Sialidase Deficiency in Porphyromonas gingivalis Increases IL-12 Secretion in Stimulated Macrophages Through Regulation of CR3, IncRNA GAS5 and miR-21
- 1Department of Periodontics, School of Stomatology, China Medical University, Shenyang, China
- 2Shenyang Medical College, Shenyang, China
- 3Department of Periodontics, Dalian Stomatology Hospital, Dalian Shi, China
- 4Liaoning Province Key Laboratory of Oral Diseases, Shenyang, China
- 5Liaoning Province Translational Medicine Research Center of Oral Diseases, Shenyang, China
Porphyromonas gingivalis (P. gingivalis) is a major periodontal pathogen that can induce an immune response leading to a destructive inflammatory process. During the inflammatory process, interleukin-12 (IL-12) is secreted, correlating with bacterial clearance by macrophages. Bacterial sialidase has recently been shown to influence the synthesis and modification of the macromolecules on its surface, and is associated with the interaction between bacteria and host cells. We have previously constructed a P. gingivalis sialidase gene mutant strain in P. gingivalis W83 (ΔPG0352) and found that ΔPG0352 showed less pathogenicity than the wild-type strain. In this study, U937-differentiated macrophages were stimulated by P. gingivalis W83, ΔPG0352, or PG0352 complemented strain (comΔPG0352). Transmission electron microscopy showed that P. gingivalis caused a loss of membrane integrity in macrophages and the intracellular bacteria were enclosed within endocytic vacuoles. The expression of both IL-12p35 and IL-12p40 genes and the levels of IL-12p70 were significantly higher in U937 stimulated by ΔPG0352 than in those with P. gingivalis W83 and comΔPG0352. In order to explain why ΔPG0352 induced more IL-12 in macrophages, immunofluorescence assays, PCR arrays, and gene silence or overexpression experiments were carried out. Immunofluorescence assays showed that ΔPG0352 induced lower expression of CR3 in macrophages. After CR3 was suppressed, there were no significant differences in the IL-12p70 levels between macrophages stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352. PCR array experiments showed that miR-21 and lncRNA GAS5 were differentially expressed between macrophages stimulated by P. gingivalis W83 and ΔPG0352, which had been identified by real-time PCR. The results of CR3 blocking and lncRNA GAS5 gene silence or overexpression showed that the difference in IL-12 levels between P. gingivalis W83 and ΔPG0352 groups was associated with CR3, lncRNA GAS5 and miR-21. Thus it can be concluded that the sialidase-deficient strain is more easily cleared by attenuating CR3 activation, reducing the inhibition of lncRNA GAS5, inducing less miR-21 and more IL-12 in macrophages. These results indicate that inhibiting the activity of sialidase in P. gingivalis will cause rapid clearing by macrophages.
Introduction
Chronic periodontitis is a type of chronic inflammatory disorder of bacterial origin that affects tooth-supporting tissue, eventually leading to tooth loss (Highfield, 2009). Plaque is the initiator of chronic periodontitis caused by more than 700 microorganism species. Among these, Porphyromonas gingivalis (P. gingivalis) has been implicated as a major etiologic agent in the pathogenesis of chronic periodontitis (Loesche and Grossman, 2001; Aas et al., 2005). P. gingivalis is a Gram-negative an aerobic coccobacillus and can produce a number of virulence factors, such as fimbriae, gingipains and lipopolysaccharide (LPS) (Bostanci and Belibasakis, 2012). High levels of P. gingivalis induce an immune response leading to a destructive inflammatory process (Garlet, 2010). It is noteworthy that P. gingivalis can escape the host's innate immune response and internalize to the host cells, as well as invade into deep tissue (Lamont et al., 1995).
During the initiation and progression of periodontal disease, macrophages in the periodontal tissue play a central role in the front-line immune response against invading pathogens (Zhou et al., 2005). In response to infection by periodontal pathogens, macrophages eliminate pathogens by phagocytosis, or secrete cytokines leading to an inflammatory process (Reichrath et al., 2016). Macrophages also have antigen-presenting functions resulting in activation of acquired immunity (Lavin et al., 2015). During the inflammatory process, many cytokines and chemokines produced by macrophages activate the immune response (Okada and Murakami, 1998; Park et al., 2014). IL-12 is a heterodimeric proinflammatory cytokine, composed of the disulfide-linked p35 and p40 subunits to form bioactive IL-12 (also known as IL-12p70; Gately et al., 1998). IL-12p70 is correlated with bacterial clearance by macrophages (Trinchieri, 2003) and has been shown to stimulate NK cell production. In addition, IL-12p70 promotes CD4+ T cell differentiated from Th1 cells to Th2 cells, and can also activate the bacterial clearance function of macrophages (Yun et al., 2001; Trinchieri, 2003).
The study of sialic acid and the sialidase gene in pathogenic bacteria has been the subject of much recent research. Sialic acids are nine-carbon sugar acids derived from N-acetylneuraminic acid (Vanterpool et al., 2006). Their presence or absence has played an important role in determining the physiological state of a cell or tissue. Many symbiotic pathogens take advantage of sialic acids in their environment (the host) as sources of carbon, nitrogen, energy, and amino acids to synthesize the cell wall (Plumbridge and Vimr, 1999). Sialic acids may also regulate the expression of other genes, such as the fimbriae gene, and may modify macromolecules on the surface of bacteria (El-Labany et al., 2003; Comstock and Kasper, 2006; Severi et al., 2007). Sialylation on the surface of bacteria on the one hand enhances the virulence of bacteria; while on the other hand leads to the host mistakenly recognizing the pathogen as host cells evading an immune response. Such a mechanism is beneficial to the pathogen's ability to escape the host's defense. There are two ways for bacteria to acquire sialic acid: endogenous synthesis and exogenous decomposition (Severi et al., 2007). Endogenous synthesis occurs in bacteria that contain enzymes associated with sialic acid synthesis. While some bacteria, such as P. gingivalis, do not contain these enzymes and cannot synthesize sialic acid; instead, they obtain sialic acid from the environment by sialidase in a process known as exogenous decomposition. Sialidase (neuraminidase) is a type of glycosyl hydrolase that can cut the connection between glycosylation and the sialic acid O-receptor substrate by external hydrolysis to obtain free sialic acid (Vimr, 1994). The sialidase gene can be expressed in a variety of pathogens, is associated with macromolecule synthesis and modification, leading to infection, tissue damage, peroxide elimination in the process of oxidative stress and host innate immune regulation (Vimr, 1994; Iijima et al., 2004; Wang et al., 2010).
As early as 1990, Moncla et al. discovered that P. gingivalis showed sialidase activity (Moncla et al., 1990). However, the roles of sialidase in P. gingivalis have not been studied further possibly due to limitations in technology and knowledge. Recently, we have constructed a mutant strain of the P. gingivalis sialidase gene in P. gingivalis W83 (ΔPG0352) by means of homologous recombination. It was found that PG0352 was the sole gene encoding sialidase in P. gingivalis W83. Sialidase deficiency did not influence P. gingivalis growth, but showed less pathogenicity than the wild-type strain in a mouse abscess model (Li et al., 2012), while the mechanism remains unknown. Our previous study and the study of Aruni W et al. showed that the existence of sialidase was associated with the expression of virulence factors in P. gingivalis, including fimbriae, gingipain, capsule, and LPS (Aruni et al., 2011; Li et al., 2017). The sialylation on the surface of pathogen made it difficult to be recognized by macrophages, allowing escape from the hosts' defense. Therefore, we hypothesized that sialidase deficiency prevented P. gingivalis from acquiring sialic acid, thus affecting the virulence factors and modification of macromolecules. As a result, sialidase-deficient P. gingivalis is easier to be recognized and activate phagocytosis in macrophages.
In this study, we stimulated U937-differentiated macrophages with P. gingivalis W83 and ΔPG0352, compared the differences in IL-12 expression, and analyzed the differences of miRNAs and lncRNAs by PCR array. In addition, gene silence or overexpression experiments were conducted on the lncRNA GAS5 in order to decipher the mechanism of the effect of sialidase on P. gingivalis subverting phagocytosis by macrophages.
Materials and Methods
Bacterial Strains and Growth Conditions
The construction of ΔPG0352 and comΔPG0352 were as previously described (Li et al., 2017). Porphyromonas gingivalis W83, ΔPG0352 and comΔPG0352 were cultured on tryptic soy blood (TSB) agar plates supplemented with 5% sheep blood, 5% yeast extract, hemin (5 μg/ml) and 0.1% vitamin K under anaerobic conditions at 37°C. Bacterial suspensions were prepared in TSB infusion broth at 37°C under anaerobic conditions (N2, 80%; H2, 10%; CO2, 10%).
Cell Culture and Macrophage Preparation
Human monocytes U937 (ATCC CRL-1593.2), a monoblastic leukemia cell line were purchased from American Type Culture Collection (Manassas, VA, USA). The cells were cultured in RPMI1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco, USA) and antibiotics (100 units/ml penicillin, 100 μg/ml streptomycin, Sigma, USA) at 37°C, 5%CO2. The cells were then incubated with 10 ng/ml phorbol myristic acid (PMA; Sigma, USA) to induce differentiation into macrophage-like cells. After 72 h, the medium was replaced with fresh medium without PMA or antibiotics, and the differentiated cells were incubated for an additional 24 h prior to use (Roberts et al., 1997).
MTS Assay
Cell proliferation assays were performed using the CellTiter 96® Aqueous One Solution Cell Proliferation Assay kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. In brief, U937 cells (1 × 104 cells/well) were seeded in 24-cell culture plates with PMA. After differentiation into macrophage-like cells, the cells were stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352 (MOI = 100) for 0, 2, 6, 8, 12, 24, and 48 h. A total of 20 μl MTS reagent was added into each well at each time point and incubated for 4 h, and finally the absorbance at 490 nm monitored using a microplate reader.
Transmission Electron Microscopy (TEM)
U937-differentiated macrophages were stimulated by P. gingivalis W83, ΔPG0352, or comΔPG0352 (MOI = 100) for 6 h at 37°C, 5%CO2. TEM was performed according to methods previously described (Giacona et al., 2004; Willingham et al., 2007). Briefly, cells were washed, scraped and centrifuged at 800 rpm for 5 min. The pellets were fixed in 2.5% glutaraldehyde at 4°C overnight. Then, cells were washed three times with sterilized water and suspended in osmium solution for 2 h. The washed cell pellets were dehydrated in a graded of series of ethanol and acetone solutions (30, 50, 70, and 80%, 90 and 100% respectively for 30 min each). Finally, the pellets were embedded in Epox 812 resin. Thin sections were cut, post-stained with uranyl acetate and lead citrate, and viewed using the transmission electron microscope (H-7650, HIACHI, Japan).
Real-Time Polymerase Chain Reaction (Real-Time PCR) for IL-12p35 and IL-12p40
U937-differentiated macrophages were stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352 (MOI = 100) for 6, 12, and 24 h at 37°C, 5%CO2. Total RNA was extracted from cells (miRNeasy Mini Kit, Qiagen, Germany) and reverse-transcribed (PrimeScript™ RT Master Mix kit, Takara, Japan) according to the manufacturer's instructions. Real-time PCR was performed with a SYBR Green approach (SYBR® Premix Ex Taq, Takara, Japan) and the expressions of IL-12p35 and IL-12p40 were measured with ABI System (Applied Biosystems, USA). The primers were as follows: GAPDH: F: 3′-GAAGGTGAAGGTCGGAGTC-5′ and R: 3′-GAAGATGGTGATGGGATTTC-5′ (Dutzan et al., 2012); IL-12p35: F: 3′-CACTCCCAAAACCTGCTGAG-5′ and R: 3′-TCTCTTCAGAAGTGCAAGGGTA-5′ (Farkas et al., 2008); IL-12p40: F: 3′-CCCTGACATTCTGCGTTCA-5′ and R: 3′-AGGTCTTGTCCGTGAAGACTCTA-5′ (Farkas et al., 2008). The amplification cycling conditions were 95°C for 30 s, 40 cycles of 5 s each at 95°C, and 34 s at 60°C. 2−ΔΔCt was used to evaluate the variability of target genes as previously described (Suzuki et al., 2005).
Enzyme-Linked Immunosorbent Assay (ELISA)
U937-differented macrophages were cultured in 6-well culture plates and a final concentration of 2 μg/ml anti-CD11b antibody was added for pretreatment (1 h) in inhibitor groups. Then macrophages were stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352 (MOI = 100) for 6, 12 and 24 h. The levels of IL-12p70 were determined using a Human IL-12A/IL-12B ELISA kit (Sigma-Aldrich, USA), according to the manufacturer's instructions. A microplate reader was used to measure optical density at 450 nm. Standard curves were generated by plotting the mean optical density and concentration of each standard dilution, and the concentrations of each indicator in samples were calculated.
Immunofluorescence Assay
U937-differentiated macrophages cultivated on 24-well chambered cover-glass slides were stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352 (MOI = 100) for 6 h and were fixed in 4% paraformaldehyde at 4°C for 60–90 min. Cells were then punched with 0.25% Triton for 30 min and blocked in PBS with 10% human serum for 20 min. Samples were incubated with CR3 specific antibody CD11b (1:100 dilution in PBS solution; Abcam, USA) overnight at 4°C, and incubated with the secondary antibody conjugated to fluorescein isothiocyanate isomer (FITC) (1:100 dilution in PBS solution; Abcam, USA) for 30 min at 37°C. The slides were incubated with DiIC18 (3) (DiI) and 4,6-diamidino-phenylindole (DAPI) for 20 and 5 min respectively. Quencher was added and observed under a fluorescence microscope. Positive-staining sections in each cell of a ×200 image were analyzed for the intensity of fluorescence emitted with National Institutes of Health (NIH) IMAGEJ analysis software.
PCR Array
U937-differentiated macrophages were stimulated by P. gingivalis W83 or ΔPG0352 (MOI = 100) for 6 h at 37°C, 5%CO2. Total RNA was extracted from macrophages with TRIzol and purified using a miRNeasy Mini Kit (Qiagen, Germany). A total of 200 ng RNA was reverse-transcribed to cDNA with miScript®II RT Kit (Qiagen, Germany). Next, 200 μl RNase-free water was added to each 20 μl cDNA solution. The reaction mixture was prepared, including 1,375 μl 2 × QuantiTect SYBR Green PCR Master Mix, 275 μl 10 × miScript Universal Primer, 1,000 μl RNase-free water and 100 μl cDNA according to the manufacturer's instructions of RT2 miRNA PCR Array (MIHS-105, Qiagen, USA). Next, 25 μl per well reaction mixture was added to the 96-well plates. Plates were placed in the real-time cycler and the amplification cycling conditions were programmed as follow: 95°C for 15 min, 40 cycles of 15 s each at 94°C, 30 s at 55°C and 30 s at 70°C. Data were analyzed using miScript miRNA PCR Array Data Analysis Tool. SNORD48 and RNU6-2 were used as references. RNA from three independent cultures was analyzed. TargetScan (http://www.targetscan.org/vert_50/) and miRDB (http://mirdb.org/miRDB/download.html.) databases were used to predict IL-12 related miRNAs for further exploration of differently expressed miRNA between P. gingivalis W83 and ΔPG0352 groups.
LncRNAs Selection
LncRNAs MEG3, GAS5, FASLG, BTG2, SPRY2, and TAGAP that correlated with miR-21 were primarily screened by referring previous reports. U937-differentiated macrophages were stimulated by P. gingivalis W83 or ΔPG0352 (MOI = 100) for 6 h at 37°C, 5%CO2. Total RNA was extracted from macrophages by using TRIzol reagent (Invitrogen, Carlsbad, CA). Real-time PCR was used to detect the expression of different lncRNAs. The sequences were as follow: MEG3: F: 5′-ATCATCCGTCCACCTCCTTGTCTTC-3′, R: 5′-GTATGAGCA TAGCAAAGGTCAGGGC-3′ (Jia et al., 2014); GAS5: F: 5′-CTTCTGGGCTCAAGTGATCCT-3′, and R: 5′-TTGTGCCATGAGACTCCATCAG-3′ (Khalil et al., 2009); BTG2: F:5′-CATCATCAGCAGGGTGGC-3′, R: 5′-CCCAATGCGGTAGGACAC-3′ (Li et al., 2015); SPRY2: 5′-GAGTCGTCTCCAGCTCCGAAC-3′, 5′-AGCTCTGGCCTCCATCAGG-3′ (Ahn et al., 2015).
Preparation of shRNA-Expressing Lentiviruses and Cell Infection
A targeted sequence designed to be homologous to lncRNA GAS5 were cloned into the LV3 (H1/GFP&Puro) and LV5 (EF-1αF/GFP&Puro) lentiviral vectors (GenePharma, Shanghai, China) for GAS5 gene silence and overexpression, respectively (See Images 1, 2 in Supplementary Material). These vectors were termed LV3-GAS5-Lentivirus and LV5-GAS5-Lentivirus, and the vectors without lncRNA GAS5 gene were LV3NC-GAS5-Lentivirus and LV5NC-GAS5-Lentivirus. The sequences were as follow: LV3-GAS5-Lentivirus: 5′-CTTGCCTGGACCAGCTTAA-3′; LV3NC-GAS5-Lentivirus: 5′-TTCTCCGAACGTGTCACGT-3′. The construction of LV3 and LV5 lentiviral vectors was as previously described (Shen et al., 2017). Briefly, the restriction enzyme cutting sites were digested by special enzyme (NotI&BamHI or BamI&EcoRI). Then the vectors were recovered using DNA gel recovery kit and the amplified fragment was subjected to recombinant cloning. The ligation products were transferred to the competent cells, and the newly generated clones were verified by double enzyme digestion. The vectors were transfected into the 293T packaging cell line with RNAi-Mate. The supernatant was collected after infection to harvest the recombinant virus. U937-differentiated macrophages cells were infected with the virus (MOI = 100) with 5 μg/ml polybrene and non-transfected cells were used as a control. The cells were divided into five groups: control group, LV3NC (control for lncRNA GAS5 silence), LV3 (lncRNA GAS5 silence), LV5NC (control for lncRNA GAS5 overexpression), and LV5 (lncRNA GAS5 overexpression) groups. After 72 h infection, the expression of green fluorescent protein (GFP) was observed using a fluorescent microscope to assess the infection efficiency. The silence efficiency of GAS5 was evaluated by real-time PCR.
U937-differentiated macrophages cells were seeded on 6-well culture plates post-lentiviral infection, and stimulated by P. gingivalis W83 or ΔPG0352 (MOI = 100) for 24 h at 37°C, 5%CO2. The levels of IL-12p70 were determined using a Human IL-12A/IL-12B ELISA kit (Sigma-Aldrich, USA).
Confirmation of Differently Expressed miR-21 and lncRNA GAS5
U937-differentiated macrophages were stimulated by P. gingivalis W83, ΔPG0352 and comΔPG0352 (MOI = 100) respectively for 6 h, 12 and 24 h at 37°C, 5%CO2 condition. RNA and protein was extracted. Real-time PCR was used to detect the changes of miR-21 and lncRNA GAS5 gene expressions. The primers were as followed: miR-21: F: 5′-AGTGGGGAGTAGCTTATCAGAC-3′, and R: 5′- CAACGGGGCATCAACATCA-3′ (Yu et al., 2012); U6: F: 5′- CTCGCTTCGGCA GCACA-3′, R: 5′- AACGCTTCACGAATT TGCGT-3′ (Jia et al., 2014).
Real-Time PCR for GAS5 After CR3 Receptor Was Suppressed
U937-differented macrophages were cultured in 6-well culture plates and a final concentration of 2 μg/ml anti-CD11b antibody was added for pretreatment (1 h). Then macrophages were stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352 (MOI = 100) for 6, 12, and 24 h. The expression of lncRNA GAS5 was detected by real-time PCR.
Statistical Analysis
One-way analysis of variance (ANOVA) was used to compare the differences between parameters in each group. Measurement data were represented as mean ± standard deviation. All tests were two-sided, with a significance level of 0.05. All statistical analyses were performed with statistical software.
Results
The Morphology of Macrophages After Various P. gingivalis Infections
The TEM assay showed that the three P. gingivalis strains could induce uptake by macrophages. In the control group, macrophages were morphologically normal and the membrane was integrated (shown in Figure 1). However, there were microvilli and coated pits on the cell surface of macrophages in the P. gingivalis W83, ΔPG0352 and comΔPG0352 groups. The membrane was discontinuous and the intracellular bacteria were enclosed within endocytic vacuoles.
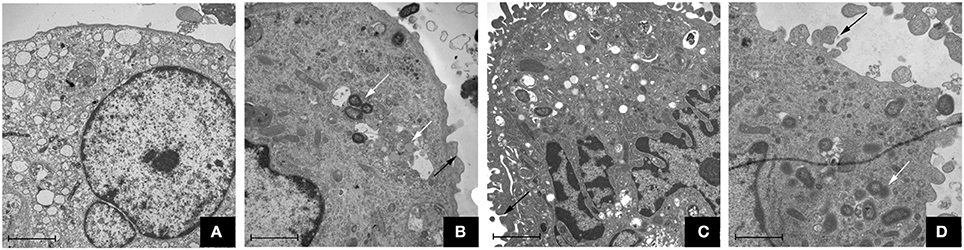
Figure 1. Transmission electron microscope observation of macrophages stimulated by P. gingivalis W83, ΔPG0352 and comΔPG0352 respectively (15,000 ×, bar: 2 μm). (A) control group; (B) P. gingivalis W83 group; (C) ΔPG0352 group; (D) comΔPG0352 group. White arrow: P. gingivalis internalized by macrophage; black arrow: P. gingivalis adhering to macrophage surface.
Compared to P. gingivalis W83, ΔPG0352 Induced More IL-12 in Macrophages
The expression of IL-12p35 and IL-12p40 genes was detected by real-time PCR. Compared with the P. gingivalis W83 group, the expression of IL-12p35 and IL-12p40 genes were higher in the ΔPG0352 group (P < 0.01) (shown in Figure 2). The results of ELISA showed that, compared with the P. gingivalis W83 and comΔPG0352 groups, the IL-12p70 level was significantly higher in the ΔPG0352 group from 6 to 24 h (P < 0.01) (shown in Figure 3). There were no significant differences between the P. gingivalis W83 and comΔPG0352 groups in the gene expression of IL-12p35 and IL-12p40 and the protein level of IL-12p70 (P > 0.05).
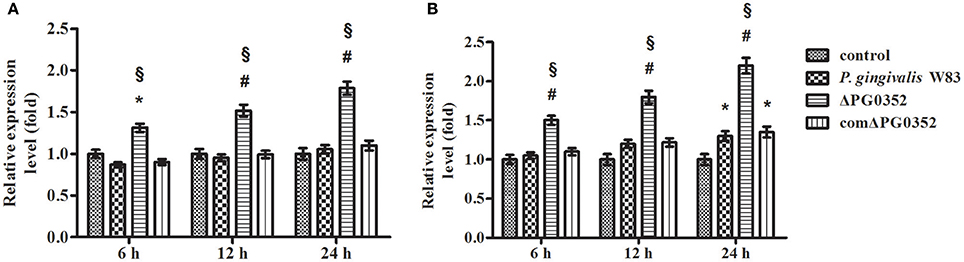
Figure 2. The expression of IL-12p35 (A) and IL-12p40 (B) genes after macrophages stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352. *P < 0.05 vs. control group, #P < 0.01 vs. control group, §P < 0.01 vs. P. gingivalis W83 group, using ANOVA.
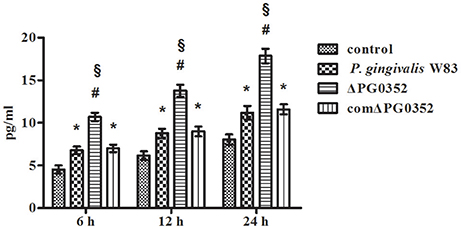
Figure 3. The levels of IL-12p70 after macrophages stimulated by P. gingivalis W83, ΔPG0352, or comΔPG0352. *P < 0.05 vs. control group, #P < 0.01 vs. control group, §P < 0.01 vs. P. gingivalis W83 group, using ANOVA.
The Difference in IL-12 Level Between P. gingivalis W83 and ΔPG0352 Groups Was Associated with CR3
As shown in Figure 4A, CR3 was expressed on almost the entire cell surface, but the fluorescence intensity was different in each group. NIH software IMAGEJ was used to analyze the fluorescence intensity of CR3 expression in each group. The fluorescence intensity levels in the P. gingivalis W83, ΔPG0352 and comΔPG0352 groups were significantly higher than that in the control group (shown in Figure 4B). The fluorescence intensity of CR3 in the P. gingivalis W83 group was about two times higher than that in the control group, and about one and a half times higher than that in the ΔPG0352 group. There were no significant differences between the P. gingivalis W83 and comΔPG0352 groups.
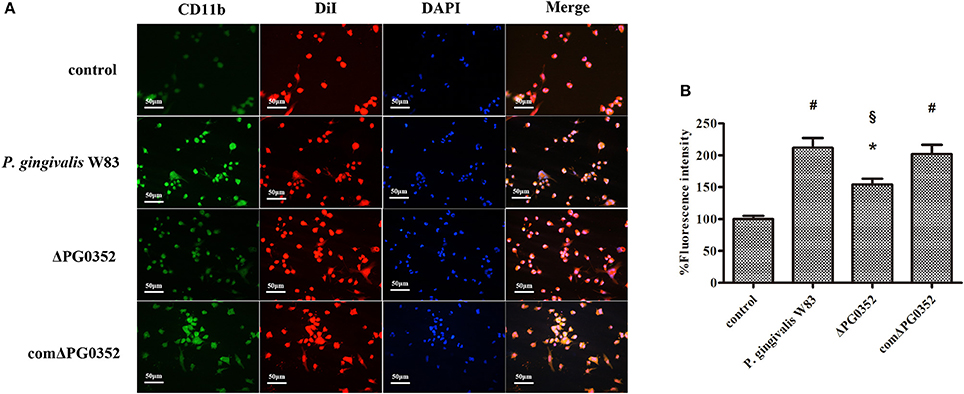
Figure 4. The CR3 expression levels after stimulated by P. gingivalis W83, ΔPG0352 and comΔPG0352. (A) The U937-differentiated macrophages membranes stained with DiI appear red, while the nuclei appeared blue (200×). (B) Analysis of fluorescent levels using IMAGEJ software revealed elevated CD11b levels in P. gingivalis W83, ΔPG0352 and comΔPG0352 groups compared with control group. After 6 h post-infection, the fluorescence intensity of CR3 in P. gingivalis W83 group was about 2 times higher than that in the control group, and about 1.5 times higher than that in ΔPG0352 group. There were no significant differences between P. gingivalis W83 and comΔPG0352 groups. *P < 0.05 vs. control group, #P < 0.01 vs. control group, §P < 0.01 vs. P. gingivalis W83 group, using ANOVA.
We then used anti-CD11b antibodies to suppress the CR3 receptor and detected the IL-12p70 level in each group. The level of IL-12p70 increased and showed no significant differences in the P. gingivalis W83, ΔPG0352 and comΔPG0352 groups after CR3 was suppressed (shown in Figure 5).
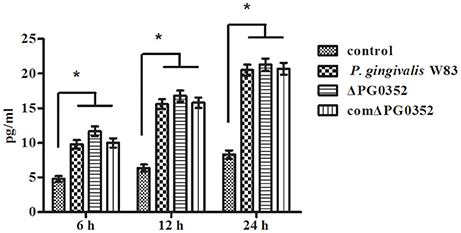
Figure 5. The levels of IL-12p70 in macrophages stimulated by different P. gingivalis strains after CR3 receptor suppressed by CD11b antibody. *P < 0.01 vs. control group, using ANOVA.
Differential Expression of miRNAs in Macrophages Stimulated by P. gingivalis W83 or ΔPG0352
The expression of miRNAs in macrophages in response to P. gingivalis stimulation was examined using a PCR array that included 84 key miRNAs predicted to regulate the expression of proinflammatory or autoimmunity genes. There were six miRNAs differently expressed between the P. gingivalis W83 and ΔPG0352 groups (shown in Table 1). To investigate why the level of IL-12 increased in the ΔPG0352 group, TargetScan and miRDB databases were used to forecast miRNAs correlated with IL-12. The results showed that IL-12A (IL-12p35) was the target gene of miR-21-5p and miR-590-5p, and IL-12B (IL-12p40) was the target gene of miR-23-3p and miR-130-5p. As shown in Table 1, it was found that there was a significant difference in miR-21-5p between the P. gingivalis W83 and ΔPG0352 groups; the fold change of expression was 0.444. The difference in miR-21 expression between the P. gingivalis W83 and ΔPG0352 groups was confirmed by real-time PCR for 6, 12, and 24 h (shown in Figure 6). The results showed that expression of the miR-21 gene increased gradually over time in macrophages stimulated by P. gingivalis and was significantly higher in U937 stimulated by P. gingivalis W83 than that in macrophages stimulated by ΔPG0352. These results were consistent with the results from PCR array.
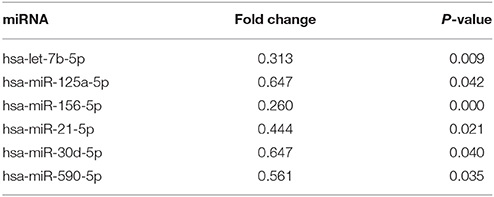
Table 1. The significant differences of miRNA expression after macrophages stimulated by ΔPG0352 compared with P. gingivalis W83.
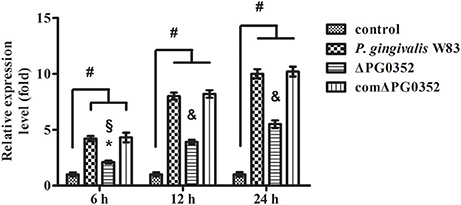
Figure 6. The expression of miR-21 after macrophages stimulated by P. gingivalis W83, ΔPG0352, or comΔPG0352. *P < 0.05 vs. control group, #P < 0.01 vs. control group, §P < 0.05 vs. P. gingivalis W83 group, &P < 0.01 vs. P. gingivalis W83 group, using ANOVA.
Different Expression of lncRNAs in Macrophages Stimulated by P. gingivalis W83 or ΔPG0352
According to previous findings, it was concluded that MEG3, GAS5, FASLG, BTG2, SPRY2, and TAGAP could regulate miR-21 expression. After 6 h stimulated by P. gingivalis W83 or ΔPG0352, the expression of GAS5, BTG2, SPRY2, and TAGAP was significantly lower in the P. gingivalis W83 or ΔPG0352 groups compared with the control group (shown in Figure 7). The expression of GAS5 in the P. gingivalis W83 and ΔPG0352 groups was significantly different and there were no significant differences between the P. gingivalis W83 and ΔPG0352 groups in MEG3, FASLG, BTG2, SPRY2, and TAGAP expression. Real-time PCR was used to detect the different expression of lncRNA GAS5 when macrophages were stimulated by different P. gingivalis strains for 6, 12, and 24 h. Compared to the control group, the expression of GAS5 decreased in macrophages stimulated by P. gingivalis in a time dependent manner. Compared to the P. gingivalis W83 group, the expression of GAS5 was significantly higher in the ΔPG0352 group (shown in Figure 8).
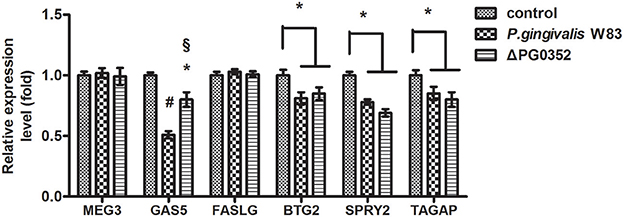
Figure 7. The expression of lncRNA MEG3, GAS5, FASLG, BTG2, SPRY2, and TAGAP genes after macrophages stimulated by P. gingivalis W83 and ΔPG0352. *P < 0.05 vs. control group, #P < 0.01 vs. control group, §P < 0.01 vs. P. gingivalis W83 group, using ANOVA.
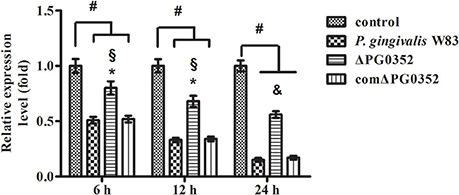
Figure 8. The expression of lncRNA GAS5 after macrophages stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352. *P < 0.05 vs. control group, #P < 0.01 vs. control group, §P < 0.05 vs. P. gingivalis W83 group, &P < 0.01 vs. P. gingivalis W83 group, using ANOVA.
The Expression of GAS5 in Macrophages After Transfected With Lentivirus-Mediated shRNA
The expression of GAS5 in LV3, LV3NC, LV5, and LV5NC groups was detected by using real-time PCR (shown in Figure 9). These results indicated that LV3 significantly down-regulated the GAS5 expression in macrophages, the silence efficiency was 78 ± 6%, and the expression of GAS5 in LV5 group was 3.45 ± 0.26 times that of the control. These results indicated that lentivirus-mediated GAS5 shRNALV3 and LV5 were able to significantly down-regulate or up-regulate GAS5 expression in macrophages, respectively.
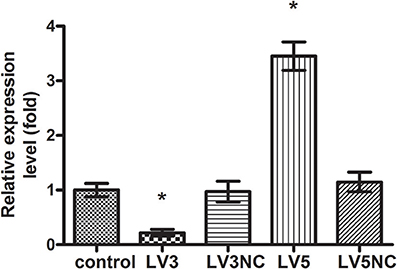
Figure 9. The expression of lncRNA GAS5 genes after transfected with Lentivirus-mediated shRNA. *P < 0.05 vs. control group, using ANOVA.
The Levels of IL-12p70 in Macrophages After Transfected With Lentivirus-Mediated shRNA
ELISA was used to detect the level of IL-12p70 in transfected macrophages stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352 after 24 h (shown in Figure 10). The results showed that, compared with U937, the levels of IL-12p70 were significantly lower in LV3, and higher in LV5 after stimulated by different P. gingivalis strains. The differences of IL-12 level between the P. gingivalis W83 and ΔPG0352 groups in LV3 were less than those in U937.
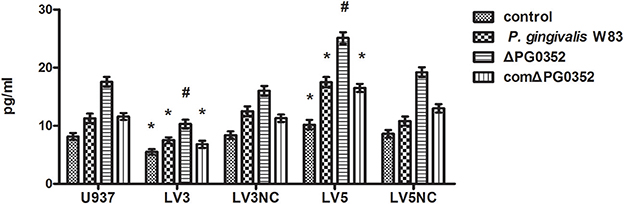
Figure 10. The levels of IL-12p70 after transfected with Lentivirus-mediated shRNA. *P < 0.05 vs. control group, #P < 0.01 vs. control group, using ANOVA.
The Difference in GAS5 Expression Between P. gingivalis W83 and ΔPG0352 Groups Was Associated With CR3
The expression of GAS5 increased in each group after CR3 receptor was suppressed by anti-CD11b antibodies (shown in Figure 11). Compared with the P. gingivalis W83 and comΔPG0352 groups, GAS5 expression was significantly higher in the ΔPG0352 group from 6 to 24 h (P < 0.01). There were no significant differences between the P. gingivalis W83 and comΔPG0352 groups in the gene expression of GAS5 (P > 0.05).
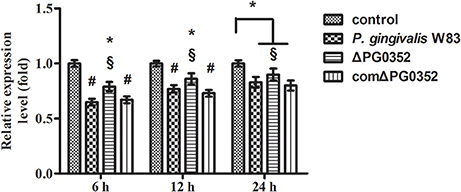
Figure 11. The expression of lncRNA GAS5 in macrophages stimulated by P. gingivalis W83, ΔPG0352 or comΔPG0352 after CR3 suppressed by CD11b antibody. *P < 0.05 vs. control group, #P < 0.01 vs. control group, §P < 0.05 vs. P. gingivalis W83 group, using ANOVA.
Discussion
Macrophages are an important part of the innate immune system. They can engulf pathogens, regulate immune function by antigen presentation, and secreting inflammatory cytokines to clear pathogens invading host tissues. The phagocytosis of macrophages includes the recognition of surface receptors of pathogens, wrapping pathogens and clearing them. In this study, we stimulated U937 with P. gingivalis and performed TEM assays to observe the appearance of P. gingivalis internalized into macrophages. The results showed that there was microvilli formation on the macrophage surface and pits surrounding the pathogens. P. gingivalis W83, ΔPG0352 and comΔPG0352 were visible within the cytoplasm of macrophages, enclosed within membrane-bound endocytic vacuoles. Therefore, we suggest that irrespective of sialidase deficiency, P. gingivalis can interact with macrophages inducing phagocytes is to clear pathogens to different extents.
The inflammatory response is essential for host defense against invading pathogens. Previous studies showed that IL-12 produced by macrophages plays an important role in resistance to pathogen infection (Trinchieri, 2003). IL-12 is described as a natural killer cell stimulatory factor (NKSF), linking the activation of innate immune cells to the induction of an effective adaptive immune response (Rossol et al., 2011). The results in this study showed that both the expression of IL-12p35 and IL-12p40 genes and the level of IL-12p70 increased in U937 stimulated by both P. gingivalis W83 and ΔPG0352. The expression of IL-12 was significantly higher in U937 stimulated by ΔPG0352 than P. gingivalis W83, indicating that sialidase deficiency in P. gingivalis W83 increases IL-12 levels in macrophages. There are two possible reasons for this observation. The first reason might be that the lack of sialidase reduces the activity of important P. gingivalis virulence factors, such as fimbriae, LPS, capsule, and gingipains (Aruni et al., 2011; Li et al., 2012, 2017). Virulence factors of P. gingivalis affect the interaction with macrophages. Gmiterek et al. (2016) found that the HmuY gene mutant in P. gingivalis affected the interaction between P. gingivalis and macrophages by decreasing cytokines and chemokines. Singh et al. compared the host inflammatory response induced by encapsulated and non-encapsulated P. gingivalis and found that the capsule of P. gingivalis reduced the host inflammatory response (Singh et al., 2011). Hou et al. showed that P. gingivalis gingipain decreased expression of IL-12 which Escherichia coli LPS induced in macrophages (Hou et al., 2017). The second reason might be that the macromolecules on the cell surface of ΔPG0352 cannot be sialylated due to sialidase deficiency. This would affect the interaction between P. gingivalis and macrophages.
Several publications reported data about IL-12 expression in macrophages stimulated by whole P. gingivalis cells. Sugano et al. and Huang et al. found that the expression of IL-12 induction in macrophages stimulated by P. gingivalis was two-fold higher than that of controls at 24 h post-infection (Sugano et al., 2004; Huang et al., 2011). These results are consistent with the data presented here. Conversely, Park et al. found that P. gingivalis LPS can increase the level of IL-12 in macrophages nine-fold (Park et al., 2011). These results demonstrated that some components in P. gingivalis, such as LPS, can increase the level of IL-12 in macrophages and some may attenuate this increase. Besides, different P. gingivalis strains induced different levels of IL-12 in macrophages. Gmiterek et al. (2016) stimulated U937 with P. gingivalis A7436, the levels of IL-12 increased much higher than that stimulated by P. gingivalis W83 the data presented here. Sugano et al. also compared cytokine induction in two strains of P. gingivalis, and the results showed that P. gingivalis ATCC 49417 showed statistically higher gene expression of IL-12 induction than that of controls at 24 h post-infection. However, P. gingivalis 381 showed no significant induction of IL-12 levels (Sugano et al., 2004). These data indicate that different P. gingivalis strains can result in different IL-12 expression levels in macrophages.
Complement receptor 3 (CR3; CD11b/CD18), which is the most common integrin expressed abundantly in monocytes and neutrophils (Bhat et al., 1999; Yakubenko et al., 2002), can recognize and interact with molecules derived from host cells and pathogens. The interaction of CD11b/CD18 and P. gingivalis is very important for P. gingivalis to internalize and subvert phagocytosis by macrophages (Hajishengallis et al., 2007). The immunofluorescence results presented here showed that the expression of CR3 was significantly lower than that of P. gingivalis W83 and comΔPG0352 groups, indicating that sialidase deficiency attenuated the ability of P. gingivalis activating CR3 in macrophages. There are three possible reasons for this observation. First, sialidase deficiency influences the synthesis and modification of the macromolecules on P. gingivalis surface, which affect P. gingivalis to bind with macrophages and induce less CR3 expression. Second, sialidase deficient mutant strain fails to produce an intact capsule. Capsule contains antigen, immature capsule in sialidase deficient mutant strain cannot activate CR3 as much as mature capsule in wild-type strain. Third, deletion of sialidase gene in P. gingivalis W83 reduces gingipain activity and the amount of LPS, thus sialidase deficient mutant strain induces less CR3 expression than wild type. CR3 was reported to regulate production of inflammatory mediators and phagocytosis of cells by inside-out signaling pathways (Ehlers, 2000; Laudanna et al., 2002). When P. gingivalis interacts with CR3 on the surface of macrophages, extracellular signal-regulated kinases (ERK) 1/2 is activated (Trinchieri, 2003). The activation of ERK1/2 reduced the level of IL-12p70, and inhibited IL-12 mediated clearance to pathogens (Trinchieri, 2003). Therefore, CR3 is thought to be exploited by P. gingivalis to undermine IL-12-mediated bacterial clearance (Hajishengallis et al., 2007). Harokopakis and Shimaoka inhibited CR3 in a mouse periodontitis model and the results showed that the survivability of P. gingivalis in host cells was reduced, thus inhibiting alveolar bone resorption (Shimaoka et al., 2002; Harokopakis et al., 2006). In this study, the levels of IL-12p70 became higher in macrophages stimulated by P. gingivalis and much higher in the ΔPG0352 group than in the P. gingivalis W83 and comΔPG0352 groups. There were no significant differences between the P. gingivalis W83, ΔPG0352 and comΔPG0352 groups when CR3 was suppressed by anti-CD11b. These results indicated that sialidase deficiency in P. gingivalis weakened the increasing level of CR3 in macrophages. Compared with P. gingivalis W83, ΔPG0352 could stimulate more IL-12 secretion in macrophages. The difference in IL-12 levels between the P. gingivalis W83 and ΔPG0352 groups were associated with CR3.
In order to explore why sialidase-deficient P. gingivalis induced more IL-12 in macrophages, microRNA PCR array was carried out. It was shown that miR-21, which could regulate the expression of IL-12, was significantly lower in macrophages stimulated by ΔPG0352. MiR-21 is one of the most highly expressed microRNAs in multiple types of mammalian cells (Lagos-Quintana et al., 2001, 2002; Landgraf et al., 2007). In addition, MiR-21 is also expressed in immune cells, including monocytes, macrophages, B/T-cells and dendritic cells (DCs) (Sheedy, 2015). Previous studies have shown that miR-21 is up-regulated in LPS-treated periodontal ligament cells (Du et al., 2016) and also in various cancers (Wang et al., 2014). Finally, miR-21 levels are thought to be a marker of immune cell activation (Kasashima et al., 2004; Landgraf et al., 2007). Up-regulated miR-21 in macrophages/monocytes is commonly related to pro-inflammatory factors by viruses, bacteria and other molecular patterns and plays an important role in the innate immune process (Sheedy, 2015). TargetScan and miRDB database analysis showed that miR-21 could negatively regulate the expression of IL-12. Lu et al. constructed a luciferase receptor vector to determine the direct effect of miR-21 on IL-12p35 (Lu et al., 2009). The results indicated that IL-12p35 is a target gene of miR-21; moreover, miR-21 has the 8-mer seed sequence that binds to the IL-12p35 3'UTR region, resulting in the repression of IL-12. Therefore, the sialidase-deficient strain of P. gingivalis secreted more IL-12 by inhibiting the level of miR-21 in macrophages.
LncRNAs are defined as a kind of long-chain RNA with transcripts longer than 200 bp. These molecules are involved in regulation of gene expression directly by participating in protein modifications or recruitment of transcription factors. In this study, we also screened six lncRNAs associated with the expression of miR-21 by referring to previous reports. Among these lncRNAs, only growth arrest-specific 5 (GAS5) showed significantly different levels between macrophages stimulated by P. gingivalis W83 and ΔPG0352. GAS5, which has an identified gene regulatory function, was originally found to accumulate in growth-arrested cells (Schneider et al., 1988). GAS5 levels are alerted in virus or bacterial infectious diseases. Mayama et al. found that GAS5 played an important roles in the regulation of immune functions and pathogenesis/pathophysiology of autoimmune, inflammatory, and infectious diseases (Mayama et al., 2016). The results in this study showed that the expression of GAS5 decreased in macrophages stimulated by P. gingivalis. However, compared to the P. gingivalis W83 group, the expression of GAS5 was significantly higher in macrophages stimulated by ΔPG0352. The levels of IL-12p70 were lower when GAS5 was silenced in macrophages and higher when GAS5 was overexpressed. Differences in IL-12p70 levels between P. gingivalis W83 and ΔPG0352 groups were reduced when GAS5 was silenced in macrophages. Therefore, compared to P. gingivalis W83, the sialidase-deficient strain of P. gingivalis reduced the inhibition of GAS5 leading to more IL-12 secretion in macrophages. Recently, reports have shown that GAS5 is a direct target of miR-21 through interaction with the putative miR-21-binding site at exon 4 of GAG5, which indicates that GAS5 negatively regulates miR-21 through the RNA-induced silencing complex (RISC) (Zhang et al., 2013; Song et al., 2014). Taken together, these results indicated that compared to P. gingivalis W83, the sialidase-deficient strain of P. gingivalis reduced the inhibition of GAS5, induced lower expression of miR-21, and led to higher IL-12 level in macrophages.
To our knowledge, there have been few reports associated with the relationship between CR3 and lncRNA GAS5. To explore the relationship between CR3 and lncRNA GAS5, we used anti-CD11b antibodies to suppress CR3 and detected the expression of GAS5 by real-time PCR. The results showed that GAS5 expression increased after CR3 suppression in all three P. gingivalis groups. GAS5 was higher in the ΔPG0352 group compared with the P. gingivalis W83 and comΔPG0352 groups. This result indicated a reciprocal interaction between CR3 and GAS5. It was noteworthy that the level of IL-12p70 showed no significant differences in P. gingivalis W83, ΔPG0352, and comΔPG0352 groups after CR3 was suppressed. However, there were differences in the IL-12p70 levels in the ΔPG0352 and the P. gingivalis W83 groups after silencing GAS5. We speculated the reason was that, in addition to GAS5, there was another pathway which could take part in the influence of CR3 on IL-12 production.
In this study, we first found that compared to P. gingivalis W83, sialidase deficiency in P. gingivalis induced more IL-12 in macrophages. Second, the mechanism was clarified by using PCR array, gene silence and overexpression experiments. It can be concluded that the sialidase-deficiency in P. gingivalis attenuates CR3 activation in macrophages, reduces the inhibition of lncRNA GAS5, induces less miR-21 and more IL-12 in macrophages. (shown in Figure 12). The results of this study indicate that inhibiting the activity of sialidase in P. gingivalis would render P. gingivalis more easily cleared by macrophages. This finding will open up a new direction in prevention and treatment of chronic periodontitis.
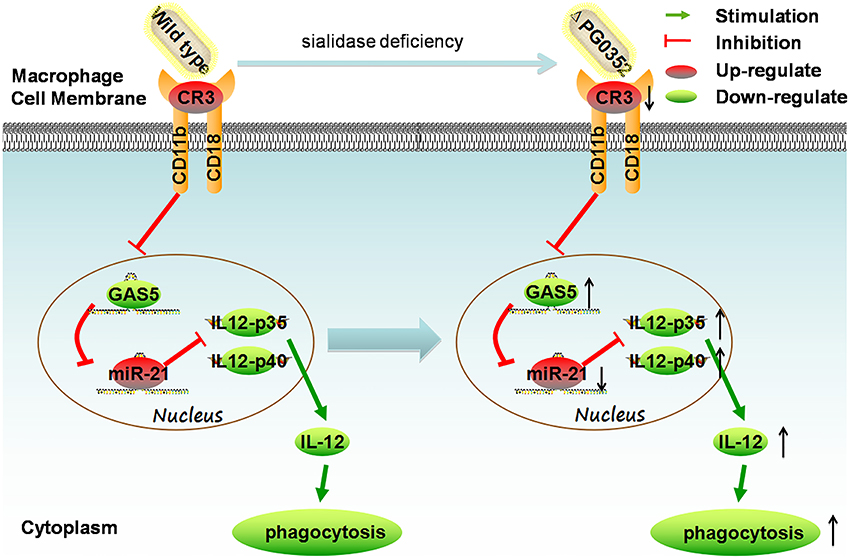
Figure 12. The mechanism of sialidase deficiency increasing IL-12 secretion in P. gingivalis-stimulated macrophages. P. gingivalis can activate CR3 in macrophages, inhibit the expression of lncRNA GAS5, increase the expression of miR-21, decrease the level of IL-12 and subvert phagocytosis by macrophages. The sialidase-deficiency in P. gingivalis attenuates CR3 activation in macrophages, reduces the inhibition of lncRNA GAS5, induces less miR-21 and more IL-12 in macrophages.
Author Contributions
CL, YP, and JL conceived and designed the experiments. XY, XX, TT, SY, LL, and YZ performed the experiments. YP, LL, and DZ analyzed the data. XY, CL, and YP wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Nature Science Foundation of China (Nos. 81200785, 81470745, and 81500869 for CL, YP, and JL, respectively).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00100/full#supplementary-material
Image 1. Map of LV3 lentiviral vector (H1/GFP&Puro).
Image 2. Map for LV5 lentiviral vector (EF-1α/GFFP&Puro).
References
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., and Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005
Ahn, J. H., Han, B. I., and Lee, M. (2015). Induction of resistance to BRAF inhibitor is associated with the inability of Spry2 to inhibit BRAF-V600E activity in BRAF mutant cells. Biomol. Ther. (Seoul). 23, 320–326. doi: 10.4062/biomolther.2015.007
Aruni, W., Vanterpool, E., Osbourne, D., Roy, F., Muthiah, A., Dou, Y., et al. (2011). Sialidase and sialoglycoproteases can modulate virulence in Porphyromonas gingivalis. Infect. Immun. 79, 2779–2791. doi: 10.1128/IAI.00106-11
Bhat, N., Perera, P. Y., Carboni, J. M., Blanco, J., Golenbock, D. T., Mayadas, T. N., et al. (1999). Use of a photoactivatable taxol analogue to identify unique cellular targets in murine macrophages: identification of murine CD18 as a major taxol-binding protein and a role for Mac-1 in taxol-induced gene expression. J. Immunol. 162, 7335–7342.
Bostanci, N., and Belibasakis, G. N. (2012). Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol. Lett. 333, 1–9. doi: 10.1111/j.1574-6968.2012.02579.x
Comstock, L. E., and Kasper, D. L. (2006). Bacterial glycans: key mediators of diverse host immune responses. Cell 126, 847–850. doi: 10.1016/j.cell.2006.08.021
Du, A., Zhao, S., Wan, L., Liu, T., Peng, Z., Zhou, Z., et al. (2016). MicroRNA expression profile of human periodontal ligament cells under the influence of Porphyromonas gingivalis LPS. J. Cell. Mol. Med. 20, 1329–1338. doi: 10.1111/jcmm.12819
Dutzan, N., Vernal, R., Vaque, J. P., Garcia-Sesnich, J., Hernandez, M., Abusleme, L., et al. (2012). Interleukin-21 expression and its association with proinflammatory cytokines in untreated chronic periodontitis patients. J. Periodontol. 83, 948–954. doi: 10.1902/jop.2011.110482
Ehlers, M. R. (2000). CR3: a general purpose adhesion-recognition receptor essential for innate immunity. Microb. Infect. 2, 289–294. doi: 10.1016/S1286-4579(00)00299-9
El-Labany, S., Sohanpal, B. K., Lahooti, M., Akerman, R., and Blomfield, I. C. (2003). Distant cis-active sequences and sialic acid control the expression of fimB in Escherichia coli K-12. Mol. Microbiol. 49, 1109–1118. doi: 10.1046/j.1365-2958.2003.03624.x
Farkas, A., Tonel, G., and Nestle, F. O. (2008). Interferon-alpha and viral triggers promote functional maturation of human monocyte-derived dendritic cells. Br. J. Dermatol. 158, 921–929. doi: 10.1111/j.1365-2133.2008.08508.x
Garlet, G. P. (2010). Destructive and protective roles of cytokines in periodontitis: a re-appraisal from host defense and tissue destruction viewpoints. J. Dent. Res. 89, 1349–1363. doi: 10.1177/0022034510376402
Gately, M. K., Renzetti, L. M., Magram, J., Stern, A. S., Adorini, L., Gubler, U., et al. (1998). The interleukin-12/interleukin-12-receptor system: role in normal and pathologic immune responses. Annu. Rev. Immunol. 16, 495–521. doi: 10.1146/annurev.immunol.16.1.495
Giacona, M. B., Papapanou, P. N., Lamster, I. B., Rong, L. L., D'agati, V. D., Schmidt, A. M., et al. (2004). Porphyromonas gingivalis induces its uptake by human macrophages and promotes foam cell formation in vitro. FEMS Microbiol. Lett. 241, 95–101. doi: 10.1016/j.femsle.2004.10.009
Gmiterek, A., Klopot, A., Wojtowicz, H., Trindade, S. C., Olczak, M., and Olczak, T. (2016). Immune response of macrophages induced by Porphyromonas gingivalis requires HmuY protein. Immunobiology 221, 1382–1394. doi: 10.1016/j.imbio.2016.07.007
Hajishengallis, G., Shakhatreh, M. A., Wang, M., and Liang, S. (2007). Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J. Immunol. 179, 2359–2367. doi: 10.4049/jimmunol.179.4.2359
Harokopakis, E., Albzreh, M. H., Martin, M. H., and Hajishengallis, G. (2006). TLR2 transmodulates monocyte adhesion and transmigration via Rac1- and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J. Immunol. 176, 7645–7656. doi: 10.4049/jimmunol.176.12.7645
Highfield, J. (2009). Diagnosis and classification of periodontal disease. Aust. Dent. J. 54(Suppl. 1), S11–S26. doi: 10.1111/j.1834-7819.2009.01140.x
Hou, Y., Yu, H., Liu, X., Li, G., Pan, J., Zheng, C., et al. (2017). Gingipain of Porphyromonas gingivalis manipulates M1 macrophage polarization through C5a pathway. In vitro Cell. Dev. Biol. Anim. 53, 593–603. doi: 10.1007/s11626-017-0164-z
Huang, C. B., Altimova, Y., Strange, S., and Ebersole, J. L. (2011). Polybacterial challenge effects on cytokine/chemokine production by macrophages and dendritic cells. Inflamm. Res. 60, 119–125. doi: 10.1007/s00011-010-0242-0
Iijima, R., Takahashi, H., Namme, R., Ikegami, S., and Yamazaki, M. (2004). Novel biological function of sialic acid (N-acetylneuraminic acid) as a hydrogen peroxide scavenger. FEBS Lett. 561, 163–166. doi: 10.1016/S0014-5793(04)00164-4
Jia, L. F., Wei, S. B., Gan, Y. H., Guo, Y., Gong, K., Mitchelson, K., et al. (2014). Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int. J. Cancer 135, 2282–2293. doi: 10.1002/ijc.28667
Kasashima, K., Nakamura, Y., and Kozu, T. (2004). Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem. Biophys. Res. Commun. 322, 403–410. doi: 10.1016/j.bbrc.2004.07.130
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672. doi: 10.1073/pnas.0904715106
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. doi: 10.1126/science.1064921
Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739. doi: 10.1016/S0960-9822(02)00809-6
Lamont, R. J., Chan, A., Belton, C. M., Izutsu, K. T., Vasel, D., and Weinberg, A. (1995). Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63, 3878–3885.
Landgraf, P., Rusu, M., Sheridan, R., Sewer, A., Iovino, N., Aravin, A., et al. (2007). A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414. doi: 10.1016/j.cell.2007.04.040
Laudanna, C., Kim, J. Y., Constantin, G., and Butcher, E. (2002). Rapid leukocyte integrin activation by chemokines. Immunol. Rev. 186, 37–46. doi: 10.1034/j.1600-065X.2002.18604.x
Lavin, Y., Mortha, A., Rahman, A., and Merad, M. (2015). Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 15, 731–744. doi: 10.1038/nri3920
Li, C., Hu, B., Bian, J., Sun, J., Zhang, W., Li, C., et al. (2012). Abrogation of neuraminidase reduces biofilm formation, capsule biosynthesis, and virulence of Porphyromonas gingivalis. Infect. Immun. 80, 3–13. doi: 10.1128/IAI.05773-11
Li, C., Yang, X., Pan, Y., Yu, N., Xu, X., Tong, T., et al. (2017). A sialidase-deficient Porphyromonas gingivalis Mutant Strain Induces Less Interleukin-1beta and Tumor Necrosis Factor-alpha in Epi4 Cells Than W83 strain through regulation of c-Jun N-Terminal Kinase pathway. J. Periodontol. 88, e129–e139. doi: 10.1902/jop.2017.160815
Li, Y. J., Dong, B. K., Fan, M., and Jiang, W. X. (2015). BTG2 inhibits the proliferation and metastasis of osteosarcoma cells by suppressing the PI3K/AKT pathway. Int. J. Clin. Exp. Pathol. 8, 12410–12418.
Loesche, W. J., and Grossman, N. S. (2001). Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clin. Microbiol. Rev. 4, 727–752, table of contents. doi: 10.1128/CMR.14.4.727-752.2001
Lu, T. X., Munitz, A., and Rothenberg, M. E. (2009). MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 182, 4994–5002. doi: 10.4049/jimmunol.0803560
Mayama, T., Marr, A. K., and Kino, T. (2016). Differential expression of glucocorticoid receptor noncoding RNA repressor Gas5 in autoimmune and inflammatory diseases. Horm. Metab. Res. 48, 550–557. doi: 10.1055/s-0042-106898
Moncla, B. J., Braham, P., and Hillier, S. L. (1990). Sialidase (neuraminidase) activity among gram-negative anaerobic and capnophilic bacteria. J. Clin. Microbiol. 28, 422–425.
Okada, H., and Murakami, S. (1998). Cytokine expression in periodontal health and disease. Crit. Rev. Oral Biol. Med. 9, 248–266. doi: 10.1177/10454411980090030101
Park, E., Na, H. S., Kim, S. M., Wallet, S., Cha, S., and Chung, J. (2014). Xylitol, an anticaries agent, exhibits potent inhibition of inflammatory responses in human THP-1-derived macrophages infected with Porphyromonas gingivalis. J. Periodontol. 85, e212–e223. doi: 10.1902/jop.2014.130455
Park, S. Y., Park, D. J., Kim, Y. H., Kim, Y., Choi, Y. W., and Lee, S. J. (2011). Schisandra chinensis alpha-iso-cubebenol induces heme oxygenase-1 expression through PI3K/Akt and Nrf2 signaling and has anti-inflammatory activity in Porphyromonas gingivalis lipopolysaccharide-stimulated macrophages. Int. Immunopharmacol. 11, 1907–1915. doi: 10.1016/j.intimp.2011.07.023
Plumbridge, J., and Vimr, E. (1999). Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 181, 47–54.
Reichrath, S., Reichrath, J., Moussa, A. T., Meier, C., and Tschernig, T. (2016). Targeting the non-neuronal cholinergic system in macrophages for the management of infectious diseases and cancer: challenge and promise. Cell Death Discov. 2:16063. doi: 10.1038/cddiscovery.2016.63
Roberts, F. A., Richardson, G. J., and Michalek, S. M. (1997). Effects of Porphyromonas gingivalis and Escherichia coli lipopolysaccharides on mononuclear phagocytes. Infect. Immun. 65, 3248–3254.
Rossol, M., Heine, H., Meusch, U., Quandt, D., Klein, C., Sweet, M. J., et al. (2011). LPS-induced cytokine production in human monocytes and macrophages. Crit. Rev. Immunol. 31, 379–446. doi: 10.1615/CritRevImmunol.v31.i5.20
Schneider, C., King, R. M., and Philipson, L. (1988). Genes specifically expressed at growth arrest of mammalian cells. Cell 54, 787–793. doi: 10.1016/S0092-8674(88)91065-3
Severi, E., Hood, D. W., and Thomas, G. H. (2007). Sialic acid utilization by bacterial pathogens. Microbiology 153, 2817–2822. doi: 10.1099/mic.0.2007/009480-0
Sheedy, F. J. (2015). Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. 6:19. doi: 10.3389/fimmu.2015.00019
Shen, C., Yuan, Y., Li, F., Hu, Y., Song, Y., Zhao, S., et al. (2017). Mechanism of genistein regulating the differentiation of vascular smooth muscle cells into osteoblasts via the OPG/RANKL pathway. Oncotarget 8, 76857–76864. doi: 10.18632/oncotarget.20167
Shimaoka, M., Takagi, J., and Springer, T. A. (2002). Conformational regulation of integrin structure and function. Annu. Rev. Biophys. Biomol. Struct. 31, 485–516. doi: 10.1146/annurev.biophys.31.101101.140922
Singh, A., Wyant, T., Anaya-Bergman, C., Aduse-Opoku, J., Brunner, J., Laine, M. L., et al. (2011). The capsule of Porphyromonas gingivalis leads to a reduction in the host inflammatory response, evasion of phagocytosis, and increase in virulence. Infect. Immun. 79, 4533–4542. doi: 10.1128/IAI.05016-11
Song, J., Ahn, C., Chun, C. H., and Jin, E. J. (2014). A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J. Orthop. Res. 32, 1628–1635. doi: 10.1002/jor.22718
Sugano, N., Ikeda, K., Oshikawa, M., Sawamoto, Y., Tanaka, H., and Ito, K. (2004). Differential cytokine induction by two types of Porphyromonas gingivalis. Oral Microbiol. Immunol. 19, 121–123. doi: 10.1046/j.0902-0055.2003.00119.x
Suzuki, N., Yoshida, A., and Nakano, Y. (2005). Quantitative analysis of multi-species oral biofilms by TaqMan real-time PCR. Clin. Med. Res. 3, 176–185. doi: 10.3121/cmr.3.3.176
Trinchieri, G. (2003). Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3, 133–146. doi: 10.1038/nri1001
Vanterpool, E., Roy, F., Zhan, W., Sheets, S. M., Sangberg, L., and Fletcher, H. M. (2006). VimA is part of the maturation pathway for the major gingipains of Porphyromonas gingivalis W83. Microbiology 152, 3383–3389. doi: 10.1099/mic.0.29146-0
Vimr, E. R. (1994). Microbial sialidases: does bigger always mean better? Trends Microbiol. 2, 271–277. doi: 10.1016/0966-842X(94)90003-5
Wang, Q., Chang, B. J., and Riley, T. V. (2010). Erysipelothrix rhusiopathiae. Vet. Microbiol. 140, 405–417. doi: 10.1016/j.vetmic.2009.08.012
Wang, Y., Gao, X., Wei, F., Zhang, X., Yu, J., Zhao, H., et al. (2014). Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene 533, 389–397. doi: 10.1016/j.gene.2013.09.038
Willingham, S. B., Bergstralh, D. T., O'connor, W., Morrison, A. C., Taxman, D. J., Duncan, J. A., et al. (2007). Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe 2, 147–159. doi: 10.1016/j.chom.2007.07.009
Yakubenko, V. P., Lishko, V. K., Lam, S. C., and Ugarova, T. P. (2002). A molecular basis for integrin alphaMbeta 2 ligand binding promiscuity. J. Biol. Chem. 277, 48635–48642. doi: 10.1074/jbc.M208877200
Yu, Y., Kanwar, S. S., Patel, B. B., Oh, P. S., Nautiyal, J., Sarkar, F. H., et al. (2012). MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis 33, 68–76. doi: 10.1093/carcin/bgr246
Yun, P. L., Decarlo, A. A., Collyer, C., and Hunter, N. (2001). Hydrolysis of interleukin-12 by Porphyromonas gingivalis major cysteine proteinases may affect local gamma interferon accumulation and the Th1 or Th2 T-cell phenotype in periodontitis. Infect. Immun. 69, 5650–5660. doi: 10.1128/IAI.69.9.5650-5660.2001
Zhang, Z., Zhu, Z., Watabe, K., Zhang, X., Bai, C., Xu, M., et al. (2013). Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 20, 1558–1568. doi: 10.1038/cdd.2013.110
Keywords: P. gingivalis, neuraminidase, macrophage, interleukin-12, microRNA-21, lncRNA GAS5
Citation: Yang X, Pan Y, Xu X, Tong T, Yu S, Zhao Y, Lin L, Liu J, Zhang D and Li C (2018) Sialidase Deficiency in Porphyromonas gingivalis Increases IL-12 Secretion in Stimulated Macrophages Through Regulation of CR3, IncRNA GAS5 and miR-21. Front. Cell. Infect. Microbiol. 8:100. doi: 10.3389/fcimb.2018.00100
Received: 13 October 2017; Accepted: 16 March 2018;
Published: 05 April 2018.
Edited by:
Jan Potempa, University of Louisville, United StatesReviewed by:
Teresa Olczak, University of Wrocław, PolandAshu Sharma, University at Buffalo, United States
Copyright © 2018 Yang, Pan, Xu, Tong, Yu, Zhao, Lin, Liu, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Li, lichen1980716@163.com
†These authors have contributed equally to this work.
 Xue Yang
Xue Yang Yaping Pan
Yaping Pan Xiaoyu Xu
Xiaoyu Xu Tong Tong
Tong Tong Shiwen Yu
Shiwen Yu Yue Zhao
Yue Zhao Li Lin
Li Lin Jingbo Liu
Jingbo Liu Dongmei Zhang
Dongmei Zhang Chen Li
Chen Li