Two Novel Salmonella Bivalent Vaccines Confer Dual Protection against Two Salmonella Serovars in Mice
- 1Research Center of Avian Diseases, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
- 3Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
Non-typhoidal Salmonella includes thousands of serovars that are leading causes of foodborne diarrheal illness worldwide. In this study, we constructed three bivalent vaccines for preventing both Salmonella Typhimurium and Salmonella Newport infections by using the aspartate semialdehyde dehydrogenase (Asd)-based balanced-lethal vector-host system. The constructed Asd+ plasmid pCZ11 carrying a subset of the Salmonella Newport O-antigen gene cluster including the wzx-wbaR-wbaL-wbaQ-wzy-wbaW-wbaZ genes was introduced into three Salmonella Typhimurium mutants: SLT19 (Δasd) with a smooth LPS phenotype, SLT20 (Δasd ΔrfbN) with a rough LPS phenotype, and SLT22 (Δasd ΔrfbN ΔpagL::T araC PBAD rfbN) with a smooth LPS phenotype when grown with arabinose. Immunoblotting demonstrated that SLT19 harboring pCZ11 [termed SLT19 (pCZ11)] co-expressed the homologous and heterologous O-antigens; SLT20 (pCZ11) exclusively expressed the heterologous O-antigen; and when arabinose was available, SLT22 (pCZ11) expressed both types of O-antigens, while in the absence of arabinose, SLT22 (pCZ11) expressed only the heterologous O-antigen. Exclusive expression of the heterologous O-antigen in Salmonella Typhimurium decreased the swimming ability of the bacterium and its susceptibility to polymyxin B. Next, the crp gene was deleted from the three recombinant strains for attenuation purposes, generating the three bivalent vaccine strains SLT25 (pCZ11), SLT26 (pCZ11), and SLT27 (pCZ11), respectively. Groups of BALB/c mice (12 mice/group) were orally immunized with 109 CFU of each vaccine strain twice at an interval of 4 weeks. Compared with a mock immunization, immunization with all three vaccine strains induced significant serum IgG responses against both Salmonella Typhimurium and Salmonella Newport LPS. The bacterial loads in the mouse tissues were significantly lower in the three vaccine-strain-immunized groups than in the mock group after either Salmonella Typhimurium or Salmonella Newport lethal challenge. All of the mice in the three vaccine-immunized groups survived the lethal Salmonella Typhimurium challenge. In contrast, SLT26 (pCZ11) and SLT27 (pCZ11) conferred full protection against lethal Salmonella Newport challenge, but SLT25 (pCZ11) provided only 50% heterologous protection. Thus, we developed two novel Salmonella bivalent vaccines, SLT26 (pCZ11) and SLT27 (pCZ11), suggesting that the delivery of a heterologous O-antigen in attenuated Salmonella strains is a prospective approach for developing Salmonella vaccines with broad serovar coverage.
Introduction
Non-typhoidal Salmonella (NTS), a gram-negative and facultative intracellular bacterium, predominantly causes enteric diarrheal disease in a broad spectrum of animal hosts and humans, which represents a major public health concern, with an estimated 93.8 million cases worldwide and 155,000 deaths each year (Majowicz et al., 2010). NTS also infrequently causes serious dysentery and septicemia with substantial mortality, particularly in young infants, the elderly and immunocompromised individuals, such as HIV patients (Arshad et al., 2008; Parry et al., 2013). Salmonella infections have been linked to a variety of sources, particularly livestock and related products (e.g., poultry, eggs, beef, and dairy products) and produce (Braden, 2006; Scallan et al., 2011; Painter et al., 2013). NTS infections often result in an asymptomatic carrier state in adult domestic animals and the establishment of persistent infections, which have serious impacts on public health due to the risks of food poisoning via consumption of contaminated products. Furthermore, the prevalence of multidrug resistance among human and animal Salmonella isolates has increased over the past 2 decades (Threlfall, 2002; Varma et al., 2005; Karon et al., 2007). Thus, safe and effective NTS vaccines are urgently needed to prevent salmonellosis in both humans and domestic animals.
The global epidemiology of NTS diseases is complex, with diverse serovars in different regions worldwide, posing a substantial challenge for vaccine development. Salmonella enterica serovar Typhimurium (Salmonella Typhimurium) and Salmonella Enteritidis have been the most prevalent serovars in people and animals for many years; however, other serovars, such as Salmonella Newport and Salmonella Infantis have increasingly been detected in many countries in association with foodborne outbreaks (Bayer et al., 2014; Angelo et al., 2015). Salmonella Newport ranks among the top six serovars in the North American, European, and Latin American regions (Hendriksen et al., 2011); appears to be the most common serotype isolated from patients with diarrhea in some areas of China (Deng et al., 2012); and exhibits a high antimicrobial resistance rate in several countries (Espie et al., 2005; Centers for Disease Prevention, 2008 and Centers for Disease and Prevention, 2013). Additionally, immune cross-protection between serovar variants of S. enterica is limited due to the expression of different immunodominant O-antigens by these pathogens (Hormaeche et al., 1996). Thus, an ideal vaccine with broad serovar coverage should be developed to prevent diverse NTS serovar infections worldwide (Mahan et al., 2012); however, no licensed human NTS vaccines currently exist, and most human NTS vaccines in preclinical or clinical stages, such as the live vaccines CVD1921 and CVD1941 and the O-antigen-conjugated vaccine O:4,12-TT, are being explored for a single serovar, mainly Typhimurium or Enteritidis (MacLennan et al., 2014; Tennant and Levine, 2015). In addition, current commercial Salmonella vaccines used in animals exhibit efficacy but have limited cross-protection (Desin et al., 2013).
Oral live NTS vaccines can confer protection against salmonellosis by inducing both cell-mediated and humoral immune responses. To overcome the limitations of Salmonella vaccines, considerable efforts have been made to develop live attenuated vaccines with cross-protection. One strategy is the construction of live vaccines by mutating global regulators, leading to the overexpression of potential antigens that may be shared among heterologous serovars. The Salmonella Typhimurium double mutant Δcya Δcrp induces cross-protection against heterologous serovars, including Salmonella Enteritidis and group C Salmonella, in chickens (Hassan and Curtiss, 1994). Salmonella Typhimurium DNA adenine methylase (Dam) mutants elicit protective immune responses to homologous and heterologous challenges in murine, avian and bovine animal models (Dueger et al., 2001; Heithoff et al., 2008; Mohler et al., 2008). However, the protection efficacies conferred by these vaccines against heterologous challenges are not complete and are not as good as their protective effects against the homologous strain (Dueger et al., 2001; Mahan et al., 2012; Heithoff et al., 2015). The other strategy is the introduction of mutations in genes required for lipopolysaccharide (LPS) or enterobacterial common antigen (ECA) biosynthesis, such as rfaH (Nagy et al., 2008), rfbB and rffG (Huang et al., 2016) and wecA (Bridge et al., 2015), resulting in the exposure of conserved outer membrane proteins (OMPs) to the host immune system. Likewise, the resulting vaccine strains, which exhibit a truncated LPS or ECA phenotype, can induce improved cross-immunity to heterologous bacteria but cannot provide full protection against heterologous serovars in most cases (Bridge et al., 2015; Huang et al., 2016). Thus, new approaches are required to develop multivalent Salmonella vaccines.
LPS O-antigens, composed of repeat-unit polysaccharides, are quite variable and determine the serogroup of Salmonella. Serovars in the same serogroup share the same dominant O-antigen epitope (Reeves et al., 2013). O-antigen-specific antibodies mediate strong protection against several homologous serovars of S. enterica, which is evidenced by O:4-specific IgG- or IgA-based adoptive transfer experiments (Colwell et al., 1984; Michetti et al., 1992; Forbes et al., 2012) and the potent efficacies displayed by O-antigen-conjugated vaccines, such as O:9-flagellin, O:4,12-TT, and O:4,5/O:9-CRM197 in animal models (Simon and Levine, 2012). Attenuated live Salmonella strains, which are ideal vaccine vectors with unparalleled merits, have been extensively used to deliver heterologous antigens from various pathogens and have been shown to induce protective responses against lethal challenges (Roland and Brenneman, 2013). These vaccine vectors are easy to handle and manipulate genetically and can be administered orally to both human and farm animals without the need for needles, inducing long-lasting systematic and mucosal immune responses against not only heterologous antigens but also the Salmonella carrier itself while causing few or no side effects when attenuated appropriately by deleting one or two key genes that are essential for virulence. In the past several decades, a variety of technologies, such as Asd (aspartate semialdehyde dehydrogenase)-based balanced-lethal vector-host systems and arabinose-dependent regulated delayed attenuation, have been exploited in Salmonella Typhimurium to enhance its efficacy as a vaccine vector (Wang et al., 2013). Asd is responsible for the synthesis of diaminopimelic acid (DAP), which is an essential component for the biosynthesis of cell walls in Salmonella; cell lysis will occur in its absence. The balanced-lethal host-vector system based on the essential bacterial gene for Asd has been developed to deliver heterologous antigens in a vaccine strain with the asd gene deleted from the chromosome using a recombinant plasmid carrying the wild-type asd gene to establish a complementation heterozygote. The absence of DAP in mammalian tissues drives Salmonella vaccine strains with an asd mutation to stably maintain Asd+ plasmids in vivo. This system possesses a safety advantage by avoiding drug-resistance gene markers in the development of recombinant vaccines. Additionally, a tightly regulated araC PBAD activator-promoter has been frequently applied to replace upstream promoter sequences for O-antigen synthesis genes, making O-antigen expression dependent on arabinose provided during in vitro growth, which achieves regulated delayed attenuation. We hypothesized that immunization with a recombinant Salmonella Typhimurium vaccine strain stably expressing heterologous O-antigens from other serovars using the balanced-lethal vector-host system could provide protection against both homologous and heterologous Salmonella infections.
To address this issue, we attempted to construct recombinant Salmonella Typhimurium strains with heterologous expression of Salmonella Newport O-antigen. The genes specific for Salmonella O-antigen synthesis are generally present as a gene cluster in the chromosome, called the O-antigen gene cluster, which maps between the galF and gnd genes and is the major determinant of differences among the diverse O-antigen forms (Liu et al., 2014). The O-antigen gene cluster of Salmonella Newport includes 19 genes, among which the wzx-wbaR-wbaL-wbaQ-wzy-wbaW-wbaZ (wzx-wbaZ) gene sequences are distinct from or absent in the O-antigen gene cluster of Salmonella Typhimurium. Thus, we constructed a recombinant low-copy Asd+ plasmid, pCZ11, containing this subset of the Salmonella Newport O-antigen gene cluster (wzx-wbaZ) for heterologous O-antigen expression and introduced it into three Salmonella Typhimurium Δasd mutant strains: SLT19 (ATCC14028 Δasd), with a smooth LPS phenotype; SLT20 (ATCC14028 ΔasdΔrfbN), with a rough LPS phenotype; and SLT22 (ATCC14028 Δasd ΔrfbN ΔpagL::T araC PBAD rfbN), with an arabinose-dependent LPS phenotype. This resulted in three recombinant strains expressing the heterologous Salmonella Newport O-antigen, SLT19 harboring the plasmid pCZ11 [herein termed SLT19 (pCZ11)], SLT20 (pCZ11), and SLT22 (pCZ11). The aim of the asd gene deletion was to ensure the stability of plasmid pCZ11 in the Salmonella Typhimurium Δasd strains. rfbN was selected for gene deletion and gene regulation by the arabinose-dependent araC PBAD promoter because it encodes a rhamnosyl transferase, which is required for the synthesis of Salmonella Typhimurium O-antigen but not Salmonella Newport O-antigen (Reeves et al., 2013). As the rfbN gene is located within the O-antigen gene cluster, the arabinose regulated gene cassette, T araC PBAD rfbN, was inserted into the pagL site to achieve expression of Salmonella Typhimurium O-antigen in an arabinose-dependent manner. The deletion of pagL does not alter Salmonella virulence (Kong et al., 2010). Since O-antigens are associated with bacterial survival and virulence (Kong et al., 2011), we determined the effects of heterologous O-antigen expression on several Salmonella Typhimurium biological activities, including swimming, sensitivities to polymyxin B and sodium deoxycholate (DOC), colonization and virulence. The cAMP-receptor protein (CRP) is a global regulator of a number of genes involved in the uptake and utilization of carbon sources, flagellum synthesis, OMPs, etc., after binding to cAMP in Escherichia coli (E. coli) and Salmonella Typhimurium (Lawson et al., 2004). Salmonella with mutations in crp alone or in combination with other genes were shown to be greatly attenuated in virulence and could serve as effective vaccine candidates against salmonellosis (Chu et al., 2007). Thus, for attenuation and vaccine construction, we deleted the crp gene from the three recombinant strains and ultimately evaluated the immunogenicity and protective efficacy of the attenuated vaccine strains in BALB/c mice.
Materials and Methods
Ethics Statement
The treatment of animals in this study was in accordance with the Guide for the Care and Use of Laboratory Animals from the Ministry of Science and Technology of China. All animal protocols were approved by the Animal Ethics Committee at Sichuan Agricultural University and the Sichuan Administration Committee of Laboratory Animals under protocol number SYXK2014-187.
Bacterial Strains, Plasmids, Media, and Growth Conditions
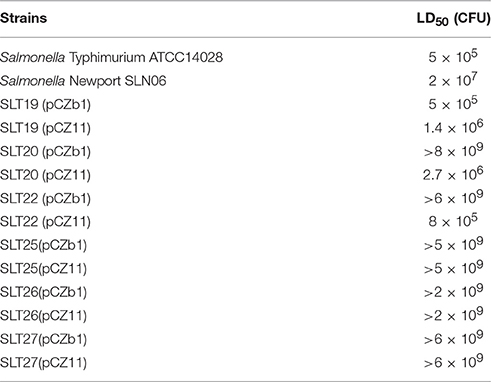
The bacterial strains and plasmids used are listed in Table 1. The recombinant Salmonella Typhimurium strains were derived from the highly virulent ATCC14028 strain. The wild-type Salmonella Newport SLN06 strain was isolated from fecal samples from a duck farm in China and was virulent, with an oral LD50 of 2 × 107 CFU in BALB/c mice (Table 2). All bacterial strains were grown in Luria-Bertani (LB) broth or on LB agar with or without 0.1% arabinose. When required, antibiotics and supplements were added at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; and diaminopimelic acid (DAP), 50 μg/ml. DAP is essential for the growth of the Salmonella Typhimurium Δasd mutant. LB agar without NaCl and containing 10% sucrose was used for sacB gene-based counterselection in the allelic exchange experiments. The transformation of E. coli and Salmonella Typhimurium was performed via electroporation. Transformants were selected on LB agar plates containing the appropriate antibiotics.
Plasmid Construction
The primers used in this study are listed in Table S1. The Asd+ plasmid pCZb1 was constructed according to the plasmid pYA3337 (Baek et al., 2009). To construct the plasmid pCZb0, the primer pair pSC101-2F and pSC101ori-R was used to amplify a DNA fragment containing the lambda t0 terminator and pSC101 origin from the plasmid pKS011 (Sekar et al., 2016), and the primers Pasd-F and pSC101-2R were used to amplify the asd gene cassette from the ATCC14028 genome. The two fragments were then joined by PCR using the primers pSC101-2F and pSC101-2R. The product was then assembled by incubating at 50°C for 1 h with a DNA fragment containing the TIT2 terminator, Ptrc promoter, sacB gene cassette and ampicillin-resistance cassette cloned from the plasmid pTRC-LIC with the primers pSC101-1F and pSC101-1R to form the plasmid pCZb0 using a Gibson Assembly Kit (NEB, Beverley, MA, USA). The pKS011 and pTRC-LIC plasmids were gifts from Keith Tyo (Addgene plasmid #65464) and Cheryl Arrowsmith (Addgene plasmid #62343), respectively. Next, to remove the sacB gene cassette from plasmid pCZb0, the DNA fragment containing the TIT2 terminator and ampicillin-resistance cassette cloned from pCZb0 with the primers TIT2-F and pSC101-1R was assembled with the DNA fragment containing the lambda t0 terminator, pSC101 origin, asd gene cassette and Ptrc promoter cloned from pCZb0 by the primers pSC101-2F and Ptrc-R using a Gibson Assembly Kit (NEB), forming the plasmid pCZb1. Furthermore, to express the Salmonella Newport O-antigen in Salmonella Typhimurium, the recombinant plasmid pCZ11 was constructed based on pCZb1. In brief, the primers C2-O-antigen-F and C2-O-antigen-R were used to amplify the gene cluster wzx-wbaR-wbaL-wbaQ-wzy-wbaW-wbaZ (7,547 bp) from the Salmonella Newport ATCC27869 genome, and the primers pCZb1-C2F and pCZb1-C2R were used to amplify the entire coding sequence of plasmid pCZb1. Finally, the two PCR products were joined using a Gibson Assembly Kit (NEB), generating the recombinant plasmid pCZ11. The plasmid construction is depicted in Figure S1.
Construction of Bacterial Mutant Strains and Recombinant Strains
Salmonella Typhimurium mutant strains were constructed by allelic exchange using the suicide vector pRE112, which carries a chloramphenicol resistance gene and the sucrose-sensitivity gene sacB (Edwards et al., 1998), a gift from Dieter Schifferli (Addgene plasmid #43828), as previously described (Zhou et al., 2014). To delete the rfbN gene, the primer pairs DrfbN-1F/DrfbN-1R and DrfbN-2F/DrfbN-2R were used to amplify the regions ~400 bp upstream and downstream of the ATCC14028 genome, respectively. The two fragments were then joined by PCR using the primers DrfbN-1F and DrfbN-2R. The resulting PCR product was digested with KpnI and XmaI and ligated into plasmid pRE112 that had been digested with the same enzymes to generate plasmid pCZ13, which carries a deletion of the entire rfbN gene sequence. pCZ13 was subsequently introduced into Salmonella Typhimurium ATCC14028 by E. coli SM10λpir (Rubires et al., 1997) via conjugation. Recipient cells were cultured on LB agar supplemented with chloramphenicol to select for a trans-conjugant strain of ATCC14028 (pCZ13) that contained pCZ13 integrated into the Salmonella genome after a single crossover. Next, a colony of ATCC14028 (pCZ13) was grown in LB to allow for a second crossover. After overnight growth, the ATCC14028 (pCZ13) culture was plated on LB agar containing 10% (w/v) sucrose for the counterselection of colonies that lost the pRE112 vector carrying the counterselectable marker, sacB. Expression of sacB in Salmonella is lethal in the presence of sucrose (Edwards et al., 1998). The colonies that grew on the LB agar containing sucrose were then tested for chloramphenicol sensitivity to ensure loss of the plasmid. The rfbN mutants were finally confirmed by PCR using the primers DrfbN-1F and DrfbN-2R. The same method was used to delete the asd, pagL, and crp genes. To construct the arabinose-regulated mutants, the primers rfbN-F and rfbN-R were used to amplify the rfbN gene from the ATCC14028 genome, and the primers T araC PBAD-F and T araC PBAD-R were used to amplify the T araC PBAD DNA fragment from the plasmid BBa_J72113-BBa_J72152 (Kittleson et al., 2012), a gift from Christopher Anderson (Addgene plasmid #40784). The two fragments were then joined by PCR using primers T araC PBAD-F and rfbN-R. The product was inserted into the NotI and Sbf I double-digested pCZ14 (pRE112-ΔpagL) plasmid, generating the plasmid pCZ15, which was introduced into the SLT21 strain (ATCC14028 Δasd ΔrfbN ΔpagL) to insert TaraCPBAD rfbN into the pagL gene site, generating the SLT22 strain. In addition, for heterologous O-antigen expression, pCZ11 and the control plasmid pCZb1 were individually transformed into Salmonella Typhimurium SLT19 (Δasd), SLT20 (Δasd ΔrfbN) and SLT22 (Δasd ΔrfbN ΔpagL::T araC PBAD rfbN), generating three recombinant strains, SLT19 (pCZ11), SLT20 (pCZ11), and SLT22 (pCZ11), and three control strains, SLT19 (pCZb1), SLT20 (pCZb1), and SLT22 (pCZb1). The Asd+ plasmid and Salmonella Typhimurium Δasd mutant constituted the balanced-lethal host-vector system. Loss of the plasmid is lethal for the Δasd mutant.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE), Silver Staining and Immunoblot Analysis
To confirm the expression of Salmonella Typhimurium and Salmonella Newport O-antigens, silver staining and Western immunoblot analyses were performed as previously described (Xu et al., 2002; Digiandomenico et al., 2004). Whole-cell lysates of Salmonella organisms were separated on 12% (w/v) acrylamide gels using a Tricine-SDS buffer system (Bio-Rad Laboratories, California, USA). The gels were then silver-stained or analyzed by Western blotting with Salmonella O:4-specific or O:8-specific antiserum diluted to 1:200 (Tianjin Biochip Corporation, Tianjin, China).
Swimming and Sensitivity Assays
Salmonella Typhimurium strains were grown to an OD600 of 0.8–0.9 in LB broth with or without 0.1% arabinose, harvested, and washed in phosphate-buffered saline (PBS). For the swimming assay, 6 μl of bacterial suspension at a concentration of 1 × 108 CFU/ml was spotted onto the middle of an LB plate solidified with 0.25% agar and supplemented with or without arabinose. The colony diameters were measured after incubation at 37°C for 6 h. For the sensitivity assays, 100 μl of a 100-fold-diluted bacterial culture (~1 × 106–5 × 106 CFU) was inoculated with or without polymyxin B at a final concentration of 0.12 μg/ml or DOC at a final concentration of 4 mg/ml for 1 h at 37°C. The bacteria were diluted to the appropriate concentration and plated onto LB plates. The survival rate was calculated as the CFU per ml of the polymyxin B- or DOC-treated group divided by the CFU per ml of the non-treated group. Each experiment was repeated three times.
Determination of Bacterial Colonization and Virulence (50% Lethal Dose, LD50) in Mice
Six-week-old female BALB/c mice were obtained from Dashuo Experimental Animal, Ltd. (Chengdu, China) and acclimated for 7 days after arrival before inoculation. The colonization ability and LD50 of Salmonella Typhimurium strains were determined as previously described (Kong et al., 2011). In brief, BALB/c mice were inoculated orally with 20 μl of buffered saline with gelatin (BSG) containing 1 × 109 CFU of bacteria. For arabinose-regulated strains, 0.1% arabinose was added to the culture medium before inoculation. Six days after oral inoculation, four animals per group were euthanized by CO2 asphyxiation, and tissues including the Peyer's patches (PPs), spleen and liver were collected. The weight of each sample was measured, and the samples were homogenized in a total volume of 1 ml of BSG. Then, appropriate dilutions were plated onto MacConkey agar or LB agar to determine the number of viable bacteria. The colonization value was calculated as CFU per gram of tissue (CFU/g). To measure the LD50 of Salmonella Typhimurium strains, stepwise increasing doses of Salmonella Typhimurium strains in 20-μl bacterial suspensions were orally inoculated into groups of BALB/c mice (6 mice per group). For arabinose-regulated strains, 0.1% arabinose was added to the culture medium before inoculation. The mice were observed for 1 month after infection, and the mortality rates were recorded to calculate the LD50 of the strains using the method of Reed and Muench. To minimize suffering, all animals that displayed extreme signs of moribundity, including shallow breathing, shaking, unresponsiveness to touch, and an inability to move and obtain food and water, were immediately euthanized by CO2. The carcasses of dead animals were sterilized, enclosed and delivered to the laboratory animal center of Sichuan Agricultural University for bio-safety disposal.
Plasmid Stability of Recombinant Salmonella Vaccine Strains
The plasmid stability in vitro was determined as previously described (Xin et al., 2012). An overnight culture of Salmonella Typhimurium strains carrying Asd+ vector pCZ11 was diluted 1:100 into fresh LB medium containing DAP (non-selective media) for 12 h of incubation with rotation at 37°C (T0). Then, serial dilutions of the culture were plated on LB plates containing DAP. The process described above was repeated every 12 h five times, and the culture from the final passage was considered T5 (~50 generations). Before each passage, a sample from the culture was diluted and plated onto LB plates containing DAP. Then, 100 single colonies from overnight growth were selected and streaked on LB agar plates without DAP (selective media) and on LB agar plates containing DAP (non-selective media). Plasmid stability was determined as the percentage of the 100 selected colonies that grew on selective media after each of the five passages.
The plasmid stability in vivo was determined as previously described (Gahan et al., 2007). Groups of BALB/c mice (12 mice/group) were inoculated orally with ~1 × 109 CFU of Salmonella Typhimurium strains carrying Asd+ vector pCZ11. Four mice in each group were euthanized by CO2 asphyxiation, and PPs were collected at 3, 6, and 9 days post-infection. The PPs were weighed and homogenized in a total volume of 1 ml of BSG. Then, to determine the number of viable bacteria, appropriate dilutions were plated onto both LB agar plates without DAP (selective media) and LB plates containing DAP (non-selective media).
Immunization and Protection in Mice
The Salmonella Typhimurium vaccine strains SLT25 (pCZb1), SLT25 (pCZ11), SLT26 (pCZb1), SLT26 (pCZ11), SLT27 (pCZb1), and SLT27 (pCZ11) were grown statically overnight in LB broth at 37°C. For arabinose-regulated strains, 0.1% arabinose was added to the culture medium. The following day, 1 ml of the overnight culture was inoculated into 100 ml of LB broth (arabinose was added to the arabinose-regulated strain cultures) and grown with shaking at 37°C to an OD600 of 0.8–0.9. The cells were harvested by centrifugation at 4,500 rpm for 10 min and resuspended in 1 ml of BSG. Groups of mice (16 mice per group) were orally inoculated with 20 μl of BSG containing 1 × 109 CFU of each strain or with BSG without bacteria and were boosted with the same dose of each strain 4 weeks later. Serum was collected from 8 mice in each group via tail-vein bleeds or retroorbital venous plexus at 3 weeks after each immunization. One month after the second immunization, the mice in each group were challenged by oral inoculation with at least 100 × LD50 of the Salmonella Typhimurium virulent strain ATCC14028 or Salmonella Newport virulent strain SLN06. PP, spleen, and liver tissues were collected from 4 mice in each group 6 days post-challenge, and the bacterial count in each tissue was measured. The mortality of the remaining 12 mice was recorded daily for 21 days.
Antigen Preparation and Quantitative Enzyme-Linked Immunosorbent Assay (ELISA)
Salmonella Typhimurium LPS was purchased from Sigma (St. Louis, MO, USA), and Salmonella Newport LPS was extracted and purified as previously described (Rezania et al., 2011). Serum IgG against Salmonella Typhimurium LPS or Salmonella Newport LPS was measured using a quantitative ELISA as previously described (Huang et al., 2016). In brief, a 96-well ELISA microplate was coated with 100 ng/well LPS or 100 ng/well goat anti-mouse Ig(H+L) (BD, San Diego, CA) in PBS and then blocked with 2% BSA (BD) for 2 h at room temperature after an overnight incubation at 4°C. The serum samples were diluted to 1:100 in PBS containing 2% BSA, and 100 μl of this solution was added to the LPS-coated wells in triplicate. Meanwhile, serial 2-fold dilutions of mouse IgG (BD) in 100 μl, starting from 0.5 mg/ml, were added to the goat anti-mouse Ig(H+L)-coated wells to generate a standard curve. After 1 h of incubation at 37°C, 100 μl of biotinylated goat anti-mouse IgG (Southern Biotech, Birmingham, AL, USA), biotinylated goat anti-mouse IgG1 (Southern Biotech) or biotinylated goat anti-mouse IgG2a (Southern Biotech) and 100 μl of a streptavidin-alkaline phosphatase conjugate (Southern Biotech) were added to each well sequentially followed by p-nitrophenyl phosphate substrate in diethanolamine buffer. Finally, the plate was read at 405 nm using a microplate reader (Bio-Rad Laboratories). The standard curve was drawn using Curve Expert (Hyams DG, Starkville, MS, USA), and the concentration of serum antibodies was calculated according to the standard curve.
Serum Bactericidal Assay
The serum collected from mice in each immunized group at week 3 post-second immunization was pooled for serum bactericidal assays as described previously (Rondini et al., 2015). Commercially available Salmonella O:4-specific and O:8-specific antisera (Tianjin Biochip Corporation) were used as positive controls. All serum samples were heated at 56°C for 30 min to inactivate endogenous complement. Salmonella Typhimurium ATCC14028 or Salmonella Newport SLN06 were grown in LB medium to log phase and were diluted in SBA buffer (50 mM phosphate, 0.041% MgCl2·6H2O, 33 mg/ml CaCl2, and 0.5% BSA) to ~3 × 103 CFU/ml. The bacteria were incubated with 25% heat-inactivated serum supplemented with or without active guinea pig complement (Sigma) for 1.5 h. The relative survival was calculated as the percent CFU counted in each pooled serum sample with active complement compared to the CFU of the same serum with no complement. Each sample and control was tested in triplicate.
Statistical Analysis
The data are shown as the means ± SD. One-way ANOVA followed by Tukey's multiple-comparison test was used to evaluate statistical significance. A probability value of P < 0.05 was considered statistically significant. The correlation between serum IgG concentrations post-immunization and bacterial loads post-challenge was analyzed using Spearman's rank correlation test in GraphPad Prism (GraphPad Software, California, USA). All in vitro experiments were repeated at least three times, and the in vivo experiments were repeated twice.
Results
Characterization of the Constructed Recombinant Salmonella Typhimurium Strains
To express the heterologous Salmonella Newport O-antigen in Salmonella Typhimurium, the control Asd+ plasmid pCZb1 and the recombinant Asd+ plasmid pCZ11 carrying the O-antigen gene cluster (wzx-wbaZ) of Salmonella Newport ATCC27869 were constructed first. Maps of the two plasmids are shown in Figure S1. To maintain the Asd+ plasmid in Salmonella Typhimurium, an ATCC14028 Δasd mutant termed SLT19 was constructed; to block expression of the homologous Salmonella Typhimurium O-antigen, the rfbN gene, which encodes the rhamnosyl transferase that is responsible for the addition of rhamnose sugar to Und-PP-Gal of the O-antigen unit (Reeves et al., 2013), was deleted from SLT19, generating SLT20 (ATCC14028 Δasd ΔrfbN); and to regulate the expression of the homologous O-antigen by exogenous arabinose provided during in vitro growth, the rfbN promoter was replaced with the araC PBAD promoter, and the gene segment T araC PBAD rfbN was inserted into the pagL site, generating SLT22 (ATCC14028 Δasd ΔrfbN ΔpagL::T araC PBAD rfbN). Then, pCZ11 or pCZb1 was introduced into the three Δasd mutants, SLT19, SLT20, and SLT22, generating three recombinant strains, SLT19 (pCZ11), SLT20 (pCZ11), and SLT22 (pCZ11), and three control strains SLT19 (pCZb1), SLT20 (pCZb1), and SLT22 (pCZb1). The Asd+ plasmid and the Δasd mutant constituted the balanced-lethal vector-host system for the stable expression of exogenous genes.
Silver staining and Western immunoblotting were used to analyze O-antigen expression. Silver staining showed that the wild-type Salmonella Newport ATCC27869 and Salmonella Typhimurium ATCC14028 produced typical LPS ladders (Figure 1A, lanes 3 and 4) representative of the smooth LPS phenotype. As expected, the SLT19 (pCZb1), SLT19 (pCZ11) and SLT20 (pCZ11) strains displayed smooth LPS phenotypes (Figure 1A, lanes 1, 2 and 6), whereas the control strain SLT20 (pCZb1) with an rfbN mutation displayed a rough LPS phenotype (Figure 1A, lane 5). Western immunoblotting demonstrated that SLT19 (pCZ11) expressed both the homologous and heterologous O-antigens (Figures 1B,C, lane 2). Conversely, the control strain SLT19 (pCZb1) only expressed the homologous O-antigen (Figures 1B,C, lane 1). The recombinant strain SLT20 (pCZ11) only expressed the heterologous O-antigen (Figures 1B,C, lane 6), while the control strain SLT20 (pCZb1) did not express either the homologous or heterologous O-antigen (Figures 1B,C, lane 5). Additionally, the recombinant strain SLT22 (pCZ11) expressed O-antigens in an arabinose-regulated manner: both types of O-antigens were expressed when the growth medium was supplemented with arabinose (Figures 1D–F, lane 4), but only the heterologous O-antigen was expressed in the absence of arabinose (Figures 1D–F, lane 5). In contrast, the control strain SLT22 (pCZb1) expressed the homologous O-antigen, but not the heterologous O-antigen, when grown with arabinose (Figures 1D–F, lane 2) and did not express either the homologous or heterologous O-antigen when grown without arabinose (Figures 1D–F, lane 3). Thus, SLT22 (pCZ11) co-expressed two types of O-antigens in the presence of arabinose, similar to SLT19 (pCZ11), and exclusively expressed the heterologous Salmonella Newport O-antigen when grown without arabinose, similar to SLT20.
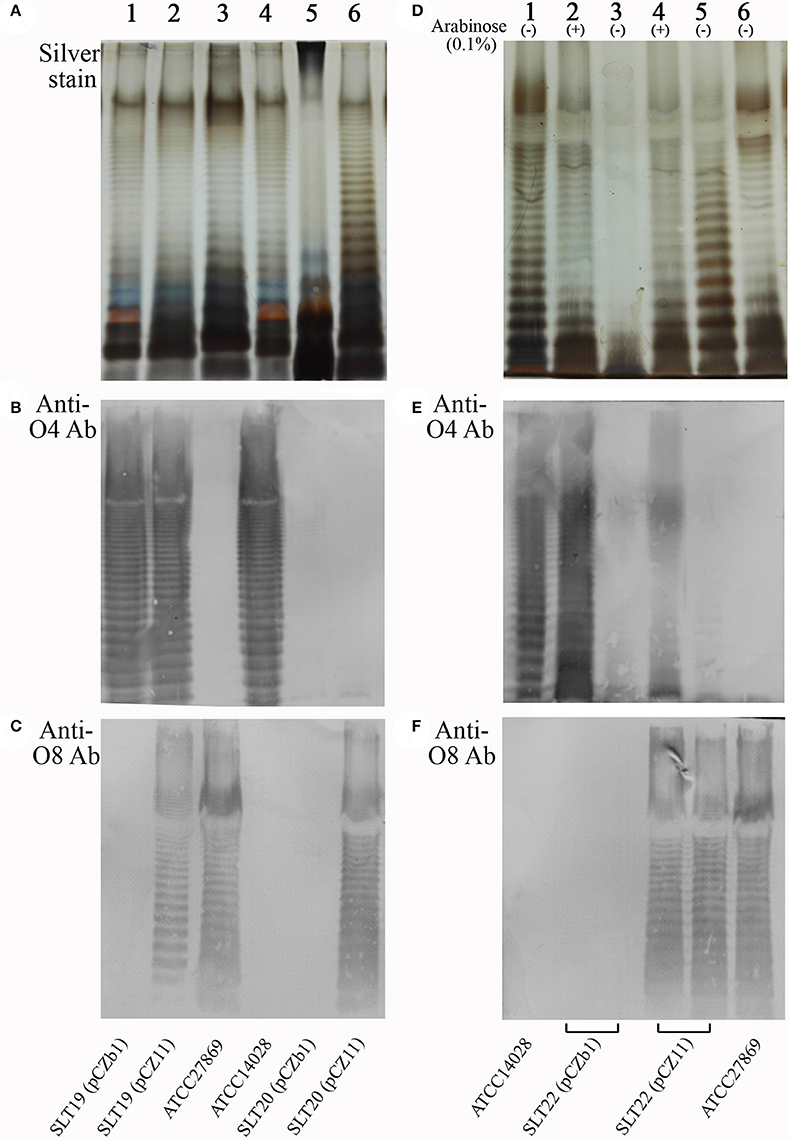
Figure 1. LPS analysis by silver staining and Western immunoblotting. (A–C) LPS extracted from SLT19 (pCZb1) (lane 1), SLT19 (pCZ11) (lane 2), ATCC27869 (lane 3), ATCC14028 (lane 4), SLT20 (pCZb1) (lane 5) and SLT20 (pCZ11) (lane 6) was subjected to SDS-PAGE followed by silver staining (A) and immunoblotting analysis using O:4-specific antiserum (B) and O:8-specific antiserum (C). (D–F) LPS extracted from ATCC14028 (lane 1), SLT22 (pCZb1) grown with arabinose (lane 2) or without arabinose (lane 3), SLT22 (pCZ11) grown with arabinose (lane 4) or without arabinose (lane 5), and ATCC27869 (Lane 6) was subjected to SDS-PAGE followed by silver staining (D) and immunoblotting using O:4-specific antiserum (E) and O:8-specific antiserum (F).
The Effects of Heterologous O-Antigen Expression on Bacterial Phenotypes and Virulence
As the O-antigen performs a key role in bacterial virulence, we evaluated whether the altered O-antigen profiles of the recombinant strains influenced the biological activities of Salmonella Typhimurium, including swimming, resistance to polymyxin B and DOC, colonization and virulence (LD50). Interestingly, compared with the parent strain SLT19 (pCZb1), SLT19 (pCZ11), SLT20 (pCZ11), and SLT22 (pCZ11) (grown with or without arabinose) exhibited decreased swimming ability, while SLT22 (pCZb1) (grown with arabinose), which had the homologous O-antigen profile, showed a similar swimming ability (Figure 2A). The control strains with the rough LPS phenotype, SLT20 (pCZb1) and SLT22 (pCZb1) (without arabinose), had very little swimming ability (Figure 2A). Compared with SLT19 (pCZb1), SLT20 (pCZ11), and SLT22 (pCZ11) (grown without arabinose), which displayed heterologous O-antigen profiles, showed slightly increased resistance to polymyxin B, but not DOC, while the strains SLT19 (pCZ11) and SLT22 (pCZ11) (grown with arabinose), which displayed chimeric O-antigen profiles, and SLT22 (pCZb1) (grown with arabinose), which displayed the homologous LPS profile, showed similar resistance to polymyxin B and DOC (Figures 2B,C). In addition, bacterial resistance to polymyxin B and DOC was dramatically reduced in SLT20 (pCZb1) and SLT22 (pCZb1) (without arabinose), which had rough LPS phenotypes, compared with that in SLT19 (pCZb1) (Figures 2B,C).
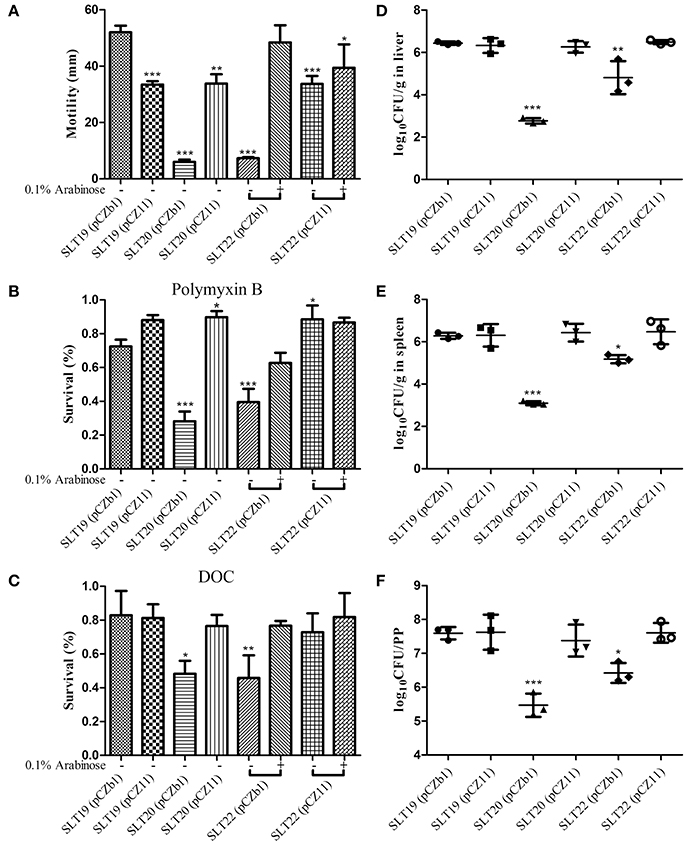
Figure 2. Biological activities and colonization of the Salmonella Typhimurium strains. (A) The swimming assay. SLT19 (pCZb1), SLT19 (pCZ11), SLT20 (pCZb1), SLT20 (pCZ11) and the two arabinose-regulated strains SLT22 (pCZb1) and SLT22 (pCZ11) were grown in LB broth with or without 0.1% arabinose. Then, 6 μl of the bacterial suspension (~1 × 108 CFU/ml) was spotted onto the middle of LB plates containing 0.25% agar with or without arabinose, and the diameter of the colonies was measured after incubation at 37°C for 6 h. (B,C) The sensitivity assays. The six Salmonella Typhimurium strains were cultured as described in (A), and 100 μl of each bacterial culture (~1 × 106–5 × 106 CFU) was inoculated with or without polymyxin B at a final concentration of 0.12 μg/ml or DOC at a final concentration of 4 mg/ml for 1 h at 37°C. The survival rate was calculated the following day. (D–F) Colonization. Groups of BALB/c mice (n = 4/group) were orally inoculated with 1 × 109 CFU of each indicated strain. Viable bacteria were recovered from the liver (D), spleen (E), and PPs (F) 6 days after infection. The number of bacteria in each tissue was calculated as log10CFU/g. The asterisk above the error bar indicates significance compared with the SLT19 (pCZb1) group. *p < 0.05, **p < 0.01, ***p < 0.001.
Regarding bacterial colonization in BALB/c mouse tissues, the recombinant strains SLT19 (pCZ11), SLT20 (pCZ11) and SLT22 (pCZ11) colonized the liver, spleen and PP at a high level that was similar to that of SLT19 (pCZb1), while the colonization level in all three tissues was significantly decreased for the SLT20 (pCZb1) and SLT22 (pCZb1) groups compared with the SLT19 group (pCZb1) (Figures 2D–F). Furthermore, the LD50 of SLT19 (pCZ11), SLT20 (pCZ11) and SLT22 (pCZ11) was 1.4 × 106, 2.7 × 106, and 8 × 105 CFU, respectively, which was similar to that of the SLT19 (pCZb1) and wild-type strain ATCC14028 (LD50 of 5 × 105 CFU). Conversely, the LD50of SLT20 (pCZb1) and SLT22 (pCZb1) declined substantially, to more than 109 CFU (Table 2).
Attenuation of the Virulence of the Recombinant Strains
The crp gene, which encodes the global regulator CRP, which is associated with the utilization of carbon sources as well as bacterial virulence (Poncet et al., 2009), was deleted from SLT19 (pCZb1), SLT19 (pCZ11), SLT20 (pCZb1), SLT20 (pCZ11), SLT22 (pCZb1), and SLT22 (pCZ11) to attenuate the virulence, generating three recombinant vaccine strains harboring the plasmid pCZ11, SLT25 (SLT19 Δcrp), SLT26 (SLT20 Δcrp), and SLT27 (SLT22 Δcrp), and three control vaccine strains, SLT25 (pCZb1), SLT26 (pCZb1), and SLT27 (pCZb1). Western immunoblotting demonstrated that SLT25 (pCZ11), SLT26 (pCZ11), and SLT27 (pCZ11) displayed similar LPS phenotypes as SLT19 (pCZ11), SLT20 (pCZ11), and SLT22 (pCZ11) (Figure S2). SLT25 (pCZ11) (Figures S2A,B, lane 3), and SLT27 (pCZ11) (when grown with arabinose) (Figures S2C,D, lane 4) co-expressed both types of O-antigens; SLT26 (pCZ11) (Figures S2A,B, lane 5), and SLT27 (pCZ11) (when grown without arabinose) (Figures S2C,D, lane 5) expressed only the heterologous O-antigen. In contrast, the control vaccine strain SLT25 (pCZb1) (Figures S2A,B, lane 2) and SLT27 (pCZb1) (when grown with arabinose) (Figures S2C,D, lane 2) only expressed the homologous O-antigen; SLT26 (pCZb1) (Figures S2A,B, lane 4) and SLT27 (pCZb1) (when grown without arabinose) (Figures S2C,D, lane 3) showed rough LPS phenotypes. Furthermore, the LD50 for each of the six strains was determined in BALB/c mice. All six strains were highly attenuated, and their LD50 values were more than 109 CFU, which was at least four orders of magnitude higher than that of the wild-type strain ATCC14028 (LD50 of 5 × 105 CFU) (Table 2).
Measurement of Plasmid Stability In vitro and In vivo
Stable maintenance of plasmid pCZ11 over some generations is an essential requirement to ensure the efficacy of recombinant vaccine strains. The stability of plasmid pCZ11 in the recombinant vaccine strains SLT25, SLT26 and SLT27 was determined in vitro by plating actively growing cultures on both selective and non-selective medium for 50 generations. As shown in Figure 3A, the percent retention of the Asd+ plasmid pCZ11 was 100% in SLT25, SLT26 and SLT27. To measure the stability in vivo, groups of BALB/c mice were orally inoculated with 109 CFU of SLT25 (pCZ11), SLT26 (pCZ11) or SLT27 (pCZ11). The plasmid stability and persistence of the bacteria in vivo were investigated in the PPs at 3, 6, and 9 days post-infection. The recovered bacteria from the PPs were diluted and plated onto LB plates and LB plates with DAP. As shown in Figures 3B–D, the bacterial loads of all the vaccine strain-infected groups calculated from the LB plates supplemented with DAP were similar to those from the plates without DAP at 3, 6, and 9 days post-infection, indicating that the plasmid pCZ11 was stably retained in the recombinant vaccine strains SLT25, SLT26, and SLT27 in vivo for 9 days.
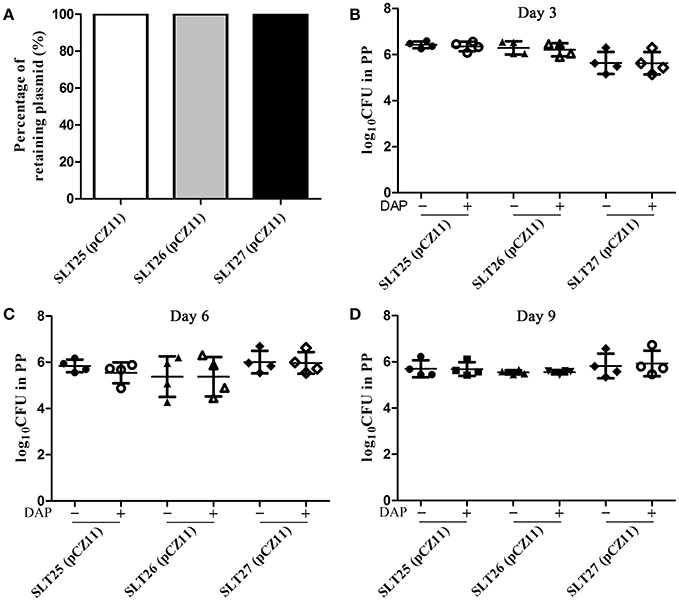
Figure 3. Stability assay of the Asd+ plasmid pCZ11 in the Salmonella vaccine strains. (A) Plasmid stability in vitro. The recombinant vaccine strains SLT25 (pCZ11), SLT26 (pCZ11), and SLT27 (pCZ11) were subjected to passage for 50 generations in LB containing DAP (non-selective medium). After the fifth passage, 100 single colonies were selected and streaked on LB agar plates without DAP (selective media) and on LB plates containing DAP (non-selective media). Plasmid stability was determined as the percentage of the 100 selected colonies that grew on the selective media. (B–D) Plasmid stability in vivo. Groups of BALB/c mice (12 mice/per group) were inoculated orally with ~1 × 109 CFU of SLT25 (pCZ11), SLT26 (pCZ11), or SLT27 (pCZ11). PPs were collected from four mice of each group at 3 days (B), 6 days (C), and 9 days (D) post-infection and were weighed and homogenized in BSG. Then, appropriate dilutions were plated onto LB agar plates without DAP (selective media) and LB plates containing DAP (non-selective media) to determine the number of viable bacteria.
Antibody Responses and Serum Bactericidal Effects after Immunization with the Generated Vaccine Strains
To evaluate the immunogenicity of the delivered O-antigens, BALB/c mice were immunized orally with ~109 CFU of each of the six vaccine strains or BSG twice at an interval of 4 weeks. ELISA results showed that compared with the control BSG group, a weak but significant serum total IgG response specific to the homologous LPS was induced in the SLT25 (pCZb1), SLT25 (pCZ11), SLT26 (pCZ11), and SLT27 (pCZb1) groups, but not the SLT26 (pCZb1) and SLT27 (pCZ11) groups, 3 weeks post-first immunization (Figure 4A). An increased total IgG response to the homologous LPS was observed in all of the vaccine groups 7 weeks post-first immunization; however, the IgG amount was much lower in the SLT26 (pCZb1) group than in the other five groups post-second immunization (p < 0.001, Figure 4A). For the heterologous LPS-specific serum IgG responses, only the vaccine strain SLT26 (pCZ11) produced significantly higher IgG levels than the BSG group 3 weeks post-first immunization, and all vaccine-immunized groups except the SLT26 (pCZb1) group produced increased IgG responses 7 weeks post-first immunization (Figure 4B). Immunization with SLT25 (pCZ11) produced a serum IgG level similar to that induced by the control SLT25 (pCZb1) strain, while the SLT27 (pCZ11) group exhibited a higher IgG level than the control SLT27 (pCZb1) group post-second immunization. Additionally, the antibody amounts detected in the SLT26 (pCZ11) and SLT27 (pCZ11) groups were higher than that detected in the SLT25 (pCZ11) group post-second immunization (Figure 4B). Furthermore, the antibody levels of serum IgG subtypes IgG2a, which is indicative of a Th1-type immune response, and IgG1, which is indicative of a Th2-type immune response, were also determined post-second immunization. Immunization with all six vaccines elicited significantly higher levels of IgG2a than IgG1 in response to the homologous O-antigens (Figure 4C). Interestingly, compared to the antibody levels in the BSG group, no significant IgG1 responses against the heterologous O-antigen were induced in the three control vaccine groups, SLT25 (pCZb1), SLT26 (pCZb1) and SLT27 (pCZb1), whereas significantly higher IgG2a levels against the heterologous O-antigen were produced in all of the vaccine-immunized groups except for the SLT26 (pCZb1) group (p < 0.001, Figure 4D). The IgG2a levels were significantly higher than the IgG1 levels in all vaccine-immunized groups except the SLT26 (pCZb1) group (Figure 4D). These results suggested that the recombinant vaccines stimulated predominantly Th1-type immune responses post-second immunization.
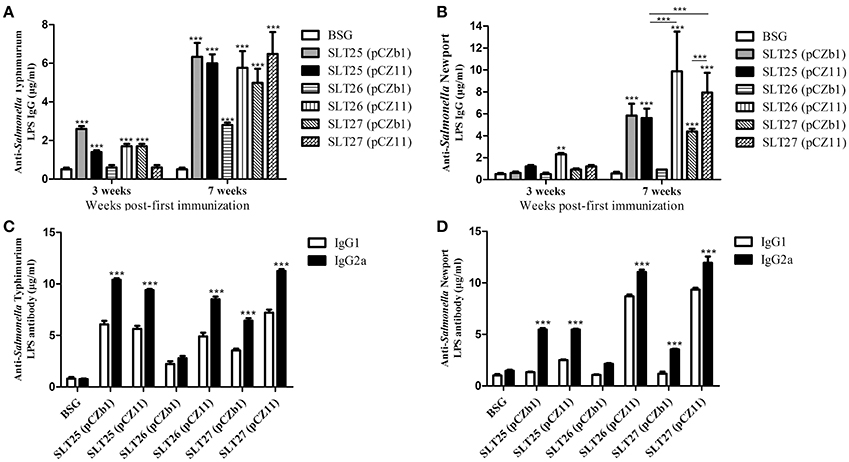
Figure 4. ELISA assay. (A,B) Serum total IgG responses. Groups of BALB/c mice were inoculated with SLT25 (pCZb1), SLT25 (pCZ11), SLT26 (pCZb1), SLT26 (pCZ11), SLT27 (pCZb1), SLT27 (pCZ11), or BSG twice at an interval of 4 weeks. Serum samples (n = 8/group) were collected at 3 weeks and 7 weeks post-first immunization. Serum IgG specific to Salmonella Typhimurium LPS (A) and Salmonella Newport LPS (B) was detected by quantitative ELISA in each group. (C,D) Serum IgG2a and IgG1 responses. Serum IgG2a and IgG1 specific to Salmonella Typhimurium LPS (C) and Salmonella Newport LPS (D) was detected by quantitative ELISA in each group at 7 weeks post-first immunization. The asterisk above the error bar indicates significance compared with the BSG control group. The asterisk above the line indicates significance between the two indicated groups. **p < 0.01, ***p < 0.001.
The serum collected from all immunized groups at 7 weeks post-first immunization were used to measure bactericidal activity against the homologous Salmonella Typhimurium strain ATCC14028 or the heterologous Salmonella Newport strain SLN06. The bacteria were incubated with 25% heat-inactivated serum plus active guinea pig complement or no complement for 1.5 h, and the relative survival was then calculated as the percent CFU in each serum with active complement compared to the CFU of the same serum with no complement. The sera of all vaccine-immunized groups elicited significant bactericidal effects against ATCC14028 compared with the BSG group (Figure 5A). The bacterial survival rates in all of the vaccine-immunized groups were much lower than that of the BSG group and were similar to that of the positive O:4-specific antiserum, and the survival rate of the SLT26 (pCZb1) group was significantly higher than that of the other five vaccine groups (p < 0.001, Figure 5A). Furthermore, the sera from all vaccine-immunized groups except for the SLT26 (pCZb1) group had significant bactericidal effects against SLN06 compared with the BSG group; however, the bactericidal levels in these vaccine groups were quite diverse. The sera from mice vaccinated with the two recombinant vaccine strains SLT26 (pCZ11) and SLT27 (pCZ11) were able to kill SLN06 at a similar level, which was comparable to that of the positive O:8-specific antiserum but much better than those of the remaining vaccine strains, SLT25 (pCZb1), SLT25 (pCZ11) and SLT27 (pCZb1) (Figure 5B).
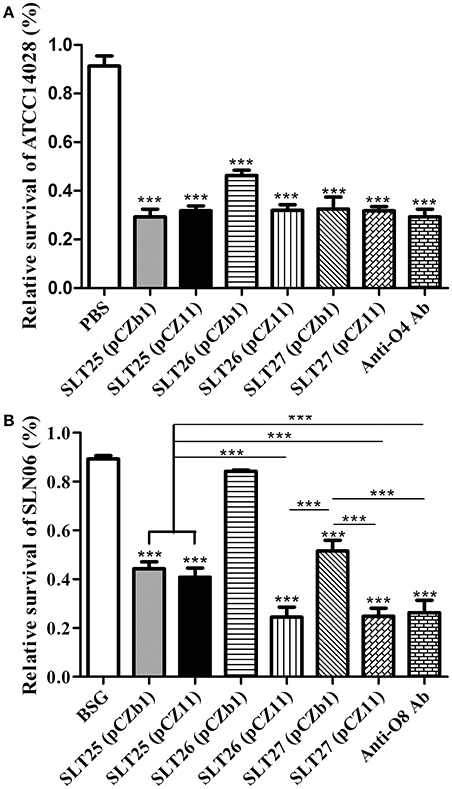
Figure 5. Serum bactericidal assay. (A,B) Salmonella Typhimurium ATCC14028 (A) or Salmonella Newport SLN06 (B) was incubated with heat-inactivated serum collected at week 7 post-first immunization from mice in each immunized group. Active guinea pig complement was added to or omitted from the mixture. The relative survival was measured at 1.5 h post-incubation. The asterisk above the error bar indicates significance compared with the BSG control group. The asterisk above the line indicates significance between the two indicated groups. ***p < 0.001.
Protection Efficacies Provided by the Vaccine Strains
The mice in each immunized group were challenged by oral inoculation with at least 100 × LD50 of the Salmonella Typhimurium virulent strain ATCC14028 or Salmonella Newport virulent strain SLN06 at 1 month post-second immunization. The bacterial loads in the mouse tissues were measured at 6 days post-challenge. After the challenge with ATCC14028, the PPs, spleen and liver from all vaccine-immunized groups had significantly fewer bacterial CFU compared with the BSG group (Figures 6A–C). The bacterial count of the SLT26 (pCZb1)-immunized group was higher than that of the other five vaccine groups in PP (Figure 6A), the SLT25 (pCZb1) and SLT27 (pCZ11) groups in the spleen (Figure 6B), and the SLT25 (pCZ11) and SLT27 (pCZb1) groups in the liver (Figure 6C). Furthermore, no mortality was observed in any of the vaccine-immunized groups, while all 12 mice in the BSG group succumbed to the challenge within 14 days (Figure 6D).
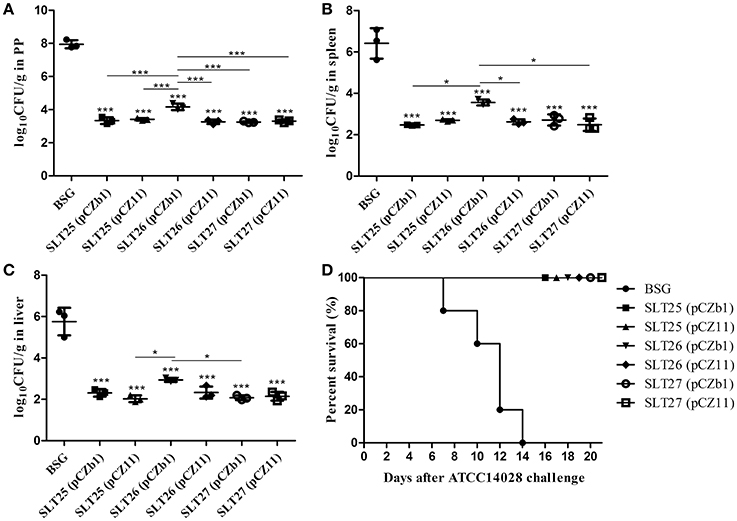
Figure 6. Bacterial loads and survival of mice post-challenge with ATCC14028. The BALB/c mice in each immunization group were orally inoculated with a lethal dose of Salmonella Typhimurium ATCC14028. Six days after oral inoculation, the PPs (A), spleen (B), and liver (C) were collected from mice in each group (n = 4/group). Then, the bacteria in the tissues were recovered and measured as log10CFU/g. Animal mortality (n = 12/group) was recorded daily for 21 days post-challenge (D). The asterisk above the error bar indicates significance compared with the BSG control group. The asterisk above the line indicates significance between the two indicated groups. *p < 0.05, ***p < 0.001.
After the SLN06 challenge, the mice immunized with SLT25 (pCZb1), SLT25 (pCZ11), SLT26 (pCZ11), SLT27 (pCZb1), and SLT27 (pCZ11), but not SLT26 (pCZb1), showed a significant reduction in the number of CFU of Salmonella Newport in the PPs, spleen and liver compared with the BSG-immunized mice. The CFU numbers in these three tissues of mice immunized with SLT25 (pCZ11) were nearly the same as those observed in the SLT25 (pCZb1)-immunized mice, while the CFU numbers in all three tissues were also significantly reduced in mice immunized with SLT27 (pCZ11) compared with the SLT27 (pCZb1)-dosed group (Figures 7A–C). In the PP, the number of CFU was lower in the SLT26 (pCZ11) group than in the SLT25 (pCZ11) and SLT27 (pCZ11) groups (Figure 7A). In the spleen and liver, the number of CFU in the SLT26 (pCZ11) group was similar to that in the SLT27 (pCZ11) group and lower than that observed in the SLT25 (pCZ11) group (Figures 7B,C). Additionally, a significant reduction in the number of CFU in the liver was observed in the group of mice immunized with SLT27 (pCZ11) compared with the group immunized with SLT25 (pCZ11) (Figure 7C). Furthermore, all 12 mice in the SLT26 (pCZ11)- and SLT27 (pCZ11)-immunized groups survived the Salmonella Newport SLN06 challenge, suggesting 100% protection. Immunization with SLT25 (pCZb1) or SLT25 (pCZ11) resulted in 50% survival, and immunization with SLT27 (pCZb1) resulted in 41.7% survival. By contrast, all of the mice in the SLT26 (pCZb1) and BSG groups succumbed to the challenge (Figure 7D).
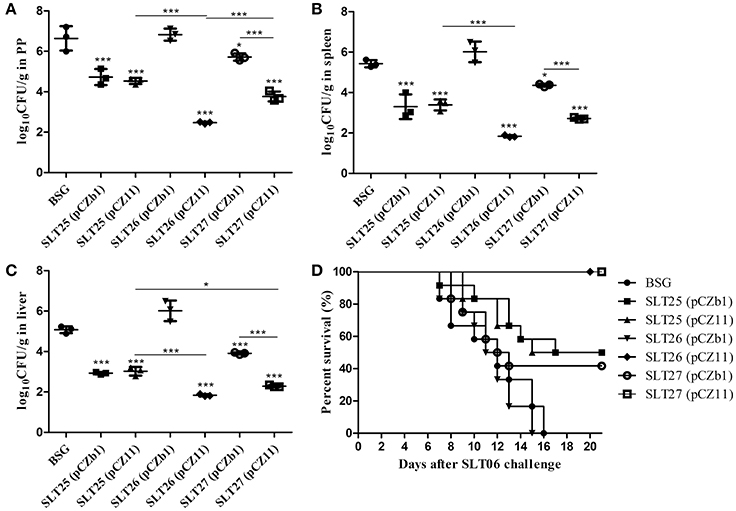
Figure 7. Bacterial load and survival of mice post-challenge with SLN06. The BALB/c mice in each immunization group were orally inoculated with a lethal dose of Salmonella Newport SLN06. Six days after oral inoculation, the PPs (A), spleen (B) and liver (C) were collected from mice in each group (n = 4/group). Then, the bacteria in the tissues were recovered and measured as log10CFU/g. Animal mortality (n = 12/group) was recorded daily for 21 days post-challenge (D). The asterisk above the error bar indicates significance compared with the BSG control group. The asterisk above the line indicates significance between the two indicated groups. *p < 0.05, ***p < 0.001.
Finally, to evaluate the role of the O-antigen-mediated antibody response in protection against lethal Salmonella Typhimurium or Salmonella Newport challenge, the correlation between the mean value of the total IgG levels against Salmonella Typhimurium LPS or Salmonella Newport LPS in each immunized group post-second immunization and the mean value of the bacterial loads of each immunized group in the PP, spleen and liver post-ATCC14028 or SLN06 challenge was analyzed using Spearman's rank correlation test. The results showed that the IgG levels against Salmonella Typhimurium LPS showed a negative but not statistically significant correlation with the bacterial load post-ATCC14028 challenge in the PP, spleen and liver (Figure 8A), with r values of −0.6429, −0.679, and −0.3571, respectively. In contrast, the IgG levels against Salmonella Newport LPS were significantly and negatively correlated to the bacterial load post-SLN06 challenge in the PP (Figure 8B, Spearman r = −0.9286, p = 0.0067), spleen (Figure 8B, r = −0.9643, p = 0.0028) and liver (Figure 8B, r = −0.9643, p = 0.0028), indicating that heterologous O-antigen-specific serum IgG acted as a functional correlate of protection against Salmonella Newport challenge.
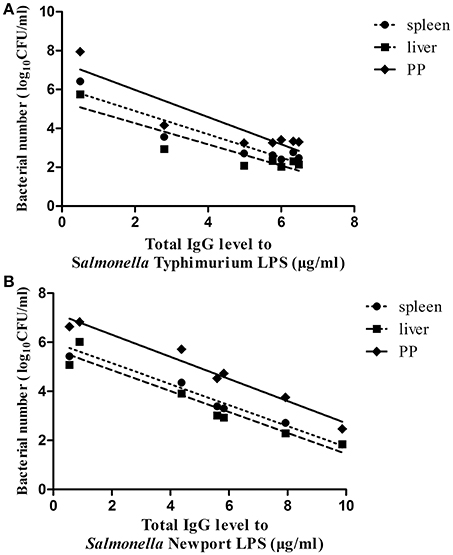
Figure 8. The correlation between serum total IgG levels post-second immunization and bacterial loads post-challenge. (A,B) Spearman's rank correlation analysis was applied to determine the correlations between the total IgG levels against Salmonella Typhimurium LPS post-second immunization and the bacterial loads in the PP, spleen and liver in all of the immunized groups post-ATCC14028 challenge (A), as well as the correlations between the total IgG levels against Salmonella Newport LPS post-second immunization and the bacterial loads in the PP, spleen and liver in all of the immunized groups post-SLN06 challenge (B). Lines represent the non-linear regression of the data, and each closed circle is the value of one of the seven immunized groups.
Discussion
NTS serovars are important causes of gastrointestinal disease in healthy people and of invasive disease in immunocompromised individuals worldwide (Majowicz et al., 2010; Feasey et al., 2012). Vaccination is one of the best forms of prophylaxis against the development of infectious disease. An effective Salmonella vaccine with broad serovar coverage is urgently needed to control infections by a variety of NTS serovars. In this study, we developed bivalent Salmonella vaccines through the recombinant expression of the heterologous Salmonella Newport O-antigen in Salmonella Typhimurium. This strategy was applied for four reasons: (1) O-antigen-specific antibodies can mediate robust protection against Salmonella Typhimurium, Salmonella Enteritidis or Salmonella Paratyphi infections (MacLennan et al., 2014), although this protective role has not been observed in Salmonella Newport infections; (2) O-antigen polymorphism is a mechanism used to evade cross-immunity between different Salmonella serovars, such as Enteritidis and Typhimurium (Hormaeche et al., 1996; Kingsley and Baumler, 2000); (3) Salmonella Typhimurium is an ideal vaccine vector for the delivery of exogenous antigens, including O-antigen, to the host immune system (Bridge et al., 2016), and a set of recombinant vaccine technologies has been developed for this serovar (Wang et al., 2013); and (4) a potential advantage of O-antigen-based vaccines is cross-protection against other serovars within the same serogroup because all serovars in one serogroup express the same dominant O-antigen epitopes, which has been demonstrated in the serogroup D serovars Salmonella Enteritidis and Salmonella Gallinarum (Chacana and Terzolo, 2006). The Asd-based balanced-lethal vector-host system was utilized to construct three bivalent Salmonella vaccines. We proved that the recombinant Asd+ plasmid pCZ11, which expressed the heterologous O-antigen, was stably retained in the three recombinant Salmonella Typhimurium vaccines in vitro and in vivo, consistent with previous reports utilizing the balanced-lethal vector-host system (Xin et al., 2012). The duration time examined in vivo for the recombinant strains carrying pCZ11 was at least 9 days, which is sufficient to stimulate protective immunity to Salmonella infection, as a previous report has demonstrated (Griffin and McSorley, 2011). Our study demonstrates that immunization with the constructed bivalent vaccine strains SLT26 (pCZ11), which has the heterologous O-antigen phenotype, and SLT27 (pCZ11), which has an arabinose-dependent O-antigen phenotype, stimulate strong serum IgG responses, including IgG2a and IgG1, against two types of O-antigen and provide full protection against lethal challenges with the homologous and heterologous Salmonella strains.
LPS O-antigen forms the outer layer of Salmonella and other gram-negative bacteria, protecting the bacteria from extreme environments, antimicrobial peptides and other host defense factors. We found that SLT20 (pCZb1) and SLT22 (pCZb1) (grown without arabinose), which displayed rough LPS phenotypes due to the rfbN mutation, were significantly defective in swimming, resistance to polymyxin B and DOC, colonization and virulence in mice, similar to findings in previous reports of a Salmonella Typhimurium ΔrfbP mutant (Kong et al., 2011) and a Salmonella Enteritidis strain with transposon insertions in the rfbN gene (Addwebi et al., 2014), confirming the essential role of O-antigen in bacterial virulence, motility, and immune evasion. Interestingly, we also found that the exclusive expression of the Salmonella Newport O-antigen in Salmonella Typhimurium decreases bacterial swimming. Flagellum-mediated motility is considered a basic function of Salmonella that contributes to its interactions with host cells during pathogenesis; however, the mechanism underlying the function of O-antigen in motility has not been fully elucidated. An early study suggested that the loss of surface O-antigen inhibits the swarming motility of Salmonella Typhimurium because it generally acts to increase the surface “wettability” required for bacterial colony expansion (Toguchi et al., 2000). Recent reports demonstrated that auto-aggregation and a defect in flagellar expression were responsible for the reduced swimming motility of a Salmonella Choleraesuis rough strain with a wzx gene deletion (Zhou et al., 2014), and the inactivation of genes involved in LPS synthesis decreases flagellar protein production in Salmonella Typhimurium (Liu et al., 2016). Thus, whether a similar mechanism for defective swimming motility occurs in the Salmonella ΔrfbN mutant and whether expression of the Salmonella Newport O-antigen has an adverse effect on flagellar expression still need to be addressed. Additionally, we demonstrated that the exclusive expression of the Salmonella Newport O-antigen in Salmonella Typhimurium has no influence on bacterial colonization and virulence in mice, indicating that O-antigen expression itself, rather than O-antigen variety, was vital for Salmonella virulence. An analogous observation was also found in a previous report in which the initial interaction between newly hatched chickens and Salmonella was found to be dependent on the presence of O-antigen but not serovar classification (Varmuzova et al., 2014).
As an ideal vaccine vector, live attenuated Salmonella vaccines have been utilized to deliver O-antigens from other pathogenic bacteria, such as Shigella sonnei (Dharmasena et al., 2013) and Burkholderia mallei (Moustafa et al., 2015), exhibiting protection against lethal challenge. However, a Salmonella vaccine strain expressing Pseudomonas aeruginosa O-antigen exhibits only partial protection against lethal challenge (Bridge et al., 2016). All of these developed Salmonella vectors possess complete homologous LPS phenotypes. The co-expression of two types of O-antigens may attenuate the immunogenicity of the heterologous O-antigen and the resulting protection potency. Therefore, to avoid this potentially negative effect in our study, in addition to the construction of SLT25 (pCZb1), which co-expressed two types of O-antigen, we also constructed two other recombinant vaccine strains, SLT26 (pCZ11), which expressed only the heterologous O-antigen, and SLT27 (pCZ11), which expressed two types of O-antigen when exogenous arabinose was supplied during in vitro growth and expressed only the heterologous O-antigen in vivo, as the expression of the homologous O-antigen ceases after Salmonella invasion due to cell division and the absence of arabinose in the gut-associated lymphoid tissues (Kong et al., 2008). Not unexpectedly, SLT26 (pCZ11) and SLT27 (pCZ11) immunization induced higher levels of serum IgG specific to the heterologous O-antigen and better bactericidal effects against Salmonella Newport than immunization with SLT25 (pCZ11). Correspondingly, in contrast to the 50% heterologous protection provided by SLT25 (pCZ11) immunization, the other two vaccines conferred full protection against Salmonella Newport infection. We speculate that the presence of the homologous O-antigen influenced the epitope exposure of the heterologous O-antigen to B-cells or decreased the amount of heterologous O-antigen expressed in SLT25 (pCZ11), resulting in its poorer immunogenicity. The results suggest that the exclusive expression of a heterologous O-antigen might be a better strategy for developing an O-antigen-based bivalent Salmonella vaccine than the co-expression of two types of O-antigen.
However, the serum IgG levels induced against the homologous LPS and their related bactericidal effects to Salmonella Typhimurium post-immunization as well as the resultant bacterial clearance levels and protection efficacies after homologous challenge were similar between the three recombinant vaccine-immunized groups. In combination with the finding that immunization with SLT26 (pCZb1), which did not express the homologous LPS, also provided full homologous protection, the results suggest that the protective role of antibodies specific to the homologous O-antigen are not indispensable for the live attenuated Salmonella vaccines. This finding is in line with previous reports based on attenuated Salmonella Typhimurium rough strains with mutations in O-antigen synthesis genes (Nagy et al., 2006) and is similar to the role of Vi-mediated antibodies in the protection against Salmonella Typhi infections, as demonstrated by the protection conferred by the commercial Vi CPS and live Ty21a vaccines without the Vi antigen (MacLennan et al., 2014). Nevertheless, the presence of O-antigen may be essential for the efficacy of inactivated Salmonella vaccines, as the sera induced by inactivated rough strain immunization were not able to kill Salmonella (Rondini et al., 2013). Immunization with live Salmonella vaccines elicits robust humoral and T cell-mediated immune responses, both of which are required for preventing Salmonella infection (Wahid et al., 2015; Zhu et al., 2017). In addition to the protective antigen O-polysaccharide, specific immune responses stimulated by other Salmonella antigens, such as flagellin proteins and OMPs are strongly associated with protection against Salmonella infection (Mastroeni and Menager, 2003; Bergman et al., 2005). Thus, these kinds of immune responses are likely responsible for the full protection conferred by the vaccine strain SLT26 (pCZb1), which had a rough LPS phenotype, or SLT26 (pCZ11), which had a heterologous O-antigen profile. The finding that the serum IgG response against homologous LPS was negatively but not significantly correlated to the bacterial loads in tissues after homologous challenge might be attributable to the same cause.
Bacterial-specific Th1 immune responses including antibodies, CD4 T cells, cytokines are essential for the clearance of disseminated Salmonella infections, and Th1-promoting vaccines are likely to help prevent these infections (Ravindran and McSorley, 2005). All three of the recombinant vaccine strains stimulated more serum IgG2a (characteristic of a Th1-type response) than IgG1 (characteristic of a Th2-type response) against the heterologous LPS post-second immunization, indicating a biased Th1-type immune response. This finding was consistent with a number of studies in which recombinant antigens expressed in oral, attenuated Salmonella vaccines induced a predominately Th1 response, as did the vector itself (VanCott et al., 1996; Pathangey et al., 2009; Moustafa et al., 2015).
Interestingly, we also found that the SLT26 (pCZ11) strain, which expressed only the heterologous O-antigen, and the SLT25 (pCZb1) and SLT27 (pCZb1) strains, which expressed only the homologous O-antigen, stimulated specific serum IgG responses to both the homologous and heterologous LPS post-second immunization, indicating that there is cross-reactivity between the antibodies specific to Salmonella Typhimurium LPS and Salmonella Newport LPS. In other words, there are epitopes common to both Salmonella Typhimurium LPS and Salmonella Newport LPS. A similar observation was also demonstrated in a previous study in which monoclonal antibody against O:5 antigen expressed by Salmonella strains from group B (O-antigenic formula, 1,4,5,12) cross-reacted with an unidentified lipopolysaccharide epitope of the Salmonella serogroup C2 (O-antigenic formula, 6,8). The cross-reactivity between antibodies against two types of LPS also resulted in cross-protection as evidenced by the finding that immunization with SLT25 (pCZb1) or SLT27 (pCZb1) expressing only the homologous O-antigen resulted in a significant reduction in the bacterial loads in mouse tissues and provided 50 or 41.7% heterologous protection efficacy upon heterologous Salmonella Newport challenge, respectively; conversely, the mice in the rough SLT26 (pCZb1)-immunized group had bacterial loads similar to those in the mice in the BSG group, and all of these mice succumbed to the heterologous challenge. A previous report showed that a Salmonella Typhimurium Δcrp Δcya mutant conferred significant but marginal protection against a challenge with group C2 serovars in chickens (Hassan and Curtiss, 1994). Our study suggests that the cross-reactivity of IgG against the LPS of Group B and Group C2 Salmonella might be partially responsible for the observed cross-protection. Finally, we also demonstrated that the amounts of serum total IgG against Salmonella Newport LPS were positively and significantly correlated to bacterial clearance after heterologous protection, proving the protective role of the Salmonella Newport O-antigen-mediated IgG response in the prevention of Salmonella Newport infection, similar to the essential role of O-antigens from extensively studied serovars such as Salmonella Typhimurium, Salmonella Enteritidis, and Salmonella Paratyphi (MacLennan et al., 2014). Therefore, the level of antibody response could become a marker for evaluating the protection efficacy of O-antigen-based vaccines against Salmonella infections.
In summary, we constructed three bivalent Salmonella vaccine strains, SLT25 (pCZ11), SLT26 (pCZ11), and SLT27 (pCZ11), with different LPS profiles by stable recombinant expression of the heterologous Salmonella Newport O-antigen in attenuated Salmonella Typhimurium. Expression of the heterologous O-antigen had no adverse effects on Salmonella Typhimurium colonization and virulence in mice but decreased bacterial swimming and susceptibility to polymyxin B. All three of the vaccine strains provided full protection against a Salmonella Typhimurium challenge. In contrast to the moderate heterologous protection provided by SLT25 (pCZ11), immunization with SLT26 (pCZ11) and SLT27 (pCZ11) conferred full protection against a Salmonella Newport challenge. Thus, the O-antigen-stimulated antibody response plays a protective role in preventing Salmonella Newport infection, and the expression of heterologous O-antigens in attenuated Salmonella Typhimurium strains is a promising strategy for the development of effective Salmonella vaccines with broad serovar coverage.
Author Contributions
XZ, QD, and AC designed the experiments, analyzed the data, and drafted the manuscript. XZ, QD, RJ, DZ, ML, MW, and SC performed the research. KS, QY, and YW participated in the animal experiments. XZ and AC edited the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31502106), the Applied Basic Research Programs of Science and Technology Department of Sichuan Province (2015JY0244) and the China Agricultural Research System (CARS-42-17).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sheng Liang, Hai-Yan Du, Xin-Yu Lei, and Yu-Xin Liu in our laboratory for generous assistance during the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fcimb.2017.00391/full#supplementary-material
References
Addwebi, T. M., Call, D. R., and Shah, D. H. (2014). Contribution of Salmonella Enteritidis virulence factors to intestinal colonization and systemic dissemination in 1-day-old chickens. Poult. Sci. 93, 871–881. doi: 10.3382/ps.2013-03710
Angelo, K. M., Chu, A., Anand, M., Nguyen, T. A., Bottichio, L., Wise, M., et al. (2015). Outbreak of Salmonella Newport infections linked to cucumbers–United States, 2014. MMWR Morb. Mortal. Wkly. Rep. 64, 144–147. doi: 10.15585/mmwr.mm655051a3
Arshad, M. M., Wilkins, M. J., Downes, F. P., Rahbar, M. H., Erskine, R. J., Boulton, M. L., et al. (2008). Epidemiologic attributes of invasive non-typhoidal Salmonella infections in Michigan, 1995–2001. Int. J. Infect. Dis. 12, 176–182. doi: 10.1016/j.ijid.2007.06.006
Baek, C. H., Wang, S., Roland, K. L., and Curtiss, III. R. (2009). Leucine-responsive regulatory protein (Lrp) acts as a virulence repressor in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191, 1278–1292. doi: 10.1128/JB.01142-08
Bayer, C., Bernard, H., Prager, R., Rabsch, W., Hiller, P., Malorny, B., et al. (2014). An outbreak of Salmonella Newport associated with mung bean sprouts in Germany and the Netherlands, October to November 2011. Euro. Surveill. 19:20665. doi: 10.2807/1560-7917.ES2014.19.1.20665
Bergman, M. A., Cummings, L. A., Alaniz, R. C., Mayeda, L., Fellnerova, I., and Cookson, B. T. (2005). CD4+-T-cell responses generated during murine Salmonella enterica serovar Typhimurium infection are directed towards multiple epitopes within the natural antigen FliC. Infect. Immun. 73, 7226–7235. doi: 10.1128/IAI.73.11.7226-7235.2005
Braden, C. R. (2006). Salmonella enterica serotype Enteritidis and eggs: a national epidemic in the United States. Clin. Infect. Dis. 43, 512–517. doi: 10.1086/505973
Bridge, D. R., Whitmire, J. M., Gilbreath, J. J., Metcalf, E. S., and Merrell, D. S. (2015). An enterobacterial common antigen mutant of Salmonella enterica serovar Typhimurium as a vaccine candidate. Int. J. Med. Microbiol. 305, 511–522. doi: 10.1016/j.ijmm.2015.05.004
Bridge, D. R., Whitmire, J. M., Makobongo, M. O., and Merrell, D. S. (2016). Heterologous Pseudomonas aeruginosa O-antigen delivery using a Salmonella enterica serovar Typhimurium wecA mutant strain. Int. J. Med. Microbiol. 306, 529–540. doi: 10.1016/j.ijmm.2016.06.005
Centers for Disease and Prevention (2008). Outbreak of multidrug-resistant Salmonella enterica serotype Newport infections associated with consumption of unPasteurized Mexican-style aged cheese–Illinois, March 2006-April 2007. MMWR Morb. Mortal. Wkly. Rep. 57, 432–435. Available online at: www.cdc.gov/mmwr/preview/mmwrhtml/mm5716a4.htm
Centers for Disease and Prevention (2013). Notes from the field: Multistate outbreak of Salmonella infantis, newport, and lille infections linked to live poultry from a single mail-order hatchery in Ohio–March-September, 2012. MMWR Morb. Mortal. Wkly. Rep. 62:213. Available online at: www.cdc.gov/mmwr/preview/mmwrhtml/mm6211a5.htm
Chacana, P. A., and Terzolo, H. R. (2006). Protection conferred by a live Salmonella Enteritidis vaccine against fowl typhoid in laying hens. Avian Dis. 50, 280–283. doi: 10.1637/7463-102705R.1
Chu, C. Y., Wang, S. Y., Chen, Z. W., Chien, M. S., Huang, J. P., Chen, J. J., et al. (2007). Heterologous protection in pigs induced by a plasmid-cured and crp gene-deleted Salmonella choleraesuis live vaccine. Vaccine 25, 7031–7040. doi: 10.1016/j.vaccine.2007.07.063
Colwell, D. E., Michalek, S. M., Briles, D. E., Jirillo, E., and McGhee, J. R. (1984). Monoclonal antibodies to Salmonella lipopolysaccharide: anti-O-polysaccharide antibodies protect C3H mice against challenge with virulent Salmonella typhimurium. J. Immunol. 133, 950–957.
Deng, X., Ran, L., Wu, S., Ke, B., He, D., Yang, X., et al. (2012). Laboratory-based surveillance of non-typhoidal Salmonella infections in Guangdong Province, China. Foodborne Pathog. Dis. 9, 305–312. doi: 10.1089/fpd.2011.1008
Desin, T. S., Koster, W., and Potter, A. A. (2013). Salmonella vaccines in poultry: past, present and future. Expert Rev. Vaccines 12, 87–96. doi: 10.1586/erv.12.138
Dharmasena, M. N., Hanisch, B. W., Wai, T. T., and Kopecko, D. J. (2013). Stable expression of Shigella sonnei form I O-polysaccharide genes recombineered into the chromosome of live Salmonella oral vaccine vector Ty21a. Int. J. Med. Microbiol. 303, 105–113. doi: 10.1016/j.ijmm.2013.01.001
Digiandomenico, A., Rao, J., and Goldberg, J. B. (2004). Oral vaccination of BALB/c mice with Salmonella enterica serovar Typhimurium expressing Pseudomonas aeruginosa O antigen promotes increased survival in an acute fatal pneumonia model. Infect. Immun. 72, 7012–7021. doi: 10.1128/IAI.72.12.7012-7021.2004
Dueger, E. L., House, J. K., Heithoff, D. M., and Mahan, M. J. (2001). Salmonella DNA adenine methylase mutants elicit protective immune responses to homologous and heterologous serovars in chickens. Infect. Immun. 69, 7950–7954. doi: 10.1128/IAI.69.12.7950-7954.2001
Edwards, R. A., Keller, L. H., and Schifferli, D. M. (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157. doi: 10.1016/S0378-1119(97)00619-7
Espie, E., De Valk, H., Vaillant, V., Quelquejeu, N., Le Querrec, F., and Weill, F. X. (2005). An outbreak of multidrug-resistant Salmonella enterica serotype Newport infections linked to the consumption of imported horse meat in France. Epidemiol. Infect. 133, 373–376. doi: 10.1017/S0950268804003449
Feasey, N. A., Dougan, G., Kingsley, R. A., Heyderman, R. S., and Gordon, M. A. (2012). Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379, 2489–2499. doi: 10.1016/S0140-6736(11)61752-2
Forbes, S. J., Martinelli, D., Hsieh, C., Ault, J. G., Marko, M., Mannella, C. A., et al. (2012). Association of a protective monoclonal IgA with the O antigen of Salmonella enterica serovar Typhimurium impacts type 3 secretion and outer membrane integrity. Infect. Immun. 80, 2454–2463. doi: 10.1128/IAI.00018-12
Gahan, M. E., Webster, D. E., Wesselingh, S. L., and Strugnell, R. A. (2007). Impact of plasmid stability on oral DNA delivery by Salmonella enterica serovar Typhimurium. Vaccine 25, 1476–1483. doi: 10.1016/j.vaccine.2006.10.042
Griffin, A. J., and McSorley, S. J. (2011). Generation of Salmonella-specific Th1 cells requires sustained antigen stimulation. Vaccine 29, 2697–2704. doi: 10.1016/j.vaccine.2011.01.078
Hassan, J. O., and Curtiss, III. R. (1994). Development and evaluation of an experimental vaccination program using a live avirulent Salmonella typhimurium strain to protect immunized chickens against challenge with homologous and heterologous Salmonella serotypes. Infect. Immun. 62, 5519–5527.
Heithoff, D. M., Enioutina, E. Y., Bareyan, D., Daynes, R. A., and Mahan, M. J. (2008). Conditions that diminish myeloid-derived suppressor cell activities stimulate cross-protective immunity. Infect. Immun. 76, 5191–5199. doi: 10.1128/IAI.00759-08
Heithoff, D. M., House, J. K., Thomson, P. C., and Mahan, M. J. (2015). Development of a Salmonella cross-protective vaccine for food animal production systems. Vaccine 33, 100–107. doi: 10.1016/j.vaccine.2014.11.012
Hendriksen, R. S., Vieira, A. R., Karlsmose, S., Lo Fo Wong, D. M., Jensen, A. B., Wegener, H. C., et al. (2011). Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog. Dis. 8, 887–900. doi: 10.1089/fpd.2010.0787
Hormaeche, C. E., Mastroeni, P., Harrison, J. A., Demarco de Hormaeche, R., Svenson, S., and Stocker, B. A. (1996). Protection against oral challenge three months after i.v. immunization of BALB/c mice with live Aro Salmonella typhimurium and Salmonella enteritidis vaccines is serotype (species)-dependent and only partially determined by the main LPS O antigen. Vaccine 14, 251–259. doi: 10.1016/0264-410X(95)00249-Z
Huang, C., Liu, Q., Luo, Y., Li, P., Liu, Q., and Kong, Q. (2016). Regulated delayed synthesis of lipopolysaccharide and enterobacterial common antigen of Salmonella Typhimurium enhances immunogenicity and cross-protective efficacy against heterologous Salmonella challenge. Vaccine 34, 4285–4292. doi: 10.1016/j.vaccine.2016.07.010
Karon, A. E., Archer, J. R., Sotir, M. J., Monson, T. A., and Kazmierczak, J. J. (2007). Human multidrug-resistant Salmonella Newport infections, Wisconsin, 2003-2005. Emerg. Infect. Dis. 13, 1777–1780. doi: 10.3201/eid1311.061138
Kingsley, R. A., and Baumler, A. J. (2000). Host adaptation and the emergence of infectious disease: the Salmonella paradigm. Mol. Microbiol. 36, 1006–1014. doi: 10.1046/j.1365-2958.2000.01907.x
Kittleson, J. T., DeLoache, W., Cheng, H. Y., and Anderson, J. C. (2012). Scalable plasmid transfer using engineered P1-based phagemids. ACS Synth. Biol. 1, 583–589. doi: 10.1021/sb300054p
Kong, Q., Liu, Q., Jansen, A. M., and Curtiss, III. R. (2010). Regulated delayed expression of rfc enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Vaccine 28, 6094–6103. doi: 10.1016/j.vaccine.2010.06.074
Kong, Q., Yang, J., Liu, Q., Alamuri, P., Roland, K. L., and Curtiss, III. R. (2011). Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect. Immun. 79, 4227–4239. doi: 10.1128/IAI.05398-11
Kong, W., Wanda, S. Y., Zhang, X., Bollen, W., Tinge, S. A., Roland, K. L., et al. (2008). Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proc. Natl. Acad. Sci. U.S.A. 105, 9361–9366. doi: 10.1073/pnas.0803801105
Lawson, C. L., Swigon, D., Murakami, K. S., Darst, S. A., Berman, H. M., and Ebright, R. H. (2004). Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 14, 10–20. doi: 10.1016/j.sbi.2004.01.012
Liu, B., Knirel, Y. A., Feng, L., Perepelov, A. V., Senchenkova, S. N., Reeves, P. R., et al. (2014). Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev. 38, 56–89. doi: 10.1111/1574-6976.12034
Liu, Q., Liu, Q., Yi, J., Liang, K., Liu, T., Roland, K. L., et al. (2016). Outer membrane vesicles derived from Salmonella Typhimurium mutants with truncated LPS induce cross-protective immune responses against infection of Salmonella enterica serovars in the mouse model. Int. J. Med. Microbiol. 306, 697–706. doi: 10.1016/j.ijmm.2016.08.004
MacLennan, C. A., Martin, L. B., and Micoli, F. (2014). Vaccines against invasive Salmonella disease: current status and future directions. Hum. Vaccin. Immunother. 10, 1478–1493. doi: 10.4161/hv.29054
Mahan, M. J., Heithoff, D. M., and House, J. K. (2012). Salmonella cross-protective vaccines: fast-forward to the next generation of food safety. Future Microbiol. 7, 805–808. doi: 10.2217/fmb.12.60
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O'Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Mastroeni, P., and Menager, N. (2003). Development of acquired immunity to Salmonella. J. Med. Microbiol. 52(Pt 6), 453–459. doi: 10.1099/jmm.0.05173-0
Michetti, P., Mahan, M. J., Slauch, J. M., Mekalanos, J. J., and Neutra, M. R. (1992). Monoclonal secretory immunoglobulin A protects mice against oral challenge with the invasive pathogen Salmonella typhimurium. Infect. Immun. 60, 1786–1792.
Mohler, V. L., Heithoff, D. M., Mahan, M. J., Walker, K. H., Hornitzky, M. A., Shum, L. W., et al. (2008). Cross-protective immunity conferred by a DNA adenine methylase deficient Salmonella enterica serovar Typhimurium vaccine in calves challenged with Salmonella serovar Newport. Vaccine 26, 1751–1758. doi: 10.1016/j.vaccine.2008.01.018
Moustafa, D. A., Scarff, J. M., Garcia, P. P., Cassidy, S. K., DiGiandomenico, A., Waag, D. M., et al. (2015). Recombinant Salmonella Expressing Burkholderia mallei LPS O Antigen Provides Protection in a Murine Model of Melioidosis and Glanders. PLoS ONE 10:e0132032. doi: 10.1371/journal.pone.0132032
Nagy, G., Danino, V., Dobrindt, U., Pallen, M., Chaudhuri, R., Emody, L., et al. (2006). Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect. Immun. 74, 5914–5925. doi: 10.1128/IAI.00619-06
Nagy, G., Palkovics, T., Otto, A., Kusch, H., Kocsis, B., Dobrindt, U., et al. (2008). “Gently rough”: the vaccine potential of a Salmonella enterica regulatory lipopolysaccharide mutant. J. Infect. Dis. 198, 1699–1706. doi: 10.1086/593069
Painter, J. A., Hoekstra, R. M., Ayers, T., Tauxe, R. V., Braden, C. R., Angulo, F. J., et al. (2013). Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998-2008. Emerg. Infect. Dis. 19, 407–415. doi: 10.3201/eid1903.111866
Parry, C. M., Thomas, S., Aspinall, E. J., Cooke, R. P., Rogerson, S. J., Harries, A. D., et al. (2013). A retrospective study of secondary bacteraemia in hospitalised adults with community acquired non-typhoidal Salmonella gastroenteritis. BMC Infect. Dis. 13:107. doi: 10.1186/1471-2334-13-107
Pathangey, L., Kohler, J. J., Isoda, R., and Brown, T. A. (2009). Effect of expression level on immune responses to recombinant oral Salmonella enterica serovar Typhimurium vaccines. Vaccine 27, 2707–2711. doi: 10.1016/j.vaccine.2009.02.072
Poncet, S., Milohanic, E., Maze, A., Nait Abdallah, J., Ake, F., Larribe, M., et al. (2009). Correlations between carbon metabolism and virulence in bacteria. Contrib. Microbiol. 16, 88–102. doi: 10.1159/000219374
Ravindran, R., and McSorley, S. J. (2005). Tracking the dynamics of T-cell activation in response to Salmonella infection. Immunology 114, 450–458. doi: 10.1111/j.1365-2567.2005.02140.x
Reeves, P. R., Cunneen, M. M., Liu, B., and Wang, L. (2013). Genetics and evolution of the Salmonella galactose-initiated set of o antigens. PLoS ONE 8:e69306. doi: 10.1371/journal.pone.0069306
Rezania, S., Amirmozaffari, N., Tabarraei, B., Jeddi-Tehrani, M., Zarei, O., Alizadeh, R., et al. (2011). Extraction, Purification and Characterization of Lipopolysaccharide from Escherichia coli and Salmonella typhi. Avicenna J. Med. Biotechnol. 3, 3–9.
Roland, K. L., and Brenneman, K. E. (2013). Salmonella as a vaccine delivery vehicle. Expert Rev. Vaccines 12, 1033–1045. doi: 10.1586/14760584.2013.825454
Rondini, S., Lanzilao, L., Necchi, F., O'Shaughnessy, C. M., Micoli, F., Saul, A., et al. (2013). Invasive African Salmonella Typhimurium induces bactericidal antibodies against O-antigens. Microb. Pathog. 63, 19–23. doi: 10.1016/j.micpath.2013.05.014
Rondini, S., Micoli, F., Lanzilao, L., Gavini, M., Alfini, R., Brandt, C., et al. (2015). Design of glycoconjugate vaccines against invasive African Salmonella enterica serovar Typhimurium. Infect. Immun. 83, 996–1007. doi: 10.1128/IAI.03079-14
Rubires, X., Saigi, F., Pique, N., Climent, N., Merino, S., Alberti, S., et al. (1997). A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179, 7581–7586. doi: 10.1128/jb.179.23.7581-7586.1997
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Sekar, K., Gentile, A. M., Bostick, J. W., and Tyo, K. E. (2016). N-Terminal-Based targeted, inducible protein degradation in Escherichia coli. PLoS ONE 11:e0149746. doi: 10.1371/journal.pone.0149746
Simon, R., and Levine, M. M. (2012). Glycoconjugate vaccine strategies for protection against invasive Salmonella infections. Hum. Vaccin. Immunother. 8, 494–498. doi: 10.4161/hv.19158
Tennant, S. M., and Levine, M. M. (2015). Live attenuated vaccines for invasive Salmonella infections. Vaccine 33(Suppl. 3), C36–C41. doi: 10.1016/j.vaccine.2015.04.029
Threlfall, E. J. (2002). Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol. Rev. 26, 141–148. doi: 10.1111/j.1574-6976.2002.tb00606.x
Toguchi, A., Siano, M., Burkart, M., and Harshey, R. M. (2000). Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182, 6308–6321. doi: 10.1128/JB.182.22.6308-6321.2000
VanCott, J. L., Staats, H. F., Pascual, D. W., Roberts, M., Chatfield, S. N., Yamamoto, M., et al. (1996). Regulation of mucosal and systemic antibody responses by T helper cell subsets, macrophages, and derived cytokines following oral immunization with live recombinant Salmonella. J. Immunol. 156, 1504–1514.
Varma, J. K., Molbak, K., Barrett, T. J., Beebe, J. L., Jones, T. F., Rabatsky-Ehr, T., et al. (2005). Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J. Infect. Dis. 191, 554–561. doi: 10.1086/427263
Varmuzova, K., Matulova, M. E., Sebkova, A., Sekelova, Z., Havlickova, H., Sisak, F., et al. (2014). The early innate response of chickens to Salmonella enterica is dependent on the presence of O-antigen but not on serovar classification. PLoS ONE 9:e96116. doi: 10.1371/journal.pone.0096116
Wahid, R., Fresnay, S., Levine, M. M., and Sztein, M. B. (2015). Immunization with Ty21a live oral typhoid vaccine elicits crossreactive multifunctional CD8+ T-cell responses against Salmonella enterica serovar Typhi, S. Paratyphi, A., and S. Paratyphi B in humans. Mucosal Immunol. 8, 1349–1359. doi: 10.1038/mi.2015.24
Wang, S., Kong, Q., and Curtiss, III. R. (2013). New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microb. Pathog. 58, 17–28. doi: 10.1016/j.micpath.2012.10.006
Xin, W., Wanda, S. Y., Zhang, X., Santander, J., Scarpellini, G., Ellis, K., et al. (2012). The Asd+-DadB+ dual-plasmid system offers a novel means to deliver multiple protective antigens by a recombinant attenuated Salmonella vaccine. Infect. Immun. 80, 3621–3633. doi: 10.1128/IAI.00620-12
Xu, D. Q., Cisar, J. O., Ambulos, N. Jr., Burr, D. H., and Kopecko, D. J. (2002). Molecular cloning and characterization of genes for Shigella sonnei form I O polysaccharide: proposed biosynthetic pathway and stable expression in a live salmonella vaccine vector. Infect. Immun. 70, 4414–4423. doi: 10.1128/IAI.70.8.4414-4423.2002
Zhou, X., Liu, B., Shi, C., and Shi, X. (2014). Mutation of a Salmonella serogroup-C1-specific gene abrogates O7-antigen biosynthesis and triggers NaCl-dependent motility deficiency. PLoS ONE 9:e106708. doi: 10.1371/journal.pone.0106708
Keywords: Salmonella typhimurium, Salmonella newport, O-antigen, bivalent vaccines, protection
Citation: Zhao X, Dai Q, Jia R, Zhu D, Liu M, Wang M, Chen S, Sun K, Yang Q, Wu Y and Cheng A (2017) Two Novel Salmonella Bivalent Vaccines Confer Dual Protection against Two Salmonella Serovars in Mice. Front. Cell. Infect. Microbiol. 7:391. doi: 10.3389/fcimb.2017.00391
Received: 21 May 2017; Accepted: 22 August 2017;
Published: 04 September 2017.
Edited by:
Alfredo G. Torres, University of Texas Medical Branch, United StatesReviewed by:
Enrique Medina-Acosta, State University of Norte Fluminense, BrazilAdam Cunningham, University of Birmingham, United Kingdom
Copyright © 2017 Zhao, Dai, Jia, Zhu, Liu, Wang, Chen, Sun, Yang, Wu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinxin Zhao, xxinzhao@sicau.edu.cn
Anchun Cheng, chenganchun@vip.163.com
†These authors have contributed equally to this work.
 Xinxin Zhao1,2,3*†
Xinxin Zhao1,2,3*†  Qinlong Dai
Qinlong Dai Dekang Zhu
Dekang Zhu Mafeng Liu
Mafeng Liu Shun Chen
Shun Chen Anchun Cheng
Anchun Cheng