- 1M1:03 Rolf Luft Center for Diabetes and Endocrinology Research, Department of Molecular Medicine and Surgery, Karolinska Institutet, Karolinska University Hospital, Stockholm, Sweden
- 2Department of Anesthesiology, Anzhen Hospital, Capital Medical University, Beijing, People’s Republic of China
- 3Viktor Rydberg Gymnasium Odenplan School, Stockholm, Sweden
Diabetes and diabetic nephropathy are complex diseases affected by genetic and environmental factors. Identification of the susceptibility genes and investigation of their roles may provide useful information for better understanding of the pathogenesis and for developing novel therapeutic approaches. Intercellular adhesion molecule 1 (ICAM1) is a cell surface glycoprotein expressed on endothelial cells and leukocytes in the immune system. The ICAM1 gene is located on chromosome 19p13 within the linkage region of diabetes. In the recent years, accumulating reports have implicated that genetic polymorphisms in the ICAM1 gene are associated with diabetes and diabetic nephropathy. Serum ICAM1 levels in diabetes patients and the icam1 gene expression in kidney tissues of diabetic animals are increased compared to the controls. Therefore, ICAM1 may play a role in the development of diabetes and diabetic nephropathy. In this review, we present genomic structure, variation, and regulation of the ICAM1 gene, summarized genetic and biological studies of this gene in diabetes and diabetic nephropathy and discussed about the potential application using ICAM1 as a biomarker and target for prediction and treatment of diabetes and diabetic nephropathy.
Introduction
Diabetes mellitus is a group of metabolic diseases in which the patients have high blood glucose levels. Its epidemic has become a national and global crisis. Based upon the figures today, at least 366 million people at the worldwide have diabetes. By the year 2030, this number is expected to be double (Bonow and Gheorghiade, 2004; Wild et al., 2004; Cornell and Dorsey, 2012; Lam and LeRoith, 2012). There are two major types of diabetes. Type 1 diabetes (T1D), previously called juvenile diabetes or insulin-dependent diabetes, develops on the basis of autoimmune destruction of pancreatic β-cells, which results in insulin deficiency. It mostly affects young people (<20 years old) but occurs also in adults (Lightfoot et al., 2012). Type 2 diabetes (T2D) is the most common form of diabetes and accounts for approximately 85–90% of all diabetic patients. In T2D, hyperglycemia results from a combination of impaired insulin secretion and insulin resistance. When the pancreatic β-cells loose the ability to compensate for insulin resistance in liver, skeletal muscle, and adipose tissues, hyperglycemia becomes manifest (Alberti and Zimmet, 1998). Diabetes patients often develop macro- and/or micro-vascular complications. Diabetic nephropathy (DN) is one of serious complications and occurs in 30–40% of diabetic patients (Heerspink and de Zeeuw, 2011; Marshall, 2012). This diabetic complication is characterized by pathophysiological changes in glomerular hyperfiltration, renal hypertrophy, tubular function and then progress to proteinuria and reduction of glomerular filtration rate (GFR). The patients with DN exhibit persistent proteinuria, hypertension, declining renal function, and increased premature mortality largely as a result of cardiovascular disease. DN is the most common single cause of end-stage renal disease (ESRD). Once overt DN occurs, it progresses slowly or rapidly to the most advanced stage of chronic kidney disease which needs dialysis or transplantation treatment (Marshall, 2004; Shields and Maxwell, 2010; Weil et al., 2010; Thomas and Groop, 2011). The treatment cost for diabetes patients has been increasing staggering in the recent decades and becomes a further burden of the healthcare system. Diabetes and DN are multi-factorial diseases, which are influenced by both genetic and environmental factors (Satko et al., 2005; Pitkäniemi et al., 2007; Ashcroft and Rorsman, 2012; Gonzalez-Bulnes and Ovilo, 2012; Morahan, 2012). Therefore, identification of the susceptibility genes in development of diabetes and diabetic complications and investigation of their roles are of importance to provide useful information for improvement of the prevention and medication programs.
Intercellular adhesion molecule 1 (ICAM1, OMIM: 147840) is a cell surface glycoprotein and expressed in endothelial cells and leukocytes in the immune system. This endothelial- and leukocyte-associated transmembrane protein has been known for its importance in stabilizing cell–cell interactions and facilitating leukocyte endothelial transmigration. Recently, the accumulating reports from genetic studies in diabetic patients with and without DN and from biological studies with diabetic animal models have implicated that ICAM1 may play a role in the pathogenesis of diabetes and DN. In this review, we will summarize the genetic and pathophysiological relevance of ICAM1 and discuss about the possible role of ICAM1 in the development of diabetes and DN as well as the perspectives of the ICAM1 research.
Genomic DNA Structure, mRNA and Protein of ICAM1
The ICAM1 gene (GeneID: 3383) is located in chromosome 19p13.2 and spans 15,775 base pairs (bp) along the short arm of this chromosome (10,381,517–10,397,291 bp from pter). Its aliases include cluster of differentiation 54 (CD54) and cell surface glycoprotein P3.58 (BB2). Figure 1 demonstrates genomic structure, regulation, and variation of the ICAM1 gene. There are seven exons and six introns in the ICAM1 gene. A relatively large un-translation region (UTRs) resides respectively at both 5′ and 3′-sequences of the gene, while the translation start point is located in exon 1. From the 2–6 exon, there is one non-synonymous single-nucleotide polymorphism (SNP) each, by which the amino acid changes of ICAM1 protein are caused. ICAM1 is a transmembrance glycoprotein molecule of the immunoglobulin superfamily and characterized by five distinct immunoglobulin-like domains, a transmembrance domain and a cytoplasmic tail (van de Stolpe and van der Saag, 1996). ICAM1 protein is 505 amino acids in the length, the molecule weights between 80 and 114 kDa depending upon the levels of glycosylation, which varies among cell types and environments (Newman et al., 1990). Interestingly, there are several transcription binding proteins including NF-kappaB, NF-kappaB1, STAT3, δCREB, STAT1, STAT1α, and STAT1β, which may up-regulate the ICAM1 gene activity (Roebuck and Finnegan, 1999). MicroRNA (miR-221) may down-regulate the gene expression in the region of 3′-UTR of ICAM1 (Gong et al., 2011). There are CpG islands for methylation in the promoter of the ICAM1 gene. A study in tumor endothelial cells has demonstrated that the ICAM1 gene activity can be epigenetically silenced by promoter histone modifications (Hellebrekers et al., 2006). All regulatory factors are of importance to control the ICAM1 activities in immune-related processes. Our research has been focused on investigating whether alterations in the ICAM1 gene structure and function are associated with the development of diabetes and DN.
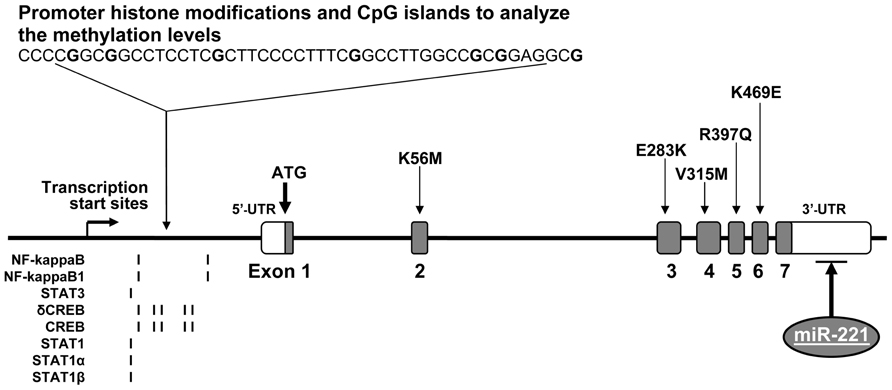
FIGURE 1. Genomic DNA structure, variation, and regulation of the ICAM1 gene. The ICAM1 gene is consisted of seven exons and six introns. There are several non-synonymous SNPs, which reside one each in exons 2–6. In the gene promoter, there are CpG islands for methylation and epigenetically silence by histone modifications as well as transcription binding sites for the proteins, including NF-kappaB, NF-kappaB1, STAT3, δCREB, STAT1, STAT1α, and STAT1β. In addition, microRNA (miR-221) in the region of 3′-UTR may down-regulate the ICAM1 gene expression.
Linkage of the ICAM1 Gene to Diabetes and Diabetic Nephropathy
Diabetes and DN are multi-factorial diseases, which are influenced by both genetic and environmental factors. To search for the susceptibility genes for the diseases, genome-wide scan linkage analysis has been used. This is a family-based approach to investigate if the genetic markers such as microsatellites or SNPs that span the whole genome and co-segregate with disease phenotypes. Microsatellites are simple repeats of 1–6 bp in genome and (CA)n is the most common form. A total of ~30,000 microsatellites present high levels of inter- and intra-specific polymorphisms and distribute in whole genome (often in intergenic DNA regions and rarely in the sequences of the genes). SNPs are the substitutions of nucleotides in genomic DNA. In general speaking, bi-allelic SNPs are the most common type. Tri-allelic ones and small insertions/deletions are also included. C/T is the most common SNP in the human genome. SNPs reside in the coding region of the genes are called cSNPs, which include non-synonymous SNPs (with amino acid changes) and synonymous SNPs (without amino acid changes). Most of SNPs are located in non-coding regions of the genes, including promoter, intron, and UTRs. SNPs in the promoter region may alter transcription binding site and thereby affect the transcriptional activity of the gene. Moreover, a number of SNPs are found in inter-genic sequences. Up to date, more than 40 million SNPs are recorded in the public SNP databases, which are freely available for research use. Using the genetic markers, the linkage analysis allows us to identify the genome that is transmitted within families along with the disease phenotypes of interest at a genome-wide scale. Based on finding a statistical signal, the probability of co-segregation of a disease with a chromosomal locus is given (Gulcher, 2012). In this approach of genome-wide scan, highly polymorphic genetic markers distributed across the genome are genotyped in large family pedigrees or in affected and discordant sibling pairs.
By using the approach of genome-wide scan and linkage analyses, several chromosomal regions including 19p13 have been predicted to link with diabetes and DN. Mein et al. (1998) have previously conducted genome wide scan analysis in 93 affected sibpair families and 263 multiplex families from the UK and indicated that loci in chromosome 19p13 are linked to T1DM. Later on, the linkage of T1DM to chromosome 19p13 is replicated by the study with 2658 affected sib-pairs in USA (Concannon et al., 2009). Interestingly, lipid-related traits such total cholesterol, triglycerides and low-density lipoprotein (LDL) concentrations in T2DM are linked to chromosome 19p13 and this finding has been replicated in Caucasians, African-American and Hispanic families (Imperatore et al., 2000; Adeyemo et al., 2005; Malhotra and Wolford, 2005). Several interesting candidate genes, including insulin receptor, resistin and ICAM1, are involved in the region of chromosome 19p13. The linkage of LDL concentrations to chromosome 19p13 has been replicated in the study with 612 individuals from 28 Amish families in USA (Pollin et al., 2004). Furthermore, Leon et al. (2007) have performed a genome-wide scan study in 1251 African Americans (AA) and 1129 European Americans (EA) hypertensive siblings from the Hypertension Genetic Epidemiology Network study and indicated that loci in chromosome 19p are linked with albumin to creatinine ratio (ACR) when both AA and EA subjects are combined in the analyses. Particularly, Kathiresan et al. (2008) have analyzed the genome-wide scan data from three studies including 8816 T2D subjects and found six new loci associated with LDL, cholesterol, HDL, and triglycerides. One of the loci is located in chromosome 19p13. Previously, Arya et al. (2006) have conducted a genome-wide scan and linkage study and suggested that loci in both chromosomal arms of 19p13.2 and 19q13.4 may be linked to birth weight in T2D families of both Mexican Americans and EA. By the analyses of the combined metabolic syndrome and echocardiographic factors, Kraja et al. (2008) have found the linkage of blood pressure in T2D patients of AA and EA with the region chromosome 19p13. Taking together with the information from above briefly described genome-wide scan studies, it is clear that there are the loci in chromosome 19p may confer the susceptibility risk to diabetes and DN.
Association of the ICAM1 Genetic Polymorphisms with Diabetes and Diabetic Nephropathy
Several research groups including ours have reported the genetic association studies of the ICAM1 gene in T1DM and DN. Guja et al. (1999) reported that the transmission of the G allele of SNP K469E (A/G) is increased in Romanian T1DM families. One year later, Nishimura et al. (2000) replicated that this K469E polymorphism is associated with adult-onset T1DM in a Japanese population. However, the association of K469E polymorphism in the ICAM1 gene with T1DM was not found in Danish, Finnish, and British Caucasian (Nejentsev et al., 2000). Furthermore, Nejentsev et al. (2003) demonstrated that another synonymous SNP G241R in the ICAM1 gene was associated with T1DM. All these previous studies were designed for analysis of one or two SNP(s). In order to ascertain whether the ICAM1 genetic polymorphisms are associated with T1DM and DN, we conducted the comprehensive genetic association studies. The studied SNPs including K469E and G241R were selected based upon the information of their position in the ICAM1 gene and their linkage disequilibrium (LD) values in the HapMap. Our data from single marker association analyses indicated that K469E and another intronic polymorphism (rs281432) were significantly associated with T1DM in Swedish Caucasians (Ma et al., 2006). Interestingly, these two SNPs are located in intron 2 and exon 6, respectively. According to the data of pair-wise LD values for SNPs in the ICAM1 gene, a relatively strong LD (/D′/ ≥ 0.7) existed to extend over the region between these two SNPs. Due to the large 5′- and 3′- UTRs in exons 1 and 7, the LD block covers almost the whole coding region of the gene. Further multiplex marker association analysis was done and the common haplotype C-A constructed by C allele from K469E and A allele from rs281432 was found to be associated with T1D (Ma et al., 2006). Later on, we found that K469E polymorphism in the ICAM1 is associated with DN in T1D patients of Americans of European descent and selected from the Genetic of Kidney Diseases in Diabetes (GoKinD) study (Mueller et al., 2006). However, no association of G241R in the ICAM1 gene with T1DM and DN in Swedish and GoKinD populations was found (Ma et al., 2008). In patients with T2D, the K469E polymorphism in the ICAM1 gene was found to associate with plasma fibrinogen levels and diabetic retinopathy (Kamiuchi et al., 2002; Yokoyama et al., 2005; Liu et al., 2006; Petrovic et al., 2008; Vinita et al., 2012). Up to date, there is, however, no report regarding the association of ICAM1 genetic polymorphism with DN in T2D.
Interestingly, we have observed that genotype distribution of K469E polymorphism in the ICAM1 gene presents a high heterozygous index (~50%; Ma et al., 2006). Figure 2 represents the genotype distribution of the ICAM1 E469K polymorphism in Swedish non-diabetic control subjects, T1D patients without and with DN. From non-diabetic control subjects, to T1D patients without DN and the patients with DN, the frequencies of the carriers with heterozygous genotype are increased, while the carriers with 469E homozygous genotype decreased. In order to avoid the possibility that high heterozygous index may be caused by genotyping errors, we confirmed the genotyping experiments with two different techniques such as dynamic allele-specific hybridization (DASH) and pyrosequencing (Ronaghi et al., 1998; Howell et al., 1999). In the human genome, there are segmental duplications (duplicons) with >90% sequence similarity between the copies, which may cause specific allelic and genotypic diversities, such as high heterozygous index in complex diseases (Venter et al., 2001; Shaw and Lupski, 2004). To ascertain whether K469E SNP is involved in a duplicon, we further performed a cloning and sequencing analysis and found that no duplication resides in the gene region (Ma et al., 2006). K469E is a non-synonymous SNP in exon 6 of the ICAM1 gene, which causes the amino acid changes of the ICAM1 protein. We have submitted ICAM1 amino acid sequences with K469 and 469E alleles respectively into SWISS-MODEL (Peitsch, 1995; Arnold et al., 2006) to understand the changes of ICAM1 protein. There are 532 amino acids in the protein sequence of ICAM1, K469 is wild-type and has 100% identified homology. Compared to the DIMER image of wild ICAM1 protein, however, the structure of ICAM with mutant 469E is significantly changed. Although the modeling analysis implicates that the K469E polymorphism in the ICAM1 gene may have functional effect, further investigation with transfection of 469E allele into cells such human embryonic kidney (HEK) 293A or with icam1 knock-out mouse model is necessary in order to further understand the pathogenic mechanism.
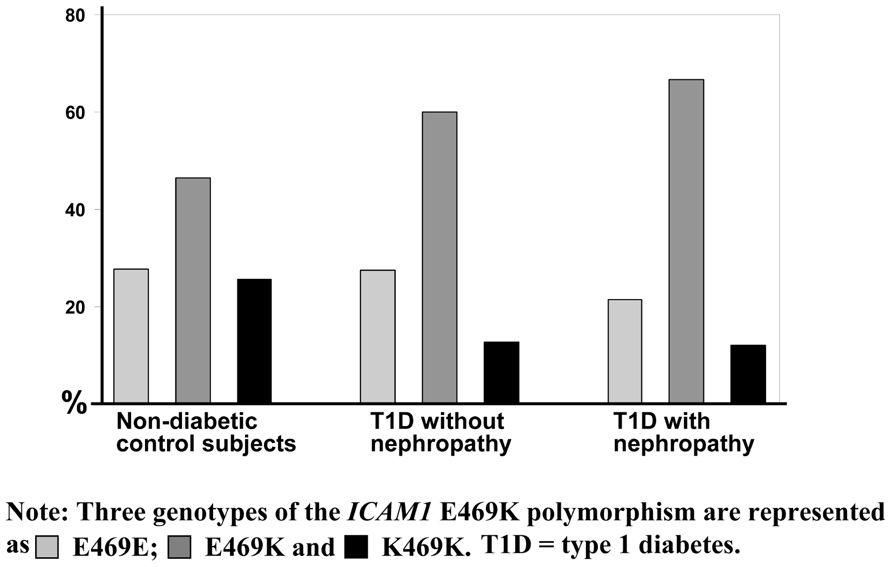
FIGURE 2. Genotype distribution of the ICAM1 K469E polymorphism. The genotype distribution of the ICAM1 K469E polymorphism is represented from a genetic association study in Swedish population (Ma et al., 2006). Three genotypes of the ICAM1 K469E polymorphism are shown in as light gray color for K469K, gray for K469E, and dark for E469E. Obviously, the heterozygous index is high compared to the percentage of homozygous and increased from the group of non-diabetic control subjects, to type 1 diabetes (T1D) patients without diabetic nephropathy and the patients with diabetic nephropathy.
Possible Role of ICAM1 in Development of Diabetes and Diabetic Nephropathy
In general speaking, ICAM1 proteins act as ligands and the primary receptors for ICAM1 are integrins, which mediate cell–cell interactions and allow signal transduction. Specifically, ICAM1, unlike most integrin-binding proteins, does not contain an RGD (Arg-Gly-Asp) motif to promote integrin binding (van de Stolpe and van der Saag, 1996), but is targeted to two integrins of the β2 subunit family, i.e., leukocyte adhesion protein-1 (LFA-1) and Mac-1 (integrin, alpha M; Janeway, 2001). Thus, based upon the interaction with these two molecules, ICAM1 has a role for two important immune-related functions: T lymphocytes activation and leukocyte–endothelial cell interaction. The role of ICAM1 in the development of diabetes and DN has not been fully explored. Recent studies, however, have provided the information to predict that ICAM1 is involved in the pathogenesis of diabetes and DN (Sahakyan et al., 2010a,b).
Diabetic nephropathy is a progress disease, which is categorized into stages based upon urinary albumin excretion (UAE) values. The early phase, which can be reversed, is microalbuminuria. The reduction of renal function begins with proteinuria. Clinical investigation has demonstrated that soluble ICAM1 levels in stored blood samples from T1D patients are higher compared to non-diabetic control subjects. High ICAM1 levels in T1D patients are associated with a relative risk of 1.67 (95 CI 0.96–2.92, P = 0.03) of developing incident sustained microalbuminuria after adjustment for baseline age, sex, duration of diabetes, and randomized treatment assignment (Lin et al., 2008). Furthermore, Astrup et al. (2008) have reported that soluble ICAM1 levels are associated with all-caused mortality and cardiovascular morbidity in T1D patients with DN. The similar findings have been observed in T2D patients. Soluble ICAM1 levels are significantly correlated with albuminuria in T2D patients (Rubio-Guerra et al., 2007). T2D patients with diabetic micro-angiopathic complications have higher soluble ICAM1 levels in comparison with diabetic group without micro-angiopathic complications and healthy control subjects (Mastej and Adamiec, 2008). The findings from clinical investigations have been supported by studies with diabetic animal models such as the db/db mice and streptozotocin-induced rats. Compared to non-diabetic rats, serum and urinary ICAM1 levels in streptozotocin-induced rats are found to be increased, which are parallel with the elevation of UAE (Qian et al., 2008). Furthermore, evidence has indicated that ICAM1 is overexpressed in glomeruli diabetic rats (Watanabe et al., 2011) and in tubular epithelial cells of kidney in T2D db/db mice (Kosugi et al., 2009). Therefore, ICAM1 may play a role in the development of diabetes DN and possible mechanism is shown in Figure 3. In a diabetic condition with hyperglycemia, the ICAM1 gene transcription in the nuclei is increased and the ICAM1 gene expression on the surface of endothelium cells is up-regulated. ICAM1 binding activity with LFA-1 is increased and more lymphocytes from blood are transferred into cells in glomeruli and peritubular capillaries of nephron in kidney. Consequently, injure of kidney glomeruli and tubular occurs and the proteins are excreted to urine. In this figure, however, two questions still remain. First, how is ICAM1 gene activity stimulated by high blood glucose levels? Second, how does ICAM1 elevation cause kidney tubular and glomeruli injury?
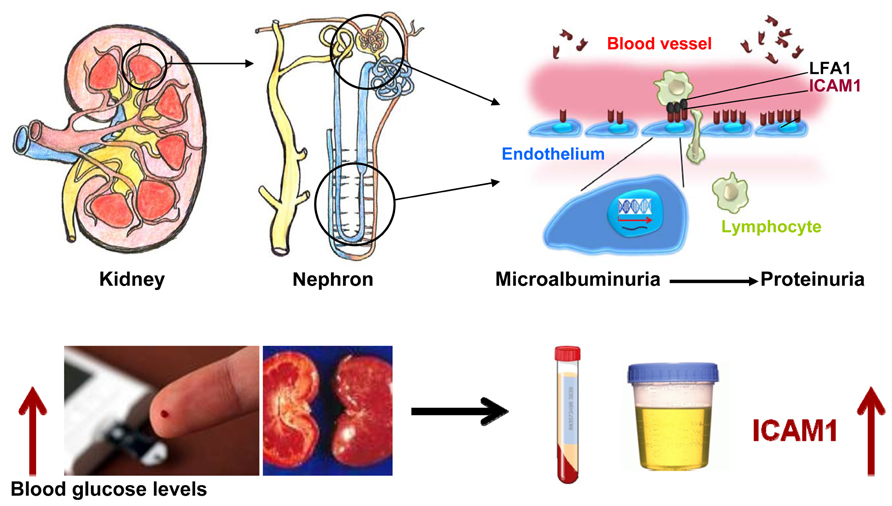
FIGURE 3. Possible role of ICAM1 in the development of diabetic nephropathy. Under a diabetic condition with hyperglycemia, the ICAM1 gene transcription in the nuclei is increased and the ICAM1 gene expression on the surface of endothelium cells is up-regulated. ICAM1 protein binding activity with leukocyte adhesion protein-1 (LFA-1) is increased and more lymphocytes from blood are transferred into cells of glomeruli and peritubular capillaries of nephron in kidney. Consequently, serum ICAM1 levels are increased. Injury in kidney glomeruli and tubular is occurred and the proteins are released to urine.
ICAM1 as a Biomarkers for Prediction of Diabetic Nephropathy
Biomarkers are substances and structures that can be measured in biological samples such as urine, blood, saliva, DNA, and protein. A biomarker can be used as an indicator of a particular disease state such as diabetes and DN (Caveney and Cohen, 2011). Currently, the biomarker used for prediction and diagnosis of DN is UAE. This biomarker, however, is less valuable for early prediction and diagnosis of DN. Therefore, researchers have searched for novel biomarkers of DN in order to improve the diagnosis approach and prevention program (Hellemons et al., 2012). As we described above, genetic and biological studies have implicated that ICAM1 plays a role in the development of diabetes and DN. First, the ICAM1 gene is located in a linkage region with diabetes and DN. Second, the K469E polymorphism in the ICAM1 gene is associated with diabetes and DN. Third, serum ICAM1 levels are gradually increased from low levels in normal albuminuria to high levels in micro-albuminuria and to even higher levels in proteinuria. Therefore, we concluded that ICAM1 is associated with diabetes and DN. This molecule is most likely a useful biomarker for prediction of endothelial dysfunction in diabetes and DN. Further evaluation of ICAM1 as a biomarker in a large cohort of T1D and T2D patients with and without DN needs to be done.
ICAM1 as a Target for Drug Development
ICAM1 is a molecule involved in many pathways including anti-inflammation. Although the picture of ICAM1 involvement and interaction is complex, experiments have indicated that inhibition of the ICAM1 gene expression may improve the progress of diabetes and DN. Glucagon-like peptide-1 (GLP-1) has various extra-pancreatic actions, in addition to its enhancement of insulin secretion from pancreatic islets. Kodera et al. (2011) have demonstrated that GLP-1 receptor agonist, exendin-4, decreases the ICAM1 gene expression and ameliorates albuminuria, glomerular hyperfiltration, glomerular hypertrophy, and mesangial matrix expansion in the diabetic rats without changing blood pressure or body weight. Furthermore, Matsui et al. (2010) have reported that nifedipine, a calcium-channel blocker, blocks the advanced glycation end product (AGE)-induced tubular damage and also inhibits ICAM1 gene activity in tubular cells, which may have benefits in treatment of DN. Liu et al. (2010) have suggested that berberine can ameliorate renal dysfunction in diabetic rats by decreasing ICAM1 gene expression and nuclear factor-kappa B (NF-kappaB) activation. Taking together, the data from these studies suggest that ICAM1 may be a good candidate as target for drug development. Inhibition of ICAM1 gene activity may benefit in treatment of diabetes and DN.
Summary and Perspectives
We have described genetic and biological studies of ICAM1 in diabetes and DN. Data implicate that ICAM1 can be used a biomarker for prediction of diabetes and DN and may also be serviced as a target for drug development. To reach these two objectives, however, we need to continuously put efforts into the study of ICAM1 in diabetes research. First, genetic polymorphisms in the ICAM1 gene are associated with T1D, T2D and DN in T1D. But, no genetic association with DN in T2D has been reported. Although it is known that methylation modification of genomic DNAs and histone proteins is involved in the regulation of ICAM1 gene expression, no epigenetic study of this gene in diabetes and DN has been done. Second, clinical observations in diabetes patients and experimental studies with diabetic animal models have demonstrated that elevation of serum ICAM1 levels and overexpression of the gene in kidney tissues. Inhibition of ICAM1 gene activity with agents may improve the kidney function, suggesting that ICAM1 may be a target for drug development in treatment of diabetes and DN.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ACR, urinary albumin/creatinine ratio; AER, albumin excretion rate; CD54, cluster of differentiation 54; DN, diabetic nephropathy; ESRD, end-stage renal disease; GFR, glomerular filtration rate; GoKinD, Genetics of Kidneys in Diabetes; HDL, high-density lipoprotein; HWE, Hardy–Weinberg equilibrium; ICAM1, intercellular adhesion molecule 1; LD, linkage disequilibrium; LDL, low-density lipoprotein; LFA, leukocyte adhesion protein; SNP, single-nucleotide polymorphism; T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus; UTR, un-translation region.
References
Adeyemo, A. A., Johnson, T., Acheampong, J., Oli, J., Okafor, G., Amoah, A., et al. (2005). A genome wide quantitative trait linkage analysis for serum lipids in type 2 diabetes in an African population. Atherosclerosis 181, 389–397.
Alberti, K. G., and Zimmet, P. Z. (1998). Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet. Med. 15, 539–553.
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201.
Arya, R., Demerath, E., Jenkinson, C. P., Göring, H. H., Puppala, S., Farook, V., et al. (2006). quantitative trait locus (QTL) on chromosome 6q influences birth weight in two independent family studies. Hum. Mol. Genet. 15, 1569–1579.
Ashcroft, F. M., and Rorsman, P. (2012). Diabetes mellitus and the β cell: the last ten years. Cell 148, 1160–1171.
Astrup, A. S., Tarnow, L., Pietraszek, L., Schalkwijk, C. G., Stehouwer, C. D., Parving, H. H., et al. (2008). Markers of endothelial dysfunction and inflammation in type 1 diabetic patients with or without diabetic nephropathy followed for 10 years: association with mortality and decline of glomerular filtration rate. Diabetes Care 31, 1170–1176.
Bonow, R. O., and Gheorghiade, M. (2004). The diabetes epidemic: a national and global crisis. Am. J. Med. 116(Suppl. 5A), 2S–10S.
Caveney, E. J., and Cohen, O. J. (2011). Diabetes and biomarkers. J. Diabetes Sci. Technol. 5, 192–197.
Concannon, P., Chen, W. M., Julier, C., Morahan, G., Akolkar, B., Erlich, H. A., et al. (2009). Type 1 Diabetes Genetics Consortium. Genome-wide scan for linkage to type 1 diabetes in 2,496 multiplex families from the Type 1 Diabetes Genetics Consortium. Diabetes 58, 1018–1022.
Cornell, S., and Dorsey, V. J. (2012). Diabetes pharmacotherapy in 2012: considerations in medication selection. Postgrad. Med. 2012 124, 84–94.
Gonzalez-Bulnes, A., and Ovilo, C. (2012). Genetic basis, nutritional challenges and adaptive responses in the prenatal origin of obesity and type-2 diabetes. Curr. Diabetes Rev. 8, 144–154.
Gong, A. Y., Hu, G., Zhou, R., Liu, J., Feng, Y., Soukup, G. A., et al. (2011). MicroRNA-221 controls expression of intercellular adhesion molecule-1 in epithelial cells in response to Cryptosporidium parvum infection. Int. J. Parasitol. 41, 397–403.
Guja, C., Todd, J. A., Welsh, K., Marshall, S., and Ionescu-Tirgoviste, C. (1999). Increased transmission of intercellular adhesion-molecule 1, 469E allele in type 1 Romanian diabetic families. Diabetologia 42, 327.
Gulcher, J. (2012). Microsatellite markers for linkage and association studies. Cold Spring Harb. Protoc. 2012, 425–432.
Heerspink, H. J., and de Zeeuw, D. (2011). The kidney in type 2 diabetes therapy. Rev. Diabet. Stud. 8, 392–402.
Hellebrekers, D. M., Castermans, K., Viré, E., Dings, R. P., Hoebers, N. T., Mayo, K. H., et al. (2006). Epigenetic regulation of tumor endothelial cell anergy: silencing of intercellular adhesion molecule-1 by histone modifications. Cancer Res. 66, 10770–10777.
Hellemons, M. E., Kerschbaum, J., Bakker, S. J., Neuwirt, H., Mayer, B., Mayer, G., et al. (2012). Validity of biomarkers predicting onset or progression of nephropathy in patients with type 2 diabetes: a systematic review. Diabet. Med. 29, 567–577.
Howell, W. M., Jobs, M., Gyllensten, U., and Brookes, A. J. (1999). Dynamic allele-specific hybridization. A new method for scoring single nucleotide polymorphisms. Nat. Biotechnol. 17, 87–88.
Imperatore, G., Knowler, W. C., Pettitt, D. J., Kobes, S., Fuller, J. H., Bennett, P. H., et al. (2000). A locus influencing total serum cholesterol on chromosome 19p: results from an autosomal genomic scan of serum lipid concentrations in Pima Indians. Arterioscler. Thromb. Vasc. Biol. 20, 2651–2656.
Janeway, C. A. Jr. (2001). How the immune system works to protect the host from infection: a personal view. Proc. Natl. Acad. Sci. U.S.A. 98, 7461–7468.
Kamiuchi, K., Hasegawa, G., Obayashi, H., Kitamura, A., Ishii, M., Yano, M., et al. (2002). Intercellular adhesion molecule-1 (ICAM-1) polymorphism is associated with diabetic retinopathy in type 2 diabetes mellitus. Diabet. Med. 19, 371–376.
Kathiresan, S., Melander, O., Guiducci, C., Surti, A., Burtt, N. P., Rieder, M. J., et al. (2008). Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40, 189–197.
Kodera, R., Shikata, K., Kataoka, H. U., Takatsuka, T., Miyamoto, S., Sasaki, M., et al. (2011). Glucagon-like peptide-1 receptor agonist ameliorates renal injury through its anti-inflammatory action without lowering blood glucose level in a rat model of type 1 diabetes. Diabetologia 54, 965–978.
Kosugi, T., Nakayama, T., Heinig, M., Zhang, L., Yuzawa, Y., Sanchez-Lozada, L. G., et al. (2009). Effect of lowering uric acid on renal disease in the type 2 diabetic db/db mice. Am. J. Physiol. Renal Physiol. 297, F481–F488.
Kraja, A. T., Huang, P., Tang, W., Hunt, S. C., North, K. E., Lewis, C. E., et al. (2008). QTLs of factors of the metabolic syndrome and echocardiographic phenotypes: the hypertension genetic epidemiology network study. BMC Med. Genet. 9:103. doi: 10.1186/1471-2350-9-103
Lam, D. W., and LeRoith, D. (2012). The worldwide diabetes epidemic. Curr. Opin. Endocrinol. Diabetes Obes. 19, 93–96.
Leon, J. M., Freedman, B. I., Miller, M. B., North, K. E., Hunt, S. C., Eckfeldt, J. H., et al. (2007). Genome scan of glomerular filtration rate and albuminuria: the HyperGEN study. Nephrol. Dial. Transplant. 22, 763–771.
Lightfoot, Y. L., Chen, J., and Mathews, C. E. (2012). Immune-mediated β-cell death in type 1 diabetes: lessons from human β-cell lines. Eur. J. Clin. Invest. 42, 1244–1251.
Lin, J., Glynn, R. J., Rifai, N., Manson, J. E., Ridker, P. M., Nathan, D. M., et al. (2008). Inflammation and progressive nephropathy in type 1 diabetes in the diabetes control and complications trial. Diabetes Care 31, 2338–2343.
Liu, L., Yu, Q., Wang, H., Zhang, S. X., Huang, C., and Chen, X. (2006). Association of intercellular adhesion molecule 1 polymorphisms with retinopathy in Chinese patients with type 2 diabetes. Diabet. Med. 23, 643–648.
Liu, W., Zhang, X., Liu, P., Shen, X., Lan, T., Li, W., et al. (2010). Effects of berberine on matrix accumulation and NF-kappa B signal pathway in alloxan-induced diabetic mice with renal injury. Eur. J. Pharmacol. 638, 150–155.
Ma, J., Möllsten, A., Prázny, M., Falhammar, H., Brismar, K., Dahlquist, G., et al. (2006). Genetic influences of the intercellular adhesion molecule 1 (ICAM-1) gene polymorphisms in development of type 1 diabetes and diabetic nephropathy. Diabet. Med. 23, 1093–1099.
Ma, J., Zhang, D., Brismar, K., Efendic, S., and Gu, H. F. (2008). Evaluation of the association between the common E469K polymorphism in the ICAM-1 gene and diabetic nephropathy among type 1 diabetic patients in GoKinD population. BMC Med. Genet. 9:47. doi: 10.1186/1471-2350-9-47.
Malhotra, A., and Wolford, J. K. (2005). American Diabetes Association GENNID Study Group. Analysis of quantitative lipid traits in the genetics of NIDDM (GENNID) study. Diabetes 54, 3007–3014.
Marshall, S. M. (2012). Diabetic nephropathy in type 1 diabetes: has the outlook improved since the 1980s? Diabetologia 55, 2301–2306.
Mastej, K., and Adamiec, R. (2008). Neutrophil surface expression of CD11b and CD62L in diabetic microangiopathy. Acta Diabetol. 45, 183–190.
Matsui, T., Yamagishi, S., Takeuchi, M., Ueda, S., Fukami, K., and Okuda, S. (2010). Nifedipine inhibits advanced glycation end products (AGEs) and their receptor (RAGE) interaction-mediated proximal tubular cell injury via peroxisome proliferator-activated receptor-gamma activation. Biochem. Biophys. Res. Commun. 398, 326–330.
Mein, C. A., Esposito, L., Dunn, M. G., Johnson, G. C., Timms, A. E., Goy, J. V., et al. (1998). A search for type 1 diabetes susceptibility genes in families from the United Kingdom. Nat. Genet. 19, 297–300.
Morahan, G. (2012). Insights into type 1 diabetes provided by genetic analyses. Curr. Opin. Endocrinol. Diabetes Obes. 19, 263–270.
Mueller, P. W., Rogus, J. J., Cleary, P. A., Zhao, Y., Smiles, A. M., Steffes, M. W., et al. (2006). Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J. Am. Soc. Nephrol. 17, 1782–1790.
Nejentsev, S., Guja, C., McCormack, R., Cooper, J., Howson, J. M., Nutland, S., et al. (2003). Association of intercellular adhesion molecule-1 gene with type 1 diabetes. Lancet 362, 1723–1724.
Nejentsev, S., Laine, A. P., Simell, O., and Ilonen, J. (2000). Intercellular adhesion molecule-1 (ICAM-1) K469E polymorphism: no association with type 1 diabetes among Finns. Tissue Antigens 55, 568–570.
Newman, P. J., Berndt, M. C., Gorski, J., White, G. C. II, Lyman, S., Paddock, C., et al. (1990). PECAM-1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily. Science 247, 1219–1222.
Nishimura, M., Obayashi, H., Maruya, E., Ohta, M., Tegoshi, H., Fukui, M., et al. (2000). Association between type 1 diabetes age-at-onset and intercellular adhesion molecule-1 (ICAM-1) gene polymorphism. Hum. Immunol. 61, 507–510.
Petrovic, M. G., Osredkar, J., Saraga-Babić, M., and Petrovic, D. (2008). K469E polymorphism of the intracellular adhesion molecule 1 gene is associated with proliferative diabetic retinopathy in Caucasians with type 2 diabetes. Clin. Exp. Ophthalmol. 36, 468–472.
Pitkäniemi, J., Moltchanova, E., Haapala, L., Harjutsalo, V., Tuomilehto, J., and Hakulinen, T. (2007). Genetic random effects model for family data with long-term survivors: analysis of diabetic nephropathy in type 1 diabetes. Genet. Epidemiol. 31, 697–708.
Pollin, T. I., Hsueh, W. C., Steinle, N. I., Snitker, S., Shuldiner, A. R., and Mitchell, B. D. (2004). A genome-wide scan of serum lipid levels in the Old Order Amish. Atherosclerosis 173, 89–96.
Qian, Y., Li, S., Ye, S., Chen, Y., Zhai, Z., Chen, K., et al. (2008). Renoprotective effect of rosiglitazone through the suppression of renal intercellular adhesion molecule-1 expression in streptozotocin-induced diabetic rats. J. Endocrinol. Invest. 31, 1069–1074.
Roebuck, K. A., and Finnegan, A. (1999). Regulation of intercellular adhesion molecule-1 (CD54) gene expression. J. Leukoc. Biol. 66, 876–888.
Ronaghi, M., Uhlén, M., and Nyrén, P. (1998). A sequencing method based on real-time pyrophosphate. Science 281, 363–365.
Rubio-Guerra, A. F., Vargas-Robles, H., Ayala, G. V., and Escalante-Acosta, B. A. (2007). Correlation between circulating adhesion molecule levels and albuminuria in type 2 diabetic normotensive patients. Med. Sci. Monit. 13, CR349–CR352.
Sahakyan, K., Klein, B. E., Lee, K. E., Tsai, M. Y., and Klein, R. (2010a). Inflammatory and endothelial dysfunction markers and proteinuria in persons with type 1 diabetes mellitus. Eur. J. Endocrinol. 162, 1101–1105.
Sahakyan, K., Klein, B. E., Myers, C. E., Tsai, M. Y., and Klein, R. (2010b). Novel risk factors in long-term hypertension incidence in type 1 diabetes mellitus. Am. Heart J. 159, 1074–1080.
Satko, S. G., Freedman, B. I., and Moossavi, S. (2005). Genetic factors in end-stage renal disease. Kidney Int. Suppl. 94, S46–S49.
Shaw, C. J., and Lupski, J. R. (2004). Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum. Mol. Genet. 13, R57–R64.
Thomas, M. C., and Groop, P. H. (2011). New approaches to the treatment of nephropathy in diabetes. Expert. Opin. Investig. Drugs 20, 1057–1071.
Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., et al. (2001). The sequence of the human genome. Science 291, 1304–1351.
Vinita, K., Sripriya, S., Prathiba, K., Vaitheeswaran, K., Sathyabaarathi, R., Rajesh, M., et al. (2012). SNDREAMS project. ICAM-1 K469E polymorphism is a genetic determinant for the clinical risk factors of T2D subjects with retinopathy in Indians: a population-based case-control study. BMJ Open 2, pii: e001036.
van de Stolpe, A., and van der Saag, P. T. (1996). Intercellular adhesion molecule-1. J. Mol. Med. (Berl.) 74, 13–33.
Watanabe, N., Shikata, K., Shikata, Y., Sarai, K., Omori, K., Kodera, R., et al. (2011). Involvement of MAPKs in ICAM-1 expression in glomerular endothelial cells in diabetic nephropathy. Acta Med. Okayama 65, 247–257.
Weil, E. J., Curtis, J. M., Hanson, R. L., Knowler, W. C., and Nelson, R. G. (2010). The impact of disadvantage on the development and progression of diabetic kidney disease. Clin. Nephrol. 74(Suppl. 1), S32–S8.
Wild, S., Roglic, G., Green, A., Sicree, R., and King, H. (2004). Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 27, 1047–1053.
Keywords: intercellular adhesion molecule 1, diabetic nephropathy, end-stage renal disease, type 1 diabetes mellitus, type 2 diabetes mellitus
Citation: Gu HF, Ma J, Gu KT and Brismar K (2013) Association of intercellular adhesion molecule 1 (ICAM1) with diabetes and diabetic nephropathy. Front. Endocrin. 3:179. doi: 10.3389/fendo.2012.00179
Received: 22 October 2012; Accepted: 17 December 2012;
Published online: 22 January 2013.
Edited by:
Gabriel Virella, Universidade de Lisboa, PortugalReviewed by:
Kay Waud, Eastern Virginia Medical School, USAShinichi Oikawa, Nippon Medical School, Japan
Melissa Irene March, Beth Israel Deaconess Medical Center, USA
Copyright: © 2013 Gu, Ma, Gu and Brismar. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Harvest F. Gu, M1:03 Rolf Luft Center for Diabetes and Endocrinology Research, Department of Molecular Medicine and Surgery, Karolinska Institutet, Karolinska University Hospital, Stockholm SE-17176, Sweden. e-mail: harvest.gu@ki.se
