Yarrowia lipolytica as an oleaginous cell factory platform for production of fatty acid-based biofuel and bioproducts
- Bioprocessing and Bioproducts Engineering Laboratory, Department of Biological Systems Engineering, Washington State University, Pullman, WA, USA
Today’s biotechnologists seek new biocatalysts to meet the growing demand for the bioproducts. This review critically evaluates the potential use of Y. lipolytica as an oleaginous cell factory platform. This yeast has undergone extensive modifications for converting a wide range of hydrophobic and hydrophilic biomass, including alkane, oil, glycerol, and sugars to fatty acid-based products. This article highlights challenges in the development of this platform and provides an overview of strategies to enhance its potential in the sustainable production of biodiesel, functional dietary lipid compounds, and other value-added oleochemical compounds. Future applications of the recombinant Y. lipolytica platform are also discussed.
Introduction
The development of sustainable microbial platforms for chemical and fuel production is becoming even more crucial as petroleum reserves are depleted and environmental problems increase throughout the world. Microbial-based bioconversion processes can be used to produce a variety of products from a vast array of biomass. The biorefining process relies on a biotransformational step under relatively mild conditions. Several strategies have been explored to enhance the productivity and competitiveness of microbial-based processes. These include genetic and metabolic engineering of microorganisms, novel fermentation technologies, low-value feedstock, and recycling byproducts (Koutinas et al., 2014).
The sustainable production of fatty acids (FAs) as energy rich molecules (Steen et al., 2010) has garnered recent attention. FAs can be obtained from plant and microbial resources. Microbial lipids offer several advantages, including faster production, less labor, more flexibility in terms of season and climate, and easier scale-up (Huang et al., 2013a). A microbial platform can be established to convert lignocellulosic sugars or low-value hydrophobic substrates such as waste oils to biofuels, functional lipids, and other value-added bioproducts. The rise in plant oil prices, introduction of novel fermentation technology, and the use of versatile engineered oleaginous strains have made microbial oil more competitive (Koutinas et al., 2014). However, the production cost of microbial oil must be reduced to increase its economic viability for use in biodiesels, drop-in biofuels, and other bioproducts (Koutinas et al., 2014).
Microbial oil is produced by a wide range of oleaginous organisms. Studies demonstrate that yeast is more effective than bacteria and microalgae in terms of lipid accumulation in the arsenal of the oleaginous microorganisms (Enshaeieh et al., 2012). Yeast is not influenced by climate, while phototrophic microalgae are. Yeast also uses more diverse sugar sources, such as xylose, at higher growth rates than do heterotrophic microalgae (Chi et al., 2010b). In addition, yeast shows more tolerance to metal ions and low oxygen availability than fungi. Unlike E. coli, yeast possesses post-translational modification, subcellular compartments, larger cellular size for easier separation, higher tolerance of potentially prohibitory byproducts such as organic acids, and a high accumulation of end-products (Beopoulos et al., 2010; Ageitos et al., 2011). Its lack of endotoxin, relatively easy genetic manipulation, low duplication time (<1 h), potential for large scale fermentation (Zhou et al., 2012), as well as its ability to grow in high concentrations of sugar and accumulate lipid in the form of polyunsaturated FAs containing triglycerides are also strong advantages (Ageitos et al., 2011).
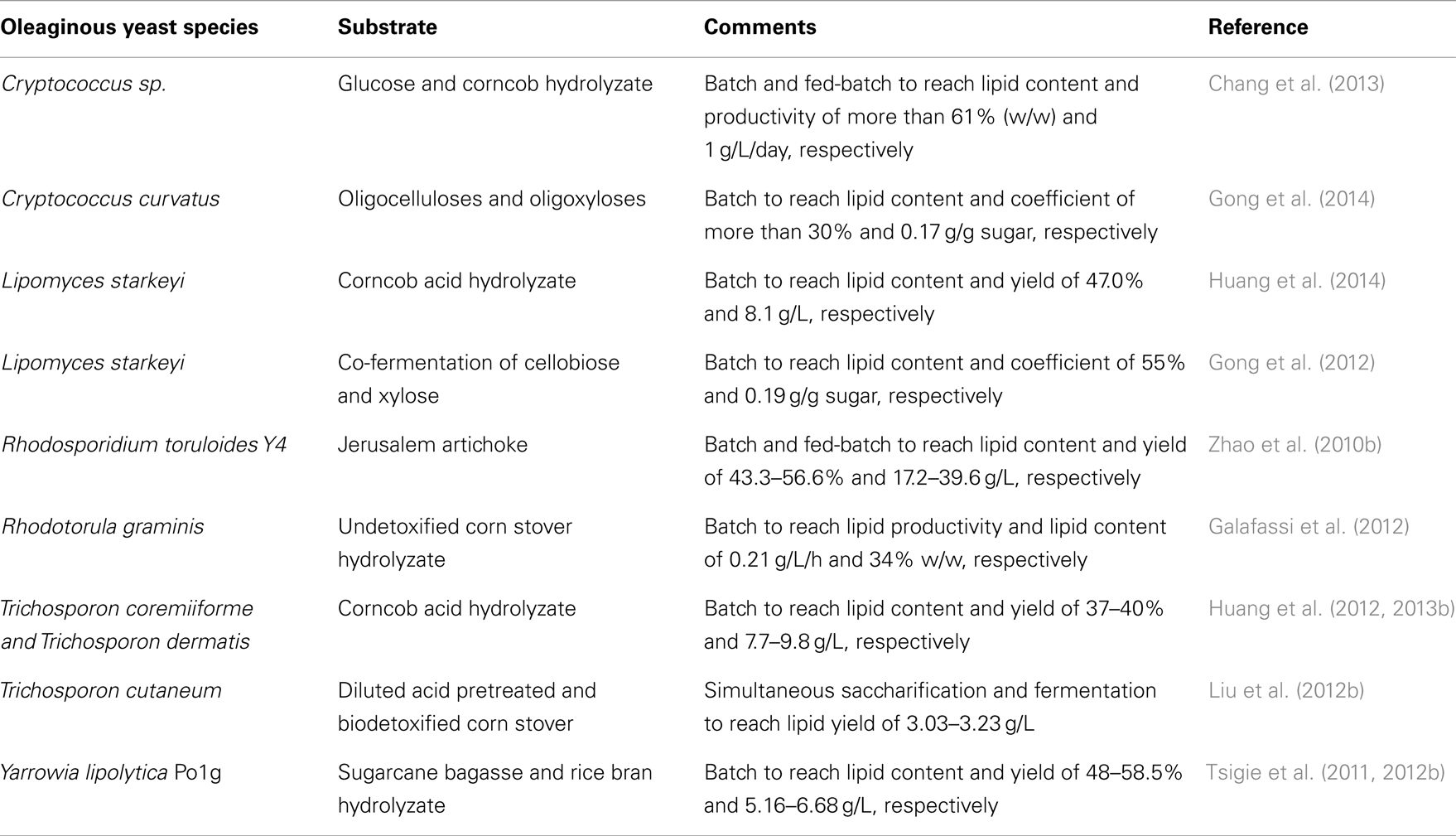
Many studies have explored the use of common oleaginous yeast species in single-cell oil (SCO) production (Breuer and Harms, 2006; Chen et al., 2009; Beopoulos and Nicaud, 2012; Magdouli et al., 2014). Table 1 shows the typical oleaginous yeast species that were used for bioconversion of cellulosic sugars to SCO.
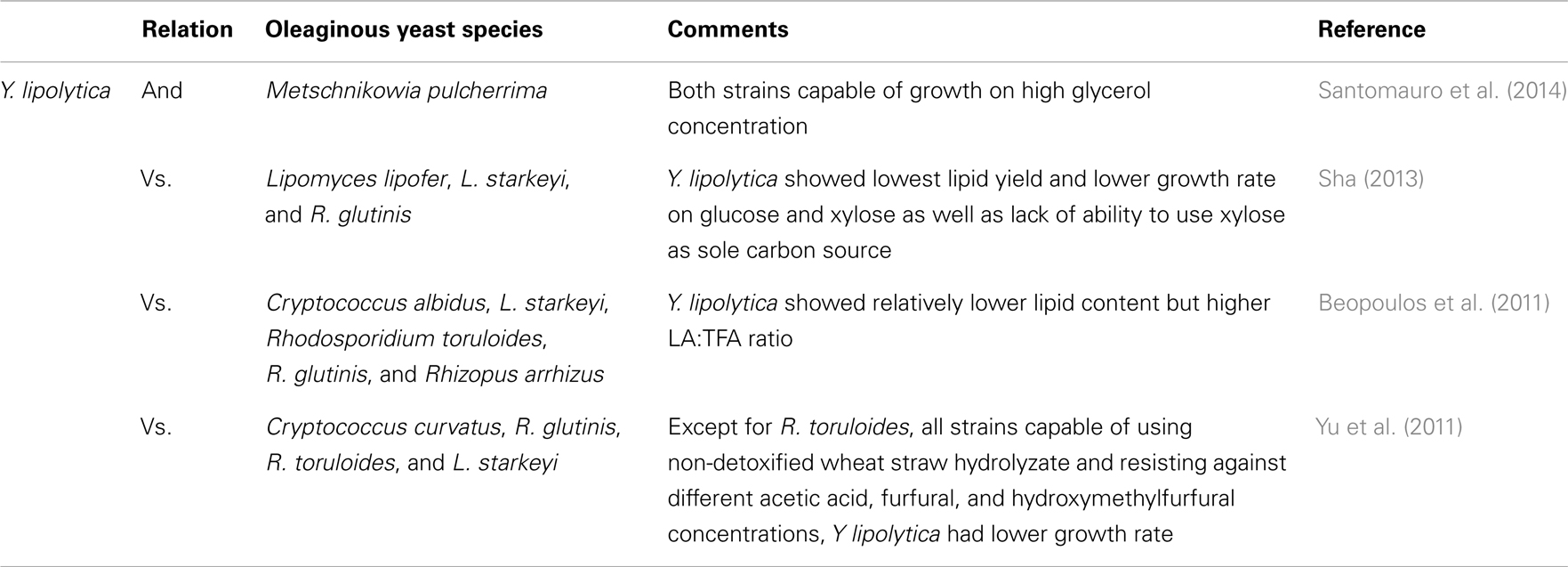
Generally, oleaginous yeasts have the capability of accumulating oil at more than 20–25% of their cell biomass (Beopoulos and Nicaud, 2012). Lipid accumulation can reach beyond 65% of dry weight in some oleaginous yeast species such as Cryptococcus albidus, Cryptococcus curvatus, Lipomyces starkeyi, Lipomyces tetrasporus, Rhodotorula glutinis, and Trichosporon pullulans under specific conditions. The importance of Y. lipolytica platform in the production of SCO and bioconversion of FA was recently discussed by Harzevili (2014). Although relatively lower level of lipid accumulation occurs in Y. lipolytica (up to 40% of the DCW), it is feasible to engineer this species for higher lipid accumulation (Beopoulos et al., 2010; Dulermo and Nicaud, 2011; Blazeck et al., 2014). Large portions of accumulated FAs in this yeast are in the form of unsaturated FAs (e.g., C18:1 = 28% and C18:2 = 51%) (Beopoulos et al., 2009; Nicaud, 2012; Sriwongchai et al., 2013). This facilitates the development of lipid-derived bioproducts that are more functional and reactive. Table 2 highlights some comparative studies on Y. lipolytica and other oleaginous yeast species.
Y. lipolytica has also been extensively used as the model oleaginous yeast in studies of heterologous protein synthesis. It has also been studied in terms of lipid accumulation and degradation steps, including the action of lipases, cytochrome pP450, acyl-CoA oxidase, and peroxisome biogenesis; salt tolerance; dimorphism; lysine metabolism and general secretion pathways. In addition, it has been studied for industrial and environmental applications to produce vast range of valuable bioproducts (Bankar et al., 2009). Y. lipolytica possesses haploid genome that is easily manipulated. This non-conventional yeast is regarded as non-pathogenic and categorized as GRAS (generally recognized as safe) by the FDA (Food and Drug Administration). It has the ability to form biofilm and possesses a robust secretory system (Beopoulos et al., 2008; Bankar et al., 2009). It is adaptable to cold, salt, and metal stresses that are common in hypersaline marine environments polluted with heavy metals and oil (Bankar et al., 2009). It is also adaptable to biphasic systems (Braun et al., 2012). Y. lipolytica is intrinsically equipped with an alkane efflux pumping system. This elevates alkane tolerance and makes it useful for the production of drop-in biofuel (Chen et al., 2013). Finally, Y. lipolytica can be more competitive than the non-oleaginous yeast, S. cerevisiae, in terms of product yield, decreased over glycosylation activity, and secretion efficiency, particularly for high molecular weight proteins. The co-translational pathway is the principal route, similar to that of mammalian cells (Madzak et al., 2004; Nicaud, 2012). Thus, many heterologous proteins have been produced by Y. lipolytica (Gellissen et al., 2005). This oleaginous platform also shows promise in industrial applications of lipid-modifying enzymes. All of these features make Y. lipolytica a suitable production platform in an array of fields and highlight its potential for use in sustainable production of second-generation biofuels.
A major point of consideration in selecting a cell factory platform is its ability to utilize a wide substrate. Using renewable materials as substrates can enhance the economy of the bioconversion process for Y. lipolytica (Chen et al., 2009; Liang and Jiang, 2013). Molasses, N-acetylglucosamine, sewage sludge, palm oil mill, olive oil mill wastewater, whey, municipal wastewater, industrial fats, and saturated spent fat are examples of such materials (Barth and Gaillardin, 1997; Papanikolaou et al., 2003; Fickers et al., 2005; Chen et al., 2009; Thevenieau and Nicaud, 2013). Y. lipolytica can also utilize multiple carbon sources such as glycerol, alkanes, ethanol (at concentration of up to 3%), sodium acetate (at concentration of up to 1%), lactic, butyric, propionic, malic, and succinic acids (Barth et al., 2003; Papanikolaou and Aggelis, 2009; Coelho et al., 2010; Fontanille et al., 2012; Zhou et al., 2012; Sriwongchai et al., 2013). For example, the capability of this yeast to convert glycerol to SCO or citric acid is important (Papanikolaou and Aggelis, 2009), since annual production of this carbon source is estimated at 1.9 million tons from the biodiesel industry (Rywinska et al., 2012). Thus, Y. lipolytica can play dual roles in the upstream and downstream of the biodiesel industry.
Recently, the microbial production of oil and oil-based bioproducts from renewable sources such as lignocellulosic biomass has received attention because they are abundant and low cost. However, the economically feasible production of cellulosic lipid has been hampered by the lack of systematic management for food waste, agro-, forest-, industrial process-, and microbial biomass residues; the lack of highly dedicated technologies; inadequate cultivation of energy crops in marginal lands; the shortage of highly engineered microbial strains for industrial application; the inadequate expansion of valuable co-product; potential environmental consequences (Yousuf, 2012; Chung, 2013; Balan, 2014). However, some of these issues can be addressed by systematically cultivating high-yielding crops, applying tailored technologies and genetically engineering plant and inhibitor-tolerant microbial strains (Pu et al., 2011). The efficient bioconversion of glucose- and xylose-containing sugars can improve biomass energy conversion and lower final costs. Y. lipolytica is not a particularly efficient sugar-utilizing platform, since it cannot use xylose as a sole source of carbon and prefers glycerol over glucose (Sestric et al., 2014). Nonetheless, upgrading this platform to enhance biomass and cellulosic lipid yields is feasible due to new genetic engineering (Blazeck et al., 2014) and co-fermentation strategies (Workman et al., 2013; Sestric et al., 2014).
Yarrowia lipolytica may be used to produce non-specific lipids for biofuel production or functional lipids for various applications (Beopoulos et al., 2008). The GRAS status of this yeast further enhances its potential for the production of polyunsaturated FAs (Xue et al., 2013) or lipids such as cocoa butter for dietary applications (Papanikolaou et al., 2003). The following sections provide general information on lipid accumulation and degradation pathways in the oleaginous yeast.
Lipid Accumulation and Degradation in Y. lipolytica
In oleaginous yeasts, lipids are found mainly in the form of neutral lipids, glycolipids, phospholipids, and free FAs (FFA) (Dey and Maiti, 2013). Neutral and non-neutral lipid fractions account for more than 90% and <10% of total lipids, respectively (Huang et al., 2013a). Triglycerides (TAG) and steryl esters (SE) are the major cellular lipids synthesized from sterol, glycerol-3-phosphate, and acyl-CoA. These lipid compounds comprise the hydrophobic core of the dynamic storage compartment called oleosome or lipid bodies (LB) (Han et al., 2013). LB can also accommodate FFA. FFAs may constitute more than 30% of the total accumulated lipids without negatively affecting the growth of the cell (Tsigie et al., 2011). Y. lipolytica has been shown to withstand high levels of intracellular FFA accumulation (Beopoulos et al., 2012). Several studies have explored the schematic pathway from hydrophilic or hydrophobic substrates to lipid accumulation and subsequent degradation (Fickers et al., 2005; Papanikolaou and Aggelis, 2009; Dulermo and Nicaud, 2011; Liang and Jiang, 2013; Tai and Stephanopoulos, 2013; Thevenieau and Nicaud, 2013; Wang et al., 2013; Garay et al., 2014; Matthäus et al., 2014). Generally, lipid accumulation takes place intracellularly through two pathways, de novo and ex novo.
The de novo pathway of FA biosynthesis requires precursors such as acetyl-CoA and malonyl-CoA, ATP, along with the reducing power of NADPH. Converting cytosolic acetyl-CoA to malonyl-CoA by acetyl-CoA carboxylase (ACC) is the first dedicated step to biosynthesizing FA (Kohlwein, 2010). This is followed by the elongation process through FA synthetase. The unsaturated FAs portion of accumulated lipid may increase during de novo FAs biosynthesis when non-fat substrates such as glycerol or sugar are used under nutrient limitation. In the ex novo pathway of lipid accumulation, the FA enters the cell and becomes activated. Then, it is absorbed as CoA-thioesters into the storage lipid. The synthesized or imported lipid may also degrade to other lipid-derived metabolites (Beopoulos et al., 2011; Beopoulos and Nicaud, 2012).
Synthesized FFAs may be stored in the LB, used in the structural lipid, degraded to provide energy or secreted into the surrounding media. Typically, the FAs of shorter chains (e.g., C12:0, C14:0, and C16:0) are more susceptible to degradation to support of growth, while saturated or unsaturated FAs of longer chains are more involved in lipid accumulation (Papanikolaou et al., 2002). Notably, the hydrolysis of TAG or FFAs formed from the de novo synthesis pathway often do not undergo metabolic degradation unless they are activated through esterification to acyl carrier protein or CoA via its carboxyl group (Scharnewski et al., 2008). Whether Acyl-CoA is used in the anabolic pathway (lipid biosynthesis) or the catabolic pathway (β-oxidation) determines the lipid content of the cell (Xue et al., 2013). This depends upon physiological and environmental conditions (e.g., the relative flux of carbon and nitrogen into the cell) and is regulated globally based on energy intake and expenditure (Coleman et al., 2002).
In an anabolic pathway, TAG is synthesized from acyl-CoA in endoplasmic reticulum (ER) or LB membrane via the Kennedy pathway. This pathway consists of three major steps through which activated FAs are esterified to the glycerol-3-phosphate (G3P) backbone (Beopoulos et al., 2008). The first acylation of G3P to lysophosphatidic acid (LPA) is followed by the second acylation to phosphatidic acid (PA). The final step of acylation takes place through action of three classes of diacylglycerol acyltransferase (DGA) (i.e., DGAT1, DGAT2, and PDAT) (Athenstaedt, 2011; Zhang et al., 2012c). This committed step of TAG synthesis is regulated at the transcriptional and translational levels (Courchesne et al., 2009).
In general, the lipid biosynthesis pathway is firmly regulated (Tai and Stephanopoulos, 2013) and the reactions occur at different organelles in the oleaginous yeasts. The general regulatory mechanism of lipid accumulation in the oleaginous microorganism has not yet been elucidated (Morin et al., 2011). The above stages at different organelles create more complexity, underscoring the need for transportation of intermediate among the organelles. The flux through the lipid pathway is intensified at specific phases of the oleaginous yeast life and under specific conditions.
Lipid accumulation is not usually growth-associated property (Ageitos et al., 2011). However, a significant amount of lipid may be produced at the early growth phase (André et al., 2009). The increase of lipid accumulation may occur during the logarithmic growth phase in response to the need for membrane biosynthesis (Sha, 2013). The stored lipid usually reaches its highest level at the start of the stationary phase when the oleaginous cells exhaust their nutrient supply and have excess carbon (Tai and Stephanopoulos, 2013). In general, the limitation of nitrogen, iron, zinc, phosphate, or inositol can stimulate de novo lipid biosynthesis (Papanikolaou et al., 2009). However, the ex novo synthesis of lipids may occur, regardless of whether nitrogen is available (Papanikolaou et al., 2007). High lipid accumulation may prevent the sugar substrate from being thoroughly consumed (Jadhav et al., 2012). The turnover of accumulated lipid takes place toward the late stationary phase to deal with shortage of external carbon sources and for maintenance purposes (Makri et al., 2010). Finally, lipid accumulation can be restricted by the repression of glycolysis under long-term nutrient limitation (Morin et al., 2011).
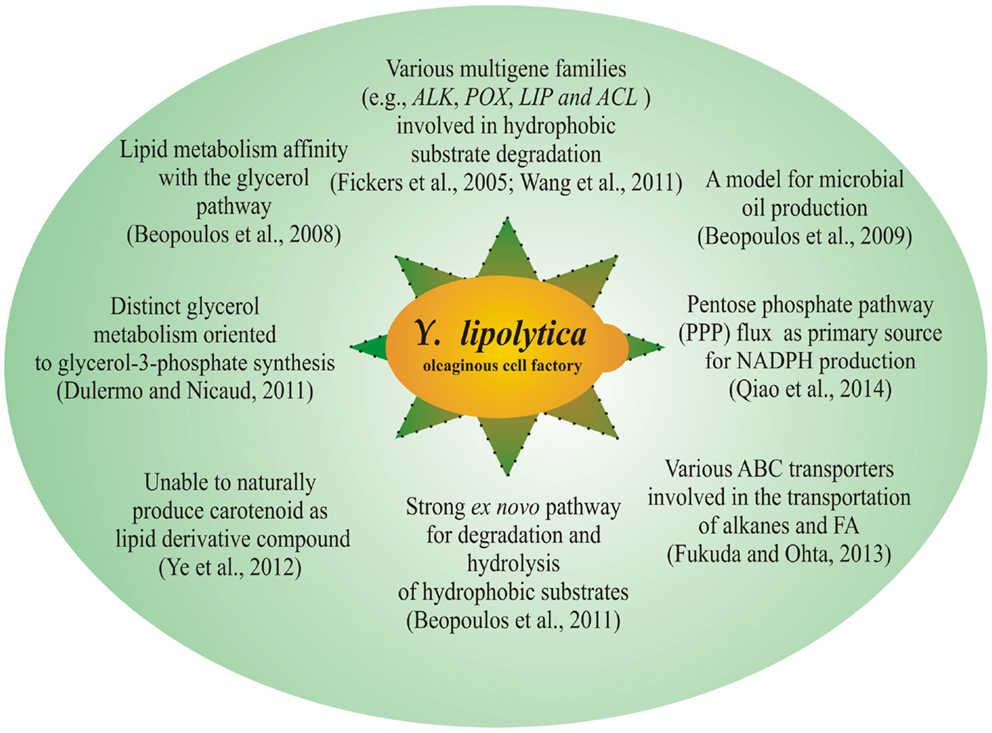
In general, three types of pathways consisting of α-oxidation, β-oxidation, and ω-oxidation contribute to the oxidative degradation of the hydrophobic substrate. Briefly, α-oxidation involves the conversion of alkanes to related alcohol; β-oxidation contributes to the FAs degradation to CO2 and water; and ω-oxidation involves the hydroxylation of alkanes and FAs. The hydroxylation of n-alkanes and FAs to fatty alcohols or hydroxy FAs takes place in the ER. These intermediate products are further degraded through either peroxisomal β-oxidation pathways or microsomal ω-oxidation pathways (Iida et al., 2000). Therefore, both ER and peroxisome involves several metabolic steps to degrade hydrophobic substrates. Several distinct features of Y. lipolytica for lipid metabolism are shown in Figure 1.
Challenges in Upgrading the Y. lipolytica Platform
The development of Y. lipolytica platform for industrial applications still poses challenges. The most important ones include obtaining more naturally versatile strains, making the platform more robust for enhanced yield and productivity especially when alternative cost–effective carbon and nitrogen sources are used (Jadhav et al., 2012), and improving fermentation and downstream processes in favor of the modified strain.
The selection of the naturally versatile Y. lipolytica strain through screening is still one of the most important needs in industrial applications. This is because Y. lipolytica strains differ in terms of biomass and bioproduct production rate under similar conditions (Juszczyk et al., 2013; Sil et al., 2013). Dependable methods should be established for quick, cost–effective, and high-throughput screening of potential strains. These strains are checked for their capability to utilize low-value substrates, biomass yield, lipid productivity, and quality.
Oleaginous yeasts typically do not have a high growth rate, so the exponential phase rate can affect the performance of lipid production by these yeasts considerably. In addition, the maximum level of intracellular lipid accumulation in resting cells is usually achieved over several days (Seip et al., 2013), making this process time-consuming. Therefore, any attempt to enhance the growth and accumulation rate will result in a higher overall productivity. On the other hand, the growth of the recombinant strain can be negatively affected by metabolic perturbation or burden caused by classical metabolic engineering approaches. This necessitates the use of new tools such as protein engineering to develop versatile mutant transporters and enzymes for better biomass utilization and growth rates (Fisher and Tullman-Ercek, 2013). This can also enhance the yield of FA-based bioproduct (Lennen and Pfleger, 2013).
One of the major challenges for commercialization is to achieve yields close to theoretical limits. A study on the economy of biofuel production from sugar-based feedstock found that, FAEE was a less efficient biofuel than ethanol, butanol, and hydrogen, because it is produced through aerobic fermentation. In this process, a higher ratio of carbohydrate is directed to biomass and enzyme production over biofuel production. On the other hand, part of the sugar combustion energy is wasted in the separation process especially when the targeted metabolite or bioproduct is not efficiently secreted out of the cell (Huang and Zhang, 2011). It has also been reported that about 1 kg of oil is typically obtained from 6 kg of sugar in the yeast culture process, while the rest of the sugars may be used in biomass and byproduct formation (Chi et al., 2011). Actual lipid yield and production rate affect feedstock and fermentation costs (Ageitos et al., 2011). Therefore, it is necessary to improve product yield. However, it poses a challenge to rely solely on selecting versatile strains and modifying environmental conditions in order to accomplish this. For instance, the level of lipid accumulation rarely rises above 20% of the dry weight in Y. lipolytica when glucose is the sole source of carbon (Papanikolaou et al., 2006). This leaves some spaces for metabolic engineering.
From an economic standpoint, the production of value-added lipids and lipid products using oleaginous yeast can be successful if substrates with nearly zero cost are used by the oleaginous yeasts (Tsigie et al., 2011). The production of cellulosic oil and bioproducts is currently under development. In lignocellulosic feedstock, the ratio of hexoses over pentose sugars is about 1.5–3 to 1, highlighting the importance of pentoses (Huang et al., 2013a). It is notable that the concentration of utilizable sugars in some lignocellulosic feedstock is not adequate for SCO production (Huang et al., 2013a). This points to the need for supplementation or co-application of different feedstocks. Successful application of these materials in the bioconversion process requires highly dedicated technology and a robust oleaginous yeast platform. The strain should be able to efficiently utilize C5 and C6 sugars, and possibly the byproducts of pretreatment. Moreover, it should show tolerance for the inhibitory byproducts in the pretreatment process, including acetic acid, formic acid, furfural, and vanillin.
Although Y. lipolytica is naturally well-adapted to the hydrophobic biomass utilization, it is not naturally well-equipped with di- or polysaccharide hydrolyzing enzymes (Barth et al., 2003; Coelho et al., 2010). These enzymes aid in better utilization of sugar-based feedstock such as sustainable lignocellulosic materials. Y. lipolytica can not utilize xylose as the sole source of carbon (Sha, 2013). On the other hand, this yeast has a lower glucose uptake rate than S. cerevisiae (Tai, 2012), despite the existence of various hexose transporters (Young et al., 2011). These add up to relatively lower efficiency of sugar uptake system and underline the need to upgrade the intrinsic membrane sugar transporter to a more versatile type. In order to achieve a higher growth rate, this yeast should be able to import and conferment the pentoses and cello oligomers. This aids in prevention of the inhibitory effect of cellobiose on the cellulase enzyme and the repressive effect of glucose on sugar uptake.
Not only the lack of sugar but also the high concentration of carbon sources may be problematic because they negatively affect yeast growth and bioproduct composition. At concentrations of 320 g/L, glucose can reduce growth in engineered strains (Blazeck et al., 2014). At lower concentrations of 150 g/L, growth remains unchanged under nitrogen-limited conditions, but unsaturated FAs content is depleted (Papanikolaou et al., 2006). In a recent study, Y. lipolytica showed a decrease in growth with higher glycerol concentration, from 50 to 300 g/L in a chemically defined medium (Sriwongchai et al., 2013). Thus, the concentration of carbon sources should be adjusted accordingly to produce high yields and desirable compositions of lipid-based bioproducts. In addition to carbon sources, the application of other nutrients should be properly managed.
Nitrogen and phosphorous deprivations are a typical strategy to induce lipid accumulation in the oleaginous cell. These conditions can increase lipid yields, since almost no carbon flux is spent on biomass formation (Huang and Zhang, 2011). However, major drawback of this strategy is that lower biomass formation also reduces overall lipid productivity (Dey and Maiti, 2013). On the other hand, the carbon assimilation rate gradually decreases in response to the nitrogen-limiting conditions (Beopoulos et al., 2009). Inactivation of methyl-citrate cycle was successfully employed to enhance lipid production by 1.6-fold independent of nitrogen availability. However, this results in lower biomass generation of Y. lipolytica (Papanikolaou et al., 2013). Thus, further strategies need to be developed to increase lipid accumulation in the presence of a nitrogen source, while maintaining a constant carbon flux to the targeted lipid-derived metabolite is maintained at high productivity.
Mass transfer is also a factor in increasing productivity during fermentation of oleaginous microorganisms. Rheological properties of the culture media can be affected by dynamic parameters such as biomass concentration, lipid accumulation, and the morphology of the Y. lipolytica (Fillaudeau et al., 2009). This effect creates complexity in heat, mass, and air transfer. Furthermore, energy and electricity demands may also fluctuate. Thus, dynamic parameters should be considered when the bioprocess is performed at a large scale. Both intrinsic cell permeability and rheological properties affect the rate of substrate uptake.
Lipid accumulation is followed by lipid extraction, one of the most costly stages of biodiesel production (Wang et al., 2012). To date, no approaches have been developed to produce 100% oil yield, due to the thick yeast cell wall and the susceptibility of the microbial lipid to intracellular lipases (Poli et al., 2013). Although chemical extraction followed by quantitative analysis of lipids with GC–FID has been successfully used, it is considered to be time-consuming and impractical for industry.
Despite the production of different heterologous bioproducts in Y. lipolytica, the permeability of the plasma membrane for secreting those products poses a challenge. Adding surfactants to the growth media (Mirbagheri et al., 2011) or engineering this yeast for higher cell permeability may allow target bioproducts to be exported to surrounding media. This facilitates the downstream process for extraction, reducing costs. This suggests that existing efflux pumping systems could be modified by protein engineering and directed evolution strategies. Additionally, more compatible heterologous transporters could be introduced into the Y. lipolytica platform to enhance bioproduct tolerance. These modifications along with integration of bioconversion with a continuous product recovery system can reduce potentially toxic bioproducts. In terms of intracellular accumulation, extraction tactics should be emphasized.
There are several major barriers to efficiently exploiting genetic and metabolic engineering strategies. In oleaginous yeast, different organelles accommodate individual steps of the metabolic pathway, requiring understanding of the function and regulation of the pathway. Also, the mechanism by which acyl-CoA is transported among different subcellular organelles is not fully understood. Similarly, the mechanisms by which acetyl-CoA is transported from mitochondria to cytosol for synthesizing the FAs has not been fully clarified (Cao et al., 2006). To understand such issues, one must identify the complex FA-dependent pathways, regulatory mechanism of key contributory genes and enzymes, contributory DNA binding proteins and the relationship between the pool of FA precursors such as acyl-CoA with the expression of different gene sets.
More research is needed to determine the importance and frequency of recombination mechanisms in oleaginous yeasts, specifically Y. lipolytica. In fact, studies show that homologous recombination (HR) and non-homologous end-joining recombination occurs in this yeast, but microhomology-mediated end-joining and further non-homologous recombinases have not yet been found (Kretzschmar et al., 2013).
Genetic engineering may be used to manipulate the lipid-associated metabolic pathway. However, engineering the lipid pathway in this yeast has several limitations. For example, the β-oxidation pathway has not been thoroughly understood in terms of the contributory enzymes, its relation with TCA cycle, and the regulatory mechanisms for managing flux through various channels. On the other hand, the set of four main reactions that are repeated several times at different levels and handled by similar sets of enzymes to breakdown the FAs can complicate the engineering of the β-oxidation pathway (Waché et al., 2006). Undoubtedly, understanding this issue will help us fully harness the potential of peroxisome to compartmentalize production of specific lipid-derived compounds. Glucose, which is typically found in pretreated lignocellulosic feedstock, can repress the proliferation of this organelle, while oleic acid induces the β-oxidation pathway and proliferation of the organelle (Kunau and Hartig, 1992). Therefore, in order to fully take advantage of peroxisome in the glucose-grown oleaginous yeast, we must identify and subsequently modify the genes involved in peroxisome biogenesis and activity. Peroxisomal acyl-CoA synthetases pose another challenge, since they may have overlapping functions and activate FAs of different chain lengths and hydrophobicity. However, they have not been clearly assigned to specific oxidative pathways and substrates thereof (Watkins and Ellis, 2012).
Strategies for Developing the Y. lipolytica Platform for Lipid Production
The Y. lipolytica platform shows promise for the production of microbial oil. This section reviews strategies for extending the adaptability of this platform to leverage a wider range of carbon sources and enhance production of lipid-based bioproducts. The section explores optimal cultivation conditions, the development of associated tools, and genetic engineering strategies.
Strategies for Cultivation and Fermentation
Exploring industrial application of oleaginous yeast strains first requires versatile measuring tools to determine the physiological characteristics of the yeast and optimize them during fermentation. From the product perspective, the Nile red stain has been used with fluorescence microscopy to analyze intracellular lipid accumulation in Y. lipolytica (Poli et al., 2013, 2014). During fermentation, the online glucose biosensor has been successfully employed to monitor substrate concentration during long-term incubation of a repeated fed-batch process (Moeller et al., 2011). In this method, the ideal time duration of targeted bioproduct production can be estimated based on the consumption rate of the substrate. Finally, flow cytometry and flow particle image analysis were used together to analyze vitality, cell shape, and lipid development in other oleaginous yeast cultures (Raschke and Knorr, 2009).
A key strategy to improve the performance of Y. lipolytica in industrial applications is to optimize fermentation conditions. As a strict aerobic yeast, Y. lipolytica culture requires efficient aeration systems during its growth in bioreactors. In fact, the dissolved oxygen should be controlled based on the requirements of targeted bioproduct formation. For instance, high oxygen can cause overproduction of cell mass and lower production of some bioproducts (Huang and Zhang, 2011). However, production of oxidized bioproducts may require a sufficient amount of oxygen. For instance, the rate of oxygen consumption decreased during the transition from growth to lipogenic phases in engineered strains (Blazeck et al., 2014). The aerated cultures can boost oil biodegradation (Martins et al., 2012) or FFA oxidation to unsaturated ones (Sha, 2013). On the other hand, high agitation and aeration may intensify lipid degradation by increasing acyl-CoA oxidase activities (Papanikolaou et al., 2007). As a control, oxygen was adjusted to increase lipid content with a specific degree of saturation and regulate ω,β-oxidation enzymes (Choi et al., 1982; Hallenbeck, 2012). Some strategies have been developed to address oxygen as a typical limiting factor in the industrial aerobic process. For instance, perfluorodecalin can be used as an oxygen transporter or pressurized bioreactor to improve the growth rate and extracellular enzyme production in Y. lipolytica cultures (Coelho et al., 2010). Adjusting the culture media is also feasible to mitigate its oxygen requirement level. This strategy was practiced by using media with high iron (3.5 mg/L) (Kamzolova et al., 2003).
In addition to dissolved oxygen, other culture conditions should also be adjusted to increase biomass and bioproduct formation. These cultivation parameters include but are not limited to oxidoreduction potential, temperature, pH, the concentration of metal traces and minerals (e.g., Mg2+, Zn2+, Mn2+, Cu2+, Ca2+), the type of carbon and nitrogen source, and the C/N ratio. For instance, the C/N ratio of 50–100 usually induces lipid accumulation (Ageitos et al., 2011; Hallenbeck, 2012). A recent study used the D-stat continuous system of cultivation to determine the appropriate ratio for lipid accumulation in glucose-grown cells. Lipid accumulation occurred at the C/N ratio of 11.7–47 when citric acid production was absent or limited (Ochoa-Estopier and Guillouet, 2014). Higher ratios may reduce lipid yield and increase citric acid and other undesired byproducts (Rossi et al., 2011; Thevenieau and Nicaud, 2013). However, another study found that total glucose amount plays a major role in lipid accumulation for engineered glucose-grown strains (Blazeck et al., 2014).
Proper fermentation modes should also be selected to improve production and cultivation conditions (Beopoulos and Nicaud, 2012). Typical fermentation modes include batch, single-stage and double-stage fed batches, and continuous culture. For continuous or fed-batch mode, the C/N ratio and dilution rate should be optimized to control lipid accumulation and acid production. The fed-batch system is a suitable mode to separate the growth and lipid accumulation phases because it offers control of the aforementioned factors (Beopoulos et al., 2009). The two-stage fed-batch system has been used to achieve high biomass for subsequent conversion of volatile FAs to the SCO by Y. lipolytica (Fontanille et al., 2012). However, this system is rather complex for industrial use. Lipid accumulation can also be achieved in a continuous culture under nitrogen-limiting conditions and with stable dilution rates (Ageitos et al., 2011). Traditional stirred-tank and air-lift bioreactors are often used to optimize and develop bioprocesses involving Y. lipolytica strains (Coelho et al., 2010). The biphasic system of fermentation can simplify and stabilize whole-cell bioconversion of the hydrophobic substrate to value-added products. For example, the aqueous and organic phases can be used to improve biotransformation of progesterone by recombinant Y. lipolytica in a stirred-tank bioreactor. In this strategy, ethyl oleate and dibutylphthalate were relatively more efficient at elevating the eventual production levels. This was facilitated by higher accessibility of the hydrophobic substrate, lower inhibitory effects of the hydrophobic compounds, and the facilitated co-uptake of the organic element and the substrate (Braun et al., 2012). Finally, the agitation rate can also affect the rate of biomass and bioproduct generation and should be adjusted accordingly (Rywinska et al., 2012). Generally, the Y. lipolytica culture should be properly maintained between 25 and 30°C and a pH range of 3–6 for lipid accumulation (Ageitos et al., 2011). Enshaeieh et al. (2013) reported that optimum temperature, pH, agitation rate, and incubation time for lipid accumulation in Y. lipolytica M7 were 25°C, 5, 200 rpm, and 72 h, respectively.
The fermentation process is typically followed by the extraction process, and various approaches to extracting oil from oleaginous microorganisms have been reviewed (Wang et al., 2012). Glass bead disruption followed by solvent extraction has been noted to extract oil from yeast. Ultrasound, microwave, and electroporation have been considered as novel methods to enhance extraction. However, these approaches are not practical at the industrial scale due to technological restrictions. Oil has also been extracted from oleaginous yeast through solvent extraction from dry biomass, vortex and glass beads, ultrasonic bath and glass beads, liquid nitrogen, and liquid nitrogen with sonication (Poli et al., 2013). The later approach was found to be the most efficient for lab use. To improve the lipid extraction from the Y. lipolytica biomass, applying subcritical water pretreatment at an optimal of 175°C and a water addition of 20 mL/g of biomass for 20 min has been suggested (Tsigie et al., 2012a). This enhances the crude lipid extraction from 51.54 to 84.79% in the subsequent solvent-assisted lipid extraction. This may be due to higher reactivity of water ions including H+ and OH− as catalysts that facilitate the chemical reactions that break the cell wall. Finally, it is noteworthy that the most suitable lipid extraction method for the industrial scale has not yet been determined.
Using mixed cultures of organisms, including oleaginous and non-oleaginous ones, can also enhance the efficiency of fermentation and substrate utilization. For example, mixed cultures of microalgae and some oleaginous yeast species on sugarcane juice have been introduced as a desirable strategy for lipid accumulation. In this strategy, yeast as a CO2 generator and microalgae as an O2 generator supported one another’s growth under mixotrophic cultivation (Papone et al., 2012). The co-culture system of fermentation may demonstrate outstanding characteristic for more efficient utilization of feedstock, higher tolerance of inhibitory compounds, and higher production of biomass and target bioproducts.
Another approach to increase productivity during fermentation is biomass immobilization. An alginate bead carrier in a batch system and a k-carrageenan carrier in an air-lift bioreactor were used to immobilize Y. lipolytica for citric acid production (Kautola, 1991). Although this yeast shows promise for biofilm generation, this approach may be inefficient for lipid production under nutrient starvation. However, the Y. lipolytica biofilm on the selective substrate can be more efficiently used in a fixed bed biofilm reactor (Lehocký, 2007). The immobilization technique may also prevent reconsumption of secreted bioproducts. Another related application of immobilization technique is to enhance the effectiveness of yeast products. This yeast produces several types of lipases, and its enzymes can also be immobilized to enhance hydrolysis, esterification, interesterification, transesterification, and ring-opening polymerization processes. These processes have potential industrial applications for biodiesel production (Hallenbeck, 2012), digestive acid production (Marova et al., 2011), and flavor modification with the aid of free FAs and esters (Waché et al., 2006; Fickers et al., 2011). Immobilization strategies may also pave the way for continuous whole-cell or enzyme-based biotransformation processes.
To offset the cost of microbial lipid-based biofuel production, yeast biomass may be recycled for various purposes. For example, the direct use of residual yeast biomass by animals can improve the economy of SCO production, since the biomass of oleaginous yeast has considerable amounts of protein, carbohydrates, and other nutrient ingredients. The potential of a marine Y. lipolytica strain as a feed component for cultured marine animals has been examined (Wang et al., 2009). The anaerobic digestion of residual biomass is another means of reducing the production costs (Meng et al., 2009). In one study, a spent medium and yeast biomass from the first step of lipid accumulation and extraction were reused in the next steps of yeast and algae cultivation in an integrated system to maximize substrate utilization and improve the economy of the process (Chi et al., 2011). In a recent study, the hydrolyzate of Y. lipolytica biomass acted as a medium for bioethanol production using S. cerevisiae (Tsigie et al., 2013). Higher yields of glucose were obtained from hydrolyzate after lipid extraction, and 98.05% of this glucose was converted to bioethanol at a yield of 0.38 g ethanol/g glucose. These studies provide evidence for maximal exploitation of this yeast.
Strategies Associated with Extension of Substrate Range
A key step for producing lipids and co-products by Y. lipolytica is to upgrade this platform to utilize pentoses from lignocellulose. There are contradictory reports on xylose utilization in this yeast. One study showed that this yeast can simultaneously utilize d-xylose and d-glucose by consuming xylose at a relatively higher rate. However, glucose could competitively inhibit the uptake of xylose (Tsigie et al., 2011). The co-fermentation of xylose and glucose may enhance biomass and lipid formation in this yeast (Sha, 2013). In another study, the xylose reductase encoding gene, XYL1, and xylitol dehydrogenase encoding gene, XYL2, were transferred from Scheffersomyces stipitis into Y. lipolytica (Tai, 2012). Actually, Y. lipolytica has the basic structure for this manipulation, since it possesses putative XYL12 genes within its genome. The combination of this strategy with several adaptation steps helped develop a strain that grows on xylose as the sole source of carbon and reach a lipid accumulation level of 42%. Further study may target this platform to enhance the uptake of pentose and oligo-sugars, which are present in pretreated lignocellulosic materials, while maintaining the appropriate growth rate.
Yu et al. (2011) used wheat straw hydrolyzate from a dilute sulfuric acid pretreatment for lipid accumulation in several oleaginous yeast strains including Y. lipolytica. This strain could use both detoxified and non-detoxified forms of the substrate to grow and reach a lipid content of about 4.6% w/w (Yu et al., 2011). Sugarcane bagasse hydrolyzate was also used to grow Y. lipolytica and achieve a maximum lipid content of 58.5% (Tsigie et al., 2011). Defatted rice bran hydrolyzate and sugarcane bagasse hydrolyzates as carbon and nitrogen source have been used to cultivate this yeast to reach a lipid content of 48.02 and 58.5%, respectively (Tsigie et al., 2011, 2012b). These studies indicate that the lipid content of this yeast varies, depending on strain and feedstock.
One promising strategy is to express heterologous polysaccharide cleaving enzymes and target them to the cell surface in order to enable Y. lipolytica to grow on lignocellulosic feedstocks. The advantage of this strategy lies in the fact that Y. lipolytica is a non-cellulose degrading yeast. On the other hand, the cost of hydrolyzing enzymes is still high, which can hamper the development of economically viable cellulosic biofuel technology. In an attempt to upgrade this platform to utilize plant polysaccharides, the gene for exo-inulinase was cloned from the yeast Kluyveromyces marxianus and expressed into Y. lipolytica. This resulted in direct bioconversion of inulin into 48.3% (w/w) oil (Zhao et al., 2010a). In fact, a remarkable lipid accumulation and small citric acid production were observed during fermentation of inulin containing a substrate by Y. lipolytica. In another study, the gene encoding exo-inulinase from the yeast K. marxianus was expressed and immobilized on Y. lipolytica cells using a surface-display plasmid (Liu et al., 2010). This strategy was used for simultaneous utilization of inulin and production of citric acid. By targeting heterologous proteins at the surface of the cell, yeast cells become more suitable for other applications such as the development of live vaccines, antibody libraries, whole-cell biocatalysts, and absorbent. The combination of fungal α-amylase gene, together with its signal peptide, successfully led to the extracellular secretion of this enzyme in Y. lipolytica (Park et al., 1997). The bacterial chitosanase is another example of a sugar-hydrolyzing enzyme whose gene, chi, was transformed into Y. lipolytica for the overexpression of bacterial chitosanase. A remarkable level of expression (i.e., 9.0 U/ml) and activity were achieved in the conversion of chitin into constituent oligosaccharides (Liu et al., 2012a). Heterologous mannanase has also been produced on the cell surface of this yeast, reaching the high activity level of 62.3 IU/g DCW during the 96 h of cultivation period (Yang et al., 2009). Molasses, a byproduct of sugar refinery, have also been used at an optimum concentration of 8% to achieve lipid accumulation of 59.9% by this yeast (Karatay and Dönmez, 2010). In order to achieve efficient utilization of molasses, the invertase-positive strain of Y. lipolytica is required. Alternatively, the heterologous invertase gene can be introduced into this yeast (Lazar et al., 2013). Recently, several heterologous cellulases including endoglucanase, cellobiohydrolase, and β-glucosidase were expressed in this yeast. This enhanced the enzymatic hydrolysis of cellulosic substance, which can be further converted to biofuels. This strategy may enable the platform to simultaneously utilize lignocellulosic sugars and produce targeted bioproducts in consolidated bioprocessing (CBP). This strategy may also improve enzyme stability and the economics of downstream processing.
To improve the economy of the microbial lipid technology, simultaneous hydrolysis of cellulosic biomass and lipid accumulation in the oleaginous yeast Cryptococcus curvatus has been suggested (Gong et al., 2013). In this integrated system, a high dosage of inoculum combined with hydrolytic enzymes could improve the level of sugar utilization and mitigate the susceptibility of oleaginous yeast culture to contamination. Additionally, the absence of supplementary nutrients aids in lipid accumulation and higher cell resistance. One study used CBP to produce cellulosic bioethanol with a non-oleaginous yeast consortium was also reported (Kim et al., 2013). Such studies can spark further research on the Y. lipolytica platform in advanced CBP to produce cellulosic lipid-based bioproducts. Therefore, overexpressing heterologous sugar-hydrolyzing enzymes in this GRAS yeast can help facilitate the use of CBP for the production of cellulosic lipid and functional oligosaccharides. These oligosaccharides can be used as prebiotics in various food products to boost proliferation of probiotic bacteria in the colon and improve human health (Abghari et al., 2011).
Yarrowia lipolytica can also be potentially used to enhance the biorefinery of lignocellulose-based feedstock and utilization of byproducts. Y. lipolytica has been used to develop a biological detoxification process, due to its natural capability to produce extracellular laccase (Lee et al., 2012). This capability was harnessed to oxidize the lignin-derived phenolic compounds for pretreated rice straw to mitigate their inhibitory impacts on cellulase. Other studies have explored the suitability of Y. lipolytica as an expression system for recombinant laccase (Madzak et al., 2005b). However, the high concentration of Y. lipolytica biomass can potentially maintain volatile FAs, the byproducts of pretreatment process, below the inhibitory level for recycling (Fontanille et al., 2012).
Yarrowia lipolytica has shown promise for converting glycerol to SCO and citric acid (André et al., 2009; Makri et al., 2010). Recently, lipid accumulation has been found in the Y. lipolytica strains grown on glycerol in the range of 37.1–54.4% of cell dry weight (Sriwongchai et al., 2013). The introduction of heterologous glycerol dehydratase genes and several alcohol oxidoreductases under the control of glycerol inducible promoter, G3P dh, gave rise to a higher growth rate and a lipid yield at13 g/L of culture volume (40% of DCW) (Celinska and Grajek, 2013). This could be enhanced by the codon optimization of the heterologous genes and the prevention of further degradation of accumulated lipid. The mixed substrate utilization of glycerol and other substrates also show promise for enhancing lipid accumulation (Papanikolaou et al., 2003). However, the inhibitory impact of glycerol on the metabolism of hydrophobic biomass and the regulatory mechanisms should be addressed in mixed carbon sources (Mori et al., 2013).
Strategies for Genetic Engineering
Selecting optimum cultivation conditions should be accompanied with the appropriate metabolic and genetic engineering of Y. lipolytica. These engineering strategies have mainly been used to intensify carbon flux to the targeted bioproduct, produce new FA-based co-products, and extend the range of utilizable feedstock. Most of the strategies can be found tabulated elsewhere (Liang and Jiang, 2013). Successful application of these engineering techniques requires understanding of the roles of contributory metabolic enzymes, regulatory mechanisms, and the mechanisms of secretion and/or re-uptake of bioproducts. Moreover, the application of next generation DNA sequencing, microarray analysis, and mass spectrometry can also help to comprehend how the Y. lipolytica platform works and how it should be manipulated.
The major engineering strategies to improve the Y. lipolytica platform include but are not limited to the expansion of the pools of G3P and acyl-CoA, the promotion of key steps in TAG biosynthesis pathway, and the prevention of lipid degradation through the competitive pathway (Courchesne et al., 2009). Some critical contributory enzymes to lipid biosynthesis are ACC, Δ 9 stearoyl-ACP desaturase (D9), and DGA, and ACL. The foregoing enzymes were ordered based on the highest to the lowest degree of contribution to lipid overproduction as DGA, ACC, D9, and ACL (Tai, 2012). The transporting enzyme can also be targeted to enhance production of lipid-derived bioproducts. This strategy was used in other oleaginous yeasts, and appears to be simpler than manipulating the central metabolic pathway (Cao et al., 2006).
Researchers have transformed the genes encoding acetyl-CoA synthase and ATP:citrate lyase from S. cerevisiae and Mus musculus into Y. lipolytica to increase the acetyl-CoA pool (Zhou et al., 2012). Upregulation of the latter gene is important, since its enzyme activity seems to increase as intracellular carbon flux increases (André et al., 2009). TAG accumulation was also enhanced in this yeast by elevating the G3P pool. This was achieved by modifying the G3P shuttle by disrupting GUT2 and the overexpression of GPD1 genes. These genes govern the reverse reaction between G3P and DHAP. Overexpression of the GPD1gene, in combination with deletion of POX1–6 genes, may cause lipid accumulation of up to 65% dry weight in Y. lipolytica (Dulermo and Nicaud, 2011). Others have disrupted the putative acyl-CoA synthetase gene YAL1 in order to identify the function of one of the fatty acyl-CoA synthetases in Y. lipolytica. This led to a 1.47-fold increase in lipid production and an elevation of saturated to unsaturated FAs. This gene may play a role in elongating and desaturating FA (Wang et al., 2011b). However, more research is needed to determine the function of various acyl-CoA synthetase genes in Y. lipolytica organelles, as well as their selectivity toward activation of specific FAs and how activated FAs are directed to lipid accumulation or degradation pathways. The disruption of TAG lipases coding genes TGL3 and TGL4 may also improve lipid content by repressing TAG hydrolysis (Dulermo and Nicaud, 2011). The POX genes and corresponding Aox proteins can be manipulated in different ways to produce various lipid bioproducts, control of FA chain length, and supplement expression of heterologous FA modifying enzymes in Y. lipolytica. In fact, POX deficiency may heighten modification to lipid-associated genes. For example, the impact of G3P shuttle modification on improvement of TAG content is intensified when modification is followed by POX deficiency. This combinational strategy promotes the pool of G3P and FA, altering gene expression in TAG homeostasis and enhanced lipid accumulation (Dulermo and Nicaud, 2011). The disruption of POX genes can also limit TAG biosynthesis thorough the feedback inhibitory effect (Mlickova et al., 2004).
Genetic modification of oleaginous microorganism is considered to be a sustainable and economical strategy for producing essential FAs, such as linoleic acid (LA), α-linolenic acid, and other functional poly unsaturated FAs that are not synthesized by the human body. Y. lipolytica and some other oleaginous yeasts are naturally capable of producing LA acid or linolenic acid. Introducing specific desaturase, elongase, hydroxylase coding genes, and their facilitator genes can convert FAs to PUFA and other functional FAs. Tuned expression of the heterologous Δ 9 pathway containing Δ-9 elongase, Δ-8 desaturase, Δ-5 desaturase, and Δ-17 desaturase, along disrupting PEX10, involved in the biogenesis of peroxisome, produces a high yield of eicosapentaenoic acid (EPA) in Y. lipolytica (Xue et al., 2013).
Some research has targeted the intrinsic regulatory system of this yeast. The inhibitory effect of glucose on transcript levels of lipid biosynthesis-associated genes should be mitigated to improve lipid content in glucose-grown Y. lipolytica. To this end, researchers have disrupted Mig1 protein as a global transcriptional repressor, which is the major constituent of the glucose repression pathway in this yeast. This disruption could enhance lipid content from 36 to 48.7% (w/w) at the cost of a lower growth rate (Wang et al., 2013). This could be due to the activation of more metabolic pathways when they are no longer repressed. Other researchers have intensified and hastened the lipid accumulation process by manipulating the SNF1 complex and some of its subunits. Ylsnf1 is one of the major negative regulators of lipid accumulation in Y. lipolytica. Deleting the Ylsnf1 gene has been found to enhance FA content 2.6 times in the absence of nutrient starvation. This is because this gene plays a role in the transcriptional regulation of key lipid metabolism-associated genes (Seip et al., 2013). The combined manipulation of the SNF1 and lipid degradation pathway may help continuously produce FAs of specific functionality during exponential growth and stationary phases. Identifying and manipulating these regulatory pathways will improve the ability of this yeast to produce lipid-based biofuel.
In addition to directing flux to the engineered pathway, a balance should be maintained between substrates and cofactors and also between the upstream and downstream flux. In a recent study on Y. lipolytica, balance was established between these two fluxes by amplifying ACC1 and overexpressing DGA (DGA1). This balance minimized the accumulation of intermediate metabolites and created a synergistic effect of gene co-expression. This co-expression, in tandem gene construction, resulted in a fivefold increase of lipid content to 41.4%, and subsequently to 61.7% of DCW during 120 h of fermentation in a 2-L bioreactor (Tai and Stephanopoulos, 2013). This diverted the flux toward the desirable product and created a driving force. A de novo synthesized lipid has recently been enhanced to 90% DCW in glucose-grown cells, through upregulation of key lipid-associated genes along with the suppression of peroxisomal FA degrading pathway and application of leucine, an intracellular inducer signal. Leucine may facilitate lipogenesis and alleviate dependency on nitrogen starvation (Blazeck et al., 2014). This rises beyond the maximum ex novo lipid accumulation of 70% DCW, which was obtained from an engineered strain fed on pure oleic acid (Beopoulos et al., 2008). This shows great promise for application of this platform for the production of cellulosic biofuel.
Another promising strategy is to compartmentalize specific metabolic pathways in one organelle. Targeting the enzymes of the Ehrlich pathway to mitochondria was recently found to be more efficient than overexpressing contributory enzymes to achieve higher yield of isobutanol (Avalos et al., 2013). This advantage is due to the concentrated availability of the precursors and enzymes, obviating the need to transport metabolites. These strategies inspire more exploration of the heterologous lipid-associated metabolic pathway in the lipid body, peroxisome, or mitochondria of this oleaginous yeast, and pave the way for purposeful and concentrated degradation of lipid and lipid precursors to lipid-derived bioproducts of higher value.
The FA biosynthesis pathway can be dynamically controlled by identifying key contributory transcriptional factors and developing tunable hybrid promoters. For instance, the synthetic regulator system is well-constructed for enhanced production of biodiesel in E. coli (Zhang et al., 2012b). IPTG has been used to activate the hybrid promoter, and the induction level was finely tuned by FA/acyl-CoA level based on naturally existing FA-sensing protein. The invention of such synthetic regulators can be inspiring for the dynamic control of FA-associated pathways in the Y. lipolytica platform.
Finally, after successful optimization of the metabolic and culture conditions for the maximal production of bioproducts, the scale-up process should be properly carried out for consistent production of targeted byproducts at a larger scale.
Genetic tools
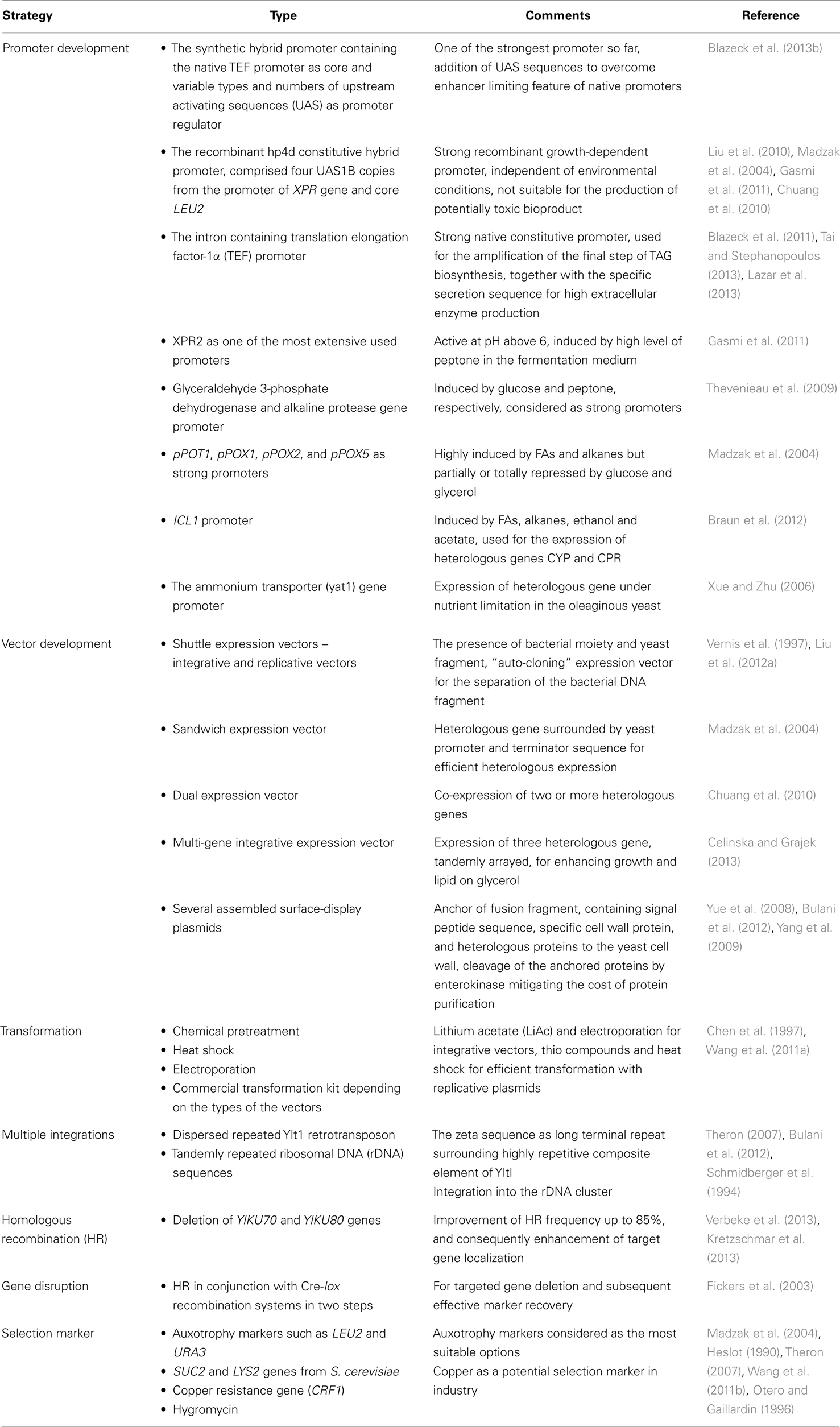
Since Y. lipolytica is a model of oleaginous yeast, many genetic tools have been developed for modification of this platform. Table 3 briefly shows some of the tools developed to genetically engineer this platform, including development of different promoters, vectors, selection markers, and so on. More information can be found elsewhere (Madzak et al., 2005a).
Genome-scale metabolic model for in silico analysis
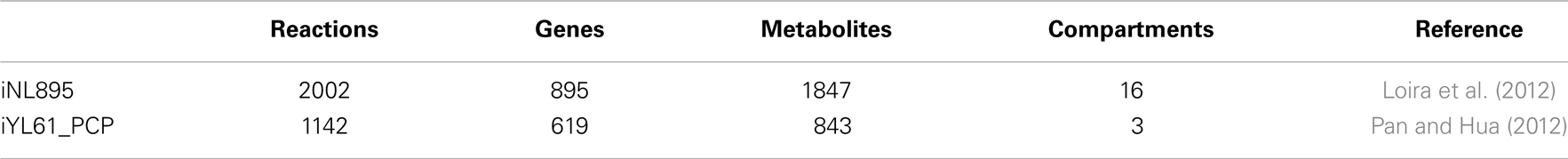
Genome-scale metabolic model (GSMM) can greatly improve the performance of efficient genetic and metabolic engineering strategies. Such models are based on known protein coding genes, gene homology data, biochemical information from databases such as KEGG (Kanehisa et al., 2008), ENZYME (http://enzyme.expasy.org/), BIGG (Schellenberger et al., 2010), and other related in silico approaches. The first functional GSMM for Y. lipolytica was proposed by Loira et al. (2012) and called iNL895, using the model of a phylogenetically distant yeast S. cerevisiae. Some reactions are included in the model that are related to alkane (i.e., cytochrome P450 oxidases and reductase) and TAG (i.e., extracellular or membrane bound lipases) degradation, FA synthesis (i.e., acyl-CoA:diacylglycerol acyl transferase), hydrophobic molecule transportation, and other interorganelle transporters (i.e., ABC). However, reactions related to utilization of sucrose and production of ethanol were removed from the model. In addition to simulating gene knockout, biomass formation was estimated using flux balance analysis by considering experimental information and limitations on the intake capacity. The major drawbacks of the model are generation of false negative or positive results, failure to account for kinetic and substrate specificity of contributory enzymes and failure to describe metabolic pathways dynamically. A second in silico model is based on conclusive data from well-known databases and has successfully simulated minimal media, growth behavior on different substrates, and single gene knockouts. However, regulatory mechanisms should be well-established and applied when upgrading these models (Pan and Hua, 2012). Table 4 shows some basic properties of the proposed models.
Despite their advantages, these models require modification using new data from functional genomics of this yeast, new discoveries on the corresponding regulatory mechanisms, and new information on the kinetic and substrate specificity of related enzymatic reactions. Undoubtedly, these improvements will result in more functional models to predict behavior of the engineered Y. liplytica by in silico analysis.
Use of Y. lipolytica Platform to Produce FA-Based Biofuel and Bioproducts
The endogenous functions of Y. lipolytica have been manipulated for biosynthesis of various bioproducts. This created a plethora of yeast products, as reviewed in the literature (Barth and Gaillardin, 1996). Generally, Y. lipolytica strains can be used to produce fuels and chemicals whose microbial productions require the carbon flux to be directed to the FA synthesis pathway. Together with GRAS status and intrinsic capability for high-level production of large heterologous proteins, this makes the engineered Y. lipolytica platform quite attractive for use in the production of various FA-based biofuel and bioproducts. For instance, using Y. lipolytica to produce a variety of lipid-derived compounds such as wax ester, isoprenoid-derived compounds, polyhydroxyalkanoates (PHA), and free hydroxylated FAs from lipid substrate has been investigated in European project, called “lipoYeasts” (Sabirova et al., 2011).
The next subsections focus on FA-based bioproducts that are derived or produced from the engineered lipid pathways in this yeast. These bioproducts are briefly presented in Figure 2.
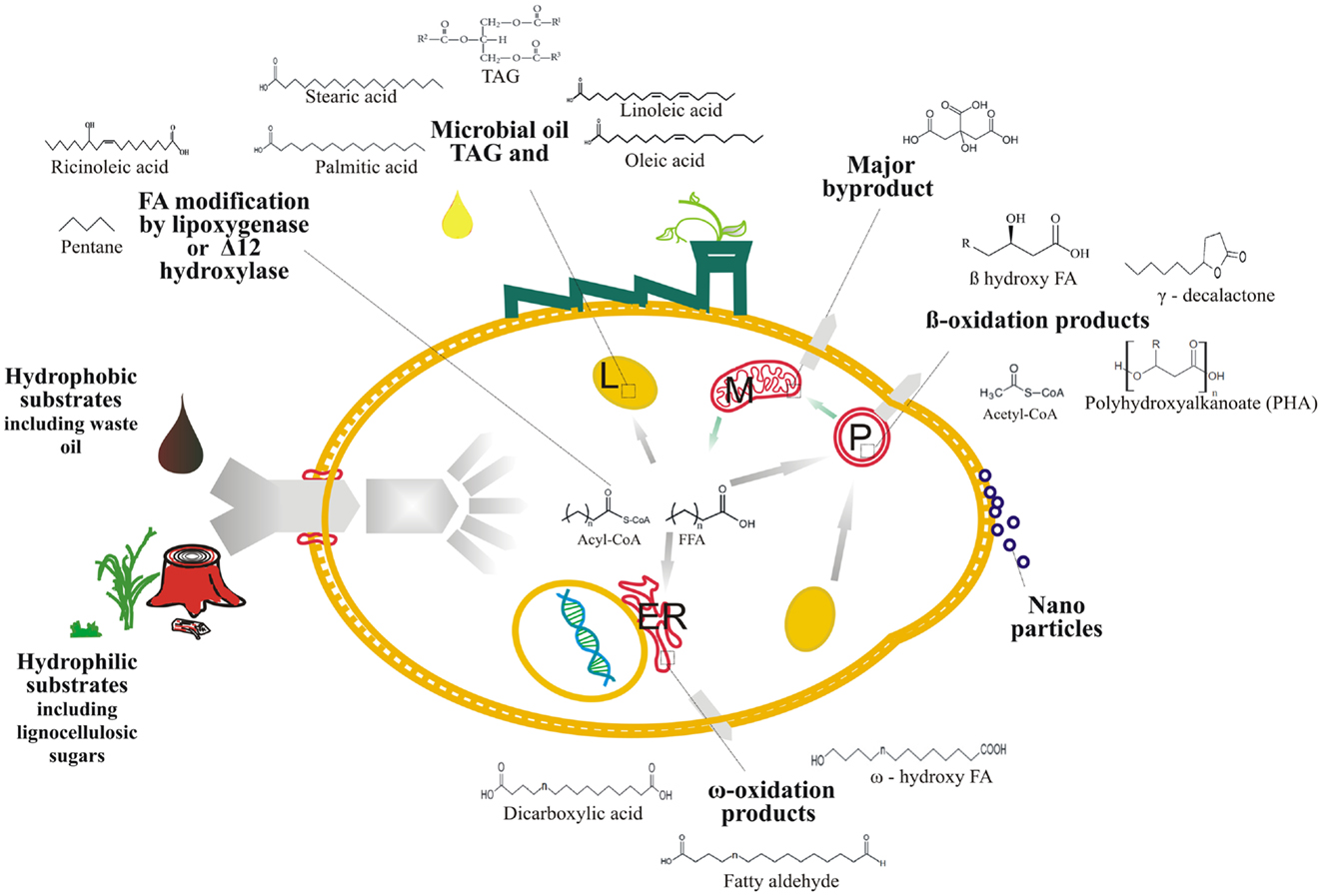
Figure 2. Overall schematic of the fatty acid-derived bioproducts production from hydrophobic and hydrophilic substrates using Y. lipolytica cell factory, ER, endoplasmic reticulum, L, lipid body, M, mitochondria, and P, peroxisome.
Production of FA-Based Advanced Biofuels
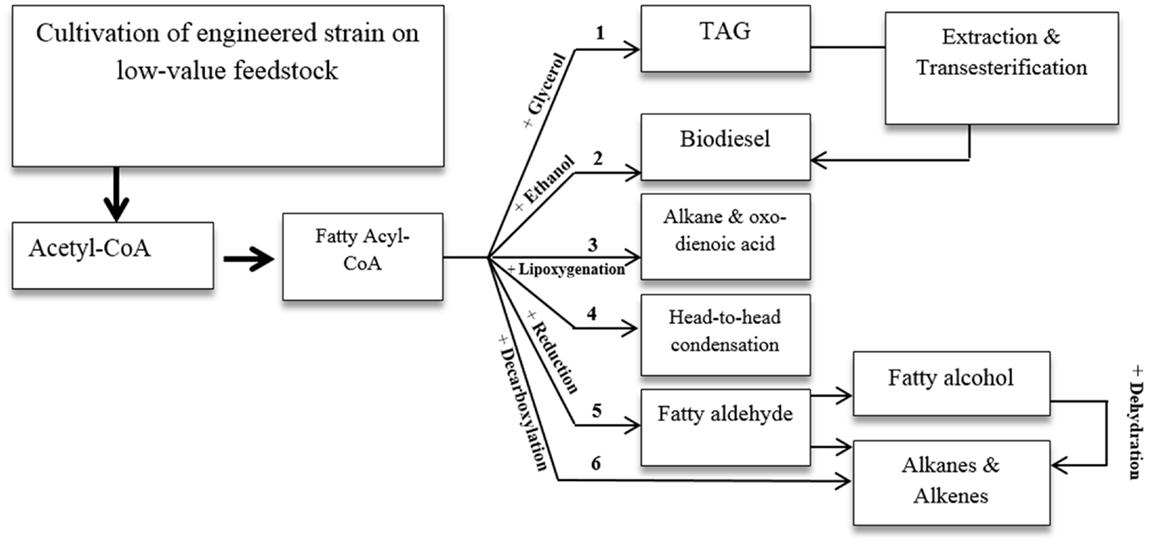
The intrinsic FA and lipid production of oleaginous yeasts make them ideal for production of lipid-derived bio fuel. Several potential routes for FA-based biofuel production are shown in Figure 3 (Brown and Shanks, 2012; Hallenbeck, 2012; Wang and Lu, 2013). Some of these routes have already been investigated for Y. lipolytica, while others need further exploration.
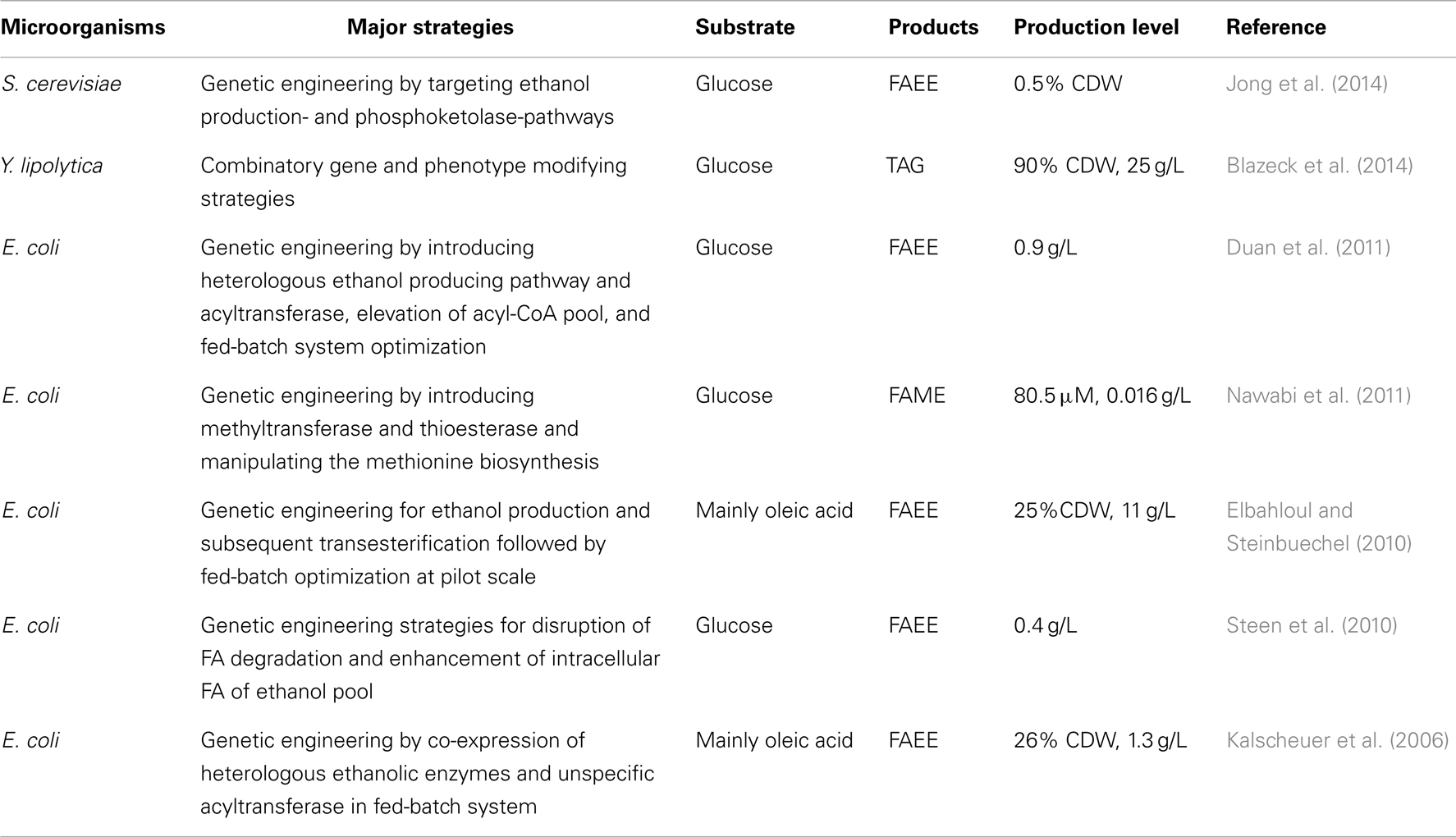
Yarrowia lipolytica shows promise in the conversion of low-cost materials to microbial oil and biodiesel, which is regarded as the first feasible resolution for the shortage of oily resources (Katre et al., 2012; Huang et al., 2013a; Blazeck et al., 2014). However, direct production of biodiesel has not yet been achieved with this yeast. However, this was achieved in recombinant E. coli and S. cerevisiae. Table 5 presents major achievements in microbial biodiesel production.
Genetic strategies for producing FAEE and FA methy ester (FAME) have typically targeted de novo or ex novo FA biosynthesis, ethanol production, and phosphoketolase pathways by introducing heterologous genes for higher production of acetyl-CoA, NADPH, ethanol, S-adenosylmethionine, acyltransferase, including FA methyltransferase and wax ester synthase, and disruption of FA degrading genes.
Another strategy was introduced by Blazeck et al. (2013a) who produced short-chain n-alkane in Y. lipolytica by introducing soybean lipoxygenase enzyme to cleave LA to pentane and tridecadienoic acid. In fact, many oxygenases can be used in specific biooxidation of FAs and lipids to functional metabolites, including alkanes and aldehydes. These membrane-associated enzymes require cofactors such as NADPH and NADH to introduce oxygen at single or double bond locations in the presence of electron transfer partner such as flavin reductase and iron sulfur protein (Urlacher and Schmid, 2006). Introducing such heterologous lipid-related enzymes into Y. lipolytica can pave the way for production of new lipid-derived biofuels. Nonetheless, complexity in the membrane-associated metabolic steps can complicate the introduction of such pathways into the yeast.
Some genes known as ole genes were found in Gram-negative and Gram-positive bacteria and encoded head-to-head FA condensation pathways. In this pathway, two FAs become connected through their carboxyl group releasing the carboxyl group and generating double bonds at the linkage point. Heterologous expression of these genes allows for long-chain olefinic hydrocarbon or monoketone production to the host cell (Sukovich et al., 2010). FA-overproducing E. coli has been used as platform for the biosynthesis of large chain alkenes. Heterologous three-gene cluster encoding for the homologous of the condensing enzymes, involved in FA biosynthesis (i.e., β-ketoacyl-ACP synthases), has been expressed in the host (Beller et al., 2010). In the suggested biosynthesis pathway, β-ketoacyl-CoA, a product of the β-oxidation pathway, and acyl-CoA, a product of FA biosynthesis, undergo condensation reaction. The product of the reaction goes through a pathway that is homologous to the FA elongation pathway in the presence of different reductases and dehydratases, resulting in long-chain alkene production (Ruffing, 2013). The suggested pathway can potentially be installed on the oleaginous yeast platform, since peroxisomal β-oxidation and FA biosynthesis are inherently and efficiently present in this platform. However, successful installation and compartmentalization requires comprehensive understanding of the peroxisome, its metabolic components, and corresponding regulatory and transformational mechanisms. These strategies may be employed in the future to produce advanced biofuels by manipulating oleaginous yeast platform.
Reducing fatty acyl-CoA to the corresponding aldehyde through eukaryotic acyl-CoA reductase followed by enzymatic decarbonylation can open the door to the production of alkane and alkenes from FAs with this oleaginous platform. However, it should be noted that fatty aldehyde decarbonylation is not well-understood. Some genes with aldehyde decarbonylase activity have been discovered in microbial species such as cyanobacteria. Codon optimization may be necessary to achieve an acceptable expression level. On the other hand, the above-mentioned multistep process may be compartmented into specific organelles to boost efficiency and conversion yield. A recent study shows that introducing specific heterologous FA reductase complex and aldehyde decarbonylase enables E. coli to directly use FFA for the production of linear and branched-chain alkane and alkene (Howard et al., 2013). The intracellular pool of FFA, and consequently the composition of the produced alkanes and alkenes were modified by supplementing media exogenously, and introducing heterologous thioesterase, branched-chain α-keto acid dehydrogenase, and β-keto acyl carrier protein synthase. FA intermediates may also undergo decarboxylation to hydrocarbon by FA decarboxylases of the P450 enzyme family (Rude et al., 2011). This is another inspiring way to make this platform a hydrocarbon producer.
Production of FA-Based Nutraceuticals and Pharmaceuticals
Yarrowia lipolytica has been engineered to produce various functional FAs. For instance, arachidonic acid, as a ω-6 polyunsaturated FA, was produced with recombinant strains. This was achieved by transferring several heterologous desaturases, elongases, and acyltransferases into the yeast (Damude et al., 2006). Y. lipolytica was also recently engineered for the production of EPA at 56.6 and 15% of lipid content and DCW, respectively, the highest titer reported so far (Xue et al., 2013). Y. lipolytica has also been used to produce up to 5.9% of trans-10, cis-12 conjugated linoleic acid (CLA) of the total FA yield from glucose. The heterologous gene encoding LA isomerase was codon optimized and introduced into the yeast platform that is capable of free LA production, to covert the free FAs to the CLA (Zhang et al., 2012a). That study showed that codon optimization, integration sites, and the gene’s copy number are major factors for modifying the expression level. Also, the manipulation of heterologous and homologous desaturase, elongase, and acyltransferase system enabled this yeast to produce docosahexaenoic acid at 5.6% of total accumulated lipid (DHA) (Damude et al., 2014).
This yeast has also been used to produce other lipid-derived neutraceuticals. Some examples of these products are sterols (Bailey et al., 2010), carotenoids, and antioxidants (Sharpe et al., 2008). In the lipoyeasts project, various heterologous carotenoid genes (crt genes) were placed under the control of pPOX2 promoter in the integration expression vector and transformed into Y. lipolytica. In spite of the disruption of β-oxidation pathway and the restriction of lipid accumulation, the modification did not yield an outstanding production level of carotenoids (Sabirova, 2011). Y. lipolytica has also been engineered to direct the products of its isoprenoid biosynthesis pathway to β-carotene as lipophilic bioproducts (Grenfell-Lee, 2009). In a recent study, Y. lipolytica β-carotene did not display a safety profile that differed from other commercial carotenoid products, including microbial and synthetic ones (Grenfell-Lee et al., 2014). However, its application as a safe food additive may require further long-term oral toxicity analysis. This yeast has also been engineered for lycopene production (Ye et al., 2012; Matthäus et al., 2014). In a recent study, disruption of POX1–6 and GUT2 genes were followed by the introduction of heterologous genes and upregulation of native genes to achieve a maximum lycopene yield of 16 mg/g DCW in a fed-batch system under oil accumulation conditions. In fact, stimulation of lipid accumulation may play a key role in accommodating such hydrophobic carotenoids.
Recombinant Y. lipolytica also shows potential for expression of various heterologous P450 systems (Obiero, 2006; Theron, 2007). This expression may aid in production of active steroids (Mauersberger et al., 2013) or other oxygenated FAs with antimicrobial properties.
Production of FA-Based Biochemicals
Several subcellular metabolic pathways of Y. lipolytica have been used to produce various lipid-derived biochemicals. For instance, the pathway of microsomal ω-oxidation, peroxisomal β-oxidation, FA synthesis, and TCA cycle have been engineered and harnessed in favor of specific bioproducts. In fact, these pathways accommodate a remarkable amount of carbon flux in this yeast.
The addition of functional groups such as hydroxy, carboxyl, or epoxy groups to FAs, can be achieved through ω-oxidation. The resultant products have a tendency to polycondense and cyclisize (Thevenieau et al., 2010). The ω-oxidation pathway can be manipulated to produce a variety of α,ω-dicarboxylic acids (DCAs), ω-hydroxy acids, and lactones. Production of these compounds using engineered Y. lipolytica has been recently discussed (Wache, 2013). Y. lipolytica has also been selected as a host for the expression of various CYP540s (Obiero, 2006). DCA production requires that a pool of FA is converted to DCA through ω-oxidation. The typical strategy for producing DCA from alkanes or FAs is to improve the flux toward ω-oxidation pathway by amplifying the first limiting step in this pathway (e.g., ALK1 and/or ALK2, and CPR). The next step is to reduce the flux toward the competitive β-oxidation pathway by partially disrupting POX genes (e.g., POX2, POX 3, POX 4, and POX 5). DCA9 and DCA11 have been produced from the β-oxidation of DCA13 when Aox1p and Aox6p are present (Thevenieau et al., 2010). Finally, it is challenging to produce DCAs with chain lengths of more than 16 carbons, since contributory oxidizing enzymes have not been fully characterized. On the other hand, it is typically challenging to purify P450 enzymes and study their functions in vitro, due to their co-factor dependency, sensitivity to oxygen species, and product inhibition (Van Beilen and Funhoff, 2007; Meng et al., 2009).
The β-oxidation pathway can potentially be engineered to produce β-hydroxyl FAs, lactones, methyl ketones, and vanillin with the applications in flavor, fragrance, and biofuel industries. To achieve this aim, directing the substrate to the correct pathway and favoring its exit at a specific level of degradation is necessary (Waché et al., 2006).
Some researchers have produced several types of lactone with aromatic characteristic from ricinoleic acid (C18:1-OH) or methyl ricinoleate using Y. lipolytica (Waché et al., 2003). In fact, γ-decalactone can reach the production level of 10 g/L. Ricinoleic acid is converted to 4-hydroxydecanoic acid through four cycles of β-oxidation, and then undergoes lactonization under acidic conditions (Swizdor et al., 2012). However, it is not clear whether lactonization occurs on acyl-CoA or on the FFAs. Intact β-oxidation genes can degrade lactone, highlighting the need to modify this pathway and avoid further degradation of the products (Thevenieau et al., 2009). The disruption of POX3, along with overexpression of POX2, can be used to continuously produce γ-decalactone from methyl ricinoleate (Guo et al., 2012). More specifically, disrupting acyl-CoA oxidase with short-chain specificity (i.e., <10 carbons) can promote the production of γ-decalactone (Waché et al., 2006; Thevenieau et al., 2009). In fact, many types of lactone can be produced from different hydroxyl FAs released at different stages of β-oxidation cycle. Y. lipolytica is one of the most promising candidates for production of γ-decalactone. This yeast was also recently engineered to produce ricinoleic acid. This was achieved through the disruption of β-oxidation, native Δ12 desaturase, and the majority of native TAG acyltransferases, except for Lro1p, followed by the heterologous expression of fungal hydroxylase. This FA reached 43% of total lipid at 15% CDW (Beopoulos et al., 2014).
β-hydroxy FAs are important chiral building blocks in the asymmetric synthesis of drugs, vitamins, fragrances, pheromones, and other bioproducts. They can be the products of partial β-oxidation cycle through the reduction of β-ketoacids, oxidation of 1,3-diol, and subsequently, hydration of α,β-unsaturated acids (Swizdor et al., 2012). When using β-oxidation, further metabolization of β-hydroxy acids to the corresponding acids should be avoided by genetic engineering to increase molar conversion yield.
Another type of lipid-derived flavor compound is green note, including a variety of aldehyde and alcohols of six to nine carbons. These flavor compounds can be produced through successive action of lipoxygenase, hydroperoxide lyase, and if necessary, alcohol dehydrogenase on LA and linolenic acids, which are naturally found in yeast lipids. Different tactics have been reviewed for the heterologous expression of the above-mentioned enzymes from plants, fruits, and conventional yeast into oleaginous yeasts such as Y. lipolytica (Waché et al., 2006).
Methyl ketones are reduced compounds with applications in the fragrance, flavor, and pharmacological industries. They can be produced from various sources, such as fungal biotransformation of medium chain FAs or oxidation of secondary alcohols by yeast cell suspension (Patel et al., 1979; Hagedorn and Kaphammer, 1994). Recently, E. coli was genetically manipulated to convert glucose to methyl ketones using 3-ketoacyl-ACP as precursor (Park et al., 2012). In another strategy, a β-oxidation pathway of E. coli was engineered for methyl ketone production. In these studies, overexpression and interruption of some native genes along with the expression of heterologous genes were employed (Goh et al., 2012). The presence of strong infrastructure for the production and degradation of FA flux makes the Y. lipolytica a good candidate to produce methyl ketone. However, the successful application of such strategies requires comprehensive understanding of contributory genes, proteins, regulatory mechanisms, and points of action in this oleaginous yeast.
Polyhydroxyalkanoate synthase can be used to polymerize the released hydroxylated acyl-CoA from β-oxidation to produce PHAs (Beopoulos and Nicaud, 2012). Although bacteria can produce PHA, the oleaginous yeast platform is also highly competitive, due to its ability to utilize a wider range of cheap feedstock, and intrinsically accumulate lipid and convert it to PHA. For example, the expression of PHA synthetase from Pseudomonas aeruginosa in this yeast under different POX genetic profiles has been reported (Haddouche et al., 2010). Based on the results, only Aox3p and Aox5p have been found to be involved in PHA biosynthesis from specific FAs. The hydroxylated FAs can also be obtained directly from hydroxy acyl-CoA and used as chiral building blocks to synthesize some chemicals (Sabirova et al., 2011).
Production of Nanoproducts
Among the oleaginous yeasts, some Y. lipolytica strains show a good tolerance to different metals and possess inherent reductive compounds (e.g., reductases, proteases, melanin, and specific protein metallothionein). These compounds improve the potential for these strains to produce metal nanoparticles as prominent building blocks in nanotechnology (Pimprikar et al., 2009). Some potential strains of Y. lipolytica may be used to generate elemental metal nanoparticles (Agnihotri et al., 2009; Apte et al., 2013). This field of research is still in its infancy. Further studies will shed light on the molecular mechanism of nanoparticle biosynthesis (Chi et al., 2010a) and engineer Y. lipolytica strains. The use of Y. lipolytica to produce nanoproducts and lipid-based bioproducts will transform this platform into an effective tool for bridging between biotechnology and nanotechnology through development of novel biocatalysts and nano-oils.
Conclusion
Yarrowia lipolytica provides a promising platform to produce a wide range of lipid-based bioproducts. Recommended aims of future efforts to upgrade this platform include selecting naturally versatile Y. lipolytica strains, appropriate genetic and metabolic manipulation of the strain, developing more efficient GSMM predictive tools, applying protein engineering strategies, and the achieving high productivity and product recovery at a larger scale. Undoubtedly, integrated information from genomics, transcriptomics, metabolomics, and lipidomics will support future studies on underlying mechanisms and new strategies to better manipulate this organism. Further exploration on regulatory and transport mechanisms, transcriptional machinery, and signal transduction pathways, involved in lipid accumulation and degradation will pave the way to better understanding and utilization of this platform. The feasibility of this platform will be greatly enhanced when highly engineered Y. lipolytica is tailored to efficiently convert different agro-industrial byproducts, food waste materials, and low-value lipids (e.g., tallow and soapstocks) to valuable lipid-derived compounds with acceptable yield and adequate productivity for industrial scale applications.
Authors Contribution
Shulin Chen suggested the outline of this review paper. Ali Abghari developed this review paper based on the comprehensive literature review. Both Ali Abghari and Shulin Chen finalized the topic of this review. Shulin Chen helped with the revision of this review paper and gave final approval of the version to be sent to the journal. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to express their sincere thanks to Dr. Xiaochao Xiong for his valuable comments.
Abbreviations
FA, fatty acid; LA, linoleic acid; SCO, single-cell oil; TFA, total fatty acid.
References
Abghari, A., Sheikh-Zeinoddin, M., and Soleimanian-Zad, S. (2011). Nonfermented ice cream as a carrier for Lactobacillus acidophilus and Lactobacillus rhamnosus. Int. J. Food Sci. Technol. 46, 84–92. doi: 10.1111/j.1365-2621.2010.02453.x
Ageitos, J. M., Vallejo, J. A., Veiga-Crespo, P., and Villa, T. G. (2011). Oily yeasts as oleaginous cell factories. Appl. Microbiol. Biotechnol. 90, 1219–1227. doi:10.1007/s00253-011-3200-z
Agnihotri, M., Joshi, S., Kumar, A. R., Zinjarde, S., and Kulkarni, S. (2009). Biosynthesis of gold nanoparticles by the tropical marine yeast Yarrowia lipolytica NCIM 3589. Mater. Lett. 63, 1231–1234. doi:10.1016/j.matlet.2009.02.042
André, A., Chatzifragkou, A., Diamantopoulou, P., Sarris, D., Philippoussis, A., Galiotou-Panayotou, M., et al. (2009). Biotechnological conversions of bio-diesel-derived crude glycerol by Yarrowia lipolytica strains. Eng. Life Sci. 9, 468–478. doi:10.1002/elsc.200900063
Apte, M., Sambre, D., Gaikawad, S., Joshi, S., Bankar, A., Kumar, A. R., et al. (2013). Psychrotrophic yeast Yarrowia lipolytica NCYC 789 mediates the synthesis of antimicrobial silver nanoparticles via cell-associated melanin. AMB Express 3, 32. doi:10.1186/2191-0855-3-32
Athenstaedt, K. (2011). YALI0E32769g (DGA1) and YALI0E16797g (LRO1) encode major triacylglycerol synthases of the oleaginous yeast Yarrowia lipolytica. Biochim. Biophys. Acta 1811, 587–596. doi:10.1016/j.bbalip.2011.07.004
Avalos, J., Fink, G. R., and Stephanopoulos, G. (2013). Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat. Biotechnol. 31, 335–341. doi:10.1038/nbt.2509
Bailey, R., Trueheart, J., and Madden, K. T. (2010). Production of Sterols in Oleaginous Yeast and Fungi. Google Patents.
Balan, V. (2014). Current challenges in commercially producing biofuels from lignocellulosic biomass. ISRN Biotechnol. 2014, 31. doi:10.1155/2014/463074
Bankar, A. V., Kumar, A. R., and Zinjarde, S. S. (2009). Environmental and industrial applications of Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 84, 847–865. doi:10.1007/s00253-009-2156-8
Barth, G., Beckerich, J.-M., Dominguez, A., Kerscher, S., Ogrydziak, D., Titorenko, V., et al. (2003). “Functional genetics of Yarrowia lipolytica,” in Functional Genetics of Industrial Yeasts, ed. J. H. D. Winde (Berlin Heidelberg: Springer), 227–271. doi:10.1007/3-540-37003-X_8
Barth, G., and Gaillardin, C. (1996). “Yarrowia lipolytica,” in Nonconventional Yeasts in Biotechnology, ed. K. Wolf (Berlin: Springer), 313–388.
Barth, G., and Gaillardin, C. (1997). Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 19, 219–237. doi:10.1111/j.1574-6976.1997.tb00299.x
Beller, H. R., Goh, E.-B., and Keasling, J. D. (2010). Genes involved in long-chain alkene biosynthesis in Micrococcus luteus. Appl. Environ. Microbiol. 76, 1212–1223. doi:10.1128/aem.02312-09
Beopoulos, A., Cescut, J., Haddouche, R., Uribelarrea, J. L., Molina-Jouve, C., and Nicaud, J. M. (2009). Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 48, 375–387. doi:10.1016/j.plipres.2009.08.005
Beopoulos, A., Desfougeres, T., Sabirova, J., Zinjarde, S., Neuvéglise, C., and Nicaud, J.-M. (2010). “The hydrocarbon-degrading oleaginous yeast Yarrowia lipolytica,” in Handbook of Hydrocarbon and Lipid Microbiology, ed. K. N. Timmis (Berlin Heidelberg: Springer), 2111–2121. doi:10.1007/978-3-540-77587-4_152
Beopoulos, A., Haddouche, R., Kabran, P., Dulermo, T., Chardot, T., and Nicaud, J.-M. (2012). Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl. Microbiol. Biotechnol. 93, 1523–1537. doi:10.1007/s00253-011-3506-x
Beopoulos, A., Mrozova, Z., Thevenieau, F., Le Dall, M.-T., Hapala, I., Papanikolaou, S., et al. (2008). Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 74, 7779–7789. doi:10.1128/aem.01412-08
Beopoulos, A., and Nicaud, J.-M. (2012). Yeast: a new oil producer? OCL 19, 22–28. doi:10.1684/ocl.2012.0426
Beopoulos, A., Nicaud, J.-M., and Gaillardin, C. (2011). An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl. Microbiol. Biotechnol. 90, 1193–1206. doi:10.1007/s00253-011-3212-8
Beopoulos, A., Verbeke, J., Bordes, F., Guicherd, M., Bressy, M., Marty, A., et al. (2014). Metabolic engineering for ricinoleic acid production in the oleaginous yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 98, 251–262. doi:10.1007/s00253-013-5295-x
Blazeck, J., Hill, A., Liu, L., Knight, R., Miller, J., Pan, A., et al. (2014). Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 5, doi:10.1038/ncomms4131
Blazeck, J., Liu, L., Knight, R., and Alper, H. S. (2013a). Heterologous production of pentane in the oleaginous yeast Yarrowia lipolytica. J. Biotechnol. 165, 184–194. doi:10.1016/j.jbiotec.2013.04.003
Blazeck, J., Reed, B., Garg, R., Gerstner, R., Pan, A., Agarwala, V., et al. (2013b). Generalizing a hybrid synthetic promoter approach in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 97, 3037–3052. doi:10.1007/s00253-012-4421-5
Blazeck, J., Liu, L., Redden, H., and Alper, H. (2011). Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl. Environ. Microbiol. 77, 7905–7914. doi:10.1128/AEM.05763-11
Braun, A., Geier, M., Buehler, B., Schmid, A., Mauersberger, S., and Glieder, A. (2012). Steroid biotransformations in biphasic systems with Yarrowia lipolytica expressing human liver cytochrome P450 genes. Microb. Cell Fact. 11:106. doi:10.1186/1475-2859-11-106
Breuer, U., and Harms, H. (2006). Debaryomyces hansenii – an extremophilic yeast with biotechnological potential. Yeast 23, 415–437. doi:10.1002/yea.1374
Brown, M., and Shanks, J. (2012). “Linear hydrocarbon producing pathways in plants, algae and microbes,” in Sustainable Bioenergy and Bioproducts, eds K. Gopalakrishnan, J. H. V. Leeuwen, and R. C. Brown (London: Springer), 1–11.
Bulani, S. I., Moleleki, L., Albertyn, J., and Moleleki, N. (2012). Development of a novel rDNA based plasmid for enhanced cell surface display on Yarrowia lipolytica. AMB Express 2, 27. doi:10.1186/2191-0855-2-27
Cao, Z., Gao, H., Liu, M., and Jiao, P. (2006). Engineering the acetyl-CoA transportation system of Candida tropicalis enhances the production of dicarboxylic acid. Biotechnol. J. 1, 68–74. doi:10.1002/biot.200500008
Celinska, E., and Grajek, W. (2013). A novel multigene expression construct for modification of glycerol metabolism in Yarrowia lipolytica. Microb. Cell Fact. 12, 1–16. doi:10.1186/1475-2859-12-102
Chang, Y.-H., Chang, K.-S., Hsu, C.-L., Chuang, L.-T., Chen, C.-Y., Huang, F.-Y., et al. (2013). A comparative study on batch and fed-batch cultures of oleaginous yeast Cryptococcus sp. in glucose-based media and corncob hydrolysate for microbial oil production. Fuel 105, 711–717. doi:10.1016/j.fuel.2012.10.033
Chen, B., Ling, H., and Chang, M. W. (2013). Transporter engineering for improved tolerance against alkane biofuels in Saccharomyces cerevisiae. Biotechnol. Biofuels 6:21. doi:10.1186/1754-6834-6-21
Chen, D. C., Beckerich, J. M., and Gaillardin, C. (1997). One-step transformation of the dimorphic yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 48, 232–235. doi:10.1007/s002530051043
Chen, X., Li, Z., Zhang, X., Hu, F., Ryu, D. D. Y., and Bao, J. (2009). Screening of oleaginous yeast strains tolerant to lignocellulose degradation compounds. Appl. Biochem. Biotechnol. 159, 591–604. doi:10.1007/s12010-008-8491-x
Chi, Z., Zheng, Y., Jiang, A., and Chen, S. (2011). Lipid production by culturing oleaginous yeast and algae with food waste and municipal wastewater in an integrated process. Appl. Biochem. Biotechnol. 165, 442–453. doi:10.1007/s12010-011-9263-6
Chi, Z.-M., Liu, G., Zhao, S., Li, J., and Peng, Y. (2010a). Marine yeasts as biocontrol agents and producers of bio-products. Appl. Microbiol. Biotechnol. 86, 1227–1241. doi:10.1007/s00253-010-2483-9
Chi, Z., Zheng, Y., Lucker, B., and Chen, S. (2010b). Integrated system for productioin of biofuel feedstock. Google Patents.
Choi, S. Y., Ryu, D. D. Y., and Rhee, J. S. (1982). Production of microbial lipid – effects of growth rate and oxygen on lipid synthesis and fatty acid composition of Rhodotorula gracilis. Biotechnol. Bioeng. 24, 1165–1172. doi:10.1002/bit.260240513
Chuang, L.-T., Chen, D.-C., Nicaud, J.-M., Madzak, C., Chen, Y.-H., and Huang, Y.-S. (2010). Co-expression of heterologous desaturase genes in Yarrowia lipolytica. N Biotechnol. 27, 277–282. doi:10.1016/j.nbt.2010.02.006
Chung, J. N. (2013). Grand challenges in bioenergy and biofuel research: engineering and technology development, environmental impact, and sustainability. Front. Energy Res. 1, doi:10.3389/fenrg.2013.00004
Coelho, M. A. Z., Amaral, P. F. F., and Belo, I. (2010). Yarrowia lipolytica: an industrial workhorse. Curr. Res. Technol. Educ. Top. Appl. Microbiol. Microb. Biotechnol. 2, 930–940.
Coleman, R. A., Lewin, T. M., Van Horn, C. G., and Gonzalez-Baro, M. R. (2002). Do long-chain Acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 132, 2123–2126.
Courchesne, N. M. D., Parisien, A., Wang, B., and Lan, C. Q. (2009). Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J. Biotechnol. 141, 31–41. doi:10.1016/j.jbiotec.2009.02.018
Damude, H., Gillies, P., Macool, D., Picataggio, S., Pollak, D., Ragghianti, J., et al. (2006). High Arachidonic Acid Producing Strains of Yarrowia lipolytica. Google Patents.
Damude, H. G., Gillies, P. J., Macool, D. J., Picataggio, S. K., Ragghianti, J. J., Seip, J. E., et al. (2014). Docosahexaenoic Acid Producing Strains of Yarrowia lipolytica. Google Patents.
Dey, P., and Maiti, M. K. (2013). Molecular characterization of a novel isolate of Candida tropicalis for enhanced lipid production. J. Appl. Microbiol. 114, 1357–1368. doi:10.1111/jam.12133
Duan, Y., Zhu, Z., Cai, K., Tan, X., and Lu, X. (2011). De novo biosynthesis of biodiesel by Escherichia coli in optimized fed-batch cultivation. PLoS ONE 6(5):e20265. doi:10.1371/journal.pone.0020265
Dulermo, T., and Nicaud, J. M. (2011). Involvement of the G3P shuttle and b-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 13, 482–491. doi:10.1016/j.ymben.2011.05.002
Elbahloul, Y., and Steinbuechel, A. (2010). Pilot-scale production of fatty acid ethyl esters by an engineered Escherichia coli strain harboring the p(Microdiesel) plasmid. Appl. Environ. Microbiol. 76, 4560–4565. doi:10.1128/aem.00515-10
Enshaeieh, M., Abdoli, A., Nahvi, I., and Madani, M. (2013). Medium optimization for biotechnological production of single cell oil using Yarrowia lipolytica M7 and Candida sp. J. Cell Mol. Res. 5, 17–23.
Enshaeieh, M., Abdoli, A., Nahvi, I., and Madani, M. (2012). Bioconversion of different carbon sources in to microbial oil and biodiesel using oleaginous yeasts. J. Biol. Todays World 1, 82–92.
Fickers, P., Benetti, P. H., Waché, Y., Marty, A., Mauersberger, S., Smit, M. S., et al. (2005). Hydrophobic substrate utilisation by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 5, 527–543. doi:10.1016/j.femsyr.2004.09.004
Fickers, P., Le Dall, M. T., Gaillardin, C., Thonart, P., and Nicaud, J. M. (2003). New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J. Microbiol. Methods 55, 727–737. doi:10.1016/j.mimet.2003.07.003
Fickers, P., Marty, A., and Nicaud, J. M. (2011). The lipases from Yarrowia lipolytica: genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol. Adv. 29, 632–644. doi:10.1016/j.biotechadv.2011.04.005
Fillaudeau, L., Cescut, J., Anne-Archard, D., Nicaud, J. M., Uribelarrea, J.-L., and Molina-Jouve, C. (2009). “Morphology and rheological behaviour of Yarrowia lipolytica during production of intra-cellular energetic molecules. Impact of lipid accumulation and genetic modifications,” in Proceeding of 8th World Congress of Chemical Engineering (Montréal, Canada: Canadian Society for Chemical Engineering).
Fisher, M. A., and Tullman-Ercek, D. (2013). Change, exchange, and rearrange: protein engineering for the biotechnological production of fuels, pharmaceuticals, and other chemicals. Curr. Opin. Biotechnol. 24, 1010–1016. doi:10.1016/j.copbio.2013.02.027
Fontanille, P., Kumar, V., Christophe, G., Nouaille, R., and Larroche, C. (2012). Bioconversion of volatile fatty acids into lipids by the oleaginous yeast Yarrowia lipolytica. Bioresour. Technol. 114, 443–449. doi:10.1016/j.biortech.2012.02.091
Galafassi, S., Cucchetti, D., Pizza, F., Franzosi, G., Bianchi, D., and Compagno, C. (2012). Lipid production for second generation biodiesel by the oleaginous yeast Rhodotorula graminis. Bioresour. Technol. 111, 398–403. doi:10.1016/j.biortech.2012.02.004
Garay, L. A., Boundy-Mills, K. L., and German, J. B. (2014). Accumulation of high-value lipids in single-cell microorganisms: a mechanistic approach and future perspectives. J. Agric. Food Chem. 62, 2709–2727. doi:10.1021/jf4042134
Gasmi, N., Ayed, A., Ammar, B. B., Zrigui, R., Nicaud, J. M., and Kallel, H. (2011). Development of a cultivation process for the enhancement of human interferon alpha 2b production in the oleaginous yeast, Yarrowia lipolytica. Microb. Cell Fact. 10, doi:10.1186/1475-2859-10-90
Gellissen, G., Kunze, G., Gaillardin, C., Cregg, J. M., Berardi, E., Veenhuis, M., et al. (2005). New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica – a comparison. FEMS Yeast Res. 5, 1079–1096. doi:10.1016/j.femsyr.2005.06.004
Goh, E.-B., Baidoo, E. E. K., Keasling, J. D., and Beller, H. R. (2012). Engineering of bacterial methyl ketone synthesis for biofuels. Appl. Environ. Microbiol. 78, 70–80. doi:10.1128/aem.06785-11
Gong, Z., Shen, H., Wang, Q., Yang, X., Xie, H., and Zhao, Z. K. (2013). Efficient conversion of biomass into lipids by using the simultaneous saccharification and enhanced lipid production process. Biotechnol. Biofuels 6:36. doi:10.1186/1754-6834-6-36
Gong, Z., Wang, Q., Shen, H., Hu, C., Jin, G., and Zhao, Z. K. (2012). Co-fermentation of cellobiose and xylose by Lipomyces starkeyi for lipid production. Bioresour. Technol. 117, 20–24. doi:10.1016/j.biortech.2012.04.063
Gong, Z., Wang, Q., Shen, H., Wang, L., Xie, H., and Zhao, Z. K. (2014). Conversion of biomass-derived oligosaccharides into lipids. Biotechnol. Biofuels 7, 13. doi:10.1186/1754-6834-7-13
Grenfell-Lee, D., Zeller, S., Cardoso, R., and Pucaj, K. (2014). The safety of β-carotene from Yarrowia lipolytica. Food Chem. Toxicol. 65, 1–11. doi:10.1016/j.fct.2013.12.010
Grenfell-Lee, D. G. (2009). A fermentation-based process for carotenoid synthesis in Yarrowia lipolytica. SIM Annual Meeting and Exhibition Industrial Microbiology and Biotechnology, Toronto.
Guo, Y., Song, H., Wang, Z., and Ding, Y. (2012). Expression of POX2 gene and disruption of POX3 genes in the industrial Yarrowia lipolytica on the γ-decalactone production. Microbiol. Res. 167, 246–252. doi:10.1016/j.micres.2011.10.003
Haddouche, R., Delessert, S., Sabirova, J., Neuveglise, C., Poirier, Y., and Nicaud, J.-M. (2010). Roles of multiple acyl-CoA oxidases in the routing of carbon flow towards beta-oxidation and polyhydroxyalkanoate biosynthesis in Yarrowia lipolytica. FEMS Yeast Res. 10, 917–927. doi:10.1111/j.1567-1364.2010.00670.x
Hagedorn, S., and Kaphammer, B. (1994). Microbial biocatalysis in the generation of flavor and fragrance chemicals. Annu. Rev. Microbiol. 48, 773–800. doi:10.1146/annurev.mi.48.100194.004013
Hallenbeck, P. C. (2012). “Microbial production of fatty-acid-based biofuels,” in Microbial Technologies in Advanced Biofuels Production, ed. P. C. Hallenbeck (New York: Springer), 213–230. doi:10.1007/978-1-4614-1208-3_12
Han, Z., Madzak, C., and Su, W. W. (2013). Tunable nano-oleosomes derived from engineered Yarrowia lipolytica. Biotechnol. Bioeng. 110, 702–710. doi:10.1002/bit.24761
Harzevili, F. D. (2014). Biotechnological Applications of the Yeast Yarrowia lipolytica. Springer International Publishing.
Heslot, H. (1990). Genetics and genetic engineering of the industrial yeast Yarrowia lipolytica. Adv. Biochem. Eng. Biotechnol. 43, 43–73.
Howard, T. P., Middelhaufe, S., Moore, K., Edner, C., Kolak, D. M., Taylor, G. N., et al. (2013). Synthesis of customized petroleum-replica fuel molecules by targeted modification of free fatty acid pools in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110, 7636–7641. doi:10.1073/pnas.1215966110
Huang, C., Chen, X.-F., Xiong, L., Chen, X.-D., and Ma, L.-L. (2012). Oil production by the yeast Trichosporon dermatis cultured in enzymatic hydrolysates of corncobs. Bioresour. Technol. 110, 711–714. doi:10.1016/j.biortech.2012.01.077
Huang, C., Chen, X.-F., Xiong, L., Chen, X.-D., Ma, L.-L., and Chen, Y. (2013a). Single cell oil production from low-cost substrates: the possibility and potential of its industrialization. Biotechnol. Adv. 31, 129–139. doi:10.1016/j.biotechadv.2012.08.010
Huang, C., Chen, X.-F., Xiong, L., Yang, X.-Y., Chen, X.-D., Ma, L.-L., et al. (2013b). Microbial oil production from corncob acid hydrolysate by oleaginous yeast Trichosporon coremiiforme. Biomass Bioenergy 49, 273–278. doi:10.1016/j.biombioe.2012.12.023
Huang, C., Chen, X.-F., Yang, X.-Y., Xiong, L., Lin, X.-Q., Yang, J., et al. (2014). Bioconversion of corncob acid hydrolysate into microbial oil by the oleaginous yeast Lipomyces starkeyi. Appl. Biochem. Biotechnol. 172, 2197–2204. doi:10.1007/s12010-013-0651-y
Huang, W.-D., and Zhang, Y. H. P. (2011). Analysis of biofuels production from sugar based on three criteria: thermodynamics, bioenergetics, and product separation. Energy Environ. Sci. 4, 784–792. doi:10.1039/c0ee00069h
Iida, T., Sumita, T., Ohta, A., and Takagi, M. (2000). The cytochrome P450ALK multigene family of an n-alkane-assimilating yeast, Yarrowia lipolytica: cloning and characterization of genes coding for new CYP52 family members. Yeast 16, 1077–1087. doi:10.1002/1097-0061(20000915)16:12<1077::AID-YEA601>E3.0.CO;2-K
Jadhav, V. V., Salunkhe, D. S., and Bhadekar, R. K. (2012). Effect of alterations in conventional medium on lipid accumulation and fatty acid content in oleaginous yeasts. Int. J. Pharm. Bio. Sci. 3, 757–769.
Jong, B. W. D., Shi, S., Siewers, V., and Nielsen, J. (2014). Improved production of fatty acid ethyl esters in Saccharomyces cerevisiae through up-regulation of the ethanol degradation pathway and expression of the heterologous phosphoketolase pathway. Microb. Cell Fact. 13, doi:10.1186/1475-2859-13-39
Juszczyk, P., Tomaszewska, L., Kita, A., and Rymowicz, W. (2013). Biomass production by novel strains of Yarrowia lipolytica using raw glycerol, derived from biodiesel production. Bioresour. Technol. 137, 124–131. doi:10.1016/j.biortech.2013.03.010
Kalscheuer, R., Stoelting, T., and Steinbuechel, A. (2006). Microdiesel: Escherichia coli engineered for fuel production. Microbiology 152, 2529–2536. doi:10.1099/mic.0.29028-0
Kamzolova, S. V., Shishkanova, N. V., Morgunov, I. G., and Finogenova, T. V. (2003). Oxygen requirements for growth and citric acid production of Yarrowia lipolytica. FEMS Yeast Res. 3, 217–222. doi:10.1016/s1567-1356(02)00188-5
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi:10.1093/nar/gkm882
Karatay, S. E., and Dönmez, G. (2010). Improving the lipid accumulation properties of the yeast cells for biodiesel production using molasses. Bioresour. Technol. 101, 7988–7990. doi:10.1016/j.biortech.2010.05.054
Katre, G., Joshi, C., Khot, M., Zinjarde, S., and Ravikumar, A. (2012). Evaluation of single cell oil (SCO) from a tropical marine yeast Yarrowia lipolytica NCIM 3589 as a potential feedstock for biodiesel. AMB Express 2, 36. doi:10.1186/2191-0855-2-36
Kautola, H., Rymowicz, W., Linko, Y.-Y., and Linko, P. (1991). Production of citric acid with immobilized Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 35, 447–449. doi:10.1007/bf00169747
Kim, S., Baek, S.-H., Lee, K., and Hahn, J.-S. (2013). Cellulosic ethanol production using a yeast consortium displaying a minicellulosome and beta-glucosidase. Microb. Cell Fact. 12:14. doi:10.1186/1475-2859-12-14
Kohlwein, S. D. (2010). Triacylglycerol homeostasis: insights from yeast. J. Biol. Chem. 285, 15663–15667. doi:10.1074/jbc.R110.118356
Koutinas, A. A., Chatzifragkou, A., Kopsahelis, N., Papanikolaou, S., and Kookos, I. K. (2014). Design and techno-economic evaluation of microbial oil production as a renewable resource for biodiesel and oleochemical production. Fuel 116, 566–577. doi:10.1016/j.fuel.2013.08.045
Kretzschmar, A., Otto, C., Holz, M., Werner, S., Huebner, L., and Barth, G. (2013). Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Curr. Genet. 59, 63–72. doi:10.1007/s00294-013-0389-7
Kunau, W. H., and Hartig, A. (1992). Peroxisome biogenesis in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 62, 63–78. doi:10.1007/bf00584463
Lazar, Z., Rossignol, T., Verbeke, J., Crutz-Le Coq, A.-M., Nicaud, J.-M., and Robak, M. (2013). Optimized invertase expression and secretion cassette for improving Yarrowia lipolytica growth on sucrose for industrial applications. J. Ind. Microbiol. Biotechnol. 40, 1273–1283. doi:10.1007/s10295-013-1323-1
Lee, K.-M., Kalyani, D., Tiwari, M. K., Kim, T.-S., Dhiman, S. S., Lee, J.-K., et al. (2012). Enhanced enzymatic hydrolysis of rice straw by removal of phenolic compounds using a novel laccase from yeast Yarrowia lipolytica. Bioresour. Technol. 123, 636–645. doi:10.1016/j.biortech.2012.07.066
Lehocký, M., Amaral, P. F. F., St’ahel, P., Coelho, M. A. Z., Barros-Timmons, A. M., and Coutinho, J. A. P. (2007). Preparation and characterization of organosilicon thin films for selective adhesion of Yarrowia lipolytica yeast cells. J. Chem. Technol. Biotechnol. 82, 360–366. doi:10.1002/jctb.1677
Lennen, R. M., and Pfleger, B. F. (2013). Microbial production of fatty acid-derived fuels and chemicals. Curr. Opin. Biotechnol. 24, 1044–1053. doi:10.1016/j.copbio.2013.02.028
Liang, M.-H., and Jiang, J.-G. (2013). Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog. Lipid Res. 52, 395–408. doi:10.1016/j.plipres.2013.05.002
Liu, G.-L., Li, Y., Zhou, H.-X., Chi, Z.-M., and Madzak, C. (2012a). Over-expression of a bacterial chitosanase gene in Yarrowia lipolytica and chitosan hydrolysis by the recombinant chitosanase. J. Mol. Catal. B Enzym. 83, 100–107. doi:10.1016/j.molcatb.2012.07.012
Liu, W., Wang, Y., Yu, Z., and Bao, J. (2012b). Simultaneous saccharification and microbial lipid fermentation of corn stover by oleaginous yeast Trichosporon cutaneum. Bioresour. Technol. 118, 13–18. doi:10.1016/j.biortech.2012.05.038
Liu, X.-Y., Chi, Z., Liu, G.-L., Wang, F., Madzak, C., and Chi, Z.-M. (2010). Inulin hydrolysis and citric acid production from inulin using the surface-engineered Yarrowia lipolytica displaying inulinase. Metab. Eng. 12, 469–476. doi:10.1016/j.ymben.2010.04.004
Loira, N., Dulermo, T., Nicaud, J. M., and Sherman, D. J. (2012). A genome-scale metabolic model of the lipid-accumulating yeast Yarrowia lipolytica. BMC Syst. Biol. 6:35. doi:10.1186/1752-0509-6-35
Madzak, C., Gaillardin, C., and Beckerich, J. M. (2004). Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J. Biotechnol. 109, 63–81. doi:10.1016/j.jbiotec.2003.10.027
Madzak, C., Nicaud, J.-M., and Gaillardin, C. (2005a). “Yarrowia lipolytica,” in Production of Recombinant Proteins: Novel Microbial and Eukaryotic Expression Systems, ed. G. Gellissen (Wiley-VCH Verlag GmbH & Co. KGaA), 163–189.
Madzak, C., Otterbein, L., Chamkha, M., Moukha, S., Asther, M., Gaillardin, C., et al. (2005b). Heterologous production of a laccase from the basidiomycete Pycnoporus cinnabarinus in the dimorphic yeast Yarrowia lipolytica. FEMS Yeast Res. 5, 635–646. doi:10.1016/j.femsyr.2004.10.009
Magdouli, S., Yan, S., Tyagi, R. D., and Surampalli, R. Y. (2014). Heterotrophic microorganisms: a promising source for biodiesel production. Crit. Rev. Environ. Sci. Technol. 44, 416–453. doi:10.1080/10643389.2012.728523
Makri, A., Fakas, S., and Aggelis, G. (2010). Metabolic activities of biotechnological interest in Yarrowia lipolytica grown on glycerol in repeated batch cultures. Bioresour. Technol. 101, 2351–2358. doi:10.1016/j.biortech.2009.11.024
Marova, I., Certik, M., and Breierova, E. (2011). “Production of enriched biomass by carotenogenic yeasts – application of whole-cell yeast biomass to production of pigments and other lipid compounds,” in Biomass – Detection, Production and Usage, ed. D. Matovic (Rijeka, Croatia: InTech), 345–384.
Martins, F. F., Ferreiraa, T. F., Azevedob, D. A., Alice, M., and Coelho, Z. (2012). Evaluation of crude oil degradation by Yarrowia lipolytica Chem. Eng. Trans. 27, 223–228. doi:10.3303/CET1227038
Matthäus, F., Ketelhot, M., Gatter, M., and Barth, G. (2014). Production of lycopene in the non-carotenoid-producing yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 80, 1660–1669. doi:10.1128/AEM.03167-13
Mauersberger, S., Novikova, L. A., and Shkumatov, V. M. (2013). “Cytochrome P450 expression in Yarrowia lipolytica and its use in steroid biotransformation,” in Yarrowia lipolytica Biotechnological Applications, ed. G. Barth (Berlin Heidelberg: Springer), 171–226. doi:10.1007/978-3-642-38583-4_7
Meng, X., Yang, J., Xu, X., Zhang, L., Nie, Q., and Xian, M. (2009). Biodiesel production from oleaginous microorganisms. Renew. Energy 34, 1–5. doi:10.1016/j.renene.2008.04.014
Mirbagheri, M., Nahvi, I., Emtiazi, G., and Darvishi, F. (2011). Enhanced production of citric acid in Yarrowia lipolytica by triton X-100. Appl. Biochem. Biotechnol. 165, 1068–1074. doi:10.1007/s12010-011-9325-9
Mlickova, K., Roux, E., Athenstaedt, K., D’Andrea, S., Daum, G., Chardot, T., et al. (2004). Lipid accumulation, lipid body formation, and acyl coenzyme A oxidases of the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 70, 3918–3924. doi:10.1128/aem.70.7.3918-3924.2004
Moeller, L., Grünberg, M., Zehnsdorf, A., Aurich, A., Bley, T., and Strehlitz, B. (2011). Repeated fed-batch fermentation using biosensor online control for citric acid production by Yarrowia lipolytica. J. Biotechnol. 153, 133–137. doi:10.1016/j.jbiotec.2011.03.013
Mori, K., Iwama, R., Kobayashi, S., Horiuchi, H., Fukuda, R., and Ohta, A. (2013). Transcriptional repression by glycerol of genes involved in the assimilation of n-alkanes and fatty acids in yeast Yarrowia lipolytica. FEMS Yeast Res. 13, 233–240. doi:10.1111/1567-1364.12025
Morin, N., Cescut, J., Beopoulos, A., Lelandais, G., Le Berre, V., Uribelarrea, J. L., et al. (2011). Transcriptomic analyses during the transition from biomass production to lipid accumulation in the oleaginous yeast Yarrowia lipolytica. PLoS ONE 6(11):e27966. doi:10.1371/journal.pone.0027966
Nawabi, P., Bauer, S., Kyrpides, N., and Lykidis, A. (2011). Engineering Escherichia coli for biodiesel production utilizing a bacterial fatty acid methyltransferase. Appl. Environ. Microbiol. 77, 8052–8061. doi:10.1128/aem.05046-11
Obiero, G. O. (2006). Evaluation of Yarrowia lipolytica as a Host for Cytochrome P450 Monooxygenase Expression. Ph.D. thesis, University of the Free State, Bloemfontein.
Ochoa-Estopier, A., and Guillouet, S. E. (2014). D-stat culture for studying the metabolic shifts from oxidative metabolism to lipid accumulation and citric acid production in Yarrowia lipolytica. J. Biotechnol. 170, 35–41. doi:10.1016/j.jbiotec.2013.11.008
Otero, R. C., and Gaillardin, C. (1996). Efficient selection of hygromycin-B-resistant Yarrowia lipolytica transformants. Appl. Microbiol. Biotechnol. 46, 143–148. doi:10.1007/s002530050796
Pan, P., and Hua, Q. (2012). Reconstruction and in silico analysis of metabolic network for an oleaginous yeast, Yarrowia lipolytica. PLoS ONE 7(12):e51535. doi:10.1371/journal.pone.0051535
Papanikolaou, S., and Aggelis, G. (2009). Biotechnological valorization of biodiesel derived glycerol waste through production of single cell oil and citric acid by Yarrowia lipolytica. Lipid Technol. 21, 83–87. doi:10.1002/lite.200900017
Papanikolaou, S., Beopoulos, A., Koletti, A., Thevenieau, F., Koutinas, A. A., Nicaud, J.-M., et al. (2013). Importance of the methyl-citrate cycle on glycerol metabolism in the yeast Yarrowia lipolytica. J. Biotechnol. 168, 303–314. doi:10.1016/j.jbiotec.2013.10.025
Papanikolaou, S., Chatzifragkou, A., Fakas, S., Galiotou-Panayotou, M., Komaitis, M., Nicaud, J.-M., et al. (2009). Biosynthesis of lipids and organic acids by Yarrowia lipolytica strains cultivated on glucose. Eur. J. Lipid Sci. Technol. 111, 1221–1232. doi:10.1002/ejlt.200900055
Papanikolaou, S., Chevalot, I., Galiotou-Panayotou, M., Komaitis, M., Marc, I., and Aggelis, G. (2007). Industrial derivative of tallow: a promising renewable substrate for microbial lipid, single-cell protein and lipase production by Yarrowia lipolytica. Electron. J. Biotechnol. 10, doi:10.2225/vol10-issue3-fulltext-8
Papanikolaou, S., Chevalot, I., Komaitis, M., Marc, I., and Aggelis, G. (2002). Single cell oil production by Yarrowia lipolytica growing on an industrial derivative of animal fat in batch cultures. Appl. Microbiol. Biotechnol. 58, 308–312. doi:10.1007/s00253-001-0897-0
Papanikolaou, S., Galiotou-Panayotou, M., Chevalot, I., Komaitis, M., Marc, I., and Aggelis, G. (2006). Influence of glucose and saturated free-fatty acid mixtures on citric acid and lipid production by Yarrowia lipolytica. Curr. Microbiol. 52, 134–142. doi:10.1007/s00284-005-0223-7
Papanikolaou, S., Muniglia, L., Chevalot, I., Aggelis, G., and Marc, I. (2003). Accumulation of a cocoa-butter-like lipid by Yarrowia lipolytica cultivated on agro-industrial residues. Curr. Microbiol. 46, 124–130. doi:10.1007/s00284-002-3833-3
Papone, T., Kookkhunthod, S., and Leesing, R. (2012). Microbial oil production by monoculture and mixed cultures of microalgae and oleaginous yeasts using sugarcane juice as substrate. World Acad. Sci. Eng. Technol. 6, 908–912.
Park, C. S., Chang, C. C., Kim, J. Y., Ogrydziak, D. M., and Ryu, D. D. Y. (1997). Expression, secretion, and processing of rice alpha-amylase in the yeast Yarrowia lipolytica. J. Biol. Chem. 272, 6876–6881. doi:10.1074/jbc.272.11.6876
Park, J., Rodriguez-Moya, M., Li, M., Pichersky, E., San, K.-Y., and Gonzalez, R. (2012). Synthesis of methyl ketones by metabolically engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 39, 1703–1712. doi:10.1007/s10295-012-1178-x
Patel, R. N., Hou, C. T., Laskin, A. I., Derelanko, P., and Felix, A. (1979). Oxidation of secondary alcohols to methyl ketones by yeasts. Appl. Environ. Microbiol. 38, 219–223.
Pimprikar, P. S., Joshi, S. S., Kumar, A. R., Zinjarde, S. S., and Kulkarni, S. K. (2009). Influence of biomass and gold salt concentration on nanoparticle synthesis by the tropical marine yeast Yarrowia lipolytica NCIM 3589. Colloids Surf. B Biointerfaces 74, 309–316. doi:10.1016/j.colsurfb.2009.07.040
Poli, J. S., Dallé, P., Senter, L., Mendes, S., Ramirez, M., Vainstein, M. H., et al. (2013). Fatty acid methyl esters produced by oleaginous yeast Yarrowia lipolytica QU21: an alternative for vegetable oils. Rev. Bras. Biosci. 11, 203–208.
Poli, J. S., Lützhøft, H.-C. H., Karakashev, D. B., Valente, P., and Angelidaki, I. (2014). An environmentally-friendly fluorescent method for quantification of lipid contents in yeast. Bioresour. Technol. 151, 388–391. doi:10.1016/j.biortech.2013.09.128
Pu, Y., Kosa, M., Kalluri, U., Tuskan, G., and Ragauskas, A. (2011). Challenges of the utilization of wood polymers: how can they be overcome? Appl. Microbiol. Biotechnol. 91, 1525–1536. doi:10.1007/s00253-011-3350-z
Raschke, D., and Knorr, D. (2009). Rapid monitoring of cell size, vitality and lipid droplet development in the oleaginous yeast Waltomyces lipofer. J. Microbiol. Methods 79, 178–183. doi:10.1016/j.mimet.2009.08.011
Rossi, M., Amaretti, A., Raimondi, S., and Leonardi, A. (2011). “Getting lipids for biodiesel production from oleaginous fungi,” in Biodiesel – Feedstocks and Processing Technologies, ed. M. Stoytcheva (Rijeka, Croatia: InTech), 71–92.
Rude, M. A., Baron, T. S., Brubaker, S., Alibhai, M., Del Cardayre, S. B., and Schirmer, A. (2011). Terminal olefin (1-alkene) biosynthesis by a novel P450 fatty acid decarboxylase from Jeotgalicoccus species. Appl. Environ. Microbiol. 77, 1718–1727. doi:10.1128/aem.02580-10
Ruffing, A. M. (2013). “Metabolic engineering of hydrocarbon biosynthesis for biofuel production,” in Liquid, Gaseous and Solid Biofuels – Conversion Techniques, ed. Z. Fang (Rijeka, Croatia: InTech), 263–299.
Rywinska, A., Musial, I., Rymowicz, W., Zarowska, B., and Boruczkowski, T. (2012). Effect of agitation and aeration on the citric acid production by Yarrowia lipolytica grown on glycerol. Prep. Biochem. Biotechnol. 42, 279–291. doi:10.1080/10826068.2012.656868
Sabirova, J. (2011). “Lipoyeasts, mobilising the enzymatic potential of hydrocarbonoclastic bacteria and the oleaginous yeast Yarrowia lipolytica to create a powerful cellular production platform for lipid-derived industrial materials," in Lipo Yeasts (213068). Ghent University.
Sabirova, J. S., Haddouche, R., Van Bogaert, I. N., Mulaa, F., Verstraete, W., Timmis, K. N., et al. (2011). The ‘LipoYeasts’ project: using the oleaginous yeast Yarrowia lipolytica in combination with specific bacterial genes for the bioconversion of lipids, fats and oils into high-value products. Microb. Biotechnol. 4, 47–54. doi:10.1111/j.1751-7915.2010.00187.x
Santomauro, F., Whiffin, F., Scott, R., and Chuck, C. (2014). Correction: low-cost lipid production by an oleaginous yeast cultured in non-sterile conditions using model waste resources. Biotechnol. Biofuels 7, 42. doi:10.1186/1754-6834-7-42
Scharnewski, M., Pongdontri, P., Mora, G., Hoppert, M., and Fulda, M. (2008). Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J. 275, 2765–2778. doi:10.1111/j.1742-4658.2008.06417.x
Schellenberger, J., Park, J. O., Conrad, T. M., and Palsson, B. O. (2010). BiGG: a biochemical genetic and genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinform. 11:213. doi:10.1186/1471-2105-11-213
Schmidberger, N., Schmid, B., and Barth, G. (1994). YLT1, a highly repetitive retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. J. Bacteriol. 176, 2477–2482.
Seip, J., Jackson, R., He, H., Zhu, Q., and Hong, SP. (2013). Snf1 is a regulator of lipid accumulation in Yarrowia lipolytica. Appl. Environ. Microbiol. 79, 7360–7370. doi:10.1128/AEM.02079-13
Sestric, R., Munch, G., Cicek, N., Sparling, R., and Levin, D. B. (2014). Growth and neutral lipid synthesis by Yarrowia lipolytica on various carbon substrates under nutrient-sufficient and nutrient-limited conditions. Bioresour. Technol. 164, 41–46. doi:10.1016/j.biortech.2014.04.016
Sha, Q. (2013). A Comparative Study on Four Oleaginous Yeasts on their Lipid Accumulating Capacity. Master’s Degree, Swedish University of Agricultural Sciences, Uppsala.
Sharpe, P. L., Ye, R. W., and Zhu, Q. Q. (2008). Carotenoid Production in a Recombinant Oleaginous Yeast. USA Patent PCT/US2007/025222.
Sil, J., Das, S., Oliveira, R. G., Amaral, P. F. F., and Coelho, M. A. Z. (2013). “Screening six potential Yarrowia lipolytica strains for best lipid, citric acid, biosurfactant and lipase production,” in 2nd International Conference on Environment, Energy and Biotechnology (Singapore: IACSIT Press).
Sriwongchai, S., Pokethitiyook, P., Kruatrachue, M., Bajwa, P. K., and Lee, H. (2013). Screening of selected oleaginous yeasts for lipid production from glycerol and some factors which affect lipid production by Yarrowia lipolytica strains. J. Microbiol. Biotechnol. Food Sci. 2, 2344–2348.
Steen, E. J., Kang, Y., Bokinsky, G., Hu, Z., Schirmer, A., Mcclure, A., et al. (2010). Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–U182. doi:10.1038/nature08721
Sukovich, D. J., Seffernick, J. L., Richman, J. E., Hunt, K. A., Gralnick, J. A., and Wackett, L. P. (2010). Structure, function, and insights into the biosynthesis of a head-to-head hydrocarbon in Shewanella oneidensis strain MR-1. Appl. Environ. Microbiol. 76, 3842–3849. doi:10.1128/aem.00433-10
Swizdor, A., Panek, A., Milecka-Tronina, N., and Kolek, T. (2012). Biotransformations utilizing b-oxidation cycle reactions in the synthesis of natural compounds and medicines. Int. J. Mol. Sci. 13, 16514–16543. doi:10.3390/ijms131216514
Tai, M. (2012). Metabolic Engineering of Oleaginous Yeast for the Production of Biofuels. Ph.D. thesis, Massachusetts Institute of Technology, Cambridge.
Tai, M., and Stephanopoulos, G. (2013). Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 15, 1–9. doi:10.1016/j.ymben.2012.08.007
Theron, C. W. (2007). Hetrologous Expression of Cytochrome P450 Monoxygenase by the Yeast Yarrowia lipolytica. University of the Free State, Bloemfontein.
Thevenieau, F., Beopoulos, A., Desfougeres, T., Sabirova, J., Albertin, K., Zinjarde, S. S., et al. (2010). “Uptake and assimilation of hydrophobic substrates by the oleaginous yeast Yarrowia lipolytica,” in Handbook of Hydrocarbon and Lipid Microbiology, ed. K. N. Timmis (Berlin: Springer), 1513–1527.
Thevenieau, F., Nicaud, J.-M., and Gaillardin, C. (2009). “Applications of the non-conventional yeast Yarrowia lipolytica,” in Yeast Biotechnology: Diversity and Applications, ed. G. K. T. Satyanarayana (Netherlands: Springer), 589–613. doi:10.1007/978-1-4020-8292-4_26
Thevenieau, F., and Nicaud, J.-M. (2013). Microorganisms as sources of oils. OCL 20, 1–8. doi:10.1051/ocl/2013034
Tsigie, Y. A., Huynh, L. H., Ahmed, I. N., and Ju, Y.-H. (2012a). Maximizing biodiesel production from Yarrowia lipolytica Po1g biomass using subcritical water pretreatment. Bioresour. Technol. 111, 201–207. doi:10.1016/j.biortech.2012.02.052
Tsigie, Y. A., Wang, C.-Y., Kasim, N. S., Quy-Do, D., Lien-Huong, H., Quoc-Phong, H., et al. (2012b). Oil production from Yarrowia lipolytica Po1g using rice bran hydrolysate. J. Biomed. Biotechnol. 2012:378384. doi:10.1155/2012/378384
Tsigie, Y. A., Wang, C.-Y., Chi-Thanh, T., and Ju, Y.-H. (2011). Lipid production from Yarrowia lipolytica Po1g grown in sugarcane bagasse hydrolysate. Bioresour. Technol. 102, 9216–9222. doi:10.1016/j.biortech.2011.06.047
Tsigie, Y. A., Wu, C.-H., Huynh, L. H., Ismadji, S., and Ju, Y.-H. (2013). Bioethanol production from Yarrowia lipolytica Po1g biomass. Bioresour. Technol. 145, 210–216. doi:10.1016/j.biortech.2012.11.091
Urlacher, V. B., and Schmid, R. D. (2006). Recent advances in oxygenase-catalyzed biotransformations. Curr. Opin. Chem. Biol. 10, 156–161. doi:10.1016/j.cbpa.2006.02.001
Van Beilen, J. B., and Funhoff, E. G. (2007). Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 74, 13–21. doi:10.1007/s00253-006-0748-0
Verbeke, J., Beopoulos, A., and Nicaud, J.-M. (2013). Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol. Lett. 35, 571–576. doi:10.1007/s10529-012-1107-0
Vernis, L., Abbas, A., Chasles, M., Gaillardin, C. M., Brun, C., Huberman, J. A., et al. (1997). An origin of replication and a centromere are both needed to establish a replicative plasmid in the yeast Yarrowia lipolytica. Mol. Cell. Biol. 17, 1995–2004.
Wache, Y. (2013). Yarrowia lipolytica biotechnological applications: production of dicarboxylic acids and flagrances by Yarrowia lipolytica. Microbiol. Monogr. 25, 151–170. doi:10.1007/s00253-012-4085-1
Waché, Y., Aguedo, M., Nicaud, J. M., and Belin, J. M. (2003). Catabolism of hydroxyacids and biotechnological production of lactones by Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 61, 393–404. doi:10.1007/s00253-002-1207-1
Waché, Y., Husson, F., Feron, G., and Belin, J. M. (2006). Yeast as an efficient biocatalyst for the production of lipid-derived flavours and fragrances. Antonie Van Leeuwenhoek 89, 405–416. doi:10.1007/s10482-005-9049-3
Wang, C., Chen, L., Rakesh, B., Qin, Y., and Lv, R. (2012). Technologies for extracting lipids from oleaginous microorganisms for biodiesel production. Front. Energy 6:266–274. doi:10.1007/s11708-012-0193-y
Wang, F., Yue, L., Wang, L., Madzak, C., Li, J., Wang, X., et al. (2009). Genetic modification of the marine-derived yeast Yarrowia lipolytica with high-protein content using a GPI-anchor-fusion expression system. Biotechnol. Prog. 25, 1297–1303. doi:10.1002/btpr.235
Wang, J.-H., Hung, W., and Tsai, S.-H. (2011a). High efficiency transformation by electroporation of Yarrowia lipolytica. J. Microbiol. 49, 469–472. doi:10.1007/s12275-011-0433-6
Wang, J., Zhang, B., and Chen, S. (2011b). Oleaginous yeast Yarrowia lipolytica mutants with a disrupted fatty acyl-CoA synthetase gene accumulate saturated fatty acid. Process Biochem. 46, 1436–1441. doi:10.1016/j.procbio.2011.03.011
Wang, W., and Lu, X. (2013). Microbial synthesis of alka(e)nes. Front. Bioeng. Biotechnol. 1, doi:10.3389/fbioe.2013.00010
Wang, Z.-P., Xu, H.-M., Wang, G.-Y., Chi, Z., and Chi, Z.-M. (2013). Disruption of the MIG1 gene enhances lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Biochim. Biophys. Acta 1831, 675–682. doi:10.1016/j.bbalip.2012.12.010
Watkins, P. A., and Ellis, J. M. (2012). Peroxisomal acyl-CoA synthetases. Biochim. Biophys. Acta 1822, 1411–1420. doi:10.1016/j.bbadis.2012.02.010
Workman, M., Holt, P., and Thykaer, J. (2013). Comparing cellular performance of Yarrowia lipolytica during growth on glucose and glycerol in submerged cultivations. AMB Express 3, 1–9. doi:10.1186/2191-0855-3-58
Xue, Z., Sharpe, P. L., Hong, S.-P., Yadav, N. S., Xie, D., Short, D. R., et al. (2013). Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat. Biotechnol. 31, 736–741. doi:10.1038/nbt.2622
Xue, Z., and Zhu, Q. Q. (2006). Ammonium Transporter Promoters for Gene Expression in Oleaginous Yeast. US 11/185,301.
Yang, X.-S., Jiang, Z.-B., Song, H.-T., Jiang, S.-J., Madzak, C., and Ma, L.-X. (2009). Cell-surface display of the active mannanase in Yarrowia lipolytica with a novel surface-display system. Biotechnol. Appl. Biochem. 54, 171–176. doi:10.1042/ba20090222
Ye, R. W., Sharpe, P. L., and Zhu, Q. (2012). “Bioengineering of oleaginous yeast Yarrowia lipolytica for lycopene production,” in Microbial Carotenoids from Fungi, ed. J.-L. Barredo (Humana Press), 153–159. doi:10.1007/978-1-61779-918-1_9
Young, E., Poucher, A., Comer, A., Bailey, A., and Alper, H. (2011). Functional survey for heterologous sugar transport proteins, using Saccharomyces cerevisiae as a host. Appl. Environ. Microbiol. 77, 3311–3319. doi:10.1128/aem.02651-10
Yousuf, A. (2012). Biodiesel from lignocellulosic biomass – prospects and challenges. Waste Manag. 32, 2061–2067. doi:10.1016/j.wasman.2012.03.008
Yu, X., Zheng, Y., Dorgan, K. M., and Chen, S. (2011). Oil production by oleaginous yeasts using the hydrolysate from pretreatment of wheat straw with dilute sulfuric acid. Bioresour. Technol. 102, 6134–6140. doi:10.1016/j.biortech.2011.02.081
Yue, L., Chi, Z., Wang, L., Liu, J., Madzak, C., Li, J., et al. (2008). Construction of a new plasmid for surface display on cells of Yarrowia lipolytica. J. Microbiol. Methods 72, 116–123. doi:10.1016/j.mimet.2007.11.011
Zhang, B., Rong, C., Chen, H., Song, Y., Zhang, H., and Chen, W. (2012a). De novo synthesis of trans-10, cis-12 conjugated linoleic acid in oleaginous yeast Yarrowia lipolytica. Microb. Cell Fact. 11, doi:10.1186/1475-2859-11-51
Zhang, F., Carothers, J. M., and Keasling, J. D. (2012b). Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 30, 354–U166. doi:10.1038/nbt.2149
Zhang, H., Damude, H. G., and Yadav, N. S. (2012c). Three diacylglycerol acyltransferases contribute to oil biosynthesis and normal growth in Yarrowia lipolytica. Yeast 29, 25–38. doi:10.1002/yea.1914
Zhao, C.-H., Cui, W., Liu, X.-Y., Chi, Z.-M., and Madzak, C. (2010a). Expression of inulinase gene in the oleaginous yeast Yarrowia lipolytica and single cell oil production from inulin-containing materials. Metab. Eng. 12, 510–517. doi:10.1016/j.ymben.2010.09.001
Zhao, X., Wu, S., Hu, C., Wang, Q., Hua, Y., and Zhao, Z. (2010b). Lipid production from Jerusalem artichoke by Rhodosporidium toruloides Y4. J. Ind. Microbiol. Biotechnol. 37, 581–585. doi:10.1007/s10295-010-0704-y
Keywords: Yarrowia lipolytica, genetic and metabolic engineering, oleaginous yeast, fatty acid-based biofuel and bioproducts, molecular tools, microbial platform, biotransformation
Citation: Abghari A and Chen S (2014) Yarrowia lipolytica as an oleaginous cell factory platform for production of fatty acid-based biofuel and bioproducts. Front. Energy Res. 2:21. doi: 10.3389/fenrg.2014.00021
Received: 27 February 2014; Accepted: 01 June 2014;
Published online: 19 June 2014.
Edited by:
Lew Paul Christopher, South Dakota School of Mines and Technology, USAReviewed by:
Zongbao K. Zhao, Chinese Academy of Sciences, ChinaBiplab Kumar Debnath, Politecnico di Torino, Italy
Yu-Shen Cheng, University of California Davis, USA
Copyright: © 2014 Abghari and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shulin Chen, Bioprocessing and Bioproducts Engineering Laboratory, Department of Biological Systems Engineering, Washington State University, L. J. Smith Hall, 1935 E Grimes Way, Pullman, WA 99164-6120, USA e-mail: chens@wsu.edu
 Ali Abghari
Ali Abghari Shulin Chen
Shulin Chen