The LuxS/AI-2 Quorum-Sensing System of Streptococcus pneumoniae Is Required to Cause Disease, and to Regulate Virulence- and Metabolism-Related Genes in a Rat Model of Middle Ear Infection
- 1Department of Otorhinolaryngology-Head and Neck Surgery, Korea University College of Medicine, Seoul, South Korea
- 2Institute for Medical Device Clinical Trials, Korea University College of Medicine, Seoul, South Korea
- 3Hubert Department of Global Health, Rollins School of Public Health, Emory University, Atlanta, GA, United States
Objective: Streptococcus pneumoniae colonizes the nasopharynx of children, and from nasopharynx it could migrate to the middle ear and causes acute otitis media (AOM). During colonization and AOM, the pneumococcus forms biofilms. In vitro biofilm formation requires a functional LuxS/AI-2 quorum-sensing system. We investigated the role of LuxS/AI-2 signaling in pneumococcal middle ear infection, and identified the genes that are regulated by LuxS/AI-2 during pneumococcal biofilm formation.
Methods: Streptococcus pneumoniae D39 wild-type and an isogenic D39ΔluxS strain were utilized to evaluate in vitro biofilm formation, and in vivo colonization and epithelial damage using a microtiter plate assay and a rat model of pneumococcal middle ear infection, respectively. Biofilm structures and colonization and epithelial damage were evaluated at the ultrastructural level by scanning electron microscopy and confocal microscopy. Microarrays were used to investigate the global genes that were regulated by LuxS/AI-2 during biofilm formation.
Results: The biofilm biomass and density of D39ΔluxS were significantly (p < 0.05) lower than those of D39 wild-type. SEM and confocal microscopy revealed that D39ΔluxS formed thin biofilms in vitro compared with D39 wild-type. The in vivo model of middle ear infection showed that D39ΔluxS resulted in ~60% less (p < 0.05) bacterial colonization than the wild-type. SEM analysis of the rat middle ears revealed dense biofilm-like cell debris deposited on the cilia in wild-type D39-infected rats. However, little cell debris was deposited in the middle ears of the D39ΔluxS-inoculated rats, and the cilia were visible. cDNA-microarray analysis revealed 117 differentially expressed genes in D39ΔluxS compared with D39 wild-type. Among the 66 genes encoding putative proteins and previously characterized proteins, 60 were significantly downregulated, whereas 6 were upregulated. Functional annotation revealed that genes involved in DNA replication and repair, ATP synthesis, capsule biosynthesis, cell division, the cell cycle, signal transduction, transcription regulation, competence, virulence, and carbohydrate metabolism were downregulated in the absence of LuxS/AI-2.
Conclusion: The S. pneumoniae LuxS/AI-2 quorum-sensing system is necessary for biofilm formation and the colonization of the ear epithelium, and caused middle ear infection in the rat model. LuxS/AI-2 regulates the expression of the genes involved in virulence and bacterial fitness during pneumococcal biofilm formation.
Introduction
Otitis media (OM) is one of the main reasons antibiotics are prescribed for children in both developing and developed countries (Grijalva et al., 2009; Arguedas et al., 2010). More than 80% of children up to the age of 3 year experience at least one episode of acute OM (AOM), and the health and economic burdens associated with AOM are significant (Pichichero, 2013; Usonis et al., 2016). Streptococcus pneumoniae (S. pneumoniae) is the most important bacteria that causes AOM in children (Bergenfelz and Hakansson, 2017). Although S. pneumoniae can cause disease, it is a commensal bacterium that quiescently and asymptomatically colonizes the mucosal surface of the nasopharynx in the form of a specialized structure called a biofilm (Bogaert et al., 2004; Simell et al., 2012). Once established as a biofilm, the bacteria can disperse to other typically sterile anatomical sites and cause pneumonia, OM, bacteremia, or meningitis (Hall-Stoodley et al., 2006; Sanchez et al., 2010; Weimer et al., 2010; Ash and Sheffield, 2013; Pichichero, 2013; Shak et al., 2013). It has been suggested that pneumococcal biofilms can also asymptomatically colonize the mucosal surfaces of the middle ear (during OM) and sinuses (during rhinosinusitis) (Hall-Stoodley et al., 2006; Sanderson et al., 2006; Hoa et al., 2009). During colonization and biofilm formation, the pneumococci replicate slowly and express low levels of virulence factors, such as the polysaccharide capsule. They also produce extracellular DNA, proteins, lipids, and polysaccharides.
The bacteria within biofilms are embedded in a self-produced extracellular polymeric substance (EPS) matrix, and are resistant to both host immune defenses and antibiotics (Donlan and Costerton, 2002). More frequent genetic transformation has also been detected in biofilms. The available DNA in the biofilm matrix serves as a substrate for transformation that can result in the evolution of resistant strains and the spread of drug-resistant genotypes (Trappetti et al., 2011c; Vidal et al., 2011; Croucher et al., 2012; Chao et al., 2014). It has been reported that the changes observed in pneumococcal transcription during biofilm formation are also associated with colonization (Sanchez et al., 2011). Furthermore, bacteria dispersed in biofilms have an increased propensity for tissue dissemination and pathogenesis compared with bacteria in free-floating, planktonic culture (Marks et al., 2013; Chao et al., 2014). The biofilm mode of growth provides an opportunity for pneumococci to colonize the upper respiratory tract and persist without causing disease; thus, pneumococcal carriage is common (Simell et al., 2012; Shak et al., 2013; Gilley and Orihuela, 2014). The persistence, pathogenesis, and drug resistance of pneumococcal biofilms is of high clinical importance.
During biofilm formation, bacteria regulate gene expression in response to changes in population density through a mechanism called quorum sensing (QS) (Hense et al., 2007). QS is mediated by secreted molecules called auto-inducers (AIs). One of these, a furanosyl borate diester called AI-2, is a metabolic byproduct of a luxS gene-encoded synthase: an enzyme involved primarily in the conversion of ribosyl-homocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione (DPD), which is the precursor of AI-2 (Chen et al., 2002; Trappetti et al., 2011a). Various studies have shown that LuxS regulates pneumococcal biofilm formation, competence, and autolysis (Trappetti et al., 2011c; Vidal et al., 2011).
Previous studies using a mouse model of pneumococcal colonization suggest that LuxS plays an important role in persistence in the nasopharynx (Joyce et al., 2004). It has also been reported that S. pneumoniae luxS mutant strain has low in vitro biofilm formation capacity, and is defective in genetic competence and iron uptake (Trappetti et al., 2011c; Vidal et al., 2011, 2013). Using a mouse model of intranasal challenge, Stroeher et al. (2003) demonstrated that the ability to spread from the nasopharynx to the lungs or blood was reduced in an S. pneumoniae D39 luxS mutant compared with the wild-type D39 strain (Stroeher et al., 2003). More recently, it has been reported that LuxS/AI-2 signaling enables pneumococci to use galactose as a carbon source, and enhances capsular polysaccharide production and the hyper-virulent phenotype (Trappetti et al., 2017). However, the role of the LuxS/AI-2 QS system in the global gene expression of pneumococcal biofilms, and in the in vivo colonization of the rat middle ear has not been reported. Therefore, in the present study we confirmed that LuxS/AI-2 is necessary for in vitro biofilm formation, analyzed its ultrastructure by electron microscopy, and assessed the effects of the absence of LuxS/AI-2 signaling on pneumococcal middle ear infection. We used a rat model of pneumococcal middle ear infection for the in vivo studies, and ultimately identified the global genes that are regulated by LuxS/AI-2 during pneumococcal biofilm formation.
Materials and Methods
Ethics Statement
The animal experiment protocol was approved by the Institute Review Board of Korea University, Guro Hospital, Seoul, South Korea. The animal experiments were carried out as per the guidelines provided by the Animal Research Committee, Korea University College of Medicine, Seoul, South Korea.
Bacterial Strains and Culture Media
Streptococcus pneumoniae D39 (NCTC 7466) was obtained from the Health Protection Agency Culture Collection (Salisbury, UK). It is Avery's Virulent Serotype 2 encapsulated strain, and is extremely virulent in animal models of infection (Avery et al., 1944). The S. pneumoniae luxS mutant strain (D39ΔluxS) has been prepared and characterized in previous studies (Vidal et al., 2011, 2013). The S. pneumoniae strains were grown on blood agar plates (BAPs) (Shin Yang Chemicals Co., Ltd., Seoul, Korea) and in brain heart infusion (BHI) broth.
In Vitro Biofilm Formation Abilities of S. pneumoniae D39 Wild-Type and D39ΔluxS
The in vitro biofilm formation abilities of S. pneumoniae D39 wild-type and an isogenic D39ΔluxS strain were evaluated using a static microtiter plate assay, as described previously (Christensen et al., 1982; Yadav et al., 2017b). Briefly, the pneumococcal strains were grown on BAPs overnight. A single colony from each plate was transferred to BHI broth and grown to the mid-exponential phase. The log-phase cells were diluted (1:200), and 1 mL of each cell suspension was seeded into a 24-well polystyrene flat-bottomed microtiter plate (BD Falcon, Sparks, MD, USA), and incubated at 37°C for various times. After incubation, the planktonic cells and medium were removed, and the biofilms remaining in the wells were washed twice with phosphate-buffered saline (PBS). The biofilms were then stained with 200 μL of 0.1% crystal violet (CV) for 15 min. After staining, the plates were washed twice with PBS and air-dried. The CV in the wells was dissolved in 1 mL of ethanol, 200 μL of the CV solution from each well was transferred to a 96-well plate, and the absorbance at 570 nm was measured using a micro-plate reader.
Alternative, the viable bacterial within the biofilms were detected by cfu counting. The biofilms were washed twice with PBS and suspended by sonication in 1 mL of PBS. The resulting biofilm suspensions were serially diluted and plated onto BAPs, and the bacteria were counted after overnight incubation at 37°C.
Effect of Incubation Time on the in Vitro Biofilm Growth of S. pneumoniae D39 Wild-Type and D39ΔluxS
It has been reported that S. pneumoniae biofilms are affected by incubation time. Therefore, to evaluate the effect of incubation time on in vitro biofilm formation, we grew S. pneumoniae D39 wild-type and D39ΔluxS biofilms for various times (6, 12, 18, and 24 h). We then determined biofilm biomass using a CV-microtiter assay, as described above.
Scanning Electron Microscopy (SEM) Analysis of in Vitro Biofilms Formed by the S. pneumoniae D39 Wild Type and D39ΔluxS
We investigated the morphologies of the in vitro-formed biofilms of the D39 wild-type and the D39ΔluxS strains using SEM. The biofilms were grown for 18 h, as described above. After incubation, the biofilms were washed with PBS, and fixed with 2% glutaraldehyde and 2.5% paraformaldehyde. The biofilms were then treated with 1% osmic acid for 2 h, and dehydrated with a graded series of ethanol (60–95%). Biofilm samples were washed thrice with t-butyl alcohol (Sigma, St. Louis, MO, USA), and preserved under freezing conditions. The biofilm samples were freeze-dried using ES-2030 equipment (Hitachi, Tokyo, Japan), and coated with platinum using an ion coater (IB-5; Eiko, Kanagawa, Japan). SEM images were captured by field emission-SEM (FE-SEM, S-4700; Hitachi, Tokyo, Japan).
Confocal Microscopy of S. pneumoniae D39 Wild-Type and D39ΔluxS Biofilms
Streptococcus pneumoniae D39 wild-type and D39ΔluxS biofilms were evaluated by confocal microscopy. The biofilms were grown on μ-slides (ibidi, Germany) for 18 h using the procedure described above. After incubation, the biofilms were stained using a LIVE/DEAD biofilm viability kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. After washing with PBS, the stained biofilms were examined using a Nikon A1 confocal microscope (Nikon Instruments, Inc., NY, USA) with fluorescein (green) and Texas red (red) band-pass filter sets.
Evaluation of the in Vivo Colonization Capability of the S. pneumoniae D39 Wild-Type and D39ΔluxS
The in vivo colonization capability of the D39 wild-type and D39ΔluxS strains was evaluated using a rat model of OM (Yadav et al., 2012b, 2017a). Twenty healthy, pathogen-free Sprague Dawley (SD) rats weighing approximately 150–200 g were purchased from Koatech (Pyeongtaek, South Korea). All rats were housed isolated under sterile conditions for 2 weeks prior to the start of the experiments, and were examined for abnormalities in the middle ear. They were then divided into four groups: the rats in group 1 (n = 7) were inoculated with S. pneumoniae D39 wild-type; the rats in group 2 (n = 7) were inoculated with S. pneumoniae D39ΔluxS; the rats in group 3 (n = 3) received the medium only (vehicle control); and the rats in group 4 (n = 3) received no treatment (no procedure control). The rats were anesthetized with a combination of Zoletil H (tiletamine-zolazepam; Virbac, Carros, France) and Rompun H (xylazine-hydrochloride; Bayer, Leverkusen, Germany) at a ratio of 1:1. The bacteria suspensions containing the S. pneumoniae wild-type or ΔluxS strain were prepared in BHI medium, and 50 μL (~1 × 107 colony-forming units (CFUs)) of the suspension (or medium only) was injected into the right middle ear of each rat in groups 1 (wild-type), 2 (ΔluxS), and 3 (medium) through the tympanic membrane (trans-tympanic membrane inoculation) using a tuberculin syringe and a 27-gauge needle. The rats were monitored daily for 1 week for abnormalities. The rats were then sacrificed, and bullae were acquired aseptically. The tympanic membranes and upper tissues were removed, and the middle ears were dissected and photographed. For the SEM analysis, representative bullae from each group were cleaned by trimming the bony parts so that the middle ear was clearly visible, and were preserved in SEM solution (glutaraldehyde and paraformaldehyde). To determine the number of viable bacteria, bullae from each group were aseptically homogenized with a mortar and pestle, serially diluted in PBS, and plated on BAP. After incubation at 37°C for 24 h, S. pneumoniae colonies were counted, and CFUs were calculated.
Differential Gene Expression Analysis of in Vitro Biofilms of S. pneumoniae D39 Wild-Type or D39ΔluxS
The global gene expression of S. pneumoniae D39 wild-type and D39ΔluxS biofilms were determined using a cDNA-microarray. For the experiment, S. pneumoniae D39 wild-type and D39ΔluxS biofilms were grown in 24-well plates for 18 h, as described above. The biofilms were washed twice with PBS, scraped, and suspended in PBS. The biofilm cells were then pelleted by centrifugation and treated with 100 μL of lysozyme [3 mg/mL in Tris-ethylenediaminetetraacetic acid (EDTA) buffer (TE); Sigma-Aldrich, St. Louis, MO, USA] for 4 min to lyse the cells. Total RNA was extracted using an RNeasy Total RNA Isolation System Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Contaminating DNA was removed by on-column RNase-free DNase (Qiagen) treatment for 10 min at 20–25°C. The quantity and quality of total RNA was detected using a NanoDrop (NanoDrop Technologies, Inc., Wilmington, DE, USA), and the integrity of the RNA was assessed using Bioanalyzer 2100 equipment (Agilent, Palo Alto, CA, USA).
RNA probe synthesis and hybridization were performed using the Agilent Low Input Quick Amp WT Labeling Kit according to the manufacturer's protocol. Briefly, 200 ng of total biofilm RNA was mixed with WT primer mix, and the samples were incubated at 65°C for 10 min. The cDNA master mix was then prepared with 5 × first strand buffer, 0.1 M dithiothreitol, 10 mM dNTP mix, and RNase Block Mix (AffinityScript), and added to the RNA + WT primer reaction mixture. The samples were incubated at 40°C for 2 h, and reverse transcription and dsDNA synthesis were terminated by incubation at 70°C for 15 min. The transcription master mix was prepared according to the manufacturer's protocol (5 × transcription buffer, 0.1 M dithiothreitol, NTP mix, T7-RNA polymerase blend, and cyanine 5-CTP in nuclease-free water). The transcription of dsDNA was performed by adding transcription master mix to the dsDNA reaction samples and incubating the mix at 40°C for 2 h. The amplified and labeled cRNA was purified on an RNase mini column (Qiagen) according to the manufacturer's protocol. The labeled complementary RNA (cRNA) target was quantified using an spectrophotometer.
After checking the labeling efficiency of the cyanine 5-labeled cRNA target, the cRNA was fragmented by adding 10 × blocking agent and 25 × fragmentation buffer, and incubating at 60°C for 30 min. The fragmented cRNA was resuspended in 2 × hybridization buffer and directly pipetted onto an assembled S.pneumoniae_6 x 7k V2 Microarray (mycroarray.com). The arrays were hybridized at 57°C for 17 h in an Agilent Hybridization oven. The hybridized microarrays were washed according to the manufacturer's washing protocol (Agilent Technology). After overnight incubation at 42°C, the slides were washed twice with washing solution 1 (containing 2 × saline-sodium citrate buffer (SSC) and 0.1% sodium dodecyl sulfate) for 5 min at 42°C, washed once with washing solution 2 (containing 0.1 × SSC and 0.1% sodium dodecyl sulfate) for 10 min at room temperature, and finally washed four times with 0.1 × SSC for 1 min at room temperature. The slides were dried by centrifugation at 650 rpm for 5 min. The hybridization image on the slide was scanned using 4000B apparatus (Axon Instruments, Union City, CA, USA).
The hybridization images were analyzed using GenePix Pro 3.0 software (Axon Instruments, Union City, CA, USA) to obtain the gene expression ratios of the D39 wild-type and D39ΔluxS biofilms. The microarray data were analyzed using Genowiz 4.0™ (Ocimum Biosolutions, Hyderabad, India), and normalized with Global LOWESS. The cutoffs for upregulated and downregulated genes were +2-fold and−2-fold, respectively. The microarray experiment was performed with three biological replicates. Statistical significance was calculated using Student's t-test, and p-values < 0.05 were considered significant. STRING version 10.5 (https://string-db.org) was used for functional annotation, and the UniProtKB database (http://www.uniprot.org/uniprot/P0A4M0) was used to search for clusters of biological processes in the gene ontology database within the two sets of differentially expressed genes (D39ΔluxS and D39 wild-type). The microarray data have been deposited in the National Center for Biotechnology Information (NCBI)'s Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/info/linking.html), and are accessible through GEO Series accession number GSE109347.
Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR) Analysis
To confirm the microarray data by real-time RT-PCR, we chose 15 genes that are differentially expressed in biofilms, and the 16S gene as a control. The primer sequences are presented in Table 1. Each 20-μL real-time RT-PCR reaction mixture included 10 μL of 2 × SYBR Green PCR Master Mix (Roche Applied Science, Indianapolis, IN, USA), 5 pmol each of the forward and reverse primers, and 2 μL of complementary DNA (cDNA). The PCR conditions were: an initial denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, and annealing and extension at 60°C for 1 min. Negative controls, which contained nuclease-free water instead of RNA, were included to confirm that the samples were free from contamination. To verify the absence of contaminating genomic DNA, each RT-PCR experiment included a no reverse transcriptase control. Relative gene expression was determined using the 2−ΔΔCT method. The reference gene was 16S, and the standard condition was the D39 wild-type biofilm.
Results
The luxS Mutant Strain Formed Less Biofilm in Vitro
It has been reported that the LuxS/AI-2 QS system regulates biofilm formation in S. pneumoniae. In the present study, we compared the planktonic growth and biofilm formation capability of S. pneumoniae D39 wild-type and the isogenic D39ΔluxS strain. No significant difference in the density of planktonic cells was observed between the D39 wild-type and D39ΔluxS strains (Figure 1A). After 18 h, the biofilm biomass of D39ΔluxS was significantly (p < 0.05) lower than that of the D39 wild-type when it was analyzed using the CV microplate assay (Figure 1B), or by bacterial counts (Figure 1C).
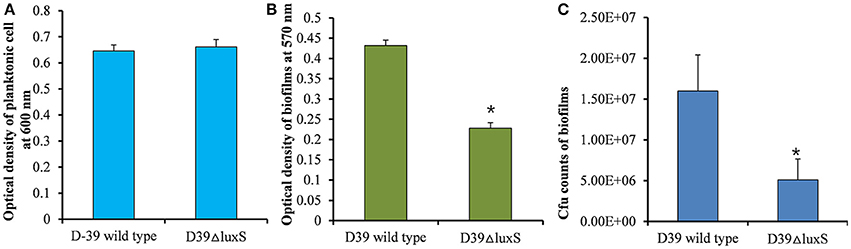
Figure 1. Streptococcus pneumoniae D39 wild-type and D39ΔluxS planktonic and biofilm growth. (A) Planktonic growth optical density at 600 nm. (B) Quantification of biomass of in vitro biofilms grown for 18 h, using a CV-microplate assay. (C) Colony-forming unit (CFU) counts of in vitro biofilms grown for 18 h. Error bars are the standard deviation from the mean. Statistical significance was calculated using the Student's t-test,*p < 0.05.
We then conducted a time-course experiment to evaluate the effect of the absence of the luxS gene at various stages of biofilm formation. The results revealed a significant (p < 0.05) decrease in biofilm biomass in D39ΔluxS at 6, 12, 18, and 24 h post-inoculation (Figure 2). The results revealed that the D39ΔluxS mutant formed significantly less biofilm biomass than the D39 wild-type at all time-points.
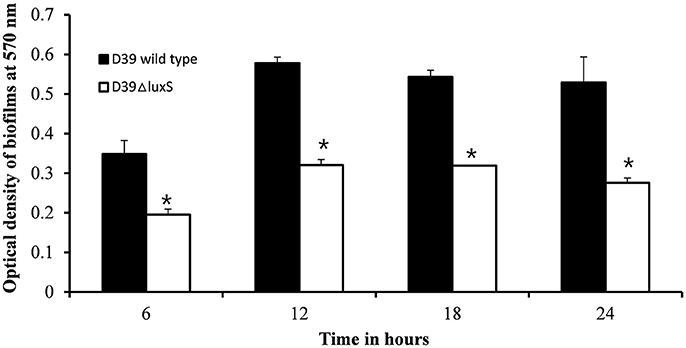
Figure 2. In vitro biofilm growth of Streptococcus pneumoniae D39 wild-type and D39ΔluxS strains at different time-points after inoculation (6, 12, 18, and 24 h). The error bars are the standard deviation from the mean. Statistical significance was calculated using the Student's t-test,*p < 0.05.
SEM Revealed Thin and Scattered Biofilms Formed by the luxS Mutant Strain
We used SEM to investigate the morphologies of the D39 wild-type and D39ΔluxS biofilms grown for 18 h. The SEM analysis revealed that the D39 wild-type strain formed thick, three-dimensionally (3D) organized heterogeneous biofilms. The cells in the D39 wild-type biofilms were surrounded by extracellular polysaccharides (EPS), and were attached to both the bottom of the plate and to each other, forming an organized 3D biofilm structure with significant depth (Figures 3A–C). In contrast, the biofilms formed by D39ΔluxS were thin and disorganized. The cells were attached only to the base of the plate, and no cell–cell adherence was observed (Figures 3D–F). The cell surfaces were smooth and devoid of EPS.
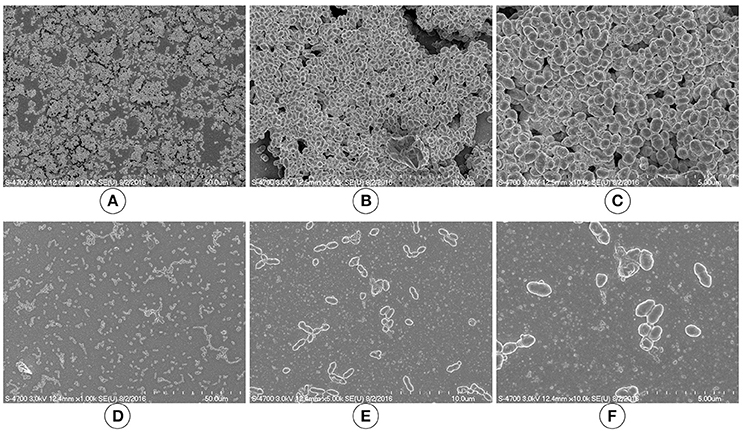
Figure 3. Scanning electron microscopy (SEM) images of Streptococcus pneumoniae in vitro biofilms grown for 18 h. (A–C) are representative SEM images of the D39 wild-type strain. The wild-type strain biofilms were thick and organized with a significant depth. (D–F) are SEM images of the D39ΔluxS strain. The D39ΔluxS biofilms were thin and disorganized, and extracellular polymeric substance (EPS) was absent.
Confocal Microscopy of in Vitro Biofilms Formed by the D39ΔluxS Mutant and D39 Wild-Type Strains
We investigated the D39ΔluxS and D39 wild-type biofilms grown for 18 h in vitro using a confocal microscope. Confocal microscopy revealed a significant difference in the morphology of the biofilms formed by the D39ΔluxS and D39 wild-type strains. The D39 wild-type biofilms were compact, thick, and had a well-organized 3D structure (Figure 4A). In contrast, the D39ΔluxS biofilms were thin with scattered pneumococci attached to the bottom of the dish. Their 3D structure was disorganized (Figure 4B).
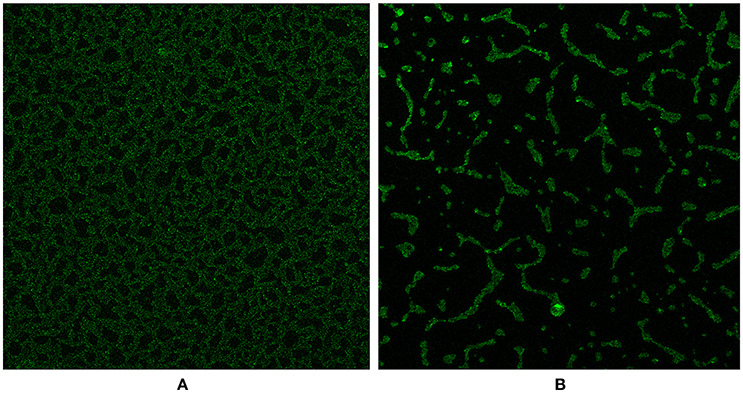
Figure 4. Confocal microscopy images of Streptococcus pneumoniae in vitro biofilms grown for 18 h. (A) Confocal microscopy image of the D39 wild-type strain biofilm. (B) Confocal microscopy image of the D39ΔluxS strain biofilm.
A Mutation in luxS Decreases Colonization of S. pneumoniae in the Rat Middle Ear
The in vivo colonization abilities of the D39 wild-type and D39ΔluxS strains were evaluated using a rat model of middle ear infection. At 1 week post-inoculation, the rats were sacrificed, and their bullae were obtained, dissected, and cleaned of unwanted tissue. As Figure 5A shows, the rat bullae inoculated with the D39 wild-type were completely filled with biofilm-like debris, and exhibited severe mucosal swelling (Figure 5A). In contrast, the rat bullae inoculated with D39ΔluxS showed signs of inflammation, with a thick mucosa, but no visible biofilm-like debris (Figure 5B). As expected, the bullae of the control animals were clean, with no signs of inflammation (Figure 5C). The bacterial density in the middle ears of the rats inoculated with D39 wild-type was ~7.04 × 104 (SD ± 26083.2) CFU/bullae, whereas that of the rats inoculated with the D39ΔluxS strain was significantly lower at ~1.85 × 104 (SD ± 8859) CFU/bullae (Figure 5D). The mean CFUs of the D39ΔluxS strain were significantly lower (~60% reduction, p < 0.05) than the CFUs of the D39 wild-type strain.
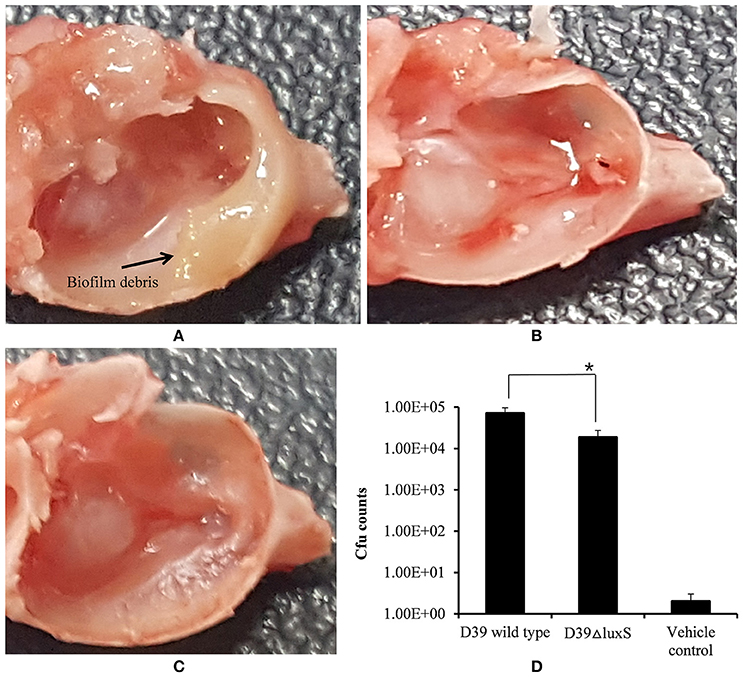
Figure 5. Digital photos of rat bullae and colony-forming unit (CFU) counts of bullae inoculated with Streptococcus pneumoniae D39 wild-type or D39ΔluxS. (A) Rat bullae inoculated with the D39 wild-type. (B) Rat bullae inoculated with the D39ΔluxS, and (C) Rat bullae inoculated with medium. (D) CFU of S. pneumoniae colonizing the rat middle ear mucosa. Error bars are the standard deviation from the mean. Statistical significance was calculated using the Student's t-test,*p < 0.05.
Histologically, the middle ear mucosa comprises both non-ciliated squamous epithelium and ciliated epithelium. The ciliated epithelium is distributed in the hypotympanum and Eustachian tube orifice, whereas the remainder of the middle ear bulla is covered with non-ciliated squamous epithelium. SEM images of a rat middle ear colonized by D39 wild-type are shown in Figures 6A–C. In these images, thick cells or biofilm debris deposits were visible (arrow). Cilia, however, were not visible but were completely covered with biofilm debris (Figures 6A–C). Conversely, in the rat middle ear colonized with D39ΔluxS, less biofilm debris was observed, although the cilia were coagulated (Figures 6D–F). The middle ears of the rats inoculated with vehicle (control) were clean, and neither biofilms nor cell debris were detected (Figures 6G–I).
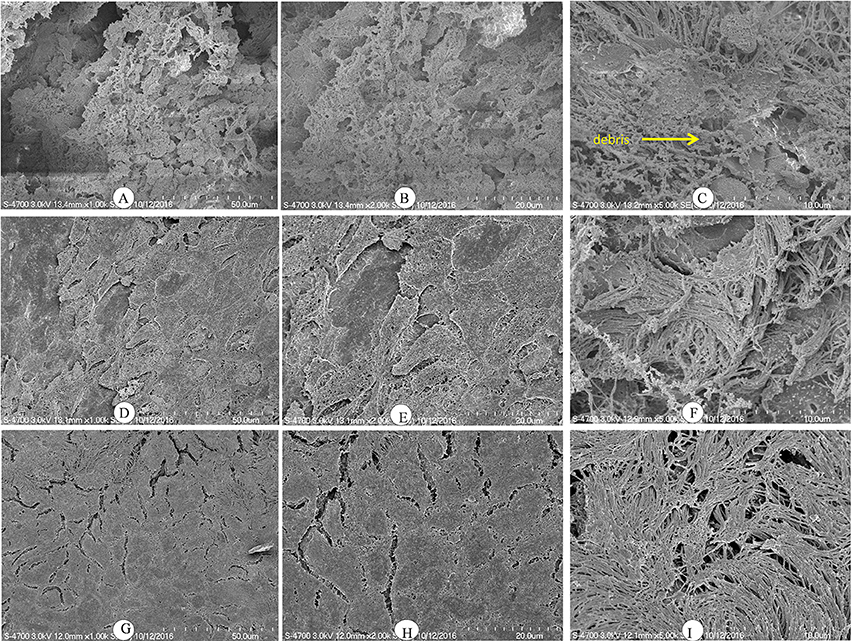
Figure 6. Scanning electron microscopy (SEM) images of rat bullae inoculated with Streptococcus pneumoniae D39 wild-type and D39ΔluxS. (A–C) are representative SEM images of rat bullae inoculated with the D39 wild-type strain. In rats colonized with the wild-type strain, dense biofilm/cell debris was deposited on the cilia, and the cilia were coagulated and completely covered with biofilm debris. (D–F) are representative SEM images of rat bullae inoculated with the D39ΔluxS strain. In rat bulla colonized with the D39ΔluxS strain, less biofilm debris was visible, although the cilia were coagulated. (G–I) are representative SEM images of rat bullae inoculated with medium (vehicle control). The vehicle control rat bulla were clean.
Differential Gene Expression in D39 Wild-Type Biofilms and Those Produced by the D39ΔluxS Mutant
The changes in the gene expression levels of the D39ΔluxS mutant biofilm with respect to D39 wild-type were evaluated using a cDNA microarray. Total RNA was extracted from 18-h-old biofilm samples grown in triplicate on a microtiter plate. The cDNA synthesis, labeling, and hybridization were performed as per standard protocols. The hybridization images were analyzed to detect the gene expression ratios in the D39 wild-type and D39ΔluxS biofilms. The fold changes in gene expression of the D39ΔluxS biofilms were detected with respect to the D39 wild-type. The ±2-fold changes in gene expression in the three biological replicate samples were included in this study. The cDNA-microarray analysis demonstrated that 117 genes were differentially expressed in D39ΔluxS compared with the D39 wild-type. According to functional group analysis, 51 of these genes were uncharacterized/hypothetical. Among the 66 genes encoding putative and previously characterized proteins, 60 were significantly downregulated and 6 were significantly upregulated (Table 2). The KEGG pathways of down-regulated genes in biofilms of S. pneumoniae D39ΔluxS with respect to D39 wild-type are shown in Figure 7.
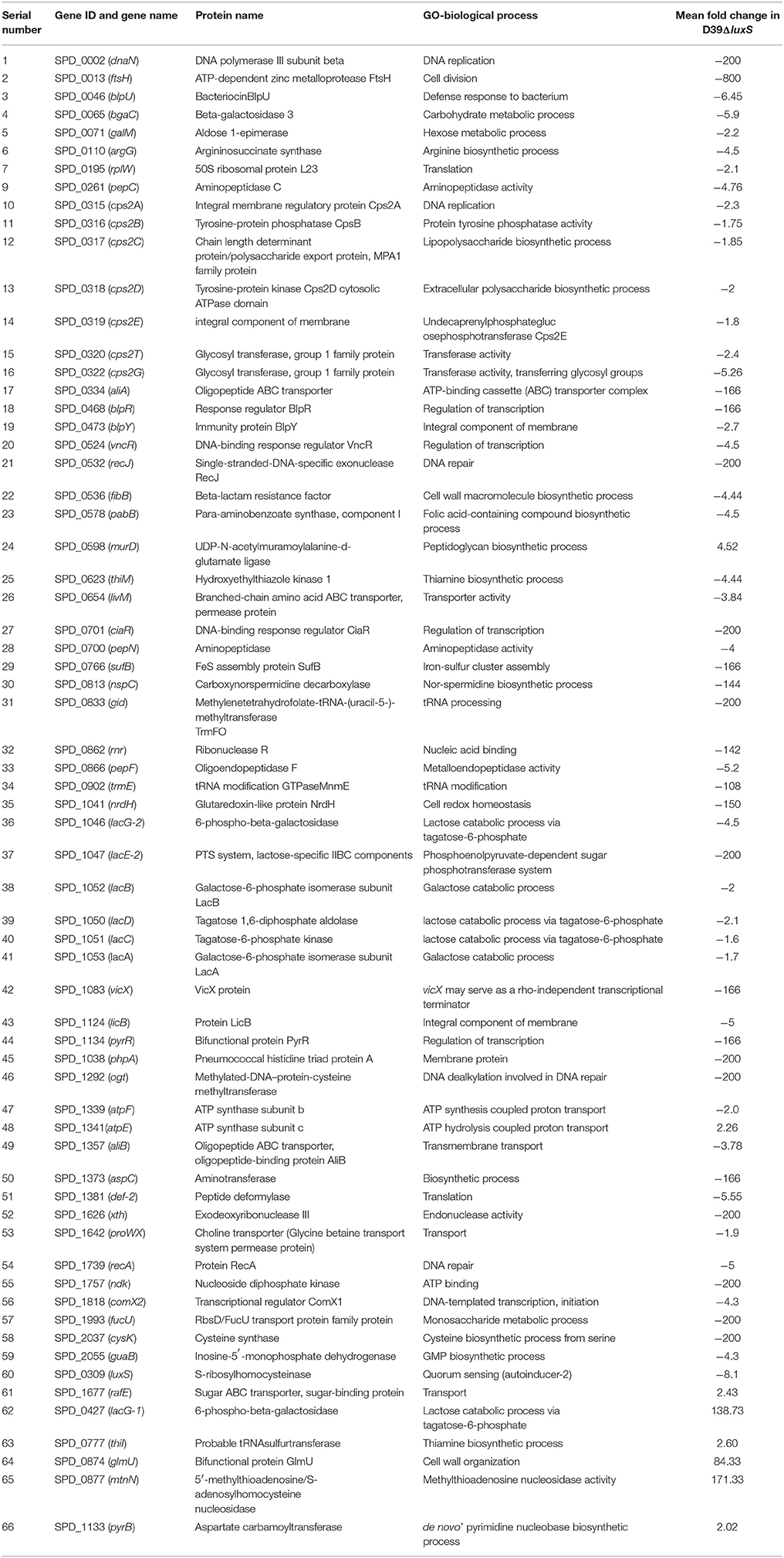
Table 2. List of differentially expressed genes in biofilms of the Streptococcus pneumoniae D39luxS mutant strain compared with the wild-type D39.
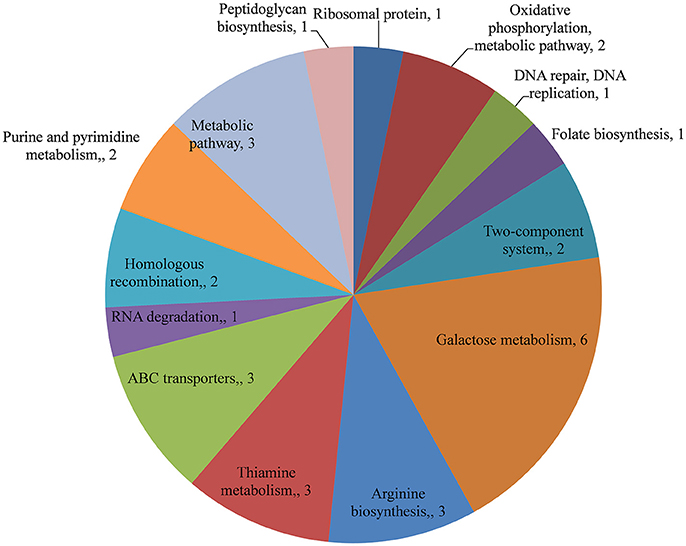
Figure 7. KEGG pathway analysis of genes downregulated in Streptococcus pneumoniae D39ΔluxS biofilms compared with the D39 wild-type biofilms.
The functional annotation of the differentially regulated genes in D39ΔluxS and D39 wild-type revealed that the genes involved in DNA replication and repair, ATP synthesis, capsule biosynthesis, cell division and the cell cycle, signal transduction, transcription regulation, competence, virulence, and fructose, lactose and galactose metabolism were down-regulated in the absence of LuxS/AI-2. The fold changes in gene expression in D39ΔluxS compared with the D39 wild-type from three independent samples are shown in Table 2.
Galactose metabolism involves the tagatose-6-phosphate and Leloir pathways. Our results revealed that the tagatose-6-phosphate pathway genes (lacA, lacB, lacC, lacD, and lacG-2), and the Leloir pathway (galM) gene were significantly downregulated in D39ΔluxS. However, the lacG-1 gene was upregulated. Furthermore, lacE2 (encoding the PTS system, lactose-specific IIBC components), bgaC (encoding beta-galactosidase 3), and fucU (encoding RbsD/FucU transport protein family) were also significantly downregulated. The cps2A, cps2B, cps2C, cps2D, cps2E, cps2T, and cps2G genes involved in pneumococcal capsule synthesis were significantly downregulated in the D39ΔluxS. Similarly, the gene expression levels of the genes encoding DNA replication (dnaN), DNA repair (recJ, recA, and ogt), and ABC transportation (aliA, aliB, livM, and rafE) were downregulated in the D39ΔluxS. The xth and rnr genes encoding exodeoxyribonuclease III and ribonuclease R were also downregulated. Furthermore, the luxS gene mutation downregulated the transcription of regulatory protein-encoding genes such as ciaR, comX2, blpR, vncR, and pyrR, and ATP synthesis genes (atpF and atpE). The ciaR gene encodes a DNA-binding response regulator protein of the two-component regulatory system known as CiaH/CiaR, which is involved in the early steps of competence regulation (Guenzi et al., 1994), and the comX2 gene encodes a sigma factor that functions as a competence-specific global transcription modulator involved in bacterial competence.
Gene Expression Analysis Using Real-Time RT-PCR
To validate the microarray results, real-time RT-PCR was performed on 15 differentially expressed genes. The 16S rDNA gene was included as a control. The fold changes in gene expression were calculated after normalization of each gene to 16S gene expression levels using the comparative threshold method. The real-time RT-PCR results agree with the microarray shown in Table 3.

Table 3. Fold changes in the gene expression of a Streptococcus pneumoniae luxS mutant strain (D39ΔluxS) during biofilm growth compared with the parental wild-type strain, detected by real-time polymerase chain reaction (PCR).
Discussion
Streptococcus pneumoniae asymptomatically colonizes the nasopharyngeal cavity in the form of structures called biofilms (Moscoso and García, 2009). However, bacteria from these biofilms can disperse to other typically sterile sites and causes diseases of the lungs (pneumonia), middle ear (OM), brain (meningitis), and blood (bacteremia) (Hava et al., 2003). The biofilm growth of S. pneumoniae is in part regulated by the LuxS/AI QS system (Vidal et al., 2011). Moreover, Trappetti et al. (2011c) reported that LuxS regulates pneumococcal biofilm formation, competence, and fratricide (Trappetti et al., 2011c). However, the effects of luxS mutation on global gene expression in pneumococcal biofilms and colonization capability in the middle ear were previously unknown. In the present study, we investigated the effect of S. pneumoniae luxS gene mutation on in vitro biofilm formation capability and colonization of the rat middle ear mucosa as well as global gene expression in biofilms.
Vidal et al. (2011) detected low in vitro biofilm capability in D39ΔluxS compared with its parental D39 wild-type strain (Vidal et al., 2011). Herein, we showed that D39ΔluxS was unable to form robust early (6–12-h) and late (18–24-h) biofilms. The D39ΔluxS biofilms had ~60% less biomass and lower viable bacterial counts. LuxS/AI-2 QS is vital for in vitro pneumococcal biofilm growth, and in vivo colonization and pathogenesis. In S. pneumoniae, AI-2 synthesis from methionine occurs in the activated methyl cycle (AMC), which involves MTA/SAH nucleosidase (Pfs) and LuxS (Parveen and Cornell, 2011). MTA/SAH nucleosidase (Pfs) produces SRH, which is further cleaved by LuxS into homocysteine and 4,5-dihydroxy-2,3-pentanedione, the precursor of AI-2. Herein, we detected increased expression levels of the MTA/SAH nucleosidase-encoding gene, indicating an imbalance due to inactivation of LuxS and high MTA/SAH nucleosidase expression. The products of AMC—such as MTA, SAH, and 5′-deoxyadenosine (5′dADO)—are toxic and product inhibitors that need to be removed for normal growth (Parveen and Cornell, 2011). Therefore, it can be assumed that the low biofilm capability of the luxS mutant strain could be caused by the attenuation of AI-2 synthesis and an imbalance in the methionine pathway or AMC due to the accumulation of product inhibitors (Halliday et al., 2010; Vidal et al., 2013).
Another interesting finding was the presence of low levels of EPS in the D39ΔluxS biofilms. Although we did not quantify the EPS present in the biofilms, the SEM images revealed that the D39ΔluxS biofilms were devoid of EPS, and the bacteria were scattered on the plate.
Pneumococci typically colonize the nasopharyngeal cavities of young children and elderly people. However, under immune-suppressed conditions, these bacteria can disperse to other sterile sites, such as the middle ear, and cause OM. Disease severity depends on the successful colonization of the middle ear mucosa by S. pneumoniae. Our in vivo results demonstrated that D39ΔluxS was less capable of colonizing the rat middle ear mucosa than the wild-type strain. Significantly (> 60%) fewer bacteria were recovered from the rat middle ears inoculated with D39ΔluxS compared with the ears inoculated with the D39 wild-type strain. These results indicate that a loss of luxS renders bacteria unfit for successful colonization of the rat mucosal membrane. The results also indicate that in S. pneumoniae, LuxS plays an important role in the colonization of the rat middle ear, causing OM. Using a pneumonia model, it has been demonstrated that a less virulent luxS mutant strain is less likely to spread to the lungs and blood (Stroeher et al., 2003). Previous studies have demonstrated that the LuxS QS system plays an important role in the persistence, virulence, and dissemination of S. pneumoniae (Stroeher et al., 2003; Joyce et al., 2004; Vidal et al., 2013). This QS system has been implicated in the persistence of pneumococci in the mouse nasopharynx (Joyce et al., 2004). In pneumococci, LuxS-mediated QS plays an important role in survival and fitness. QS is mediated by a small molecule called auto-inducer-2, which is synthesized by the product of the luxS gene in an AMC. The LuxS catalyzes the conversion of SRH to AI-2. Moreover, the LuxS enzyme is absent in humans, and could be an attractive target for novel therapeutic agents against S. pneumoniae. Our previous studies showed that blocking the auto-inducer synthesis pathway decreased pneumococcal colonization in the rat middle ear, as well as in vitro biofilm formation (Yadav et al., 2012a, 2014). Furthermore, it has been suggested that quorum sensing or auto-inducer activity inhibition may increase the success of antibiotic treatment by increasing the susceptibility of bacterial biofilms and/or by increasing host survival following infection (Wnuk et al., 2009; Brackman et al., 2011).
To further investigate the effect of luxS mutation on global gene expression in biofilms, we performed a global gene expression analysis using a cDNA microarray. The expression analysis revealed 117 genes that were differentially expressed in D39ΔluxS compared with the D39 wild-type. A large number of genes (60) encoding putative proteins were significantly downregulated, whereas only 6 genes were significantly upregulated. The downregulation of genes encoding DNA replication, repair, cell division, and cell wall protein biosynthesis in the luxS mutant indicates that cell division may be perturbed, which results in reduced in vitro biofilm formation and in vivo colonization capability. Interestingly, a large number of genes encoding ribosomal proteins were downregulated; however, the expression levels were <2-fold (data not shown).
In S. pneumoniae, the CSP-mediated QS system plays an important role in biofilm growth, genetic competence, and pathogenesis. The CSP-mediated QS competence system includes the two-component regulatory system CiaH-CiaR, early competence genes (comAB and comCD), and the late competence gene comX (which encodes a global transcription modulator; Ishii et al., 2017). In the present study, we detected a significant reduction in the expression levels of the ciaR gene (which encodes the DNA-binding response regulator protein of the two-component regulatory system CiaH/CiaR), and the late competence comX2 gene. Recently, it has been reported that a ciaR gene mutant of Streptococci sanguinis was unable to form robust biofilms (Zhu et al., 2017). In S. pneumoniae, the ComX alternative sigma factor plays an important role in the initiation of the transcription of the late competence-specific operon, which facilitates DNA uptake and the recombination of DNA (Lee and Morrison, 1999; Luo and Morrison, 2003). Moreover, it has been reported that competence QS plays an important role in biofilm formation, and a ciaR/H gene mutant strain is unable to form biofilms (Trappetti et al., 2011b).
The two ABC transporter genes aliA and aliB were downregulated in the luxS mutant (which encodes the Ami-AliA/AliB oligopeptide permease, an ATP-binding cassette transporter that is involved in nutrient uptake). It has been reported that the expression of aliA and aliB genes is required for nasopharyngeal cavity colonization, and the mutant strain is unable to colonize in vivo (Kerr et al., 2004).
The pneumococcal capsule is a major virulence factor, and protects bacteria by interfering with the phagocytic activity of the host (Hyams et al., 2010). The pneumococcal capsule is composed of immunogenic capsular polysaccharides (CPSs) that are encoded by the cpsABCD genes (Guidolin et al., 1994; Feldman and Anderson, 2014). In the present study, microarray analysis revealed significantly reduced expression levels of cps2A, cps2B, cps2C, cps2D, cps2E, cps2T, and cps2G in the D39ΔluxS strain compared with the corresponding levels in the D39 wild-type strain. It has been suggested that mutation in each of the cpsABCD genes results in a significant reduction in capsule synthesis, as well as reduced virulence and decreased bacterial colonization of the nasopharyngeal cavity (Bender et al., 2003; Morona et al., 2004, 2006). Therefore, blocking LuxS activity probably reduces virulence and renders the bacteria vulnerable to the opsonophagocytic activity of the host (Kim et al., 1999).
The ATP synthesis genes atpF and atpE were downregulated in the D39ΔluxS strain. Membrane-associated F0F1 H+-ATPase is essential for bacteria, and in S. pneumoniae, the primary roles of this enzyme are to create a proton gradient using the energy provided by ATP hydrolysis, and to maintain intracellular pH via proton extrusion (Martín-Galiano et al., 2001). In S. pneumoniae, the activity of the F0F1 ATPase increases as the pH of the growth medium decreases. Regulation of this pH-inducible phenotype occurs at the level of transcription initiation (Martín-Galiano et al., 2001). In streptococci, the F0F1 H+-ATPase-encoding operon is atpEBFHAGDC, and the F0 gene order is atpEBF (Shabayek and Spellerberg, 2017).
In the present study our results demonstrated the downregulation of galactose pathway genes, such as lacB, lacC, lacD, and lacA (of the T6P pathway), and galM (of the Leloir pathway) in the D39ΔluxS strain (Figure 8). In addition, lacE2 (encoding the PTS system, lactose-specific IIBC components), bgaC (encoding beta-galactosidase 3), and fucU (encoding the RbsD/FucU transport protein family) were also downregulation in D39ΔluxS. Pneumococci can utilize various carbohydrate sources during colonization of the nasopharyngeal cavity (Buckwalter and King, 2012; Yadav et al., 2013). It has been reported that galactose catabolic route genes such as lacAB and lacD (of the T6P pathway) and galM (of the Leloir pathway) are required for pneumococcal colonization, and were upregulated during bacteria growth on mucin (Paixão et al., 2015). Furthermore, it has been suggested that lacD (T-6-p) or the Leloir pathway mutant D39 are less capable of colonizing the murine nasopharynx and have reduced virulence (Paixão et al., 2015). Recently, Trappetti et al. (2017) suggested that LuxS/AI-2 signaling enables pneumococci to utilize galactose as a carbon source, and enhances capsular polysaccharide production and the hyper-virulent phenotype (Trappetti et al., 2017). Present and previous study results demonstrate that the loss of functional LuxS hinders carbohydrate utilization, leading to the reduced colonization capability of pneumococci in vivo.
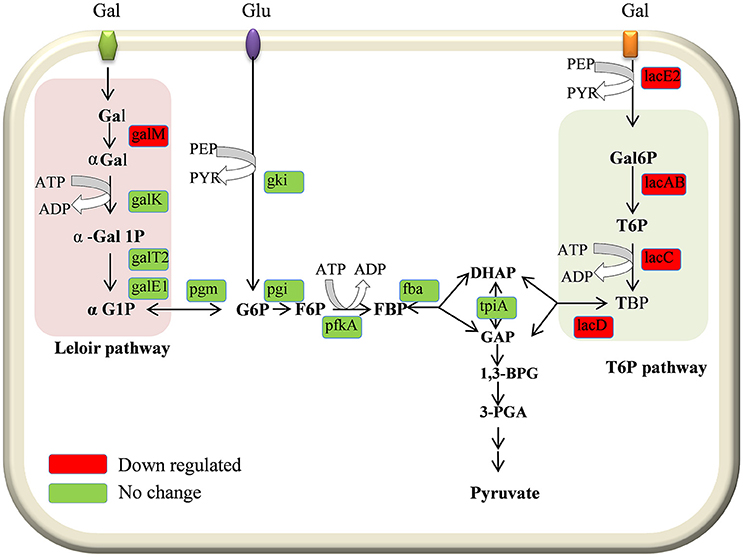
Figure 8. Schematic representation of galactose metabolism pathway in Streptococcus pneumoniae D39. In S. pneumoniae, lactose, and galactose are metabolized by the tagatose-6-phosphate pathway (light gray box) and the Leloir pathway (left; light pink box). The lacA, lacB, lacC, and lacD genes are involved in the tagatose-6-phosphate pathway, and the galM, galK, galT-1, and galE-2 genes are involved in the Leloir pathway. The lacE2 gene (which encodes the PTS system, lactose-specific IIBC component) is required for galactose transport. The lacA, lacB, lacC, lacD, galM, and lacE-2 genes were downregulated in the present study.
Conclusion
The results of this study demonstrate that the S. pneumoniae D39ΔluxS strain has a reduced ability to form early and late in vitro biofilm, and is less capable of colonizing the rat middle ear mucosa. LuxS/AI-2 regulates the expression of the genes involved in cell division and growth, capsule synthesis, carbohydrate metabolism, competence, virulence, and bacterial fitness during colonization.
Author Contributions
MY, J-JS, S-WC, YG, SK and JV conceived and designed the experiments. MY and SK performed the experiments. MY, J-JS, S-WC, YG, and JV analyzed the data; S-WC, YG, J-JS, SK, and JV contributed reagents, materials, analysis tools. MY, J-JS, and JV wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (NFR-2017R1A2B40005163) and Korea University Grant. JV was supported by a grant from the National Institutes of Health (NIH; 5R21AI112768-02), the content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH.
References
Arguedas, A., Kvaerner, K., Liese, J., Schilder, A. G., and Pelton, S. I. (2010). Otitis media across nine countries: disease burden and management. Int. J. Pediatr. Otorhinolaryngol. 74, 1419–1424. doi: 10.1016/j.ijporl.2010.09.022
Ash, S. Y., and Sheffield, J. V. L. (2013). Pneumococcus. Med. Clin. North Am. 97, 647–666. doi: 10.1016/j.mcna.2013.03.005
Avery, O. T., MacLeod, C. M., and McCarty, M. (1944). Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J. Exp. Med. 79, 137–158.
Bender, M. H., Cartee, R. T., and Yother, J. (2003). Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J. Bacteriol. 185, 6057–6066. doi: 10.1128/JB.185.20.6057-6066.2003
Bergenfelz, C., and Hakansson, A. P. (2017). Streptococcus pneumoniae otitis media pathogenesis and how it informs our understanding of vaccine strategies. Curr. Otorhinolaryngol. Rep. 5, 115–124. doi: 10.1007/s40136-017-0152-6
Bogaert, D., de Groot, R., and Hermans, P. W. (2004). Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4, 144–154. doi: 10.1016/S1473-3099(04)00938-7
Brackman, G., Cos, P., Maes, L., Nelis, H. J., and Coenye, T. (2011). Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 55, 2655–2661. doi: 10.1128/AAC.00045-11
Buckwalter, C. M., and King, S. J. (2012). Pneumococcal carbohydrate transport: food for thought. Trends Microbiol. 20, 517–522. doi: 10.1016/j.tim.2012.08.008
Chao, Y., Marks, L. R., Pettigrew, M. M., and Hakansson, A. P. (2014). Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front. Cell. Infect. Microbiol. 4:194. doi: 10.3389/fcimb.2014.00194
Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L., et al. (2002). Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549. doi: 10.1038/415545a
Christensen, G. D., Bisno, A. L., Parisi, J. T., McLaughlin, B., Hester, M. G., and Luther, R. W. (1982). Nosocomial septicemia due to multiply antibiotic-resistant staphylococcus epidermidis. Ann. Intern. Med. 96, 1–10. doi: 10.7326/0003-4819-96-1-1
Croucher, N. J., Harris, S. R., Barquist, L., Parkhill, J., and Bentley, S. D. (2012). A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 8:e1002745. doi: 10.1371/journal.ppat.1002745
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Feldman, C., and Anderson, R. (2014). Review: current and new generation Pneumococcal vaccines. J. Infect. 69, 309–325. doi: 10.1016/j.jinf.2014.06.006
Gilley, R. P., and Orihuela, C. J. (2014). Pneumococci in biofilms are non-invasive: implications on nasopharyngeal colonization. Front. Cell. Infect. Microbiol. 4:163. doi: 10.3389/fcimb.2014.00163
Grijalva, C. G., Nuorti, J. P., and Griffin, M. R. (2009). Antibiotic prescription rates for acute respiratory tract infections in us ambulatory settings. JAMA 302, 758–766. doi: 10.1001/jama.2009.1163
Guenzi, E., Gasc, A. M., Sicard, M. A., and Hakenbeck, R. (1994). A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12, 505–515. doi: 10.1111/j.1365-2958.1994.tb01038.x
Guidolin, A., Morona, J. K., Morona, R., Hansman, D., and Paton, J. C. (1994). Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect. Immun. 62, 5384–5396.
Halliday, N. M., Hardie, K. R., Williams, P., Winzer, K., and Barrett, D. A. (2010). Quantitative liquid chromatography–tandem mass spectrometry profiling of activated methyl cycle metabolites involved in LuxS-dependent quorum sensing in Escherichia coli. Anal. Biochem. 403, 20–29. doi: 10.1016/j.ab.2010.04.021
Hall-Stoodley, L., Hu, F. Z., Gieseke, A., Nistico, L., Nguyen, D., Hayes, J., et al. (2006). Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296, 202–211. doi: 10.1001/jama.296.2.202
Hava, D. L., LeMieux, J., and Camilli, A. (2003). From nose to lung: the regulation behind Streptococcus pneumoniae virulence factors. Mol. Microbiol. 50, 1103–1110. doi: 10.1046/j.1365-2958.2003.03764.x
Hense, B. A., Kuttler, C., Müller, J., Rothballer, M., Hartmann, A., and Kreft, J. U. (2007). Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5, 230–239. doi: 10.1038/nrmicro1600
Hoa, M., Tomovic, S., Nistico, L., Hall-Stoodley, L., Stoodley, P., Sachdeva, L., et al. (2009). Identification of adenoid biofilms with middle ear pathogens in otitis-prone children utilizing SEM and FISH. Int. J. Pediatr. Otorhinolaryngol. 73, 1242–1248. doi: 10.1016/j.ijporl.2009.05.016
Hyams, C., Camberlein, E., Cohen, J. M., Bax, K., and Brown, J. S. (2010). The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect. Immun. 78, 704–715. doi: 10.1128/IAI.00881-09
Ishii, S., Fukui, K., Yokoshima, S., Kumagai, K., Beniyama, Y., Kodama, T., et al. (2017). High-throughput screening of small molecule inhibitors of the Streptococcus quorum-sensing signal pathway. Sci. Rep. 7:4029. doi: 10.1038/s41598-017-03567-2
Joyce, E. A., Kawale, A., Censini, S., Kim, C. C., Covacci, A., and Falkow, S. (2004). LuxS is required for persistent pneumococcal carriage and expression of virulence and biosynthesis genes. Infect. Immun. 72, 2964–2975. doi: 10.1128/IAI.72.5.2964-2975.2004
Kerr, A. R., Adrian, P. V., Estevão, S., de Groot, R., Alloing, G., Claverys, J. P., et al. (2004). The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect. Immun. 72, 3902–3906. doi: 10.1128/IAI.72.7.3902-3906.2004
Kim, J. O., Romero-Steiner, S., Sørensen, U. B., Blom, J., Carvalho, M., Barnard, S., et al. (1999). Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect. Immun. 67, 2327–2333.
Lee, M. S., and Morrison, D. A. (1999). Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181, 5004–5016.
Luo, P., and Morrison, D. A. (2003). Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185, 349–358. doi: 10.1128/JB.185.1.349-358.2003
Marks, L. R., Davidson, B. A., Knight, P. R., and Hakansson, A. P. (2013). Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio 4, e00438-13. doi: 10.1128/mBio.00438-13
Martín-Galiano, A. J., Ferrándiz, M. J., and De La Campa, A. G. (2001). The promoter of the operon encoding the F0F1 ATPase of Streptococcus pneumoniae is inducible by pH. Mol. Microbiol. 41, 1327–1338. doi: 10.1046/j.1365-2958.2001.02597.x
Morona, J. K., Miller, D. C., Morona, R., and Paton, J. C. (2004). The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 189, 1905–1913. doi: 10.1086/383352
Morona, J. K., Morona, R., and Paton, J. C. (2006). Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc. Nat. Acad. Sci. U.S.A. 103, 8505–8510. doi: 10.1073/pnas.0602148103
Moscoso, M., and García, E. (2009). Transcriptional regulation of the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae: a bioinformatic analysis. DNA Res. 16, 177–186. doi: 10.1093/dnares/dsp007
Paixão, L., Oliveira, J., Veríssimo, A., Vinga, S., Lourenço, E. C., Ventura, M. R., et al. (2015). Host glycan sugar-specific pathways in streptococcus pneumonia: galactose as a key sugar in colonisation and infection. PLoS ONE 10:e0121042. doi: 10.1371/journal.pone.0121042
Parveen, N., and Cornell, K. A. (2011). Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol. Microbiol. 79, 7–20. doi: 10.1111/j.1365-2958.2010.07455.x
Pichichero, M. E. (2013). Otitis media. Pediatr. Clin. North Am. 60, 391–407. doi: 10.1016/j.pcl.2012.12.007
Sanchez, C. J., Hurtgen, B. J., Lizcano, A., Shivshankar, P., Cole, G. T., and Orihuela, C. J. (2011). Biofilm and planktonic pneumococci demonstrate disparate immunoreactivity to human convalescent sera. BMC Microbiol. 11:245. doi: 10.1186/1471-2180-11-245
Sanchez, C. J., Shivshankar, P., Stol, K., Trakhtenbroit, S., Sullam, P. M., Sauer, K., et al. (2010). The Pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 6:e1001044. doi: 10.1371/journal.ppat.1001044
Sanderson, A. R., Leid, J. G., and Hunsaker, D. (2006). Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope 116, 1121–1126. doi: 10.1097/01.mlg.0000221954.05467.54
Shabayek, S., and Spellerberg, B. (2017). Acid stress response mechanisms of group B Streptococci. Front. Cell. Infect. Microbiol. 7:395. doi: 10.3389/fcimb.2017.00395
Shak, J. R., Vidal, J. E., and Klugman, K. P. (2013). Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol. 21, 129–135. doi: 10.1016/j.tim.2012.11.005
Simell, B., Auranen, K., Käyhty, H., Goldblatt, D., Dagan, R., and O'Brien, K. L. (2012). The fundamental link between pneumococcal carriage and disease. Expert Rev. Vaccines 11, 841–855. doi: 10.1586/erv.12.53
Stroeher, U. H., Paton, A. W., Ogunniyi, A. D., and Paton, J. C. (2003). Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 71, 3206–3212. doi: 10.1128/IAI.71.6.3206-3212.2003
Trappetti, C., Gualdi, L., Di Meola, L., Jain, P., Korir, C. C., Edmonds, P., et al. (2011a). The impact of the competence quorum sensing system on Streptococcus pneumoniae biofilms varies depending on the experimental model. BMC Microbiol. 11:75. doi: 10.1186/1471-2180-11-75
Trappetti, C., McAllister, L. J., Chen, A., Wang, H., Paton, A. W., Oggioni, M. R., et al. (2017). Autoinducer 2 signaling via the phosphotransferase FruA drives galactose utilization by Streptococcus pneumoniae, resulting in hypervirulence. MBio 8, e02269-16. doi: 10.1128/mBio.02269-16
Trappetti, C., Ogunniyi, A. D., Oggioni, M. R., and Paton, J. C. (2011b). Extracellular matrix formation enhances the ability of Streptococcus pneumoniae to cause invasive disease. PLoS ONE 6:e19844. doi: 10.1371/journal.pone.0019844.
Trappetti, C., Potter, A. J., Paton, A. W., Oggioni, M. R., and Paton, J. C. (2011c). LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect. Immun. 79, 4550–4558. doi: 10.1128/IAI.05644-1
Usonis, V., Jackowska, T., Petraitiene, S., Sapala, A., Neculau, A., Stryjewska, I., et al. (2016). Incidence of acute otitis media in children below 6 years of age seen in medical practices in five East European countries. BMC Pediatr. 16:108. doi: 10.1186/s12887-016-0638-2
Vidal, J. E., Howery, K. E., Ludewick, H. P., Nava, P., and Klugman, K. P. (2013). Quorum-sensing systems LuxS/Autoinducer 2 and com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect. Immun. 81, 1341–1353. doi: 10.1128/IAI.01096-12
Vidal, J. E., Ludewick, H. P., Kunkel, R. M., Zähner, D., and Klugman, K. P. (2011). The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect. Immun. 79, 4050–4060. doi: 10.1128/IAI.05186-11
Weimer, K. E., Armbruster, C. E., Juneau, R. A., Hong, W., Pang, B., and Swords, W. E. (2010). Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J. Infect. Dis. 202, 1068–1075. doi: 10.1086/656046
Wnuk, S. F., Robert, J., Sobczak, A. J., Meyers, B. P., Malladi, V. L. A., Zhu, J., et al. (2009). Inhibition of S-ribosylhomocysteinase (LuxS) by substrate analogues modified at the ribosyl C-3 position. Bioorg. Med. Chem. 17, 6699–6706. doi: 10.1016/j.bmc.2009.07.057
Yadav, M. K., Chae, S. W., Go, Y. Y., Im, G. J., and Song, J. J. (2017a). In vitro multi-species biofilms of methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa and their host interaction during in vivo colonization of an otitis media rat model. Front. Cell. Infect. Microbiol. 7:125. doi: 10.3389/fcimb.2017.00125
Yadav, M. K., Chae, S. W., Park, K., and Song, J. J. (2013). Hyaluronic acid derived from other streptococci supports Streptococcus pneumoniae in vitro biofilm formation. Biomed Res. Int. 2013:690217. doi: 10.1155/2013/690217.
Yadav, M. K., Chae, S. W., and Song, J. J. (2012a). Effect of 5-azacytidine on in vitro biofilm formation of Streptococcus pneumoniae. Microb. Pathog. 53, 219–226. doi: 10.1016/j.micpath.2012.08.003
Yadav, M. K., Chae, S.-W., and Song, J.-J. (2012b). In vitro Streptococcus pneumoniae biofilm formation and in vivo middle ear mucosal biofilm in a rat model of acute otitis induced by S. pneumoniae. Clin. Exp. Otorhinolaryngol. 5, 139–144. doi: 10.3342/ceo.2012.5.3.139
Yadav, M. K., Go, Y. Y., Kim, S. H., Chae, S. W., and Song, J. J. (2017b). Antimicrobial and antibiofilm effects of human amniotic/chorionic membrane extract on Streptococcus pneumoniae. Front. Microbiol. 8:1948. doi: 10.3389/fmicb.2017.01948
Yadav, M. K., Park, S. W., Chae, S. W., and Song, J. J. (2014). Sinefungin, a natural nucleoside analogue of S-adenosylmethionine, inhibits Streptococcus pneumoniae biofilm growth. Biomed. Res. Int. 2014:156987. doi: 10.1155/2014/156987
Keywords: Streptococcus pneumoniae, LuxS/AI-2, quorum-sensing, luxS mutation, biofilm, in vivo colonization
Citation: Yadav MK, Vidal JE, Go YY, Kim SH, Chae S-W and Song J-J (2018) The LuxS/AI-2 Quorum-Sensing System of Streptococcus pneumoniae Is Required to Cause Disease, and to Regulate Virulence- and Metabolism-Related Genes in a Rat Model of Middle Ear Infection. Front. Cell. Infect. Microbiol. 8:138. doi: 10.3389/fcimb.2018.00138
Received: 23 January 2018; Accepted: 18 April 2018;
Published: 04 May 2018.
Edited by:
W. Edward Swords, University of Alabama at Birmingham, United StatesReviewed by:
Justin A. Thornton, Mississippi State University, United StatesGuangchun Bai, Albany Medical College, United States
Copyright © 2018 Yadav, Vidal, Go, Kim, Chae and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae-Jun Song, jjsong23@gmail.com
 Mukesh K. Yadav
Mukesh K. Yadav Jorge E. Vidal
Jorge E. Vidal Yoon Y. Go1
Yoon Y. Go1  Shin H. Kim
Shin H. Kim Jae-Jun Song
Jae-Jun Song