt) of bacteria [post-infection (pi)].
Reverse Transcription Quantitative PCR (qRT-PCR)
THP-1 cells infected with Etf-1 or CTL PNA-transfected E. chaffeensis were harvested at 36 h pi. Cells were washed once with phosphate buffer saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) and total cellular RNA was extracted from ~106 cells per sample using an RNeasy kit (Qiagen, Valencia, CA). RNA concentrations and quality were determined by NanoDrop (Thermo Fisher). Total RNA (2 μg) was reverse transcribed using a Maxima H Minus First Strand cDNA Synthesis Kit and random hexamers (Thermo Fisher). The qPCR reaction mixture (20 μl) included 2 μl cDNA (corresponding to 0.2–0.3 μg of total RNA), 0.25 μM each primer and 10 μl SYBR Green qPCR master mix (Thermo Fisher). PCR was performed in an Mx3000P instrument (Stratagene). Primer sequences are described elsewhere for Etf-1(Liu et al., 2012) and E. chaffeensis 16S rRNA, and human G3PDH (Miura et al., 2011).
Etf-1 Protein Expression Analysis
Host cell-free E. chaffeensis were transfected with CTL or Etf-1 PNA and used to infect THP-1 or HEK293 cells. The infected cells were harvested by centrifugation at 48 h pi. The cell pellet was washed with PBS and solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Proteins were separated by 10% SDS-PAGE, and transferred to a nitrocellulose membrane (Bio-Rad). Western blot analyses were performed using rabbit anti-Etf-1 (anti-ECH0825) antibody (Liu et al., 2012), rabbit anti-E. chaffeensis P28 major outer membrane protein (Ohashi et al., 1998) and rabbit anti-human LC3 (Cell Signaling Technology, Danvers, MA) at 1:1,000. To normalize protein loading, the membrane was washed with Restore ™ Western Blot Stripping buffer (Thermo Fisher) and probed with mouse monoclonal anti-human tubulin (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:1,000. Primary antibodies were detected with horseradish peroxidase-labeled goat anti-rabbit IgG/goat anti-mouse IgG (KPL, Gaithersburg, MD). Blots were developed with Pierce ™ ECL Western Blotting Substrate (Thermo Fisher). Chemiluminescent bands were visualized using an LAS3000 image documentation system (FUJIFILM Medical Systems USA, Stamford, CT) and band intensities were determined by densitometry using Multi Gauge software (FUJIFILM).
Trans-Complementation of E. chaffeensis Transfected with Etf-1 PNA-Transfected and Etf-1-Expressing Human Cells
HEK293 cells at 1 × 106 cells/100 μl were mixed with 5 μg (1 μg/μl) of pEGFPN1 plasmid (Clontech, Mountain View, CA) or pEGFPN1-Etf-1 (pEGFN1-ECH0825 codon-optimized) plasmid (Liu et al., 2012) in a 0.2 cm electroporation cuvette (Bio-Rad). Electroporation was performed using a Gene PulserXcell Microbial System (Bio-Rad) by applying a ~50 ms pulse (100 V, 1,000 μF, ∞ Ω) as described earlier (Niu et al., 2008; Liu et al., 2012). Transfected host cells were grown in DMEM medium (Corning Cellgro) supplemented with 10% FBS and 2 mM L-glutamine in 6-well plate at 1 × 106 cells/well at 37°C in 5% CO2 and 95% air in a humidified atmosphere for 24 h. Then host cell-free E. chaffeensis transfected with 3 μg of CTL or Etf-1 PNA as described above was added to HEK293 cells that were pre-transfected with pEGFP/pEtf-1-EGFP at approximate MOI of 100:1. The MOI was estimated as follows: GFP- or Etf-1-GFP-transfected uninfected HEK293 cells were cultured at 1 × 106 cells/well for 24 h and the host cell-free bacteria were added; at the time of bacterial addition, number of HEK293 cells were ~2 × 106 cells/well. Host cell-free ehrlichia derived from 4 × 107 infected cells (4 × 109 bacteria) were divided into 4 groups = 1 × 109 bacteria/group; however, after sonication to liberate bacteria, filtration, and electroporation, the rate of final recovery of viable bacteria was ~20%. Infected cells were harvested at 48 h pi (48 h CTL/Etf-1 PNA pt of ehrlichia, and 72 h Etf-1-GFP/GFP pt of HEK293 cells). RNA extraction, cDNA synthesis, qPCR, and western blot analysis were performed as described above.
Statistical Analyses
Experiments were repeated at least three times. Comparison of each treatment was performed relative to samples with CTL PNA. Statistical analysis was performed by 2-tailed Student's t-test and P < 0.05 was considered significant. For experiments involving more than two groups, analysis of variance was performed and P < 0.05 was considered significant. All statistical analyses were performed using Microsoft Excel 2010.
Results
E. chaffeensis Etf-1 PNA Specifically Hybridized with Its Target Sequence
Although, the consensus Shine-Dalgarno (SD) sequence is GGAGG, most E. coli SD sequences are slight variations of GGAGG (Schurr et al., 1993). The optimal spacing between SD sequence and AUG translational start codon is about 8 to 10 bases for E. coli genes (Chen et al., 1994). Ma et al. (2002) reported only 18.6% of Rickettsia prowazeki genes has the SD consensus sequence (GGAGG). Similarly, we found most E. chaffeensis genes lack the typical SD sequence. Therefore, we assumed 8 to 10 bases upstream of AUG is the putative SD region and designed anti-sense PNA complementary to this region of E. chaffeensis Etf-1, and confirmed the PNA sequence specificity by BLASTn analysis against the entire genome of E. chaffeensis Arkansas (Dunning Hotopp et al., 2006) (Figure 1A). Scrambled CTL PNA was designed by confirming the absence of annealing sequences by BLASTn analysis against the entire genome of E. chaffeensis. To verify that Etf-1-PNA could specifically hybridize with the respective target sequence, the PNA was biotinylated, and biotinylation of PNA was confirmed via dot blot assay (Figure 1B). To demonstrate specific hybridization, equal amounts of the target ssRNA were incubated with biotinylated Etf-1 PNA or different ratios of labeled to non-labeled Etf-1 PNA, and the mixture was analyzed by electrophoresis migration shift assay. Biotinylated Etf-1 PNA showed delayed migration in the presence of the target ssRNA compared to the absence of ssRNA, which indicated hybridization between the target ssRNA and the biotinylated PNA; whereas, without biotinylated PNA, no signal was detected (Figure 1C). Hybridized biotinylated PNA was decreased in a dose-dependent manner in the presence of increasing concentrations of unlabeled Etf-1 PNA, which indicated competitive inhibition of binding (Figure 1C). The formation of ssRNA-PNA pairing established the ability of our Etf-1 PNA to hybridize with the specific nucleotide target sequence.
PNA Treatment Impaired Ehrlichial Etf-1 Expression and Infection
Knowing that the PNA was specific for its target gene, we evaluated ability of Etf-1 PNA to suppress Etf-1 mRNA expression in ehrlichiae and inhibit its infection of human monocytes. E. chaffeensis was purified to homogeneity, electroporated with non-targeting CTL or Etf-1 PNA, and incubated with the human monocyte cell line THP-1. Etf-1 mRNA and E. chaffeensis 16S rRNA (to measure live bacteria) levels were analyzed by qRT-PCR. Human GAPDH mRNA levels (to measure live host cells) were also analyzed to normalize the sample. Etf-1 mRNA/16S rRNA in E. chaffeensis and 16S rRNA/human GAPDH mRNA in host cells were significantly reduced at 36 h pt (pi) compared to CTL PNA-treated E. chaffeensis (Figures 2A,B). Western blot analysis revealed profound reduction of E. chaffeensis P28 major outer membrane proteins (to measure bacteria at protein level; encoded by a multigene family and major two bands in human cells) (Ohashi et al., 1998) and Etf-1/human tubulin (to normalize host cells at protein level) in Etf-1 PNA-treated E. chaffeensis at 48 h pt (pi) compared with CTL PNA-treated E. chaffeensis in THP-1 cells (Figure 2C). The result indicated that Etf-1 mRNA and protein expression was effectively knocked down with the Etf-1 PNA, Etf-1 was required for E. chaffeensis infection, and thus, the PNA approach was useful for examining the requirement of target ehrlichial genes in infection.
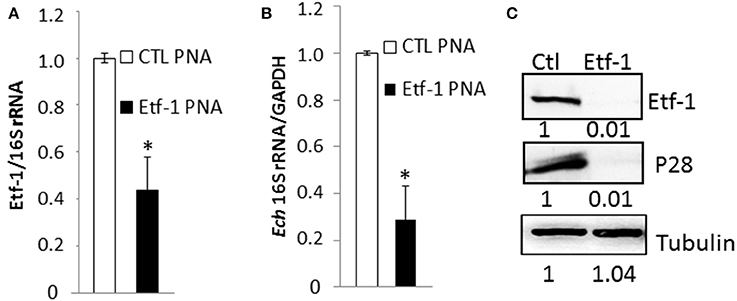
Figure 2. E. chaffeensis Etf-1 PNA impaired Etf-1 expression and ehrlichial infection. (A,B) Etf-1 PNA inhibited E. chaffeensis Etf-1 expression (A) and infection (B). RNA samples prepared from THP-1 cells incubated with Etf-1 or CTL PNA-transfected E. chaffeensis were harvested at 36 h pi and subjected to qRT-PCR analysis. The values reflect bacterial 16S rRNA normalized against human GAPDH mRNA, relative to the amount determined for the CTL PNA treatment. Data indicate the mean ± standard deviation from three independent experiments performed in triplicate. *Significantly different (P < 0.05; Student's t-test) compared to the value of the CTL PNA. (C) Western blot analysis of Etf-1 and E. chaffeensis outer membrane protein P28 normalized against human tubulin in THP-1 cells incubated with Etf-1 and CTL PNA-transfected E. chaffeensis at 48 h pi. The ratios of Etf-1: tubulin and p28:tubulin were quantified and compared to CTL PNA, which was set to 1.
Reduced Ehrlichial Growth by Etf-1 PNA Knockdown Was Complemented with Ectopically Expressed Etf-1-GFP
To rule out the possibility that the observed phenotypes of E. chaffeensis were caused by disruption of off-target sites, we performed an intracellular rescue assay by ectopic expression of Etf-1-GFP in host cells. To negate the effect of foreign plasmid, as control, host cells were transfected with GFP plasmid. Because THP-1 cells are difficult to transfect with plasmids, we used HEK293 cells that can be easily transfected and infected with E. chaffeensis (Miura et al., 2011). Our previous studies showed that E. chaffeensis behaved similarly in human monocytes and HEK293 cells (Liu et al., 2012; Lin et al., 2016). HEK293 cells were transfected with Etf-1-GFP or GFP plasmid. At 24 h pt, i.e., the time point when Etf-1-GFP expression and Etf-1-GFP-induced autophagy is clearly visible in the host cells (Lin et al., 2016), Etf-1 PNA or CTL PNA-treated E. chaffeensis were added to the pre-transfected HEK293 cells and cells were harvested at 48 h pi. The results showed that ehrlichial Etf-1 mRNA/human GAPDH mRNA and E. chaffeensis 16S rRNA/human GAPDH mRNA were significantly reduced in Etf-1 PNA than in CTL PNA-treated E. chaffeensis in HEK293 cells (Figure 3A). As we previously reported with non-PNA-treated E. chaffeensis (Lin et al., 2016), based on the levels of E. chaffeensis 16S rRNA/human GAPDH mRNA, Etf-1-GFP-transfection of HEK293 cells enhanced infection of CTL PNA-treated E. chaffeensis compared to the GFP-transfection CTL (Figure 3A). Etf-1-GFP-transfection of HEK293 cells enhanced infection by Etf-1 PNA-treated E. chaffeensis compared to the GFP-transfection CTL as well (Figure 3A). At protein level, western blot analysis revealed significant reduction of ehrlichial Etf-1 and reduction of E. chaffeensis P28 major outer membrane proteins with Etf-1 PNA-treated E. chaffeensis at 48 h pi compared with CTL PNA-treated E. chaffeensis in HEK293 cells (GFP-transfection CTL in Figures 3B,C). Etf-1-GFP transfection of HEK 293 cells resulted in a significant increase in ehrlichial Etf-1 and P28 protein levels compared to GFP-transfection CTL, in both Etf-1 PNA and CTL PNA-treated E. chaffeensis (Figures 3B,C). Taken together, these data indicate that exogenous Etf-1-GFP expression intracellularly complemented Etf-1 PNA-mediated inhibition of ehrlichial Etf-1 expression and E. chaffeensis infection.
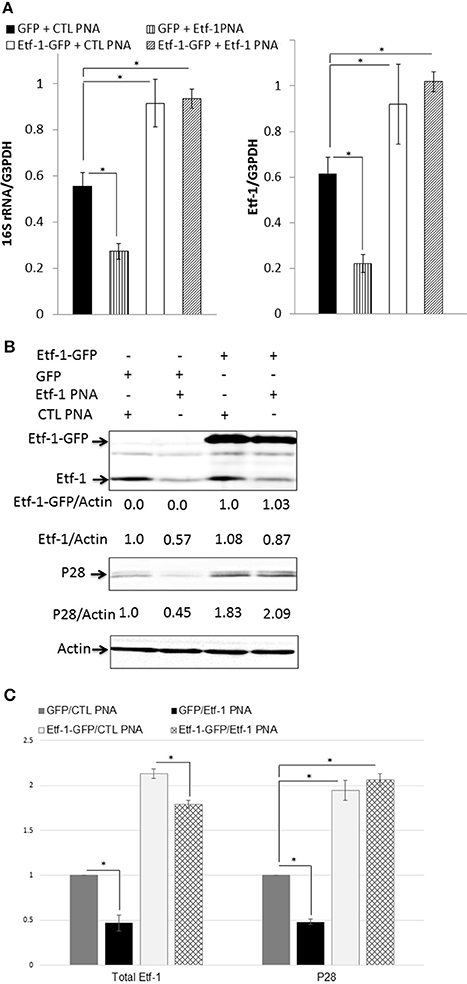
Figure 3. Intracellular complementation of Etf-1 PNA-reduced ehrlichial infection by ectopic expression of Etf-1-GFP. HEK293 cells expressing Etf-1 complemented the Etf-1 PNA knockdown and rescued E. chaffeensis infection. RNA samples prepared from HEK293 cells expressing Etf-1-GFP or GFP incubated with Etf-1 or CTL PNA-transfected E. chaffeensis were harvested at 48 h pi and subjected to qRT-PCR analysis (A) and protein analysis (B,C). The values reflect bacterial 16S rRNA and total Etf-1 normalized against human GAPDH mRNA, relative to the amount determined for the Etf-1 PNA treatment of HEK293 cells expressing Etf-1-GFP. Data indicate the mean ± standard deviation from three independent experiments performed in triplicate. *Significantly different (P < 0.05; analysis of variance) (A). Western blot analysis using anti-P28, actin, and Etf-1. The values under the bands show relative ratios of band intensities vs. actin, with the ratios of those from GFP plasmid and/or CTL PNA set to 1. (B) Results are presented as means ± standard deviations from three independent experiments. The asterisk indicates a significant difference compared with CTLs by analysis of variance (P < 0.01).
Etf-1 PNA Knockdown Impairs E. chaffeensis-Induced Host Cell Autophagy
Autophagy is a catabolic process for the autophagosome-lysosomal degradation of bulk cytoplasmic contents (Yorimitsu and Klionsky, 2005; Klionsky, 2007). Autophagy is an important innate immune mechanism to clear intracellular infection (Deretic, 2011; Levine et al., 2011). However, intracellular E. chaffeensis is not only resistant to host cell autophagy, but rather ehrlichial infection is enhanced by host cell autophagy, and ehrlichial Etf-1 secreted into host cell cytoplasm or ectopically expressed Etf-1-GFP induces host cell autophagy and promote ehrlichial growth (Lin et al., 2016). Mammalian light Chain 3 (LC3, MAP1LC3, the homolog of yeast ATG8) undergoes post-translational modifications during autophagy: the cytosolic form LC3-I is converted to LC3-II through cleavage and lipidation that allows LC3 to become associated with autophagic vesicles (Kabeya et al., 2000). The presence of LC3 in autophagosomes and the conversion of LC3-I to the lower migrating form LC3-II have been used as indicators of autophagy (Klionsky et al., 2008). To investigate whether Etf-1 PNA treatment of E. chaffeensis inhibited ehrlichial Etf-1-induced cellular autophagy, HEK293 cells were infected with Etf-1 PNA or CTL PNA-treated E. chaffeensis and the cells were harvested at 48 h pi. Western blot analysis revealed significant reduction of ehrlichial Etf-1 protein expression, and decrease in LC3-II levels with Etf-1 PNA-treated E. chaffeensis at 48 h pi compared with CTL PNA-treated E. chaffeensis in HEK293 cells (Figures 4A,B). Taken together, these data indicate that Etf-1 PNA treatment reduces ehrlichial Etf-1 levels and consequently diminishes Etf-1-mediated host cell autophagy induced by E. chaffeensis infection.
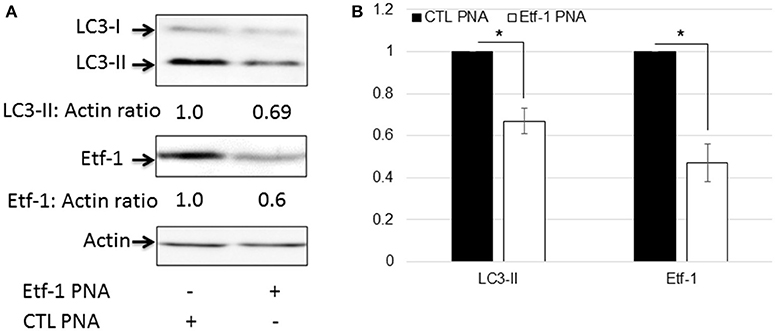
Figure 4. Etf-1 PNA knockdown impedes E. chaffeensis-induced host cell autophagy. HEK293 cells were infected with Etf-1 or CTL PNA-transfected E. chaffeensis and harvested at 48 h pi and subjected to western blot analysis using anti-LC3, actin, and Etf-1 (A,B). The values under the bands show relative ratios of band intensities vs. actin, with the ratios of those from CTL PNA set to 1. (B) Results are presented as means ± standard deviations from three independent experiments. The asterisk indicates a significant difference compared with CTLs by Student's t-test (P < 0.01).
Discussion
Significant progresses have been made in generating mutants in rickettsial organisms including Rickettsia, Anaplasma, and Ehrlichia by random transposon mutagenesis and by transformation with rickettsial or other plasmids, and these procedures generated stable florescent bacteria that allows to track bacterial growth and motility, and study in vitro and in vivo functions of knockout mutants isolated using the cell culture system (Felsheim et al., 2006, 2010; Kleba et al., 2010; Noh et al., 2011; Chen et al., 2012; Cheng et al., 2013; Crosby et al., 2014; Oliver et al., 2014; Oliva Chavez et al., 2015; Hauptmann et al., 2017). However, so far stable targeted mutagenesis including conditional mutagenesis that allows functional studies have not been achieved. This also precludes complementation studies using the knockout mutants. One of unique features of obligatory intracellular bacteria is that they have evolved genes essential for survival and proliferation within the hostile eukaryotic cell environment. However, unlike facultative intracellular bacteria, knockout mutants for genes required for intracellular survival cannot be obtained for obligatory intracellular bacteria. Furthermore, obligatory intracellular bacteria have evolved unique genes (e.g., ~50% of E. chaffeensis ORFs encode “hypothetical proteins,” which have no homology to any other known proteins (Dunning Hotopp et al., 2006; Rikihisa, 2015), which preclude studies based on conserved motifs and orthologous genes in other bacteria. Indeed, many of novel Ehrlichia and Anaplasma genes so far discovered as critical for infection encode previously “hypothetical proteins” (Rikihisa, 2010, 2011, 2015). In addition, surrogate systems in model bacteria such as E. coli do not provide needed key information on obligatory intracellular survival/infection. When E. chaffeensis was treated with PNA specific to Etf-1, host cell autophagy induction and E. chaffeensis infection were significantly reduced, which validated the critical role of the T4SS effector for autophagy induction and E. chaffeensis infection. Our study demonstrated the utility of PNA delivery as an effective method to study E. chaffeensis gene function, which is more broadly applicable for obligatory intracellular bacteria to overcome limited availability of genetictools.
Off target effects are a concern when working with antisense oligos to interrupt gene function. However, advantages of PNAs are that (1) both affinity and specificity of a PNA/DNA duplex is higher than a DNA/DNA duplex (Nielsen and Egholm, 1999), and (2) a mismatch in a PNA/DNA duplex is generally more destabilizing than a mismatch in a DNA/DNA duplex (Nielsen and Egholm, 1999), thus less likely causes knockdown of the mismatched gene. When designing PNA oligos, we blasted the PNA sequences against the entire E. chaffeensis genome sequence to ensure alignment with only our target sequence: Etf-1 PNA designed complementary to the putative SD region. To negate the non-specific effect of PNA transfection in Ehrlichia, all the experiments were performed in parallel with scrambled CTL PNA. Our competitive hybridization assay results with ssRNA, cell culture infections, and mRNA and protein suppression experiments pointed to the specificity of the PNA approach for the target. Thus, appropriately designed PNA, well-controlled experiments, and objective evaluation as used in the present study (qRT-PCR and densitometry analysis of protein expression) would be expected to facilitate the discovery of unique genes important for obligatory intracellular bacteria. Our study showed that, for secreted target proteins such as Etf-1, deficiency can be complemented by ectopic expression of Etf-1 in the host cells, offering a new approach to affirm the specificity of PNA knockdown. One of the limitations of the PNA approach is the cost of custom PNA synthesis. Although, almost all published papers on PNA use a cell-permeable peptide (CPP) linked to the N-terminus of the PNA, for our study without CPP, PNA was effectively delivered into infectious live Ehrlichia, which reduced the cost of PNA synthesis.
The field needs additional tools of genetic manipulation to better scrutinize the lifestyle and virulence of obligate intracellular bacteria, and allow molecular Koch's postulates to be fulfilled. Our ability to inhibit protein expression in pathogenic E. chaffeensis indicates the broad spectrum of uses for this technology. PNA offers an advantage in that reduction of protein expression is achieved in 2 days, no antibiotic selection is required, and partial reduction can be achieved allowing study of essential genes, where investigations of a phenotype imposed by a true knockout would be impossible due to bacterial death. Given that anti-sense oligos have been developed as FDA-approved drugs for certain diseases (Jiang, 2013), by modifying PNAs to improve their targeting of specific mammalian cell types and intracellular bacteria, it is reasonable to envision enhanced bioavailability in vivo. The work presented herein represents a necessary first step in the path of developing PNAs as a drug to effectively block E. chaffeensis infection.
Author Contributions
Conceived and designed the experiments: YR. Performed experiments and analyzed data: PS, OT, and YR. Wrote the paper: YR and PS.
Funding
This work was in part supported by a National Institutes of Health grant, R01AI054476.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate Drs. Mingqun Lin and Qingming Xiong for their help in various experimental procedures.
References
Abushahba, M. F., Mohammad, H., Thangamani, S., Hussein, A. A., and Seleem, M. N. (2016). Impact of different cell penetrating peptides on the efficacy of antisense therapeutics for targeting intracellular pathogens. Sci. Rep. 6:20832. doi: 10.1038/srep20832
Alvarez-Martinez, C. E., and Christie, P. J. (2009). Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73, 775–808. doi: 10.1128/MMBR.00023-09
Bao, W., Kumagai, Y., Niu, H., Yamaguchi, M., Miura, K., and Rikihisa, Y. (2009). Four VirB6 paralogs and VirB9 are expressed and interact in Ehrlichia chaffeensis-containing vacuoles. J. Bacteriol. 191, 278–286. doi: 10.1128/JB.01031-08
Barnewall, R. E., and Rikihisa, Y. (1994). Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron-transferrin. Infect. Immun. 62, 4804–4810.
Barnewall, R. E., Rikihisa, Y., and Lee, E. H. (1997). Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect. Immun. 65, 1455–1461.
CDC (2015). RE: Table II Provisonal Cases of Selected Notifiable Diseases. Morbidity and Mortality Weekly Report 63: NO-737. Atlanta, GA: CDC.
Chen, G., Severo, M. S., Sakhon, O. S., Choy, A., Herron, M. J., Felsheim, R. F., et al. (2012). Anaplasma phagocytophilum dihydrolipoamide dehydrogenase 1 affects host-derived immunopathology during microbial colonization. Infect. Immun. 80, 3194–3205. doi: 10.1128/IAI.00532-12
Chen, H., Bjerknes, M., Kumar, R., and Jay, E. (1994). Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 22, 4953–4957. doi: 10.1093/nar/22.23.4953
Cheng, C., Nair, A. D., Indukuri, V. V., Gong, S., Felsheim, R. F., Jaworski, D., et al. (2013). Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog. 9:e1003171. doi: 10.1371/journal.ppat.1003171
Crosby, F. L., Wamsley, H. L., Pate, M. G., Lundgren, A. M., Noh, S. M., Munderloh, U. G., et al. (2014). Knockout of an outer membrane protein operon of Anaplasma marginale by transposon mutagenesis. BMC Genomics 15:278. doi: 10.1186/1471-2164-15-278
Dawson, J. E., Anderson, B. E., Fishbein, D. B., Sanchez, J. L., Goldsmith, C. S., Wilson, K. H., et al. (1991). Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29, 2741–2745.
Deretic, V. (2011). Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240, 92–104. doi: 10.1111/j.1600-065X.2010.00995.x
Dunning Hotopp, J. C., Lin, M., Madupu, R., Crabtree, J., Angiuoli, S. V., Eisen, J., et al. (2006). Comparative genomics of emerging human Ehrlichiosis agents. PLoS Genet. 2:e21. doi: 10.1371/journal.pgen.0020021
Felsheim, R. F., Chavez, A. S., Palmer, G. H., Crosby, L., Barbet, A. F., Kurtti, T. J., et al. (2010). Transformation of Anaplasma marginale. Vet. Parasitol. 167, 167–174. doi: 10.1016/j.vetpar.2009.09.018
Felsheim, R. F., Herron, M. J., Nelson, C. M., Burkhardt, N. Y., Barbet, A. F., Kurtti, T. J., et al. (2006). Transformation of Anaplasma phagocytophilum. BMC Biotechnol. 6:42. doi: 10.1186/1472-6750-6-42
Good, L., and Nielsen, P. E. (1998). Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 16, 355–358. doi: 10.1038/nbt0498-355
Hauptmann, M., Burkhardt, N., Munderloh, U., Kuehl, S., Richardt, U., Krasemann, S., et al. (2017). GFPuv-expressing recombinant Rickettsia typhi: a useful tool for the study of pathogenesis and CD8+ T cell immunology in Rickettsia typhi infection. Infect. Immun. 85:e00156–17. doi: 10.1128/IAI.00156-17
Janowski, B. A., Kaihatsu, K., Huffman, K. E., Schwartz, J. C., Ram, R., Hardy, D., et al. (2005). Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nat. Chem. Biol. 1, 210–215. doi: 10.1038/nchembio724
Jiang, K. (2013). Biotech comes to its 'antisenses' after hard-won drug approval. Nat. Med. 19:252. doi: 10.1038/nm0313-252
Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., et al. (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728. doi: 10.1093/emboj/19.21.5720
Kleba, B., Clark, T. R., Lutter, E. I., Ellison, D. W., and Hackstadt, T. (2010). Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect. Immun. 78, 2240–2247. doi: 10.1128/IAI.00100-10
Klionsky, D. J. (2007). Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937. doi: 10.1038/nrm2245
Klionsky, D. J., Abeliovich, H., Agostinis, P., Agrawal, D. K., Aliev, G., Askew, D. S., et al. (2008). Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175. doi: 10.4161/auto.5338
Levine, B., Mizushima, N., and Virgin, H. W. (2011). Autophagy in immunity and inflammation. Nature 469, 323–335. doi: 10.1038/nature09782
Lin, M., Kikuchi, T., Brewer, H. M., Norbeck, A. D., and Rikihisa, Y. (2011). Global proteomic analysis of two tick-borne emerging zoonotic agents: Anaplasma phagocytophilum and Ehrlichia chaffeensis. Front. Microbiol. 2:24. doi: 10.3389/fmicb.2011.00024
Lin, M., Liu, H., Xiong, Q., Niu, H., Cheng, Z., Yamamoto, A., et al. (2016). Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy 12, 2145–2166. doi: 10.1080/15548627.2016.1217369
Liu, H., Bao, W., Lin, M., Niu, H., and Rikihisa, Y. (2012). Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell. Microbiol. 14, 1037–1050. doi: 10.1111/j.1462-5822.2012.01775.x
Ma, J., Campbell, A., and Karlin, S. (2002). Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184, 5733–5745. doi: 10.1128/JB.184.20.5733-5745.2002
Miura, K., Matsuo, J., Rahman, M. A., Kumagai, Y., Li, X., and Rikihisa, Y. (2011). Ehrlichia chaffeensis induces monocyte inflammatory responses through MyD88, ERK, and NF-kappaB but Not through TRIF, interleukin-1 Receptor 1 (IL-1R1)/IL-18R1, or toll-like receptors. Infect. Immun. 79, 4947–4956. doi: 10.1128/IAI.05640-11
Nielsen, P. E., and Egholm, M. (1999). An introduction to peptide nucleic acid. Curr. Issues Mol. Biol. 1, 89–104.
Nielsen, P. E., Egholm, M., Berg, R. H., and Buchardt, O. (1991). Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254, 1497–1500. doi: 10.1126/science.1962210
Niu, H., Yamaguchi, M., and Rikihisa, Y. (2008). Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell. Microbiol. 10, 593–605. doi: 10.1111/j.1462-5822.2007.01068.x
Noh, S. M., Ueti, M. W., Palmer, G. H., Munderloh, U. G., Felsheim, R. F., and Brayton, K. A. (2011). Stability and tick transmission phenotype of gfp-transformed Anaplasma marginale through a complete in vivo infection cycle. Appl. Environ. Microbiol. 77, 330–334. doi: 10.1128/AEM.02096-10
Ohashi, N., Zhi, N., Zhang, Y., and Rikihisa, Y. (1998). Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect. Immun. 66, 132–139.
Oliva Chavez, A. S., Fairman, J. W., Felsheim, R. F., Nelson, C. M., Herron, M. J., Higgins, L., et al. (2015). An O-Methyltransferase is required for infection of tick cells by Anaplasma phagocytophilum. PLoS Pathog. 11:e1005248. doi: 10.1371/journal.ppat.1005248
Oliver, J. D., Burkhardt, N. Y., Felsheim, R. F., Kurtti, T. J., and Munderloh, U. G. (2014). Motility characteristics are altered for Rickettsia bellii transformed to overexpress a heterologous rickA gene. Appl. Environ. Microbiol. 80, 1170–1176. doi: 10.1128/AEM.03352-13
Paddock, C. D., and Childs, J. E. (2003). Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16, 37–64. doi: 10.1128/CMR.16.1.37-64.2003
Paddock, C. D., and Yabsley, M. J. (2007). Ecological havoc, the rise of white-tailed deer, and the emergence of Amblyomma americanum-associated zoonoses in the United States. Curr. Top. Microbiol. Immunol. 315, 289–324. doi: 10.1007/978-3-540-70962-6_12
Pelc, R. S., McClure, J. C., Kaur, S. J., Sears, K. T., Rahman, M. S., and Ceraul, S. M. (2015). Disrupting protein expression with peptide nucleic acids reduces infection by obligate intracellular Rickettsia. PLoS ONE 10:e0119283. doi: 10.1371/journal.pone.0119283
Rikihisa, Y. (2010). Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat. Rev. Microbiol. 8, 328–339. doi: 10.1038/nrmicro2318
Rikihisa, Y. (2011). Mechanisms of obligatory intracellular infection with anaplasma phagocytophilum. Clin. Microbiol. Rev. 24, 469–489. doi: 10.1128/CMR.00064-10
Rikihisa, Y. (2015). Molecular pathogenesis of Ehrlichia chaffeensis infection. Annu. Rev. Microbiol. 69, 283–304. doi: 10.1146/annurev-micro-091014-104411
Schurr, T., Nadir, E., and Margalit, H. (1993). Identification and characterization of E.coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 21, 4019–4023. doi: 10.1093/nar/21.17.4019
Keywords: PNA, Ehrlichia chaffeensis, type IV secretion, complementation, autophagy, effector, Etf-1
Citation: Sharma P, Teymournejad O and Rikihisa Y (2017) Peptide Nucleic Acid Knockdown and Intra-host Cell Complementation of Ehrlichia Type IV Secretion System Effector. Front. Cell. Infect. Microbiol. 7:228. doi: 10.3389/fcimb.2017.00228
Received: 25 March 2017; Accepted: 17 May 2017;
Published: 07 June 2017.
Edited by:
Jean Celli, Washington State University, United StatesReviewed by:
Joao Pedra, University of Maryland, United StatesBurt Anderson, University of South Florida, United States
Jere W. McBride, University of Texas Medical Branch, United States
Jason A. Carlyon, Virginia Commonwealth University School of Medicine, United States
Copyright © 2017 Sharma, Teymournejad and Rikihisa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasuko Rikihisa, rikihisa.1@osu.edu
 Pratibha Sharma
Pratibha Sharma Omid Teymournejad
Omid Teymournejad Yasuko Rikihisa
Yasuko Rikihisa