Ironing Out the Unconventional Mechanisms of Iron Acquisition and Gene Regulation in Chlamydia
- 1School of Molecular Biosciences, College of Veterinary Medicine, Washington State University, Pullman, WA, United States
- 2Jefferiss Trust Laboratories, Faculty of Medicine, Imperial College London, St. Mary's Hospital, London, United Kingdom
The obligate intracellular pathogen Chlamydia trachomatis, along with its close species relatives, is known to be strictly dependent upon the availability of iron. Deprivation of iron in vitro induces an aberrant morphological phenotype termed “persistence.” This persistent phenotype develops in response to various immunological and nutritional insults and may contribute to the development of sub-acute Chlamydia-associated chronic diseases in susceptible populations. Given the importance of iron to Chlamydia, relatively little is understood about its acquisition and its role in gene regulation in comparison to other iron-dependent bacteria. Analysis of the genome sequences of a variety of chlamydial species hinted at the involvement of unconventional mechanisms, being that Chlamydia lack many conventional systems of iron homeostasis that are highly conserved in other bacteria. Herein we detail past and current research regarding chlamydial iron biology in an attempt to provide context to the rapid progress of the field in recent years. We aim to highlight recent discoveries and innovations that illuminate the strategies involved in chlamydial iron homeostasis, including the vesicular mode of acquiring iron from the intracellular environment, and the identification of a putative iron-dependent transcriptional regulator that is synthesized as a fusion with a ABC-type transporter subunit. These recent findings, along with the noted absence of iron-related homologs, indicate that Chlamydia have evolved atypical approaches to the problem of iron homeostasis, reinvigorating research into the iron biology of this pathogen.
Introduction
Being the fourth most abundant element in the Earth's crust, iron (Fe) has developed a rich evolutionary history with life on Earth (Frey and Reed, 2012). Iron is an essential micronutrient for nearly every organism with very few notable exceptions. One such exception is Borrelia burgdorferi, which is thought to substitute iron for other metals like manganese (Mn) in its metabolism (Posey and Gherardini, 2000). The requirement for iron is no less significant for the wide array of prokaryotic human pathogens, where the limitation of iron is generally lethal. This suggests biochemical intervention in iron-related pathways may be a promising medical avenue for the treatment of infectious disease. Pathogenic bacteria, like many other organisms, utilize iron in a wide array of conserved biochemical pathways, including prominent examples such as the tricarboxcylic acid (TCA) cycle and the electron transport chain (Oexle et al., 1999; Kim et al., 2012). However, in all organisms, iron dependency comes with significant problems that must be overcome. Biologically available iron is commonly in one of either two oxidative states: ferric (Fe3+) or ferrous (Fe2+) iron. Ferric iron is notorious for its insolubility at physiological pH, and therefore its biological utility is restricted unless reduced to Fe2+. Moreover, free Fe2+ can react with hydrogen peroxide in the Fenton reaction and generate hydroxyl radicals associated with cellular oxidative stress (e.g., DNA damage; Winterbourn, 1995). Indeed, perturbation of iron-dependent regulons has been demonstrated to facilitate the acquisition of antimicrobial resistance in E. coli, likely by dysregulating iron acquisition and promoting the accumulation of free iron that can participate in DNA mutagenesis (Mehi et al., 2014). Accordingly, iron-dependent organisms are presented with a tripartite challenge of acquiring iron that is in the appropriate oxidative state, in homeostatic amounts, and properly liganded to avoid collateral damage to the cell. For bacterial pathogens, this is complicated further by the important task of acquiring iron from the host. As such, pathogenic bacteria have evolved an impressive array of iron acquisition systems and iron-responsive regulatory networks to enhance their success when competing for iron. Iron acquisition and homeostasis have been extensively researched for many important prokaryotic organisms including Escherichia coli, Salmonella typhimurium, and Mycobacterium tuberculosis (Andrews et al., 2003). It is well-documented in the literature that the Chlamydiaceae are strictly iron-dependent (Raulston, 1997; Al-Younes et al., 2001; Freidank et al., 2001), but the long-standing genetic intractability of the organism has rendered many benchmark approaches to iron biology ineffectual in past chlamydial research. Consequently, our understanding of this fundamental biological process is incomplete in Chlamydiae.
The Chlamydiaceae includes many pathogens of humans and economically important animals, including Chlamydia trachomatis (genital and ocular human infections), Chlamydia pneumoniae (pulmonary human infection), Chlamydia pecorum (cattle/sheep) and Chlamydia suis (swine). Chlamydiae are Gram-negative obligate intracellular pathogens typified by a unique biphasic developmental cycle that interconverts an infectious and metabolically dormant elementary body (EB) to a vegetative and metabolically active reticulate body (RB) (AbdelRahman and Belland, 2005). Interestingly, Chlamydiae can deviate from their normal developmental cycle under stress conditions, resulting in the formation of aberrantly enlarged RB forms that fail to divide or differentiate and are distinguished by dysregulated gene expression (Wyrick, 2010). This aberrant state has been termed “persistence.” Importantly, abatement of the stress condition can rescue Chlamydia from a persistent state. Limiting the availability of iron to Chlamydiae has long been appreciated as an inducer of persistence, though the exact mechanism that facilitates this development is not understood (Raulston, 1997). Chlamydial persistence may contribute to the development of sub-acute pathologies and Chlamydia-associated chronic diseases. Only recently has bona fide in vivo evidence provided a clear example of the development of aberrant chlamydial forms in infected women, pigs and mouse models (Pospischil et al., 2009; Phillips Campbell et al., 2012; Lewis et al., 2014). These studies are far from definitive, as they represent small sample sizes and only note the presence of enlarged RB forms among inclusions of mixed populations. Regardless, it remains plausible to hypothesize that the limitation of iron, which induces persistence in vitro, may also stimulate aberrant development in vivo. Multiple lines of evidence suggest that Chlamydia may be exposed to fluctuating iron availability during infection. Specifically for C. trachomatis serovars responsible for genital infections (D-K, L1-3), fluctuations in lactoferrin concentrations in the female genital tract in response to estrogen signaling during menstruation could significantly alter iron availability to Chlamydia (Cohen et al., 1987; Kelver et al., 1996). Sequestration of iron in the mononuclear phagocyte system (MPS) may represent a host protective mechanism against Chlamydia infection by preventing iron trafficking to mucosal epithelia (Nairz et al., 2014; Ganz and Nemeth, 2015; Soares and Weiss, 2015). Iron limitation through the MPS could be especially important for Chlamydia species that invade monocytic cells, such as C. pneumoniae or C. trachomatis lymphogranuloma venereum (LGV) seorvars (L1-3) that presumably invade monocytes to proliferate to the local lymph nodes.
How Chlamydia might respond to these nutritive insults remains unclear. In the absence of genetic tools, past studies have relied on the chlamydial genome for clues in this regard. However, the chlamydial genome is relatively small, measuring only 1.04 Mb in C. trachomatis (Stephens et al., 1998). Correspondingly, only 894 open reading frames (ORFs) are predicted to be coded on the C. trachomatis chromosome. The small genome size of Chlamydiae is accounted for by their long history of co-evolution with specific host cells. This has resulted in severe reductive evolution with respect to the size and functionality of the chlamydial genomic repertoire (Clarke, 2011). Chlamydia consequently lack or possess truncated versions of many highly conserved metabolic pathways, such as amino acid and nucleotide biosynthesis. Obligate intracellular parasites are especially susceptible to the accumulation of deleterious mutations, sequence deletions and inversions as they discard functionality from their genome that can be compensated by their hosts (Andersson and Kurland, 1998). This perhaps helps to explain the fact that roughly 32% of the ORFs predicted in the genome of C. trachomatis are annotated as hypothetical, with no assigned function based on homology (Stephens et al., 1998). It is therefore no surprise that Chlamydia lack many highly conserved components of iron-related systems found in other pathogens.
In this review, we will detail our current understanding of chlamydial iron homeostasis, focusing on both acquisition and regulation, from an historical perspective contextualized with respect to what is understood in other pathogens. This will serve to starkly contrast Chlamydia against some of its most similar contemporaries. We also hope to highlight some of the primary difficulties that have stalled progress with respect to our understanding of iron homeostasis in Chlamydia. Furthermore, we will discuss some of the recent breakthroughs that we believe will facilitate a more in-depth understanding of iron-related processes in Chlamydia.
Mammalian Iron Metabolism
Iron metabolism in mammals has been extensively studied and this work has been condensed into comprehensive reviews (for reference see Andrews, 2000; Hentze et al., 2010; Winter et al., 2014). It will suffice here to briefly recount the primary mechanisms by which iron is trafficked to cells, further disseminated throughout the body and how these processes are regulated. Each of these steps represent opportunities for pathogens to siphon iron from their host or for the host to limit iron to pathogens. In humans and all other mammals, ferric iron, obtained through diet, must be reduced before transport across the intestinal epithelium. This process is dependent on a cytochrome b-like ferrireductase (Dcytb) for reduction to the ferrous state (McKie et al., 2001), and on Divalent metal transporter-1 (DMT-1; AKA Nramp2) for transport of the divalent cation into the cytoplasm of the epithelial cell (Fleming et al., 1997, 1998). DMT-1 was shown to transport several different divalent cations, including Mn2+ and Zn2+, in a proton-coupled symport mechanism (Gunshin et al., 1997). Export across the basolateral membrane of the intestinal epithelium is accomplished via ferroportin-1 (Donovan et al., 2000). Two ferroxidases, ceruloplasmin and hephaestin, are responsible for the re-oxidation of ferrous iron to the ferric form (Osaki et al., 1966; Vulpe et al., 1999; Hellman and Gitlin, 2002). This re-oxidation is important for binding by the serum carrier protein, transferrin (Tf). Tf is a high affinity Fe3+ binding protein, with an association constant of approximately 1036 for the trivalent metal at a near neutral pH (Ratledge and Dover, 2000). Approximately, 30% of serum transferrin is saturated with iron at any given time, which drives a near immediate sequestration of ferric iron in plasma (Crichton, 2016). Conformational changes occur upon the diferric saturation of transferrin, which, at neutral pH, allows the specific recognition of the diferric-holotransferrin (dfTf) over apo-transferrin at the cell surface by transferrin receptors (TfR) (Cheng et al., 2004). Once bound, these dfTf-TfR complexes localize to clathrin-coated pits and are endocytosed (Harding et al., 1983). In the early endosome, proton pumps cause the decrease of vesicular pH, destabilizing the iron-transferrin complex. While the apo-transferrin-TfR complex is not stable at a neutral pH, at an acidic pH, this complex remains intact and is recycled back to the plasma membrane, where the return to neutral pH causes the dissociation of the apo-Tf, starting the cycle anew (Dautry-varsat et al., 1983). The ferric iron released in the early endosome must be reduced before its transport into the cytosol by DMT-1 (Fleming et al., 1998). This reduction is accomplished by another membrane bound ferrireductase, STEAP3 (Ohgami et al., 2005).
Once in the cytosol, iron is either immediately incorporated into iron utilizing proteins (termed the labile iron pool) or stored by the ferritin complex. Ferritin storage complexes exist as multimers, which are composed of two subunits, designated the heavy (H) and light (L) chains. These subunits surround an iron core that may contain up to 4,500 iron atoms (Harrison and Arosio, 1996; Hentze et al., 2004). The H-subunit possesses ferroxidase activity and can convert the Fe2+ molecules into the Fe3+ form (Lawson et al., 1989). Little is known about how iron is released from ferritin complexes, although it is assumed that complex degradation functions in this process (Hentze et al., 2004). In erythroid cells, cytosolic iron is used predominantly in the biosynthesis of hemoglobin, which utilizes iron to bind oxygen, and circulate it throughout the body. Indeed, approximately 25 mg of iron per day is devoted to hemoglobin biosynthesis (Hentze et al., 2010). Reduced Fe2+ is trafficked to the mitochondrial matrix, the site of heme biosynthesis, where at the last enzymatic step ferrochelatase inserts Fe2+ into protoporphyrin IX to form heme (Chung et al., 2012). Importantly, senescent erythroid cells (primarily red blood cells) can be phagocytosed by macrophages which then degrade heme and incorporate the liberated Fe2+ into their labile iron pool or direct it to transferrin-mediated trafficking pathways.
Iron homeostasis in mammals is predominantly regulated by one of two mechanisms: The IRP/IRE system or the small peptide hepcidin (Hentze et al., 2010). There are two iron regulatory proteins (IRP1 and IRP2) which function as regulators by binding iron-responsive elements (IRE) in the untranslated regions (UTRs) of mRNA (Rouault, 2006). Notably, IRP1 has a dual function as a cytosolic aconitase when it contains an intact Fe-S cluster, which has led to the “Fe-S switch” model for IRP activation, whereby the regulator is activated in the absence of Fe due to degradation of the Fe-S cluster. Several mRNAs contain IREs, and the regulation of these IREs by IRPs varies. For TfR mRNA, binding of IREs by multiple IRPs stabilizes the mRNA and protects it from endonucleolytic degradation, thus increasing TfR expression (Casey et al., 1989). For ferritin L- and H-chain mRNA, however, IRE binding by a single IRP impedes translation by a steric hindrance mechanism, decreasing ferritin content in the cell (Rouault et al., 1988). IRP1 and IRP2 function redundantly to some extent, as independent knock out models demonstrate significant iron-related phenotypes but double-knockouts are embryonic lethal (Wilkinson and Pantopoulos, 2014). Hepcidin was first thought to be an antimicrobial peptide but was soon found to be critical for proper iron metabolism. Hepcidin was shown to be induced in response to iron overload and hepcidin knock-out mice suffer an iron-overload phenotype, connecting the function of hepcidin to iron homeostasis (Bennoun et al., 2001; Pigeon et al., 2001). It was later shown that hepcidin exerts its effect on iron metabolism mechanistically by binding to and internalizing the iron exporter ferroportin, leading to its degradation (Nemeth et al., 2004). By preventing the transport of iron through the basolateral membrane and thus preventing the dissemination of dietary iron throughout the body, hepcidin has been termed the “master iron regulator.” Interestingly, innate immune signaling induces hepcidin expression and consequently much work has shown that hepcidin is expressed in response to infection by some pathogens, potentially limiting iron from pathogens (Drakesmith and Prentice, 2012). Conversely, in other infection models such as chronic hepatitis C virus infection, hepcidin expression is repressed, and it is thought this may promote iron overload conditions such as hepatic iron accumulation. In total, the circulation of iron through the human body presents many opportunities for pathogens to intervene in their pursuit for iron. Some prominent examples will be discussed below.
Iron Acquisition in Intracellular Bacterial Pathogens
Intracellular pathogens compete for iron sequestered in host proteins, often by intercepting or interrupting mammalian iron trafficking pathways. Iron acquisition follows a general paradigm in intracellular pathogens: Iron, in one form or another, is recognized by the bacteria and translocated across the bacterial membrane(s), generally by a permease system, where it can then be utilized. There are four predominant mechanisms by which this occurs: the biosynthesis of siderophores, transferrin/lactoferrin capture, heme capture, or direct acquisition of ferrous iron. We will briefly recount these mechanisms, but the reader is directed to several detailed reviews on the subject for further reading (Marx, 2002; Andrews et al., 2003; Caza and Kronstad, 2013).
The biosynthesis of small iron-chelating molecules, known as siderophores, represent a primary mechanism of ferric iron acquisition among bacteria. Model siderophores, such as enterobactin, have a notably high affinity for Fe3+, (Ka = 1051), which allows them to directly chelate iron bound in host proteins such as transferrin (Carrano and Raymond, 1979). Siderophores are then transported across the outer membrane by a TonB-dependent receptor protein, bound in the periplasm by a siderophore-binding protein and trafficked through an inner membrane permease system where the siderophore can then be dissociated from Fe3+ to facilitate reduction to Fe2+. These systems are represented by homologs of E. coli fepABCDEG and fhuABCDEF, the respective permease systems of enterochelin and ferrichrome. (Fecker and Braun, 1983; Pierce et al., 1983; Ozenberger et al., 1987). The mechanism of TonB-dependent energy transduction from the inner membrane to the outer membrane remains a hotly debated subject of research. What can be concluded from the available data is that the association of TonB with the accessory proteins ExbB and ExbD results in the charging of TonB with potential energy as a function of the proton motive force across the inner membrane (Gresock et al., 2015). Several hypotheses have existed for how the “charged” TonB then delivers energy to the outer membrane receptor proteins which depend on it for the translocation of important nutrients into the periplasm. Recent studies suggest that in contrast to models that posit the shuttling of TonB through the periplasm to the outer membrane, TonB is stationary at the inner membrane and confers energy transduction from this position (Gresock et al., 2011). Most of the iron acquisition systems in Gram-negative bacteria utilize TonB-dependent outer membrane receptors.
In addition to siderophore-mediated acquisition of ferric iron, many Gram-negatives possess mechanisms for the acquisition of ferrous iron, which is presumed to freely flow through the outer membrane, possibly via porins. This strategy is dependent upon ferrireductase activity, which is thought to be either cell-associated or translocated into the extracellular space. Several bacteria have been shown to display ferrireductase activity, but few ferrireductases have been directly identified. Notably, riboflavin has been implicated as a ferrireductase in the Gram-negative pathogens Campylobacter jejuni and Helicobacter pylori (Worst et al., 1998; Crossley et al., 2007). It was recently shown that Legionella pneumophila secretes a pylomelanin pigment that indeed reduces Fe3+ to Fe2+, and some fungal ferrous iron permease homologs have associated ferrireductases (Kosman, 2003; Chatfield and Cianciotto, 2007). Ferrous iron import systems are generally represented by homologs of E. coli feoABC and Yersinia pestis yfeABCD (Kammler et al., 1993; Bearden and Perry, 1999). Intriguingly, it has been demonstrated in Shigella flexneri that homologs of both ferrisiderophore and ferrous iron uptake systems are induced during host cell invasion and intracellular proliferation (Runyen-Janecky and Payne, 2002; Pieper et al., 2013). This suggests not only that intracellular bacteria experience iron deprivation likely as a protective mechanism of the host, but additionally that direct acquisition of iron ions may represent a “path-of-least-resistance” for intracellular pathogens in their quest for sufficient iron. Moreover, these mechanisms are redundant in S. flexneri, indicating that intracellular pathogens have access to both oxidative states of iron in the host cell (Runyen-Janecky et al., 2003). However, because Shigella replicates within the host cell cytoplasm (Mellouk and Enninga, 2016), it is not clear if pathogens that reside within intracellular vacuoles could similarly or as effectively utilize these sources of iron.
The serum iron-binding protein transferrin (Tf) is abundant in mammalian blood, and represents an iron resource for extracellular pathogens. Tf is endocytosed by mammalian cells, via holo-Tf recognition by the transferrin receptor (TfR; Harding et al., 1983). Upon endocytosis, TfR-holo-Tf vacuoles are trafficked through the endocytic recycling pathways. This pathway provides an opportunity for intracellular pathogens to obtain holo-Tf and thus iron. Neisseria species possess sophisticated mechanisms for the acquisition of transferrin. Neisseria meningitidis access host Tf by manipulating Tf-trafficking in the cell, redirecting it to sites of bacterial colonization (Barrile et al., 2015). Transferrin can then be acquired by Neisseria via fusion of Tf-containing endosomes with the pathogen-containing vacuole. Two transferrin binding proteins, TbpA and TbpB, are involved in binding and transporting transferrin across the outer membrane in N. gonorrhoeae (Cornelissen et al., 1992; Anderson et al., 1994). The two proteins function coordinately, where the outer membrane lipoprotein TbpB acts as a co-receptor to bind transferrin and help direct it to transmembrane TbpA where it can then be transported across the membrane (Ostberg et al., 2013). TbpA is TonB-dependent, the energy from which aids in initiating a conformational change that facilitates the dissociation of apo-transferrin from iron (Noinaj et al., 2012a,b). Free iron can then be transported into the periplasm and bound by the periplasmic ferric binding protein FbpA. The fbpABC operon encodes the periplasmic FbpA in conjunction with the inner membrane permease FbpB and the nucleotide-binding FbpC, which facilitate the translocation of Fe3+ across the inner membrane (Adhikari et al., 1996).
Most iron in the mammalian body is sequestered in heme molecules. Due to the cytotoxicity of free heme, most heme is bound in hemoproteins such as hemoglobin. Hemoproteins are ubiquitously expressed in mammalian cells, as they play crucial functions in metabolism (e.g., electron transport chain). Thus, hemoproteins and heme represent an ample pool of iron to intracellular pathogens. The acquisition of heme from the host is well-characterized in Yersinia species, and is representative of systems found in other pathogens such as Neisseria and Bartonella (Rohde and Dyer, 2004; Parrow et al., 2009). The hmuRSTUV operon encodes the full complement of gene products necessary to receive heme at the outer membrane (HmuR), shuttle it through the periplasm (HmuT), transport it across the inner membrane (HmuUV) and traffic it through the bacterial cytoplasm (HmuS; Thompson et al., 1999). Some pathogens, such as Brucella abortus, N. meningitidis and Bradyrhizobium japonicum encode heme oxygenases that facilitate the use of heme as an iron source by degrading the tetrapyrrole ring and liberating the coordinated iron (Zhu et al., 2000; Puri and O'Brian, 2006; Ojeda et al., 2012). Yersinia enterocolitica encodes an hmuRSTUV analog, hemRSTUV (Stojiljkovic and Hantke, 1992). Interestingly, Y. pestis has a secondary hemophore-dependent heme-protein acquisition system encoded by hasRADEB (Carniel et al., 2001). This system functions similarly to both hmuRSTUV and hemRSTUV with the exception that HasA encodes a secreted heme-binding protein, or hemophore, that can coordinate hemin and shuttle it to the receptor HasR (Lefèvre et al., 2008; Kumar et al., 2013). Intriguingly, essentially none of these receptor mechanisms are identifiable by homology in obligate intracellular pathogens.
Very little is presently understood about iron acquisition and homeostasis in obligate intracellular pathogens. Interestingly, many do not appear to have strict dependencies on iron availability. For example, Coxiella burnetii seem to prefer a low-iron environment, potentially as a mechanism to avoid the oxidative stress associated with free iron (Mertens and Samuel, 2012). This is supported by the fact that iron-depleted cell culture systems and animal models actively promote the growth and replication of C. burnetii (Briggs et al., 2008). Additionally, only three putative iron-regulated genes appear to be regulated by C. burnetii Fur transcriptional repression as determined by a two-plasmid reporter assay (Briggs et al., 2008). Correspondingly, Rickettsia rickettsii demonstrate inhibited growth in response to iron-depletion only at very high (500 μM) concentrations of the iron chelator desferrioxamine mesylate (DFO), and only 12 genes appear differentially expressed in response to this stimulus, most of them being hypothetical (Ellison et al., 2009). Moreover, whereas expression of Nramp1 in macrophages affects significant killing of Salmonella, Coxiella are resistant to the depletion of metal ions by this transporter (Cockrell et al., 2017). Neither Coxiella nor Rickettsia have any apparent functional homologs to siderophore biosynthesizing enzymes, but it is interesting to note that Coxiella possesses a frameshifted ORF for a siderophore synthase, suggesting that reductive evolution of the genome is actively remodeling the iron requirement of this pathogen (Briggs et al., 2008; Ellison et al., 2008). Contrastingly, Ehrlichia species are known to require iron. Both E. chaffeensis and E. sennetsu fail to proliferate in the presence of only 15 μM DFO and co-localize with TfR to possibly acquire iron (Barnewall et al., 1999). Moreover, supplementation of holo-TfR to Ehrlichia-infected THP-1 monocytes treated with the inflammatory cytokine gamma-interferon (IFN-γ) abrogates IFN-γ killing of E. chaffeensis, demonstrating the importance of iron availability to Ehrlichia pathogenesis (Barnewall and Rikihisa, 1994). Still, despite evidence that iron availability may modulate Ehrlichia pathogenesis, the only known iron acquisition protein in Ehrlichia is a homolog of FbpA (Doyle et al., 2005). Chlamydia species are somewhat distinguished from other obligate intracellular pathogens insofar as the iron-dependency of the organism has been an active subject of research within the field for nearly two decades. Like studies in other bacteria, attempts to dissect iron acquisition in Chlamydia began with investigations into chlamydial responses to iron deprivation.
Chlamydial Response to Iron Starvation
The interest in chlamydial iron biology originated from work done in the laboratory of Jane Raulston, which demonstrated that restricting iron to C. trachomatis serovar E-infected epithelial cells by DFO treatment resulted in a significant reduction in infectious progeny and the development of aberrant, morphologically enlarged RBs that exhibited a delayed maturation (Raulston, 1997). Importantly, the addition of holo-Tf to the cell culture medium resulted in the recovery of infectious progeny, indicating that the effect was reversible. These results closely mirrored what had been known for some time with respect to the development of chlamydial persistence in response to IFN-γ treatment (Byrne et al., 1986; Thomas et al., 1993; Beatty et al., 1994). Therefore, based on similarity to IFN-γ-mediated persistence, iron depletion became recognized as an inducer of chlamydial persistence. This discovery prompted research into how exactly Chlamydia responds to iron starvation, with the hopes of uncovering molecular mechanisms that function in iron acquisition and homeostasis, and perhaps even persistence. Studies were thus conducted in several other chlamydial species and serovars. Depriving C. pneumoniae of iron inhibited growth and resulted in the depletion of infectious progeny and the development of abnormal inclusion and chlamydial morphology (Al-Younes et al., 2001; Freidank et al., 2001). Interestingly, iron chelation seemed to have a more pronounced impact on C. pneumoniae than on C. trachomatis, as demonstrated by smaller inclusions and markedly fewer recoverable infectious particles following equimolar treatment with DFO. This prompted the investigation of whether iron availability was predictive of C. pneumoniae-associated heart disease (Sullivan and Weinberg, 1999). Indeed, samples from patients operated on for stenotic aortic heart valves who also tested positive for C. pneumoniae infection showed a greater than 20-fold enrichment for iron, implicating iron availability as a factor in C. pneumoniae-associated development of aortic stenosis (Nystrom-Rosander et al., 2003).
Attempts have been made to limit iron to C. trachomatis without the use of a chemical chelator, namely by the over-expression of the iron exporter ferroportin (Dill and Raulston, 2007; Paradkar et al., 2008). In Dill and Raulston's report, ponasterone A-inducible ferroportin was over-expressed in HEK293 cells, and no changes in recoverable infectious progeny or morphology were observed in C. trachomatis serovar E. In contrast, Paradkar et al. were able to demonstrate that over-expression of ferroportin in HEK293 cells leads to smaller chlamydial inclusions in both C. psittaci and C. trachomatis infection models. Upon treatment of the infected cells with the major iron regulator hepcidin, inclusion development was rescued, implicating the over-expression of ferroportin as the primary mediator of the phenotype. The authors further demonstrated that this phenotype was reproduced in primary bone marrow macrophages derived from flatiron mutant mice, which possess a missense mutation in one allele of ferroportin, resulting in improperly sorted ferrorportin that is retained in intracellular compartments (Zohn et al., 2007). It is not entirely clear why Paradkar et al. obtained such different results from Dill and Raulston's study. However, Paradkar et al. note that in contrast with Dill and Raulston, they included a 24 or 48 h iron-loading step (supplementation of the media with 10 μM ferric citrate) prior to both ferroportin over-expression in HEK293 cells and C. pisttaci infection of the flatiron mouse-derived macrophages, respectively. Because a non-iron loaded control was not reported in their study, it is not clear the effect that iron-loading may have had on chlamydial growth or host iron homeostasis.
Several studies have investigated changes in gene and protein expression in models of iron limitation for Chlamydia. Differential transcript expression analyses revealed significant down-regulation of genes associated with cell wall morphology (ompA, omcB) and nucleoid condensation (hctB) in C. pneumoniae, thought to represent part of the genetic foundation for the hallmark aberrant morphology observed during persistence (Timms et al., 2009). This study also revealed a significant induction of generic stress-response genes in C. pneumoniae, such as htrA (21.14-fold) and ahpC (8.56-fold), encoding a DO serine protease and thioredoxin peroxidase, respectively. In a parallel study on differential protein expression in iron-starved C. pneumoniae, marked discrepancies were apparent, such as the 2.3-fold induction of OmpA and unchanged expression of OmcB (Mukhopadhyay et al., 2006). Notable changes were also observed between the transcriptional responses of C. pneumoniae and C. psittaci, where the transcriptional repressor of developmentally late genes, euo, was induced 2.94-fold in C. pneumoniae, but repressed nearly 5-fold in C. psittaci (Goellner et al., 2006; Timms et al., 2009). It has been proposed that the induction of euo late in infection in response to persistence contributes significantly to the inability of Chlamydia to continue its normal development (Belland et al., 2003a). It is not entirely clear where the discrepancies between these studies originate. Some amount of disagreement is expected between species, as well as between transcript and protein expression studies. However, these studies only examined the expression of select genes from defined gene ontology groups, as opposed to examining global transcriptomic changes, possibly excluding similarities, and differences. Importantly, the transcriptional studies, predominantly relying on qRT-PCR, normalized their expression data to an internal reference gene, commonly 16S rRNA. It has been demonstrated that this is a less informative methodology for Chlamydia, as 16S rRNA expression levels are observed to decrease dramatically in response to global stresses such as those that induce persistence (e.g., IFN-γ; Ouellette et al., 2006). Iron limitation studies in C. trachomatis serovar E wherein transcript abundance was normalized to genomic DNA reported no significant difference in the expression of euo, ompA, or omcB (Dill et al., 2009). However, ahpC transcript expression was still found to be moderately up-regulated, and expression was substantially induced at the protein level.
Investigation of the global transcriptomic response of C. pneumoniae to an iron starvation model of persistence by microarray analysis revealed a broad down-regulation of transcription machinery, suggesting that a stalled transcriptome may facilitate the development of persistence (Mäurer et al., 2007). This study reported the down-regulation of omcB and hctB, however euo expression was unaffected, contributing further confusion to the body of transcriptomic data. Global proteomic studies in iron-limited C. trachomatis models have identified between 19 and 25 proteins induced in response to iron limitation, however it remains unclear whether the majority of these proteins participate directly in iron acquisition or homeostasis (Raulston, 1997; Dill et al., 2009). The identification of chlamydial Hsp60 as iron-regulated did however raise the possibility that iron availability plays a crucial role in sub-acute pathologies of C. trachomatis, such as tubal factor infertility, being that patients are often seropositive for chlamydial Hsp60 (Toye et al., 1993; LaRue et al., 2007). Unfortunately, this hypothesis has remained largely unaddressed. An analogous study in an iron-limited C. pneumoniae model only identified differential expression among six proteins, most notably the chromosome partitioning protein ParB and the thioredoxin reductase TrxB (Wehrl et al., 2004). Again, the participation of any of the identified iron-responsive proteins in iron homeostasis was unclear.
A potential contributor to the general lack of clarity produced by past studies on chlamydial responses to iron limitation has been the decision to monitor differential gene/protein expression late in infection following prolonged treatments with iron chelators. These extended treatments meant that most comparisons have been made between highly abnormal, persistent chlamydiae and their untreated, replicative and differentiating counterparts. As such, the results from these studies have been harder to interpret given that it is unclear if the phenotypes observed are a true response to iron limitation or if the organisms are simply abnormal. In part this may have been judged to be necessary, as DFO is a relatively inefficient iron chelator. DFO displays a discriminatory binding affinity for the different valence states of iron, having specific preference for ferric over ferrous iron (Keberle, 1964). Moreover, DFO is relatively membrane impermeable, effectively chelating iron only from the extracellular environment and potentially from within lysosomes into which it can be pinocytosed (Lloyd et al., 1991; Richardson et al., 1994; Cable and Lloyd, 1999; Persson et al., 2003; Glickstein et al., 2005). Thus, for an intracellular pathogen such as Chlamydia, extensive DFO treatments would be required to elicit sufficient iron starvation. Other iron chelators have been explored in Chlamydia infection models, such as desferasirox and deferriprone, which are more membrane permeable than DFO and thus have greater access to intracellular iron pools (Neufeld, 2006; Paradkar et al., 2008). While both chelators display moderate advantages over DFO regarding their ability to reduce host ferritin levels and inhibit growth of chlamydial inclusions, neither chelator was investigated thoroughly enough to suggest that they induced a more robust or rapid response to iron limitation in Chlamydia.
The shortcomings of DFO with respect to iron starvation in Chlamydia prompted the investigation of a more efficient chelator of iron, leading to the discovery that the membrane permeable 2,2-bipyridyl (Bpdl) was substantially more effective at depriving Chlamydia of iron (Thompson and Carabeo, 2011). First, Bpdl is more efficient at promoting an iron-starved “persistent” phenotype as 48 h treatment starting at the time of infection produces enlarged RBs accompanied by the noticeable lack of EBs in the inclusion lumen, whereas an identical treatment of DFO produces inclusions that resemble untreated 24 h post infection (hpi) inclusions. This is reinforced by the observation that Bpdl treatment predictably modulates the expression of persistence markers, euo and omcB, where euo expression remains elevated late in infection and omcB expression is suppressed throughout a 48 h time course (Belland et al., 2003b; Ouellette et al., 2006; Timms et al., 2009). DFO, however, fails to significantly alter the expression of either marker for persistence, demonstrating that Bpdl is much more effective at producing a relevant iron starvation phenotype. This agreed with reports from Dill et al. (2009) which showed that DFO treatment did not modulate the transcript abundance of persistence markers. Second, the reduction in recoverable infectious progeny from Bpdl treatment can be reversed not only by the addition of exogenous ferric iron (Fe(III)Cl3), but also by the addition of exogenous ferrous iron (Fe(II)SO4), demonstrating that Bpdl, unlike DFO, can be saturated, and thus chelate both ferric and ferrous species of iron. Finally, Bpdl induces a much more robust iron starvation transcriptional response than DFO. Treatment with Bpdl significantly induces the expression of two previously identified iron-responsive transcripts, ahpC and devB, while equimolar DFO treatment does not affect ahpC expression and only marginally induces devB expression following 30 h of treatment (Dill et al., 2009). In sum, the utility of Bpdl in chlamydial iron starvation studies was demonstrated by its potent induction of persistence, its broader spectrum of iron chelation, and its ability to modulate the expression of known iron-responsive transcripts.
Contrasting research in other bacteria, such as E. coli and Salmonella, where investigating iron-deprived responses yielded the discovery of many components of iron acquisition and homeostasis, results from the analogous chlamydial studies did not provide the same degree of clarity. This may have been a result of the general disagreement between studies, serovars and species. Alternatively, as the characterization of Bpdl has shown, this may have resulted from non-ideal methodologies. It has therefore required more indirect approaches to begin to understand the molecular basis of iron acquisition in Chlamydia.
Iron Acquisition in Chlamydia
Analysis of the genome sequence of C. trachomatis serovar D reveals that C. trachomatis does not possess homologs for heme capture mechanisms, transferrin or lactoferrin receptors, siderophore biosynthesis pathways, or TonB (Stephens et al., 1998). The notable lack of iron-acquisition homologs is a common feature amongst the C. trachomatis serovars and species within the Chlamydiaceae (Kalman et al., 1999; Read et al., 2000, 2003; Thomson et al., 2008; Grinblat-huse et al., 2011; Mojica et al., 2011; Donati et al., 2014). A protein estimated to be 30 kDa in size with an isoelectric point of 5.5 was originally identified by Jane Raulston in the initial set of 19 iron-regulated proteins differentially expressed in response to extended DFO treatment (Raulston, 1997). However, this was overlooked as a participator in iron acquisition being that another protein which had a molecular weight of roughly 37 kDa and an unusually basic isoelectric point (>9.0) appeared to closely resemble the biochemical properties of the periplasmic Fe3+-binding protein FbpA in Neisseria (Mietzner et al., 1984). However, a subsequent study addressing the actual identity and activity of this protein was never reported. It wasn't until a subsequent study that aimed to understand which antigenic chlamydial proteins are iron-responsive that the 30 kDa protein was identified as YtgA, the first ORF in the ytgABCD operon (Raulston et al., 2007). Based on sequence analysis, YtgA is a 37 kDa substrate-binding periplasmic protein, involved in the trafficking of divalent metals from the outer membrane to an ABC permease system in the inner membrane, closely resembling the Zn2+-binding TroA from Treponema pallidum (Lee et al., 1999). YtgA migrated with an apparent molecular weight of 30 kDa in both of Raulston's studies, despite its predicted size of 37 kDa. Subsequent investigation regarding the function of YtgA demonstrated that YtgA had a specific affinity for Fe3+ in vitro, where radiolabeled 59Fe3+ could not be competed off purified recombinant YtgA when either Mn2+ or Zn2+ were added to the system (Miller et al., 2009). However, the affinity of YtgA for Fe2+ was not reported. Additionally, prolonged exposure to DFO demonstrated that YtgA was more highly expressed 36 hpi and onward compared to untreated controls, but exposure to other insults such as penicillin treatment did not alter YtgA protein expression. It was also observed that YtgA localized predominantly to chlamydial membranes, with a substantial proportion localized within the periplasm, as determined by immunogold labeling. Interestingly, this localization was disrupted in response to DFO treatment, with a pronounced reorganization of YtgA to the cytoplasm, suggesting that iron transport is disrupted during persistence. These localization data confirmed previous experiments that had observed YtgA localized to chlamydial membranes around the periphery of the inclusion by immunofluorescent microscopy (Bannantine and Rockey, 1999). Importantly, in validating Bpdl as a more efficient iron chelator for Chlamydia, Thompson and Carabeo observed the induction of ytgA transcripts in response to Bpdl treatment (Thompson and Carabeo, 2011). This demonstrated for the first time that ytgA was iron-regulated at the transcript level. While these studies suggested a role for YtgA in the acquisition of iron in C. trachomatis, it remained unclear from where Chlamydia siphoned iron from the host.
Early studies regarding the establishment of the chlamydial inclusion upon invasion monitored the association of classical endosomal markers—such as TfR—with the inclusion as it developed. Endocytosed TfR follows two distinct recycling pathways back to the plasma membrane that are distinguished by their association with particular Rab family GTPases (Mayle et al., 2012). Upon internalization into the early endosome, TfR-containing vesicles are Rab5-positive. These Rab5-positive TfR-containing endosomal vesicles acquire both Rab4 and Rab7 and are promptly recycled to the plasma membrane within 30 min following internalization. However, TfR-containing vesicles can be routed to the sorting endosome, where they acquire Rab11 GTPases. These TfR-containing Rab11-positive vesicles are trafficked back to the plasma membrane with notably slower kinetics, potentially in a Rab4-dependent manner, and represent the slow-recycling pathway of TfR (van der Sluijs et al., 1992; Sönnichsen et al., 2000). Landmark studies in the laboratories of Ted Hackstadt, Joanne Engle and Thomas Meyer demonstrated that TfR-containing vesicles localize around the chlamydial inclusion in both C. trachomatis and C. pneumoniae-infected epithelial cells (Scidmore et al., 1996; van Ooij et al., 1997; Al-younes et al., 1999). Interestingly, these studies did not conclusively demonstrate that Chlamydia infection modulates the recycling of TfR, suggesting the inclusion association with endocytic vesicles may be fluid and dynamic. Moreover, Scidmore et al. reported that Tf localized to the inclusion periphery but never within the inclusion lumen, eliminating the possibility that Chlamydia directly acquire Tf. However, the relation of TfR localization around the chlamydial inclusion to the success of the pathogen was not addressed in these studies.
A later study identified a small molecule that specifically inhibits the slow-recycling pathway of transferrin by preventing Tf-containing Rab11-positive vesicular fusion with Rab4-positive vesicles on their way back to the plasma membrane (Ouellette and Carabeo, 2010). This molecule was identified initially because of its potent anti-chlamydial activity. The inhibition of Rab11-Rab4 hybrid vesicle formation effectively stalled Tf-recycling at the chlamydial inclusion, leading to the accumulation of Tf-containing vesicles around the inclusion membrane. Chlamydial killing was likely caused by exposure to inhibitory amounts of free iron, as removing the transferrin-containing fraction from the serum alleviated inhibition of chlamydial growth. While this study did not directly observe iron transport into the chlamydial inclusion, it determined that normal development of Chlamydia is strictly dependent upon the uninhibited recycling of Tf through the slow-recycling endosomal pathway. This finding coincides with the result of an earlier study that observed knocking down expression of Rab11, and thus disrupting Tf recycling, blocks the development of infectious progeny in C. trachomatis (Lipinski et al., 2009). Therefore, normal chlamydial development is dependent upon association with Rab11-positive endosomes, which are critical for intracellular trafficking of Tf. Mycobacterium avium-containing vacuoles also interact with Rab11-positive organelles, including the endocytic recycling compartments (ERC), but the biological significance of this interaction is unknown (Halaas et al., 2010). M. tuberculosis binds holo-transferrin on its surface via a number of proteins, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH). After binding, transferrin is internalized into the bacterial cell (Boradia et al., 2014). The interaction of the Mtb-containing vacuole with ERC-derived Rab11-positive vesicles would be consistent with this transferrin capture strategy. Indeed, such an association was reported during infection of macrophages (Tailleux et al., 2003).
The requirement of Rab11-positive vesicles may help explain why Dill and Raulston's attempt to phenocopy iron starvation in Chlamydia by ferroportin over-expression failed (Dill and Raulston, 2007). While ferroportin exports cytosolic iron, Chlamydia utilizes intravesicular iron prior to their transport to the cytosol. Chlamydial iron acquisition is a process independent, at least in part, of the dissemination of Fe2+ from the endosome into the cytosol (e.g., by the activity of STEAP3 ferrireductase and DMT1 transporter). If so, it would not be expected that increased iron export would impact Chlamydia in the absence of unaltered endocytic TfR internalization. These data help support the notion that the chlamydial inclusion functions as a pathogen-specified parasitic organelle, siphoning resources in homeostasis with the subcellular trafficking pathways of the host (Moore and Ouellette, 2014). It is worth mentioning here that it has been reported that Chlamydia can propagate in serum-free media, i.e., media lacking transferrin (Maass et al., 1993; Ouellette and Carabeo, 2010). However, the biological significance of this is unclear given that iron is predominantly sequestered in host proteins such as transferrin and ferritin in vivo, and eukaryotic host cells uptake iron not in the free elemental form, but bound in macromolecular complexes. Therefore, the concentration of free iron under normal conditions is already likely quite low. This may however indicate that other mechanisms are at work beyond the acquisition of iron via vesicular trafficking of transferrin.
Taken together, the present data allow the construction of a model by which Chlamydia may acquire iron (Figure 1). This model proposes that upon receptor-mediated endocytosis of Tf bound to TfR, a small portion of Tf is trafficked in Rab11-positive vesicles to the periphery of the chlamydial inclusion, possibly where a “kiss-and-run”-type mechanism may facilitate the entry of free iron into the inclusion lumen. In the absence of a ferrireductase homolog in the chlamydial genome, it is possible that both Fe2+ (reduced by STEAP3) and Fe3+ diffuse into the inclusion lumen upon vesicular fusion with TfR-containing Rab11-positive endosomes. However, it has been suggested that riboflavin may act to reduce Fe3+, as riboflavin biosynthetic genes (ribBA, ribC, and ribH) have been reported to be induced by 1 h post-infection (Humphrys et al., 2013). However, no additional experimental data exists to support this hypothesis. Iron transport across the outer-membrane could be mediated by an as yet unidentified siderophore/receptor mechanism that funnels Fe3+ to YtgA. The existence of unidentified siderophores is in part supported by the observation that neutrophil gelatinase-associated lipocalin (Lcn-2) is involved in host-mediated suppression of chlamydial growth (Bellmann-Weiler et al., 2013), possibly via sequestering iron-loaded bacterial siderophores, although siderophore candidates would have to be novel or non-traditional (Flo et al., 2004; Berger et al., 2006; Nairz et al., 2009). Siderophore uptake may be dependent upon a system of energy transduction, as Chlamydia possess both ExbB and ExbD, the canonical TonB accessory proteins. However, the evidence for such a system is highly circumstantial at present, as no homolog for siderophore biosynthesizing enzymes or TonB have been identified (Stephens et al., 1998). Ferrous iron may passively diffuse through outer membrane porins into the periplasm. YtgA can then traffic Fe3+, and possibly Fe2+, to the inner membrane heterodimeric permease likely formed by YtgC and YtgD. The ATP-binding YtgB provides the energy to transport iron across the inner membrane by ATP hydrolysis to ADP. If transported, Fe3+ must be reduced by an unidentified chlamydial cell-associated ferrireductase analog to become biologically utilizable. An alternate scenario may provide Chlamydia with ferrous iron, exported by DMT1 from the TfR-containing endosomes to the host cell cytosol. This cytosolic pool of unliganded iron could be passively transported across the inclusion membrane and chlamydial outer membrane to be sequestered by YtgA or some other divalent metal binding protein. However, as the ferroportin over-expression system demonstrated, this alternate pathway may not provide the bulk of iron to Chlamydia. In addition, cytosolic Fe2+ would likely be captured by host ferritins, intensifying competition for this iron pool. Further, Chlamydia may access the intracellular labile iron pool. For example, it is understood that the chlamydial inclusion associates with lysosomes to acquire nutrients such as oligopeptides, and Chlamydia may further interact with mitochondria and autophagosomes for acquisition of nutrients (Matsumoto et al., 1991; Al-Younes et al., 2004; Derré et al., 2007; Ouellette et al., 2011). However, without properly identifying the subcellular location of the labile iron pool accessed by Chlamydia, and in the absence of straight-forward tools to interrogate the interaction between Chlamydia and the labile iron pool, questions regarding chlamydial access of the labile iron pool remain difficult to address. The available data thus provides a testable framework within which to continue investigations into chlamydial iron acquisition and begin to delineate which model of acquisition predominates in Chlamydia.
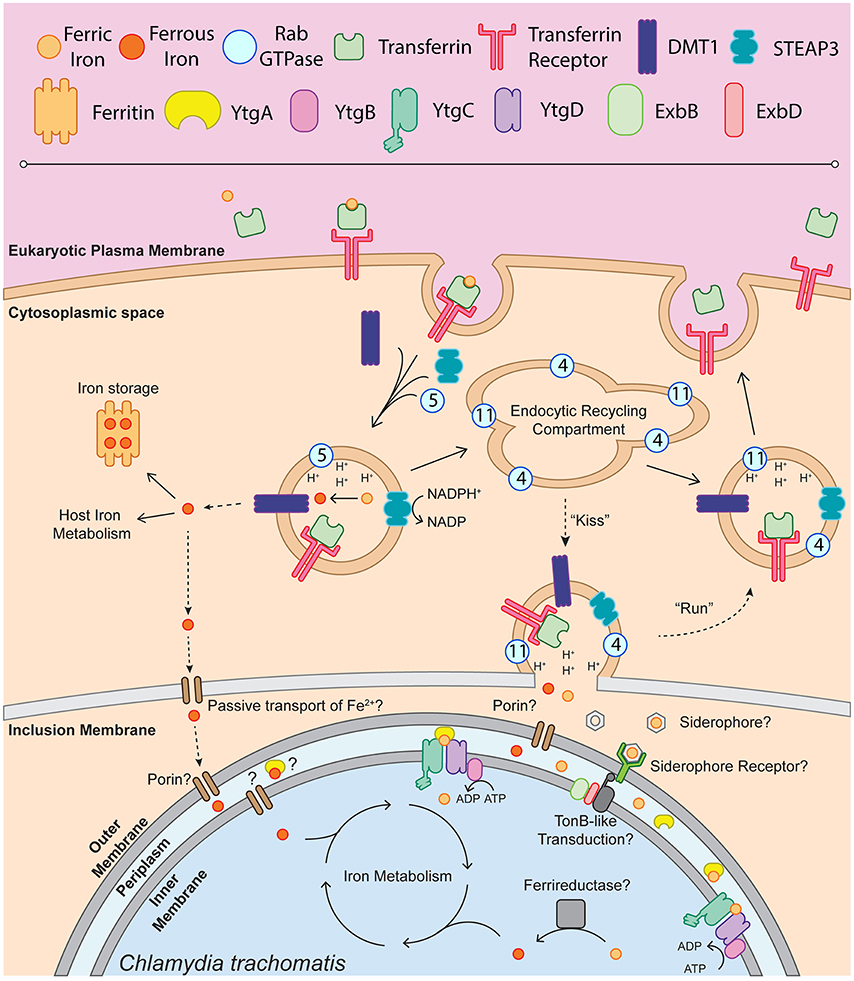
Figure 1. Schematic of chlamydial iron acquisition. Solid black arrows denote iron trafficking pathways of the host whereas dashed-line arrows indicate possible mechanisms of iron acquisition by Chlamydia. The depiction of endosome-associated Rab-family GTPases is not intended to be exhaustive and is based on the findings of Ouellette and Carabeo (2010). Ferric iron (Fe3+) is bound by apo-transferrin (Tf) and endocytosed upon recognition by the transferrin receptor (TfR). The holo-Tf-TfR complex proceeds into the early endosome, upon which TfR-containing endosomes acquire DMT1, STEAP3, and Rab5. Acidification of the early endosome potentiates the dissociation of Fe3+ from Tf, which can then be reduced to ferrous iron (Fe2+) by STEAP3. Fe2+, exported from the endosome by DMT1 into the cytoplasm, is shunted to ferritin for iron storage or incorporated into host iron metabolic processes such as the electron transport chain or TCA cycle. It is possible that labile Fe2+ is passively transported through the inclusion membrane and the chlamydial outer membrane to be sequestered in the periplasm and trafficked into the bacterial cell. A fraction of the TfR-containing endosomes will acquire Rab11 and Rab4, proceeding through the slow recycling endosomal pathway to be trafficked back to the eukaryotic plasma membrane. In an Chlamydia-infected cell, slow recycling Tf-containing endosomes associate with the chlamydial inclusion, where a “kiss & run” mechanism may deliver free ferric or ferrous iron into the inclusion lumen. Novel siderophores, siderophore receptors or porins may function to traffic iron to the periplasm. This outer membrane transport may be dependent upon TonB-like energy transduction, given that Chlamydia possess ExbB and ExbD homologs. Once inside the periplasm, Fe3+ can be bound by YtgA and shuttled to the membrane permease likely formed by YtgC and YtgD. ATP hydrolysis mediated by YtgB likely provides the energy to import iron into the chlamydial cytosol. The reduction of Fe3+ to biologically useful Fe2+ may be performed by an unidentified ferrireductase. Upon acquisition of Fe2+, Chlamydia can then incorporate iron into the electron transport chain via porphyrin biosynthesis, divert iron to Fe-S cluster biosynthesis or utilize iron in other metabolic processes.
Bacterial Iron-Dependent Transcriptional Regulation
That iron availability represents a critical developmental parameter for most organisms implies that the pathways that utilize it must be under tight regulation. Iron-dependent regulation, therefore, has been widely researched in many organisms, including pathogenic bacteria. Following the isolation of mutant strains of E. coli and S. typhimurium that constitutively expressed iron acquisition machinery, it has been appreciated that bacteria globally regulate iron acquisition in response to iron availability (Ernst et al., 1978; Hantke, 1981). The strains isolated in these studies were classified as fur mutants, reflecting the observation that ferric iron uptake was dysregulated. It was later concluded, based upon complementation studies, that the fur locus operated as a negative regulator of iron acquisition (Hantke, 1982). The specific repressor activity of Fur is dependent upon Fe2+ availability and even early studies were elucidating the role of Fur in E. coli virulence (Bagg and Neilands, 1987; Calderwood and Mekalanos, 1987). Fur is now recognized as an iron-dependent master regulator in E. coli and many other pathogens, the diverse roles of which have been reviewed extensively (Hantke, 2001; Lee and Helmann, 2007; Carpenter et al., 2009). The Fur family of repressors includes many members for which the repressor activity is dependent upon another divalent cation, such as the Zn2+-dependent Zur in E. coli and B. subtilis or the Ni2+-dependent Nur in S. coelicolor (Gaballa and Helmann, 1998; Outten et al., 2001; Ahn et al., 2006). Due to the instability of Fe2+ in aerobic environments, many other divalent cations, including Mn2+, Co2+ and Cu2+ have been used in biochemical studies of Fur (de Lorenzo et al., 1987). Fur is known to bind to a consensus “Fur-box” sequence of 19 nucleotide bases forming a palindrome (de Lorenzo et al., 1987). Fur has been shown to act both as a repressor and an activator, in both Fe2+-dependent and independent ways. However, these activities are not strictly or uniformly dependent on the consensus Fur box sequence. The general mechanism for Fur repression is based on competition with the RNA Polymerase holoenzyme (RNAP), where bound Fur prevents RNAP from associating with a given promoter region (Escolar et al., 1997, 1998; Pohl et al., 2003). Alternatively, when activating transcription, Fur can presumably facilitate the association of RNAP at a target promoter by binding upstream of the promoter element, thus leaving the association of RNAP uninhibited (Alamuri et al., 2006; Gancz et al., 2006). It has also been established that Fur autoregulates its own expression, directly by binding its own promoter region and indirectly by regulating the expression of the small non-coding RNA RyhB, which prevents Fur translation (Vecerek et al., 2007). Notably, Fur negatively regulates RyhB expression and thus indirectly regulates RyhB targets, which are predominantly positively regulated, including components of the TCA cycle and bacterial ferritin proteins (Gottesman and Masse, 2002). The biological insight gained from studies on Fur inspired investigations into other metal dependent regulators in bacteria, of which the iron-dependent DtxR has emerged as the opposing family-defining member.
The existence of the DtxR regulator was postulated as far back as 1974 as a possible mechanism for controlling the iron-dependent expression of the diphtheria toxin in Corynebacterium diphtheriae (Murphy et al., 1974). Isolation of the dtxR locus and characterization of its activity against the diphtheria toxin (tox) promoter demonstrated that DtxR functions as a negative regulator of tox expression in an iron-dependent manner, while being unable to functionally complement repression of Fur-regulated promoters (Boyd et al., 1990). Accordingly, Fur is incapable of repressing tox promoter-driven reporter expression, suggesting that Fur and DtxR are not complementary repressors. This coincides with the low nucleotide and amino acid sequence homology observed between the two repressors. Purified DtxR binds the tox promoter region in the presence of either Fe2+, Ni2+, Co2+, or Mn2+ and is dependent upon a nine base-pair interrupted palindrome (Schmitt et al., 1992; Tao et al., 1992). DtxR additionally regulates siderophore biosynthesis in C. diphtheriae along with several other iron-regulated promoter regions (Schmitt and Holmes, 1991, 1994; Lee et al., 1997; Kunkle and Schmitt, 2005). Importantly, a 19 bp consensus sequence for DtxR regulation has been defined, and exhibits limited sequence homology to the Fur consensus sequence (Schmitt and Holmes, 1994). Functional studies have demonstrated that DtxR has two important metal binding sites, with metal site 2 being essential for activation (Spiering et al., 2003). Interestingly, metal binding appears to facilitate homo-dimer stability, demonstrating how DtxR may function as an iron-sensor protein via an innate dependence on iron for dimerization and thus repressor activity. DtxR dimers aggregate around DNA in groups of two and, similarly to Fur, prevent RNAP association at the promoter region. M. tuberculosis encodes an iron-dependent DtxR-homolog annotated IdeR that appears to function analogously to DtxR in coordinating iron homeostasis via transcriptional repression, but appears to additionally act as a transcriptional activator (Gold et al., 2001). TroR in T. pallidum is a Mn2+-dependent transcriptional repressor, which autoregulates its own operon, but little else is understood about its broad regulatory function (Posey et al., 1999). Similarly, both C. diphtheriae and B. subtilis encode Mn2+-dependent MntR, which regulates manganese homeostasis by transcriptional repression under high manganese conditions (Que and Helmann, 2000; Schmitt, 2002). Interestingly, B. subtilis MntR also activates transcription of the mntABCD operon under low manganese conditions. Importantly, Fur- and DtxR-like family members can function cooperatively, such as in E. coli where both Fur and MntR coordinately regulate the expression of manganese uptake (Patzer and Hantke, 2001).
Unlike Fur and DtxR, the activity of which is modulated by coordination of Fe2+, many α-proteobacteria such as B. japonicum possess iron response regulators (Irr) that are instead regulated by the availability of heme (Hamza et al., 1998). The association of ferrocheletase with Irr inactivates the protein after which heme binding facilitates the degradation of Irr (Qi et al., 1999; Qi and O'Brian, 2002). Irr is expressed alongside a Fur homolog in B. japonicum, which represses irr expression in a Mn2+-dependent manner, while Irr itself acts as an iron-dependent antirepressor of irr expression (Hamza et al., 2000; Hohle and O'Brian, 2010). Irr has been shown to function both as an iron-dependent transcriptional repressor and activator in B. japonicum (Sangwan et al., 2008; Small et al., 2009). Irr regulation is generally dependent on a cis-acting iron control element (ICE) in the promoter region of target genes, consisting of an AT-rich 21 bp incomplete inverted repeat (Nienaber et al., 2001; Rudolph et al., 2006). The putative Irr-dependent regulon includes many genes associated with iron homeostasis, including heme-uptake (hmuR, hmuT, etc.) and iron receptor proteins (fcuA, blr3904; Rudolph et al., 2006). In sum, bacterial metallo-regulation is remarkably diverse, functioning in multiple capacities to control a wide array of pathways. Correspondingly, it is important to thoroughly interrogate putative metallo-regulators with respect to these various functions, including the examination of multiple possible metal cofactors, regulatory activities (i.e., negative vs. positive regulation, direct vs. indirect regulation) and prospective regulons. With respect to chlamydial metallo-regulation, we are only just beginning to tackle each of these criteria.
Chlamydial Iron-Dependent Regulation
The early studies of chlamydial responses to iron deprivation invigorated interest in identifying the underlying modes of gene regulation. However, sequence homology did not indicate the existence of an obvious homolog to known iron-dependent regulators such as Fur, DtxR or Irr in the chlamydial genome. In fact, to date only eight transcriptional regulators have been characterized in Chlamydia, many of which were not immediately identified by sequence homology (Wyllie and Raulston, 2001; Wilson and Tan, 2002; Carlson et al., 2006; Koo et al., 2006; Schaumburg and Tan, 2006; Case et al., 2011; Rosario and Tan, 2012; Thompson et al., 2012). Moreover, no iron-dependent post-transcriptional mechanisms, such as RyhB homologs, have been identified. Technologies to mutagenize or transform Chlamydia to screen for iron-related phenotypes have only recently been developed and as such were not viable research strategies to address iron regulation initially. Despite these hurdles, Wyllie and Raulston endeavored to identify candidate iron-dependent regulators by more rigorously surveying the available sequence data. CLUSTAL sequence alignment revealed five ORFs that demonstrated limited sequence homology to Fur in other Gram-negative bacteria (Wyllie and Raulston, 2001). Among these five ORFs, CT296 emerged as a strong candidate Fur homolog following several lines of evidence: CT296 was specifically recognized by Pseudomonas aeruginosa Fur-specific polyclonal antiserum, ectopic expression of recombinant CT296 complemented a Fur-repression phenotype in E. coli, and CT296 bound a consensus Fur-box sequence in an electrophoretic mobility shift assay (EMSA). CT296 co-migrated with DNA in the presence of both Zn2+ and Mn2+; however, Fe2+ was not assayed in this study due to precipitation of the protein upon incubation with the biometal. Together, these data prompted the renaming of CT296 to divalent cation-dependent regulator (DcrA), and represented the first characterized transcriptional repressor in Chlamydia. These studies were then extended when a subsequent investigation utilized a Fur titration assay (FURTA) screen to identify regions of the chlamydial genome recognized by E. coli Fur (Rau et al., 2005). Selected candidate Fur-binding sequences were then screened for DcrA interaction by EMSA, identifying CT733, mreB, glgP, and tolR as possible DcrA regulatory targets. This work held the promise of beginning to define the scope of metallo-regulation in Chlamydia. However, later structural modeling, both by ab initio computational modeling and X-Ray crystallography, revealed that CT296 displayed no structural similarity to Fur and moreover contained no conserved DNA-binding motifs (Kemege et al., 2011). Furthermore, this study could not reproduce both the EMSA and the Fur-complementation assays previously reported by Wyllie and Raulston. Unfortunately, the function of CT296 has not been explored in further depth.
Perhaps unsurprisingly, the identification of a subsequent metallo-regulator in Chlamydia again required the implementation of a more specific and tailored examination of the available sequence data. By analyzing 240 predicted ORFs from the C. trachomatis serovar D genome using an in silico functional assignment algorithm, CT069 was identified as a candidate transcriptional regulator containing a C-terminal DtxR-like HTH domain (Akers et al., 2011). This was a surprising discovery given that CT069 encodes YtgC, annotated as an integral membrane permease that functions in concert with the ytgABCD operon. More careful inspection of the YtgC sequence revealed that this assignment was based on sequence homology of the N-terminus of YtgC to TroC in T. pallidum, but interestingly, the C-terminus possesses high identity to TroR, the Mn2+-dependent transcriptional repressor of the troABCDR operon (Posey et al., 1999). Akers et al. characterized the function of this C-terminal domain, YtgR, by demonstrating that YtgR could bind and repress the ytg promoter region in a Zn2+-dependent fashion in vitro and in a heterologous E. coli system. Curiously, this study reported on the activity of YtgR in the presence of Zn2+ and Mn2+, but not Fe2+. Given that the expression of YtgA was previously demonstrated to be iron-responsive and functioned to specifically bind Fe3+ over both Mn2+ and Zn2+, there existed strong evidence that the ytg system functioned in iron transport (Miller et al., 2009). The Zn2+-dependent function of YtgR did not appear to correspond with what was known about the ytgABCD operon and the authors did not provide an explanation as to how zinc availability may influence iron acquisition. It is plausible that the ytgABCD permease functions as a broad-spectrum divalent metal importer, and thus may be regulated by Zn2+ and import various divalent cations, but this was not addressed in this study. Regardless, the decision not to address the potential iron-dependency of YtgR left questions unanswered, especially considering the recognized variety of metal corepressors documented in Fur and DtxR activity. In addition, Akers et al. did not directly address how this YtgC-YtgR fusion could function as a transcriptional repressor if YtgC was indeed sequestered in the chlamydial inner membrane.
Independently, it was observed that the expression of ytgA at the transcript level was significantly induced in response to iron chelation by 2,2-bipyridyl, which was discussed in detail above (Thompson and Carabeo, 2011). This observation added more evidence to support the hypothesis that A.) the ytgABCD operon functions in iron transport and B.) that the ytgABCD operon was controlled by an iron-dependent system of regulation. Therefore, Thompson et al. conducted independent in silico analyses and confirmed the presence of the C-terminal DtxR-like domain within the YtgC coding sequence (Thompson et al., 2012). Importantly, critical DtxR metal-coordinating residues were highly conserved in the YtgC repressor domain (Pohl et al., 1999). Accordingly, the authors re-evaluated YtgR repressor function, to address both the ability of the repressor to utilize Fe2+ as a corepressor and act as a transcriptional repressor when physically fused with an integral membrane domain. Thompson et al. demonstrated that YtgR specifically bound the ytg promoter region in a Fe2+-dependent manner by in vitro repressor-DNA interaction assays. Importantly, YtgR was screened for DNA-binding activity against an assortment of divalent cations, including zinc, manganese, copper and cobalt, and observed no YtgR binding to the promoter with any of these possible alternative corepressors. This was in clear disagreement with the Zn2+-dependent activation of YtgR reported by Akers et al. It is possible that Zn2+ confers activation of YtgR at a higher concentration, and thus the DNA-binding assay employed by Thompson et al. simply failed to detect this activation. This would partly agree with the observation that equimolar concentrations of Zn2+ only partially activate DtxR DNA-binding in comparison with Fe2+ (Tao and Murphy, 1992). On that point, it is worth noting that Akers et al. relied on an E. coli Zn2+-efflux mutant (zntA zitB) to demonstrate Zn2+-dependent YtgR repression in vivo, concentrating Zn2+ in their heterologous experimental system. Given that some metallo-regulators, such as B. japonicum Mur and Fur, alter their metal affinity and thus repressor activity in different cellular environments (Hohle and O'Brian, 2016), it is unclear how artificially increased Zn2+ concentrations may affect YtgR metal specificity and activity. Ultimately, the new data reported by Thompson et al. suggested a strong preference for Fe2+ in YtgR activation.
Thompson et al. additionally observed that YtgR undergoes cleavage from YtgC during infection, as CT069 (YtgC) antisera detects two bands which migrate at approximately 49 and 28 kDa, likely representing the uncleaved YtgC and the YtgR cleavage product, respectively. Intriguingly, in purified EB preparations, only the 28 kDa species is detectable, suggesting that the EB is primed for iron-dependent regulation upon invasion. In agreement with the results obtained by Akers et al. the authors demonstrated that ectopically expressed recombinant YtgR can repress transcription from the ytg promoter in a heterologous E. coli system. Importantly, however, it was also demonstrated that ectopically expressed recombinant full-length YtgC displayed the same phenotype, suggesting that functional YtgR was cleaved from YtgC in this heterologous system. Indeed, this hypothesis was supported by immunoblot analysis of ectopically expressed recombinant YtgC in E. coli, which revealed several C-terminal cleavage products, a likely consequence of non-specific cleavage. Despite non-specific cleavage events, the E. coli reporter assay suggests that at least one of the cleaved products acted to repress transcription from the ytg promoter. It is possible that this data indicates that full-length YtgC can also function as a transcriptional repressor, perhaps similarly to membrane-anchored LacI (Görke et al., 2005). However, given that the C-terminal domain is liberated endogenously, cleavage likely represents a critical step in the processing of the repressor domain. Intriguingly, the authors identified the presence of orthologous permease-repressor fusion proteins in 11 bacterial phyla, suggesting that this mechanism of transcriptional regulation may exist throughout the bacterial kingdom.
Thus, the present model for iron-dependent regulation in Chlamydia involves first the import of iron through the Ytg permease system, which allows, upon the accumulation of sufficient iron, cleaved YtgR to coordinate Fe2+ and subsequently autoregulate the expression of the iron uptake machinery likely by out-competing RNAP for DNA binding (Figure 2). While cleavage of YtgR from YtgC was observed, a protease responsible for this cleavage has not yet been identified. However, the observation that YtgC is also cleaved heterologously in E. coli may indicate that the culprit protease is represented by a chlamydial homolog of a known E. coli protease. The stimulus that induces YtgC cleavage and thus liberation of YtgR has not been described. It is worth noting, however, that regardless of the exact stimulus, the requirement of cleavage imposes an additional level of regulation on chlamydial iron homeostasis at the level of the protease. The protease in turn may be regulated, further extending the tight control Chlamydia exerts on iron acquisition. At present, it is not clear how YtgR binding of DNA accomplishes transcriptional repression. As a DtxR homolog, YtgR may similarly complex around operator sites as a couplet of homodimers (Pohl et al., 1999). However, this model will require direct investigation of YtgR binding ratios and structural analyses to be verified. The identification of YtgR as an iron-dependent transcriptional repressor should potentiate subsequent studies of the iron regulon in Chlamydia, allowing researchers to address both functional and categorical aspects of iron-dependent regulation in a more systematic manner.
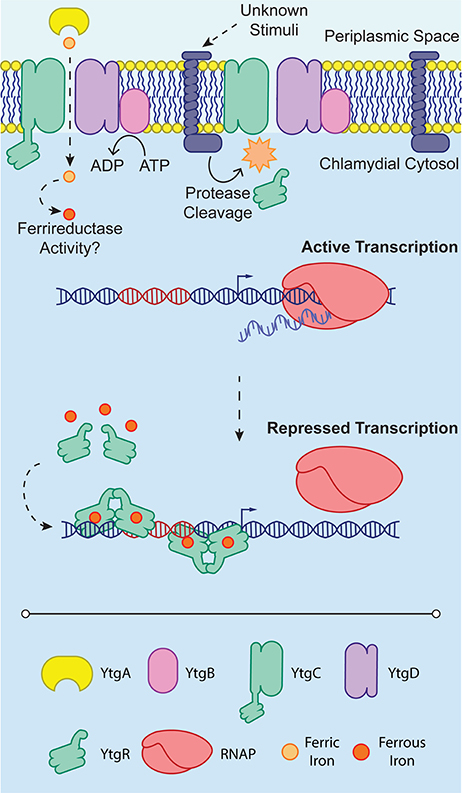
Figure 2. YtgR-mediated transcriptional regulation. To enable regulation, YtgR must be cleaved from the integral membrane YtgC domain. This cleavage is poorly understood and is depicted here as an integral membrane protease only for convenience. The stimuli that induce cleavage of YtgC in Chlamydia are not known. Under low iron conditions, expression of the ytgABCD operon is uninhibited, and the RNA polymerase holoenzyme (RNAP) can associate with the promoter region and initiate transcription. In contrast, when iron concentrations are sufficiently high, presumably following high expression of the ytgABCD operon and thus increased iron uptake, Fe2+ can activate YtgR-dependent transcriptional repression. This may occur by the association of pairs of YtgR homodimers binding to the operator sequence (depicted in red) as has been observed for DtxR. Transcriptional repression is thus accomplished by occlusion of RNAP from the promoter region of the operon.
Concluding Remarks
For 20 years it has been understood that iron represents a critical nutrient for the proper development of Chlamydia. Only in the latter half of this period have we begun to cement our understanding of chlamydial iron homeostasis in terms of molecular mechanisms. Thus, we can now place the ytgABCD operon squarely at the center of our knowledge regarding iron acquisition in Chlamydia. This agrees with the iron-dependent autoregulatory function of the cleavage product of YtgC, the YtgR repressor domain. Recent data also strongly suggests that iron is trafficked to the chlamydial inclusion via the slow recycling pathway of transferrin. Moreover, we now have more adequate tools with which to address important questions surrounding chlamydial iron biology. The iron chelator 2,2-bipyridyl should facilitate more robust studies on the responses of Chlamydia to iron starvation. The advantages of bipyridyl over previous methods may provide the opportunity to begin to truly understand the primary response of the organism to iron deprivation, as compared to the response of fully persistent aberrant chlamydial forms. In addition, as the golden age of chlamydial genetics has now dawned upon the field, we can ask more prying questions into the molecular basis underlying chlamydial iron responses by manipulating chlamydial, as well as host proteins, and understand in greater detail the iron-dependent regulon of Chlamydia and the ways in which Chlamydia acquires and traffics iron away from their host. Fully elucidating the regulatory function of YtgR and the mechanism of the ytg iron acquisition system will likely require the introduction of subtle mutations, such as base substitutions, given the potential indispensability of these systems for proper chlamydial development. Technologies such as CRISPR/Cas9 will be invaluable in these studies. Chlamydia appears to defy stereotypes at every turn, and we can no longer make assumptions on mechanisms or simply rely on referencing systems present in other organisms if we are to understand the basic biology of this pathogen.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Dr. Amanda Brinkworth, Dr. António Pedrosa and Ana Nogueira for their valuable critique in review of this manuscript. This is supported by funding from the NIH (AI065545) and start-up funds from the WSU College of Veterinary Medicine to RC, NP is supported by the NIH Protein Biotechnology Training Grant awarded to WSU and an ARCS Fellowship. CT was supported by the Jefferiss Trust Fellowship.
References
AbdelRahman, Y. M., and Belland, R. J. (2005). The chlamydial developmental cycle. FEMS Microbiol. Rev. 29, 949–959. doi: 10.1016/j.femsre.2005.03.002
Adhikari, P., Berish, S. A., Nowalk, A. J., Veraldi, K. L., Morse, S. A., and Mietzner, T. A. (1996). The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J. Bacteriol. 178, 2145–2149. doi: 10.1128/jb.178.7.2145-2149.1996
Ahn, B., Cha, J., Lee, E., Han, A., Thompson, C. J., and Roe, J. (2006). Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59, 1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x
Akers, J. C., HoDac, H., Lathrop, R. H., and Tan, M. (2011). Identification and functional analysis of CT069 as a novel transcriptional regulator in Chlamydia. J. Bacteriol. 193, 6123–6131. doi: 10.1128/JB.05976-11
Alamuri, P., Mehta, N., Burk, A., and Maier, R. J. (2006). Regulation of the Helicobacter pylori Fe-S cluster synthesis protein NifS by iron, oxidative stress conditions, and Fur. J. Bacteriol. 188, 5325–5330. doi: 10.1128/JB.00104-06
Al-Younes, H. M., Brinkmann, V., and Meyer, T. F. (2004). Interaction of Chlamydia trachomatis serovar L2 with the host autophagic pathway. Infect. Immun. 72, 4751–4762. doi: 10.1128/IAI.72.8.4751-4762.2004
Al-younes, H. M., Rudel, T., and Meyer, T. F. (1999). Characterization and intracellular trafficking pattern of vacuoles containing Chlamydia pneumoniae in human epithelial cells. Cell. Microbiol. 1, 237–247. doi: 10.1046/j.1462-5822.1999.00024.x
Al-Younes, H. M., Rudel, T., Brinkmann, V., Szczepek, A. J., and Meyer, T. F. (2001). Low iron availability modulates the course of Chlamydia pneumoniae infection. Cell. Microbiol. 3, 427–437. doi: 10.1046/j.1462-5822.2001.00125.x
Anderson, J. E., Sparling, P. F., and Cornelissen, C. N. (1994). Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J. Bacteriol. 176, 3162–3170. doi: 10.1128/jb.176.11.3162-3170.1994
Andersson, S. G. E., and Kurland, C. G. (1998). Reductive evolution of resident genomes. Trends Microbiol. 6, 263–268. doi: 10.1016/S0966-842X(98)01312-2
Andrews, N. C. (2000). Iron homeostasis: insights from genetics and animal models. Nat. Rev. Genet. 1, 208–217. doi: 10.1038/35042073
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Bagg, A., and Neilands, J. B. (1987). Ferric Uptake Regulation protein acts as a repressor, employing iron (ii) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26, 5471–5477. doi: 10.1021/bi00391a039
Bannantine, J. P., and Rockey, D. D. (1999). Use of a primate model system to identify Chlamydia trachomatis protein antigens recognized uniquely in the context of infection. Microbiology 145, 2077–2085. doi: 10.1099/13500872-145-8-2077
Barnewall, R. E., and Rikihisa, Y. (1994). Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron transferrin. Infect. Immun. 62, 4804–4810.
Barnewall, R. E., Ohashi, N., and Rikihisa, Y. (1999). Ehrlichia chaffeensis and E. sennetsu, but Not the human granulocytic ehrlichiosis agent, colocalize with transferrin receptor and up-regulate transferrin receptor mRNA by activating iron-responsive protein 1. Infect. Immun. 67, 2258–2265.
Barrile, R., Kasendra, M., Rossi-Paccani, S., Merola, M., Pizza, M., Baldari, C., et al. (2015). Neisseria meningitidis subverts the polarized organization and intracellular trafficking of host cells to cross the epithelial barrier. Cell. Microbiol. 17, 1365–1375. doi: 10.1111/cmi.12439
Bearden, S. W., and Perry, R. D. (1999). The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32, 403–414. doi: 10.1046/j.1365-2958.1999.01360.x
Beatty, W. L., Belanger, T. A., Desai, A. A., Morrison, R. P., and Byrne, G. I. (1994). Tryptophan depletion as a mechanism of gamma interferon-mediated chlamydial persistence. Infect. Immun. 62, 3705–3711.
Belland, R. J., Nelson, D. E., Virok, D., Crane, D. D., Hogan, D., Sturdevant, D., et al. (2003a). Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. U.S.A. 100, 15971–15976. doi: 10.1073/pnas.2535394100
Belland, R. J., Zhong, G., Crane, D. D., Hogan, D., Sturdevant, D., Sharma, J., et al. (2003b). Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U.S.A. 100, 8478–8483. doi: 10.1073/pnas.1331135100
Bellmann-Weiler, R., Schroll, A., Engl, S., Nairz, M., Talasz, H., Seifert, M., et al. (2013). Neutrophil gelatinase-associated lipocalin and interleukin-10 regulate intramacrophage Chlamydia pneumoniae replication by modulating intracellular iron homeostasis. Immunobiology 218, 969–978. doi: 10.1016/j.imbio.2012.11.004
Bennoun, M., Devaux, I., Beaumont, C., Grandchamp, B., Kahn, A., and Vaulont, S. (2001). Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc. Natl. Acad. Sci. U.S.A. 98, 8780–8785. doi: 10.1073/pnas.151179498
Berger, T., Togawa, A., Duncan, G. S., Elia, A. J., You-Ten, A., Wakeham, A., et al. (2006). Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U.S.A. 103, 1834–1839. doi: 10.1073/pnas.0510847103
Boradia, V. M., Malhotra, H., Thakkar, J. S., Tillu, V. A., Vuppala, B., Patil, P., et al. (2014). Mycobacterium tuberculosis acquires iron by cell-surface sequestration and internalization of human holo-transferrin. Nat. Commun. 5:5730. doi: 10.1038/ncomms5730
Boyd, J., Oza, M. N., and Murphy, J. R. (1990). Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc. Natl. Acad. Sci. U.S.A. 87, 5968–5972. doi: 10.1073/pnas.87.15.5968
Briggs, H. L., Pul, N., Seshadri, R., Wilson, M. J., Tersteeg, C., Russell-Lodrigue, K. E., et al. (2008). Limited role for iron regulation in Coxiella burnetii Pathogenesis. Infect. Immun. 76, 2189–2201. doi: 10.1128/IAI.01609-07
Byrne, G. I., Lehmann, L. K., and Landry, G. J. (1986). Induction of tryptophan catabolism is the mechanism for gamma-interferon-mediated inhibition of intracellular Chlamydia psittaci replication in T24 cells. Infect. Immun. 53, 347–351.
Cable, H., and Lloyd, J. B. (1999). Cellular uptake and release of two contrasting iron chelators. J. Pharm. Pharmacol. 51, 131–134. doi: 10.1211/0022357991772231
Calderwood, S. B., and Mekalanos, J. J. (1987). Iron regulation of shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169, 4759–4764. doi: 10.1128/jb.169.10.4759-4764.1987
Carlson, J. H., Wood, H., Roshick, C., Caldwell, H. D., and McClarty, G. (2006). In vivo and in vitro studies of Chlamydia trachomatis TrpR:DNA interactions. Mol. Microbiol. 59, 1678–1691. doi: 10.1111/j.1365-2958.2006.05045.x
Carniel, E., Rossi, M., Fetherston, J. D., Le, S., Perry, R. D., and Ghigo, J. (2001). Identification and characterization of the hemophore-dependent heme acquisition system of Yersinia pestis. Infect. Immun. 69, 6707–6717. doi: 10.1128/IAI.69.11.6707-6717.2001
Carpenter, B. M., Whitmire, J. M., and Merrell, D. S. (2009). This is not your mother's repressor: the complex role of fur in pathogenesis. Infect. Immun. 77, 2590–2601. doi: 10.1128/IAI.00116-09
Carrano, C. J., and Raymond, K. N. (1979). Ferric Iron Sequestering agents. 2. kinetics and mechanism of iron removal from transferrin by enterobactin and synthetic tricatechols. J. Am. Chem. Soc. 101, 5401–5404. doi: 10.1021/ja00512a047
Case, E. D. R., Akers, J. C., and Tan, M. (2011). CT406 encodes a chlamydial ortholog of NrdR, a repressor of ribonucleotide reductase. J. Bacteriol. 193, 4396–4404. doi: 10.1128/JB.00294-11
Casey, J. L., Koeller, D. M., Ramin, C., Klausner, R. D., and Harford, J. B. (1989). Iron regulation of transferrin receptor mRNA levels requires iron-responsive elements and a rapid turnover determinant in the 3' untranslated region of the mRNA. EMBO J. 8, 3693–3699.
Caza, M., and Kronstad, J. W. (2013). Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 3:80. doi: 10.3389/fcimb.2013.00080
Chatfield, C. H., and Cianciotto, N. P. (2007). The secreted pyomelanin pigment of Legionella pneumophila confers ferric reductase activity. Infect. Immun. 75, 4062–4070. doi: 10.1128/IAI.00489-07
Cheng, Y., Zak, O., Aisen, P., Harrison, S. C., Walz, T., and York, N. (2004). Structure of the human transferrin receptor-transferrin complex albert einstein college of medicine. Cell 116, 565–576. doi: 10.1016/S0092-8674(04)00130-8
Chung, J., Chen, C., and Paw, B. H. (2012). Heme metabolism and erythropoiesis. Curr. Opin. Hematol. 19, 156–162. doi: 10.1097/MOH.0b013e328351c48b
Clarke, I. N. (2011). Evolution of Chlamydia trachomatis. Ann. N. Y. Acad. Sci. 1230, 11–18. doi: 10.1111/j.1749-6632.2011.06194.x
Cockrell, D. C., Long, C. M., Robertson, S. J., Shannon, J. G., Miller, E., Myers, L., et al. (2017). Robust growth of a virulent phase II Coxiella burnetii in bone marrow-derived murine macrophages. PLoS ONE 12:173528. doi: 10.1371/journal.pone.0173528
Cohen, M., Britigan, B., French, M., and Bean, K. (1987). Preliminary observations on lactoferrin secretion in human vaginal mucus: variation during the menstrual cycle, evidence of hromonal regulation, and implications for infection with Neisseria gonorrhoeae. Am. J. Obstet. Gynecol. 157, 1122–1125. doi: 10.1016/S0002-9378(87)80274-0
Cornelissen, C. N. A. U., Biswas, G. D., Tsai, J., Paruchuri, D. K., Thompson, S. A., and Sparling, P. F. (1992). Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to tonB-dependent outer membrane receptors. J. Bacteriol. 174, 5788–5797. doi: 10.1128/jb.174.18.5788-5797.1992
Crichton, R. R. (ed.). (2016). “Cellular iron uptake and export in mammals,” in Iron Metabolism: From Molecular Mechanisms to Clinical Consequences (Chichester: John Wiley & Sons, Inc.), 364–428.
Crossley, R. A., Gaskin, D. J. H., Holmes, K., Mulholland, F., Wells, J. M., Kelly, D. J., et al. (2007). Riboflavin biosynthesis is associated with assimilatory ferric reduction and iron acquisition by Campylobacter jejuni. Appl. Environ. Microbiol. 73, 7819–7825. doi: 10.1128/AEM.01919-07
Dautry-varsat, A., Ciechanover, A., and Lodish, H. F. (1983). pH and the recycling of transferrin during receptor- mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 80, 2258–2262. doi: 10.1073/pnas.80.8.2258
de Lorenzo, V., Wee, S., Herrero, M., and Neilands, J. B. (1987). Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169, 2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987
Derré, I., Pypaert, M., Dautry-Varsat, A., and Agaisse, H. (2007). RNAi screen in Drosophila cells reveals the involvement of the tom complex in Chlamydia infection. PLoS Pathog. 3:30155. doi: 10.1371/journal.ppat.0030155
Dill, B. D., and Raulston, J. E. (2007). Examination of an inducible expression system for limiting iron availability during Chlamydia trachomatis infection. Microbes Infect. 9, 947–953. doi: 10.1016/j.micinf.2007.03.017
Dill, B. D., Dessus-Babus, S., and Raulston, J. E. (2009). Identification of iron-responsive proteins expressed by Chlamydia trachomatis reticulate bodies during intracellular growth. Microbiology 155, 210–219. doi: 10.1099/mic.0.022731-0
Donati, M., Huot-Creasy, H., Humphrys, M., Paolo, M., Di Francesco, A., and Myersb, G. S. A. (2014). Genome sequence of Chlamydia suis MD56, isolated from the conjunctiva of a weaned piglet. Genome Announc. 2:2147. doi: 10.1128/genomeA.00425-14
Donovan, A., Brownlie, A., Zhou, Y., Shepard, J., Pratt, S. J., Moynihan, J., et al. (2000). Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403, 776–781. doi: 10.1038/35001596
Doyle, C. K., Zhang, X., Popov, V. L., and McBride, J. W. (2005). An immunoreactive 38-kilodalton protein of Ehrlichia canis shares structural homology and iron-binding capacity with the ferric ion-binding protein family. Infect. Immun. 73, 62–69. doi: 10.1128/IAI.73.1.62-69.2005
Drakesmith, H., and Prentice, A. M. (2012). Hepcidin and the iron-infection axis. Science 338, 768–772. doi: 10.1126/science.1224577
Ellison, D. W., Clark, T. R., Sturdevant, D. E., Virtaneva, K., and Hackstadt, T. (2009). Limited transcriptional responses of Rickettsia rickettsii exposed to environmental stimuli. PLoS ONE 4:5612. doi: 10.1371/journal.pone.0005612
Ellison, D. W., Clark, T. R., Sturdevant, D. E., Virtaneva, K., Porcella, S. F., and Hackstadt, T. (2008). Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect. Immun. 76, 542–550. doi: 10.1128/IAI.00952-07
Ernst, J. F., Bennert, R. L., and Rothfield, L. I. (1978). Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J. Bacteriol. 135, 928–934.
Escolar, L., de Lorenzo, V., and Perez-Martin, J. (1997). Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol. Microbiol. 26, 799–808. doi: 10.1046/j.1365-2958.1997.6211987.x
Escolar, L., Pérez-Martín, J., and De Lorenzo, V. (1998). Coordinated repression in vitro of the divergent fepA-fes promoters of Escherichia coli by the iron uptake regulation (Fur) protein. J. Bacteriol. 180, 2579–2582.
Fecker, L., and Braun, V. (1983). Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J. Bacteriol. 156, 1301–1314.
Fleming, M. D., Romano, M. A., Su, M. A., Garrick, L. M., Garrick, M. D., and Andrews, N. C. (1998). Nramp 2 is mutated in the anemic Belgrade (b) rat : Evidence of a role for Nramp2 in endosomal iron transport. Proc. Natl. Acad. Sci. U.S.A. 95, 1148–1153. doi: 10.1073/pnas.95.3.1148
Fleming, M., Trenor, C. III, Su, M., Foernzler, D., Beier, D., Dietrich, W., et al. (1997). Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat. Genet. 16, 383–386. doi: 10.1038/ng0897-383
Flo, T. H. T., Smith, K. D. K., Sato, S., Rodriguez, D. J. D. J., Holmes, M. A., Strong, R. K., et al. (2004). Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921. doi: 10.1038/nature03104
Freidank, H. M., Billing, H., and Wiedmann-Al-Ahmad, M. (2001). Influence of iron restriction on Chlamydia pneumoniae and C. trachomatis. J. Med. Microbiol. 50, 223–227. doi: 10.1099/0022-1317-50-3-223
Frey, P. A., and Reed, G. H. (2012). The ubiquity of iron. ACS Chem. Biol. 7, 1477–1481. doi: 10.1021/cb300323q
Gaballa, A., and Helmann, J. D. (1998). Identification of a zinc-specific metalloregulatory protein, zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180, 5815–5821.
Gancz, H., Censini, S., and Merrell, D. S. (2006). Iron and pH homeostasis intersect at the level of fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74, 602–614. doi: 10.1128/IAI.74.1.602-614.2006
Ganz, T., and Nemeth, E. (2015). Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 15, 500–510. doi: 10.1038/nri3863
Glickstein, H., El, R., Ben, M., and Cabantchik, Z. I. (2005). Intracellular labile iron pools as direct targets of iron chelators: a fluorescence study of chelator action in living cells. Blood 106, 3242–3251. doi: 10.1182/blood-2005-02-0460
Goellner, S., Schubert, E., Liebler-Tenorio, E., Hotzel, H., Saluz, H. P., and Sachse, K. (2006). Transcriptional response patterns of Chlamydophila psittaci in different in vitro models of persistent infection. Infect. Immun. 74, 4801–4808. doi: 10.1128/IAI.01487-05
Gold, B., Rodriguez, G. M., Salvatore, A., Marras, E., Pentecost, M., and Smith, I. (2001). The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol. 42, 851–865. doi: 10.1046/j.1365-2958.2001.02684.x
Görke, B., Reinhardt, J., and Rak, B. (2005). Activity of Lac repressor anchored to the Escherichia coli inner membrane. Nucleic Acids Res. 33, 2504–2511. doi: 10.1093/nar/gki549
Gottesman, S., and Masse, E. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. doi: 10.1073/pnas.032066599
Gresock, M. G., Kastead, K. A., and Postle, K. (2015). From homodimer to heterodimer and back: elucidating the TonB energy transduction cycle. J. Bacteriol. 197, 3433–3445. doi: 10.1128/JB.00484-15
Gresock, M. G., Savenkova, M. I., Larsen, R. A., Ollis, A. A., and Postle, K. (2011). Death of the TonB shuttle hypothesis. Front. Microbiol. 2:206. doi: 10.3389/fmicb.2011.00206
Grinblat-huse, V., Drabek, E. F., Creasy, H. H., Daugherty, S. C., Jones, K. M., Santana-cruz, I., et al. (2011). Genome sequences of the zoonotic pathogens Chlamydia psittaci. J. Bacteriol. 193, 4039–4040. doi: 10.1128/JB.05277-11
Gunshin, H., Mackenzie, B., Berger, U. V., Gunshin, Y., Romero, M. F., Boron, W. F., et al. (1997). Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388, 482–488. doi: 10.1038/41343
Halaas, O., Steigedal, M., Haug, M., Awuh, J. A., Ryan, L., Brech, A., et al. (2010). Intracellular Mycobacterium avium intersect transferrin in the Rab11+ recycling endocytic pathway and avoid lipocalin 2 trafficking to the lysosomal pathway. J. Infect. Dis. 201, 783–792. doi: 10.1086/650493
Hamza, I., Chauhan, S., Hassett, R., and O'Brian, M. R. (1998). The bacterial irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273, 21669–21674.
Hamza, I., Qi, Z., King, N. D., and O'Brian, M. R. (2000). Fur-independent regulation of iron metabolism by Irr in Bradyrhizobium japonicum. Microbiology 146, 669–676. doi: 10.1099/00221287-146-3-669
Hantke, K. (1981). Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182, 288–292. doi: 10.1007/BF00269672
Hantke, K. (1982). Negative control of iron uptake systems in Escherichia coli. FEMS Microbiol. Lett. 15, 83–86. doi: 10.1111/j.1574-6968.1982.tb00043.x
Hantke, K. (2001). Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4, 172–177. doi: 10.1016/S1369-5274(00)00184-3
Harding, C., Heuser, J., and Stahl, P. (1983). Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes biochemical approaches to transferrin. J. Cell Biol. 97, 329–339. doi: 10.1083/jcb.97.2.329
Harrison, P. M., and Arosio, P. (1996). The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1275, 161–203. doi: 10.1016/0005-2728(96)00022-9
Hellman, N. E., and Gitlin, J. D. (2002). Ceruloplasmin metabolsim and function. Annu. Rev. Nutr. 22, 439–458. doi: 10.1146/annurev.nutr.22.012502.114457
Hentze, M. W., Muckenthaler, M. U., and Andrews, N. C. (2004). Balancing acts : molecular control of mammalian iron metabolism. Cell 117, 285–297. doi: 10.1016/S0092-8674(04)00343-5
Hentze, M. W., Muckenthaler, M. U., Galy, B., and Camaschella, C. (2010). Two to tango: regulation of mammalian iron metabolism. Cell 142, 24–38. doi: 10.1016/j.cell.2010.06.028
Hohle, T. H., and O'Brian, M. R. (2010). Transcriptional control of the Bradyrhizobium japonicum irr gene requires repression by fur and antirepression by irr. J. Biol. Chem. 285, 26074–26080. doi: 10.1074/jbc.M110.145979
Hohle, T. H., and O'Brian, M. R. (2016). Metal-specific control of gene expression mediated by Bradyrhizobium japonicum mur and Escherichia coli fur is determined by the cellular context. Mol. Microbiol. 101, 152–166. doi: 10.1111/mmi.13381
Humphrys, M. S., Creasy, T., Sun, Y., Shetty, A. C., Chibucos, M. C., Drabek, E. F., et al. (2013). Simultaneous transcriptional profiling of bacteria and their host cells. PLoS ONE 8:80597. doi: 10.1371/journal.pone.0080597
Kalman, S., Mitchell, W., Marathe, R., Lammel, C., Fan, J., Hyman, R. W., et al. (1999). Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21, 385–389. doi: 10.1038/7716
Kammler, M., Schon, C., and Hantke, K. (1993). Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175, 6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993
Keberle, H. (1964). The biochemistry of desferrioxamine and its relation to iron metabolism. Ann. N.Y. Acad. Sci. 119, 758–768. doi: 10.1111/j.1749-6632.1965.tb54077.x
Kelver, M. E., Kaul, A., Nowicki, B., Findley, W. E., Hutchens, T. W., and Nagamani, M. (1996). Estrogen regulation of lactoferrin expression in human endometrium. Am. J. Reprod. Immunol. 36, 243–247. doi: 10.1111/j.1600-0897.1996.tb00171.x
Kemege, K. E., Hickey, J. M., Lovell, S., Battaile, K. P., Zhang, Y., and Hefty, P. S. (2011). Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes. J. Bacteriol. 193, 6517–6528. doi: 10.1128/JB.05488-11
Kim, H. J., Khalimonchuk, O., Smith, P. M., and Winge, D. R. (2012). Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta 1823, 1604–1616. doi: 10.1016/j.bbamcr.2012.04.008
Koo, I. C., Walthers, D., Hefty, P. S., Kenney, L. J., and Stephens, R. S. (2006). ChxR is a transcriptional activator in Chlamydia. Proc. Natl. Acad. Sci. U.S.A. 103, 750–755. doi: 10.1073/pnas.0509690103
Kosman, D. J. (2003). Molecular mechanisms of iron uptake in fungi. Mol. Microbiol. 47, 1185–1197. doi: 10.1046/j.1365-2958.2003.03368.x
Kumar, R., Lovell, S., Matsumura, H., Battaile, K. P., Moenne-Loccoz, P., and Rivera, M. (2013). The hemophore hasa from Yersinia pestis (HasAyp) coordinates hemin with a single residue, Tyr75, and with minimal conformational change. Biochemistry 52, 2705–2707. doi: 10.1021/bi400280z
Kunkle, C. A., and Schmitt, M. P. (2005). Analysis of a DtxR-regulated iron transport and siderophore biosynthesis gene cluster in Corynebacterium diphtheriae. J. Bacteriol. 187, 422–433. doi: 10.1128/JB.187.2.422-433.2005
LaRue, R. W., Dill, B. D., Giles, D. K., Whittimore, J. D., and Raulston, J. E. (2007). Chlamydial Hsp60-2 is iron responsive in Chlamydia trachomatis serovar E-infected human endometrial epithelial cells in vitro. Infect. Immun. 75, 2374–2380. doi: 10.1128/IAI.01465-06
Lawson, D. M., Treffry, A., Artyrniuk, P. J., Harrison, P. M., Yewdall, S. J., Luzzago, A., et al. (1989). Identification of the ferroxidase centre in ferritin. FEBS Lett. 254, 207–210. doi: 10.1016/0014-5793(89)81040-3
Lee, J. H., Wang, T., Ault, K., Liu, J., Schmitt, M. P., and Holmes, R. K. (1997). Identification and characterization of three new promoter/operators from Corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. Infect. Immun. 65, 4273–4280.
Lee, J. W., and Helmann, J. D. (2007). Functional specialization within the fur family of metalloregulators. Biometals 20, 485–499. doi: 10.1007/s10534-006-9070-7
Lee, Y., Deka, R. K., Michael, V., Radolf, J. D., and Hasemann, C. A. (1999). Treponema pallidum TroA is a periplasmic zinc-binding protein with a helical backbone. Nat. Struct. Biol. 6, 628–633. doi: 10.1038/10677
Lefèvre, J., Delepelaire, P., Delepierre, M., and Izadi-pruneyre, N. (2008). Modulation by substrates of the interaction between the HasR outer membrane receptor and its specific. J. Mol. Biol. 378, 840–851. doi: 10.1016/j.jmb.2008.03.044
Lewis, M. E., Belland, R. J., AbdelRahman, Y. M., Beatty, W. L., Aiyar, A. A., Zea, A. H., et al. (2014). Morphologic and molecular evaluation of Chlamydia trachomatis growth in human endocervix reveals distinct growth patterns. Front. Cell. Infect. Microbiol. 4:71. doi: 10.3389/fcimb.2014.00071
Lipinski, A. R., Heymann, J., Meissner, C., Karlas, A., Brinkmann, V., Meyer, T. F., et al. (2009). Rab6 and Rab11 regulate Chlamydia trachomatis development and golgin-84-dependent golgi fragmentation. PLoS Pathog. 5:e1000615. doi: 10.1371/journal.ppat.1000615
Lloyd, J. B., Cable, H., and Rice-Evans, C. (1991). Evidence that desferrioxamine cannot enter cells by passive diffusion. Biochem. Pharmacol. 41, 1361–1363. doi: 10.1016/0006-2952(91)90109-I
Maass, M., Essig, A., Marre, R., and Henkel, W. (1993). Growth in serum-free medium improves isolation of Chlamydia pneumoniae. J. Clin. Microbiol. 31, 3050–3052.
Marx, J. J. M. (2002). Iron and infection: competition between host and microbes for a precious element. Best Pract. Res. Clin. Haematol. 15, 411–426. doi: 10.1053/beha.2002.0001
Matsumoto, A., Bessho, H., Uehira, K., and Suda, T. (1991). Morphological studies of the association of mitochondria with chlamydial inclusions and the fusion of chlamydial inclusions. J. Electron. Microsc. 40, 356–363. doi: 10.1093/oxfordjournals.jmicro.a050908
Mäurer, A. P., Mehlitz, A., Mollenkopf, H. J., and Meyer, T. F. (2007). Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 3, 0752–0769. doi: 10.1371/journal.ppat.0030083
Mayle, K. M., Le, A. M., and Kamei, D. T. (2012). The intracellular trafficking pathway of transferrin. Biochim. Biophys. Acta 1820, 264–281. doi: 10.1016/j.bbagen.2011.09.009
McKie, A. T., Barrow, D., Latunde-dada, G. O., Rolfs, A., Sager, G., Mudaly, E., et al. (2001). An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 291, 1755–1759. doi: 10.1126/science.1057206
Mehi, O., Bogos, B., Csorgo, B., Pal, F., Nyerges, A., Papp, B., et al. (2014). Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol. Biol. Evol. 31, 2793–2804. doi: 10.1093/molbev/msu223
Mellouk, N., and Enninga, J. (2016). Cytosolic access of intracellular bacterial pathogens: the shigella paradigm. Front. Cell. Infect. Microbiol. 6:35. doi: 10.3389/fcimb.2016.00035
Mertens, K., and Samuel, J. E. (2012). “Defense mechanisms against oxidative stress in Coxiella burnetii: adaptation to a unique intracellular niche,” in Coxiella burnetii: Recent Advances and New Perspectives in Research of the Q Fever Bacterium, eds R. Toman, R. A. Heinzen, J. E. Samuel, and J. L. Mege (Dordrecht: Springer Netherlands), 39–63.
Mietzner, T. A., Luginbuhl, G. H., Sandstrom, E., and Morse, S. A. (1984). Identification of an iron-regulated 37,000-dalton protein in the cell envelope of Neisseria gonorrhoeae. Infect. Immun. 45, 410–416.
Miller, J. D., Sal, M. S., Schell, M., Whittimore, J. D., and Raulston, J. E. (2009). Chlamydia trachomatis YtgA is an iron-binding periplasmic protein induced by iron restriction. Microbiology 155, 2884–2894. doi: 10.1099/mic.0.030247-0
Mojica, S., Creasy, H. H., Daugherty, S., Read, T. D., Kim, T., Kaltenboeck, B., et al. (2011). Genome sequence of the obligate intracellular animal pathogen Chlamydia pecorum E58. J. Bacteriol. 193, 9193. doi: 10.1128/JB.00454-11
Moore, E. R., and Ouellette, S. P. (2014). Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Front. Cell. Infecit. Microbiol. 4:157. doi: 10.3389/fcimb.2014.00157
Mukhopadhyay, S., Miller, R. D., Sullivan, E. D., Theodoropoulos, C., Mathews, S. A., Timms, P., et al. (2006). Protein expression profiles of Chlamydia pneumoniae in models of persistence versus those of heat shock stress response. Infect. Immun. 74, 3853–3863. doi: 10.1128/IAI.02104-05
Murphy, J. R., Pappenheimer, A. M. Jr., and Tayart de Borms, S. (1974). Synthesis of diphtheria tox-gene products in Eacherichia coli Extracts. Proc. Natl. Acad. Sci. U.S.A. 71, 11–15. doi: 10.1073/pnas.71.1.11
Nairz, M., Haschka, D., Demetz, E., and Weiss, G. (2014). Iron at the interface of immunity and infection. Front. Pharmacol. 5:152. doi: 10.3389/fphar.2014.00152
Nairz, M., Theurl, I., Schroll, A., Theurl, M., Fritsche, G., Lindner, E., et al. (2009). Absence of functional Hfe protects mice from invasive Salmonella enterica serovar typhimurium infection via induction of lipocalin-2. Blood 114, 3642–3651. doi: 10.1182/blood-2009-05-223354
Nemeth, E., Tuttle, M. S., Powelson, J., Vaughn, M. B., Donovan, A., Ward, D. M., et al. (2004). Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306, 2090–2093. doi: 10.1126/science.1104742
Neufeld, E. J. (2006). Oral chelators deferasirox and defefiprone for transfusional iron overload in thalassen a major: new data, new questions. Blood 107, 3436–3441. doi: 10.1182/blood-2006-02-002394
Nienaber, A., Hennecke, H., and Fischer, H. M. (2001). Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol. Microbiol. 41, 787–800. doi: 10.1046/j.1365-2958.2001.02555.x
Noinaj, N., Buchanan, S. K., and Cornelissen, C. N. (2012a). The transferrin—iron import system from pathogenic Neisseria species. Mol. Microbiol. 86, 246–257. doi: 10.1111/mmi.12002
Noinaj, N., Easley, N. C., Oke, M., Mizuno, N., Gumbart, J., Boura, E., et al. (2012b). Structural basis for iron piracy by pathogenic Neisseria. Nature 482, 53–58. doi: 10.1038/nature10823
Nystrom-Rosander, C., Lindh, U., Ilback, N.-G., Hjelm, E., Thelin, S., Lindqvist, O., et al. (2003). Interactions between Chlamydia pneumoniae and trace elements: a possible link to aortic valve sclerosis. Biol. Trace Elem. Res. 91, 97–110. doi: 10.1385/BTER:91:2:97
Oexle, H., Gnaiger, E., and Weiss, G. (1999). Iron-dependent changes in cellular energy metabolism: influence on citric acid cycle and oxidative phosphorylation. Biochim. Biophys. Acta 1413, 99–107. doi: 10.1016/S0005-2728(99)00088-2
Ohgami, R. S., Campagna, D. R., Greer, E. L., Antiochos, B., McDonald, A., Chen, J., et al. (2005). Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat. Genet. 37, 1264–1269. doi: 10.1038/ng1658
Ojeda, J. F., Martinson, D. A., Menscher, E. A., and Roop, R. M. (2012). The bhuQ gene encodes a heme oxygenase that contributes to the ability of Brucella abortus 2308 to use heme as an iron source and is regulated by Irr. J. Bacteriol. 194, 4052–4058. doi: 10.1128/JB.00367-12
Osaki, S., Johnson, D. A., and Frieden, E. (1966). The Possible Significance of the ferrous oxidase activity of ceruloplasmin in normal human serum. J. Biol. Chem. 241, 2746–2751.
Ostberg, K. L., DeRocco, A. J., Mistry, S. D., Dickinson, M. K., and Cornelissen, C. N. (2013). Conserved regions of gonococcal TbpB are critical for surface exposure and transferrin iron utilization. Infect. Immun. 81, 3442–3450. doi: 10.1128/IAI.00280-13
Ouellette, S. P., and Carabeo, R. A. (2010). A functional slow recycling pathway of transferrin is required for growth of Chlamydia. Front. Microbiol. 1:112. doi: 10.3389/fmicb.2010.00112
Ouellette, S. P., Dorsey, F. C., Moshiach, S., Cleveland, J. L., and Carabeo, R. A. (2011). Chlamydia species-dependent differences in the growth requirement for lysosomes. PLoS ONE 6:16783. doi: 10.1371/journal.pone.0016783
Ouellette, S. P., Hatch, T. P., AbdelRahman, Y. M., Rose, L. A., Belland, R. J., and Byrne, G. I. (2006). Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol. Microbiol. 62, 1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x
Outten, C. E., Tobin, D. A., Penner-Hahn, J. E., and O'Halloran, T. V. O. (2001). Characterization of the metal receptor sites in Escherichia coli zur, an ultrasensitive Zinc(II) metalloregulatory protein. Biochemistry 40, 10417–10423. doi: 10.1021/bi0155448
Ozenberger, B. A., Nahlik, M. S., and McIntosh, M. A. (1987). Genetic organization of multiple fep genes encoding ferric enterobactin transport functions in Escherichia coli. J. Bacteriol. 169, 3638–3646. doi: 10.1128/jb.169.8.3638-3646.1987
Paradkar, P. N., Domenico, I., De Durchfort, N., Zohn, I., Kaplan, J., and Ward, D. M. (2008). Iron depletion limits intracellular bacterial growth in macrophages. Blood 112, 866–874. doi: 10.1182/blood-2007-12-126854
Parrow, N. L., Abbott, J., Lockwood, A. R., Battisti, J. M., and Minnick, M. F. (2009). Function, regulation, and transcriptional organization of the hemin utilization locus of Bartonella quintana. Infect. Immun. 77, 307–316. doi: 10.1128/IAI.01194-08
Patzer, S. I., and Hantke, K. (2001). Dual repression by Fe(2+)-Fur and Mn(2+)-MntR of the mntH Gene, encoding an NRAMP-like Mn(2+) transporter in Escherichia coli. J. Bacteriol. 183, 4806–4813. doi: 10.1128/JB.183.16.4806-4813.2001
Persson, H. L., Yu, Z., Tirosh, O., Eaton, J. W., and Brunk, U. T. (2003). Prevention of oxidant-induced cell death by lysosomotropic iron chelators. Free Radic. Biol. Med. 34, 1295–1305. doi: 10.1016/S0891-5849(03)00106-0
Phillips Campbell, R., Kintner, J., Whittimore, J., and Schoborg, R. V. (2012). Chlamydia muridarum enters a viable but non-infectious state in amoxicillin-treated BALB/c mice. Microbes Infect. 14, 1177–1185. doi: 10.1016/j.micinf.2012.07.017
Pieper, R., Fisher, C. R., Suh, M. J., Huang, S. T., Parmar, P., and Payne, S. M. (2013). Analysis of the proteome of intracellular Shigella flexneri reveals pathways important for intracellular growth. Infect. Immun. 81, 4635–4648. doi: 10.1128/IAI.00975-13
Pierce, J. R., Pickett, C. L., and Earhart, C. F. (1983). Two fep genes are required for Ferrienterochelin Uptake in Escherichia coli K-12. J. Bacteriol. 155, 330–336.
Pigeon, C., Ilyin, G., Courselaud, B., Leroyer, P., Turlin, B., Brissot, P., et al. (2001). A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J. Biol. Chem. 276, 7811–7819. doi: 10.1074/jbc.M008923200
Pohl, E., Haller, J. C., Mijovilovich, A., Meyer-Klaucke, W., Garman, E., and Vasil, M. L. (2003). Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 47, 903–915. doi: 10.1046/j.1365-2958.2003.03337.x
Pohl, E., Holmes, R. K., and Hol, W. G. J. (1999). Crystal Structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain. J. Mol. Biol. 292, 653–667. doi: 10.1006/jmbi.1999.3073
Posey, J. E., and Gherardini, F. C. (2000). Lack of a role for iron in the Lyme disease pathogen. Science 288, 1651–1653. doi: 10.1126/science.288.5471.1651
Posey, J. E., Hardham, J. M., Norris, S. J., and Gherardini, F. C. (1999). Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc. Natl. Acad. Sci. U.S.A. 96, 10887–10892. doi: 10.1073/pnas.96.19.10887
Pospischil, A., Borel, N., Chowdhury, E. H., and Guscetti, F. (2009). Aberrant chlamydial developmental forms in the gastrointestinal tract of pigs spontaneously and experimentally infected with Chlamydia suis. Vet. Microbiol. 135, 147–156. doi: 10.1016/j.vetmic.2008.09.035
Puri, S., and O'Brian, M. R. (2006). The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. J. Bacteriol. 188, 6476–6482. doi: 10.1128/JB.00737-06
Qi, Z., and O'Brian, M. R. (2002). Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol. Cell 9, 155–162. doi: 10.1016/S1097-2765(01)00431-2
Qi, Z., Hamza, I., and O'Brian, M. R. (1999). Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. U.S.A. 96, 13056–13061. doi: 10.1073/pnas.96.23.13056
Que, Q., and Helmann, J. D. (2000). Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35, 1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x
Ratledge, C., and Dover, L. G. (2000). Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54, 881–941. doi: 10.1146/annurev.micro.54.1.881
Rau, A., Wyllie, S., Whittimore, J., and Raulston, J. E. (2005). Identification of Chlamydia trachomatis genomic sequences recognized by chlamydial divalent cation-dependent regulator A (DcrA). J. Bacteriol. 187, 443–448. doi: 10.1128/JB.187.2.443-448.2005
Raulston, J. E. (1997). Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect. Immun. 65, 4539–4547.
Raulston, J. E., Miller, J. D., Davis, C. H., Schell, M., Baldwin, A., Ferguson, K., et al. (2007). Identification of an iron-responsive protein that is antigenic in patients with Chlamydia trachomatis genital infections. FEMS Immunol. Med. Microbiol. 51, 569–576. doi: 10.1111/j.1574-695X.2007.00336.x
Read, T. D., Brunham, R. C., Shen, C., Gill, S. R., Heidelberg, J. F., White, O., et al. (2000). Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28, 1397–1406. doi: 10.1093/nar/28.6.1397
Read, T. D., Myers, G. S. A., Brunham, R. C., Nelson, W. C., Paulsen, I. T., Heidelberg, J., et al. (2003). Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31, 2134–2147. doi: 10.1093/nar/gkg321
Richardson, D., Ponka, P., and Baker, E. (1994). The effect of the iron(III) chelator, desferrioxamine, on iron and transferrin. Cancer Res. 54, 685–689.
Rohde, K. H., and Dyer, D. W. (2004). Analysis of haptoglobin and hemoglobin-haptoglobin interactions with the Neisseria meningitidis TonB-dependent receptor HpuAB by flow cytometry. Infect. Immun. 72, 2494–2506. doi: 10.1128/IAI.72.5.2494-2506.2004
Rosario, C. J., and Tan, M. (2012). The early gene product EUO is a transcriptional repressor that selectively regulates promoters of Chlamydia late genes. Mol. Microbiol. 84, 1097–1107. doi: 10.1111/j.1365-2958.2012.08077.x
Rouault, T. A. (2006). The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2, 406–415. doi: 10.1038/nchembio807
Rouault, T. A., Hentze, M. W., Caughman, S. W., Harford, J. B., and Klausner, R. D. (1988). Binding of a cytosolic protein to the iron-responsive element of human ferritin messenger, R. N. A. Science 241, 1207–1210. doi: 10.1126/science.3413484
Rudolph, G., Semini, G., Hauser, F., Lindemann, A., Friberg, M., Hennecke, H., et al. (2006). The iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J. Bacteriol. 188, 733–744. doi: 10.1128/JB.188.2.733-744.2006
Runyen-Janecky, L. J., and Payne, S. M. (2002). Identification of chromosomal Shigella flexneri genes induced by the eukaryotic intracellular environment. Infect. Immun. 70, 4379–4388. doi: 10.1128/IAI.70.8.4379-4388.2002
Runyen-Janecky, L. J., Reeves, S. A., Gonzales, E. G., and Payne, S. M. (2003). Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect. Immun. 71, 1919–1928. doi: 10.1128/IAI.71.4.1919-1928.2003
Sangwan, I., Small, S. K., and O'Brian, M. R. (2008). The Bradyrhizobium japonicum Irr protein is a transcriptional repressor with high-affinity DNA-binding activity. J. Bacteriol. 190, 5172–5177. doi: 10.1128/JB.00495-08
Schaumburg, C. S., and Tan, M. (2006). Arginine-dependent gene regulation via the argr repressor is species specific in Chlamydia. J. Bacteriol. 188, 919–927. doi: 10.1128/JB.188.3.919-927.2006
Schmitt, M. P. (2002). Analysis of a DtxR-Like Metalloregulatory protein, MntR, from Corynebacterium diphtheriae that controls expression of an ABC metal transporter by an Mn2+-dependent mechanism. J. Bacteriol. 184, 6882–6892. doi: 10.1128/JB.184.24.6882-6892.2002
Schmitt, M. P., and Holmes, R. K. (1991). Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect. Immun. 59, 1899–1904.
Schmitt, M. P., and Holmes, R. K. (1994). Cloning, sequence, and footprint analysis of two promoter/operators from corynebacterium diphtheriae that are regulated by the diphtheria toxin repressor (DtxR) and iron. J. Bacteriol. 176, 1141–1149. doi: 10.1128/jb.176.4.1141-1149.1994
Schmitt, M. P., Twiddy, E. M., and Holmes, R. K. (1992). Purification and characterization of the diphtheria toxin repressor. Proc. Natl. Acad. Sci. U.S.A. 89, 7576–7580. doi: 10.1073/pnas.89.16.7576
Scidmore, M. A., Fischer, E. R., and Hackstadt, T. (1996). Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J. Cell Biol. 134, 363–374. doi: 10.1083/jcb.134.2.363
Small, S. K., Puri, S., Sangwan, I., and Brian, M. R. (2009). Positive control of ferric siderophore receptor gene expression by the irr protein in Bradyrhizobium japonicum. J. Bacteriol. 191, 1361–1368. doi: 10.1128/JB.01571-08
Soares, M. P., and Weiss, G. (2015). The Iron age of host—microbe interactions. EMBO Rep. 16, 1–19. doi: 10.15252/embr.201540558
Sönnichsen, B., Renzis, S., De Nielsen, E., Rietdorf, J., and Zerial, M. (2000). Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 149, 901–913. doi: 10.1083/jcb.149.4.901
Spiering, M. M., Ringe, D., Murphy, J. R., and Marletta, M. A. (2003). Metal stoichiometry and functional studies of the diphtheria toxin repressor. Proc. Natl. Acad. Sci. U.S.A. 100, 1–6. doi: 10.1073/pnas.0737977100
Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., et al. (1998). Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759. doi: 10.1126/science.282.5389.754
Stojiljkovic, I., and Hantke, K. (1992). Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1, 4359–4367.
Sullivan, J. L., and Weinberg, E. D. (1999). Iron and the role of Chlamydia pneumoniae in heart disease. Emerging Infect. Dis. 5, 724–726. doi: 10.3201/eid0505.990519
Tailleux, L., Neyrolles, O., Honore-Bouakline, S., Perret, E., Sanchez, F., Abastado, J.-P., et al. (2003). Constrained intracellular survival of Mycobacterium tuberculosis in human dendritic cells. J. Immunol. 170, 1939–1948. doi: 10.4049/jimmunol.170.4.1939
Tao, X., and Murphy, J. R. (1992). Binding of the metalloregulatory protein dtxr to the diphtheria operator requires a divalent heavy metal ion and protects the palindromic sequence from DNase. J. Biol. Chem. 267, 21761–21764.
Tao, X., Boydt, J., and Murphy, J. R. (1992). Specific binding of the diphtheria tox regulatory element DtxR to the tox operator requires divalent heavy metal ions and a 9-base-pair interrupted palindromic sequence. Proc. Natl. Acad. Sci. U.S.A. 89, 5897–5901. doi: 10.1073/pnas.89.13.5897
Thomas, S. M., Garrity, L. F., Brandt, C. R., Schobert, C. S., Feng, G.-S., Taylor, M. W., et al. (1993). IFN-y-mediated antimicrobial response: indoleamine 2,3-dioxygenase-deficient mutant host cells no longer inhibit intracellular Chlamydia spp. or toxoplasma growth. J. Immunol. 150, 5529–5534.
Thompson, C. C., and Carabeo, R. A. (2011). An optimal method of iron starvation of the obligate intracellular pathogen, Chlamydia trachomatis. Front. Microbiol. 2:20. doi: 10.3389/fmicb.2011.00020
Thompson, C. C., Nicod, S. S., Malcolm, D. S., Grieshaber, S. S., and Carabeo, R. A. (2012). Cleavage of a putative metal permease in Chlamydia trachomatis yields an iron-dependent transcriptional repressor. Proc. Natl. Acad. Sci. U.S.A. 109, 10546–10551. doi: 10.1073/pnas.1201398109
Thompson, J. M., Jones, H. A., and Perry, R. D. (1999). Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect. Immun. 67, 3879–3892.
Thomson, N. R., Holden, M. T. G., Carder, C., Lennard, N., Lockey, S. J., Marsh, P., et al. (2008). Chlamydia trachomatis : genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18, 161–171. doi: 10.1101/gr.7020108
Timms, P., Good, D., Wan, C., Theodoropoulos, C., Mukhopadhyay, S., Summersgill, J. T., et al. (2009). Differential transcriptional responses between the interferon-γ–induction and iron-limitation models of persistence for Chlamydia pneumoniae. J. Microbiol. Immunol. Infect. 42, 27–37. doi: 10.1520/D0850-11.1
Toye, B., Laferriere, C., Claman, P., Jessamine, P., and Peeling, R. (1993). Association between antibody to the chlamydial heat-shock protein and tubal infertility. J. Infect. Dis. 168, 1236–1240. doi: 10.1093/infdis/168.5.1236
van der Sluijs, P., Hull, M., Webster, P., Mâle, P., Goud, B., and Mellman, I. (1992). The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 70, 729–740. doi: 10.1016/0092-8674(92)90307-X
van Ooij, C., Apodaca, G., and Engel, J. (1997). Characterization of the Chlamydia trachomatis vacuole and its interaction with the host endocytic pathway in hela cells. Infect. Immun. 65, 758–766.
Vecerek, B., Moll, I., and Blasi, U. (2007). Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. EMBO J. 26, 965–975. doi: 10.1038/sj.emboj.7601553
Vulpe, C. D., Kuo, Y., Murphy, T. L., Cowley, L., Askwith, C., Libina, N., et al. (1999). Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat. Genet. 21, 195–199. doi: 10.1038/5979
Wehrl, W., Meyer, T. F., Jungblut, P. R., Müller, E. C., and Szczepek, A. J. (2004). Action and reaction: Chlamydophila pneumoniae proteome alteration in a persistent infection induced by iron deficiency. Proteomics 4, 2969–2981. doi: 10.1002/pmic.200400917
Wilkinson, N., and Pantopoulos, K. (2014). The IRP/IRE system in vivo: Insights from mouse models. Front. Pharmacol. 5:176. doi: 10.3389/fphar.2014.00176
Wilson, A. C., and Tan, M. (2002). Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J. Bacteriol. 184, 6566–6571. doi: 10.1128/JB.184.23.6566-6571.2002
Winter, W., Bazydlo, L., and Harris, N. (2014). The molecular biology of human iron metabolism. Lab. Med. 45, 92–102. doi: 10.1309/LMF28S2GIMXNWHMM
Winterbourn, C. C. (1995). Toxicity of iron and hydrogen peroxide: the fenton reaction. Toxicol. Lett. 82, 969–974. doi: 10.1016/0378-4274(95)03532-X
Worst, D. J., M., Gerrits, M., Vandenbroucke-Grauls, C. M. J. E., and Kusters, J. G. (1998). Helicobacter pylori ribBA-mediated riboflavin production is involved in iron acquisition. J. Bacteriol. 180, 1473–1479.
Wyllie, S., and Raulston, J. E. (2001). Identifying regulators of transcription in an obligate intracellular pathogen: a metal-dependent repressor in Chlamydia trachomatis. Mol. Microbiol. 40, 1027–1036. doi: 10.1046/j.1365-2958.2001.02453.x
Wyrick, P. B. (2010). Chlamydia trachomatis persistence in vitro: an overview. J. Infect. Dis. 201, 88–95. doi: 10.1086/652394
Zhu, W., Hunt, D. J., Richardson, A. R., and Stojiljkovic, I. (2000). Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 182, 439–447. doi: 10.1128/JB.182.2.439-447.2000
Keywords: ABC-type permease-repressor fusion, vesicular iron, intracellular pathogen, persistence, iron homeostasis
Citation: Pokorzynski ND, Thompson CC and Carabeo RA (2017) Ironing Out the Unconventional Mechanisms of Iron Acquisition and Gene Regulation in Chlamydia. Front. Cell. Infect. Microbiol. 7:394. doi: 10.3389/fcimb.2017.00394
Received: 19 July 2017; Accepted: 23 August 2017;
Published: 08 September 2017.
Edited by:
Alfredo G. Torres, University of Texas Medical Branch, United StatesReviewed by:
Laura Runyen-Janecky, University of Richmond, United StatesAlan Paul Hudson, Wayne State University School of Medicine, United States
Amanda G. Oglesby-Sherrouse, University of Maryland, Baltimore, United States
Copyright © 2017 Pokorzynski, Thompson and Carabeo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rey A. Carabeo, rcarabeo@vetmed.wsu.edu
 Nick D. Pokorzynski
Nick D. Pokorzynski Christopher C. Thompson
Christopher C. Thompson Rey A. Carabeo
Rey A. Carabeo