Role of Low-Molecular-Mass Penicillin-Binding Proteins, NagZ and AmpR in AmpC β-lactamase Regulation of Yersinia enterocolitica
- 1Department of Pathogenic Biology, School of Medical Science, Jiangsu University, Zhenjiang, China
- 2National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, State Key Laboratory for Infectious Disease Prevention and Control, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Beijing, China
- 3Qinghai Institute for Endemic Diseases Prevention and Control, Xining, China
Yersinia enterocolitica encodes a chromosomal AmpC β-lactamase under the regulation of the classical ampR-ampC system. To obtain a further understanding to the role of low-molecular-mass penicillin-binding proteins (LMM PBPs) including PBP4, PBP5, PBP6, and PBP7, as well as NagZ and AmpR in ampC regulation of Y. enterocolitica, series of single/multiple mutant strains were systematically constructed and the ampC expression levels were determined by luxCDABE reporter system, reverse transcription-PCR (RT-PCR) and β-lactamase activity test. Sequential deletion of PBP5 and other LMM PBPs result in a continuously growing of ampC expression level, the β-lactamse activity of quadruple deletion strain YEΔ4Δ5Δ6Δ7 (pbp4, pbp5, pbp6, and pbp7 inactivated) is approached to the YEΔD123 (ampD1, ampD2, and ampD3 inactivated). Deletion of nagZ gene caused two completely different results in YEΔD123 and YEΔ4Δ5Δ6Δ7, NagZ is indispensable for YEΔ4Δ5Δ6Δ7 ampC derepression phenotype but dispensable for YEΔD123. AmpR is essential for ampC hyperproduction in these two types of strains, inactivation of AmpR notable reduced the ampC expression level in both YEΔD123 and YEΔ4Δ5Δ6Δ7.
Introduction
Yersinia enterocolitica, a member of Enterobacteriaceae, is a zoonotic pathogen widely distributed in nature (Wang et al., 2011; Liang et al., 2012). Most Y. enterocolitica exhibits intrinsic resistance to β-lactm antibiotics by the production of chromosomally encoded β-lactamases called BlaA (a class A enzyme showing constitutive expression) and BlaB (an inducible AmpC-type β-lactamase), respectively (Cornelis and Abraham, 1975; Bent and Young, 2010).
The process of ampC (blaB) regulation is tightly linked to the peptidoglycan recycling and controlled by AmpG, AmpD, AmpR, and NagZ (Vollmer et al., 2008; Zeng and Lin, 2013). Briefly, peptidoglycan degradation products including GlcNAc-1,6-anhydromuropeptide is transported into the cytoplasm by AmpG and further hydrolyzedcosaminidase) to yielding 1,6-anhydromuropeptides, which is the AmpR activator ligand for ampC derepression (Zamorano et al., 2010; Huang et al., 2012; Yang et al., 2014). On the other hand, the stem peptides of GlcNAc-1,6-anhydromuropeptide and 1,6-anhydromuropeptides can be removed by AmpD (N-acetylmuramyl-l-alanine amidase) and eventually recycled into UDP-MurNAc-pentapeptide, which is the AmpR repressor ligand to repress ampC expression level (Juan et al., 2006; Balasubramanian et al., 2015; Liu et al., 2016). Penicillin-binding proteins (PBPs) also play an important role in ampC regulation (Sanders et al., 1997; Pfeifle et al., 2000). Recent studies have found that in P. aeruginosa, PBP4 (DacB), PBP5 (DacC), and PBP7 (PbpG) are involved in ampC regulation, and PBP4 is the major cause of ampC derepressed in clinical strains (Moya et al., 2009; Ropy et al., 2015).
Theoretically, NagZ is indispensable in chromosomal ampC derepression. In P. aeruginosa, nagZ inactivation dramatically reduces the β-lactam resistance of both PAOΔampD (ampD inactivation) and PAOΔdacB (pbp4 inactivation; Zamorano et al., 2010). However, although nagZ inactivation nearly abolished the basal-level derepressed β-lactamase activity of KJΔampDI (ampD inactivation), it did not affect the β-lactamase activity of KJΔmrcA (pbp1a inactivation) in Stenotrophomonas maltophilia (Huang et al., 2012).
Since the effects of the above-mentioned genes in Y. enterocolitica were seldom reported, we elucidated the role of low-molecular-mass penicillin-binding proteins (LMM PBPs) (PBP4, PBP5, PBP6, and PBP7), NagZ and AmpR in the Y. enterocolitica ampC regulation. Firstly, we investigated the effects of each LMM PBP on the expression of AmpC β-lactamase by monitoring the ampC promoter activity from a series of LMM PBPs mutant strains and confirmed by quantitative reverse transcription-PCR (qRT-PCR). Secondly, nagZ gene was deleted in two ampC derepressed strains YEΔD123 and YEΔ4Δ6Δ5Δ7 to determine the role for ampC expression.
Materials and Methods
Bacterial Strains, Plasmids, Primers, and Growth Conditions
Strains and plasmids used in this study were listed in Table 1. Individual genes were deleted initially from Y. enterocolitica subsp. palearctica 105.5R(r) (Wang et al., 2011). Luria-Bertani (LB) agar plates and broth were used as culture media for Y. enterocolitica (28°C) and Escherichia coli (37°C). For induction assay, cefoxitin was used according to the references (Guerin et al., 2015; Liu et al., 2016).
Construction of Y. enterocolitica Mutant Strains
Knockout mutant strains were constructed using the method described previously (Chen et al., 2015; Liang et al., 2016; Liu et al., 2016). Briefly, the deletion mutants were constructed by double-crossover homologous recombination between wild-type strain chromosome and plasmids pΔNagZ, pΔAmpR, pΔPBP4, pΔPBP5, pΔPBP6l, and pΔPBP7. To evaluate the role of PBP4 (WP_005175403.1), PBP5 (WP_005158391.1) PBP6 (WP_023160783.1), and PBP7 (WP_005158897.1) in Y. enterocolitica 105.5R(r) ampC regulation, we constructed four single mutant strains: YEΔ4 (pbp4 inactivation), YEΔ5 (pbp5 inactivation), YEΔ6 (pbp6 inactivation), and YEΔ7 (pbp7 inactivation); six double mutant strains: YEΔ4Δ5, YEΔ4Δ6, YEΔ4Δ7, YEΔ5Δ6, YEΔ5Δ7, and YEΔ6Δ7; four triple mutant strains: YEΔ4Δ5Δ6, YEΔ4Δ5Δ7, YEΔ4Δ6Δ7, and YEΔ5Δ6Δ7; and one quadruple mutant strain: YEΔ4Δ5Δ6Δ7 (Table 1). The deletion mutants were identified by colony PCR firstly and then sequenced to confirm the in-frame deletion. Multiple deletion strains were sequentially constructed from the single mutant by use of the same procedure.
Measurement of the ampC Promoter Activity
The method of measuring the ampC promoter activity with the luxCDABE reporter system was reported previously (Liu et al., 2016). The reporter plasmid pLUXampC was transferred into the tested strains, and the luminescence was measured by using an Infinite M200 Pro spectrophotometer. The value of luminescence/OD600 was used to assess the ampC promoter activity.
Determination of β-Lactamase Activity and Antibiotic Susceptibility Testing
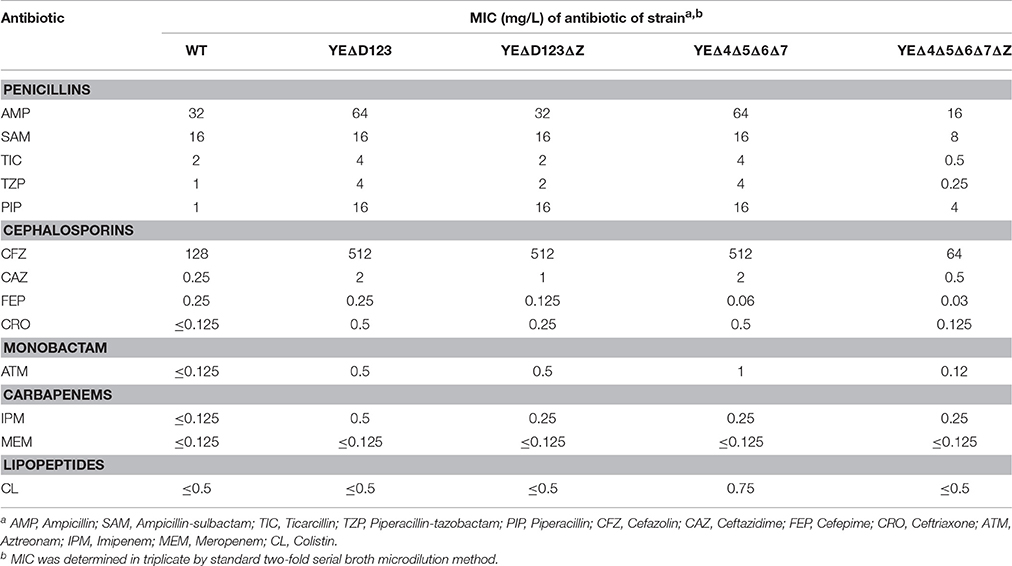
Specific β-lactamase activities were spectrophotometrically determined with nitrocefin (Oxoid) as a substrate as previously described (Liu et al., 2016). One unit of β-lactamase activity (U/mg) was defined as the number of nanomoles of nitrocefin hydrolyzed per minute per milligram of protein. Antibiotic susceptibility was determined using the standard 2-fold serial broth microdilution method according to the Guidelines of the Clinical Laboratory Standards Institute (CLSI, 2015).
N-Acetyl-β-Glucosaminidase Activity Assay
The N-acetyl-glucosaminidase activity of the whole cell lysates of wild-type strain 105.5R(r) and YEΔZ were measured using 4-nitrophenyl N-acetyl-β-D-glucosaminide as a chromogenic substrate (Sigma). The presence of p-nitrophenol were detected by monitoring the optical density at 405 nm by 10 h continuously.
Complementation Assay
The ORF of nagZ was amplified and cloned into the broad-host-range expression vector pSRKTc to construct plasmid pNagZ. Transformants were selected on 10 μg/ml tetracycline Yersinia selective LB plates, acquisition of the appropriate plasmid was confirmed by colony PCR.
Results
Role of LMM PBPs in the Expression of AmpC β-Lactamase
After a series of LMM PBPs mutant strains were constructed, reporter plasmid pLUXampC was used to monitor the ampC expression level (Liu et al., 2016). As shown in Figure 1, deletion pbp5 caused a visible increase in the ampC promoter activity under both basal and induced conditions; but deletion of pbp4, pbp6, and pbp7 did not affect the AmpC expression obviously. In the group of double and triple mutant strains, ampC derepression only appeared in Δpbp5 background, the ampC promoter activity of YEΔ4Δ5, YEΔ5Δ6, and YEΔ5Δ7 exhibited a marked rise compared with YEΔ4Δ6, YEΔ4Δ7, or YEΔ6Δ7. The level of ampC expression keep increasing in triple mutant strains YEΔ4Δ5Δ6, YEΔ4Δ5Δ7, and YEΔ5Δ6Δ7, but not in YEΔ4Δ6Δ7. Finally, the quadruple deletion strain YEΔ4Δ5Δ6Δ7 displayed the highest level of ampC promoter activity. These results suggested that PBP5 plays the most important roles in Y. enterocolitica ampC regulation. The qRT-PCR assay reconfirmed the results observed from ampC promoter activity assay (Table 2).
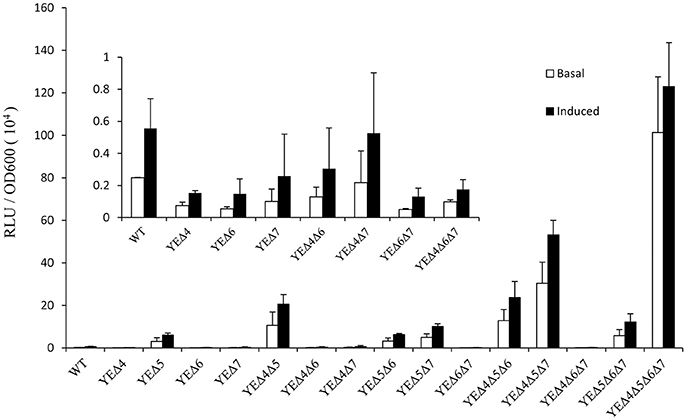
Figure 1. Analysis of the ampC promoter activities in Y. enterocolitica 105.5R(r) wild-type strain and pbp mutants. The induction group was incubated with 40 μg/ml cefoxitin for 1 h. The error bars represent the standard deviations of triplicate tests.
Role of NagZ in AmpC Derepression of Y. enterocolitica
In agreement with our previous data (Liu et al., 2016), AmpD deletion strain YEΔD123 exhibit a derepression phenotype, and the β-lactamase activity of YEΔD123 is slightly higher than YEΔ4Δ5Δ6Δ7 (Figure 2). To evaluate the role of NagZ in AmpC derepression, nagZ gene was deleted in both derepression strains to construct YEΔD123ΔZ and YEΔ4Δ5Δ6Δ7ΔZ. As shown in Figure 2, nagZ was indispensable for ampC over expression of YEΔ4Δ5Δ6Δ7, the β-lactamase activity of nagZ deletion strain YEΔ4Δ5Δ6Δ7ΔZ was decreased significantly, closed to the wild-type strain level. In complementation assay, YEΔ4Δ5Δ6Δ7ΔZ (pNagZ) restored the β-lactamase activity to the level of YEΔ4Δ5Δ6Δ7. However, NagZ was dispensable in YEΔD123, the β-lactamase activity of nagZ deletion strain YEΔD123ΔZ was nearly as high as YEΔD123 (Figure 2). These results suggested that NagZ was needed in ΔPBPs-driven AmpC derepression, but did not perform its expected function in AmpD mutation strains. Antibiotic susceptibility test was also performed, as shown in Table 3, the MIC values of YEΔ4Δ5Δ6Δ7ΔZ were slightly below the wild-type strain 105.5R(r), far from its parent strain YEΔ4Δ5Δ6Δ7 for almost all tested β-lactams; but only a marginal distinction between YEΔD123 and YEΔD123ΔZ was found. These results illustrated that AmpD/PBPs regulate AmpC expression through NagZ dispensable/indispensable ways in Y. enterocolitica.
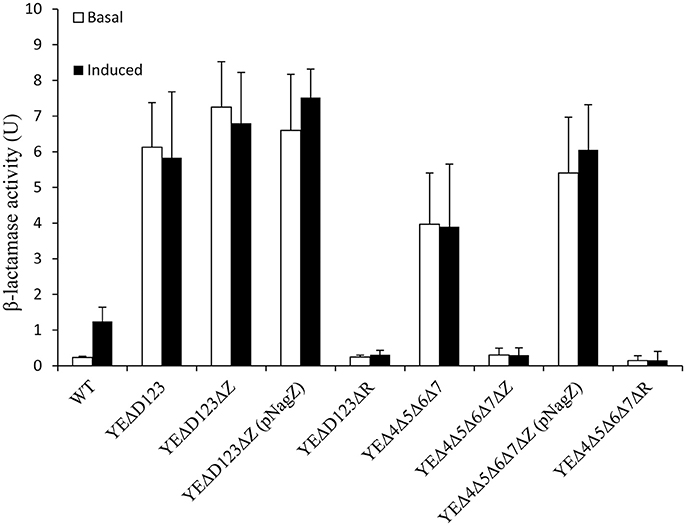
Figure 2. The role of AmpD, PBPs, NagZ, and AmpR in the β-lactamase expression of Y. enterocolitica by measuring the β-lactamase activity. These data are the average of three repeat experiments. The induction group was incubated with 40 μg/ml cefoxitin for 1 h. Error bars indicate the standard deviations of triplicate tests.
N-Acetyl-β-Glucosaminidase Activity Assay
The nagZ mutation strain YEΔZ was constructed, and determined by the enzyme activity of the both wild-type strain and YEΔZ for 10 h using N-acetyl-β-D-glucosaminide as substrate. As shown in Figure 3, YEΔZ abolished the N-acetyl-β-glucosaminidase activity completely, it was suggested that NagZ is the only enzyme that with N-acetyl-β-glucosaminidase activity in Y. enterocolitica.
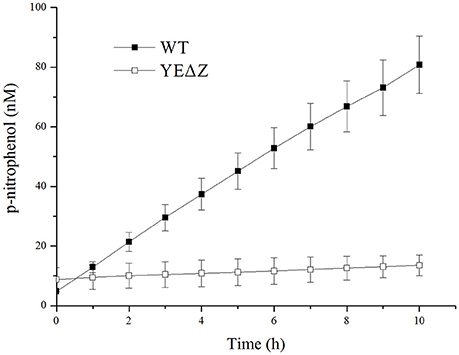
Figure 3. The N-acetyl-β-glucosamididase activity of wild-type Y. enterocolitica and nagZ deletion mutants was tested using 4-nitrophenyl N-acetyl-β-D-glucosaminide as a chromogenic substrate, and the p-nitrophenol present in the supernatant was measured at 405 nm.
Role of AmpR in ampC Expression of in Y. enterocolitica
In the paradigm of the ampR-ampC system, the ampR gene is located immediately adjacent to ampC, and AmpR plays a pivotal role in the regulation of AmpC (Seoane et al., 1992). To assess the role of AmpR in Y. enterocolitica, we compared the β-lactamase activity of YEΔD123ΔR, YEΔ4Δ5Δ6Δ7ΔR with their parent strains YEΔD123, YEΔ4Δ5Δ6Δ7, respectively. As a result, ampR inactivation dramatically reduced the β-lactamase activity of both YEΔD123ΔR and YEΔ4Δ5Δ6Δ7ΔR, regardless of adding cefoxitin or not (Figure 2).
Discussion
The ampR-ampC system from Citrobacter freundii and Enterobacter cloacae has been well studied in the early 1990s (Lindberg et al., 1987; Peter et al., 1988). However, newly discovered ampC regulators such as, PBP4 (DacB) or NagZ in Enterobacteriaceae was not yet understood. A deep study in Y. enterocolitica ampR-ampC system would be helpful to improve the comprehensive understanding of Enterobacteriaceae ampC regulation.
PBPs are a group of enzymes involved in cell-wall recycling and the processes of AmpC β-lactamases regulation. In E. coli model, deletion of three or four PBPs and the concomitant inhibition of PBP 1a, 1b, and/or 2 results in an increased level of β-lactamase induction (Pfeifle et al., 2000). However, since E. coli lacks the chromosomal ampR gene, the result may be inconsistent with other members of the Gram-negative bacteria which have a chromosome encoding the ampR-ampC system. In 2009, Moya et al. demonstrated the inactivation of DacB (PBP4), a nonessential low-molecular mass PBPs is the principal reason for one-step high-level ampC expression in clinical strains of P. aeruginosa (Moya et al., 2009). Interestingly, inactivation of PBP4 in E. cloacae triggered a significant increase of β-lactams resistance, but without an obvious upregulation of ampC gene, it may be suggested that PBP4 regulates AmpC at a post-transcriptional level (Guerin et al., 2015). In this study, we found deletion of pbp4 did not elevate the ampC expression level, this result is accordance with E. cloacae. After that, we deleted all four LMM PBPs one after another, and found that PBP5 is the most effective PBP involved in the regulation of ampC in Y. enterocolitica. Of the single-mutation strains, only the pbp5 deletion strain YEΔ5 showed an obvious rise in ampC expression level. Likewise, for multi-mutation strains, the function of PBP4, PBP6, and PBP7 in ampC regulation were detected only if in Δpbp5 background. According to the results shown in Figure 1 and Table 2, we deduced the hierarchy of the role of PBPs genes in ampC derepression: PBP5 > PBP4 > PBP7 > PBP6. Although DacB may regulates AmpC at a post-transcriptional level (Guerin et al., 2015), but no trace of post-transcriptional mechanism has been found in Y. enterocolitica.
Along with the popular research of ampC regulation, there is growing evidence that some bacteria may regulate the expression of ampC through at least two different ways, one of which was NagZ-dependent, while the other worked without the participation of NagZ (Huang et al., 2012; Guerin et al., 2015). In the study on P. aeruginosa, nagZ inactivation was shown to attenuate ampC expression and was critical for basal-level ampC derepression in both PAΔD (ampD inactivation) and PAΔdB (pbp4 inactivation) mutants (Asgarali et al., 2009; Zamorano et al., 2010). However, ΔnagZ had little effect on the cefoxitin-induced ampC expression level in both PAΔD and PAΔdB, which indicated that an unidentified non-NagZ product at work in this induction process. Furthermore, two different regulation ways of β-lactamase have been found in S. maltophilia, on one hand NagZ was essential for KJΔDI (ampD inactivation) ampC overexpression, on the other hand, nagZ inactivation hardly influenced the ampC expression level of KJΔmrcA (pbp1a inactivation; Huang et al., 2012). In this study, we also found two different ampC regulation ways exist in Y. enterocolitica, the patterns of which were just the reverse of that in S. maltophilia (Huang et al., 2012). The β-lactamase activity of YEΔD123 was not affected by the inactivation of the nagZ gene, whereas the introduction of ΔnagZ into the PBP mutation strain YEΔ4Δ5Δ6Δ7 dramatically reduced the β-lactamase activities at both the basal and induced level (Figure 2). As shown in Table 3, the antibiotic resistance of YEΔ4Δ5Δ6Δ7 and YEΔD123 were marked improved compare with wild-type strain, the MIC value of these two strains in TZP, PIP, CFZ, CAZ, CRO, and ATM is rising sharply. While after inactivation of nagZ gene simultaneously, only a marginal distinction between YEΔD123 and YEΔD123ΔZ was found, but the MIC values of YEΔ4Δ5Δ6Δ7ΔZ has shifted down significantly, far from its parent strain YEΔ4Δ5Δ6Δ7 for almost all tested β-lactams. To further confirm the function of NagZ, we constructed a nagZ deletion strain YEΔZ, and detected the N-acetyl-β-glucosaminidase activity of it to compare with the wild-type strain Y. enterocolitica 105.5R(r), the results showed that the ability of hydrolysis chromogenic substrate was completely lost in nagZ mutation strain YEΔZ (Figure 3), suggesting that NagZ (YE105_RS06670) was the only enzyme that possessed N-acetyl-β-glucosaminidase activity in Y. enterocolitica 105.5R(r). However, even though there is no readable N-acetyl-β-glucosaminidase activity in YEΔZ, we also did the bioinformatic search to look for possible NagZ homologs in genome to find the protein worked in YEΔD123ΔZ. According to the gene function annotation of 105.5R(r), we considered the YE105_RS13000 may have similar function with NagZ, but it was not clear if this protein participated the ampC regulation or not. Therefore, further studies needed to performed to elucidate the function of YE105_RS13000 in Y. enterocolitica ampC regulation.
In Y. enterocolitica, the function of AmpR was roughly the same as other members of Enterobacteriaceae or P. aeruginosa. The introduction of ΔampR into the AmpC hyperproduction strains YEΔD123 and YEΔ4Δ5Δ6Δ7 resulted in a sharp decline in the ampC expression (Figure 2). The inducibility of YEΔD123ΔR and YEΔ4Δ5Δ6Δ7ΔR also disappeared completely (Lindberg et al., 1985; Lindberg and Normark, 1987).
In conclusion, in terms of AmpC β-lactamase regulation, Y. enterocolitica shared some common characteristics with P. aerugiosa and other members of Enterobacteriaceae, but it also had its own features. This was the first investigation to the characterization of Y. enterocolitica ampC regulation. It provided a more comprehensive understanding of the AmpC β-lactamase regulation in Gram-negative bacteria.
Author Contributions
CL, CCL, SS, HJ, and XW designed the experiment together. YC and HH performed data analysis. JL and RD participated in the manuscript translation. ZG, JZ, and ZZ contributed to finish the work. All authors contributed to writing of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (General Project, no. 81470092) and the National Sci-Tech Key Project (2012ZX10004201, 2013ZX10004203-002).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Liuying Tang and American Journal Experts for their critical reading and helpful comments on our manuscript (Sub ID F3Y8N2N7).
References
Asgarali, A., Stubbs, K. A., Oliver, A., Vocadlo, D. J., and Mark, B. L. (2009). Inactivation of the glycoside hydrolase NagZ attenuates antipseudomonal β-lactam resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 2274–2282. doi: 10.1128/AAC.01617-08
Balasubramanian, D., Kumari, H., and Mathee, K. (2015). Pseudomonas aeruginosa AmpR: an acute-chronic switch regulator. Pathog. Dis. 73, 1–14. doi: 10.1111/2049-632X.12208
Bent, Z. W., and Young, G. M. (2010). Contribution of BlaA and BlaB β-lactamases to antibiotic susceptibility of Yersinia enterocolitica biovar 1B. Antimicrob. Agents Chemother. 54, 4000–4002. doi: 10.1128/AAC.01754-09
Chen, Y., Duan, R., Li, X., Li, K., Liang, J., Liu, C., et al. (2015). Homology analysis and cross-immunogenicity of OmpA from pathogenic Yersinia enterocolitica, Yersinia pseudotuberculosis, and Yersinia pestis. Mol. Immunol. 68(2 Pt A), 290–299. doi: 10.1016/j.molimm.2015.09.016
CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement (M100-S24). Wayne, PA.
Cornelis, G., and Abraham, E. P. (1975). Beta-lactamases from Yersinia enterocolitica. J. Gen. Microbiol. 87, 273–284. doi: 10.1099/00221287-87-2-273
Guerin, F., Isnard, C., Cattoir, V., and Giard, J. C. (2015). Complex regulation pathways of AmpC-mediated β-lactam resistance in Enterobacter cloacae complex. Antimicrob. Agents Chemother. 59, 7753–7761. doi: 10.1128/AAC.01729-15
Huang, Y. W., Hu, R. M., Lin, C. W., Chung, T. C., and Yang, T. C. (2012). NagZ-dependent and NagZ-independent mechanisms for β-lactamase expression in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 56, 1936–1941. doi: 10.1128/AAC.05645-11
Juan, C., Moya, B., Perez, J. L., and Oliver, A. (2006). Stepwise upregulation of the Pseudomonas aeruginosa chromosomal cephalosporinase conferring high-level β-lactam resistance involves three AmpD homologues. Antimicrob. Agents Chemother. 50, 1780–1787. doi: 10.1128/AAC.50.5.1780-1787.2006
Liang, J., Li, X., Zha, T., Chen, Y., Hao, H., Liu, C., et al. (2016). DTDP-rhamnosyl transferase RfbF, is a newfound receptor-related regulatory protein for phage phiYe-F10 specific for Yersinia enterocolitica serotype O:3. Sci. Rep. 6:22905. doi: 10.1038/srep22905
Liang, J., Wang, X., Xiao, Y., Cui, Z., Xia, S., Hao, Q., et al. (2012). Prevalence of Yersinia enterocolitica in pigs slaughtered in Chinese abattoirs. Appl. Environ. Microbiol. 78, 2949–2956. doi: 10.1128/AEM.07893-11
Lindberg, F., and Normark, S. (1987). Common mechanism of ampC beta-lactamase induction in enterobacteria: regulation of the cloned Enterobacter cloacae P99 beta-lactamase gene. J. Bacteriol. 169, 758–763. doi: 10.1128/jb.169.2.758-763.1987
Lindberg, F., Lindquist, S., and Normark, S. (1987). Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii β-lactamase. J. Bacteriol. 169, 1923–1928. doi: 10.1128/jb.169.5.1923-1928.1987
Lindberg, F., Westman, L., and Normark, S. (1985). Regulatory components in Citrobacter freundii ampC beta-lactamase induction. Proc. Natl. Acad. Sci. U.S.A. 82, 4620–4624. doi: 10.1073/pnas.82.14.4620
Liu, C., Wang, X., Chen, Y., Hao, H., Li, X., Liang, J., et al. (2016). Three Yersinia enterocolitica AmpD homologs participate in the multi-step regulation of chromosomal Cephalosporinase, AmpC. Front. Microbiol. 7:1282. doi: 10.3389/fmicb.2016.01282
Moya, B., Dotsch, A., Juan, C., Blazquez, J., Zamorano, L., Haussler, S., et al. (2009). β-lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 5:e1000353. doi: 10.1371/journal.ppat.1000353
Peter, K., Korfmann, G., and Wiedemann, B. (1988). Impact of the ampD gene and its product on beta-lactamase production in Enterobacter cloacae. Rev. Infect. Dis. 10, 800–805. doi: 10.1093/clinids/10.4.800
Pfeifle, D., Janas, E., and Wiedemann, B. (2000). Role of penicillin-binding proteins in the initiation of the AmpC β-lactamase expression in Enterobacter cloacae. Antimicrob. Agents Chemother. 44, 169–172. doi: 10.1128/AAC.44.1.169-172.2000
Philippe, N., Alcaraz, J. P., Coursange, E., Geiselmann, J., and Schneider, D. (2004). Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51, 246–255. doi: 10.1016/j.plasmid.2004.02.003
Ropy, A., Cabot, G., Sanchez-Diener, I., Aguilera, C., Moya, B., Ayala, J. A., et al. (2015). Role of Pseudomonas aeruginosa low-molecular-mass penicillin-binding proteins in AmpC expression, β-lactam resistance, and peptidoglycan structure. Antimicrob. Agents Chemother. 59, 3925–3934. doi: 10.1128/AAC.05150-14
Sanders, C. C., Bradford, P. A., Ehrhardt, A. F., Bush, K., Young, K. D., Henderson, T. A., et al. (1997). Penicillin-binding proteins and induction of AmpC β-lactamase. Antimicrob. Agents Chemother. 41, 2013–2015.
Seoane, A., Francia, M. V., and Garcia Lobo, J. M. (1992). Nucleotide sequence of the ampC-ampR region from the chromosome of Yersinia enterocolitica. Antimicrob. Agents Chemother. 36, 1049–1052. doi: 10.1128/AAC.36.5.1049
Simon, R., Priefer, U., and Puhler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791. doi: 10.1038/nbt1183-784
Vollmer, W., Joris, B., Charlier, P., and Foster, S. (2008). Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286. doi: 10.1111/j.1574-6976.2007.00099.x
Wang, X., Li, Y., Jing, H., Ren, Y., Zhou, Z., Wang, S., et al. (2011). Complete genome sequence of a Yersinia enterocolitica “Old World” (3/O:9) strain and comparison with the “New World” (1B/O:8) strain. J. Clin. Microbiol. 49, 1251–1259. doi: 10.1128/JCM.01921-10
Yang, T. C., Chen, T. F., Tsai, J. J., and Hu, R. M. (2014). NagZ is required for beta-lactamase expression and full pathogenicity in Xanthomonas campestris pv. campestris str. 17. Res. Microbiol. 165, 612–619. doi: 10.1016/j.resmic.2014.08.008
Zamorano, L., Reeve, T. M., Deng, L., Juan, C., Moya, B., Cabot, G., et al. (2010). NagZ inactivation prevents and reverts β-lactam resistance, driven by AmpD and PBP 4 mutations, in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 54, 3557–3563. doi: 10.1128/AAC.00385-10
Keywords: Yersinia enteocolitica, AmpC β-lactamase, AmpD, PBPs, NagZ, AmpR
Citation: Liu C, Li C, Chen Y, Hao H, Liang J, Duan R, Guo Z, Zhang J, Zhao Z, Jing H, Wang X and Shao S (2017) Role of Low-Molecular-Mass Penicillin-Binding Proteins, NagZ and AmpR in AmpC β-lactamase Regulation of Yersinia enterocolitica. Front. Cell. Infect. Microbiol. 7:425. doi: 10.3389/fcimb.2017.00425
Received: 20 July 2017; Accepted: 14 September 2017;
Published: 27 September 2017.
Edited by:
Dongsheng Zhou, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Fang Huang, Beijing Center for Disease Prevention and Control, ChinaL. F. Wu, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2017 Liu, Li, Chen, Hao, Liang, Duan, Guo, Zhang, Zhao, Jing, Wang and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Wang, wangxin@icdc.cn
Shihe Shao, shaoshihe2006@163.com
†These authors have contributed equally to this work.
 Chang Liu
Chang Liu Chuchu Li1,2†
Chuchu Li1,2†  Huaiqi Jing
Huaiqi Jing Xin Wang
Xin Wang