Heterologous Complementation Studies With the YscX and YscY Protein Families Reveals a Specificity for Yersinia pseudotuberculosis Type III Secretion
- 1Department of Molecular Biology, Umeå University, Umeå, Sweden
- 2Umeå Centre for Microbial Research, Umeå University, Umeå, Sweden
- 3Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology, Chinese Academy of Sciences Wuhan, Wuhan, China
- 4Department of Chemistry, University of Turku, Turku, Finland
Type III secretion systems harbored by several Gram-negative bacteria are often used to deliver host-modulating effectors into infected eukaryotic cells. About 20 core proteins are needed for assembly of a secretion apparatus. Several of these proteins are genetically and functionally conserved in type III secretion systems of bacteria associated with invertebrate or vertebrate hosts. In the Ysc family of type III secretion systems are two poorly characterized protein families, the YscX family and the YscY family. In the plasmid-encoded Ysc-Yop type III secretion system of human pathogenic Yersinia species, YscX is a secreted substrate while YscY is its non-secreted cognate chaperone. Critically, neither an yscX nor yscY null mutant of Yersinia is capable of type III secretion. In this study, we show that the genetic equivalents of these proteins produced as components of other type III secretion systems of Pseudomonas aeruginosa (PscX and PscY), Aeromonas species (AscX and AscY), Vibrio species (VscX and VscY), and Photorhabdus luminescens (SctX and SctY) all possess an ability to interact with its native cognate partner and also establish cross-reciprocal binding to non-cognate partners as judged by a yeast two-hybrid assay. Moreover, a yeast three-hybrid assay also revealed that these heterodimeric complexes could maintain an interaction with YscV family members, a core membrane component of all type III secretion systems. Despite maintaining these molecular interactions, only expression of the native yscX in the near full-length yscX deletion and native yscY in the near full-length yscY deletion were able to complement for their general substrate secretion defects. Hence, YscX and YscY must have co-evolved to confer an important function specifically critical for Yersinia type III secretion.
Introduction
Type III secretion (T3S) is an effective means for many different Gram-negative bacteria to deliver proteins into diverse eukaryotic cell types. This is a common virulence mechanism of many harmful pathogens (Buttner, 2012; Portaliou et al., 2016), but is also useful for bacteria relying on a symbiotic lifestyle or for survival in the environment (Pallen et al., 2005), and for the biosynthesis of the flagella apparatus (Erhardt et al., 2010). Upon artificial induction, a T3S system (T3SS) can also secrete substrates into laboratory culture media and this requires a complex of ~20 proteins that all together span both bacterial membranes (inner and outer), the peptidoglycan layer and which also protrudes out from the surface. This apparatus has been purified from a few different bacteria, taking on the appearance of a syringe equipped with needle (often termed the “needle complex”) (Kubori et al., 1998; Blocker et al., 1999, 2001; Kimbrough and Miller, 2000; Tamano et al., 2000, 2002; Sekiya et al., 2001; Sukhan et al., 2001; Journet et al., 2003; Marlovits et al., 2004; Mueller et al., 2005). Generally, protein cargo is thought to be secreted through the needle complex (Radics et al., 2014), and these can be divided into three categories—“early” substrates that make up the outer needle, “middle” substrates comprising the pore-forming translocon proteins and “late” host-modulating effector proteins that with assistance from the translocon are targeted to the eukaryotic cell interior (Osborne and Coombes, 2011; Buttner, 2012; Dewoody et al., 2013).
About 10 proteins exist in the needle complex that are common to all known non-flagella and flagella T3SSs (Francis et al., 2004; Buttner, 2012; Portaliou et al., 2016). Moreover, thorough analysis of sequence similarities can classify all T3SSs into evolutionary distinct nodes (Troisfontaines and Cornelis, 2005; Abby and Rocha, 2012). Among the non-flagella T3SSs, a major node is the inv/spa system encoded by the Salmonella pathogenicity island 1 (SPI-1) of Salmonella enterica and including systems from Shigella sp. (mxi/spa), Escherichia coli (evi/epa) and Burkholderia sp. (inv/spa). Another major node is characterized by the ysc system, encoded on a common virulence plasmid of human pathogenic Yersinia sp., and includes systems found in Pseudomonas aeruginosa (psc), selected Aeromonas sp. (asc), Photorhabdus luminescens (lsc/sct) and certain Vibrio sp. (vsc). Unique to this node are a number of genetically distinct T3SS components not represented in other nodes. Two examples of this are the components YscX and YscY.
The functional role of YscX and YscY in T3S remains enigmatic. In Yersinia, a mutant lacking either yscX or yscY allele poorly synthesizes Yop substrates and their secretion is abolished (Iriarte and Cornelis, 1999; Day and Plano, 2000; Bröms et al., 2005). Moreover, YscX is a T3SS substrate that prior to secretion is stabilized in the cytoplasm by the YscY chaperone (Iriarte and Cornelis, 1999; Day and Plano, 2000). Interestingly, an in silico analysis revealed that YscY possesses three tandom tetratricopeptide repeats (Pallen et al., 2003) that are important for function of the translocator class of T3S chaperones (Bröms et al., 2006; Edqvist et al., 2006; Buttner et al., 2008; Lunelli et al., 2009; Job et al., 2010; Singh et al., 2013; Kim et al., 2014). These studies taken together indicate that YscX could be a structural component of the needle complex, while YscY would be necessary to stabilize pre-made pools of YscX and to ensure correct temporal YscX secretion. However, a recent study could find no support for YscX association with the needle, but rather together with YscY was found associated with YscV (alternatively known as LcrD), a core structural inner membrane component of all T3SSs (Diepold et al., 2012).
In a study of P. aeruginosa T3S, it was reported that the YscY homolog, Pcr4 (from here on termed PscY for consistency), is actually the secreted component, and not the YscX homolog, Pcr3 (here on termed PscX) (Yang et al., 2007). Despite this anomaly, the YscX/PscX and YscY/PscY components otherwise appear to serve somewhat analogous functions in their respective T3SSs (Bröms et al., 2005; Yang et al., 2007). Moreover, expression of yscX and yscY in the corresponding P. aeruginosa mutants can efficiently restore T3S (Bröms et al., 2005). Yet specific differences must exist since neither pscX nor pscY alone, or even when expressed in combination, could complement the T3S defects observed in Yersinia mutants lacking the corresponding alleles (Bröms et al., 2005). A known difference is the YscY interaction with the LcrH chaperone (also known as SycD) that has an undisclosed role in T3SS regulation in Yersinia (Francis et al., 2001; Bröms et al., 2005); the equivalent interaction between PscY and the PcrH chaperone in P. aeruginosa has not been detected (Broms et al., 2003; Bröms et al., 2005). Moreover, it is not yet known whether PscX and PscY complex with PscV (the YscV homolog) as do the three equivalent Yersinia components (Diepold et al., 2012).
Hence, in this study we further investigated the notion that an YscX-YscY complex has evolved molecular attributes unique to T3SS function in Yersinia. We analyzed T3S from Yersinia yscX or yscY mutants ectopically expressing in trans homologous alleles from Aeromonas salmonicida and A. hydrophila (termed ascXAs/ascXAh and ascYAs /ascYAh), P. luminescens (termed sctX or sctY), V. harveyi and V. parahaemolyticus (termed vscXVh /vscXVp and vscYVh /vscYVp) or P. aeruginosa (PscX and PscY). No combination resulted in restoration of T3S, despite confirmation of reciprocal interactions giving rise to the equivalent of YscX-YscY bipartite and YscX-YscY-YscV tripartite complexes. This suggested that the YscX-YscY complex serves an exclusive T3SS function in Yersinia.
Materials and Methods
Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table S1. Bacteria were routinely cultivated in Lysogeny broth (LB) (Bertani, 2004) or LB agar at either 26°C (Yersinia pseudotuberculosis) or 37°C (E. coli) with aeration. When required, the antibiotics carbenicillin (Cb), kanamycin (Km) and gentamicin (Gm) were added to laboratory culture media at the final concentrations of 100 μg per ml, 50 μg per ml and 20 μg per ml, respectively. Analysis of T3SS by Y. pseudotuberculosis occurred at 37°C in Brain Heart Infusion (BHI) broth. Media containing Ca2+ ions was the non-inducing condition (BHI supplemented with 2.5 mM CaCl2), while media devoid of Ca2+ ions was the inducing condition (BHI supplemented with 20 mM MgCl2 and 5 mM Ethylene glycol-bis-(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid). To stimulate promoter activity from the expression vectors pMMB67EHgm and/or pMMB208, isopropyl β-D-1-thiogalactopyranoside (IPTG) at a final concentration of 0.4 mM was added.
The Saccharomyces cerevisiae reporter strain AH109 or Y190 was maintained by growth at 30°C in YEP broth [2% (w/v) peptone, 1% (w/v) yeast extract, 2% (v/v) glucose] or agar [YEP broth with 2% (w/v) agar]. pGBKT7 derived plasmids were sustained in yeast by growth on SD synthetic minimal medium (0.67% (w/v) Yeast nitrogen base without amino acids, 300 μgml−1 L-isoleucine, 1.5 mgml−1 L-valine, 200 μgml−1 L-adenine hemisulphate salt, 200 μgml−1 L-arginine HCl, 200 μgml−1 L-histidine HCl monohydrate, 1 mgml−1 L-leucine, 300 μgml−1 L-lysine HCl, 200 μgml−1 L-methionine, 500 μgml−1 L-phenylalanine, 2 mgml−1 L-threonine) while pBridge derived plasmids were grown on the same media minus methionine. Yeast containing pGADT7 derivatives were cultured on similar SD media with the exception that leucine was replaced with 200 μgml−1 L-tryptophan.
PCR Amplification and Sequence Analysis
Amplified DNA fragments were obtained by PCR using the appropriate oligonucleotide combinations listed as online Supplementary Information (Table S2). These were synthesized by either DNA Technology A/S (Aarhus, Denmark), TAG Copenhagen A/S (Copenhagen, Denmark) or Sigma-Aldrich Sweden AB (Stockholm, Sweden). Amplified fragments were confirmed to be mutation free by first cloning into pCR®4-TOPO TA (Invitrogen AB, Stockholm, Sweden) or pTZ57R using the InsTAclone PCR cloning kit (Thermo Fisher Scientific, Gothenburg, Sweden) and then via commercial sequencing (Eurofins MWG Operon, Ebersberg, Germany or GATC Biotech AB, Solna, Sweden).
Phylogenetic Analysis of YscX and YscY Protein Families
Protein sequences corresponding to YscX and YscY protein family were mined from National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/sites/entrez) and phylogenetic trees obtained by the Neighbor-Joining method using MEGA6 (Molecular Evolutionary Genetics Analysis) (Tamura et al., 2013). Briefly, proteins were aligned by ClustalW using default settings and the unaligned regions and gaps were trimmed. The significance of the phylogenetic grouping was assessed using confidence level for 1,000 replicates. The trees were rooted with genetically related Bordetella pertusis I475 YscX-like or YscY-like protein as the respective out-groups.
Generation of Bacterial Protein Expression Constructs
Constructs containing the yscX and yscY alleles from Y. pseudotuberculosis and pscX and pscY alleles from P. aeruginosa have been described previously (Bröms et al., 2005). ascX and ascY were amplified from both A. salmonicida subsp. salmonicida JF2267 (a gift from Joachim Frey, Universität Bern, Switzerland) and A. hydrophila AH-3 (Juan Tomás, Universidad de Barcelona, Spain). vscX and vscY were amplified from both Vibrio harveyi BB120 (Debra Milton, Umeå University, Sweden) and V. parahaemolyticus RIMD2210633 (Tetsuya Iida, Osaka University, Japan). Finally, sctX and sctY were amplified from P. luminescens TT01 (RifR) (David Clarke, University of Bath, United Kingdom). For the complementation assay in Y. pseudotuberculosis, with one exception all alleles were cloned individually or together as a dual expression construct into EcoRI-BamHI digested pMMB67EHgm. (The vscY allele from V. harveyi was digested by BamHI-PstI.) To examine stable protein expression in Y. pseudotuberculosis, with two exceptions all PCR amplified alleles with a 5-prime FLAG™ epitope were cloned individually into BamHI-EcoRI digested pMMB208. (The yscY allele from Y. pseudotuberculosis and the vscY allele from V. harveyi was digested by PstI-BamHI.) The same approach was utilized to generate expression constructs of codon-optimized gene variants.
Construction of Yeast Plasmids
To investigate reciprocal binding between the products of yscY, yscX and related alleles, a series of constructs were generated in the yeast two-hybrid vectors pGBKT7 and pGADT7 (Clontech Laboratories, Palo Alto, CA). Constructs expressing the yscY and pscY alleles fused to the GAL4 DNA binding domain in pGBKT7 are already described (Francis et al., 2001; Bröms et al., 2005). PCR amplified ascY, vscY, and sctY alleles were fused to the GAL4 DNA binding domain by cloning into EcoRI-BamHI digested pGBKT7. Plasmids expressing the yscX and pscX alleles fused to the GAL4 activation domain in pGADT7 are already established (Bröms et al., 2005). PCR amplified ascX, vscX, and sctX alleles were fused to the GAL4 activation domain by cloning into EcoRI-BamHI digested pGADT7.
Reciprocal interaction between YscX, YscY, YscV, and related family members were assessed with a yeast three hybrid approach by utilizing a three hybrid GAL4 DNA binding domain pBridge vector (Clontech Laboratories, Palo Alto, CA) and Gal4 activation domain pGADT7 vector. Constructs expressing yscY and related alleles were expressed as Gal4 DNA binding fusion protein by introducing into EcoRI-PstI digested pBridge downstream of ADH1 promoter. Additionally, the coding sequences of yscX and related alleles were cloned into NotI-BglII sites of pBridge downstream of MET25 promoter to finally yield pBridge-yscY-yscX and its variants. Likewise, yscV and related members were expressed as GAL4 activation fusion protein from pGADT7. While PCR-amplified yscV, ascV and sctV alleles were cloned into EcoRI-XhoI sites, PCR-amplified pscV and vscV were inserted into NdeI-BamHI sites of pGADT7.
Yeast Transformation and the n-Hybrid Assays
For yeast two-hybrid assay, transformation of the S. cerevisiae reporter strain AH109 was performed as described earlier (Francis et al., 2000; Bröms et al., 2005; Amer et al., 2016). Protein interactions from multiple independent transformations were determined by measuring the activation of the ADE2 reporter gene activation and the HIS3 reporter gene during growth on tryptophan and leucine minus SD synthetic minimal medium also lacking either adenine or histidine, respectively. The latter also required the addition of 4 mM 3-aminotriazole in the growth media to overcome any risk of false positives (James et al., 1996). Analysis of protein stability in yeast was performed as previously described (Francis et al., 2000).
The yeast three-hybrid assay was performed in S. cerevisiae reporter strain Y190 as described previously (Carlo et al., 2007; Glass et al., 2015). Briefly, yeast were co-transformed with pBridge and pGADT7-based vectors and a master plate established by initial growth at 30°C on SD synthetic minimal medium lacking leucine, tryptophan and methionine. The extent of protein interactions were then measured via the activation of the HIS3 reporter gene upon growth on the above mentioned medium also lacking histidine, but supplemented with 40 mM 3-aminotriazole (3-AT) to neutralize the inherent leakiness of the reporter. Equal amount of colony from each of the transformants were resuspended in water and subjected to 5-fold serial dilutions. An aliquot of each serial dilution was then transferred onto two sets of plates: (1) master plate (SD -Leu, -Trp, -Met) and (2) selective plate (SD −Leu, −Trp, −Met, −His + 40 mM 3-AT). The growth of yeast on the selective plate indicated a positive interaction. As a control, 1 mM methionine was used for near complete repression of MET25 repressible promoter (Her et al., 2003).
Analysis of Gene Transcription by Qualitative RT-PCR
Total RNA was isolated from T3SS-induced Yersinia cultures as previously described (Carlsson et al., 2007a,b). Briefly, overnight cultures of Yersinia grown in a secretion permissive condition (BHI minus Ca2+) were diluted into 5 ml fresh media to an optical density (OD) at 600 nm of 0.1. After incubating at 26°C for 30 min, cultures were shifted to 37°C and grown for approximately 90 min to an OD at 600 nm of 0.4 to 0.8. RNA was immediately stabilized by mixing two volumes of RNA protect bacterial reagent (QIAGEN GmbH, Hilden, Germany) with one volume of the bacterial culture. Total RNA was isolated using the NucleoSpin RNA II method (Macherey Nagel, Düren, Germany) that included an on-column DNase treatment. For reverse transcription, 0.2 μg of total RNA was used to generate cDNA by RevertAid H Minus Reverse Transcriptase (RT) system (Thermo Scientific, Vilnius, Lithuania). RNA samples that were not treated with RT were used as negative controls to confirm the absence of contaminating DNA. PCR to confirm the presence of specific transcript was performed using gene-specific primers as listed in Table S2 and the cDNA generated above as template (Carlsson et al., 2007a,b).
Qualitative RT-PCR to determine the stability of mRNA transcript was performed as described elsewhere (Okan et al., 2006; Jeters et al., 2009; Chen and Anderson, 2011) with some modifications. Briefly, Yersinia cultures were grown in BHI media lacking Ca2+ at 37°C to an OD at 600 nm of 0.4 to 0.8. At time point zero, one volume of bacterial culture was removed and rifampicin to a final concentration of 100 μg/ml was added to the bacterial culture to inhibit de novo RNA synthesis. Thereafter, one volume of bacterial cultures was taken at 5, 10, and 20 min time points. RNA was always stabilized by the addition of two volumes of RNA protect bacterial reagent (QIAGEN GmbH, Hilden, Germany). Isolation of total RNA, synthesis of cDNA and qualitative presence of transcript by PCR was performed as described above.
Synthesis and Secretion of Type III-Secreted Substrates
Induction of type III substrate synthesis and secretion from Y. pseudotuberculosis was performed as previously described (Francis et al., 2000, 2001). All protein samples were normalized to the amount of bacterial cells at an optical density of 600 nm. Protein associated with whole bacteria was assessed by sampling from pelleted bacterial cultures. Total fraction contained proteins associated within intact bacteria and secreted to the culture medium. Sampling of the cell-free supernatant assessed the secreted protein levels. All protein fractions were separated by SDS-PAGE and subjected to immunoblotting. Detection of Yersinia substrates used rabbit polyclonal antisera raised against secreted YopE, YopD, and YopB (AgriSera AB, Vännäs, Sweden).
Quantification of Protein Production in Y. pseudotuberculosis
Relative protein levels were quantified from protein bands on scanned western blot X-ray films using the gel analysis tool in ImageJ (Schneider et al., 2012). In every case, the lane profile plot area of each protein band of interest was normalized to the corresponding protein band appearing in the same lane in the loading control blot.
Results
Amino Acid Identity Among the YscX and YscY Protein Families
In silico analysis has identified up to 9 core structural components in all T3SSs (Francis et al., 2004). Additional common components restricted to genetically related T3SS sub-families can also be found. A genetically related T3SSs exists in the notable human and animal pathogens Y. pestis, Yersinia enterocolitica, Y. pseudotuberculosis, and P. aeruginosa, the marine pathogens Aeromonas hydrophilia, A. salmonicida, V. harveyi, and V. parahaemolyticus, and the insect pathogen P. luminescens. Components common only to these systems include the YscX and YscY protein families. To ascertain genetic relatedness between representatives of the YscX and YscY protein families, amino acid sequences were retrieved from the sequenced genomes at http://www.ncbi.nlm.nih.gov/sites/entrez of the above mentioned bacteria and then aligned with ClustalW (http://www.ebi.ac.uk/Tools/clustalw/index.html). The evolutionary pattern was then examined by generating a phylogenetic tree rooted against the genetically related YscX-like and/or YscY-like protein from B. pertusis I475 by using the neighbor-joining method (Tamura et al., 2013; Bhattacharyya et al., 2017).
Based on amino acid sequence, the percent amino acid identity to YscX (122 aa) ranged from 54.1% over a 122 residue overlap (A. salmonicida) to 36.5% across a 126 residue overlap (V. parahaemolyticus) (Figure 1A). Apart from the near complete identity of YscX among the three Yersinia species, most relatedness occurred between the AscX proteins produced by the two Aeromonas species (~97% identical) and the VscX proteins produced by the two Vibrio species (~75%) (Figure 1A). Consistent with this, a phylogenetic tree analysis revealed that they each formed a separate clade distinct from other homologs (Figure 1C).
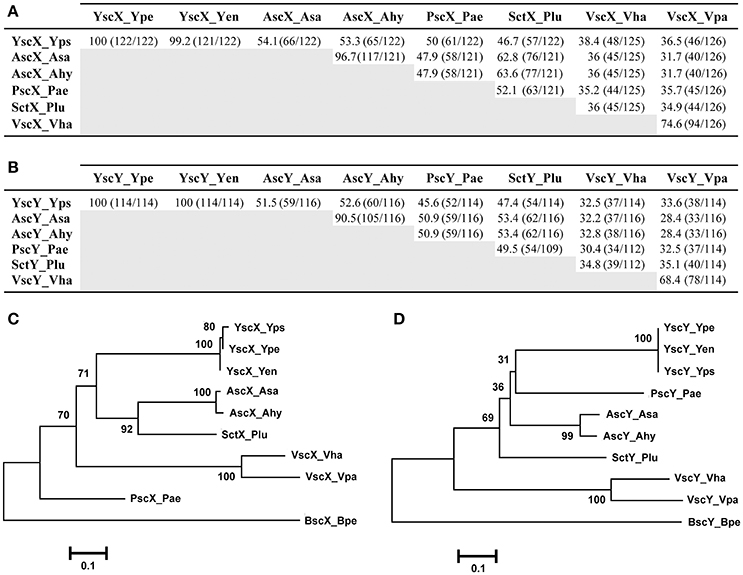
Figure 1. Amino acid sequence relatedness between the YscX and YscY protein families. Amino acid sequence identity was determined by BLASTP analysis (BLASTP 2.8.0+) (Altschul et al., 1997, 2005) for the YscX protein (A) and YscY protein (B) families. Numbers outside of parenthesis indicate percent amino acid identity. This is calculated from a ratio of the numbers inside parentheses; the left number indicating the number of identical residues, and the right number indication the total number of overlapping residues in a pairwise alignment. Representative sequences were retrieved from the NCBI genome database archived with the following GI reference numbers shown in parentheses: Yps, Yersinia pseudotuberculosis (YscX, 51593904 and YscY, 51593903); Ype, Yersinia pestis (YscX, 16082724 and YscY, 16082723); Yen, Yersinia enterocolitica (YscX, 4324343 and YscY, 4324342); Asa, Aeromonas salmonicida (AscX, 66947966 and AscY, 66947967); Ahy, Aeromonas hydrophilia (AscX, 46398260 and AscY, 46398261); Pae, Pseudomonas aeruginosa (PscX, 62865832 and PscY, 62865833); Plu, Photorhabdus luminescens (SctX, 36787060 and SctY, 36787059); Vha, Vibrio harveyi (VscX, 41834176 and VscY, 41834175); Vha, Vibrio parahaemolyticus (VscX, 28898438 and VscY, 28898437). Alignments of these sequences were used to generate phylogenetic trees depicting relationships between amino acid sequences of YscX-like (C) and YscY-like (D) family members. The neighbor-joining phylogenetic tree was generated using full length amino acid sequences of YscX and YscY homologs. Genetically related YscX-like and YscY-like protein sequences from Bordetella pertusis I475 were used as an outgroup. Values next to each branch node represent bootstrap percentages for 1,000 replicates. The scale bar represents the number of amino acid substitutions per site.
Similarly, amino acid sequence identity to YscY (114 aa) from Yersinia ranged between 52.6% (A. hydrophila) over a 116 residue overlap down to 32.5% (V. harveyi) over a 114 residue overlap (Figure 1B). Apart from the YscY proteins, the AscY proteins displayed most similarity to each other (~91% identical), followed by the VscY proteins (~68%), consistent with them forming separate phylogenetic clades (Figure 1D). In addition, the P. luminescens proteins SctX and SctY were most similar to the AscX and AscY counterparts from Aeromonas species (~63 and ~61%, respectively) (Figures 1A,B) and they all grouped close together on the phylogenetic tree (Figures 1C,D). Finally, among the three human pathogenic Yersinia species, YscY amino acid sequence was identical, whereas in YscX the amino acid glutamine at position 29 in both Y. pseudotuberculosis and Y. pestis existed as an aspartate residue in Y. enterocolitica (Figure S1). This subtle sequence divergence of YscX from Y. enterocolitica is reflected in the phylogenetic tree (Figure 1C).
In summary, this analysis reveals that YscX and YscY generally share most genetic relatedness with equivalent proteins from the two Aeromonas sp. Moreover, the YscX and YscY homologs that have diverged the most stem from the two Vibrio sp. This is in concordance with the phylogeny of non-flagella-T3SS of other core Ysc structural components (Troisfontaines and Cornelis, 2005; Romano et al., 2016).
Reciprocal Chaperone-Substrate Interactions
A cornerstone of YscX and YscY function appears to be their ability to interact with each other (Day and Plano, 2000; Bröms et al., 2005), and this seems true also of PscX and PscY function in P. aeruginosa T3S (Bröms et al., 2005; Yang et al., 2007). The yeast two-hybrid system has proven to be a reliable tool to demonstrate reciprocal interactions of YscX/PscX with YscY/PscY (Francis et al., 2001; Bröms et al., 2005). Thus, we utilized this approach to examine the interaction reciprocity between the representative YscX and YscY family members detailed in Figure 1. The PCR amplified yscX-related alleles were all separately cloned into the pGADT7 vector to establish fusions to the C-terminus of the GAL4 activation domain (AD). Additionally, the PCR amplified yscY-related alleles were all individually cloned into the pGBKT7 vector to establish fusions to the C-terminus of the GAL4 DNA binding domain (BD). The various pair-wise combinations of vectors expressing a AD and BD fusion were established in S. cerevisiae AH109 containing an ADE2 and HIS3 reporter gene. Interactions between YscX-like and YscY-like proteins were determined by the ability of transformed yeast to grow on either minimal media lacking histidine (but supplemented with 3-aminotriazole at a final concentration of 4 mM) or adenine. Interaction specificity was confirmed by the failure of a particular strain to grow on this same media after being cured of either the AD or BD expressing plasmid (data not shown). Reciprocal chaperone (YscY-like)-substrate (YscX-like) interactions could be observed for every possible combination (Figure 2, Figure S2). However, YscY-like variants derived from Y. pseudotuberculosis, P. aeruginosa, Aeromonas species, and P. luminescens all interacted better with YscX-like proteins also derived from these same bacteria. In contrast, these proteins consistently displayed weaker interactions with their more distantly related counterparts originating from Vibrio species (Figure 2, Figure S2). Importantly however, this was not a reflection of reduced protein expression, since cognate proteins derived from V. harveyi and V. parahaemolyticus demonstrated strong reciprocal binding with each other (Figure 2, Figure S2). Hence, reciprocal interactions between YscY- and YscX-like proteins are all generated in a yeast two-hybrid system. Conservation of these interactions is suggestive of a common functional requirement in T3S.
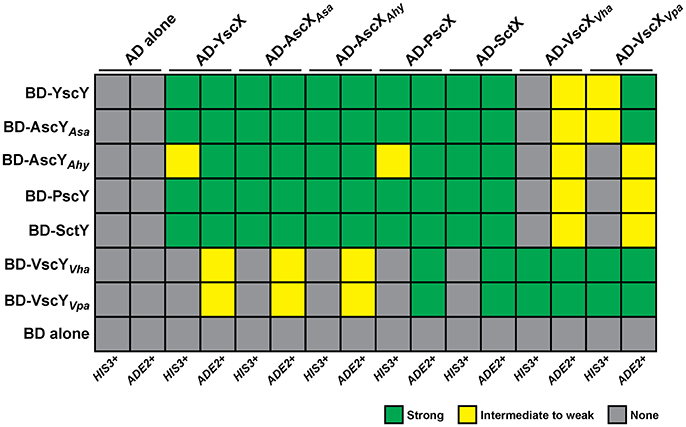
Figure 2. Summary of the reciprocal binding between members of the YscX and YscY protein families. Protein–protein interactions were determined using the yeast two-hybrid assay. YscX family members were fused to the GAL4 activation domain (in pGADT7), whereas YscY family members were fused to the GAL4 DNA binding domain (in pGBKT7). Pairwise transformations were performed in S. cerevisiae AH109 (Clontech Laboratories) that contained the HIS3 and ADE2 reporter genes. Strength of interactions were determined by the extent of growth on minimal medium devoid of histidine or adenine and recorded after day 4. Green shade indicates robust yeast growth (strong binary interaction), yellow shade reflects modest growth (moderate interaction) and gray shade specifies no growth (no interaction). Due to an intrinsic leakiness with the HIS3 reporter, 4 mM 3-aminotriazole was added to histidine dropout media to suppress false positives (James et al., 1996).
Reciprocal Ternary Interactions Between YscX, YscY and the Inner Membrane Component YscV Indicate Formation of a Conserved Substrate Secretion Sorting Complex
Together YscX and YscY co-interact with YscV, a highly conserved T3SS component that makes up part of the inner membrane export apparatus (Diepold et al., 2012). This YscX-YscY-YscV tripartite complex helps to facilitate the temporal export of early T3S substrates (Diepold et al., 2012). We utilized the yeast three hybrid approach to verify that YscX and YscY together form a ternary complex with YscV (Figure 3). Y190 yeast competent cells were co-transformed with pBridge vector carrying PADH1-BD-YscY/pMET25-YscX and pGADT7 carrying pADH1-AD-YscV. Transformed yeast cells were grown on SD minimal medium lacking histidine (but supplemented with 40 mM 3-aminotriazole) either in the absence or presence of methionine as a means to control output from the leaky MET25 promoter (Carlo et al., 2007). Expression of yscX under the control of MET25 promoter can be repressed in the presence of 1 mM methionine and activated in methionine-free media. Based on the extent of yeast growth in the absence of methionine, we could verify the formation of a YscY-YscX-YscV complex (Figure 3, panel 7). Importantly, the interaction was comparatively diminished upon growing yeast transformants on media containing 1 mM methionine. As expected, YscX did not bind to YscV in the absence of YscY (unpublished data), nor did YscY bind to YscV in the absence of YscX (Figure 3, panel 5). Moreover, transformation of empty vectors, YscV or YscX-YscY alone did not permit auto-activation of HIS3 reporter gene. These data demonstrated the suitability of the yeast three hybrid system for detecting the YscY-YscX-YscV interaction. Hence, we then tested if YscY-like, YscX-like, and YscV-like proteins from P. aeruginosa, A. hydrophila, V. parahaemolyticus, and P. luminescens have evolved to maintain the tripartite interaction with their specific counterparts. A significant amount of interaction between the cognate partners was evident in all YscY, YscX, YscV homologs tested (Figure 3, panel 16–19). Given the ability of YscY, YscX, and YscV homologs to maintain tripartite interactions, we then tested the possibility of reciprocal interactions involving the different combinations of family members. A reciprocal interaction between the members of ternary complex was observed in all combinations (Figure 3, panel 8–15). Thus, conservation of this interaction suggests that YscY-like, YscX-like, and YscV-like proteins have coevolved to perform a common T3S function, which is to facilitate the temporal export of early T3S substrates (Diepold et al., 2012).
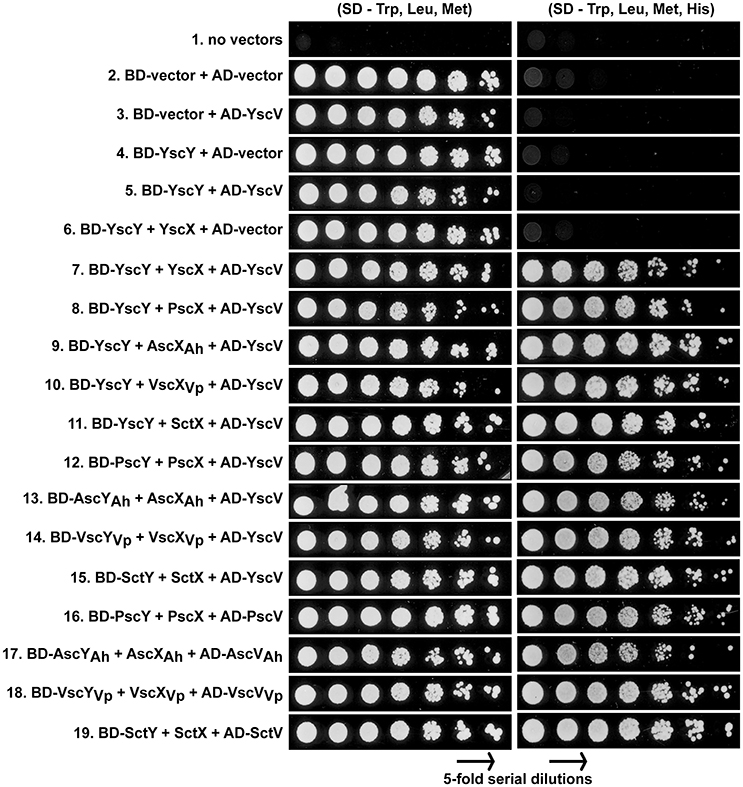
Figure 3. Reciprocal interactions between members of YscX, YscY, and YscV protein families. Different protein members of the YscY family were produced as recombinant GAL4 binding domain fusions from the pBridge vector. From this vector were also produced native forms of YscX protein family members to act as bridging proteins. Different protein members of the YscV family were produced as recombinant GAL4 activation domain fusions from the pGADT7 vector. As indicated, pairwise combinations of pBridge and pGADT7 derivatives were co-transformed into the yeast strain S. cerevesiae Y190. Five-fold serial dilutions of transformed yeast were grown on a synthetic dropout (SD) agar plates lacking tryptophan (Trp) and leucine (Leu) for maintenance of the plasmid pairs and methionine (Met) to induce production of YscX-like proteins. The strength of interaction was studied by growing yeast in equivalent media lacking Histidine (His). A concentration of 40 mM 3-aminotriazole was added to reduce leaky HIS3 expression. The result shown is a representative of three independent experiments.
Production of YscX and YscY Is a Strict Requirement for T3SS Assembly and Function in Y. pseudotuberculosis
Next we asked whether members of the YscX and YscY protein families could restore T3S function to a respective ΔyscX or ΔyscY null mutant of Y. pseudotuberculosis. To achieve this, the various PCR-amplified yscX- and yscY-like alleles were cloned either separately or together under the control of an IPTG inducible promoter in the expression vector pMMB67EHgm and then conjugated into the appropriate Y. pseudotuberculosis background. Following standardization against bacterial cell number, the level of T3S substrates associated with the total bacteria (a mixture of proteins contained within intact bacteria and secreted to the culture supernatant) and secreted free into the culture media was examined from bacteria grown in both T3S restrictive (BHI plus Ca2+) and permissive (BHI minus Ca2+) conditions. Expectantly, YscX when expressed in a ΔyscX null mutant (Figure 4A) and YscY when expressed in a ΔyscY null mutant (Figure 4B) restored functional T3S in Y. pseudotuberculosis. Moreover, YscX derived from Y. enterocolitica could readily complement the T3S defect in the ΔyscX null mutant (data not shown). Furthermore, dual expression of both YscX and YscY together in a ΔyscX, yscY double mutant also restored functional T3S (Figure 5). On the other hand, no expressed YscX-like or YscY-like protein could restore functional T3S to a ΔyscX null mutant (Figure 4A) or to a ΔyscY null mutant (Figure 4B), respectively. Additionally, co-expression of the cognate substrate-chaperone pair did not help to restore T3S to a ΔyscX, yscY double mutant (Figure 5). These data are consistent with an earlier study which concluded that PscX and PscY from P. aeruginosa could not complement the Yop synthesis and secretion defect apparent in a Y. pseudotuberculosis mutant lacking the yscX and yscY alleles respectively (Bröms et al., 2005). Taken all together, these data suggest that YscX-YscY interplay has functional consequences unique to T3S by Yersinia.
Figure 4. Assessing complementation of YscX and/or YscY function in Yersinia type III secretion. Bacteria were grown in BHI medium under secretion-permissive conditions (absence of Ca2+). Proteins contained within intact bacteria and secreted to the culture medium (Total) or secreted free to the extracellular medium (Supernatant) were fractionated on a 12% SDS-PAGE and analyzed by immunoblotting using polyclonal rabbit anti-YopB, anti-YopD and anti-YopE antiserum. IPTG was added to a final concentration of 0.4 mM where indicated. (A) Parent Y. pseudotuberculosis and ΔyscX complemented with pMMB67EHgm or pMMB67EHgm-encoded YscX family members. Strains: Parent (YPIII/pIB102); Complemented YPIII/pIB102, pMMB67EHgm (Vector);ΔyscX (YPIII/pIB880); complemented YPIII/pIB880, pMMB67EHgm (Vector); complemented YPIII/pIB880, pJEB291 (YscX+); complemented YPIII/pIB880, pJEB295 (PscX+); complemented YPIII/pIB880, pMF720 (); complemented YPIII/pIB880, pMF722 (); complemented YPIII/pIB880, pMF724 (); complemented YPIII/pIB880, pMF725 (); complemented YPIII/pIB880, pMF727 (SctX+). (B) Parent Y. pseudotuberculosis and ΔyscY complemented with pMMB67EHgm or pMMB67EHgm-encoded YscY family members. Strains: Parent (YPIII/pIB102); ΔyscY (YPIII/pIB890); complemented YPIII/pIB890, pMMB67EHgm (Vector); complemented YPIII/pIB890, pJEB292 (YscY+); complemented YPIII/pIB890, pJEB296 (PscY+); complemented YPIII/pIB890, pMF721 (); complemented YPIII/pIB890, pMF723 (); complemented YPIII/pIB890, pMF796 (); complemented YPIII/pIB890, pMF726 (); complemented YPIII/pIB890, pMF728 (SctY+). Molecular mass values shown in parentheses were deduced from primary amino acid sequences.
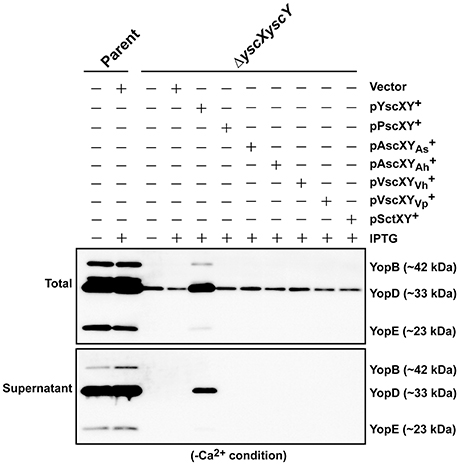
Figure 5. Assessing functional complementation of YscX and YscY by dual expression of substrate and chaperone homologs. Bacteria were grown in BHI medium under secretion-permissive conditions (absence of Ca2+). Proteins contained within intact bacteria and secreted to the culture medium (Total) or secreted free into the extracellular medium (Supernatant) were fractionated on a 12% SDS-PAGE and analyzed by immunoblotting using polyclonal rabbit anti-YopB, anti-YopD, and anti-YopE antiserum. IPTG was added to a final concentration of 0.4 mM where indicated. Strains: Parent (YPIII/pIB102); ΔyscXyscY (YPIII/pIB881); complemented YPIII/pIB881, pMMB67EHgm (Vector);complemented YPIII/pIB881, pJEB340 (YscX, YscY+); complemented YPIII/pIB881, pJEB335 (PscX, PscY+); complemented YPIII/pIB881, pMF733 (AscXAs, ); complemented YPIII/pIB881, pMF734 (AscXAh, ); complemented YPIII/pIB881, pMF797 (VscXVh, ); complemented YPIII/pIB881, pMF735 (VscXVp, ); complemented YPIII/pIB881, pMF736 (SctX, SctY+). Molecular mass values shown in parentheses were deduced from primary amino acid sequences.
Codon Optimization Fails to Improve the Complementation Efficiency of Non-endogenous yscX- and yscY-Like Alleles
Complementation may have failed because the non-cognate alleles were not sufficiently expressed. Due to the absence of available antibodies, production of YscX-like and YscY-like proteins in Y. pseudotuberculosis was assessed by appending an N-terminal FLAG™ epitope, followed by immunoblotting using an anti-FLAG™ monoclonal antibody. Accumulated levels of recombinant PscX, VscXVp, SctX and to a much lesser extent VscXVh could be detected (Figure S3A). Additionally, accumulated levels of recombinant PscY, VscXVh, VscXVp, and to a lesser extent SctY could be detected (Figure S3B). However, accumulated levels of all four recombinant Asc proteins were undetectable under these assay conditions (Figure S3). A number of reasons could contribute to the wide disparity in heterologous protein production in Yersinia, although it can often be attributed to species-specific GC content and species-specific codon usage (Gustafsson et al., 2004; Yu et al., 2015; Zhou et al., 2016). Indeed, the percent GC content of the individual genomes targeted here do vary considerably—Y. pseudotuberculosis (47.6% GC content), Aeromonas sp. (61.6%), P. aeruginosa (66.6%), Vibrio sp. (45.4%), and P. luminescens (42.8%) (http://archaea.ucsc.edu/) (Schneider et al., 2006; Chan et al., 2012), and this variance can be assumed to tailor the codon usage within each individual allele that may be incompatible with optimal translation in Y. pseudotuberculosis (Gustafsson et al., 2004; Yu et al., 2015; Zhou et al., 2016).
Interestingly, an analysis of codon usage scores in the yscX and yscY-like gene families based on the codon adaption index (CAI) revealed the presence of codon usage biases of different genes (Figure S4). Hence, we synthesized commercially a series of synthetic alleles that were optimized for production in Y. pseudotuberculosis by actively considering (i) codon usage bias, (ii) GC content, and (iii) mRNA secondary structure. In all cases the decoded amino acid sequence remained identical to that encoded by the native gene sequences. In the optimized synthetic genes, the number of unfavorable codons was reduced by upgrading the CAI value to ~0.9 and by adjusting the GC content more closely resembling that of Yersinia (Figures S4, S5). Predicted mRNA secondary structure was not affected by optimization (data not shown). To assess protein production, we cloned both native and synthetically optimized gene sequences as N-terminal FLAG™-tagged variants into pMMB208 vector under the control of an IPTG-inducible promoter. Following introduction into Y. pseudotuberculosis and subsequent growth in BHI minus Ca2+, bacterial pellets standardized against bacterial cell number were examined for accumulation of FLAG™-tagged protein. Except for the SctX and VscYVh variants, codon optimization led to improved production of all the YscX-like and YscY-like variants with accumulated levels ranging between 1.3- and 7.0-fold more abundant then their native (non-optimized) counterparts (Figure S6). Interestingly, despite the successful efforts to codon optimize, accumulated levels of some recombinant proteins such as those originally derived from Aeromonas sp. still failed to reach a level close to that achieved for the native Yersinia proteins (Figure 6).
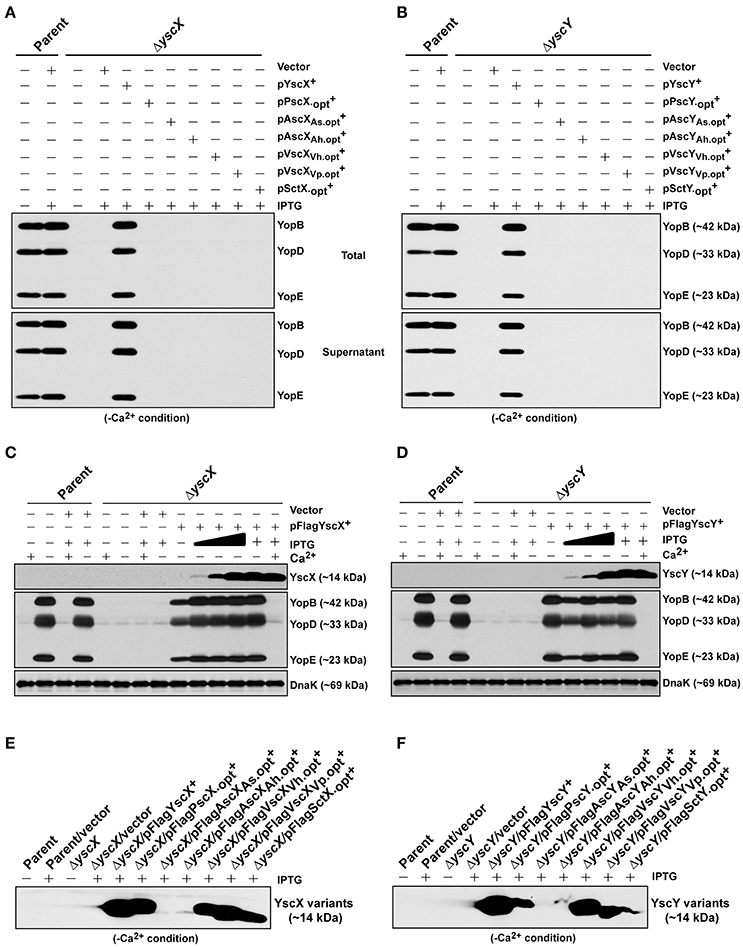
Figure 6. Assessing functional complementation by codon-optimized yscX- and yscY-like alleles. Synthesized Yop's associated with total fractions (proteins contained within intact bacteria and secreted to the culture medium) or with supernatant (secreted free to the extracellular medium) were analyzed in ΔyscX null-mutant complemented with YscX and codon-optimized YscX-like protein family members (A) or in ΔyscY null-mutant complemented with YscY and codon-optimized YscY-like protein family (B). Polyclonal anti-YopB, anti-YopD and anti-YopE were used for detection of Yops. The “+” symbol indicates the addition of IPTG to a final concentration of 0.4 mM. Proteins associated with bacterial pellet from Y. pseudotuberculosis cultures harboring either pFLAG-YscX (C) or pFLAG-YscY (D) grown in secretion-permissive condition (absence of Ca2+) and supplemented with increasing concentration of IPTG were analyzed by Western immunoblot using anti-FLAG™, anti-YopB, anti-YopD, anti-YopE, and anti-DnaK. The “−” symbol indicates absence of a particular component, while the “+” symbol indicates the presence of a particular component. In the case of IPTG, the “+” symbol indicates a final concentration of 0.4 mM IPTG was added, while the filled in graduation symbol reflects an incremental increase of IPTG according to the following final concentrations of 0.01, 0.02, and 0.1 mM. Steady state accumulation in the bacterial pellet of YscX protein family (E) and YscY protein family (F) produced from codon-optimized alleles following growth of Y. pseudotuberculosis in secretion-permissive conditions (absence of Ca2+). Protein was fractionated by SDS-PAGE and detected by immunoblot using monoclonal anti-FLAG™ antiserum. The “−” symbol indicates bacterial growth in the absence of IPTG, while “+” symbol indicates a final concentration of 0.4 mM was added. Strains: Parent (YPIII/pIB102); Complemented YPIII/pIB102, pMMB208 (Vector);ΔyscX (YPIII/pIB880); complemented YPIII/pIB880, pMMB208 (Vector); complemented YPIII/pIB880, pJMG242 (FLAG™-YscX+); complemented YPIII/pIB880, pJMG261(FLAG™-); complemented YPIII/pIB880, pJMG262 (FLAG™-); complemented YPIII/pIB880, pJMG263 (FLAG™-); complemented YPIII/pIB880, pJMG264 (FLAG™-); complemented YPIII/pIB880, pJMG265 (FLAG™-); complemented YPIII/pIB880, pMF266 (FLAG™-); ΔyscY (YPIII/pIB890); complemented YPIII/pIB890, pMMB208 (Vector); complemented YPIII/pIB890, pMF800 (FLAG™-YscY+); complemented YPIII/pIB890, pJMG267 (FLAG™-PscY.opt+); complemented YPIII/pIB890, pJMG268 (FLAG™-); complemented YPIII/pIB890, pJMG269 (FLAG™-); complemented YPIII/pIB890, pJMG270 (FLAG™-); complemented YPIII/pIB890, pJMG271 (FLAG™-); complemented YPIII/pIB890, pJMG272 (FLAG™-); complemented YPIII/pIB880, pJEB291 (YscX+); complemented YPIII/pIB880, pJMG293 (); complemented YPIII/pIB880, pJMG294 (); complemented YPIII/pIB880, pJMG295 (); complemented YPIII/pIB880, pJMG296 (); complemented YPIII/pIB880, pJMG297 (); complemented YPIII/pIB880, pJMG298 (); YPIII/pIB890, pJEB292 (YscY+); YPIII/pIB890, pJMG286 (); YPIII/pIB890, pJMG287 (AscYAs); YPIII/pIB890, pJMG288 (AscYAh); YPIII/pIB890, pJMG289 (VscYVh); YPIII/pIB890, pJMG290 (VscYVp); YPIII/pIB890, pJMG291 (). Molecular mass values shown in parentheses were deduced from primary amino acid sequences.
Significantly, this low level protein production was not due to an inferior transcriptional output because accumulated mRNA levels transcribed from all yscX-like (Figure S7A) and yscY-like (Figure S7B) genes was similar, which is consistent with transcription being driven by an identical promoter and transcriptional start site architecture in every case. Furthermore, the stability of accumulated ascX mRNA transcripts (Figure S7C) and ascY mRNA transcripts (Figure S7D) was equivalent to yscX and yscY mRNA transcripts. Thus, additional hitherto unknown regulatory elements must be limiting AscX and AscY production in Y. pseudotuberculosis.
Nevertheless, given that in most cases elevated production arose from codon optimization, we assessed if these codon optimized YscX-like and YscY-like protein variants could restore T3S to the ΔyscX null mutant and ΔyscY null-mutant, respectively. The optimized yscX-like and yscY-like genes were cloned individually into pMMB67EHgm and Yops production associated with total bacterial culture and Yops secretion associated with bacterial supernatants were monitored in secretion permissive growth conditions. Once again however, Yops production and secretion could be restored only when the ΔyscX null mutant was complemented with native YscX protein (Figure 6A) and ΔyscY null mutant with native YscY protein (Figure 6B).
To be sure that all other codon-optimized homologs were sufficiently produced to permit complementation, we compared these levels to the minimal amount of YscX and YscY sufficient to restore T3S to the ΔyscX and ΔyscY null mutants respectively. Critically, an undetectable amount of native YscX and YscY ectopically produced during growth in the presence of 0.02 mM of IPTG was already sufficient to restore high capacity T3S to the ΔyscX null mutant (Figure 6C) and ΔyscY null mutant (Figure 6D), respectively. These levels were quite noticeably much less than accumulated amounts detected for all of the non-complementing YscX-like (Figure 6E) and YscY-like (Figure 6F) variants when bacteria were grown in the presence of 0.4 mM IPTG. From this data we infer that insufficient expression of non-native YscX- and YscY-like family members cannot explain their inability to function in Y. pseudotuberculosis.
Codon Optimized Alleles Do Not Suppress a Functional Yersinia T3SS
Despite the YscX- and YscY-like homologs being unable to substitute for the respective loss of yscX or yscY in Y. pseudotuberculosis, we still wondered if they possess any activity at all when expressed in Y. pseudotuberculosis. To assess this, we examined for the ability of produced homologs to exert a dominant negative effect on Yersinia T3SS. All of the codon-optimized homologs of YscX and YscY under control of the IPTG inducible promoter of pMMB67EHgm were introduced in parental Y. pseudotuberculosis harboring an intact and fully functional T3SS. Firstly, we examined the protein expression profile of YscX and YscY family members in parental Yersinia background and found them to be comparable to what we had previously observed in the knockout strain backgrounds (data not shown). We then assessed the impact of this production on T3SS function as measured by Yops production and secretion. Significantly, neither the production of codon-optimized YscX-like (Figure 7A) nor YscY-like (Figure 7B) homologs interfered with T3SS activity by parental bacteria. The complete absence of any dominant negative effect implies that none of the YscX-like or YscY-like proteins are recognized by the Yersinia T3SS, which underscores that these proteins are truly non-functional in Yersinia bacteria. Taken altogether, our data indicates that an undisclosed feature(s) inherent in the native YscX and YscY products is necessary for proper T3S function in Yersinia, and one or more of these features cannot be substituted for by a genetically related gene from another bacterial source.
Figure 7. Assessing YscX-like and YscY-like homolog function in the presence of a fully intact type III secretion system. Bacteria were grown in BHI medium under secretion-permissive condition (absence of Ca2+). Proteins contained within intact bacteria and secreted to the culture medium (Total) or secreted free into the extracellular medium (Supernatant) were fractionated on a 12% SDS-PAGE and analyzed by immunoblotting using polyclonal rabbit anti-YopB, anti-YopD, and anti-YopE antiserum. IPTG was added to a final concentration of 0.4 mM where indicated. Strains: Parent (YPIII/pIB102); ΔyscX (YPIII/pIB880); Complemented YPIII/pIB102, pMMB67EHgm (Vector); Complemented YPIII/pIB102, pJEB291 (YscX+); complemented YPIII/pIB102, pJMG293 (); complemented YPIII/pIB102, pJMG294 (); complemented YPIII/pIB102, pJMG295 (); complemented YPIII/pIB102, pJMG296 (); complemented YPIII/pIB102, pJMG297 (); complemented YPIII/pIB102, pJMG298 (); ΔyscY (YPIII/pIB890); YPIII/pIB102, pJEB292 (YscY+); YPIII/pIB102, pJMG286 (); YPIII/pIB102, pJMG287 (AscYAs); YPIII/pIB102, pJMG288 (AscYAh); YPIII/pIB102, pJMG289 (VscYVh); YPIII/pIB102, pJMG290 (VscYVp); YPIII/pIB102, pJMG291 (). Molecular mass values shown in parentheses were deduced from primary amino acid sequences.
Discussion
On the basis that a number of core T3SS basal body components associated with bacterial inner and outer membranes are highly conserved, these have been the focus of studies to establish at least eight phylogenetically distinct T3SS sub-families among sequenced bacteria (Troisfontaines and Cornelis, 2005; Abby and Rocha, 2012). In addition, there exist critical components that are present only in a restricted T3SS sub-family. For example, the poorly studied proteins YscX and YscY exist only in the Ysc T3SS sub-family, and the prototype of this family is the plasmid encoded Ysc-Yop system of human pathogenic species of Yersinia. In this study, we have reported on the evolutionary conservation among the YscX and YscY protein families. We also explored their function in two ways. The first approach examined the binary interactions between all YscX-like and YscY-like family members, and the ability of this YscX-YscY complex to interact with the core basal body component YscV. The second approach examined the ability of all YscX-like and YscY-like family members to complement a type III secretion defect in Y. pseudotuberculosis brought about by a deletion of yscX and/or yscY. Our focus was on six different homologs sourced from P. aeruginosa, A. hydrophila, A. salmonicida, V. harveyi, V. parahaemolyticus and P. luminescens. Despite showing robust and interchangeable binary YscX-YscY interactions and ternary YscX-YscY-YscV interactions, none of the homologs could restore T3S to Y. pseudotuberculosis lacking functional YscX and/or YscY. Thus, it seems that this YscX-YscY-YscV system is functionally conserved in Ysc-like T3SSs, but that each set exhibits functional specificity to assemble their own apparatus, and this could include recognition of their own set of substrates.
Events allowing YscX to form stable complexes with the known cognate chaperone YscY, and for the YscX-YscY complex to interact with the core T3SS structural component YscV, are considered to constitute integral elements of T3SS assembly and function (Day and Plano, 2000; Diepold et al., 2012). Our n-hybrid analyses suggests that these interactions are a key attribute of all YscX-like, YscY-like and YscV-like homologs, serving an important T3SS assembly function that is evolutionarily conserved. In fact, the need to form a bipartite and a tripartite interaction is most likely an ancient role of these protein families, and maybe the reason why YscX and YscY could functionally substitute for the absence of PscX and PscY in mutants of P. aeruginosa (Bröms et al., 2005). However, merely having the ability to establish these known reciprocal interactions was clearly not enough to permit any of the homologs to mediate functional exchange in Y. pseudotuberculosis. Indeed this suggests that both YscX and YscY possess other molecular targets that are arguably unique to Ysc-Yop T3SS function. An approach to identify these putative targets within soluble bacterial lysate or solubilized bacterial membrane fractions would be to combine pull down technology using the Flag-YscX or Flag-YscY as a bait with mass spectroscopy detection. We can only assume that this need for additional molecular interactions arose as a consequence of Yersinia evolving the ability to thrive in a particular ecological niche. This notion is consistent with previous findings that correlate a diversification of bacterial T3SSs with known bacterial-host interactions (Abby and Rocha, 2012).
Sequence alignments revealed several obvious regions of uniqueness within the YscX and YscY amino acid sequence, respectively. At first glance, these are too numerous to perform targeted mutagenesis to screen for loss-of-function in the Yersinia proteins or gain-of-function in the heterologous proteins. Hence, we are now attempting to first map potentially critical functional domains unique to YscX and YscY, prior to embarking on a site-directed mutagenesis strategy. A series of chimeric alleles between yscX and yscX-like homologs as well as yscY and yscY-like homologs are being created. We proposed that by generating large N-terminal and C-terminal chimeric sets that contain progressively more native YscX sequence and native YscY sequence presents an alternative tool to define a minimal genetic content that confers to the chimera a functional competence for Y. pseudotuberculosis Ysc-Yop T3SS activity, which can then be subsequently confirmed using site-directed mutagenesis.
It is likely that such sequence would serve as an interaction interface for engaging with one or more unknown targets, making it useful as a bait to identify this unique contributor to T3SS activity in Y. pseudotuberculosis. Some unique non-reciprocal interactions are already known—YscY interacts with the translocator class chaperone LcrH/SycD in Y. pseudotuberculosis (Francis et al., 2001; Bröms et al., 2005), while PscY (termed Pcr4) interacts with Pcr1 (the equivalent of TyeA in Yersinia) in P. aeruginosa (Yang et al., 2007). However, neither of these two interactions are prerequisites for the physical assembly of their respective T3SSs, but rather function primarily in the fine-tuning of substrate secretion control. In particular, LcrH/SycD influences the ability of YopD to bind both yop mRNA and the 30S ribosomal subunit to modulate translation initiation of yop mRNA and accelerate its turnover (Anderson et al., 2002; Chen and Anderson, 2011; Kopaskie et al., 2013). The consequence of this LcrH-dependent YopD RNA binding activity is to post-transcriptionally regulate T3SS genes to impart a level of hierarchal secretion control. Significantly, a strain defective in LcrH does not loose capacity to secrete substrates (Wattiau et al., 1994; Francis et al., 2001; Edqvist et al., 2006), and neither do defined mutants in LcrH that are unable to interact with YscY (Francis et al., 2001; Bröms et al., 2005). Thus, LcrH-YscY complex formation is likely to feed into the post-transcriptionally active complex of YopD, LcrH, and yop mRNA with the sole purpose to further fine-tune temporal and spatial control of substrate secretion. It is not anticipated to assist in assembly of a secretion-competent apparatus per se. On this basis, we do not believe that the specificity of LcrH-YscY interaction in Yersinia is the reason why heterologous protein expression cannot support T3S. Hence, we will continue to pursue evidence for a unique non-reciprocal interaction involving YscX and YscY that is essential for the physical assembly of the Ysc-Yop T3SS, rather than one involved in substrate secretion control.
Through sequence alignments we did observe that only native YscX contained two cysteine residues in codon positions that were absolutely conserved. At this stage we do not know if these cysteine residues are reduced or participate in inter- or intra-disulfide bond formation. If they do participate in disulfide bond formation, this would be a unique feature of native YscX, and gives a reason as to why heterologous YscX-like proteins that lack these two conserved cysteines are not functional in Yersinia. It is well-established that disulfide bonds are an important structural element of the secretome of bacterial pathogens (De Geyter et al., 2016; Bocian-Ostrzycka et al., 2017). This is true also for human pathogenic Yersinia sp., where a role for thiol bonding is reported for the Ysc-Yop T3SS, the Caf1 chaperone-usher system, and the invasin autotransporter (Leong et al., 1993; Zav'yalov et al., 1997; Jackson and Plano, 1999; Mitchell et al., 2017). Thus, a logical next step is to use site-directed mutagenesis to investigate possible inter- or intra-disulfide bond formation in YscX.
Following completion of the needle assembly in Y. enterocolitica, YscX secretion into the extracellular milieu occurs (Diepold et al., 2012). However, the purpose of YscX secretion is enigmatic. Our ongoing studies have so far revealed that secretion of YscX is absolutely required for a functional T3SS in Y. pseudotuberculosis (unpublished data). One reason for YscX secretion might be to free up a complex of YscY-YscV to recognize and secrete other later subsets of T3S substrates, namely the translocators that are necessary to trigger the next stage of injectisome assembly and the host-modulating effectors that are injected into eukaryotic cells (Dewoody et al., 2013). In parallel, secreted YscX might comprise a minor component of the external needle, although experimental evidence to support this is completely lacking. Alternatively, secreted YscX may comprise a structural component within the needle assembly platform, such as via a cooperation with the YscI inner rod. Hence, to understand the role of YscX secretion in T3SS function is important. This pursuit is made all the more intriguing on the basis that PscY is the secreted component of the T3SS of P. aeruginosa, not PscX as one may have expected (Yang et al., 2007).
Given that all indicators point to the fact that YscX secretion is essential for T3SS function in Yersinia, it follows that a functional requirement for complementation of the ΔyscX deletion mutant would likely be secretion of the corresponding YscX-like homologs. Taking account of the already stated PscY secretion exception (Yang et al., 2007), we are aware of only one other YscX-like homolog that has been proven experimentally to be a T3SS substrate—the AscX substrate of A. salmonicida (Vanden Bergh et al., 2013). We attempted to examine secretion of all YscX-like proteins by parental Y. pseudotuberculosis containing a fully functional Ysc-Yop T3SS. However, this was complicated by the need to produce all YscX-like variants as a FLAG™ fusion to facilitate their immune-detection, for this tag may impact on fusion protein secretion efficiency. Indeed, the amount of FLAG™-YscX secreted was only a fraction of the amount that was produced (data not shown). Furthermore, we could not corroborate the findings of Vanden Bergh et al. (2013), for the yields of AscX were routinely too low to assess its secretion. Consistent with this, most other FLAG™-tagged variants were not even detected in the secretion fraction, including PscX that is not a T3S substrate (Yang et al., 2007) (data not shown). The exception to this was the observation that FLAG™-VscXVp and FLAG™-SctX were active substrates of a fully intact Ysc-T3SS of Y. pseudotuberculosis (data not shown). Hence, at present we cannot establish a clear correlation between lack of secretion and lack of functional complementation.
One final observation of interest stemming from this study concerned the poor production of the two AscX proteins and the two AscY proteins in the Y. pseudotuberculosis background. This occurred despite (1) YscX and AscX as well as YscY and AscY sharing a sequence identity above the 50 percentile, (2) an abundance of comparatively stable mRNA being initiated from an identical promoter used to support the expression of all other genes studied in this report, and (3) the use of synthetic genes that had been codon optimized to maximize efficient translation in Y. pseudotuberculosis. Hence, some hitherto unknown phenomenon is responsible for the lack of heterologous production of these proteins in Yersinia. A straightforward reason for this is that they are normally stabilized by species-specific protein–protein interactions. The YscY chaperone is thought to function as an YscX stabilizer (Iriarte and Cornelis, 1999; Day and Plano, 2000). Indeed, this motivated the dual expression complementation experiment performed in this study. However, co-expression of AscX with cognate AscY did not increase the production of either protein. A conclusion from this work is that AscY alone is probably insufficient for AscX stabilization, meaning that the complex of AscX-AscY requires stabilizing influence from additional protein–protein interactions not supported by expression in a heterologous host like Yersinia. Another reason for poor AscX and AscY yield could also be that some aspect of mRNA sequence and/or structure defined by the ascX and ascY open reading frames influences that rate of their translation in Yersinia. As a means to pin-point the reason for low yield, we are currently analyzing allelic chimeras between ascX and yscX as well as ascY and yscY. This approach has the possibility to locate the genetic regions in AscX and AscY that are either (1) the interface for species-specific protein–protein interactions that are necessary for protein stability, or (2) the site of novel posttranscriptional control mechanisms that have potential to influence the spatial and temporal control of T3SS.
In summary, on the basis of complementation assays measuring the capacity for in vitro synthesis and secretion of Yops, it appears that the function of native YscX and YscY is evolutionary optimized to contribute unique and essential information for the physical assembly of the Ysc-Yop T3SS by pathogenic Yersinia sp. Thus, the challenge that lies ahead is to define the function of both YscX and YscY, and to decipher whether this function requires the recruitment of additional molecular components. In contrast, from this work we now know that the established bipartite YscX-YscY interaction and the tripartite YscX-YscY-YscV interaction is not a unique feature restricted only to native components of Yersinia. Rather, these interactions are preserved among all heterologous YscX-like and YscY-like family members, and this probably reflects an ancient and evolutionarily conserved universal function of these proteins.
Author Contributions
JG, AA, MKF, TC, and MSF conceived and planned the experiments. SC and MSF contributed essential materials. JG, AA, MKF, and TC carried out the experiments and contributed to sample preparation. JG, AA, MKF, TC, SC, AZ, and MSF contributed to the interpretation of the results. JG and MSF took the lead in writing the manuscript. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Funding
This work has been supported by grants from the Swedish Research Council (2009-5628 and 2014-2105), the Foundation for Medical Research at Umeå University and the Faculty of Science and Technology at Umeå University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was performed within the framework of the Umeå Centre for Microbial Research at Umeå University. We appreciate the contributions made by undergraduate students of the Umeå University Life science and Biomedicine programs. Andreas Diepold is acknowledged for valuable discussion surrounding YscX and YscY function. We express gratitude to Hans Wolf-Watz for antibodies to YopE, YopD, and YopB. We are also grateful to Joachim Frey, Juan Tomás, Debra Milton, Tetsuya Iida, and David Clarke for the gift of strains and/or chromosomal DNA.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00080/full#supplementary-material
References
Abby, S. S., and Rocha, E. P. (2012). The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 8:e1002983. doi: 10.1371/journal.pgen.1002983
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Altschul, S. F., Wootton, J. C., Gertz, E. M., Agarwala, R., Morgulis, A., Schaffer, A. A., et al. (2005). Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x
Amer, A. A., Gurung, J. M., Costa, T. R., Ruuth, K., Zavialov, A. V., Forsberg, A., et al. (2016). YopN and TyeA hydrophobic contacts required for regulating Ysc-Yop Type III secretion activity by Yersinia pseudotuberculosis. Front. Cell. Infect. Microbiol. 6:66. doi: 10.3389/fcimb.2016.00066
Anderson, D. M., Ramamurthi, K. S., Tam, C., and Schneewind, O. (2002). YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J. Bacteriol. 184, 1287–1295. doi: 10.1128/JB.184.5.1287-1295.2002
Bertani, G. (2004). Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186, 595–600. doi: 10.1128/JB.186.3.595-600.2004
Bhattacharyya, C., Bakshi, U., Mallick, I., Mukherji, S., Bera, B., and Ghosh, A. (2017). Genome-guided insights into the plant growth promotion capabilities of the physiologically versatile Bacillus aryabhattai strain AB211. Front. Microbiol. 8:411. doi: 10.3389/fmicb.2017.00411
Blocker, A., Gounon, P., Larquet, E., Niebuhr, K., Cabiaux, V., Parsot, C., et al. (1999). The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J. Cell Biol. 147, 683–693. doi: 10.1083/jcb.147.3.683
Blocker, A., Jouihri, N., Larquet, E., Gounon, P., Ebel, F., Parsot, C., et al. (2001). Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol. Microbiol. 39, 652–663. doi: 10.1046/j.1365-2958.2001.02200.x
Bocian-Ostrzycka, K. M., Grzeszczuk, M. J., Banas, A. M., and Jagusztyn-Krynicka, E. K. (2017). Bacterial thiol oxidoreductases - from basic research to new antibacterial strategies. Appl. Microbiol. Biotechnol. 101, 3977–3989. doi: 10.1007/s00253-017-8291-8
Bröms, J. E., Edqvist, P. J., Carlsson, K. E., Forsberg, Å., and Francis, M. S. (2005). Mapping of a YscY binding domain within the LcrH chaperone that is required for regulation of Yersinia type III secretion. J. Bacteriol. 187, 7738–7752. doi: 10.1128/JB.187.22.7738-7752.2005
Bröms, J. E., Edqvist, P. J., Forsberg, Å., and Francis, M. S. (2006). Tetratricopeptide repeats are essential for PcrH chaperone function in Pseudomonas aeruginosa type III secretion. FEMS Microbiol. Lett. 256, 57–66. doi: 10.1111/j.1574-6968.2005.00099.x
Broms, J. E., Forslund, A. L., Forsberg, A., and Francis, M. S. (2003). PcrH of Pseudomonas aeruginosa is essential for secretion and assembly of the type III translocon. J. Infect. Dis. 188, 1909–1921. doi: 10.1086/379898
Buttner, C. R., Sorg, I., Cornelis, G. R., Heinz, D. W., and Niemann, H. H. (2008). Structure of the Yersinia enterocolitica type III secretion translocator chaperone SycD. J. Mol. Biol. 375, 997–1012. doi: 10.1016/j.jmb.2007.11.009
Buttner, D. (2012). Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. doi: 10.1128/MMBR.05017-11
Carlo, J. M., Osman, A., Niles, E. G., Wu, W., Fantappie, M. R., Oliveira, F. M., et al. (2007). Identification and characterization of an R-Smad ortholog (SmSmad1B) from Schistosoma mansoni. FEBS J. 274, 4075–4093. doi: 10.1111/j.1742-4658.2007.05930.x
Carlsson, K. E., Liu, J., Edqvist, P. J., and Francis, M. S. (2007a). Extracytoplasmic-stress-responsive pathways modulate type III secretion in Yersinia pseudotuberculosis. Infect. Immun. 75, 3913–3924. doi: 10.1128/IAI.01346-06
Carlsson, K. E., Liu, J., Edqvist, P. J., and Francis, M. S. (2007b). Influence of the Cpx extracytoplasmic-stress-responsive pathway on Yersinia sp.-eukaryotic cell contact. Infect. Immun. 75, 4386–4399. doi: 10.1128/IAI.01450-06
Chan, P. P., Holmes, A. D., Smith, A. M., Tran, D., and Lowe, T. M. (2012). The UCSC archaeal genome browser: 2012 update. Nucleic Acids Res. 40, D646–D652. doi: 10.1093/nar/gkr990
Chen, Y., and Anderson, D. M. (2011). Expression hierarchy in the Yersinia type III secretion system established through YopD recognition of RNA. Mol. Microbiol. 80, 966–980. doi: 10.1111/j.1365-2958.2011.07623.x
Day, J. B., and Plano, G. V. (2000). The Yersinia pestis YscY protein directly binds YscX, a secreted component of the type III secretion machinery. J. Bacteriol. 182, 1834–1843. doi: 10.1128/JB.182.7.1834-1843.2000
De Geyter, J., Tsirigotaki, A., Orfanoudaki, G., Zorzini, V., Economou, A., and Karamanou, S. (2016). Protein folding in the cell envelope of Escherichia coli. Nat Microbiol 1, 16107. doi: 10.1038/nmicrobiol.2016.107
Dewoody, R. S., Merritt, P. M., and Marketon, M. M. (2013). Regulation of the Yersinia type III secretion system: traffic control. Front. Cell. Infect. Microbiol. 3:4. doi: 10.3389/fcimb.2013.00004
Diepold, A., Wiesand, U., Amstutz, M., and Cornelis, G. R. (2012). Assembly of the Yersinia injectisome: the missing pieces. Mol. Microbiol. 85, 878–892. doi: 10.1111/j.1365-2958.2012.08146.x
Edqvist, P. J., Bröms, J. E., Betts, H. J., Forsberg, Å., Pallen, M. J., and Francis, M. S. (2006). Tetratricopeptide repeats in the type-III-secretion chaperone, LcrH: their role in substrate binding and secretion. Mol. Microbiol. 59, 31–44. doi: 10.1111/j.1365-2958.2005.04923.x
Erhardt, M., Namba, K., and Hughes, K. T. (2010). Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb. Perspect. Biol. 2:a000299. doi: 10.1101/cshperspect.a000299
Francis, M. S., Aili, M., Wiklund, M. L., and Wolf-Watz, H. (2000). A study of the YopD-LcrH interaction from Yersinia pseudotuberculosis reveals a role for hydrophobic residues within the amphipathic domain of YopD. Mol. Microbiol. 38, 85–102. doi: 10.1046/j.1365-2958.2000.02112.x
Francis, M. S., Lloyd, S. A., and Wolf-Watz, H. (2001). The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol. Microbiol. 42, 1075–1093. doi: 10.1046/j.1365-2958.2001.02702.x
Francis, M. S., Schesser, K., Forsberg, Å., and Wolf-Watz, H. (2004). “Type III secretion systems in animal- and plant-interacting bacteria,” in Cellular Microbiology, 2nd Edn., eds P. Cossart, P. Boquet, S. Normark, and R. Rappuoli (Washington, DC: American Society for Microbiology Press), 361–392.
Glass, F., Hartel, B., Zehrmann, A., Verbitskiy, D., and Takenaka, M. (2015). MEF13 requires MORF3 and MORF8 for RNA editing at eight targets in mitochondrial mRNAs in Arabidopsis thaliana. Mol. Plant 8, 1466–1477. doi: 10.1016/j.molp.2015.05.008
Gustafsson, C., Govindarajan, S., and Minshull, J. (2004). Codon bias and heterologous protein expression. Trends Biotechnol. 22, 346–353. doi: 10.1016/j.tibtech.2004.04.006
Her, C., Wu, X., Griswold, M. D., and Zhou, F. (2003). Human MutS homologue MSH4 physically interacts with von Hippel-Lindau tumor suppressor-binding protein 1. Cancer Res. 63, 865–872. Available online at: http://cancerres.aacrjournals.org/content/63/4/865
Iriarte, M., and Cornelis, G. R. (1999). Identification of SycN, YscX, and YscY, three new elements of the Yersinia yop virulon. J. Bacteriol. 181, 675–680.
Jackson, M. W., and Plano, G. V. (1999). DsbA is required for stable expression of outer membrane protein YscC and for efficient Yop secretion in Yersinia pestis. J. Bacteriol. 181, 5126–5130.
James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436.
Jeters, R. T., Wang, G. R., Moon, K., Shoemaker, N. B., and Salyers, A. A. (2009). Tetracycline-associated transcriptional regulation of transfer genes of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 191, 6374–6382. doi: 10.1128/JB.00739-09
Job, V., Mattei, P. J., Lemaire, D., Attree, I., and Dessen, A. (2010). Structural basis of chaperone recognition of type III secretion system minor translocator proteins. J. Biol. Chem. 285, 23224–23232. doi: 10.1074/jbc.M110.111278
Journet, L., Agrain, C., Broz, P., and Cornelis, G. R. (2003). The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302, 1757–1760. doi: 10.1126/science.1091422
Kim, J. S., Kim, B. H., Jang, J. I., Eom, J. S., Kim, H. G., Bang, I. S., et al. (2014). Functional insight from the tetratricopeptide repeat-like motifs of the type III secretion chaperone SicA in Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 350, 146–153. doi: 10.1111/1574-6968.12315
Kimbrough, T. G., and Miller, S. I. (2000). Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U.S.A. 97, 11008–11013. doi: 10.1073/pnas.200209497
Kopaskie, K. S., Ligtenberg, K. G., and Schneewind, O. (2013). Translational regulation of Yersinia enterocolitica mRNA encoding a type III secretion substrate. J. Biol. Chem. 288, 35478–35488. doi: 10.1074/jbc.M113.504811
Kubori, T., Matsushima, Y., Nakamura, D., Uralil, J., Lara-Tejero, M., Sukhan, A., et al. (1998). Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605. doi: 10.1126/science.280.5363.602
Leong, J. M., Morrissey, P. E., and Isberg, R. R. (1993). A 76-amino acid disulfide loop in the Yersinia pseudotuberculosis invasin protein is required for integrin receptor recognition. J. Biol. Chem. 268, 20524–20532.
Lunelli, M., Lokareddy, R. K., Zychlinsky, A., and Kolbe, M. (2009). IpaB-IpgC interaction defines binding motif for type III secretion translocator. Proc. Natl. Acad. Sci. U.S.A. 106, 9661–9666. doi: 10.1073/pnas.0812900106
Marlovits, T. C., Kubori, T., Sukhan, A., Thomas, D. R., Galan, J. E., and Unger, V. M. (2004). Structural insights into the assembly of the type III secretion needle complex. Science 306, 1040–1042. doi: 10.1126/science.1102610
Mitchell, A., Tam, C., Elli, D., Charlton, T., Osei-Owusu, P., Fazlollahi, F., et al. (2017). Glutathionylation of Yersinia pestis LcrV and its effects on plague pathogenesis. MBio 8:e00646-17. doi: 10.1128/mBio.00646-17
Mueller, C. A., Broz, P., Muller, S. A., Ringler, P., Erne-Brand, F., Sorg, I., et al. (2005). The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310, 674–676. doi: 10.1126/science.1118476
Okan, N. A., Bliska, J. B., and Karzai, A. W. (2006). A Role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2:e6. doi: 10.1371/journal.ppat.0020006
Osborne, S. E., and Coombes, B. K. (2011). Expression and secretion hierarchy in the nonflagellar type III secretion system. Future Microbiol. 6, 193–202. doi: 10.2217/fmb.10.172
Pallen, M. J., Beatson, S. A., and Bailey, C. M. (2005). Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a Darwinian perpective. FEMS Microbiol. Rev. 29, 201–229. doi: 10.1016/j.femsre.2005.01.001
Pallen, M. J., Francis, M. S., and Futterer, K. (2003). Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol. Lett. 223, 53–60. doi: 10.1016/S0378-1097(03)00344-6
Portaliou, A. G., Tsolis, K. C., Loos, M. S., Zorzini, V., and Economou, A. (2016). Type III secretion: building and operating a remarkable nanomachine. Trends Biochem. Sci. 41, 175–189. doi: 10.1016/j.tibs.2015.09.005
Radics, J., Konigsmaier, L., and Marlovits, T. C. (2014). Structure of a pathogenic type 3 secretion system in action. Nat. Struct. Mol. Biol. 21, 82–87. doi: 10.1038/nsmb.2722
Romano, S., Fernandez-Guerra, A., Reen, F. J., Glockner, F. O., Crowley, S. P., O'sullivan, O., et al. (2016). Comparative genomic analysis reveals a diverse repertoire of genes involved in prokaryote-eukaryote interactions within the Pseudovibrio genus. Front. Microbiol. 7:387. doi: 10.3389/fmicb.2016.00387
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schneider, K. L., Pollard, K. S., Baertsch, R., Pohl, A., and Lowe, T. M. (2006). The UCSC archaeal genome browser. Nucleic Acids Res. 34, D407–410. doi: 10.1093/nar/gkj134
Sekiya, K., Ohishi, M., Ogino, T., Tamano, K., Sasakawa, C., and Abe, A. (2001). Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure. Proc. Natl. Acad. Sci. U.S.A. 98, 11638–11643. doi: 10.1073/pnas.191378598
Singh, S. K., Boyle, A. L., and Main, E. R. (2013). LcrH, a class II chaperone from the type three secretion system, has a highly flexible native structure. J. Biol. Chem. 288, 4048–4055. doi: 10.1074/jbc.M112.395889
Sukhan, A., Kubori, T., Wilson, J., and Galán, J. E. (2001). Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183, 1159–1167. doi: 10.1128/JB.183.4.1159-1167.2001
Tamano, K., Aizawa, S., Katayama, E., Nonaka, T., Imajoh-Ohmi, S., Kuwae, A., et al. (2000). Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19, 3876–3887. doi: 10.1093/emboj/19.15.3876
Tamano, K., Aizawa, S., and Sasakawa, C. (2002). Purification and detection of Shigella type III secretion needle complex. Methods Enzymol. 358, 385–392. doi: 10.1016/S0076-6879(02)58104-0
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Troisfontaines, P., and Cornelis, G. R. (2005). Type III secretion: more systems than you think. Physiology 20, 326–339. doi: 10.1152/physiol.00011.2005
Vanden Bergh, P., Heller, M., Braga-Lagache, S., and Frey, J. (2013). The Aeromonas salmonicida subsp. salmonicida exoproteome: determination of the complete repertoire of Type-Three Secretion System effectors and identification of other virulence factors. Proteome Sci. 11, 42. doi: 10.1186/1477-5956-11-42
Wattiau, P., Bernier, B., Deslee, P., Michiels, T., and Cornelis, G. R. (1994). Individual chaperones required for Yop secretion by Yersinia. Proc. Natl. Acad. Sci. U.S.A. 91, 10493–10497. doi: 10.1073/pnas.91.22.10493
Yang, H., Shan, Z., Kim, J., Wu, W., Lian, W., Zeng, L., et al. (2007). Regulatory role of PopN and its interacting partners in type III secretion of Pseudomonas aeruginosa. J. Bacteriol. 189, 2599–2609. doi: 10.1128/JB.01680-06
Yu, C. H., Dang, Y., Zhou, Z., Wu, C., Zhao, F., Sachs, M. S., et al. (2015). Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell 59, 744–754. doi: 10.1016/j.molcel.2015.07.018
Zav'yalov, V. P., Chernovskaya, T. V., Chapman, D. A., Karlyshev, A. V., MacIntyre, S., Zavialov, A. V., et al. (1997). Influence of the conserved disulphide bond, exposed to the putative binding pocket, on the structure and function of the immunoglobulin-like molecular chaperone Caf1M of Yersinia pestis. Biochem J. 324 (Pt 2), 571–578. doi: 10.1042/bj3240571
Keywords: T3S chaperone, secretion hierarchy, substrate sorting, LcrH/SycD, YscV, protein–protein interaction
Citation: Gurung JM, Amer AAA, Francis MK, Costa TRD, Chen S, Zavialov AV and Francis MS (2018) Heterologous Complementation Studies With the YscX and YscY Protein Families Reveals a Specificity for Yersinia pseudotuberculosis Type III Secretion. Front. Cell. Infect. Microbiol. 8:80. doi: 10.3389/fcimb.2018.00080
Received: 16 December 2017; Accepted: 28 February 2018;
Published: 16 March 2018.
Edited by:
Sophie Bleves, Aix-Marseille Université, FranceReviewed by:
Luís Jaime Mota, Faculdade de Ciências e Tecnologia da Universidade Nova de Lisboa, PortugalEric Faudry, CEA Grenoble, France
Copyright © 2018 Gurung, Amer, Francis, Costa, Chen, Zavialov and Francis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew S. Francis, matthew.francis@umu.se
†Present Address: Tiago R. D. Costa, MRC Centre for Molecular Bacteriology and Infection, Department of Life Sciences, Imperial College London, London, United Kingdom
 Jyoti M. Gurung
Jyoti M. Gurung Ayad A. A. Amer
Ayad A. A. Amer Monika K. Francis
Monika K. Francis Tiago R. D. Costa
Tiago R. D. Costa Shiyun Chen
Shiyun Chen Anton V. Zavialov
Anton V. Zavialov Matthew S. Francis
Matthew S. Francis