Brucella Rough Mutant Induce Macrophage Death via Activating IRE1α Pathway of Endoplasmic Reticulum Stress by Enhanced T4SS Secretion
- 1Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
- 2Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonosis, Yangzhou, China
Brucella is a Gram-negative facultative intracellular pathogen that causes the worldwide zoonosis, known as brucellosis. Brucella virulence relies mostly on its ability to invade and replicate within phagocytic cells. The type IV secretion system (T4SS) and lipopolysaccharide are two major Brucella virulence factors. Brucella rough mutants reportedly induce the death of infected macrophages, which is T4SS dependent. However, the underlying molecular mechanism remains unclear. In this study, the T4SS secretion capacities of Brucella rough mutant and its smooth wild-type strain were comparatively investigated, by constructing the firefly luciferase fused T4SS effector, BPE123 and VceC. In addition, quantitative real-time PCR and western blotting were used to analyze the T4SS expression. The results showed that T4SS expression and secretion were enhanced significantly in the Brucella rough mutant. We also found that the activity of the T4SS virB operon promoter was notably increased in the Brucella rough mutant, which depends on quorum sensing-related regulators of VjbR upregulation. Cell infection and cell death assays revealed that deletion of vjbR in the Brucella rough mutant absolutely abolished cytotoxicity within macrophages by downregulating T4SS expression. This suggests that up-regulation of T4SS promoted by VjbR in rough mutant ΔrfbE contribute to macrophage death. In addition, we found that the Brucella rough mutant induce macrophage death via activating IRE1α pathway of endoplasmic reticulum stress. Taken together, our study provide evidence that in comparison to the Brucella smooth wild-type strain, VjbR upregulation in the Brucella rough mutant increases transcription of the virB operon, resulting in overexpression of the T4SS gene, accompanied by the over-secretion of effecter proteins, thereby causing the death of infected macrophages via activating IRE1α pathway of endoplasmic reticulum stress, suggesting novel insights into the molecular mechanisms associated with Brucella rough mutant-induced macrophage cytotoxicity.
Introduction
Brucella is a Gram-negative facultative intracellular bacterial species that causes zoonotic brucellosis, characterized by reproductive disease in domestic animals and chronic debilitating disease in humans (Boschiroli et al., 2001; Franco et al., 2007; Whatmore, 2009). Brucellosis in animals is endemic in most areas of the world, and it can become a serious public health problem that results in significant morbidity and economic losses (Boschiroli et al., 2001; Atluri et al., 2011).
Brucella virulence relies mainly on its ability to invade and replicate within professional and non-professional phagocytes, among which macrophages are major target cells in infected mammals (Gorvel and Moreno, 2002; Celli, 2006). To date, many virulence factors have been identified, such as lipopolysaccharide (LPS), the type IV secretion system (T4SS), a two-component regulatory system (BvrS/BvrR), and cyclic β-1,2-glucan (CβG) (Byndloss and Tsolis, 2016). Two major Brucella virulence factors are LPS and T4SS. Brucella LPS is composed of lipid A, a core oligosaccharide, and the O-antigen. It is characterized by low stimulatory activity and toxicity to cells, and mediates lower superoxide and lysozyme production in infected cells (Goldstein et al., 1992; Rasool et al., 1992). Furthermore, Brucella LPS is critical in the inhibition of programmed cell death (apoptosis) and enhances the bacterium′s ability to survive within macrophages (Fernandez-Prada et al., 2003). Rough mutants of Brucella abortus that lack the O-antigen, induce infected macrophage death, and are taken up in greater numbers by macrophages than the smooth wild-type strains (Pei and Ficht, 2003; Bronner et al., 2013; Tian et al., 2014). In previous reports, the rough mutant VTRS1 of B. suis induced proinflammatory, caspase-2- and nuclear factor kappa B (NF-κB)- mediated macrophage cell death (Chen et al., 2011). Bronner and colleagues subsequently reported that the rough mutant of B. abortus RB51 induces a hybrid cell death, mediated by caspase-2 activation, with features of apoptosis and pyroptosis (Bronner et al., 2013). In further study, Bronner found that endoplasmic reticulum (ER) stress induced by RB51 activates the inflammasome via NLRP3- and caspase-2- driven mitochondrial damage (Chen et al., 2011; Bronner et al., 2015). However, the molecular mechanism underlying Brucella rough mutant modulation of ER stress to induce macrophage death remains unclear.
Cytotoxicity in macrophages that have been infected by Brucella rough mutants is reportedly T4SS dependent (Pei et al., 2008). Overexpression of T4SS in the Brucella smooth strain enhances its ability to induce macrophage death (Zhong et al., 2009). The T4SS encoded by the virB operon in Brucella comprises multiprotein complexes that translocate specific protein substrates across the bacterial cell envelope to the host cell, and guides trafficking of the Brucella containing vacuole to the ER-associated compartment within macrophages (Zechner et al., 2012; Byndloss et al., 2016). The Brucella virB operon is induced by lysosomal acidification and nutritional deprivation within macrophages, which is tightly controlled by several regulation-associated genes, such as the LuxR family VjbR regulator and integration host factor IHF (Porte et al., 1999; Boschiroli et al., 2002; Sieira, 2013). The T4SS plays a significant role in Brucella trafficking, and is essential for Brucella to trigger a mild inflammatory response (Rolan et al., 2009). The effector protein VceC is translocated by T4SS to the ER, where it binds the ER chaperone BiP (binding immunoglobulin protein) and induces inositol-requiring enzyme 1α (IRE1α)-dependent ER stress during B. abortus infection, and this in turn effects the recruitment of the NOD-like receptors, NOD1 and NOD2, to induce NF-κB activation and expression of proinflammatory genes (de Jong et al., 2013; Keestra-Gounder et al., 2016). However, the manner in which the Brucella rough mutant regulates T4SS function has not been definitively determined.
Brucella LPS is an important virulence factor, and the O-antigen is a crucial component that is synthesized in the cytoplasmic face of the bacterial inner membrane, and then exported to the periplasmic face of the inner membrane, based on an ATP-binding cassette (ABC) transporter system that is encoded by rfbE and rfbD genes (Godfroid et al., 2000; Tian et al., 2014). In a previous study, we found that the B. abortus rough mutant ΔrfbE, induced the death of infected macrophages (Tian et al., 2014), which is in line with the findings of other reports (Pei and Ficht, 2003; Chen et al., 2011). In Shigella, shortening of the LPS molecule by O-antigen glucosylation enhances the secretion of effector proteins and function of the type III secretion system (West et al., 2005). In view of these findings, we hypothesized that Brucella smooth LPS is crucial for T4SS function, which plays a vital role in intracellular survival of Brucella and its interaction with host cells. In this study, we demonstrated that VjbR upregulation in the Brucella rough mutant ΔrfbE enhances T4SS expression and secretion, both of which contribute to the death of infected macrophages via activation of the IRE1α pathway of ER stress.
Materials and Methods
Strains, Plasmids, Macrophages, and Culture Conditions
All strains and plasmids used in this study are listed in Table 1. Brucella abortus S2308 and its derivatives were grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA) (Difco, Franklin Lakes, NJ, USA) plates at 37°C with 5% CO2. Manipulation of Brucella was performed in a biosafety level 3 laboratory facility at the Chinese Academy of Agricultural Sciences. Escherichia coli strains were cultured at 37°C in Luria Broth. When appropriate, 100 μg/mL ampicillin or 20 μg/mL chloramphenicol (Sigma–Aldrich Inc., St. Louis, MO, USA) was added. Mouse macrophage RAW264.7 (ATCC, Manassas, VA, USA) was cultured at 37°C with 5% CO2, in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, ThermoScientific, Grand Island, NY, USA).
Antibodies
The primary antibodies used in this study were: rabbit anti-firefly luciferase monoclonal antibody (Abcam, Cambridge, MA, USA); rabbit anti-Brucella VirB5 polyclonal antibody (prepared in our lab); rabbit anti-Brucella GAPDH polyclonal antibody (prepared in our lab); rabbit anti-IRE1 polyclonal antibody (phospho S724, Abcam); and rabbit anti-β-actin monoclonal antibody (Cell Signaling Technology, Danvers, MA, USA). The secondary antibodies used for western blotting were: IRDye 800CW-conjugated donkey anti-Rabbit IgG polyclonal antibody (LI-COR Biosciences, Lincoln, NE, USA) and horseradish peroxidase -conjugated goat anti-rabbit IgG (Life Technologies, Eugene, OR, USA).
Plasmid Construction
All primers used in this study are listed in Table 2. Suicide plasmids were constructed, using an overlap PCR assay, as previously reported (Tian et al., 2014). Briefly, the upstream and downstream fragments of virB123 (containing the virB promoter, virB1, virB2, and virB3 genes) and vjbR were amplified by independent PCRs and extracted from agarose gels that were used as templates for a second round of PCR. The resultant product that contained joined flanking sequences was purified by gel extraction and cloned into a pSC plasmid, after being digested with XbaI, to generate the suicide plasmids pSCΔvirB123 and pSCΔvjbR.
Luciferase reporter plasmids of pBCSP31-BPE123-Luc, pBCSP31-Luc-VceC, pBCSP31-GST-Luc, and pVirB-Luc were constructed using conventional methods. Firstly, the lacZ promoter region of the broad-host-range cloning plasmid pBBR1-MCS was replaced by a terminator sequence of rrnB from the plasmid pBAD24, using overlap PCR with the primer rrnB-F/R. The linearized plasmid was digested by the restriction enzyme Kpn I and self-ligated using T4 DNA ligase to construct a pMCR plasmid. The promoter regions of the bcsp31 gene or virB operon from B. abortus strain 2308 were obtained by PCR using the primers Pbcsp31-F/R or PvirB-F/R, cloned into the pMCR plasmid and digested by KpnI and XhoI enzymes to generate the plasmids pBCSP31 and pVirB, respectively. The luciferase gene (luc) was amplified by PCR from the pNFκB-Luc plasmid (Beyotime, Jiangsu, China), using the primers Luc-F and Luc-R, and cloned into the XhoI- and PstI-digested pBCSP31 plasmids, to generate pBCSP31-Luc-N, and facilitate the generation of N-terminal in-frame fusions of the Luc protein. The luc gene was cloned into the pBCSP31 plasmid by BamHI and XbaI digestion to generate pBCSP31-Luc-C, in which the stop codon in the luc open reading frame (ORF) was removed, to allow C-terminal fusion of the Luc protein. Furthermore, the luc gene was cloned into the pVirB plasmid by XhoI and PstI digestion, to construct the pVirB-Luc plasmid and effect promoter activation of the virB operon. The ORF of BPE123 was amplified from S2308 by PCR using the primer BPE123-F/R, and cloned into the pBCSP31-Luc-N plasmid to generate pBCSP31-BPE123-Luc. At the C-terminal, 116 amino acids of the VceC protein that are necessary for secretion by T4SS was amplified by PCR using the primer vceC-F/R, and cloned into the pBCSP31-Luc-C plasmid to construct the pBCSP31-Luc-VceC plasmid. In addition, the glutathione transferase (GST) gene was amplified from the pGEX-4T-1 plasmid (Takara, Dalian, China) by PCR using the primer GST-F/R and cloned into the pBCSP31-Luc-N plasmid, to generate the negative control plasmid pBCSP31-GST-Luc.
Overexpression plasmids of pMdrA and pBlxR were constructed using conventional methods. The mdrA and blxR genes containing the promoter and terminator regions were amplified by independent PCRs using the primers mdrA-F/R and blxR-F/R, respectively, and then cloned into the plasmid pBBR1-MCS, to generate the plasmids pMdrA and pBlxR, respectively.
All recombinant plasmids were propagated in E. coli DH5α cells (Invitrogen Corp., Carlsbad, CA, USA) and then extracted to construct recombinant Brucella strains.
Mutant Construction
The ΔrfbEΔvirB and ΔrfbEΔvjbR mutants were constructed by allelic replacement, using a two-step strategy as previously reported (Kahl-McDonagh and Ficht, 2006; Tian et al., 2014). The suicide plasmids pSCΔvirB123 and pSCΔvjbR (0.5–1.0 μg) were transferred to the ΔrfbE strain by electroporation. The first exchanged recombinants were selected by plating on TSA containing ampicillin. The second round of exchanged recombinants was selected by plating on TSA containing 5% sucrose. Analyses of PCRs were carried out to identify clones.
Luciferase reporter strains and overexpression strains were also constructed by electroporation. The recombinants were then selected by plating on TSA containing chloramphenicol. The PCR or western blotting analyses were carried out to identify recombinants. The recombinant strains constructed in this study are listed in Table 1.
Cell Infection Assay
Monolayers of RAW264.7 cells were cultured in six- or 24-well plates and infected with B. abortus S2308 or its derivatives at a multiplicity of infection (MOI) of 100 or 1,000 colony forming units (CFU) per cell. To synchronize the infection, the infected plates were centrifuged at 400× g for 5 min, and cells were then incubated at 37°C with 5% CO2 for 1 h. The monolayers were washed twice with phosphate buffered saline (PBS) (HyClone, GE Lifesciences, Logan, UT, USA) to remove extracellular nonadherent bacteria, and then incubated with DMEM containing gentamicin (100 μg/mL) for 1 h to kill extracellular bacteria. To maintain survival of the infected cells, the monolayers were incubated with DMEM containing gentamicin (20 μg/mL) and 2% FBS after being washed thrice with PBS.
Cell Death Analysis
Macrophage death was detected, using two approaches. In the first approach, infected cells were stained with annexin V and propidium iodide (PI) at 3, 5, 8, and 12 h post infection (p.i.), using the annexin V-FITC/PI staining kit (Beyotime, Shanghai, China). In the second approach, the release of lactate dehydrogenase (LDH) in the supernatant of Brucella-infected RAW264.7 cells both with and without 4μ8c (IRE1α inhibitor, 100 μM, Selleck, Houston, TX, USA) treatment was determined at 3, 5, 8, and 12 h p.i., using a CytoTox 96 nonradioactive cytotoxicity assay (Promega, Fitchburg, WI, USA). Cell death was expressed as a percentage of maximum LDH release. The percentage was calculated as follows: (optical density at 490 nm [OD490] of infected cells—OD490 of uninfected cells)/(OD490 of lysed uninfected cells—OD490 of uninfected cells) × 100%.
Determination of Luciferase Activity
For the determination of luciferase activity in the media culture, luciferase reporter strains of S2308(pVirB-Luc) and ΔrfbE(pVirB-Luc) were cultured to exponential phase (OD600 = 1.0), and then centrifuged at 8,000 × g for 5 min to precipitate bacteria. The pellets of luciferase reporter strains were resuspended in 200 μL PBS and lysed by adding 200 μL B-PER® Bacterial Protein Extraction Reagent (Thermo Scientific). The lysate was centrifuged at 15,000 × g for 5 min to separate soluble proteins, after which it was incubated for 15 min at room temperature. The luciferase activity (relative light units, RLUs) of lysate supernatants were measured using the Luc-Screen® reporter gene assay system (Abcam). Moreover, 100 μL of the luciferase reporter strains were serially diluted 10-fold with PBS and spread onto TSA plates to determine the bacterial CFU. All samples were analyzed in triplicate.
For the determination of luciferase activity in the cell culture, macrophage RAW264.7 cells were infected with luciferase reporter strains at a MOI of 1,000, as described previously. Infected cells were washed three times with PBS and lysed with 500 μL of 0.2% Triton X-100 in sterile water for 15 min at 3, 5, and 8 h p.i. The infected cell lysate (400 μL) was centrifuged at 12,000× g for 5 min. For cells infected with luciferase reporter strains of S2308(BPE123-Luc), ΔrfbE(BPE123-Luc), ΔrfbEΔvirB(BPE123-Luc), S2308(Luc-VceC), ΔrfbE(Luc-VceC), ΔrfbEΔvirB(Luc-VceC), S2308(GST-Luc), ΔrfbE(GST-Luc), and ΔrfbEΔvirB(GST-Luc), the RLUs of lysate supernatants were measured by the Luc-Screen® reporter gene assay system (Abcam). For the cells infected with luciferase reporter strains of S2308(pVirB-Luc) and ΔrfbE(pVirB-Luc), lysate pellets were resuspended in 200 μL PBS, after which 200 μL B-PER® Bacterial Protein Extraction Reagent (Thermo Scientific) was added to lyse the intracellular strains. The lysate of intracellular strains was centrifuged at 15,000× g for 5 min, after which it was incubated for 15 min at room temperature. The RLUs of lysate supernatants were also measured using the Luc-Screen® reporter gene assay system (Abcam). Moreover, the remaining 100 μL of infected cell lysates were serially diluted 10-fold with PBS and spread onto TSA plates to determine the bacterial CFU. All samples were analyzed in triplicate.
RNA Extraction and Real-Time PCR
Total RNA was extracted from bacteria using the TRIzol® RNA Isolation Reagent (Invitrogen) according to the manufacturer's protocol. Genomic DNA contamination was removed through treatment with a Turbo DNA-free kit (Ambion). The RNA quantity and quality were evaluated using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc.). The RNA integrity was assessed by standard denaturing agarose gel electrophoresis, and RNA (1 μg) was reverse transcribed into cDNA, using a PrimeScript RT-PCR kit (Takara) according to the manufacturer's instructions. A 20 μL RT-PCR mixture was made comprising 10 μL 2 × GoTaq qPCR master mix (Promega), 1 μL cDNA, 0.5 μL (each) forward and reverse primers (10 μM each), and 8 μL double-distilled water (ddH2O). The mixture was incubated at 95°C for 2 min, and then subjected to 40 cycles at 95°C for 15 s, followed by 60°C for 1 min using a Mastercycler ep Realplex system (Eppendorf). All samples were analyzed in triplicate and relative transcription levels of each gene were determined by the 2−ΔΔCt method, using 16S RNA as an internal control for data normalization.
Western Blotting
Sediments of the bacteria were collected, following centrifugation at 1,000 × g for 5 min and culture for various durations. The pellets were resuspended in Laemmli sample buffer and boiled for 10 min. The RAW264.7 cells were scraped into radioimmunoprecipitation assay buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP 40, 0.25% sodium deoxycholate, and 1 mM EDTA) that contained a protease inhibitor cocktail (Roche). The cell lysates were mixed with Laemmli sample buffer and boiled for 10 min. The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto nitrocellulose membranes (Millipore) using a semidry transfer procedure. The membranes were blocked overnight at 4°C in Tris-buffered saline containing 5% skim milk or 5% Bovine Serum Albumin (BSA). Immunodetection of proteins in total cell lysates was performed with the respective primary antibody for 2 h at room temperature. After washing three times with Tris-buffered saline and Tween 20, the membrane was incubated with the respective secondary antibody for 1 h at room temperature. After washing three times with Tris-buffered saline and Tween 20, an Odyssey two-color infrared imaging system (LI-COR Biosciences) was used to develop the fluorescence for visualization. The gray intensity of the bands was quantified using the ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Statistical Analysis
Statistical analysis was performed using the GraphPad Prism 6.0 software (GraphPad Software Inc., La Jolla, CA, USA). All p-values between identified samples were generated using unpaired two-tailed Student's t-tests, or in the case of groups, two-way analysis of variance, followed by the Tukey's test. All experiments were repeated at least three times and the results were presented as means ± SD from ≥3 replicates per condition.
Results
Rough Mutant ΔrfbE Induced Macrophage Death Is T4SS Dependent
Brucella rough mutant induces macrophage death, a process that is T4SS dependent (Pei et al., 2008). Our previous studies have shown that rough mutant ΔrfbE also induces the death of macrophages (Tian et al., 2014). To identify the role of T4SS on ΔrfbE mutant-induced cytotoxicity for RAW264.7 macrophages, virB1, virB2, and virB3 genes were deleted from the ΔrfbE mutant, thereby generating a double-knockout strain (ΔrfbEΔvirB). Firstly, morphology of the RAW264.7 cells infected with S2308, ΔrfbE, ΔrfbE(pBBR-rfbE), and ΔrfbEΔvirB were observed via light microscopy. As shown in Figure 1, ΔrfbE-infected cells exhibited obvious cell swelling and deformation at 8 and 12 h p.i.; however, the ΔrfbE(pBBR-rfbE)- and ΔrfbEΔvirB-infected cells showed no cell lesions, which is consistent with our observations of the S2308-infected cells and mock cells. Furthermore, the death of Brucella-infected RAW264.7 macrophages was analyzed following annexin V-FITC and PI staining, which was used to detect translocation of phosphatidylserine from the inner cell membrane to the outer cell membrane during the early stages of apoptosis. The PI stains the DNA of necrotic cells and/or cells at the late stage of apoptosis (Tian et al., 2014). The results showed that macrophages infected with the ΔrfbE mutant exhibited some characteristics of necrosis and late apoptosis, accompanied by cellular membrane damage and PI staining of the nucleus at 5, 8, and 12 h p.i. Further disruption of the virB operon in the ΔrfbE mutant reduced its ability to induce cell death. Certainly, the S2308 and ΔrfbE(pBBR-rfbE) with the smooth phenotype did not induce infected macrophage death in a similar manner to the mock cells (Figure 2A). These results are consistent with those of previous reports (Pei et al., 2008; Tian et al., 2014).
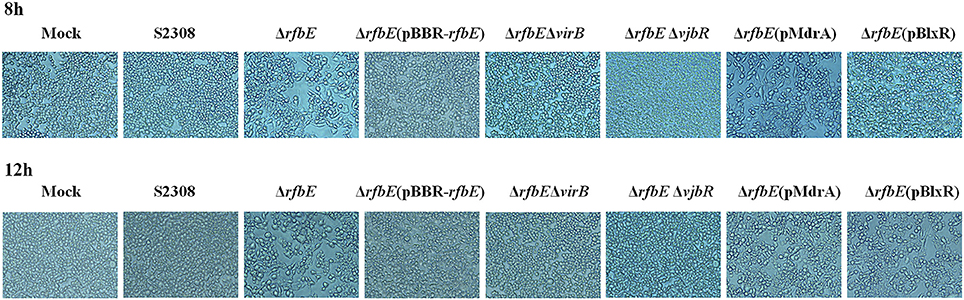
Figure 1. Phase-contrast microscopy. RAW264.7 cells cultured in a 24-well plate were infected with S2308, ΔrfbE, ΔrfbE(pBBR-rfbE), ΔrfbEΔvirB, ΔrfbEΔvjbR, ΔrfbE(pMdrA), and ΔrfbE(pBlxR) mutants at a MOI of 100. The cells were observed at a magnification of ×200 at 8 and 12 hpi. Uninfected RAW264.7 cells were used as negative controls (Mock).
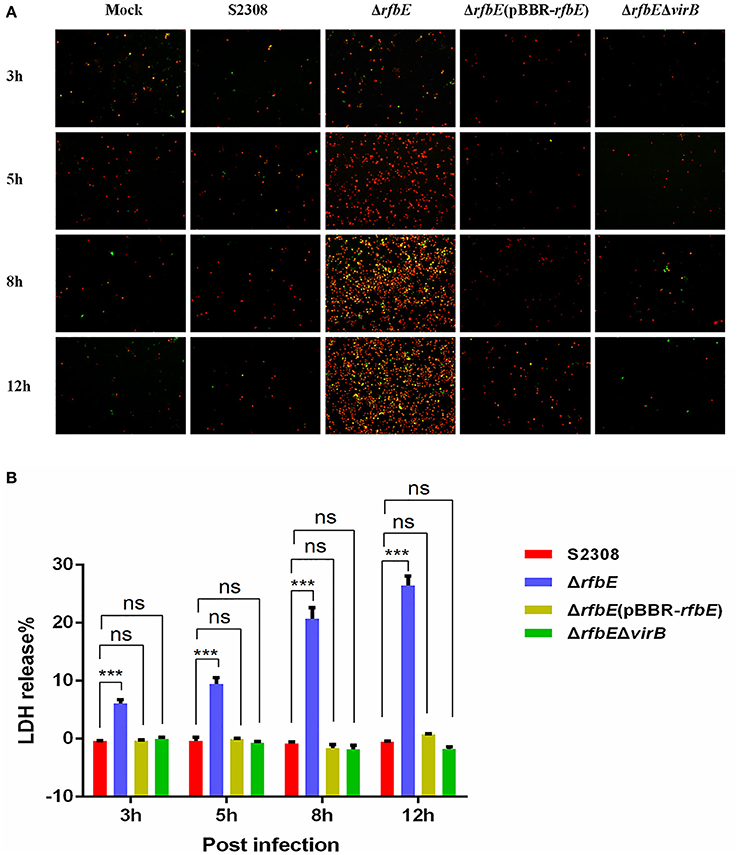
Figure 2. Macrophage death induced by Brucella rough mutant infection is T4SS dependent. RAW264.7 cells cultured in a 24-well plate were infected with S2308, ΔrfbE, ΔrfbE(pBBR-rfbE), or ΔrfbEΔvirB strains at a MOI of 100, and cell death was determined at 3, 5, 8, and 12 hpi. (A) Annexin V-FITC/PI staining. The cells were stained with FITC-annexin (green) and PI (red), and observed using fluorescence microscopy at a magnification of ×100. Uninfected RAW264.7 cells were used as negative controls (Mock). (B) LDH detection. The supernatants were collected and LDH release was detected using the CytoTox 96 nonradioactive cytotoxicity assay. The supernatants of uninfected RAW264.7 cells were used as negative controls (medium). ns, no significant difference, ***p < 0.0001.
To evaluate cell death quantitatively, the release of LDH was determined for S2308-, ΔrfbE-, ΔrfbE(pBBR-rfbE)-, and ΔrfbEΔvirB-infected cells. The levels of LDH released from the ΔrfbE-infected cells were significantly higher than those released from S2308-infected cells at 3, 5, 8, and 12 h p.i. (Figure 2B). However, the ΔrfbE(pBBR-rfbE)- and ΔrfbEΔvirB-infected cells released similar levels of LDH as the S2308-infected cells at 3, 5, 8, and 12 h p.i. (Figure 2B). These results further confirmed that the rough mutant ΔrfbE induced macrophage death is T4SS dependent.
T4SS Secretion Is Enhanced in the Brucella Rough Mutant ΔrfbE
The T4SS translocates effectors across the bacterial cell envelope to the host cell, and plays a central role in intracellular survival and replication of Brucella within the host (Ke et al., 2015). As macrophage death induced by the rough mutant ΔrfbE is T4SS dependent, we hypothesized that the capacity of the rough mutant ΔrfbE, to secrete T4SS could be altered. For this purpose, we used the previously reported T4SS effector, BPE123 and VceC as target proteins, and GST as a negative control protein, to construct the luciferase reporter strains S2308(BPE123-Luc), ΔrfbE(BPE123-Luc), ΔrfbEΔvirB(BPE123-Luc), S2308(Luc-VceC), ΔrfbE(Luc-VceC), ΔrfbEΔvirB(Luc-VceC), S2308(GST-Luc), ΔrfbE(GST-Luc), and ΔrfbEΔvirB(GST-Luc). The expression of luciferase fusion proteins was detected by western blotting analysis, indicating that luciferase was successfully expressed in the luciferase reporter strains, and the expression levels of luciferase fusion proteins in S2308, ΔrfbE, and ΔrfbEΔvirB were similar (Figure 3A).
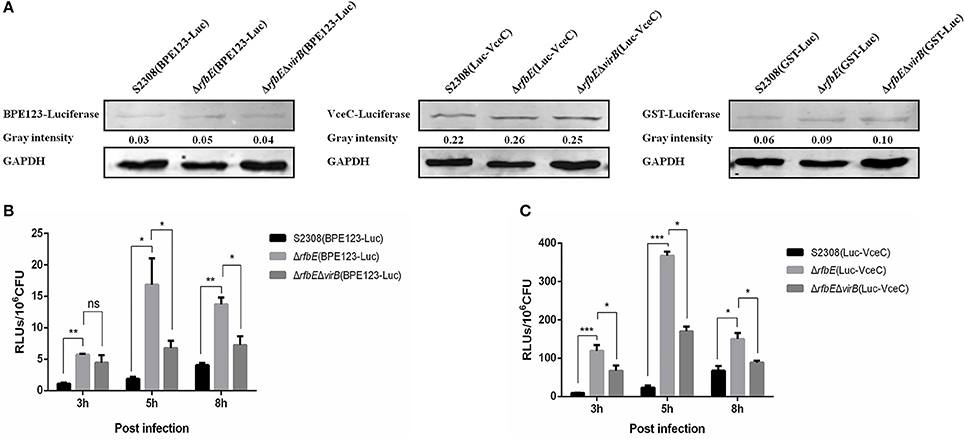
Figure 3. T4SS secretion is enhanced in Brucella rough mutant. (A) Determination of the T4SS effectors BPE123 and VceC in luciferase reporter strains. No significant difference on BPE123 or VceC expression among S2308(BPE123-Luc), ΔrfbE(BPE123-Luc), and ΔrfbEΔvirB(BPE123-Luc), or S2308(Luc-VceC), ΔrfbE(Luc-VceC), and ΔrfbEΔvirB(Luc-VceC), as determined using western blotting analysis. Protein levels are indicated by gray-scanning intensity values. GAPDH was used for normalization. (B) Determination of the BPE123 secretion on cell culture. RAW264.7 cells cultured in a 24-well plate were infected with S2308(BPE123-Luc), ΔrfbE(BPE123-Luc), or ΔrfbEΔvirB(BPE123-Luc) at a MOI of 1,000, and the cells were lysed with 500 μl of 0.2% Triton X-100 in sterile water for 15 min at 3, 5, and 8 hpi. The RLUs of lysate supernatants was measured by the Luc-Screen® reporter gene assay system. ns, no significant difference, *p < 0.05 and **p < 0.001. RAW264.7 cells infected with S2308(GST-Luc), ΔrfbE(GST-Luc), and ΔrfbEΔvirB(GST-Luc) were used as negative controls, respectively. (C) Determination of the VceC secretion on cell culture. RAW264.7 cells cultured in a 24-well plate were infected with S2308(Luc-VceC), ΔrfbE(Luc-VceC), or ΔrfbEΔvirB(Luc-VceC) at a MOI of 1000, and the cells were lysed with 500 μl of 0.2% Triton X-100 in sterile water for 15 min at 3, 5, and 8 hpi. The RLUs of lysate supernatants was measured by the Luc-Screen® reporter gene assay system. *p < 0.05 and ***p < 0.0001. RAW264.7 cells infected with S2308(GST-Luc), ΔrfbE(GST-Luc), and ΔrfbEΔvirB(GST-Luc) were used as negative controls, respectively.
Furthermore, the T4SS secretion capacity of S2308, ΔrfbE, and ΔrfbEΔvirB strains within host cells were determined. The RAW264.7 cells were infected with the luciferase reporter strains at a MOI of 1,000, and the secretion of BPE123 and VceC per 106 CFU of intracellular live Brucella were determined. Results showed that the rough mutant strain ΔrfbE translocated significantly higher levels of BPE123 and VceC to the infected cells than its smooth wild-type strain S2308 at 3, 5, and 8 h p.i., indicating an increased T4SS secretion capacity of the rough mutant ΔrfbE under the conditions of intracellular infection (Figures 3B,C). The increased T4SS secretion of the ΔrfbE mutant was partially recovered by further deletion of virB123 genes (Figures 3B,C), indicating that BPE123 and VceC oversecretion in the ΔrfbE mutant was indeed dependent on T4SS function. Taken together, the T4SS secretion capacity of the rough mutant, ΔrfbE was higher than that of the smooth wild-type strain, S2308.
T4SS Overexpression in the Brucella Rough Mutant Contributes to Its Enhanced Secretion
To confirm that the enhanced T4SS secretion is associated with enhanced T4SS expression in the Brucella rough mutant, we evaluated the expression of the T4SS components, VirB4 and VirB5 in the S2308 and ΔrfbE mutants at exponential phase in TSB, using qRT-PCR and western blotting. Results showed that virB4 and virB5 expression of the rough mutant ΔrfbE was significantly upregulated at the exponential phase, compared to that of its smooth wild-type strain, S2308 (Figures 4A,B). Based on previous reports, it is evident that the virB operon of Brucella induced expression within host cells under the conditions of nutritional deprivation and an acidic environment (Boschiroli et al., 2002). To determine the T4SS expression under acidic conditions, the smooth wild-type strain S2308 and rough mutant ΔrfbE were grown to exponential phase and exposed to TSB at pH 4.5 for 1 h. The qRT-PCR and western blotting analyses showed that virB4 and virB5 expression was induced in both strains at pH 4.5. Furthermore, much higher levels of virB4 and virB5 expression were induced in the rough mutant ΔrfbE, compared to those of the smooth wild-type strain S2308 (Figures 4A,B). To determine T4SS expression in nutritional deprivation, the S2308 and rough mutant ΔrfbE were grown to log phase and exposed to RPMI 1640 for 3 h. The results showed that virB4 and virB5 expression was also induced in both strains, and the expression level of the ΔrfbE mutant was significantly higher than that of the S2308 mutant (Figures 4A,B).
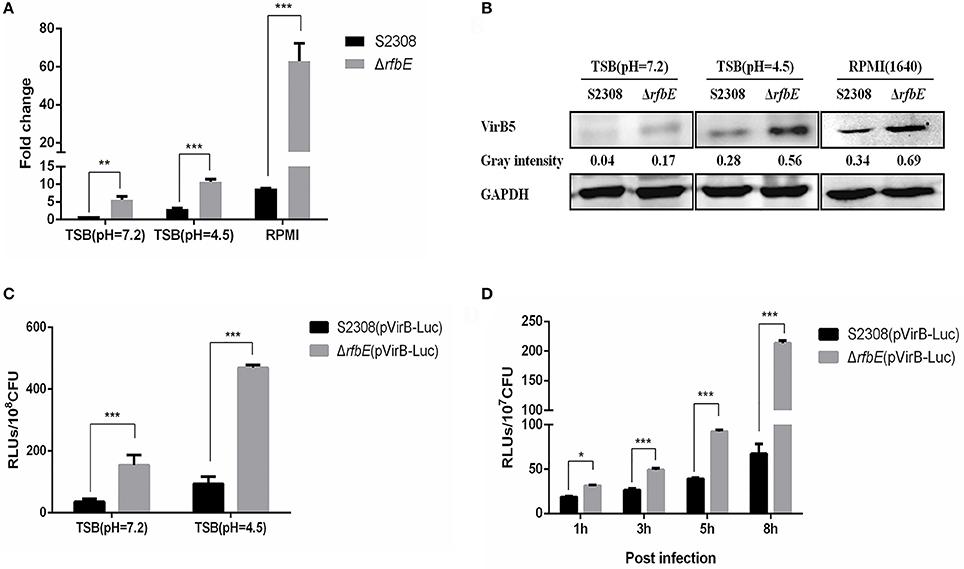
Figure 4. T4SS gene over-expressed in the Brucella rough mutant. (A) The qRT-PCR analysis. VirB4 expression in the rough mutant ΔrfbE is significantly upregulated under TSB (pH = 7.2), TSB (pH = 4.5), and RPMI 1640, compared to that in the smooth wild-type strain S2308. **p < 0.001 and ***p < 0.0001. (B) Western blotting analysis. VirB5 protein level in the rough mutant ΔrfbE is significantly enhanced under TSB (pH = 7.2), TSB (pH = 4.5), and RPMI 1640, compared to that in the smooth wild-type strain S2308. GAPDH was used for normalization and protein levels are indicated by gray-scanning intensity values. (C) Luciferase activity in TSB culture. Luciferase reporter strains S2308(pVirB-Luc) and ΔrfbE(pVirB-Luc) were cultured to exponential phase (OD600 = 1.0) under vegetative conditions, and the RLUs of culture supernatants was measured by the Luc-Screen® reporter gene assay system. ***p < 0.0001. (D) Luciferase activity in cell culture. RAW264.7 cells cultured in a 24-well plate were infected with S2308(pVirB-Luc) and ΔrfbE(pVirB-Luc) at a multiplicity of infection of 1,000, and the cells were lysed with 500 μl of 0.2% Triton X-100 in sterile water for 15 min at 1, 3, 5, and 8 hpi, and the intracellular bacteria were lysed with B-PER® Bacterial Protein Extraction Reagent. The RLUs of bacterial lysate supernatants was measured by the Luc-Screen® reporter gene assay system. *p < 0.05 and ***p < 0.0001.
To further determine T4SS upregulation in the ΔrfbE mutant, the promoter region of the virB operon was cloned and fused to the reporter luc gene, to generate luciferase reporter strains S2308(pVirB-Luc) and ΔrfbE(pVirB-Luc). The promoter activity of the virB operon was assessed in both strains during the exponential phase in TSB, indicating that the ΔrfbE(pVirB-Luc) strain displayed higher levels of luciferase activity than the S2308(pVirB-Luc) strain (Figure 4C). Furthermore, when exposed to TSB at pH 4.5 for 1 h, luciferase activity in both strains was significantly increased; however, the ΔrfbE(pVirB-Luc) strain showed much higher levels of luciferase activity than the S2308(pVirB-Luc) strain (b). To compare promoter activity of both stains within host cells, RAW264.7 cells were infected with S2308(pVirB-Luc) and ΔrfbE(pVirB-Luc), at 1, 3, 5, and 8 h p.i. The ΔrfbE(pVirB-Luc) strain showed significantly enhanced levels of luciferase activity in comparison to the S2308(pVirB-Luc) strain (Figure 4D). All these data suggest that T4SS expression in the rough mutant ΔrfbE was upregulated, which may have contributed to enhanced T4SS secretion.
Up-Regulation of T4SS Promoted by VjbR in Rough Mutant ΔrfbE Contribute to Macrophage Death
We proved that the virB operon was upregulated at the transcriptional level in the rough mutant, and further investigated whether T4SS overexpression in the rough mutant is associated with transcriptional regulators that directly bind to the virB operon. Thus, we evaluated the transcriptional expression of Brucella regulatory proteins that were found to be directly involved in transcriptional regulation of virB expression, including VjbR, IHF, HutC, BlxR, BvrR, and MdrA (Sieira, 2013). The qRT-PCR demonstrated that expression of vjbR was significantly upregulated, and that of the mdrA and blxR were evidently downregulated in the ΔrfbE mutant, compared to the S2308 strain (Figure 5A).
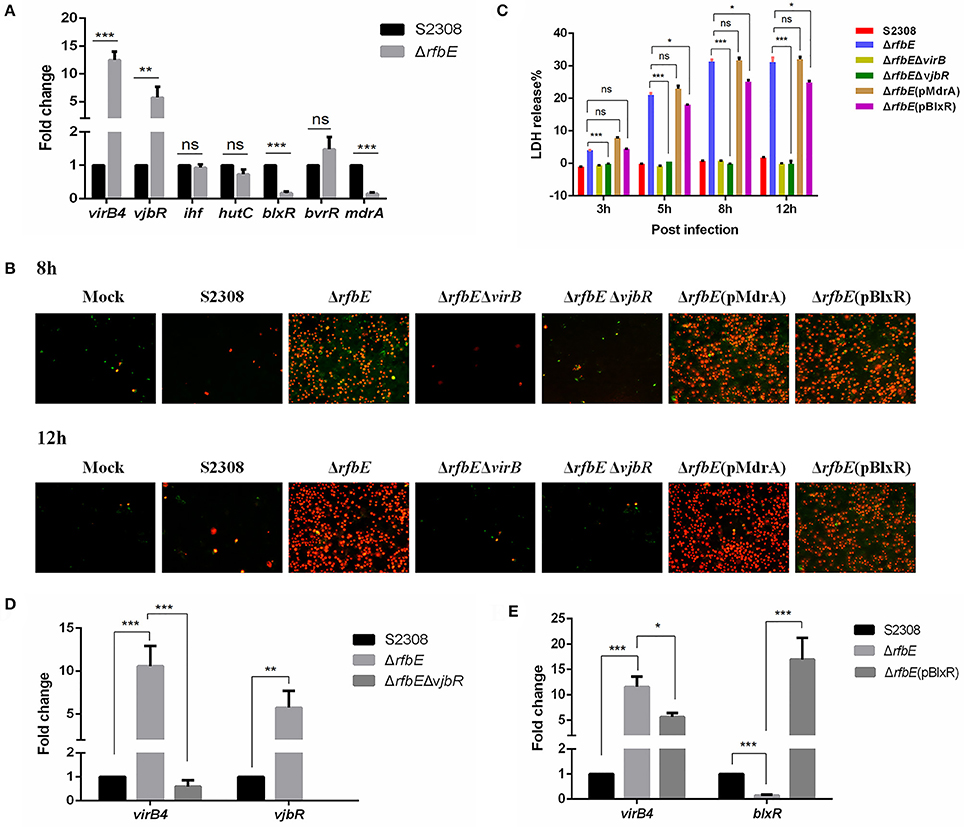
Figure 5. Brucella VjbR regulates T4SS expression in the rough mutant to induce macrophage death. (A) The qPCR analysis. Compared to the smooth wild-type strain S2308, upregulation of vjbR and downregulation of mdrA and blxR were evident in the rough mutant ΔrfbE. ns, no significant difference, **p < 0.001 and ***p < 0.0001. (B) Annexin V-FITC/PI staining. RAW264.7 cells cultured in a 24-well plate were infected with S2308, ΔrfbE, ΔrfbEΔvirB, ΔrfbEΔvjbR, ΔrfbE(pMdrA), or ΔrfbE(pBlxR) at a multiplicity of infection (MOI) of 100. The cells were stained at 8 and 12 hpi with FITC-annexin (green) and PI (red) and observed by fluorescence microscopy at a magnification of ×200. Uninfected RAW264.7 cells were used as negative controls (Mock). (C) Determination of LDH release. RAW264.7 cells were infected with S2308, ΔrfbE, ΔrfbEΔvirB, ΔrfbEΔvjbR, ΔrfbE(pMdrA), and ΔrfbE(pBlxR) at an MOI of 100. The supernatants were collected at 8 and 12 hpi, and LDH release was detected using the CytoTox 96 nonradioactive cytotoxicity assay. The supernatants of uninfected RAW264.7 cells were used as negative controls (medium). ns, no significant difference, *p < 0.05 and ***p < 0.0001. (D) VirB expression in the ΔrfbEΔvjbR was recovered to the similar level of the smooth wild-type strain S2308, as determined by qRT-PCR. **p < 0.001 and ***p < 0.0001. (E) VirB expression in the ΔrfbE(pBlxR) was partly recovered, compared to the ΔrfbE mutant, as determined by qRT-PCR. *p < 0.05 and ***p < 0.0001.
To determine whether macrophage death caused by infection with the ΔrfbE mutant is associated with VjbR, MdrA, and BlxR, we constructed ΔrfbEΔvjbR, ΔrfbE(pMdrA), and ΔrfbE(pBlxR) strains, respectively, to infect RAW264.7 macrophages. Under light microscopy, we observed no morphological changes in ΔrfbEΔvjbR-infected macrophages; however, obvious cell swelling and deformation were observed in ΔrfbE(pMdrA)- and ΔrfbE(pBlxR)-infected cells at 8 and 12 h p.i. (Figure 1). Furthermore, cell death was analyzed following annexin V-FITC and PI staining. The results showed that the ΔrfbEΔvjbR mutant was no longer cytotoxic to macrophages; however, the ΔrfbE(pMdrA) and ΔrfbE(pBlxR) strains induced macrophage death at 8 and 12 h p.i. (Figure 5B). In addition, the LDH release assay was performed to assess quantitatively the death of macrophages infected with the ΔrfbEΔvjbR, ΔrfbE(pMdrA), and ΔrfbE(pBlxR) strains. The results showed that the ΔrfbEΔvjbR mutant-infected macrophages reduced LDH release, compared to the ΔrfbE mutant infected cells, but similar to the smooth wild-type strain S2308 infected cells at 3, 5, 8, and 12 h p.i. Furthermore, the ΔrfbE(pBlxR)-infected cells also reduced LDH release at 5, 8, and 12 h p.i. compared to the ΔrfbE mutant; however, the LDH levels were much higher than those released from the S2308 infected cells (Figure 5C). The LDH release from ΔrfbE(pMdrA) infected cells showed no difference with those from the ΔrfbE mutant infected cells (Figure 5C). Taken together, these results indicated that VjbR upregulation was the key cause of ΔrfbE mutant-induced macrophage death. The BlxR downregulation played a partial role in macrophage death, but MdrA downregulation was not necessary for the ΔrfbE mutant to induce macrophage death.
To determine whether vjbR and blxR are essential for virB-upregulated expression in the ΔrfbE mutant, ΔrfbEΔvjbR, and ΔrfbE(pBlxR) strains were evaluated for virB expression using qRT-PCR. Results demonstrated that deletion of vjbR in the ΔrfbE mutant restored virB4 transcription to a level similar to that of the smooth strain S2308 (Figure 5D). In comparison to the ΔrfbE mutant, virB4 was significantly downregulated when blxR was robustly over-expressed in the ΔrfbE(pBlxR) mutant (Figure 5E), suggesting that virB-upregulated expression in the ΔrfbE mutant was associated with the regulatory proteins, VjbR and BlxR.
Taken together, T4SS overexpression induced by VjbR regulation in the Brucella rough mutant plays a key role in macrophage death.
Rough Mutant ΔrfbE Induces Macrophage Death via Activating IRE1α Pathway of ER Stress
The Brucella T4SS effector protein, VceC, is associated with triggering ER stress by activating the unfolded protein response (UPR) sensor, inositol-requiring enzyme 1α (IRE1α) (de Jong et al., 2013; Keestra-Gounder et al., 2016). To investigate the ER stress induced by Brucella rough mutant infection, activation of the UPR sensor, IRE1α was analyzed using western blotting. The results showed that compared to the Brucella smooth wild-type strain, the levels of P-IRE1α in the Brucella rough mutant were significantly increased at 3, 5, and 8 h p.i. (Figure 6A), indicating that the Brucella rough mutant induced stronger ER stress. To determine whether P-IRE1α is involved in macrophage death caused by the ΔrfbE mutant, the inhibitor of IRE1α, 4μ8c, was used to treat the macrophages before infection, which blocks the access of the substrate to the active site of IRE1α, and selectively inactivates both Xbp1 splicing and IRE1α-mediated mRNA degradation (Cross et al., 2012). We infected 4μ8c-treated macrophages with S2308 and the ΔrfbE mutant, and then evaluated cell death quantitatively, using the LDH release assay. The results demonstrated that in comparison to macrophages that had not been subjected to 4μ8c treatment, LDH levels were diminished in 4μ8c-treated macrophages infected with the ΔrfbE mutant at 3, 5, and 8 h p.i. (Figure 6B), indicating that IRE1α inhibition reduced macrophage death caused by the ΔrfbE mutant. The 4μ8c treatment did not affect LDH release from the macrophages infected with S2308 at 3, 5, and 8 h p.i. (Figure 6B).
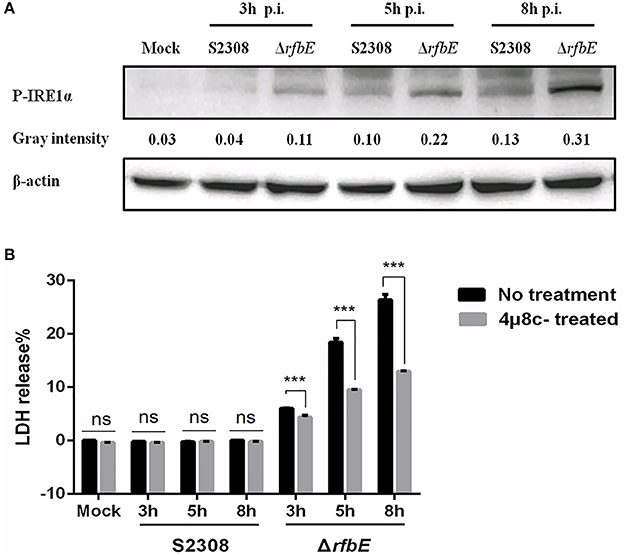
Figure 6. The Brucella rough mutant induces stronger ER stress, which plays a role in macrophage death. (A) Western blotting analysis. RAW264.7 cells were infected with S2308 or ΔrfbE at a MOI of 100. Cell lysates were collected at 3, 5, and 8 hpi, and representative immunoblots for P-IRE1α were analyzed. β-actin was used for normalization. Uninfected RAW264.7 cells were used as negative controls (Mock). The intensity of the bands was quantified using the ImageJ software. (B) Determination of LDH release. RAW264.7 cells both with and without 4μ8c (IRE1α inhibitor, 100 μM) treatment were infected with either S2308 or the ΔrfbE mutant at an MOI of 100. The supernatants were collected at 3, 5, and 8 hpi, and LDH release was detected using the CytoTox 96 nonradioactive cytotoxicity assay. The supernatants of uninfected RAW264.7 cells were used as negative controls (medium). ns, no significant difference, ***p < 0.0001.
Discussion
The cytotoxicity induced by Brucella rough mutants within macrophages was originally described more than 50 years ago (Freeman et al., 1961; Freeman and Rumack, 1964). The T4SS is essential for cytotoxic death of macrophages induced by Brucella infection (Pei et al., 2008). Brucella T4SS is tightly regulated by various regulatory proteins under specific conditions, such as acidification and nutritional deprivation. Deletion or overexpression of virB is detrimental to intracellular survival of Brucella (Zhong et al., 2009). In addition, shortening of the LPS molecule enhances the type III secretion system in Shigella (West et al., 2005). In this study, we confirmed that the capacity for T4SS secretion and the effectors being translocated to macrophages were highly increased in the ΔrfbE mutant.
The virB mRNA level has been shown to be very low when Brucella is grown in a rich medium at neutral pH, and virB transcription is upregulated when cultured in acidic conditions or minimal medium (Boschiroli et al., 2002). However, the use of lacZ reporter gene fusions has shown that the virB operon of B. abortus S2308 is expressed during the stationary phase, without the requirement for acidic induction conditions (Sieira et al., 2000). According to the analysis of mRNA levels, our results demonstrated that in comparison to the smooth wild-type strain S2308, T4SS expression of the rough mutant ΔrfbE was significantly upregulated at the exponential phase under conditions of both a rich medium at neutral pH and nutrient-deprived or acidic conditions. In the rich medium at neutral pH, the parental B. abortus and B. melitensis strains constitutively produced virB5 and virB8 (Rouot et al., 2003). However, in this study, the smooth wild-type strain, S2308, produced a low level of the virB5 protein in a rich medium at neutral pH; whereas virB5 expression levels of the rough mutant, ΔrfbE, were significantly increased. After exposure to acidic minimal medium, Brucella easily produces detectable levels of virB8 (Rouot et al., 2003). Under nutrient-deprived or acidic conditions, we confirmed that the virB5 protein is easily detected, and that expression of virB5 in the rough mutant ΔrfbE, was higher than that in the S2308 strain. Thus, enhanced expression of T4SS in the ΔrfbE mutant might account for its increased capacity for T4SS secretion.
Intracellular induction of virB expression has been observed to be transient, and the translocation and activity of VirB-secreted effectors within the host cell might be determined by the timing of expression of the virB operon (Sieira, 2013). In this work, we analyzed the activity of the virB promoter in the smooth wild-type strain S2308, and its rough mutant ΔrfbE, using the luciferase reporter assay in a rich medium and in an intracellular environment. In compared to the wild-type strain S2308, the virB promoter activity of the rough mutant ΔrfbE, was significantly increased in the rich medium and in acidic conditions, both of which enhanced T4SS expression and secretion in the rough mutant ΔrfbE. Once Brucella is internalized in macrophages, the transcriptional activity of the virB promoter reaches a maximum level at 5 h p.i., and the promoter is then turned off, when Brucella reaches its replicative niche (Sieira et al., 2004). Our results demonstrated that the activity of the virB promoter was increased in the intracellular environment of both the smooth wild-type strain S2308, and the rough mutant ΔrfbE, at an early stage of infection. Furthermore, T4SS expression and secretion of the ΔrfbE mutant was notably upregulated in comparison to that of the S2308 strain within macrophages. However, the virB promoter activity in wild-type strain S2308 did not stop at 8 h p.i. in this study, which may be due to different setting up of the time point in the cell infection assays.
On further study, we investigated the expression of vjbR, blxR, and mdrA genes that have been proven to regulate T4SS expression directly in the smooth Brucella strain. We found that vjbR expression was upregulated, and the expression of both blxR and mdrA were downregulated in the ΔrfbE mutant. The VjbR protein belongs to the LuxR family, a group of transcriptional regulators involved in the cell-to-cell communication process referred to as quorum sensing (QS), This process allows bacteria to sense changes in population density and coordinate adaptive responses, and acts as the main regulator of expression of the virB operon (Miller and Bassler, 2001; Uzureau et al., 2010; Weeks et al., 2010). A vjbR mutant of B. melitensis exhibits downregulated expression of both the virB operon and flagellar genes, either during vegetative growth or during intracellular infection, and is strongly attenuated in a mouse model of infection (Delrue et al., 2005). In addition, VjbR regulates exopolysaccharide synthesis or export, as well as the production of several outer membrane proteins, some of which are involved in virulence (Uzureau et al., 2007). In the present study, we found that deletion of vjbR in the ΔrfbE mutant significantly reduced its cytotoxicity in macrophages. The BlxR protein is the second QS-related regulator of Brucella that contains both the DNA- and AHL-binding domains characteristic of the LuxR-type proteins (Rambow-Larsen et al., 2008; Sieira, 2013). Deletion of blxR affects virulence and intracellular survival of Brucella, but to a lesser extent than deletion of vjbR (Rambow-Larsen et al., 2008). A previous report suggests that BlxR negatively modulates activity of the virB promoter in B. abortus (Caswell et al., 2012). Our results confirmed that BlxR negatively modulates the activity of the virB promoter in B. abortus, and overexpression of blxR in the ΔrfbE mutant reduces to some extent, the cytotoxicity within macrophages. However, overexpression of mdrA in the ΔrfbE mutant did not reduce cytotoxicity within macrophages. The significance of mdrA downregulation in the ΔrfbE mutant requires further study. Thus, it is evident that a QS-related transcriptional regulator plays important roles in Brucella rough mutant-induced macrophage death. The QS-related transcriptional regulators might function in sensing environmental changes, such as cell density, acidification, and nutritional deprivation. One possible explanation is that loss of LPS in Brucella makes it sensitive to environmental stress that dysregulates the QS-related transcriptional regulators and upregulates T4SS to secrete a greater number of effectors. This probably accounts for the cytotoxicity in macrophages infected by Brucella rough mutants.
During Brucella interaction with host cells, the Brucella T4SS effector protein VceC, is involved in the induction of inflammatory responses by binding chaperone BiP, to trigger ER stress (de Jong et al., 2013). The ER stress induces the UPR in macrophages, and activates IRE1α, which in turn, recruits the NOD-like receptors NOD1 and NOD2, to induce activation of NF-κB and expression of pro-inflammatory genes (Keestra-Gounder et al., 2016). Brucella abortus inhibits cell death of infected macrophages (Fernandez-Prada et al., 2003; He et al., 2006), and chronically persists under conditions of a mild inflammatory response that leads to granuloma formation (Silva et al., 2011). However, the ER stress sensor IRE1α, induced by the rough mutant RB51, induces ROS-dependent NLRP3 translocation to mitochondria, and NLRP3 stimulates the caspase-2-Bid mitochondrial damage pathway, thereby leading to the release of mitochondrial danger signals that activate the inflammasome (Bronner et al., 2015). In this study, we found that the rough mutant ΔrfbE, secreted more effector proteins and induced stronger IRE1α pathways of ER stress, in comparison to the smooth wild-type strain S2308. These actions might excessively activate the IRE1α pathway and further activate the inflammasome via NLRP3- and caspase-2- driven mitochondrial damage, and result in cell death of macrophages. The crucial components associated with activation of the IRE1α pathway of ER stress to promote macrophage death in rough mutants remain to be identified.
Taken together, this study provided evidence that VjbR upregulation in the Brucella rough mutant ΔrfbE increases transcription of the virB operon, resulting in T4SS overexpression, accompanied by over-secretion of T4SS effector proteins. This in turn, strongly activates the IRE1α pathway of ER stress to cause the death of infected macrophages. This study provides novel insights into molecular mechanisms of Brucella rough mutant ΔrfbE-induced macrophage cytotoxicity.
Author Contributions
SY, MT, and CD conceived and designed the experiments; PL and MT mainly performed the experiments and analyzed the data; YB, HH, JL, YY, and SW helped to perform some experiments; PL wrote the paper, SY revised the manuscript and coordinated the research. All authors have read and approved the manuscript.
Funding
This work was supported by funds from the Scientific and Technical Innovation Project of the Chinese Academy of Agricultural Sciences (SHVRI-ASTIP-2014-8), the National Natural Science Foundation of China (31602070), the Shanghai Sailing Program (16YF1414600), and the National Basic Fund for Research Institutes, which is supported by the Chinese Academy of Agricultural Sciences (2016JB06).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Atluri, V. L., Xavier, M. N., de Jong, M. F., den Hartigh, A. B., and Tsolis, R. M. (2011). Interactions of the human pathogenic Brucella species with their hosts. Annu. Rev. Microbiol. 65, 523–541. doi: 10.1146/annurev-micro-090110-102905
Boschiroli, M. L., Foulongne, V., and O'Callaghan, D. (2001). Brucellosis: a worldwide zoonosis. Curr. Opin. Microbiol. 4, 58–64. doi: 10.1016/S1369-5274(00)00165-X
Boschiroli, M. L., Ouahrani-Bettache, S., Foulongne, V., Michaux-Charachon, S., Bourg, G., Allardet-Servent, A., et al. (2002). The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. U.S.A. 99, 1544–1549. doi: 10.1073/pnas.032514299
Bronner, D. N., Abuaita, B. H., Chen, X., Fitzgerald, K. A., Nunez, G., He, Y., et al. (2015). Endoplasmic reticulum stress activates the inflammasome via NLRP3- and caspase-2-driven mitochondrial damage. Immunity 43, 451–462. doi: 10.1016/j.immuni.2015.08.008
Bronner, D. N., O'Riordan, M. X., and He, Y. (2013). Caspase-2 mediates a Brucella abortus RB51-induced hybrid cell death having features of apoptosis and pyroptosis. Front. Cell. Infect. Microbiol. 3:83. doi: 10.3389/fcimb.2013.00083
Byndloss, M. X., and Tsolis, R. M. (2016). Brucella spp. virulence factors and immunity. Annu. Rev. Anim. Biosci. 4, 111–127. doi: 10.1146/annurev-animal-021815-111326
Byndloss, M. X., Rivera-Chavez, F., Tsolis, R. M., and Baumler, A. J. (2016). How bacterial pathogens use type III and type IV secretion systems to facilitate their transmission. Curr. Opin. Microbiol. 35, 1–7. doi: 10.1016/j.mib.2016.08.007
Caswell, C. C., Gaines, J. M., and Roop, R. M. II. (2012). The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J. Bacteriol. 194, 3–14. doi: 10.1128/JB.05623-11
Celli, J. (2006). Surviving inside a macrophage: the many ways of Brucella. Res. Microbiol. 157, 93–98. doi: 10.1016/j.resmic.2005.10.002
Chen, F., Ding, X., Ding, Y., Xiang, Z., Li, X., Ghosh, D., et al. (2011). Proinflammatory caspase-2-mediated macrophage cell death induced by a rough attenuated Brucella suis strain. Infect. Immun. 79, 2460–2469. doi: 10.1128/IAI.00050-11
Cross, B. C., Bond, P. J., Sadowski, P. G., Jha, B. K., Zak, J., Goodman, J. M., et al. (2012). The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. U.S.A. 109, E869–E878. doi: 10.1073/pnas.1115623109
de Jong, M. F., Starr, T., Winter, M. G., den Hartigh, A. B., Child, R., Knodler, L. A., et al. (2013). Sensing of bacterial type IV secretion via the unfolded protein response. MBio 4, e00418–e00512. doi: 10.1128/mBio.00418-12
Delrue, R. M., Deschamps, C., Leonard, S., Nijskens, C., Danese, I., Schaus, J. M., et al. (2005). A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7, 1151–1161. doi: 10.1111/j.1462-5822.2005.00543.x
Fernandez-Prada, C. M., Zelazowska, E. B., Nikolich, M., Hadfield, T. L., Roop Ii, R. M., Robertson, G. L., et al. (2003). Interactions between Brucella melitensis and human phagocytes: bacterial surface O-polysaccharide inhibits phagocytosis, bacterial killing, and subsequent host cell apoptosis. Infect. Immun. 71, 2110–2119. doi: 10.1128/IAI.71.4.2110-2119.2003
Franco, M. P., Mulder, M., Gilman, R. H., and Smits, H. L. (2007). Human brucellosis. Lancet Infect. Dis. 7, 775–786. doi: 10.1016/S1473-3099(07)70286-4
Freeman, B. A., and Rumack, B. H. (1964). Cytopathogenic effect of Brucella spheroplasts on monocytes in tissue culture. J. Bacteriol. 88, 1310–1315.
Freeman, B. A., Kross, D. J., and Circo, R. (1961). Host-parasite relationships in brucellosis. II. Destruction of macrophage cultures by Brucella of different virulence. J. Infect Dis. 108, 333–338. doi: 10.1093/infdis/108.3.333
Godfroid, F., Cloeckaert, A., Taminiau, B., Danese, I., Tibor, A., de Bolle, X., et al. (2000). Genetic organisation of the lipopolysaccharide O-antigen biosynthesis region of Brucella melitensis 16M (wbk). Res. Microbiol. 151, 655–668. doi: 10.1016/S0923-2508(00)90130-X
Goldstein, J., Hoffman, T., Frasch, C., Lizzio, E. F., Beining, P. R., Hochstein, D., et al. (1992). Lipopolysaccharide (LPS) from Brucella abortus is less toxic than that from Escherichia coli, suggesting the possible use of B. abortus or LPS from B. abortus as a carrier in vaccines. Infect. Immun. 60, 1385–1389.
Gorvel, J. P., and Moreno, E. (2002). Brucella intracellular life: from invasion to intracellular replication. Vet. Microbiol. 90, 281–297. doi: 10.1016/S0378-1135(02)00214-6
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995
He, Y., Reichow, S., Ramamoorthy, S., Ding, X., Lathigra, R., Craig, J. C., et al. (2006). Brucella melitensis triggers time-dependent modulation of apoptosis and down-regulation of mitochondrion-associated gene expression in mouse macrophages. Infect. Immun. 74, 5035–5046. doi: 10.1128/IAI.01998-05
Kahl-McDonagh, M. M., and Ficht, T. A. (2006). Evaluation of protection afforded by Brucella abortus and Brucella melitensis unmarked deletion mutants exhibiting different rates of clearance in BALB/c mice. Infect. Immun. 74, 4048–4057. doi: 10.1128/IAI.01787-05
Ke, Y., Wang, Y., Li, W., and Chen, Z. (2015). Type IV secretion system of Brucella spp. and its effectors. Front Cell Infect Microbiol. 5:72. doi: 10.3389/fcimb.2015.00072
Keestra-Gounder, A. M., Byndloss, M. X., Seyffert, N., Young, B. M., Chavez-Arroyo, A., Tsai, A. Y., et al. (2016). NOD1 and NOD2 signalling links ER stress with inflammation. Nature 532, 394–397. doi: 10.1038/nature17631
Kovach, M. E., Phillips, R. W., Elzer, P. H., Roop, R. M. II., and Peterson, K. M. (1994). pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16, 800–802.
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Pei, J., and Ficht, T. A. (2003). Brucella abortus rough mutants are cytopathic for macrophages in culture. Infect. Immun. 72, 440–450. doi: 10.1128/IAI.72.1.440-450.2004
Pei, J., Wu, Q., Kahl-McDonagh, M., and Ficht, T. A. (2008). Cytotoxicity in macrophages infected with rough Brucella mutants is type IV secretion system dependent. Infect. Immun. 76, 30–37. doi: 10.1128/IAI.00379-07
Porte, F., Liautard, J. P., and Kohler, S. (1999). Early acidification of phagosomes containing Brucellasuis is essential for intracellular survival in murine macrophages. Infect. Immun. 67, 4041–4047.
Rambow-Larsen, A. A., Rajashekara, G., Petersen, E., and Splitter, G. (2008). Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J. Bacteriol. 190, 3274–3282. doi: 10.1128/JB.01915-07
Rasool, O., Freer, E., Moreno, E., and Jarstrand, C. (1992). Effect of Brucella abortus lipopolysaccharide on oxidative metabolism and lysozyme release by human neutrophils. Infect. Immun. 60, 1699–1702.
Rolan, H. G., Xavier, M. N., Santos, R. L., and Tsolis, R. M. (2009). Natural antibody contributes to host defense against an attenuated Brucella abortus virB mutant. Infect. Immun. 77, 3004–3013. doi: 10.1128/IAI.01114-08
Rouot, B., Alvarez-Martinez, M. T., Marius, C., Menanteau, P., Guilloteau, L., Boigegrain, R. A., et al. (2003). Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect. Immun. 71, 1075–1082. doi: 10.1128/IAI.71.3.1075-1082.2003
Sieira, R. (2013). Regulation of virulence in Brucella: an eclectic repertoire of transcription factors defines the complex architecture of the virB promoter. Future Microbiol. 8, 1193–1208. doi: 10.2217/fmb.13.83
Sieira, R., Comerci, D. J., Pietrasanta, L. I., and Ugalde, R. A. (2004). Integration host factor is involved in transcriptional regulation of the Brucella abortus virB operon. Mol. Microbiol. 54, 808–822. doi: 10.1111/j.1365-2958.2004.04316.x
Sieira, R., Comerci, D. J., Sanchez, D. O., and Ugalde, R. A. (2000). A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 182, 4849–4855. doi: 10.1128/JB.182.17.4849-4855.2000
Silva, T. M., Costa, E. A., Paixao, T. A., Tsolis, R. M., and Santos, R. L. (2011). Laboratory animal models for brucellosis research. J. Biomed. Biotechnol. 2011:518323. doi: 10.1155/2011/518323
Tian, M., Qu, J., Han, X., Ding, C., Wang, S., Peng, D., et al. (2014). Mechanism of Asp24 upregulation in Brucella abortus rough mutant with a disrupted O-antigen export system and effect of Asp24 in bacterial intracellular survival. Infect. Immun. 82, 2840–2850. doi: 10.1128/IAI.01765-14
Uzureau, S., Godefroid, M., Deschamps, C., Lemaire, J., De Bolle, X., and Letesson, J. J. (2007). Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J. Bacteriol. 189, 6035–6047. doi: 10.1128/JB.00265-07
Uzureau, S., Lemaire, J., Delaive, E., Dieu, M., Gaigneaux, A., Raes, M., et al. (2010). Global analysis of quorum sensing targets in the intracellular pathogen Brucella melitensis 16 M. J. Proteome Res. 9, 3200–3217. doi: 10.1021/pr100068p
Weeks, J. N., Galindo, C. L., Drake, K. L., Adams, G. L., Garner, H. R., and Ficht, T. A. (2010). Brucella melitensis VjbR and C12-HSL regulons: contributions of the N-dodecanoyl homoserine lactone signaling molecule and LuxR homologue VjbR to gene expression. BMC Microbiol. 10:167. doi: 10.1186/1471-2180-10-167
West, N. P., Sansonetti, P., Mounier, J., Exley, R. M., Parsot, C., Guadagnini, S., et al. (2005). Optimization of virulence functions through glucosylation of Shigella LPS. Science 307, 1313–1317. doi: 10.1126/science.1108472
Whatmore, A. M. (2009). Current understanding of the genetic diversity of Brucella, an expanding genus of zoonotic pathogens. Infect. Genet. Evol. 9, 1168–1184. doi: 10.1016/j.meegid.2009.07.001
Zechner, E. L., Lang, S., and Schildbach, J. F. (2012). Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1073–1087. doi: 10.1098/rstb.2011.0207
Zhang, M., Han, X., Liu, H., Tian, M., Ding, C., Song, J., et al. (2013). Inactivation of the ABC transporter ATPase gene in Brucella abortus strain 2308 attenuated the virulence of the bacteria. Vet. Microbiol. 164, 322–329. doi: 10.1016/j.vetmic.2013.02.017
Keywords: Brucella, Type IV secretion system, lipopolysaccharide, VjbR, endoplasmic reticulum stress
Citation: Li P, Tian M, Bao Y, Hu H, Liu J, Yin Y, Ding C, Wang S and Yu S (2017) Brucella Rough Mutant Induce Macrophage Death via Activating IRE1α Pathway of Endoplasmic Reticulum Stress by Enhanced T4SS Secretion. Front. Cell. Infect. Microbiol. 7:422. doi: 10.3389/fcimb.2017.00422
Received: 18 May 2017; Accepted: 14 September 2017;
Published: 27 September 2017.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Clayton Caswell, Virginia Tech, United StatesZeliang Chen, Shenyang Agricultural University, China
Copyright © 2017 Li, Tian, Bao, Hu, Liu, Yin, Ding, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengqing Yu, yus@shvri.ac.cn
†These authors have contributed equally to this work.
 Peng Li
Peng Li Mingxing Tian
Mingxing Tian Yanqing Bao1
Yanqing Bao1  Chan Ding
Chan Ding Shengqing Yu
Shengqing Yu