Inorganic Polyphosphate Is Essential for Salmonella Typhimurium Virulence and Survival in Dictyostelium discoideum
- 1Laboratorio de Microbiología de Sistemas, Departamento de Biología, Facultad de Ciencias, Universidad de Chile, Santiago, Chile
- 2Laboratorio de Microbiología, Departamento de Bioquímica y Biología Molecular, Facultad de Ciencias Químicas y Farmacéuticas, Universidad de Chile, Santiago, Chile
- 3Laboratorio de Biología Estructural y Molecular, Departamento de Biología, Facultad de Ciencias, Universidad de Chile, Santiago, Chile
Inorganic polyphosphate (polyP) deficiency in enteric bacterial pathogens reduces their ability to invade and establish systemic infections in different hosts. For instance, inactivation of the polyP kinase gene (ppk) encoding the enzyme responsible for polyP biosynthesis reduces invasiveness and intracellular survival of Salmonella enterica serovar Typhimurium (S. Typhimurium) in epithelial cells and macrophages in vitro. In addition, the virulence in vivo of a S. Typhimurium Δppk mutant is significantly reduced in a murine infection model. In spite of these observations, the role played by polyP during the Salmonella-host interaction is not well understood. The social amoeba Dictyostelium discoideum has proven to be a useful model for studying relevant aspects of the host-pathogen interaction. In fact, many intracellular pathogens can survive within D. discoideum cells using molecular mechanisms also required to survive within macrophages. Recently, we established that S. Typhimurium is able to survive intracellularly in D. discoideum and identified relevant genes linked to virulence that are crucial for this process. The aim of this study was to determine the effect of a polyP deficiency in S. Typhimurium during its interaction with D. discoideum. To do this, we evaluated the intracellular survival of wild-type and Δppk strains of S. Typhimurium in D. discoideum and the ability of these strains to delay the social development of the amoeba. In contrast to the wild-type strain, the Δppk mutant was unable to survive intracellularly in D. discoideum and enabled the social development of the amoeba. Both phenotypes were complemented using a plasmid carrying a copy of the ppk gene. Next, we simultaneously evaluated the proteomic response of both S. Typhimurium and D. discoideum during host-pathogen interaction via global proteomic profiling. The analysis of our results allowed the identification of novel molecular signatures that give insight into Salmonella-Dictyostelium interaction. Altogether, our results indicate that inorganic polyP is essential for S. Typhimurium virulence and survival in D. discoideum. In addition, we have validated the use of global proteomic analyses to simultaneously evaluate the host-pathogen interaction of S. Typhimurium and D. discoideum. Furthermore, our infection assays using these organisms can be exploited to screen for novel anti-virulence molecules targeting inorganic polyP biosynthesis.
Introduction
The ability of Dictyostelium discoideum cells to feed on bacteria has prompted the development of virulence assays for identifying host defense mechanisms and deciphering bacterial virulence factors (Cosson et al., 2002; Froquet et al., 2009). Basic cellular processes such as phagocytosis, phagosomal development and autophagy, are evolutionarily well conserved between Dictyostelium and macrophages (Hägele et al., 2000; Bozzaro and Eichinger, 2011; Dunn et al., 2018). Consequently, D. discoideum has been established as a model to study host-pathogen interaction in a wide range of pathogenic bacteria such as Legionella, Salmonella, Francisella, Mycobacterium, and Pseudomonas, among others (Pukatzki et al., 2002; Hagedorn and Soldati, 2007; Weber et al., 2014; Lampe et al., 2015; Bravo-Toncio et al., 2016; Riquelme et al., 2016; Cardenal-Muñoz et al., 2017). Unlike mammalian phagocytes, D. discoideum is amenable to a diverse array of genetic manipulations facilitating the in vivo identification of host susceptibility determinants and pathogen virulence factors (Carilla-Latorre et al., 2008; Hasselbring et al., 2011; Pan et al., 2011; Tosetti et al., 2014; Zhang et al., 2016). However, in vivo host-pathogen interaction during bacterial infection in D. discoideum remains poorly understood.
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a foodborne pathogen causative of gastroenteritis in a variety of warm-blooded animals that largely relies on its ability to survive inside host cells. Relevant genes required for this process are located in pathogenicity islands such as SPI-1 and SPI-2, which encode two independent type III secretion systems (T3SS-1 and T3SS-2, respectively) that inject effector proteins into host cells and are critical during different stages of infection (reviewed in Haraga et al., 2008). In a previous study, we showed that S. Typhimurium genes linked to virulence are required to survive in D. discoideum, including those encoding factors involved in the biosynthesis of aromatic compounds, the production of a lipopolysaccharide containing a complete O-antigen, T3SS-1, T3SS-2, the type VI secretion system (T6SS) encoded in SPI-6 and the PhoP/PhoQ two-component system (Riquelme et al., 2016). Hence, S. Typhimurium exploits a common set of molecular mechanisms to survive within amoeba and animal host cells, supporting the use of D. discoideum as a model for host-pathogen interactions and to study the cellular processes that are affected during infection.
We are particularly interested in inorganic polyphosphate (polyP) metabolism because this biopolymer is important for D. discoideum development and predation, and for virulence in many bacterial pathogens (Zhang et al., 2005; Brown and Kornberg, 2008). In fact, we have demonstrated that polyP biosynthesis is essential for P. aeruginosa PAO1 virulence toward this amoeba (Bravo-Toncio et al., 2016). Inorganic polyP is an abundant and ubiquitous biopolymer that has been conserved in every cell in nature. In the last decades, an increasing number of physiological functions have been reported for polyP in bacteria (Brown and Kornberg, 2008). Due to their phosphoanhydride bonds similar to those in ATP and their properties as polyanions, polyP serve as microbial phosphagen in a variety of biochemical reactions, as a buffer against alkalis, and as a metal storage and metal-chelating agent. In addition, recent studies have revealed the importance of polyP metabolism in signaling and regulatory processes, cell viability and proliferation, and as modulator of microbial stress response (Gray and Jakob, 2015). In numerous pathogenic bacteria, inactivation of the polyP kinase gene (ppk) encoding the enzyme responsible for polyP biosynthesis causes defects in biofilm formation, quorum sensing, motility, general stress and stringent responses, and production of virulence factors (Rao et al., 1998; Rashid and Kornberg, 2000; Rashid et al., 2000a,b; Brown and Kornberg, 2008; Varela et al., 2010; Varas et al., 2017). In S. Typhimurium, inorganic polyP is essential for long-term survival and virulence factors production (Kim et al., 2002). However, the exact mechanism that links polyP metabolism and Salmonella virulence remains to be elucidated.
In this study, we used D. discoideum as a host model to study the link between polyP biosynthesis and virulence in S. Typhimurium. To this end, we assessed the intracellular survival of S. Typhimurium wild-type and Δppk strains in the amoeba, and the effect of these strains in the social development of the host. Our results indicate that inorganic polyP is essential during S. Typhimurium infection of D. discoideum. Also, we used global proteomic profiling to get a global view of host cellular responses toward infection that gave insight into Salmonella-Dictyostelium interaction.
Materials and Methods
Bacterial Strains and Culture Conditions
The bacterial strains used in this study are listed in Table 1. All S. Typhimurium strains are derivatives of the wild-type, virulent strain 14028s (Fields et al., 1986). Bacteria were routinely grown at 37°C with agitation in Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl). When required, LB medium was supplemented with ampicillin (Amp, 100 mg/L), chloramphenicol (Cam, 20 mg/L) or kanamycin (Kan, 75 mg/L). LB medium was solidified by the addition of agar (15 g/L). All procedures involving the use of pathogenic organisms were conducted following the guidelines in the Biosafety Manual of the National Commission of Scientific and Technological Research (CONICYT), and were approved by the Institutional Biosafety Committee of Universidad de Chile, Campus Norte.
Construction of Mutant Strains
All S. Typhimurium mutants were generated by the Lambda Red recombination method (Datsenko and Wanner, 2000) with modifications (Santiviago et al., 2009), using plasmid pCLF4 (KanR, GenBank accession number EU629214) or pCLF2 (CamR, GenBank accession number HM047089) as template. Correct allelic replacement in these mutants was confirmed by PCR amplification using primers flanking the substitution site. All primers for PCR amplifications are listed in Table 2.
Construction of Complementing Plasmid pPPK
A DNA fragment containing the ppk gene (including its promoter region) was amplified from the genome of S. Typhimurium strain 14028s using Taq DNA polymerase (Invitrogen) and primers ppk_Out5 and ppk_Out3 (Table 2). The PCR product was purified from 1% agarose gels using the “QIAquick Gel Extraction Kit” (QIAGEN) and cloned into pBAD-TOPO using the “pBAD-TOPO TA Expression Kit” (Invitrogen). The presence and orientation of the insert in the recombinant plasmid generated (pPPK) was confirmed by PCR amplification using combinations of primers ppk_Out5, ppk_Out3, pBAD_Forward and pBAD_Reverse (Table 2). Finally, S. Typhimurium Δppk was transformed by electroporation with plasmid pPPK for complementation assays.
Dictyostelium Culture Conditions
D. discoideum strain AX4 was obtained from Dicty Stock Center (Kreppel et al., 2004; Basu et al., 2013; Fey et al., 2013), and cultured according to standard protocols (Fey et al., 2007). Briefly, amoebae were maintained at 22°C in SM medium (10 g/L glucose, 10 g/L peptone, 1 g/L yeast extract, 1 g/L MgSO4 × 7H2O, 1.9 g/L KH2PO4, 0.6 g/L K2HPO4, 20 g/L agar), growing on a confluent lawn of Escherichia coli B/r. Before development and intracellular survival assays, amoebae were grown in the absence of bacteria (axenic cultures) at 22°C with agitation (180 rpm) in liquid HL5 medium (14 g/L tryptone, 7 g/L yeast extract, 0.35 g/L Na2HPO4, 1.2 g/L KH2PO4, 14 g/L glucose, pH 6.3). When required, media were supplemented with streptomycin (Stp; 300 mg/L) and Amp (100 mg/L). Amoebae were harvested in early exponential phase (1–1.5 × 106 cells/mL) and centrifuged at 500 × g for 10 min at 4°C. The supernatant was discarded and the pellet was adjusted to 1 × 106 cells/mL in HL5 medium for development assays or washed three times using Soerensen buffer (2 g/L KH2PO4, 0.36 g/L Na2HPO4 × 2H2O, pH 6.0) for intracellular survival assays. The population of viable amoebae was evaluated by Trypan blue exclusion and counting in a Neubauer chamber.
Development Assay
Individual wells of a 24-well plate containing N agar (Soerensen buffer supplemented with 1 g/L peptone, 1 g/L glucose and 20 g/L agar) were inoculated with 30 μL of a stationary-phase culture from each bacterial strain to be evaluated. The plate was incubated overnight at 22°C to generate bacterial lawns. The next day, 10 μL of a cellular suspension containing 1 × 104 D. discoideum AX4 cells in HL5 was spotted in the middle of each well and the plate was further incubated at 22°C for 6 days. Amoebae were monitored daily and the developmental phase reached (“aggregation,” “elevation,” and “culmination”) was scored. A score of “1” was assigned when amoebae aggregated forming a phagocytosis plaque. A score of “2” was assigned when elevated structures, such as “slugs” or “fingers”, were observed all across the surface of the agar in the well. A score of “3” was assigned when fruiting bodies were formed all across the surface of the agar in the well. Intermediate states among two developmental phases were recorded as average values of the corresponding scores. In addition, representative images of D. discoideum development were obtained at days 2 and 4 using an Olympus MVX10 stereomicroscope.
Intracellular Survival Assay
Intracellular survival assays were performed as described previously (Riquelme et al., 2016). Briefly, each bacterial strain to be assessed was grown to stationary phase, harvested and washed twice with Soerensen buffer. Next, ~2 × 107 D. discoideum AX4 cells were mixed with each bacterial strain until reaching a multiplicity of infection (MOI) of 100 bacteria/amoeba in 10 mL of Soerensen buffer. After 1 h of co-incubation at 22°C with agitation (180 rpm), the extracellular bacteria were removed by three sequential washing steps using Soerensen buffer. The infected amoebae were suspended in 10 mL of Soerensen buffer (t = 0) and further incubated at 22°C with agitation (180 rpm). Aliquots were obtained at 0, 1, 3, 4.5, and 6 h post infection. The population of viable amoebae were determined at each time point. In parallel, infected amoebae recovered at each time point were lysed with 0.2% Triton X-100 and loads of intracellular bacteria were estimated by serial dilutions and plating on LB agar. Statistical significance was determined using a one-way ANOVA and two-way ANOVA with Fisher's LSD post-test.
Global Proteomic Profiling Using Q-Exactive Mass Spectrometry
For proteomic analyses, ~1 × 106 amoeba cells were obtained from individual intracellular survival assays after 6 h of infection with the wild-type strain or the Δppk mutant. Uninfected amoebae were used as control condition. Amoebae from each experimental condition were concentrated by centrifugation at 500 × g for 10 min, quick-frozen, and kept at −80°C until further use. Global proteomic profiles from samples representing the different experimental conditions were obtained from Bioproximity, LLC (USA). In each case, a unique proteomic analysis was performed using a pool of cells from three independent assays. Protein denaturation, digestion and desalting of samples were prepared using the filter-assisted sample preparation (FASP) method (Wiśniewski et al., 2009). Briefly, the samples were digested using trypsin, and each digestion mixture was analyzed by ultra-high pressure liquid chromatography (UHPLC-MS/MS) coupled to a high resolution, high mass accuracy quadrupole-Orbitrap mass spectrometer (Q-Exactive, Thermo Fisher). For protein quantification, intensity measurements derived from the area-under-the-curve of the MS/MS scan for each peptide ion identification were summed for each sample. These values were averaged across all samples and a normalization factor determined and applied for each sample (Zhang et al., 2010). MS/MS data were compared with the most recent protein sequence libraries available from UniProtKB. Proteins were required to have one or more unique peptides detected across the analyzed samples with an e-value ≤ 0.0001.
Proteomes from each experimental condition were compared using an online tool that generates Venn diagrams and lists of proteins detected in any given condition (http://bioinformatics.psb.ugent.be/webtools/Venn/). The proteins detected in 2 experimental conditions were analyzed by calculating log2 values of condition_1/condition_2 detection ratios. Enrichment for a protein in a particular condition was considered when the corresponding calculated value was >0.6 (~1.5 fold enrichment).
Analysis of D. discoideum Proteins Detected by Global Proteomic Profiling
D. discoideum proteins were identified from Q-proteomics data using library ID: 44689 (PubMed Taxonomy database), which include proteomes of strains AX2, AX3 y AX4. Proteins were classified according to the predicted functions annotated in the Clusters of Orthologous Groups of Proteins (COGs) database (Tatusov et al., 2000). The UniProtKB ID of each protein was mapped to the COGs database of the Social Amoeba Comparative Genome Browser (SACGB, http://sacgb.leibniz-fli.de/cgi/cog.pl?ssi=free), and the EggNOG database of orthologous groups and functional annotation (Jensen et al., 2008). To do this, both databases were downloaded, and mappings were performed using custom Python scripts. Using this approach we were able to assign COG categories for ~80% of all proteins detected in each experimental condition.
In addition, overrepresentation analyses were performed using the Protein Annotation Through Evolutionary Relationship (PANTHER) tool (http://www.pantherdb.org/), a comprehensive system that combines gene function, ontology, pathways and statistical analysis tools that enable the analysis of large-scale, genome-wide data from sequencing, proteomics or gene expression experiments (Mi et al., 2013, 2016). To do this, the UniProtKB IDs from the different sets of detected protein were used as input in PANTHER. This tool was able to map the UniProtKB ID for ~96% of all proteins detected in each experimental condition. The results of overrepresentation analyses were filtered according to three annotation data sets: “Biological process”, “Cellular component”, and “Reactome pathways”. The cut-off for the analyses was set to P < 0.05.
Analysis of Bacterial Proteins Detected by Global Proteomic Profiling
S. Typhimurium proteins were identified from Q-proteomics data using the libraries ID: 588858 and ID: 99287 (PubMed Taxonomy database), which include proteomes of strains 14028s and LT2. Virulence-related proteins were assigned following inspections of databases PATRIC_VF, VFDB and VICTORS, using tools available at the Pathosystems Resource Integration Center web site (PATRIC; www.patricbrc.org). Also, proteins encoded in genes located in close proximity to known virulence determinants, as well as genes located within Salmonella pathogenicity islands (SPIs) or prophages, were identified and analyzed using Artemis V.14, IslandViewer 3, Pathogenicity Island DataBase (PAIDB v2.0), PHAST and Islander (Rutherford et al., 2000; Zhou et al., 2011; Dhillon et al., 2015; Hudson et al., 2015; Yoon et al., 2015).
Results and Discussion
Inorganic Polyphosphate is Essential for S. Typhimurium Virulence in D. discoideum
To evaluate the role played by polyP biosynthesis in the virulence of S. Typhimurium in D. discoideum, we constructed a Δppk derivative of the wild-type, virulent strain 14028s. This mutant is impaired in the synthesis of polyP. In addition, the mutant was transformed with a plasmid harboring a wild-type version of ppk (i.e., pPPK) to confirm the specificity of the effects attributed to the inactivation of this gene in our assays.
Previous reports indicate that virulent pathogenic bacteria delay the social development of D. discoideum, while attenuated or non-pathogenic bacteria allow its rapid progression (within 3–4 days in our experimental conditions). Thus, assessing the effect of a given bacterial strain on the social development of D. discoideum can be used to evaluate its virulence (Sillo et al., 2011; Bravo-Toncio et al., 2016; Ouertatani-Sakouhi et al., 2017). Notably, this host-pathogen interaction model has been recently used to identify compounds that inhibit bacterial virulence (Bravo-Toncio et al., 2016; Ouertatani-Sakouhi et al., 2017). Therefore, we compared the effect of feeding D. discoideum with the wild-type strain or its Δppk derivative on the social development of the amoeba. In addition, a ΔaroA derivative of strain 14028s (known to be attenuated in murine infection models Sebkova et al., 2008), and Escherichia coli B/r (routinely used to feed D. discoideum during growth under laboratory conditions Fey et al., 2007) were included in our assay as attenuated and non-pathogenic controls, respectively.
The different bacterial strains were inoculated on N agar and incubated overnight to generate bacterial lawns. Then, D. discoideum cells were deposited on top of each bacterial lawn and the plates were monitored for 6 days to follow the progression of D. discoideum development, which mainly involves three sequential stages: aggregation, elevation, and culmination (Figure 1). E. coli B/r allowed the rapid progression of the development, culminating within 3 to 4 days, where mature fruiting bodies were predominant (Figure 1). A similar phenotype was observed in the case of the ΔaroA mutant, indicating that this strain is not virulent for D. discoideum. This observation is in agreement with previous reports indicating that an aroA mutant of S. Typhimurium is unable to survive intracellularly in this amoeba (Riquelme et al., 2016). On the contrary, the wild-type strain produced a delay in the development of the amoeba where only the aggregation phase was reached after 6 days of co-incubation (Figure 1). This phenotype was not observed in the case of the Δppk mutant, which allowed the development of the amoebae until reaching the elevation phase at 6 days of co-incubation. Of note, the Δppk mutant harboring plasmid pPPK showed a wild-type phenotype (Figure 1). These results indicate that polyP synthesis is essential for S. Typhimurium virulence in D. discoideum.
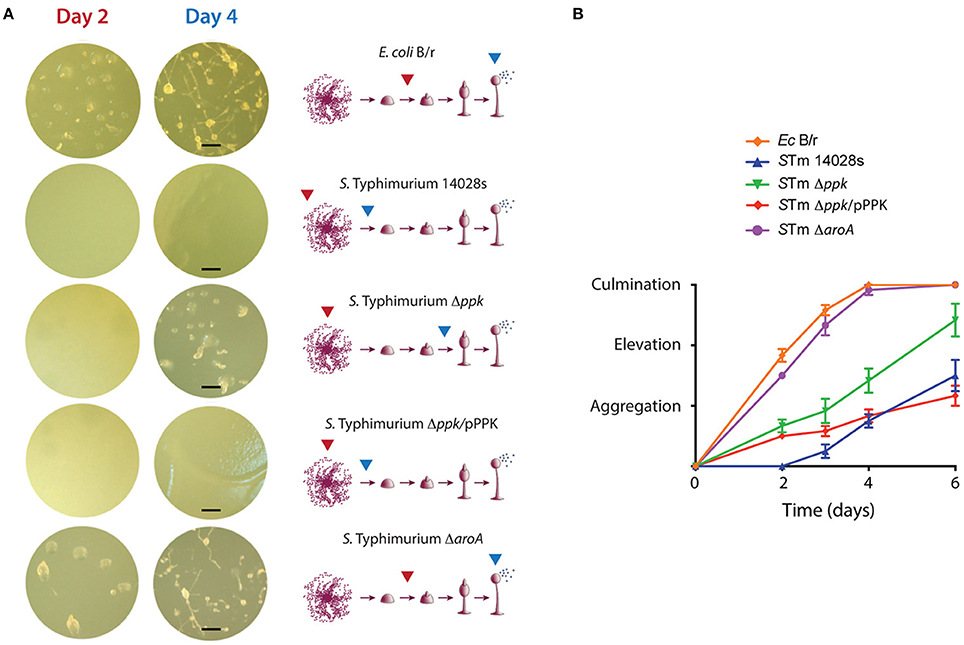
Figure 1. Development of D. discoideum co-incubated with S. Typhimurium 14028s derivatives and E. coli B/r. (A) Representative pictures of amoebae development in presence of each bacterial strain. The development stages reached at days 2 and 4 are indicated with red and blue arrows, respectively. Scale bars, 100 μm. (B) Development progress evaluated using a numerical scale defined according to the developmental phase reached at each time point (see Materials and Methods section). Graph shows mean values ± SD from 10 independent assays.
Our observations on the interaction of S. Typhimurium with D. discoideum are consistent with a previous study that evaluated the virulence of the pathogen by assessing its effect on the social development of the amoeba. The authors reported that Dictyostelium growth on a lawn of wild-type S. Typhimurium causes the aberrant development of the amoeba, or even resulted in cell death depending on nutrient conditions of the medium used for the assay. Both phenotypes required a functional T3SS-2 (Sillo et al., 2011). Additionally, the effect observed for a polyP deficiency on the social development of the amoeba has been also reported in the case of P. aeruginosa (Zhang et al., 2005; Bravo-Toncio et al., 2016) highlighting the essential role played by polyP biosynthesis in bacterial virulence using this model organism.
Inorganic Polyphosphate Is Essential for S. Typhimurium Survival in D. discoideum
Recently, we reported that wild-type S. Typhimurium can survive within D. discoideum and requires relevant genes linked to virulence for this process (Riquelme et al., 2016). To evaluate the role played by polyP in the intracellular survival of S. Typhimurium in D. discoideum, we performed infection assays where vegetative amoebae were co-incubated with the wild-type strain or its Δppk derivative. At different times post infection, intracellular bacteria were recovered from infected amoebae and titrated. The attenuated ΔaroA mutant was included as a control in our assays.
First, we evaluated the internalization of each mutant strain after 1 h of co-incubation with the amoebae, and observed that Δppk and ΔaroA mutants were internalized at higher levels than the wild-type strain. In contrast, the Δppk mutant harboring plasmid pPPK was internalized at wild-type levels (Figure 2A). Then, we evaluated the intracellular survival of each strain at different times post infection and observed that the wild-type strain was able to survive and replicate in the amoebae. On the contrary, the Δppk mutant was defective for intracellular survival at all time points evaluated. The same phenotype was observed in the case of the attenuated ΔaroA mutant. The intracellular survival of the Δppk mutant harboring plasmid pPPK was comparable to that shown by the wild-type strain (Figure 2B). It is worth mentioning that no effect in amoeba viability was observed during the course of these experiments (Figure 2C), indicating that the differences observed in the titers of intracellular bacteria are not attributable to changes in the number of viable amoebae.
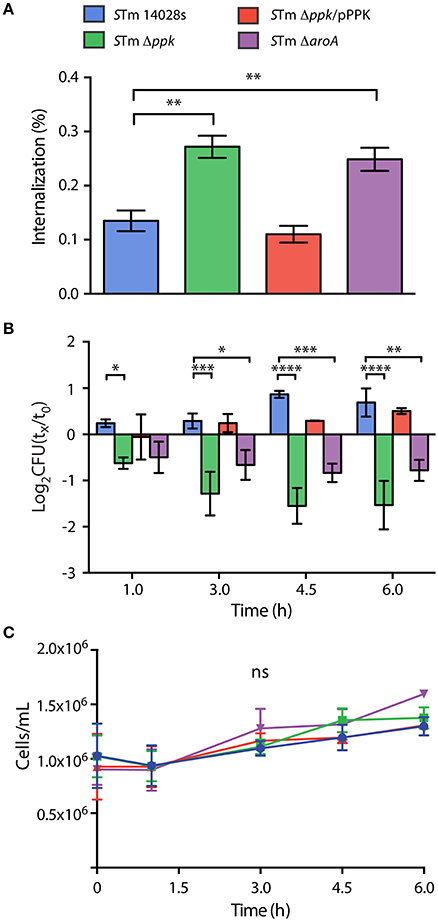
Figure 2. Internalization and intracellular survival of S. Typhimurium 14028s derivatives in D. discoideum. (A) Percentage of internalized bacteria after 1 hour of co-incubation, calculated as 100 × (CFUt=0/CFUinoculum). Statistical significance was determined using a one-way ANOVA with Fisher's LSD post-test (** = p <0.005). (B) Intracellular survival at different times post infection, expressed as log2(CFUt=x/CFUt=0). Statistical significance was determined using a two-way ANOVA with Fisher's LSD post-test (* = p < 0.05, ** = p < 0.005, *** = p < 0.001, **** = p <0.0001). (C) Population of viable amoebae at each time point expressed as cells/mL. Statistical significance was determined using a two-way ANOVA with Fisher's LSD post-test (ns = not significant). Graphs in panels (A–C) show mean values ± SD from 5 independent assays. The color code in panel (A) is also valid for panels (B,C).
Overall, our results indicate that polyP synthesis is essential for S. Typhimurium to survive intracellularly in D. discoideum. These observations are in line with a previous study indicating that a Δppk mutant of S. Typhimurium is deficient for intracellular survival in RAW 264.7 murine macrophages (Kim et al., 2002). In addition, several studied indicate that S. Typhimurium ppk mutants present a variety of phenotypes, including defective long-term survival in vitro, defective responses to oxidative stress and starvation, sensitivity to polymyxin, intolerance to acid and heat, impaired invasiveness in HEp-2 epithelial cells, and loss of swimming motility, all of which strongly influence virulence (Kim et al., 2002; McMeechan et al., 2007; Cheng and Sun, 2009). Accordingly, it has been reported that a ppk mutant of S. Typhimurium was attenuated in orally-infected Rhode Island Red chickens and BALB/c mice (McMeechan et al., 2007).
Global Proteomic Profiling of Dictyostelium-Salmonella Interaction
In order to determine the global response of D. discoideum to infections with S. Typhimurium wild type or its Δppk mutant derivative, we performed a global proteomic profiling of such interactions. To achieve this, the amoebae were co-incubated with each bacterial strain until reaching 6 h of infection. This time was chosen because the intracellular survival of both strains in D. discoideum showed the highest differences (Figure 2B). Non-infected amoebae were used as control condition. Next, shotgun proteomic profiling of infected and control amoebae were performed by UHPLC-MS/MS (Q-proteomics). Thus, 1779, 1950, and 1850 proteins were detected in samples of amoebae infected with the wild-type strain, amoebae infected with the Δppk mutant, and uninfected amoebae, respectively (Table S1).
A total of 258, 250, and 336 proteins were exclusively detected in non-infected amoebae, in amoebae infected with the wild-type strain, or in amoebae infected with the Δppk mutant, respectively. Additionally, 1277 proteins were detected in all three experimental conditions tested (Figure 3A). Considering that these proteins could be interesting if they show significant differences in expression levels, we carried out a comparative analysis of proteins detected in pairs of experimental conditions (i.e., non-infected amoebae vs. infected with the wild-type strain; non infected amoebae vs. infected with the Δppk mutant; and amoebae infected with the wild-type strain vs. infected with the Δppk mutant) to determine enrichment in a particular condition (cut-off: ~1.5 fold change). Proteins found exclusively or enriched in a given experimental condition were classified according to COG categories, which in turn were grouped in three main classes: “Cellular processes and signaling”, “Information storage and processing”, and “Metabolism” (Table S2). For most COG categories, the total number of proteins detected from non-infected amoebae was similar to those detected from amoebae infected with the Δppk mutant, in contrast to proteins detected in amoebae infected with the wild-type strain (Figure 3B). This was particularly evident in the case of COG categories “Posttranslational modification, protein turnover, chaperones”, “Signal transduction mechanisms”, “Intracellular trafficking, secretion, and vesicular transport” (associated with “Cellular processes and signaling”), “Chromatin structure and dynamics”, “Translation, ribosomal structure, and biogenesis”, “Mobilome, prophages, and transposons” (associated with “Information storage and processing”), “Energy production and conversion”, “Nucleotide transport and metabolism”, “Carbohydrate transport and metabolism”, “Coenzyme transport and metabolism”, “Lipid transport an metabolism”, and “Secondary metabolites biosynthesis, transport and catabolism” (associated with “Metabolism”; Figure 3B).
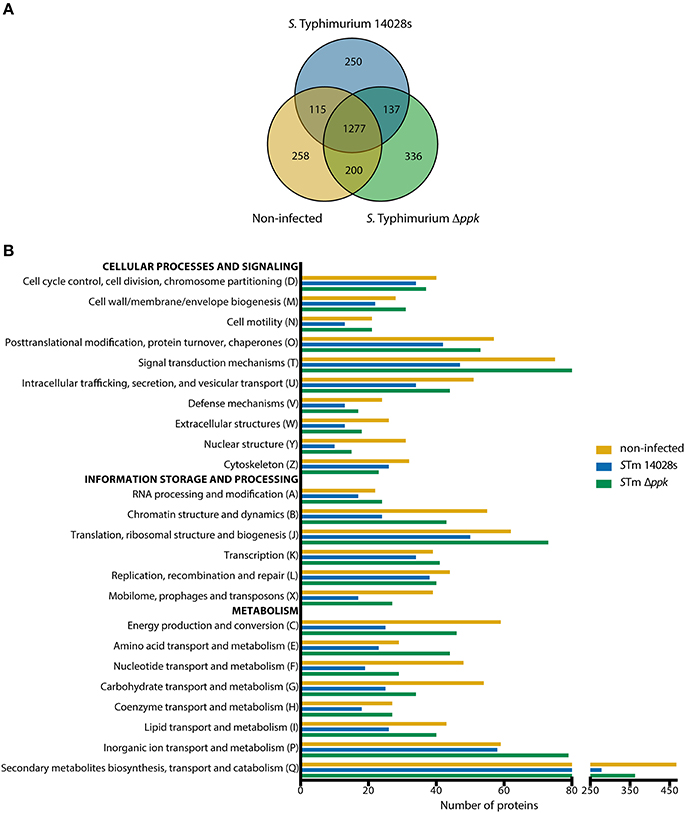
Figure 3. COG functional categorization of D. discoideum proteins detected during infection with S. Typhimurium strains. (A) Venn diagram of D. discoideum proteins detected in uninfected amoebae or in amoebae infected with S. Typhimurium wild-type or its Δppk derivative. (B) Graph showing number of proteins detected in each experimental condition and classified according to COG functional categories (see Materials and Methods section). COG categories were further grouped in three main classes: “Cellular processes and signaling”, “Information storage and processing”, and “Metabolism”.
Additionally, proteins that were exclusive or significantly enriched in each experimental condition were identified and used to perform overrepresentation analyses using the PANTHER tool (Table S3). The results were filtered according to three annotation data sets: “Biological process”, “Cellular component”, and “Reactome pathways” (Table 3). Overrepresented groups of proteins detected in D. discoideum infected with either S. Typhimurium strain include those involved in endomembrane trafficking, actin cytoskeleton organization, social development, chemotaxis and response to cAMP, immune system, response to bacteria, ubiquitination and proteasome degradation.
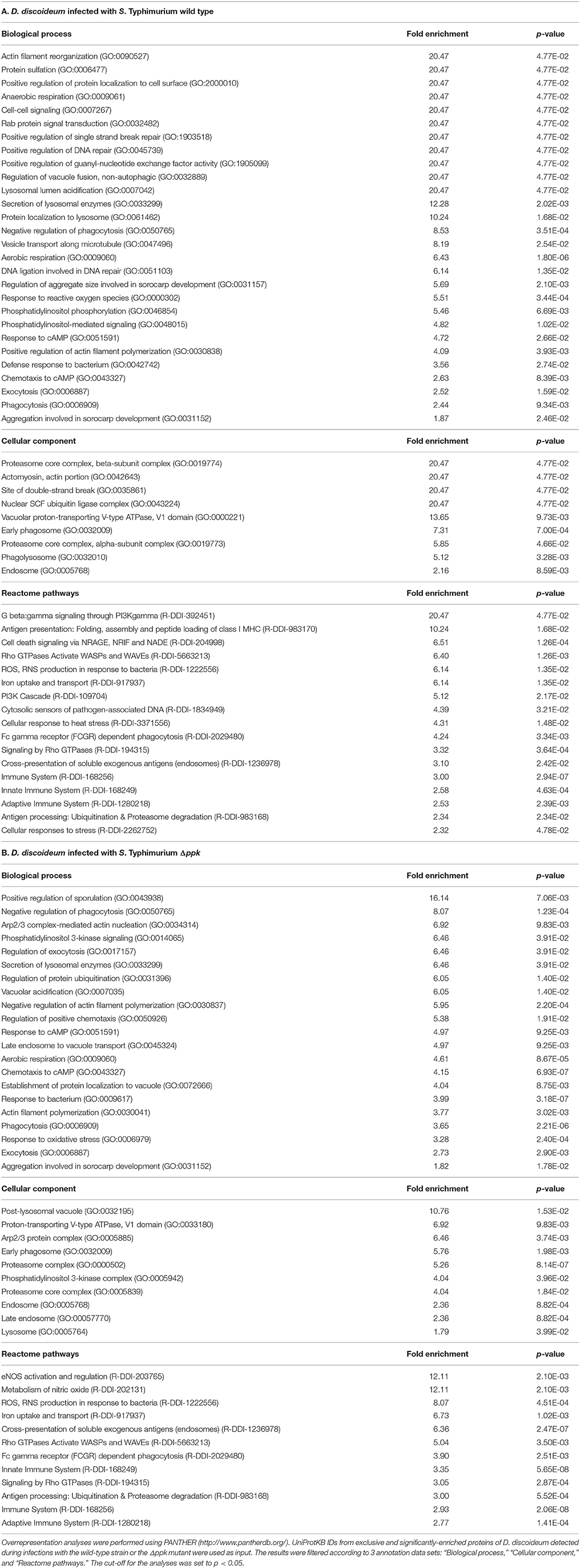
Table 3. Overrepresentation analysis of D. discoideum proteins detected during infections with S. Typhimurium wild type and Δppk.
Regarding endomembrane trafficking, common overrepresented proteins include those involved in Secretion of lysosomal enzymes (GO:0033299), Exocytosis (GO:0006887), Phagocytosis (GO:0006909), Early phagosome (GO:0032009), Endosome (GO:0005768), Signaling by Rho GTPases (R-DDI-194315), Rho GTPases Activate WASPs and WAVEs (R-DDI-5663213), and Fc gamma receptor (FCGR) dependent phagocytosis (R-DDI-2029480). Overrepresented proteins in amoebae infected with the wild-type strain include those involved in Rab protein signal transduction (GO:0032482), Positive regulation of guanyl-nucleotide exchange factor activity (GO:1905099), Regulation of vacuole fusion, non-autophagic (GO:0032889), Lysosomal lumen acidification (GO:0007042), Protein localization to lysosome (GO:0061462), Vesicle transport along microtubule (GO:0047496), Phosphatidylinositol phosphorylation (GO:0046854), Phosphatidylinositol-mediated signaling (GO:0048015), G beta:gamma signaling through PI3Kgamma (R-DDI-392451), and PI3K Cascade (R-DDI-109704). On the other hand, overrepresented proteins in amoebae infected with the Δppk mutant include those linked to Phosphatidylinositol 3-kinase signaling (GO:0014065), Phosphatidylinositol 3-kinase complex (GO:0005942), Regulation of exocytosis (GO:0017157), Vacuolar acidification (GO:0007035), Late endosome to vacuole transport (GO:0045324), Establishment of protein localization to vacuole (GO:0072666), Post-lysosomal vacuole (GO:0032195), Late endosome (GO:00057770), and Lysosome (GO:0005764). It is well known that S. Typhimurium delivers T3SS-2 effector proteins that interfere with the maturation of the endocytic route in eukaryotic host cells in order to avoid phagolysosomal fusion. This process results in a unique vacuolar compartment referred to as the Salmonella-containing vacuole (SCV), where this pathogen resides (Haraga et al., 2008; LaRock et al., 2015). We have described that inactivation of T3SS-2 abolishes the intracellular survival of S. Typhimurium in D. discoideum (Riquelme et al., 2016). Accordingly, our analysis suggest that wild-type S. Typhimurium resides in an intracellular compartment of D. discoideum comparable to an early endosome, while the Δppk mutant resides in a compartment that ultimately fuses with the lysosome, explaining the defective intracellular survival phenotype shown by this strain in the amoeba (Figure 2).
Most overrepresented proteins associated with the response to bacterial infection were detected in amoeba infected with either strain of S. Typhimurium. These proteins include those linked to Immune System (R-DDI-168256), Innate Immune System (R-DDI-168249), Adaptive Immune System (R-DDI-1280218), Cross-presentation of soluble exogenous antigens (endosomes) (R-DDI-1236978), Antigen processing: Ubiquitination & Proteasome degradation (R-DDI-983168), and ROS, RNS production in response to bacteria (R-DDI-1222556). No overrepresented proteins were exclusively detected in infections with the wild-type strain, while overrepresented proteins only detected during infections with the Δppk mutant include those associated with Metabolism of nitric oxide (R-DDI-202131) and eNOS activation and regulation (R-DDI-203765). These proteins (SprA/Q54GP3, PtsA/Q1ZXI0, and GchA/Q94465) are required for the de novo biosynthesis of tetrahydrobiopterin, an essential co-factor for the aromatic amino acid hydroxylases and nitric oxide synthases in mammals (Thöny et al., 2000; Choi et al., 2006; Vásquez-Vivar, 2009). These results indicate that D. discoideum infected with either wild-type or Δppk S. Typhimurium generates a robust response that includes production of ROS and RNS in order to eliminate the pathogen.
Proteins exclusively detected in D. discoideum infected with the wild-type strain indicate that the pathogen induces DNA damage in the host. This is revealed by a number of proteins involved in Positive regulation of single strand break repair (GO:1903518), Positive regulation of DNA repair (GO:0045739), DNA ligation involved in DNA repair (GO:0051103), Site of double-strand break (GO:0035861), and Cell death signaling via NRAGE, NRIF, and NADE (R-DDI-204998), the latter being a process associated with apoptotic cell death. It is tempting to speculate that this DNA damage is the result of excessive ROS/RNS production in response to the pathogen. Furthermore, this DNA damage needs to be repaired by the amoeba in order to resume growth and development. This is in agreement with the ability of S. Typhimurium to delay the social development of D. discoideum according to our development assay (Figure 1). In addition, two proteins linked to Protein sulfation (GO:0006477) (i.e., Kil1/Q556K8 and Phg1a/Q55FP0) were only detected in amoeba infected with the wild-type strain. Both proteins have been implicated in D. discoideum killing of intracellular K. pneumoniae (Benghezal et al., 2006; Cosson and Soldati, 2008; Le Coadic et al., 2013). Of note, protein sulfation has been implicated in decreased bacterial adherence to eukaryotic cells and reduced T3SS-dependent cytotoxicity (Blondel et al., 2016). Thus, variations in sulfation levels on the surface of D. discoideum generated during S. Typhimurium infection may explain differences in internalization observed between the wild-type and attenuated strains Δppk and ΔaroA in our infection assays (Figure 2A).
Intracellular pathogens need to cope with a hostile environment inside the host during infection. Thus, our global proteomic profiles of D. discoideum proteins suggest that the amoeba recognizes wild-type S. Typhimurium and elicits a strong response that includes production of toxic ROS and RNS. Nevertheless, the pathogen manipulates the endocytic pathway of the host to generate an intracellular replicative niche to survive this cell-autonomous defense response. On the contrary, after recognition the Δppk mutant appears to be unable to subvert the amoebal autonomous defense mechanism and to survive the unfavorable conditions within the host. This idea is in agreement with the phenotypes reported for null mutants of ppk in several bacterial pathogens (Rao et al., 1998; Rashid and Kornberg, 2000; Rashid et al., 2000a,b; Kim et al., 2002; McMeechan et al., 2007; Brown and Kornberg, 2008; Varela et al., 2010; Gray and Jakob, 2015; Varas et al., 2017). In addition the mutant seems to be incapable to modify the endocytic pathway, ending in a degradative intracellular compartment. Consequently, the Δppk mutant is unable to survive within the amoeba and to subvert the social development of this host.
In addition to D. discoideum proteins, we attempted to identify S. Typhimurium proteins expressed during the infection of the amoeba with the wild-type strain or the Δppk mutant. Using our Q-proteomics approach we were able to detect a limited number of bacterial proteins, most probably due to their low relative abundance in each sample in comparison to D. discoideum proteins. Thus, a total of 54 and 34 proteins were identified in infections of amoeba cells with the wild-type strain and the Δppk mutant, respectively. A group of seven proteins was detected in amoebae infected with either strain (Figure 4A). The identities of all S. Typhimurium proteins detected are listed in Table 4. The location of genes encoding all these proteins in the genome of S. Typhimurium 14028s is shown in Figure 4B. The list of detected proteins was compared with a list of 469 classic virulence-related proteins included in databases PATRIC_VF, VFDB and VICTORS using tools implemented in PATRIC (www.patricbrc.org). We identified 11 S. Typhimurium proteins linked to virulence in amoebae infected with the wild-type strain, and 4 of these proteins in amoebae infected with the Δppk mutant, respectively (proteins highlighted in bold type in Table 4).
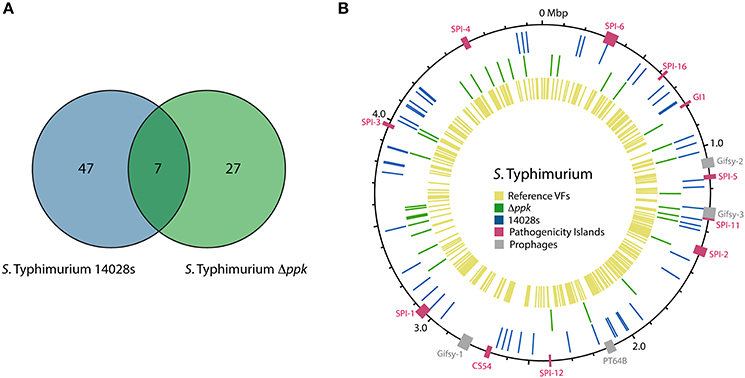
Figure 4. S. Typhimurium proteins detected by global proteomic profiling during infection of D. discoideum. (A) Venn diagram of S. Typhimurium proteins detected during infection of D. discoideum with the wild-type strain or its Δppk derivative. (B) Circular representation of S. Typhimurium 14028s chromosome. Boxes in magenta and gray indicate regions corresponding to known Salmonella pathogenicity islands (SPIs) and prophages, respectively. Blue and green lines indicate the location of genes encoding proteins detected during the infection with the wild-type strain or its Δppk derivative, respectively. Yellow lines indicate the location of genes encoding virulence-associated proteins listed in databases PATRIC_VF, VFDB, and VICTORS (see Materials and Methods section).
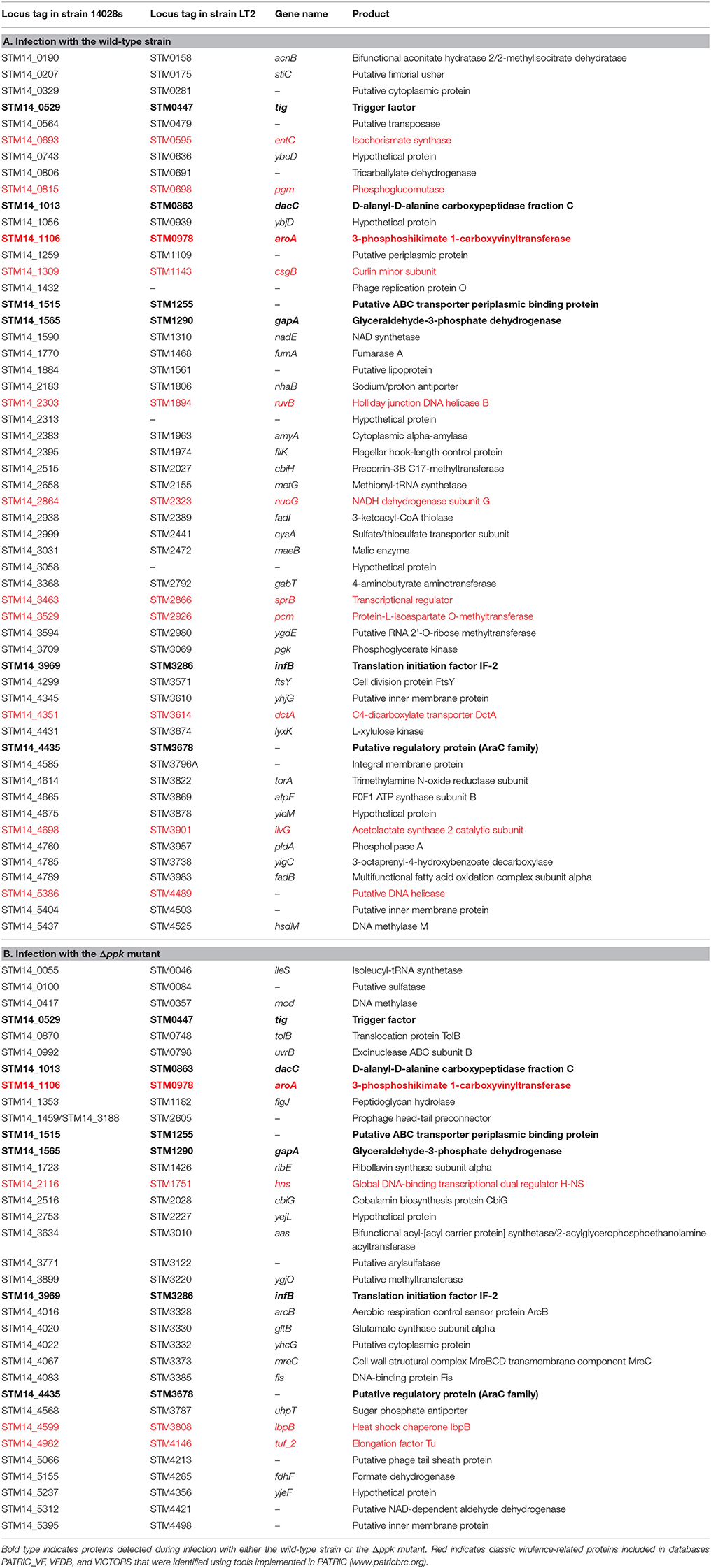
Table 4. S. Typhimurium proteins detected during infections of D. discoideum with the wild-type strain or the Δppk mutant.
The limited amount of S. Typhimurium proteins detected in our global proteomic profiling impeded us conducting an insightful comparative analysis in order to understand differences between infections of D. discoideum with the wild-type strain and the Δppk mutant. However, it is worth mentioning a particular group of S. Typhimurium proteins detected during amoebae infections that includes AroA, SprB, STM14_0329, H-NS, Fis, ArcB, NuoG, EntC, IbpB, CsgB, StiC, and FliK.
AroA is a 3-enolpyruvylshikimate-5-phosphate synthetase that is crucial for biosynthesis of aromatic compounds. As a result, Salmonella aroA null-mutants are highly attenuated in different infection models (Hoiseth and Stocker, 1981; Stocker et al., 1983; Cooper et al., 1990) and present strong defects in survival within macrophages (Fields et al., 1986; Lowe et al., 1999) and in D. discoideum (Riquelme et al., 2016). This protein was detected in amoebae infected with both S. Typhimurium wild-type and Δppk strains.
SprB is a transcription factor from the LuxR/UhaP family that is encoded in SPI-1. This protein regulates the coordinate expression of SPI-1 and SPI-4 genes during Salmonella infection (Saini and Rao, 2010). STM14_0329 (also known as SciO and TssK) is one of the 13 core components of the T6SS encoded in SPI-6 (Blondel et al., 2009; Journet and Cascales, 2016). T6SS are versatile weapons exploited by numerous bacterial pathogens to target either eukaryotic host cells or competitor bacteria (Cianfanelli et al., 2016; Hachani et al., 2016; Journet and Cascales, 2016). Noteworthy, we have recently described that null mutations in S. Typhimurium genes encoding essential components of T3SS-1 and SPI-6 T6SS cause intracellular survival defects in D. discoideum (Riquelme et al., 2016).
The nucleoid-associated protein H-NS selectively silences horizontally-acquired genes by direct binding to DNA sequences with high TA content in the genome, including all major SPIs in S. Typhimurium (Lucchini et al., 2006; Navarre et al., 2006). Accordingly, hns mutations are highly pleiotropic in S. Typhimurium (Hinton et al., 1992) and produce attenuated strains (Harrison et al., 1994). A recent study indicates that the fitness defects presented by hns mutants of S. Typhimurium are mainly due to a misregulation of SPI-1 genes (Ali et al., 2014). The factor for inversion stimulation (Fis) is a nucleoid-associated protein that influences the topological state of DNA in the cell by direct binding to DNA and by modulating DNA gyrase and topoisomerase I gene expression. As in the case of H-NS, Fis acts as a key regulator of virulence in S. Typhimurium mainly by controlling the coordinate expression of genes located in several SPIs, as well as genes involved in motility (reviewed in Duprey et al., 2014). Therefore, fis mutants of S. Typhimurium are defective for intracellular survival in macrophages (O Cróinín et al., 2006; Wang et al., 2013). ArcB is the sensor component of the master regulatory two-component system ArcA/ArcB, that controls the expression of several genes and operons encoding proteins linked to the metabolic shift from anaerobic to aerobic conditions, and the enzymatic defenses of bacteria against ROS (Evans et al., 2011). It is worth mentioning that H-NS, Fis and ArcB were only detected in amoebae infected with the Δppk mutant, perhaps reflecting adjustments required to cope with the pleiotropic phenotypes presented by this kind of mutant.
NuoG is a subunit of the NADH dehydrogenase I complex. S. Gallinarum ΔnuoG mutants are attenuated in chicken and show reduced survival and multiplication in the reticuloendothelial system of this host (Zhang-Barber et al., 1998; Turner et al., 2003). Noteworthy, it has been reported that NuoG is involved in detoxification of ROS produced by macrophages during M. tuberculosis infection (Miller et al., 2010). Thus, it is tempting to speculate that NuoG can play a similar role during S. Typhimurium infection of D. discoideum.
EntC is an isochorismate synthase involved in the biosynthesis of catecholate siderophores enterobactin and salmochelin, produced by Salmonella (and other bacterial pathogens) to capture iron from the host during infection (reviewed in Fischbach et al., 2006). It has been established that production of salmochelin is essential for full virulence of S. Typhimurium in mice (Crouch et al., 2008). Furthermore, the production of enterobactin and salmochelin is required for S. Typhimurium to survive in macrophages at early stages of infection, and these siderophores protect the pathogen against reactive oxygen species produced by macrophages during the infective process (Achard et al., 2013). Our results suggest that S. Typhimurium produces catecholate siderophores inside D. discoideum, perhaps contributing to the intracellular survival of this pathogen.
IbpB is a small heat-shock protein (sHSP) being member of a widely conserved family of ATP-independent molecular chaperones that bind to misfolded proteins and protect them from irreversible aggregation (Laskowska et al., 1996; Lee et al., 1997). It has been reported that E. coli chaperones IbpB and IbpA are substrates for the ATP-dependent Lon protease (Bissonnette et al., 2010). In addition, polyP forms a complex with Lon and stimulates the degradation of selected proteins (Kuroda et al., 2001, 2006; Nomura et al., 2004; Kuroda, 2006). Thus, there is a functional link between IbpB, Lon and the biosynthesis of polyP during the bacterial stress response.
Among the bacterial proteins detected in amoebae infected with wild-type S. Typhimurium we found CsgB, StiC, and FliK, which are associated to the assembly of proteinaceous structures such as fimbriae and flagellum, respectively. This is noteworthy because it is generally accepted that this kind of surface structures are repressed upon invasion of host cells. CsgB participates in the assembly of the curli fimbriae, favoring the polymerization of its major component CsgA (reviewed in Evans and Chapman, 2014). Curli fimbriae are amyloid fibers that act as scaffolding agents in biofilms of E. coli and Salmonella, providing increased resistance to desiccation and to sodium hypochlorite, and being extremely resistant to proteolysis and chemical denaturation (Chapman et al., 2002; White et al., 2006). These fimbriae have been linked to cell-cell contacts, and to adherence to various eukaryotic cells, tissues, and abiotic surfaces, thus promoting community behavior and host colonization, playing an important role in the initial stages of the infection process (Barnhart and Chapman, 2006). To our knowledge, there are no previous reports on the expression of CsgB when Salmonella resides inside host cells. StiC is an usher protein encoded in one of the 11 chaperone-usher fimbrial gene clusters in the genome of S. Typhimurium (Jarvik et al., 2010). Chaperone-usher fimbriae normally have one or more structural subunits, which are exported and assembled on the bacterial surface by cognate periplasmic chaperone proteins and an outer-membrane usher protein. Some of them have demonstrated roles in binding to different receptors, persisting in specific niches, promoting infections, or forming biofilms (Weening et al., 2005; Clayton et al., 2008; Yue et al., 2012). The stiABCH gene cluster encodes a class γ-fimbriae, characterized for harboring subunits comprising the domains PFAM00419 or COG3539 (Nuccio and Bäumler, 2007). To our knowledge, no previous studies have addressed the specific contribution of this fimbrial operon to S. Typhimurium virulence. However, a previous study showed that the chaperone-usher SEF14 fimbriae are essential for an efficient uptake and survival of S. Enteritidis in murine macrophages, suggesting that they may be required at stages beyond the initial host colonization (Edwards et al., 2000). FliK is the protein that controls the length of the flagellar hook, and is encoded in one of the 17 operons composing the flagellar regulon of S. Typhimurium (Chilcott and Hughes, 2000). The flagellum is required for bacterial access to the intestinal epithelium, adherence to several tissues, and immune modulation (Rossez et al., 2015). Although flagellum assembly is normally prevented inside host cells, it was previously reported that intracellular Salmonella triggers swelling of macrophages (referred to as “oncotic macrophages”) in a process where flagellated bacilli intermittently escape from infected host cells (Sano et al., 2007).
Conclusions
Overall, our results indicate that polyP biosynthesis is crucial for virulence and intracellular survival of S. Typhimurium in D. discoideum. In addition, we have validated the use of global proteomic analyses to gain insight into the host-pathogen interaction of D. discoideum and S. Typhimurium. The analysis of host proteins related to endocytic pathway, immune response, cell death, cytoskeleton dynamics, and developmental process revealed mechanisms that may explain the phenotypes shown by a S. Typhimurium strain lacking polyP during D. discoideum infection. Thus, our work demonstrates that unbiased high-throughput proteomics can be used as a powerful approach to provide new perspectives on host-pathogen interactions. Furthermore, our infection and development assays using these organisms can be exploited to screen for novel anti-virulence molecules targeting inorganic polyP biosynthesis.
Author Contributions
MV, SR-B, CV, CS, and FC: Conceived and designed the experiments; MV, SR-B, CV, and AM: Performed the experiments; MV, SR-B, CV, AM, CB-P, CS, and FC: Analyzed the data; MV, CB-P, CS, and FC: Contributed with reagents/animals/materials/analysis tools; MV, AM, CS, and FC: Wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported in part by FONDECYT grants 1120209 (to FC), 1140754 and 1171844 (to CS). MV was supported by CONICYT fellowship 21120431 and FONDECYT grant 3170449. SR-B and CV were supported by CONICYT fellowships 21160818 and 21140615, respectively.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are indebted to Nicole Molina for technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00008/full#supplementary-material
Table S1. List of all proteins detected in uninfected amoebae, and in amoebae infected with S. Typhimurium wild type or Δppk, and their corresponding PANTHER annotation.
Table S2. Proteins found exclusively or enriched in a given experimental condition, classified according to COG categories.
Table S3. Overrepresentation analysis for proteins found exclusively or enriched in amoebae infected with S. Typhimurium wild type or Δppk.
References
Achard, M. E., Chen, K. W., Sweet, M. J., Watts, R. E., Schroder, K., Schembri, M. A., et al. (2013). An antioxidant role for catecholate siderophores in Salmonella. Biochem. J. 454, 543–549. doi: 10.1042/BJ20121771
Ali, S. S., Soo, J., Rao, C., Leung, A. S., Ngai, D. H., Ensminger, A. W., et al. (2014). Silencing by H-NS potentiated the evolution of Salmonella. PLoS Pathog. 10:e1004500. doi: 10.1371/journal.ppat.1004500
Barnhart, M. M., and Chapman, M. R. (2006). Curli biogenesis and function. Annu. Rev. Microbiol. 60, 131–147. doi: 10.1146/annurev.micro.60.080805.142106
Basu, S., Fey, P., Pandit, Y., Dodson, R., Kibbe, W. A., and Chisholm, R. L. (2013). DictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res. 41, D676–D683. doi: 10.1093/nar/gks1064.
Benghezal, M., Fauvarque, M. O., Tournebize, R., Froquet, R., Marchetti, A., Bergeret, E., et al. (2006). Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 8, 139–148. doi: 10.1111/j.1462-5822.2005.00607.x
Bissonnette, S. A., Rivera-Rivera, I., Sauer, R. T., and Baker, T. A. (2010). The IbpA and IbpB small heat-shock proteins are substrates of the AAA+ Lon protease. Mol. Microbiol. 75, 1539–1549. doi: 10.1111/j.1365-2958.2010.07070.x
Blondel, C. J., Jiménez, J. C., Contreras, I., and Santiviago, C. A. (2009). Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10:354. doi: 10.1186/1471-2164-10-354
Blondel, C. J., Park, J. S., Hubbard, T. P., Pacheco, A. R., Kuehl, C. J., Walsh, M. J., et al. (2016). CRISPR/Cas9 screens reveal requirements for host cell sulfation and fucosylation in bacterial type III secretion system-mediated cytotoxicity. Cell Host Microbe 20, 226–237. doi: 10.1016/j.chom.2016.06.010
Bozzaro, S., and Eichinger, L. (2011). The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr. Drug Targets 12, 942–954. doi: 10.2174/138945011795677782
Bravo-Toncio, C., Álvarez, J. A., Campos, F., Ortíz-Severín, J., Varas, M., Cabrera, R., et al. (2016). Dictyostelium discoideum as a surrogate host-microbe model for antivirulence screening in Pseudomonas aeruginosa PAO1. Int. J. Antimicrob. Agents 47, 403–409. doi: 10.1016/j.ijantimicag.2016.02.005
Brown, M. R., and Kornberg, A. (2008). The long and short of it - polyphosphate, PPK and bacterial survival. Trends Biochem. Sci. 33, 284–290. doi: 10.1016/j.tibs.2008.04.005
Cardenal-Muñoz, E., Arafah, S., López-Jiménez, A. T., Kicka, S., Falaise, A., Bach, F., et al. (2017). Mycobacterium marinum antagonistically induces an autophagic response while repressing the autophagic flux in a TORC1- and ESX-1-dependent manner. PLoS Pathog. 13:e1006344. doi: 10.1371/journal.ppat.1006344
Carilla-Latorre, S., Calvo-Garrido, J., Bloomfield, G., Skelton, J., Kay, R. R., Ivens, A., et al. (2008). Dictyostelium transcriptional responses to Pseudomonas aeruginosa: common and specific effects from PAO1 and PA14 strains. BMC Microbiol. 8:109. doi: 10.1186/1471-2180-8-109
Chapman, M. R., Robinson, L. S., Pinkner, J. S., Roth, R., Heuser, J., Hammar, M., et al. (2002). Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855. doi: 10.1126/science.1067484
Cheng, Y., and Sun, B. (2009). Polyphosphate kinase affects oxidative stress response by modulating cAMP receptor protein and rpoS expression in Salmonella Typhimurium. J. Microbiol. Biotechnol. 19, 1527–1535. doi: 10.4014/jmb.0903.03030
Chilcott, G. S., and Hughes, K. T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708. doi: 10.1128/MMBR.64.4.694-708.2000
Choi, Y. K., Kong, J. S., and Park, Y. S. (2006). Functional role of sepiapterin reductase in the biosynthesis of tetrahydropteridines in Dictyostelium discoideum Ax2. Biochim. Biophys. Acta 1760, 877–882. doi: 10.1016/j.bbagen.2005.11.017
Cianfanelli, F. R., Monlezun, L., and Coulthurst, S. J. (2016). Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62. doi: 10.1016/j.tim.2015.10.005
Clayton, D. J., Bowen, A. J., Hulme, S. D., Buckley, A. M., Deacon, V. L., Thomson, N. R., et al. (2008). Analysis of the role of 13 major fimbrial subunits in colonisation of the chicken intestines by Salmonella enterica serovar Enteritidis reveals a role for a novel locus. BMC Microbiol. 8:228. doi: 10.1186/1471-2180-8-228
Cooper, G. L., Nicholas, R. A., Cullen, G. A., and Hormaeche, C. E. (1990). Vaccination of chickens with a Salmonella enteritidis aroA live oral Salmonella vaccine. Microb. Pathog. 9, 255–265. doi: 10.1016/0882-4010(90)90014-H
Cosson, P., and Soldati, T. (2008). Eat, kill or die: when amoeba meets bacteria. Curr. Opin. Microbiol. 11, 271–276. doi: 10.1016/j.mib.2008.05.005
Cosson, P., Zulianello, L., Join-Lambert, O., Faurisson, F., Gebbie, L., Benghezal, M., et al. (2002). Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184, 3027–3033. doi: 10.1128/JB.184.11.3027-3033.2002
Crouch, M. L., Castor, M., Karlinsey, J. E., Kalhorn, T., and Fang, F. C. (2008). Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 67, 971–983. doi: 10.1111/j.1365-2958.2007.06089.x
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dhillon, B. K., Laird, M. R., Shay, J. A., Winsor, G. L., Lo, R., Nizam, F., et al. (2015). IslandViewer 3: more flexible, interactive genomic island discovery, visualization and analysis. Nucleic Acids Res. 43, W104–W108. doi: 10.1093/nar/gkv401
Dunn, J. D., Bosmani, C., Barisch, C., Raykov, L., Lefrançois, L. H., Cardenal-Mu-oz, E., et al. (2018). Eat prey, live: Dictyostelium discoideum as a model for cell-autonomous defenses. Front. Immunol. 8:1906. doi: 10.3389/fimmu.2017.01906
Duprey, A., Reverchon, S., and Nasser, W. (2014). Bacterial virulence and Fis: adapting regulatory networks to the host environment. Trends Microbiol. 22, 92–99. doi: 10.1016/j.tim.2013.11.008
Edwards, R. A., Schifferli, D. M., and Maloy, S. R. (2000). A role for Salmonella fimbriae in intraperitoneal infections. Proc. Natl. Acad. Sci. U.S.A. 97, 1258–1262. doi: 10.1073/pnas.97.3.1258
Evans, M. L., and Chapman, M. R. (2014). Curli biogenesis: order out of disorder. Biochim. Biophys. Acta 1843, 1551–1558. doi: 10.1016/j.bbamcr.2013.09.010
Evans, M. R., Fink, R. C., Vazquez-Torres, A., Porwollik, S., Jones-Carson, J., McClelland, M., et al. (2011). Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol. 11:58. doi: 10.1186/1471-2180-11-58
Fey, P., Dodson, R. J., Basu, S., and Chisholm, R. L. (2013). One stop shop for everything Dictyostelium: dictyBase and the Dicty Stock Center in 2012. Methods Mol. Biol. 983, 59–92. doi: 10.1007/978-1-62703-302-2_4
Fey, P., Kowal, A. S., Gaudet, P., Pilcher, K. E., and Chisholm, R. L. (2007). Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2, 1307–1316. doi: 10.1038/nprot.2007.178
Fields, P. I., Swanson, R. V., Haidaris, C. G., and Heffron, F. (1986). Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U.S.A. 83, 5189–5193. doi: 10.1073/pnas.83.14.5189
Fischbach, M. A., Lin, H., Liu, D. R., and Walsh, C. T. (2006). How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2, 132–138. doi: 10.1038/nchembio771
Froquet, R., Lelong, E., Marchetti, A., and Cosson, P. (2009). Dictyostelium discoideum: a model host to measure bacterial virulence. Nat. Protoc. 4, 25–30. doi: 10.1038/nprot.2008.212
Gray, M. J., and Jakob, U. (2015). Oxidative stress protection by polyphosphate - new roles for an old player. Curr. Opin. Microbiol. 24, 1–6. doi: 10.1016/j.mib.2014.12.004
Hachani, A., Wood, T. E., and Filloux, A. (2016). Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29, 81–93. doi: 10.1016/j.mib.2015.11.006
Hagedorn, M., and Soldati, T. (2007). Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell Microbiol. 9, 2716–2733. doi: 10.1111/j.1462-5822.2007.00993.x
Hägele, S., Köhler, R., Merkert, H., Schleicher, M., Hacker, J., and Steinert, M. (2000). Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell Microbiol. 2, 165–171. doi: 10.1046/j.1462-5822.2000.00044.x
Haraga, A., Ohlson, M. B., and Miller, S. I. (2008). Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6, 53–66. doi: 10.1038/nrmicro1788
Harrison, J. A., Pickard, D., Higgins, C. F., Khan, A., Chatfield, S. N., Ali, T., et al. (1994). Role of hns in the virulence phenotype of pathogenic salmonellae. Mol. Microbiol. 13, 133–140. doi: 10.1111/j.1365-2958.1994.tb00408.x
Hasselbring, B. M., Patel, M. K., and Schell, M. A. (2011). Dictyostelium discoideum as a model system for identification of Burkholderia pseudomallei virulence factors. Infect. Immun. 79, 2079–2088. doi: 10.1128/IAI.01233-10
Hinton, J. C., Santos, D. S., Seirafi, A., Hulton, C. S., Pavitt, G. D., and Higgins, C. F. (1992). Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol. Microbiol. 6, 2327–2337. doi: 10.1111/j.1365-2958.1992.tb01408.x
Hoiseth, S. K., and Stocker, B. A. (1981). Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239. doi: 10.1038/291238a0
Hudson, C. M., Lau, B. Y., and Williams, K. P. (2015). Islander: a database of precisely mapped genomic islands in tRNA and tmRNA genes. Nucleic Acids Res. 43, D48–D53. doi: 10.1093/nar/gku1072
Jarvik, T., Smillie, C., Groisman, E. A., and Ochman, H. (2010). Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J. Bacteriol. 192, 560–567. doi: 10.1128/JB.01233-09
Jensen, L. J., Julien, P., Kuhn, M., von Mering, C., Muller, J., Doerks, T., et al. (2008). eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36, D250–D254. doi: 10.1093/nar/gkm796
Journet, L., and Cascales, E. (2016). The Type VI secretion system in Escherichia coli and related species. EcoSal Plus. doi: 10.1128/ecosalplus.ESP-0009-2015
Kim, K. S., Rao, N. N., Fraley, C. D., and Kornberg, A. (2002). Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. U.S.A. 99, 7675–7680. doi: 10.1073/pnas.112210499
Kreppel, L., Fey, P., Gaudet, P., Just, E., Kibbe, W. A., Chisholm, R. L., et al. (2004). dictyBase: a new Dictyostelium discoideum genome database. Nucleic Acids Res. 32, D332–D333. doi: 10.1093/nar/gkh138
Kuroda, A. (2006). A polyphosphate-lon protease complex in the adaptation of Escherichia coli to amino acid starvation. Biosci. Biotechnol. Biochem. 70, 325–331. doi: 10.1271/bbb.70.325
Kuroda, A., Nomura, K., Ohtomo, R., Kato, J., Ikeda, T., Takiguchi, N., et al. (2001). Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293, 705–708. doi: 10.1126/science.1061315
Kuroda, A., Nomura, K., Takiguchi, N., Kato, J., and Ohtake, H. (2006). Inorganic polyphosphate stimulates Lon-mediated proteolysis of nucleoid proteins in Escherichia coli. Cell Mol. Biol. 52, 23–29. doi: 10.1170/T723
Lampe, E. O., Brenz, Y., Herrmann, L., Repnik, U., Griffiths, G., Zingmark, C., et al. (2015). Dissection of Francisella-host cell interactions in Dictyostelium discoideum. Appl. Environ. Microbiol. 82, 1586–1598. doi: 10.1128/AEM.02950-15
LaRock, D. L., Chaudhary, A., and Miller, S. I. (2015). Salmonellae interactions with host processes. Nat. Rev. Microbiol. 13, 191–205. doi: 10.1038/nrmicro3420
Laskowska, E., Wawrzynów, A., and Taylor, A. (1996). IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie 78, 117–122. doi: 10.1016/0300-9084(96)82643-5
Le Coadic, M., Froquet, R., Lima, W. C., Dias, M., Marchetti, A., and Cosson, P. (2013). Phg1/TM9 proteins control intracellular killing of bacteria by determining cellular levels of the Kil1 sulfotransferase in Dictyostelium. PLoS ONE 8:e53259. doi: 10.1371/journal.pone.0053259
Lee, G. J., Roseman, A. M., Saibil, H. R., and Vierling, E. (1997). A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 16, 659–671. doi: 10.1093/emboj/16.3.659
Lowe, D. C., Savidge, T. C., Pickard, D., Eckmann, L., Kagnoff, M. F., Dougan, G., et al. (1999). Characterization of candidate live oral Salmonella typhi vaccine strains harboring defined mutations in aroA, aroC, and htrA. Infect. Immun. 67, 700–707.
Lucchini, S., Rowley, G., Goldberg, M. D., Hurd, D., Harrison, M., and Hinton, J. C. (2006). H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2:e81. doi: 10.1371/journal.ppat.0020081
McMeechan, A., Lovell, M. A., Cogan, T. A., Marston, K. L., Humphrey, T. J., and Barrow, P. A. (2007). Inactivation of ppk differentially affects virulence and disrupts ATP homeostasis in Salmonella enterica serovars Typhimurium and Gallinarum. Res. Microbiol. 158, 79–85. doi: 10.1016/j.resmic.2006.10.008
Mi, H., Muruganujan, A., Casagrande, J. T., and Thomas, P. D. (2013). Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566. doi: 10.1038/nprot.2013.092
Mi, H., Poudel, S., Muruganujan, A., Casagrande, J. T., and Thomas, P. D. (2016). PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44, D336–D342. doi: 10.1093/nar/gkv1194
Miller, J. L., Velmurugan, K., Cowan, M. J., and Briken, V. (2010). The type I NADH dehydrogenase of Mycobacterium tuberculosis counters phagosomal NOX2 activity to inhibit TNF-alpha-mediated host cell apoptosis. PLoS Pathog. 6:e1000864. doi: 10.1371/journal.ppat.1000864
Navarre, W. W., Porwollik, S., Wang, Y., McClelland, M., Rosen, H., Libby, S. J., et al. (2006). Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238. doi: 10.1126/science.1128794
Nomura, K., Kato, J., Takiguchi, N., Ohtake, H., and Kuroda, A. (2004). Effects of inorganic polyphosphate on the proteolytic and DNA-binding activities of Lon in Escherichia coli. J. Biol. Chem. 279, 34406–34410. doi: 10.1074/jbc.M404725200
Nuccio, S. P., and Bäumler, A. J. (2007). Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71, 551–575. doi: 10.1128/MMBR.00014-07
O Cróinín, T., Carroll, R. K., Kelly, A., and Dorman, C. J. (2006). Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 62, 869–882. doi: 10.1111/j.1365-2958.2006.05416.x
Ouertatani-Sakouhi, H., Kicka, S., Chiriano, G., Harrison, C. F., Hilbi, H., Scapozza, L., et al. (2017). Inhibitors of Mycobacterium marinum virulence identified in a Dictyostelium discoideum host model. PLoS ONE 12:e0181121. doi: 10.1371/journal.pone.0181121
Pan, Y. J., Lin, T. L., Hsu, C. R., and Wang, J. T. (2011). Use of a Dictyostelium model for isolation of genetic loci associated with phagocytosis and virulence in Klebsiella pneumoniae. Infect. Immun. 79, 997–1006. doi: 10.1128/IAI.00906-10
Pukatzki, S., Kessin, R. H., and Mekalanos, J. J. (2002). The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 99, 3159–3164. doi: 10.1073/pnas.052704399
Rao, N. N., Liu, S., and Kornberg, A. (1998). Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 180, 2186–2193.
Rashid, M. H., and Kornberg, A. (2000). Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 4885–4890. doi: 10.1073/pnas.060030097
Rashid, M. H., Rao, N. N., and Kornberg, A. (2000a). Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182, 225–227. doi: 10.1128/JB.182.1.225-227.2000
Rashid, M. H., Rumbaugh, K., Passador, L., Davies, D. G., Hamood, A. N., Iglewski, B. H., et al. (2000b). Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 9636–9641. doi: 10.1073/pnas.170283397
Riquelme, S., Varas, M., Valenzuela, C., Velozo, P., Chahin, N., Aguilera, P., et al. (2016). Relevant genes linked to virulence are required for Salmonella Typhimurium to survive intracellularly in the social amoeba Dictyostelium discoideum. Front. Microbiol. 7:1305. doi: 10.3389/fmicb.2016.01305
Rossez, Y., Wolfson, E. B., Holmes, A., Gally, D. L., and Holden, N. J. (2015). Bacterial flagella: twist and stick, or dodge across the kingdoms. PLoS Pathog. 11:e1004483. doi: 10.1371/journal.ppat.1004483
Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945. doi: 10.1093/bioinformatics/16.10.944
Saini, S., and Rao, C. V. (2010). SprB is the molecular link between Salmonella pathogenicity island 1 (SPI1) and SPI4. J. Bacteriol. 192, 2459–2462. doi: 10.1128/JB.00047-10
Sano, G., Takada, Y., Goto, S., Maruyama, K., Shindo, Y., Oka, K., et al. (2007). Flagella facilitate escape of Salmonella from oncotic macrophages. J. Bacteriol. 189, 8224–8232. doi: 10.1128/JB.00898-07
Santiviago, C. A., Reynolds, M. M., Porwollik, S., Choi, S. H., Long, F., Andrews-Polymenis, H. L., et al. (2009). Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. doi: 10.1371/journal.ppat.1000477
Sebkova, A., Karasova, D., Crhanova, M., Budinska, E., and Rychlik, I. (2008). aro mutations in Salmonella enterica cause defects in cell wall and outer membrane integrity. J. Bacteriol. 190, 3155–3160. doi: 10.1128/JB.00053-08
Sillo, A., Matthias, J., Konertz, R., Bozzaro, S., and Eichinger, L. (2011). Salmonella typhimurium is pathogenic for Dictyostelium cells and subverts the starvation response. Cell Microbiol. 13, 1793–1811. doi: 10.1111/j.1462-5822.2011.01662.x
Stocker, B. A., Hoiseth, S. K., and Smith, B. P. (1983). Aromatic-dependent “Salmonella sp.” as live vaccine in mice and calves. Dev. Biol. Stand 53, 47–54.
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Thöny, B., Auerbach, G., and Blau, N. (2000). Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 347(Pt 1), 1–16. doi: 10.1042/bj3470001
Tosetti, N., Croxatto, A., and Greub, G. (2014). Amoebae as a tool to isolate new bacterial species, to discover new virulence factors and to study the host-pathogen interactions. Microb. Pathog. 77, 125–130. doi: 10.1016/j.micpath.2014.07.009
Turner, A. K., Barber, L. Z., Wigley, P., Muhammad, S., Jones, M. A., Lovell, M. A., et al. (2003). Contribution of proton-translocating proteins to the virulence of Salmonella enterica serovars Typhimurium, Gallinarum, and Dublin in chickens and mice. Infect. Immun. 71, 3392–3401. doi: 10.1128/IAI.71.6.3392-3401.2003
Varas, M., Valdivieso, C., Mauriaca, C., Ortíz-Severín, J., Paradela, A., Poblete-Castro, I., et al. (2017). Multi-level evaluation of Escherichia coli polyphosphate related mutants using global transcriptomic, proteomic and phenomic analyses. Biochim. Biophys. Acta 1861, 871–883. doi: 10.1016/j.bbagen.2017.01.007
Varela, C., Mauriaca, C., Paradela, A., Albar, J. P., Jerez, C. A., and Chávez, F. P. (2010). New structural and functional defects in polyphosphate deficient bacteria: a cellular and proteomic study. BMC Microbiol. 10:7. doi: 10.1186/1471-2180-10-7
Vásquez-Vivar, J. (2009). Tetrahydrobiopterin, superoxide, and vascular dysfunction. Free Radic. Biol. Med. 47, 1108–1119. doi: 10.1016/j.freeradbiomed.2009.07.024
Wang, H., Liu, B., Wang, Q., and Wang, L. (2013). Genome-wide analysis of the Salmonella Fis regulon and its regulatory mechanism on pathogenicity islands. PLoS ONE 8:e64688. doi: 10.1371/journal.pone.0064688
Weber, S., Wagner, M., and Hilbi, H. (2014). Live-cell imaging of phosphoinositide dynamics and membrane architecture during Legionella infection. MBio 5, e00839–e00813. doi: 10.1128/mBio.00839-13
Weening, E. H., Barker, J. D., Laarakker, M. C., Humphries, A. D., Tsolis, R. M., and Bäumler, A. J. (2005). The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 73, 3358–3366. doi: 10.1128/IAI.73.6.3358-3366.2005
White, A. P., Gibson, D. L., Kim, W., Kay, W. W., and Surette, M. G. (2006). Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188, 3219–3227. doi: 10.1128/JB.188.9.3219-3227.2006
Wiśniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Yoon, S. H., Park, Y. K., and Kim, J. F. (2015). PAIDB v2.0: exploration and analysis of pathogenicity and resistance islands. Nucleic Acids Res. 43, D624–D630. doi: 10.1093/nar/gku985
Yue, M., Rankin, S. C., Blanchet, R. T., Nulton, J. D., Edwards, R. A., and Schifferli, D. M. (2012). Diversification of the Salmonella fimbriae: a model of macro- and microevolution. PLoS ONE 7:e38596. doi: 10.1371/journal.pone.0038596
Zhang, G., Ueberheide, B. M., Waldemarson, S., Myung, S., Molloy, K., Eriksson, J., et al. (2010). Protein quantitation using mass spectrometry. Methods Mol. Biol. 673, 211–222. doi: 10.1007/978-1-60761-842-3_13
Zhang, H., Gómez-García, M. R., Brown, M. R., and Kornberg, A. (2005). Inorganic polyphosphate in Dictyostelium discoideum: influence on development, sporulation, and predation. Proc. Natl. Acad. Sci. U.S.A. 102, 2731–2735. doi: 10.1073/pnas.0500023102
Zhang, X., Zhuchenko, O., Kuspa, A., and Soldati, T. (2016). Social amoebae trap and kill bacteria by casting DNA nets. Nat. Commun. 7:10938. doi: 10.1038/ncomms10938
Zhang-Barber, L., Turner, A. K., Dougan, G., and Barrow, P. A. (1998). Protection of chickens against experimental fowl typhoid using a nuoG mutant of Salmonella serotype Gallinarum. Vaccine 16, 899–903. doi: 10.1016/S0264-410X(97)00300-9
Keywords: Salmonella, Dictyostelium, polyphosphate, ppk, virulence, intracellular survival, proteomics
Citation: Varas MA, Riquelme-Barrios S, Valenzuela C, Marcoleta AE, Berríos-Pastén C, Santiviago CA and Chávez FP (2018) Inorganic Polyphosphate Is Essential for Salmonella Typhimurium Virulence and Survival in Dictyostelium discoideum. Front. Cell. Infect. Microbiol. 8:8. doi: 10.3389/fcimb.2018.00008
Received: 03 November 2017; Accepted: 09 January 2018;
Published: 30 January 2018.
Edited by:
Thierry Soldati, Université de Genève, SwitzerlandReviewed by:
Francisco Ramos-Morales, Universidad de Sevilla, SpainSalvatore Bozzaro, Università degli Studi di Torino, Italy
Richard H. Gomer, Texas A&M University College Station, United States
Copyright © 2018 Varas, Riquelme-Barrios, Valenzuela, Marcoleta, Berríos-Pastén, Santiviago and Chávez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlos A. Santiviago, csantiviago@ciq.uchile.cl
Francisco P. Chávez, fpchavez@uchile.cl
 Macarena A. Varas
Macarena A. Varas Sebastián Riquelme-Barrios
Sebastián Riquelme-Barrios Camila Valenzuela
Camila Valenzuela Andrés E. Marcoleta
Andrés E. Marcoleta Camilo Berríos-Pastén
Camilo Berríos-Pastén Carlos A. Santiviago
Carlos A. Santiviago Francisco P. Chávez
Francisco P. Chávez