Isolation of F. novicida-Containing Phagosome from Infected Human Monocyte Derived Macrophages
- 1Department of Microbiology and Parasitology, Faculty of Medicine, University of Rijeka, Rijeka, Croatia
- 2Department of Microbiology, Institut für Mikrobiologie, Technische Universität Braunschweig and Helmholtz Center for Infection Research, Braunschweig, Germany
- 3Department of Biotechnology, University of Rijeka, Rijeka, Croatia
- 4Department of Microbiology and Immunology and Center for Predictive Medicine, Louisville, KY, United States
Francisella is a gram-negative bacterial pathogen, which causes tularemia in humans and animals. A crucial step of Francisella infection is its invasion of macrophage cells. Biogenesis of the Francisella-containing phagosome (FCP) is arrested for ~15 min at the endosomal stage, followed by gradual bacterial escape into the cytosol, where the microbe proliferates. The crucial step in pathogenesis of tularemia is short and transient presence of the bacterium within phagosome. Isolation of FCPs for further studies has been challenging due to the short period of time of bacterial residence in it and the characteristics of the FCP. Here, we will for the first time present the method for isolation of the FCPs from infected human monocytes-derived macrophages (hMDMs). For elimination of lysosomal compartment these organelles were pre-loaded with dextran coated colloidal iron particles prior infection and eliminated by magnetic separation of the post-nuclear supernatant (PNS). We encountered the challenge that mitochondria has similar density to the FCP. To separate the FCP in the PNS from mitochondria, we utilized iodophenylnitrophenyltetrazolium, which is converted by the mitochondrial succinate dehydrogenase into formazan, leading to increased density of the mitochondria and allowing separation by the discontinuous sucrose density gradient ultracentrifugation. The purity of the FCP preparation and its acquisition of early endosomal markers was confirmed by Western blots, confocal and transmission electron microscopy. Our strategy to isolate highly pure FCPs from macrophages should facilitate studies on the FCP and its biogenesis.
Introduction
Intracellular bacteria invade eukaryotic cells, followed by subversion of endocytic pathway, which results in formation of membrane-bound phagosomes. They are cable to modulate the membrane protein and lipid composition of phagosomes. This modulation is crucial for bacterial survival within the host cell because it either promotes the establishment of an intact replicative niche or allow the pathogen to escape to replication-permissive cytosol. Many intracellular bacterial pathogens have unique life cycles. Cytosolic bacteria, like Shigella (Ray et al., 2010) and Listeria (Camejo et al., 2011) modulate the endosomal-lysosomal membrane-bound compartments and escape into the cytosol, which provides environment rich in nutrients. Intracellular bacteria, Salmonella (Steele-Mortimer, 2008; Malik-Kale et al., 2011), Legionella (Kagan and Roy, 2002; Shin and Roy, 2008; Isberg et al., 2009), and Mycobacterium spp. (Vergne et al., 2004) are intracellular bacterial pathogens that reside and replicate within host endomembrane system. These bacteria overcome the stressful conditions in membrane-bound vacuoles.
Francisella tularensis is a gram-negative, highly infectious bacterium. The bacterium causes the zoonotic disease tularemia. F. tularensis type A is a dangerous pathogen that constantly raises attention due to potential use as biological weapon. Interestingly, Francisella novicida shares many similarities to type A strain due to genome sequence, intracellular life cycle and infectivity.
F. tularensis can invade and multiply within a range of cell types (Buddingh and Womack, 1941; Shepard, 1959; Anthony L. S. et al., 1991; Ben Nasr et al., 2006; Lindemann et al., 2007), but in vivo its primary target are macrophages (Fortier et al., 1994). Within mammalian and arthropod-derived cells, Francisella resides in acidic vacuole prior to escape to the cytosol, where it replicates (Santic et al., 2009; Llewellyn et al., 2011). In contrast, within amoeba cells bacterium resides and replicates within non-acidified, membrane-bound vacuoles (Lauriano et al., 2004; Santic et al., 2008).
After entry, it is enclosed within a unique compartment. Intracellular proliferation is essential for Francisella virulence, and a lot of effort has been made on understanding of specific steps in the intracellular cycle of this bacterium. Francisella survival and proliferation strategies rely on entering in the initial phagosome along the endocytic pathway and physical escape to the cell cytosol, making this bacterium a typical cytosol-dwelling pathogen (Celli and Zahrt, 2013). Despite the fact that Francisella replicates in the cytosol of infected cells, short presence of the Francisella in the phagosome is necessary for productive multiplication.
Macrophage infection by Francisella begins with initial bacterial recognition at the cell membrane (Clemens et al., 2005). Francisella enters into macrophages by looping phagocytosis through cholesterol-rich membrane domains called “lipid rafts” with caveolin-1 (Clemens et al., 2004; Tamilselvam and Daefler, 2008; Moreau and Mann, 2013). Following uptake, Francisella resides within an initial vacuolar compartment, the Francisella-containing phagosome (FCP). Lipid raft-associated components are incorporated into the FCP during entry and the initial phase of intracellular infection of host cells (Tamilselvam and Daefler, 2008). Cholesterol, as a key structural and regulatory element for the integrity of lipid rafts, has an important role in Francisella internalization into macrophages (Tamilselvam and Daefler, 2008). FCP matures to an early endosome state regulated by Rab5, a protein that is critical for endosome-phagosome tethering and fusion (Alvarez-Dominguez et al., 1996; Jahraus et al., 1998; Duclos et al., 2000). The FCP consequently acquires late endosomal markers including CD63, LAMP-1, LAMP-2, and Rab7 (Clemens et al., 2004, 2009; Santic et al., 2005; Checroun et al., 2006; Chong et al., 2008; Wehrly et al., 2009). Eventually the late endosome becomes acidified upon acquisition of the proton vATPase pump that imports hydrogen protons into the vacuole (Chong et al., 2008; Santic et al., 2008). The FCP does not accumulate any lysosomal markers, such as Cathepsin D, or lysosomal tracers (Anthony L. D. et al., 1991; Clemens et al., 2004; Santic et al., 2005; Bonquist et al., 2008). In order to evade lysosome-mediated killing, Francisella escapes from the FCP to the cytosol. Vacuolar escape by various strains of F. tularensis and F. novicida in macrophages and other cells types has been described (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005, 2008; Chong et al., 2008). However, the study of Francisella vacuolar escape kinetics has brought controversy to the field varying from the 15 min to 8 h post-infection (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005, 2008; Checroun et al., 2006; McCaffrey and Allen, 2006). However, these differences are likely due to variation in the host cells used, the Francisella species and the methodological approaches used by various studies (Golovliov et al., 2003; Clemens et al., 2004; Santic et al., 2005, 2008; Checroun et al., 2006; McCaffrey and Allen, 2006).
The survival and replication of Francisella in host cells depends on the expression of the Francisella pathogenicity island (FPI) proteins. FPI-deficient mutants fail in formation and maturation of FCP (Nano et al., 2004; Telepnev et al., 2005; McCaffrey and Allen, 2006; Qin and Mann, 2006; Chong et al., 2008, 2013; Broms et al., 2010; Napier et al., 2012; Steele et al., 2013). Moreover, inactivated F. tularensis, as well as FPI mutants iglC and pdpC are not capable to avoid the FCP suggesting that vacuolar escape is a Francisella mediated process. This was clearly demonstrated when vacuolar escape deficient mutant pdpC was paired on magnetic bead with wild type bacteria. Wild type F. novicida secreted effector proteins, which allowed both wild type and pdpC to escape the phagosome. Studies have shown that pathogen secrete the IglA-J, PdpA, C, E, DotU, and VgrG into the macrophage cytosol during the infection (Hare and Hueffer, 2014). However, another study shown that IglC, IglI and PdpE, but not IglA and IglG are secreted in a T6SS-dependent manner during infection (Broms et al., 2012). Another importance of phagosome formation is shown by the reduced ability of Francisella LVS (live vaccine strain) strain to grow in host cell cytosol after microinjection (Meyer et al., 2015). The brief time spent in the phagosome is a dynamic step during which Francisella must actively evade host antimicrobial defenses (Jones et al., 2012). Francisella phagosomal escape is requisite to intracellular proliferation and its essential in the Francisella intracellular life cycle (Lindgren et al., 2004; Santic et al., 2005; Bonquist et al., 2008; Barker et al., 2009; Wehrly et al., 2009; Broms et al., 2012).
Techniques for the isolation and analysis of phagosomes are important experimental tools in endocytosis and apoptosis research. Since 1969, most of the available methods are based on density gradient ultracentrifugation (Wetzel and Korn, 1969). Here we present a method for isolation of FCP from infected human monocyte-derived macrophages (hMDMs). The method is based on infection of human macrophages with F. novicida, followed by mechanical lysis and separation of intracellular organelles. Several adaptations of previously described method are included (Shevchuk et al., 2009; Shevchuk and Steinert, 2013). For elimination of lysosomal compartment these organelles were loaded with dextran coated colloidal iron particles prior infection and eliminated by magnetic separation of post nuclear supernatant (PNS). The treatment of PNS with iodophenylnitrophenyltetrazolium (INT) salt was necessary to increase the density of mitochondria and fractionate it from FCP in a discontinuous sucrose density gradient.
Materials and Methods
Cultivation of F. novicida
F. novicida (Birdsell et al., 2009) was grown on buffered charcoal-yeast extract (BCYE) agar at 37°C with 5% CO2 atmosphere for 24 h.
Preparation of hMDMs from Blood of Healthy Human Donors
Human monocyte derived macrophages were differentiated from peripheral blood monocytes of healthy volunteers with no history of tularemia. Blood was diluted in ratio 1:2 with 0.9% saline, and 15 ml was applied on top of 7.5 ml of Ficoll-Hypaque (Ficoll-Paque; Pharmacia Fine Chemicals, USA). After 25 min of centrifugation at 300 × g, at room temperature (RT) the layer containing the mononuclear cell fraction was aspirated, transferred to a new tubes and centrifuged for additional 10 min at 300 × g at RT. Obtained monocytes were washed twice with 25 ml of 0.9% saline, resuspended in RPMI with glutamine (Bio Whittaker, Lonza, USA) supplemented with 20% FBS (Invitrogen, USA), and distributed in 6-well ultra-low attachment plates (Cornig Life Sciences, USA). Serum starvation was performed to promote monocyte differentiation to macrophages (Santic et al., 2005; Ozanic et al., 2016). After 3 days of incubation at 37°C in 5% CO2 the medium was replaced with 10% FBS RPMI, followed by replacement with 5% FBS RPMI (at day 6). At day 7 cells were scraped, collected and resuspended to desired concentration in RPMI with 1% FBS.
Preparation of Colloidal Iron Particles
Dextran—coated colloidal iron particles were prepared as follows. Equal volumes of 1.2 M FeCl2 (10 ml) and 1.8 M FeCl3 (10 ml) were mixed and agitated extensively while adding the same volume (10 ml) of 25% NH3 dropwise. The suspension was divided in 5 ml aliquots and placed on a magnetic unit (Dynal, Thermo Fisher, USA). Precipitate was than collected on the bottom of the tubes and washed once with 5% NH3, twice with ddH2O and resuspend in 80 ml of 0.3 M HCl. Solution was magnetically stirred for 30 min. Dextran (4 g, 64 to 76 kDa, Sigma-Aldrich, USA) was added and solution was stirred for further 30 min. In study of distribution of colloidal iron particles within endosomal compartments, small aliquot of prepared colloidal iron was incubated with dextran-tetramethylrhodamine (1 mg/ml, 70 kDa, Sigma-Aldrich, USA) and stirred for 30 min. The samples were dialyzed against 5 l of cold water, changing water four times during 2 days. The final suspension was filtered through filter paper and was used immediately or stored at 4°C for maximum 3 months. The concentration of obtained iron solution was ~10 mg/ml (Rodriguez-Paris et al., 1993).
Preparation of Optiprep™ Density Gradients
OptiPrep gradients were prepared by mixing of two working solutions, 10 and 45% of OptiPrep (Sigma-Aldrich, USA) in HB buffer (0.5 mM Na2EGTA, 20 mM HEPES, 250 mM Sucrose) in gradient mixer (Model #GM-40; C.B.S. Scientific Co, USA). Gradients were poured into polyallomer centrifuge tubes (9/16 × 3-3/4″; 14 × 95 mm; Beckman Coulter, USA) and used immediately or kept at 4°C overnight.
Infection of hMDMs with F. novicida
A total of 5·107 hMDMs were seeded in 30 ml of RPMI supplemented with 1% FBS in 75 cm2 cell culture flasks (TPP, Switzerland). Colloidal iron particles were added to a final concentration of 1 mg/ml, gently distributed 15 min before infection and left in the medium. The cells were infected with F. novicida at a multiplicity of infection (MOI) 10. In order to achieve synchronized infection, the cells were centrifuged immediately after infection at 100 × g for 3 min at RT. After 15 min of incubation at 37°C the cells were scraped, transferred to a 50 ml tube and centrifuged at 230 × g for 7 min at 4°C. Cells were washed twice in 30 ml of ice cold PBS and once in 10 ml of ice cold HB buffer.
Isolation of F. novicida-Containing Phagosome
For the isolation of FCP pellet of infected hMDMs was resuspended in 2 ml of cold HB buffer supplemented with EDTA-free protease inhibitor cocktail (Roche Diagnostic, Penzberg, Germany) according to manufacture protocol and with 5 mg/ml INT (Sigma-Aldrich, USA). The cells were mechanically disrupted in a Dura Grind stainless-steel homogenizer (Dounce Dura-Grind® Tissue Grinder; Braintree, Scientific, Inc.), transferred to a new tube and incubated with Benzonase (50 units/ml, Sigma-Aldrich, USA) for 7 min at 37°C. The nuclear and cell debris were removed by centrifugation at 110 × g for 5 min at 4°C. Obtained PNS was transferred to a new tubes and additional 2 ml of HB buffer with protease inhibitor cocktail was added to remaining pellet, carefully mixed and centrifuged at 100 × g for 3 min at 4°C. PNS was run through the MiniMACS column (OctoMACS™ Separation Unit; Miltenyi Biotec, Germany) to eliminate the lysosomal compartments loaded with colloidal iron. The flow through fraction was carefully applied on top of 8 ml of 10 to 45% OptiPrep gradient and centrifuged for 2 h in SW40 swing Rotor (Beckman Coulter, USA) at 100,000 × g at 4°C. After centrifugation, about 800 μl fractions were carefully collected from the top of gradient with cutted 1 ml tip. To analyze distribution of bacteria in gradient fractions, 10 μl of each fraction was diluted in 190 μl of ddH2O and plated on BCYE square plates 120 × 120 mm (Greiner, Sigma-Aldrich, USA). After 2 days of incubation at 37°C the CFU of F. novicida were calculated.
Confocal Laser Scanning Microscopy
The hMDMs were infected with F. novicida at MOI 10. At 15 min after infection the cells were washed with PBS, fixed using 4% paraformaldehyde (PFA, Sigma-Aldrich, USA) for 30 min at 4°C and permeabilized with 0.5% Triton X-100 (Sigma-Aldrich, USA). The coverslips were incubated with mouse monoclonal anti Francisella antibodies (1:5,000), washed with PBS and incubated with Alexa Fluor 555 (1:4,000, Molecular probes, USA) secondary antibodies for 1 h at RT.
To study the integrity of isolated FCP, equal fractions of phagosomes were seeded onto 24-well coverslips and centrifuged at 200 × g for 10 min at 4°C, followed by fixation with 4% PFA for 15 min at RT. Prepared samples were stained with 1 μl/ml of propidium iodide (PI) (Serva, Germany) for 25 min in the dark. Control samples were permeabilized with methanol at −20°C for 5 min.
To study the labeling of endosomal compartments with dextran-tetramethylrhodamine coated colloidal iron, the hMDMs were seeded on coverslips. The cells were loaded with dextran-tetramethylrhodamine colloidal iron for 15 min. After 15 min the colloidal iron particles were left in the medium and the cells were additionally infected with F. novicida for 15 min at MOI 10 followed by centrifugation at 100 × g for 3 min at RT. At 15 min after infection, and 30 min of colloidal iron particles load. the cells were washed, fixed and permeabilized as described above. The coverslips were incubated with mouse monoclonal anti Francisella antibodies (1:5,000), mouse monoclonal early endosome antigen (EEA1,1:1,000, Bio Rad, USA), mouse monoclonal lysosome associated membrane protein 1 (Lamp-1, 1:1,000, Bio Rad, USA) and mouse monoclonal anti Cathepsin-D (1:1,000, BD Biosciences, USA). The coverslips were washed with PBS and incubated with donkey anti-goat Alexa Fluor 488 and goat anti-mouse Alexa Fluor 647 (1:4,000, Molecular probes, USA) secondary antibodies for 1 h at RT. All samples were mounted in Mowiol 4-88 (Sigma-Aldrich, USA) and analyses were performed on FV 1000 Olympus confocal microscope.
SDS-PAGE and Western Blot
For Western blot analysis, equal amount of fraction proteins was applied onto 10% SDS-PAGE. After separation, proteins were transferred to nitrocellulose membrane in Transfer Buffer (Tris Base, Glycine, Methanol, ddH2O) and blocked for 1 h at room temperature in 1x Tris Buffered Saline (TBS, Sigma-Aldrich, USA) with 0.1% (w/v) Tween-20 (TBST, Sigma-Aldrich, USA) and 3% (m/v) Bovine Serum Albumine (BSA, Sigma-Aldrich, USA). Monoclonal rabbit antibody against human Rab5 (1:1,000, Cell Signaling Technology, USA), rabbit antibody against mitochondrial apoptosis-inducing factor (AIF, 1:1,000, Cell Signaling Technology, USA), mouse monoclonal KDEL antibody (1:100, Santa Cruz Biotechnology, USA), antibody against Golgi matrix protein of 130 kDa (gm130, BD Biosciences, USA), mouse monoclonal EEA1 and Lamp-1 were used for overnight incubation in staining buffer (3% BSA in TBST). After washing three times for 10 min in TBST, secondary anti-rabbit IgG and anti-mouse IgG conjugated horseradish peroxidase antibodies (1:1,000, Cell Signaling Technology, USA) were added for 1 h at RT. Membrane was again washed three times for 10 min in TBST. Enhanced chemiluminescence detection reagents Luminal Enhancer Solution (GE Healthcare, UK) and Peroxide Solution (GE Healthcare, UK) were used for visualization of the detected proteins by Bio Rad Chemi Doc XRR+ (Bio Rad Laboratories, USA).
Transmission Electron Microscopy
For transmission electron microscopy, the samples were transferred in 12-well cell culture plates (TPP, Switzerland). The samples were washed with 1x Sorensen buffer (TCS Biosciences Ltd., UK) and fixed using 2.5% glutaraldehyde (SPI Supplies, USA) for 45 min at 4°C. The post fixation was performed with 1% OsO4 (SPI Supplies, USA) for 45 min at 4°C. The sample was dehydrated by ethanol series with increased concentration, embedded in epoxy resin (SPI Supplies, USA) and polymerized for 24–48 h at 60°C. Ultra-thin sections were cut and examined by Phillips Morgany transmission electron microscope.
Statistics
Statistical significances were determined using two-tailed Student's t-test. Statistical analyses were performed using Statistica (Statsoft) software version 12 or with GraphPad Prizm version 6.0 software. P < 0.001 were accepted as significantly different and were denoted by *.
Ethics Statement
This study was carried out in accordance with the recommendations of Health Care Act Republic of Croatia (NN 158/08, 71/10, 139/10, 22/11, 84/11, 12/12, 35/12, 70/12 i 82/13), Act on the Protection of Patient's rights Republic of Croatia (NN 169/04, 37/08), was approved by the Ethical committee of Clinical Hospital Centre Rijeka as well as Ethical committee of Faculty of Medicine, with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki.
Results
F. novicida-Containing Phagosome Isolated from Infected hMDMs
Following phagocytic uptake, Francisella resides within special vacuole-FCP and its formation is absolutely required for intracytoplasmic replication of bacteria (Checroun et al., 2006). Because of apparent importance of this organelle during establishment of infection we optimized the method of FCP isolation (Figure 1). Human macrophages were infected with F. novicida at multiplicity of infection 10 resulting in 70% of hMDMs infected with bacteria at 15 min after infection (Figure 2). Macrophages, free of extracellular bacteria were disrupted in a Dura Grind stainless-steel homogenizer by optimized number of strokes. The unbroken cells and nuclei were removed by centrifugation. Obtained PNS was treated with Benzonase, an enzyme mixture for nucleic acid degradation, which allows reduction of sample viscosity and allows the separation of FCP from other organelles in PNS. The distribution of F. novicida in the gradient after ultracentrifugation was determined by plating fractions onto BCYE agar plates and counting bacterial CFU/ml (Figure 3). Our results showed that the highest number of F. novicida was in fraction 8 and it reached 6.5 × 105 CFU/ml (Figure 3). The fractions with highest number of bacteria were routinely proceeded for further analysis.
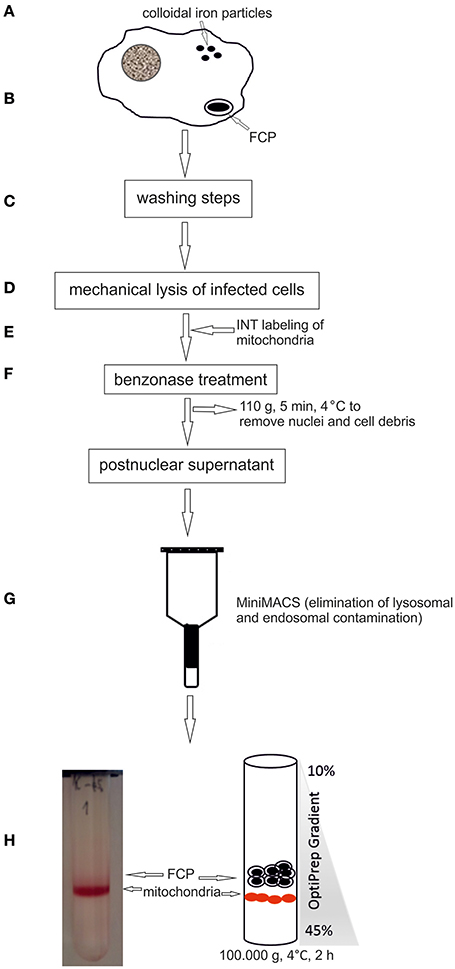
Figure 1. Schematic illustration of the FCPs isolation procedure. (A) hMDMs were loaded with colloidal iron particles. (B) Cells were infected with F. novicida, centrifuged to synchronize the infection, and the infection was allowed to proceed for 15 min. (C) Several washing steps were performed. (D) The cells were lysed mechanically. (E) Mitochondria were labeled with INT. (F) For reducing viscosity the suspension was treated with benzonase and centrifuged at low speed to remove cell debris and nuclei. (G) Post nuclear supernatant was run through a MiniMACS separation column to eliminate lysosomes. (H) The FCP were purified by OptiPrep density gradient ultracentrifugation.
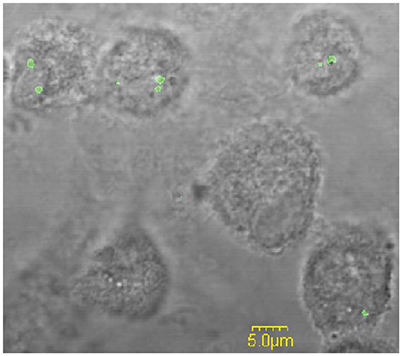
Figure 2. The analysis of the percentage of infected hMDMs. Representative fluorescence microscopy images of hMDMs infected with F. novicida at MOI 10 for 15 min. The examination of 100 hMDMs from three different coverslips shows that around 70% of the cells were infected.
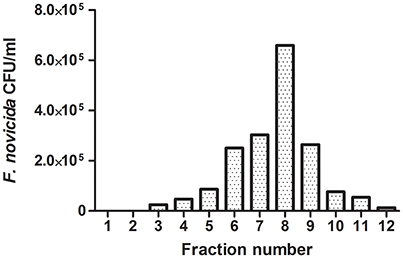
Figure 3. Representative distribution of FCPs in 10–45% OptiPrep gradient fractions. An aliquot of each OptiPrep fraction was plated onto a BCYE-agar plate and CFU/ml of F. novicida were counted. The fraction with the highest bacterial number corresponds to FCP fractions.
Separation of FCP from Subcellular Organelles
During isolation of bacterial vacuole, it is crucial to minimalize artifacts caused by other organelles. The successful separation of FCP from subcellular organelles was assessed by Western blot and transmission electron microscopy.
For separation of FCP from mitochondria we treated the PNS with INT, which results in formation of formazan, a product of activity of mitochondrial succinate dehydrogenase. This step was necessary to increase the density of mitochondria and separate two organelles by ultracentrifugation. The distribution of early endosome markers Rab5 and EEA1, lysosomal marker Lamp-1 as well and mitochondrial marker AIF, Golgi marker gm130 and ER marker KDEL were assessed by WB. Rab5 and EEA1 were enriched in fractions with the highest number of bacteria of the OptiPrep gradient, consistent with the accumulation of FCP in this fraction (Figure 4). AIF was enriched in fraction 10 of the OptiPrep gradient, confirming the presence of mitochondria in these fractions (Figure 4). Obtained results confirmed the separation of FCP from mitochondria by ultracentrifugation. Additionally, in gradient fractions formazan was visible after ultracentrifugation, and could be used as a marker for estimation of localization of bacterial fractions that were above this formazan circle (Figure 1). Further, our results showed that Golgi apparatus and endoplasmic reticulum are eliminated during the purification of the FCP and are not present in gradient fractions (Figure 4). The distribution of Lamp-1 in PNS before and after the magnetic separation, as well as in gradient fractions, was tested by Western blot (Figure 4). The results showed that lysosomes were present in PNS before magnetic separation and eliminated with this procedure.
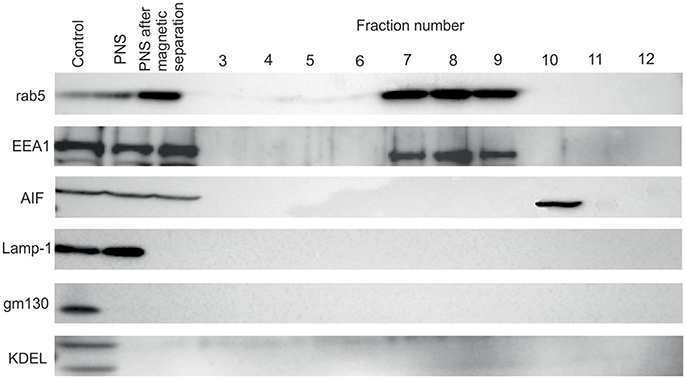
Figure 4. Western blot analysis of OptiPrep fractions. Fractions 3 to 12 as well as PNS before and after magnetic separation were tested using the markers for the following compartments: early endosome (Rab5 and EEA1), lysosomes (Lamp-1), Golgi (gm130), mitochondria (AIF) and ER (KDEL). hMDMs lysate was used as a control.
To investigate the trafficking of the dextran coated colloidal iron particles within the endocytic pathways after infection of hMDMs with F. novicida the confocal microscopy was used. Our results showed that around 30% of dextran-tetramethylrhodamine colloidal iron particles colocolized with Lamp-1 and ~65% with Cathepsin-D (Figure 5B). In contrast, F. novicida colocalized with EEA1 (Figure 5A), indicating that most of colloidal iron does not interfere with early F. novicida phagosome.
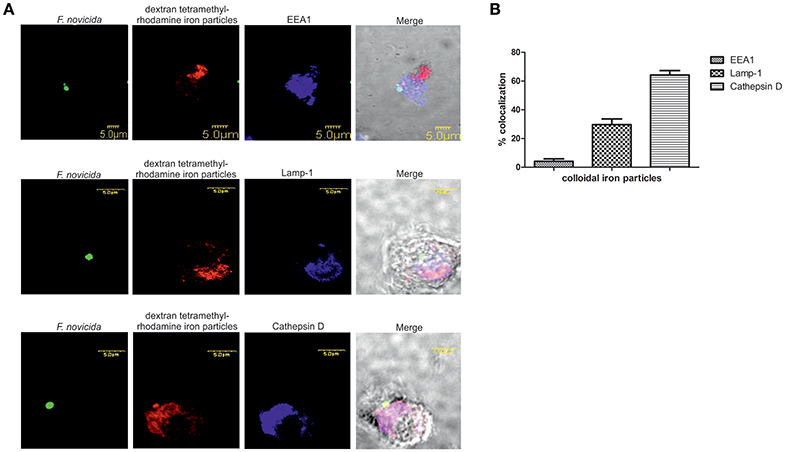
Figure 5. The distribution of dextran-tetramethylrhodamine iron particles within the endocytic pathway in F. novicida infection (A). Representative confocal laser scanning microscopy images of colocalization of dextran-tetramethylrhodamine iron particles with Lamp-1, Cathepsin-D, and EEA1 in F. novicida infected hMDMs. The images are representatives of 100 infected cells examined from three different cover slips. (B) Quantification of colocalization of the colloidal iron particles with EEA1, Lamp-1, and Cathepsin D. The results shown are representative of three independed experiments, and error bars represent standard deviation of triplicate samples.
In addition, human macrophages infected with F. novicida at MOI 10 at 15 min after infection and FCP within fractions were analyzed by transmission electron microscopy. At 15 min after infection bacteria were enclosed in intact phagosomes of infected hMDMs (Figure 6A). Low magnification TEM image of the FCP enriched fraction demonstrate the purity and small vesicle free fraction (Figure 6B). High magnification TEM image of the FCP enriched fraction revealed that single bacteria surrounded by per one-layer membrane were present (Figure 6C).
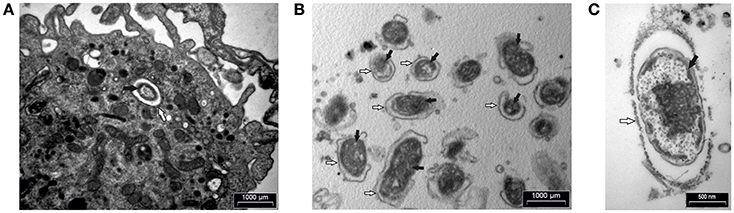
Figure 6. TEM analyses of hMDMs infected with F. novicida at MOI 10 at 15 min after infection (A) and isolated FCP (B,C). Samples were washed and fixed with glutaraldehyde and post-fixed using osmium tetroxide. Ultra-thin sections were cut and observed using TEM. The white arrows show vacuolar membrane and black arrows indicate bacteria. One representative micrographs out of three independent preparations.
Integrity of the FCP Membrane after Isolation
The integrity of phagosomal membrane of isolated FCP was tested using PI by fluorescence microscopy. The fractions with the highest number of bacteria were used for this study. As a control sample, the isolated phagosomal fraction was permeabilized in order to allow the PI to penetrate inside the phagosome and stain the bacteria. This was considered as 100% of stained bacteria. Three coverslips were analyzed and the total stained bacteria in each sample were counted. This approach provided valuable information about the quality of the isolated FCP. Our results show that FCP membrane is intact on ~70% of the isolated phagosomal fractions (P = 0.000001; Figure 7).
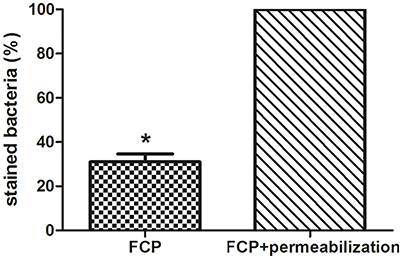
Figure 7. The analysis of FCP membrane integrity. Quantification of integrity of phagosomal membrane in the fraction with the highest number of bacteria. The percent of bacteria after staining with PI and analyses by fluorescence microscopy. Three coverslips were analyzed per fraction and STDEV were calculated. Student's t-test was used to determine statistical significance with *P < 0.001.
Discussion
It is essential for Francisella to replicate within host cells to successfully establish an infection and cause disease. Escape from the phagosome is an important step in Francisella life cycle since mutants deficient in escape are unable to cause productive infection (Chong et al., 2012). After invasion of the host cell, Francisella forms an endocytic membrane-bound phagosome. Bacteria must disrupt this phagosome and dislocate to the cytosol in order to replicate and spread from cell to cell (Clemens et al., 2004; Santic et al., 2005). However, these short-lived vacuoles can interact with the host vesicular trafficking, and can have important role for virulence and pathogenesis of Francisella. Due to importance of the phagosome in pathogenesis of tularemia we established a method for purification of FCP from infected human monocyte derived macrophages. To perform the separation of phagosome from infected cells, large cell number is necessary, making it challenging to use these approaches with human primary cells. For that reason, we pulled together the blood from different donors to obtain the necessary number of human macrophages. To ensure that we do not co-isolate the extracellular bacteria, intensive washing steps were included. In contrast to previously published protocols, the dextran coated iron particle were used for elimination of lysosomal and endosomal compartments prior infection (Shevchuk et al., 2009; Shevchuk and Steinert, 2013). With the use of confocal microscopy, we followed the trafficking of dextran-tetramethylrhodamine iron particles within hMDMs. The markers for early and late endosomes as well as lysosomes were used to document which endosomal compartments are labeled by colloidal iron. This is due to the specificity and rapidity of infection in comparison to other intracellular pathogens (Chong and Celli, 2010; Santic et al., 2010). Some of the most interesting aspects of phagosome maturation depend on the ability of intracellular pathogens to bypass the normal maturation process. Attempts to purify these compartments represents a challenge when classical organelle enrichment techniques are used. To resolve this problem a combination of classical and improved methods for enrichment and pre-fractionation must be used. In previous published methods for isolation of Legionella-containing vacuole the 5–30% OptiPrep gradient was used (Shevchuk et al., 2009; Shevchuk and Steinert, 2013). During the establishment of phagosome isolation from Dictyostelium discoideum cells infected with F. novicida, different concentration of OptiPrep were tested (5–30, 5–35, 10–40, and 10–45%, data not shown). Optimization of OptiPrep gradient for successful isolation of FCP showed that the best separation of FCP was when OptiPrep gradient 10–45% was used. In addition, the efficient removal of contaminants in the method of vacuole isolation is very important to achieve. Therefore, in order to separate two organelles with close density, mitochondria and FCP, we utilized an enzyme of the citrate cycle, the succinate dehydrogenase, located in the inner mitochondrial membrane. The INT added to the PNS is converted to formazan and increased the density of those organelles (Munujos et al., 1993). This phenomenon was used for separation of mitochondria from FCPs by discontinues ultracentrifugation method. In addition, Rab5 and EEA1, as markers for early endosomal compartment, were used to indicate the presence of FCP in OptiPrep fractions that was void of mitochondrial marker, AIF.
Isolation of bacteria-containing vacuoles (BCV) is of key importance for the understanding of these compartments, but technically is very challenging. During recent years, different groups have developed different protocols for isolation of BCV (reviewed in Herweg et al., 2015). The bacteria subvert endomembrane trafficking around the BCV and the communication around BCV and other host cell organelles has been described (Gagnon et al., 2002; Touret et al., 2005; Santos et al., 2015; Santos and Enninga, 2016).
Protocols for isolation of pathogen-containing vacuoles are based on subcellular/organelle fractionation based on physicochemical properties (Howe and Heinzen, 2008; He et al., 2012; Cheng et al., 2014). These protocols combine confocal fluorescence microscopy, Western blot and electron microscopy techniques providing the characterization of host cell compartments after infection with different intracellular pathogens. In addition, isolation of BCV can be based on immuno-affinity purification (Urwyler et al., 2010; Hoffmann et al., 2013; Vorwerk et al., 2015) or on FACS single cell enrichment by sorting bacteria and lysed host cells organelles (Becker et al., 2006; Pfortner et al., 2013; Surmann et al., 2014). Separation principles have been applied for isolation of latex bead-phagosomes from macrophages (Desjardins et al., 1994) and Dictyostelium (Gotthardt et al., 2002). In the protocol for purification of Legionella-containing vacuole (LCV) from infected D. discoideum (Shevchuk et al., 2009; Urwyler et al., 2010; Finsel et al., 2013; Shevchuk and Steinert, 2013), or murine macrophage-like RAW 264.7 (Hoffmann et al., 2014) the immuno-magnetic separation using an anti-SidC antibody was performed, followed by 10–35% Histodenz density gradient centrifugation. Others established protocol for LCV isolation from U937 macrophages using 55–65% gradient (Bruckert and Abu Kwaik, 2015). Others and our studies show that the integrity of phagosome membrane is often compromised during the early time point of infection with Francisella (Santic et al., 2008; Chong et al., 2012; Ozanic et al., 2015; Rowe and Huntley, 2015). Besides the electron microscopy methods, fluorescence microscopy could be valuable method to check the integrity of the phagosomal membrane after its isolation (Lonnbro et al., 2008; Hoffmann et al., 2013, 2014; Bruckert and Abu Kwaik, 2015). Results from this study show that the phagosomal membrane is highly conserved 15 min after infection of hMDMs and only 30% of the analyzed fraction show some damage of the FCP. The FCP is presumably intact within 30 min after infection.
Many intracellular bacteria reside and replicate inside phagosomal compartment making protocol for vacuolar isolation easy to apply. In contrast, some intracellular pathogens show ability to escape from phagosome to directly use the cytoplasm as their replicative habitat (Ray et al., 2009). Francisella resides in phagosomes ~5–30 min after infection, making it more challenging for isolation of the phagosome from infected macrophages. After cytoplasmic replication, Francisella re-enters the endocytic pathways by autophagy (Checroun et al., 2006; Jones et al., 2012), and bacteria are found in autophagosomes by 24 h after infection. Our established method for isolation of FCP could be applied for the isolation of autophagosomes as well. The protocol presented here will enable future proteomic analyses analysis of those delicate intracellular compartments.
Author Contributions
VM, OS, and MiS contributed in isolation of the vacuole and writing. MM and AJ participated in Western blot analyses. MO participated in electron microscopy analyses. MaS and YA participated in fluorescence analyses and writing.
Funding
This work is supported by a University Grants (No. 13.06.1.1.11. and No. 13.11.1.2.07), National Science Foundation (HRZZ, IP-2016-06-9003) and project RISK financed by European Regional Development Fund (ERDF). YA is supported by Public Health Service Award 1R01AI120244 and R21AI116517 from the National Institute of Health and by the Commonwealth of Kentucky Research Challenge Trust Fund.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alvarez-Dominguez, C., Barbieri, A. M., Beron, W., Wandinger-Ness, A., and Stahl, P. D. (1996). Phagocytosed live Listeria monocytogenes influences Rab5-regulated in vitro phagosome-endosome fusion. J. Biol. Chem. 271, 13834–13843. doi: 10.1074/jbc.271.23.13834
Anthony, L. D., Burke, R. D., and Nano, F. E. (1991). Growth of Francisella spp. in rodent macrophages. Infect. Immun. 59, 3291–3296.
Anthony, L. S., Gu, M. Z., Cowley, S. C., Leung, W. W., and Nano, F. E. (1991). Transformation and allelic replacement in Francisella spp. J. Gen. Microbiol. 137, 2697–2703. doi: 10.1099/00221287-137-12-2697
Barker, J. R., Chong, A., Wehrly, T. D., Yu, J. J., Rodriguez, S. A., Liu, J., et al. (2009). The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74, 1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x
Becker, D., Selbach, M., Rollenhagen, C., Ballmaier, M., Meyer, T. F., Mann, M., et al. (2006). Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307. doi: 10.1038/nature04616
Ben Nasr, A., Haithcoat, J., Masterson, J. E., Gunn, J. S., Eaves-Pyles, T., and Klimpel, G. R. (2006). Critical role for serum opsonins and complement receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in phagocytosis of Francisella tularensis by human dendritic cells (DC): uptake of Francisella leads to activation of immature DC and intracellular survival of the bacteria. J. Leukoc. Biol. 80, 774–786. doi: 10.1189/jlb.1205755
Birdsell, D. N., Stewart, T., Vogler, A. J., Lawaczeck, E., Diggs, A., Sylvester, T. L., et al. (2009). Francisella tularensis subsp. novicida isolated from a human in Arizona. BMC Res. Notes 2:223. doi: 10.1186/1756-0500-2-223
Bonquist, L., Lindgren, H., Golovliov, I., Guina, T., and Sjostedt, A. (2008). MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 76, 3502–3510. doi: 10.1128/IAI.00226-08
Broms, J. E., Meyer, L., Sun, K., Lavander, M., and Sjostedt, A. (2012). Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS ONE 7:e50473. doi: 10.1371/journal.pone.0050473
Broms, J. E., Sjostedt, A., and Lavander, M. (2010). The role of the Francisella Tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front. Microbiol. 1:136. doi: 10.3389/fmicb.2010.00136
Bruckert, W. M., and Abu Kwaik, Y. (2015). Complete and ubiquitinated proteome of the Legionella-containing vacuole within human macrophages. J. Proteome Res. 14, 236–248. doi: 10.1021/pr500765x
Buddingh, G. J., and Womack, F. C. (1941). Observations on the infection of chick embryos with Bacterium Tularense, Brucella, and Pasteurella Pestis. J. Exp. Med. 74, 213–222. doi: 10.1084/jem.74.3.213
Camejo, A., Carvalho, F., Reis, O., Leitao, E., Sousa, S., and Cabanes, D. (2011). The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2, 379–394. doi: 10.4161/viru.2.5.17703
Celli, J., and Zahrt, T. C. (2013). Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010314. doi: 10.1101/cshperspect.a010314
Checroun, C., Wehrly, T. D., Fischer, E. R., Hayes, S. F., and Celli, J. (2006). Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U.S.A. 103, 14578–14583. doi: 10.1073/pnas.0601838103
Cheng, Y., Liu, Y., Wu, B., Zhang, J. Z., Gu, J., Liao, Y. L., et al. (2014). Proteomic analysis of the Ehrlichia chaffeensis phagosome in cultured DH82 cells. PLoS ONE 9:e88461. doi: 10.1371/journal.pone.0088461
Chong, A., and Celli, J. (2010). The francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Microbiol. 1:138. doi: 10.3389/fmicb.2010.00138
Chong, A., Child, R., Wehrly, T. D., Rockx-Brouwer, D., Qin, A., Mann, B. J., et al. (2013). Structure-function analysis of DipA, a Francisella tularensis virulence Factor required for intracellular replication. PLoS ONE 8:e67965. doi: 10.1371/journal.pone.0067965
Chong, A., Wehrly, T. D., Child, R., Hansen, B., Hwang, S., Virgin, H. W., et al. (2012). Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy 8, 1342–1356. doi: 10.4161/auto.20808
Chong, A., Wehrly, T. D., Nair, V., Fischer, E. R., Barker, J. R., Klose, K. E., et al. (2008). The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76, 5488–5499. doi: 10.1128/IAI.00682-08
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72, 3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2005). Francisella tularensis enters macrophages via a novel process involving pseudopod loops. Infect. Immun. 73, 5892–5902. doi: 10.1128/IAI.73.9.5892-5902.2005
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2009). Francisella tularensis phagosomal escape does not require acidification of the phagosome. Infect. Immun. 77, 1757–1773. doi: 10.1128/IAI.01485-08
Desjardins, M., Huber, L. A., Parton, R. G., and Griffiths, G. (1994). Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J. Cell Biol. 124, 677–688. doi: 10.1083/jcb.124.5.677
Duclos, S., Diez, R., Garin, J., Papadopoulou, B., Descoteaux, A., Stenmark, H., et al. (2000). Rab5 regulates the kiss and run fusion between phagosomes and endosomes and the acquisition of phagosome leishmanicidal properties in RAW 264.7 macrophages. J. Cell Sci. 113(Pt 19), 3531–3541.
Finsel, I., Hoffmann, C., and Hilbi, H. (2013). Immunomagnetic purification of fluorescent Legionella-containing vacuoles. Methods Mol. Biol. 983, 431–443. doi: 10.1007/978-1-62703-302-2_24
Fortier, A. H., Green, S. J., Polsinelli, T., Jones, T. R., Crawford, R. M., Leiby, D. A., et al. (1994). Life and death of an intracellular pathogen: Francisella tularensis and the macrophage. Immunol. Ser. 60, 349–361.
Gagnon, E., Duclos, S., Rondeau, C., Chevet, E., Cameron, P. H., Steele-Mortimer, O., et al. (2002). Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110, 119–131. doi: 10.1016/S0092-8674(02)00797-3
Golovliov, I., Baranov, V., Krocova, Z., Kovarova, H., and Sjostedt, A. (2003). An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71, 5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003
Gotthardt, D., Warnatz, H. J., Henschel, O., Bruckert, F., Schleicher, M., and Soldati, T. (2002). High-resolution dissection of phagosome maturation reveals distinct membrane trafficking phases. Mol. Biol. Cell 13, 3508–3520. doi: 10.1091/mbc.E02-04-0206
Hare, R. F., and Hueffer, K. (2014). Francisella novicida pathogenicity island encoded proteins were secreted during infection of macrophage-like cells. PLoS ONE 9:e105773. doi: 10.1371/journal.pone.0105773
He, Y., Li, W., Liao, G., and Xie, J. (2012). Mycobacterium tuberculosis-specific phagosome proteome and underlying signaling pathways. J. Proteome Res. 11, 2635–2643. doi: 10.1021/pr300125t
Herweg, J. A., Hansmeier, N., Otto, A., Geffken, A. C., Subbarayal, P., Prusty, B. K., et al. (2015). Purification and proteomics of pathogen-modified vacuoles and membranes. Front. Cell. Infect. Microbiol. 5:48. doi: 10.3389/fcimb.2015.00048
Hoffmann, C., Finsel, I., and Hilbi, H. (2013). Pathogen vacuole purification from legionella-infected amoeba and macrophages. Methods Mol. Biol. 954, 309–321. doi: 10.1007/978-1-62703-161-5_18
Hoffmann, C., Finsel, I., Otto, A., Pfaffinger, G., Rothmeier, E., Hecker, M., et al. (2014). Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell. Microbiol. 16, 1034–1052. doi: 10.1111/cmi.12256
Howe, D., and Heinzen, R. A. (2008). Fractionation of the Coxiella burnetii parasitophorous vacuole. Methods Mol. Biol. 445, 389–406. doi: 10.1007/978-1-59745-157-4_25
Isberg, R. R., O'connor, T. J., and Heidtman, M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24. doi: 10.1038/nrmicro1967
Jahraus, A., Tjelle, T. E., Berg, T., Habermann, A., Storrie, B., Ullrich, O., et al. (1998). In vitro fusion of phagosomes with different endocytic organelles from J774 macrophages. J. Biol. Chem. 273, 30379–30390. doi: 10.1074/jbc.273.46.30379
Jones, C. L., Napier, B. A., Sampson, T. R., Llewellyn, A. C., Schroeder, M. R., and Weiss, D. S. (2012). Subversion of host recognition and defense systems by Francisella spp. Microbiol. Mol. Biol. Rev. 76, 383–404. doi: 10.1128/MMBR.05027-11
Kagan, J. C., and Roy, C. R. (2002). Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4, 945–954. doi: 10.1038/ncb883
Lauriano, C. M., Barker, J. R., Yoon, S. S., Nano, F. E., Arulanandam, B. P., Hassett, D. J., et al. (2004). MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. U.S.A. 101, 4246–4249. doi: 10.1073/pnas.0307690101
Lindemann, S. R., McLendon, M. K., Apicella, M. A., and Jones, B. D. (2007). An in vitro model system used to study adherence and invasion of Francisella tularensis live vaccine strain in nonphagocytic cells. Infect. Immun. 75, 3178–3182. doi: 10.1128/IAI.01811-06
Lindgren, H., Golovliov, I., Baranov, V., Ernst, R. K., Telepnev, M., and Sjostedt, A. (2004). Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53, 953–958. doi: 10.1099/jmm.0.45685-0
Llewellyn, A. C., Jones, C. L., Napier, B. A., Bina, J. E., and Weiss, D. S. (2011). Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS ONE 6:e24201. doi: 10.1371/journal.pone.0024201
Lonnbro, P., Nordenfelt, P., and Tapper, H. (2008). Isolation of bacteria-containing phagosomes by magnetic selection. BMC Cell Biol. 9:35. doi: 10.1186/1471-2121-9-35
Malik-Kale, P., Jolly, C. E., Lathrop, S., Winfree, S., Luterbach, C., and Steele-Mortimer, O. (2011). Salmonella - at home in the host cell. Front. Microbiol. 2:125. doi: 10.3389/fmicb.2011.00125
McCaffrey, R. L., and Allen, L. A. (2006). Francisella tularensis LVS evades killing by human neutrophils via inhibition of the respiratory burst and phagosome escape. J. Leukoc. Biol. 80, 1224–1230. doi: 10.1189/jlb.0406287
Meyer, L., Broms, J. E., Liu, X., Rottenberg, M. E., and Sjostedt, A. (2015). Microinjection of Francisella tularensis and Listeria monocytogenes reveals the importance of bacterial and host factors for successful replication. Infect. Immun. 83, 3233–3242. doi: 10.1128/IAI.00416-15
Moreau, G. B., and Mann, B. J. (2013). Adherence and uptake of Francisella into host cells. Virulence 4, 826–832. doi: 10.4161/viru.25629
Munujos, P., Coll-Canti, J., Gonzalez-Sastre, F., and Gella, F. J. (1993). Assay of succinate dehydrogenase activity by a colorimetric-continuous method using iodonitrotetrazolium chloride as electron acceptor. Anal. Biochem. 212, 506–509. doi: 10.1006/abio.1993.1360
Nano, F. E., Zhang, N., Cowley, S. C., Klose, K. E., Cheung, K. K., Roberts, M. J., et al. (2004). A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186, 6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004
Napier, B. A., Meyer, L., Bina, J. E., Miller, M. A., Sjostedt, A., and Weiss, D. S. (2012). Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc. Natl. Acad. Sci. U.S.A. 109, 18084–18089. doi: 10.1073/pnas.1206411109
Ozanic, M., Marecic, V., Abu Kwaik, Y., and Santic, M. (2015). The divergent intracellular lifestyle of Francisella tularensis in evolutionarily distinct host cells. PLoS Pathog. 11:e1005208. doi: 10.1371/journal.ppat.1005208
Ozanic, M., Marecic, V., Lindgren, M., Sjostedt, A., and Santic, M. (2016). Phenotypic characterization of the Francisella tularensis DeltapdpC and DeltaiglG mutants. Microbes Infect. 18, 768–776. doi: 10.1016/j.micinf.2016.07.006
Pfortner, H., Wagner, J., Surmann, K., Hildebrandt, P., Ernst, S., Bernhardt, J., et al. (2013). A proteomics workflow for quantitative and time-resolved analysis of adaptation reactions of internalized bacteria. Methods 61, 244–250. doi: 10.1016/j.ymeth.2013.04.009
Qin, A., and Mann, B. J. (2006). Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6:69. doi: 10.1186/1471-2180-6-69
Ray, K., Bobard, A., Danckaert, A., Paz-Haftel, I., Clair, C., Ehsani, S., et al. (2010). Tracking the dynamic interplay between bacterial and host factors during pathogen-induced vacuole rupture in real time. Cell. Microbiol. 12, 545–556. doi: 10.1111/j.1462-5822.2010.01428.x
Ray, K., Marteyn, B., Sansonetti, P. J., and Tang, C. M. (2009). Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7, 333–340. doi: 10.1038/nrmicro2112
Rodriguez-Paris, J. M., Nolta, K. V., and Steck, T. L. (1993). Characterization of lysosomes isolated from Dictyostelium discoideum by magnetic fractionation. J. Biol. Chem. 268, 9110–9116.
Rowe, H. M., and Huntley, J. F. (2015). From the outside-in: the Francisella tularensis envelope and virulence. Front. Cell. Infect. Microbiol. 5:94. doi: 10.3389/fcimb.2015.00094
Santic, M., Akimana, C., Asare, R., Kouokam, J. C., Atay, S., and Kwaik, Y. A. (2009). Intracellular fate of Francisella tularensis within arthropod-derived cells. Environ. Microbiol. 11, 1473–1481. doi: 10.1111/j.1462-2920.2009.01875.x
Santic, M., Al-Khodor, S., and Abu Kwaik, Y. (2010). Cell biology and molecular ecology of Francisella tularensis. Cell. Microbiol. 12, 129–139. doi: 10.1111/j.1462-5822.2009.01400.x
Santic, M., Asare, R., Skrobonja, I., Jones, S., and Abu Kwaik, Y. (2008). Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect. Immun. 76, 2671–2677. doi: 10.1128/IAI.00185-08
Santic, M., Molmeret, M., and Abu Kwaik, Y. (2005). Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell. Microbiol. 7, 957–967. doi: 10.1111/j.1462-5822.2005.00529.x
Santos, J. C., and Enninga, J. (2016). At the crossroads: communication of bacteria-containing vacuoles with host organelles. Cell. Microbiol. 18, 330–339. doi: 10.1111/cmi.12567
Santos, S. B., Carvalho, C., Azeredo, J., and Ferreira, E. C. (2015). Correction: population dynamics of a salmonella lytic phage and its host: implications of the host bacterial growth rate in modelling. PLoS ONE 10:e0136007. doi: 10.1371/journal.pone.0136007
Shepard, C. C. (1959). Nonacid-fast bacteria and HeLa cells: their uptake and subsequent intracellular growth. J. Bacteriol. 77, 701–714.
Shevchuk, O., Batzilla, C., Hagele, S., Kusch, H., Engelmann, S., Hecker, M., et al. (2009). Proteomic analysis of Legionella-containing phagosomes isolated from Dictyostelium. Int. J. Med. Microbiol. 299, 489–508. doi: 10.1016/j.ijmm.2009.03.006
Shevchuk, O., and Steinert, M. (2013). Isolation of pathogen-containing vacuoles. Methods Mol. Biol. 983, 419–429. doi: 10.1007/978-1-62703-302-2_23
Shin, S., and Roy, C. R. (2008). Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell. Microbiol. 10, 1209–1220. doi: 10.1111/j.1462-5822.2008.01145.x
Steele, S., Brunton, J., Ziehr, B., Taft-Benz, S., Moorman, N., and Kawula, T. (2013). Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog. 9:e1003562. doi: 10.1371/journal.ppat.1003562
Steele-Mortimer, O. (2008). The Salmonella-containing vacuole - Moving with the times. Curr. Opin. Microbiol. 11, 38–45. doi: 10.1016/j.mib.2008.01.002
Surmann, K., Michalik, S., Hildebrandt, P., Gierok, P., Depke, M., Brinkmann, L., et al. (2014). Comparative proteome analysis reveals conserved and specific adaptation patterns of Staphylococcus aureus after internalization by different types of human non-professional phagocytic host cells. Front. Microbiol. 5:392. doi: 10.3389/fmicb.2014.00392
Tamilselvam, B., and Daefler, S. (2008). Francisella targets cholesterol-rich host cell membrane domains for entry into macrophages. J. Immunol. 180, 8262–8271. doi: 10.4049/jimmunol.180.12.8262
Telepnev, M., Golovliov, I., and Sjostedt, A. (2005). Francisella tularensis LVS initially activates but subsequently down-regulates intracellular signaling and cytokine secretion in mouse monocytic and human peripheral blood mononuclear cells. Microb. Pathog. 38, 239–247. doi: 10.1016/j.micpath.2005.02.003
Touret, N., Paroutis, P., Terebiznik, M., Harrison, R. E., Trombetta, S., Pypaert, M., et al. (2005). Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell 123, 157–170. doi: 10.1016/j.cell.2005.08.018
Urwyler, S., Finsel, I., Ragaz, C., and Hilbi, H. (2010). Isolation of Legionella-containing vacuoles by immuno-magnetic separation. Curr. Protoc. Cell Biol. Chapter 3, Unit 3.34. doi: 10.1002/0471143030.cb0334s46
Vergne, I., Chua, J., Singh, S. B., and Deretic, V. (2004). Cell biology of mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 20, 367–394. doi: 10.1146/annurev.cellbio.20.010403.114015
Vorwerk, S., Krieger, V., Deiwick, J., Hensel, M., and Hansmeier, N. (2015). Proteomes of host cell membranes modified by intracellular activities of Salmonella enterica. Mol. Cell. Proteomics 14, 81–92. doi: 10.1074/mcp.M114.041145
Wehrly, T. D., Chong, A., Virtaneva, K., Sturdevant, D. E., Child, R., Edwards, J. A., et al. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x
Keywords: phagocytosis, organelle purification, pathogen-containing phagosomes, Francisella, human macrophages
Citation: Marecic V, Shevchuk O, Ozanic M, Mihelcic M, Steinert M, Jurak Begonja A, Abu Kwaik Y and Santic M (2017) Isolation of F. novicida-Containing Phagosome from Infected Human Monocyte Derived Macrophages. Front. Cell. Infect. Microbiol. 7:303. doi: 10.3389/fcimb.2017.00303
Received: 24 March 2017; Accepted: 20 June 2017;
Published: 05 July 2017.
Edited by:
Albert Descoteaux, Institut National de la Recherche Scientifique, CanadaReviewed by:
Mikhail A. Gavrilin, The Ohio State University Columbus, United StatesLee-Ann H. Allen, University of Iowa, United States
Copyright © 2017 Marecic, Shevchuk, Ozanic, Mihelcic, Steinert, Jurak Begonja, Abu Kwaik and Santic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marina Santic, marina.santic@medri.uniri.hr
 Valentina Marecic
Valentina Marecic Olga Shevchuk
Olga Shevchuk Mateja Ozanic
Mateja Ozanic Mirna Mihelcic
Mirna Mihelcic Michael Steinert
Michael Steinert Antonija Jurak Begonja
Antonija Jurak Begonja Yousef Abu Kwaik
Yousef Abu Kwaik Marina Santic
Marina Santic