Synthetic Biology and Metabolic Engineering Approaches and Its Impact on Non-Conventional Yeast and Biofuel Production
- 1Biotechnology Division, National Institute for Interdisciplinary Science and Technology, Council of Scientific and Industrial Research, Trivandrum, India
- 2Rajiv Gandhi Centre for Biotechnology, Trivandrum, India
- 3Center for Innovative and Applied Bioprocessing, Mohali, Punjab, India
- 4Dpt. Ingeniería Química, Ambiental y de los Materiales Edificio, Universidad de Jaén, Jaén, Spain
The increasing fossil fuel scarcity has led to an urgent need to develop alternative fuels. Currently microorganisms have been extensively used for the production of first-generation biofuels from lignocellulosic biomass. Yeast is the efficient producer of bioethanol among all existing biofuels option. Tools of synthetic biology have revolutionized the field of microbial cell factories especially in the case of ethanol and fatty acid production. Most of the synthetic biology tools have been developed for the industrial workhorse Saccharomyces cerevisiae. The non-conventional yeast systems have several beneficial traits like ethanol tolerance, thermotolerance, inhibitor tolerance, genetic diversity, etc., and synthetic biology have the power to expand these traits. Currently, synthetic biology is slowly widening to the non-conventional yeasts like Hansenula polymorpha, Kluyveromyces lactis, Pichia pastoris, and Yarrowia lipolytica. Herein, we review the basic synthetic biology tools that can apply to non-conventional yeasts. Furthermore, we discuss the recent advances employed to develop efficient biofuel-producing non-conventional yeast strains by metabolic engineering and synthetic biology with recent examples. Looking forward, future synthetic engineering tools’ development and application should focus on unexplored non-conventional yeast species.
Introduction
The price for non-renewable fuels as well as the level of CO2 in the atmosphere is increasing constantly. Hence, the production of bio-based fuels has attracted the attention of researchers as an alternative source of energy. Biofuels can be used as fuel for combustion engines which should be compatible. Lignocellulosic biomass is the major source of biofuel since it is economical and easily available. The yeast cell factory Saccharomyces cerevisiae is a well-known producer of ethanol and fatty acids (Tsai et al., 2015).
The topmost challenge in the marketing of microbial fuels and chemicals is the inability to cross the gap between the laboratory and the commercial market. This is primarily due to the fact that engineered strains do not meet the standard of commercialization. Synthetic biology is the fusion of biological parts and designs which evolved from the huge data of transcriptomics, proteomics, metabolomics, and fluxomics that lead to the design of novel synthetic circuits (Peralta-Yahya et al., 2012; Nielsen, 2015). The challenge in synthetic biology is to design synthetic circuits from genetic constructs toward the development of host systems that can accommodate several complex metabolic regulatory circuits. Rapid advent of DNA sequencing technologies makes the design and construction of synthetic biological devices into a reality (Unkles et al., 2014). Synthetic biology, its application in the production of fuels and chemicals, is a rapidly growing field, and synthetic microbial host cell design is the most complex area in microbial genetic engineering. Various technical platforms for assembling a library of DNA segments into synthetic pathways or circuits, which in turn inserted into the host genome while manipulating existing genes in the host, have recently been developed. Synthetic biology and metabolic engineering techniques have been widely used in Escherichia coli, S. cerevisiae, and Zymomonas mobilis to enhance ethanol production (Chubukov et al., 2016). So investigation and findings in synthetic biology hold extreme importance and need to accelerate the new design and optimization of pathways.
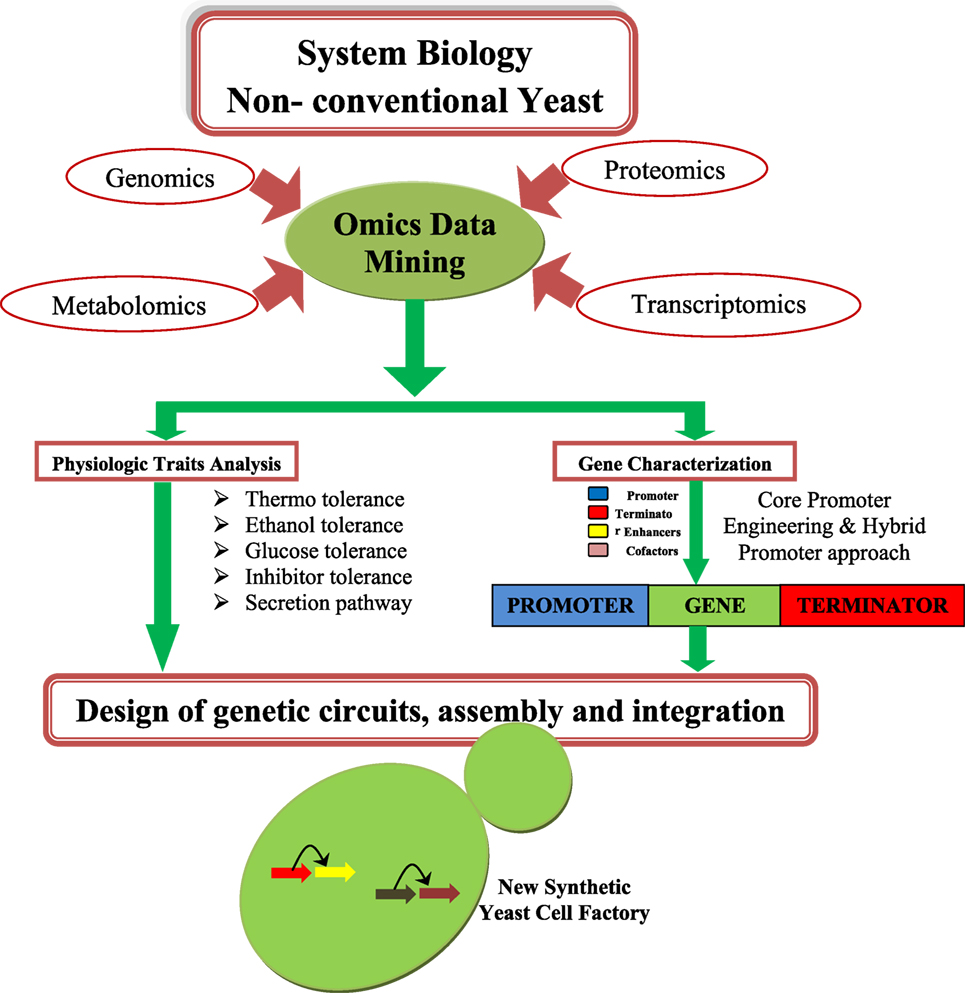
Saccharomyces cerevisiae have been considered as a model system for studying cell and molecular biology and is a well-studied organism (Blazeck et al., 2012; Blount et al., 2012a,b; Bao et al., 2015). Many synthetic biology and metabolic engineering efforts have been already established in S. cerevisiae for improving biofuel production (Tsai et al., 2015). An overview of synthetic biology design is depicted in the Figure 1. Thus, yeast is a good platform for developing synthetic biology techniques, and this system can be used for improving the status of biofuel production. The main disadvantage of S. cerevisiae is its inability to consume a wide range of substrates (e.g., xylose, arabinose) (Garcia Sanchez et al., 2010), and glycerol (Swinnen et al., 2013). S. cerevisiae is also not good for use in biofuel application that requires high temperatures (>34°C) (Caspeta et al., 2014).
Non-conventional yeast species possess several advantages over S. cerevisiae in terms of its physiology and metabolic pathways and regulation (Wolf, 2012). Non-conventional yeasts, like Yarrowia lipolytica, Hanensula polymorpha, Pichia pastoris, and Kluyveromyces lactis, are extensively studied yeast species and attractive production platforms. Most of these yeast systems developed extremely efficient mechanisms to withstand under harsh environmental conditions (Wagner and Alper, 2016). Several yeast species are diverged by evolution from S. cerevisiae and possess several unique genes and growth characteristics to withstand different stress conditions (Souciet et al., 2009). Still, several non-conventional yeast species are yet to be characterized and the advent of next-generation sequencing strategies and the other genomics and proteomics tools delineates the mechanism behind this stress tolerance strategies.
Constructing microorganisms toward desired fuel production should take into account several considerations, including increasing the yield, utilization of a wide range of substrates, and should be economical and highly efficient and simple downstream processes. In addition to these, fuel tolerance, inhibitor tolerance, thermotolerance, etc., deserve attention for enhanced fuel production. Control of redox balance is also one of the most important obstacles in strain development for the production of fuels at high yields. Non-conventional yeasts with these advantageous characters than S. cerevisiae have been utilized as industrial microorganisms for biofuel production (Ruyters et al., 2015).
The present review addresses the current status of essential synthetic biology tools being applied to construct synthetic circuits in yeast and furthermore, how these techniques can be applicable to non-conventional yeast system in order to efficiently engineer them for improving biofuel production.
Non-Conventional Yeast Systems
There are several non-conventional yeast species (e.g., Y. lipolytica) which offer many potential advantages over S. cerevisiae with respect to metabolic pathway requirements, recombinant product profile, and physiological responses (Gellissen and Hollenberg, 1997). The availability of superior quality genome sequences in public domain (Ramezani-Rad et al., 2003; Sherman et al., 2009), development of transformation vectors, gene transformation strategies (Faber et al., 1994), and metabolic engineering tools may change this scenario. Each of these organisms presents with different advantages, similarities, and differences when compared with S. cerevisiae. Most of the non-conventional yeast, like K. lactis, P. pastoris, etc., possess a broad range of substrates which is superior to S. cerevisiae and reduce the cost of industrial biofuel production (Gellissen et al., 2005). Y. lipolytica and K. lactis secrete high titers of secretory proteins extracellularly which is better than the model yeast S. cerevisiae (Dominguez et al., 1998).
The main obstacle of non-conventional yeast genetic modification is its non-homologous end-joining pathway compared to S. cerevisiae which favors homologous recombination. This results in the ectopic integration of targeted constructs which hampers synthetic biology applications (Vogl et al., 2013). Disruption of the genes involved in the NHEJ pathway Ku70 (Näätsaari et al., 2012; Verbeke et al., 2013) or Ku80 (Kooistra et al., 2004; Saraya et al., 2012) enhanced the efficiency of homologous recombination. The frequency of homologous recombination in S. cerevisiae is high achieved with 40 bp, while in the case of non-conventional yeast 500–3,000 bp of flanking sequence is required (Blazeck et al., 2014; Horwitz et al., 2015). Construction of expression cassettes for such long adaptor sequences requires a large number of PCR reactions, cloning steps, etc. Advanced synthetic biology tools may alleviate these drawbacks. In summary, non-conventional yeasts have several unique growth characteristics and serve as a potent cell factory for biofuel production with the help of synthetic biology tools.
Basic Tools for Metabolic Engineering of Non-Conventional Yeast
This section focuses on the characterized promoters from different non-conventional and previously attempted metabolic engineering strategies and its future implications in biofuel production using synthetic biology tools.
Available Promoters in Non-Conventional Yeast and Promoter Engineering
To develop a microbial cell factory, the selection of the promoter elements required for driving heterologous protein expression is crucial. At present, only very few promoters have been identified in non-conventional yeast and their metabolic regulation is not fully elucidated. Engineered promoter elements are an integral part of synthetic biology and metabolic engineering and are used to enhance the titers of homologous and heterologous proteins. Eukaryotic core promoter elements are the crucial region for the binding of transcription factors and other regulatory factors involved in transcriptional control. So the development of suitably engineered promoters in organisms with poorly defined genetic tools is extremely important for making these organisms as industrial production hosts for fuels and chemicals. The synthetic promoter approach has been developed quickly in S. cerevisiae compared to other non-conventional yeast. However, recently a large number of genome sequences of non-conventional yeasts are available (e.g., Y. lipolytica) and have boosted both basic and applied research to understand the genotypic and phenotypic features and further development of metabolic engineering tools for biofuel production. Most of the promoter engineering work has been concentrated on upstream regulatory sequences (Hartner et al., 2008; Xuan et al., 2009), engineering core promoter elements (Blazeck et al., 2012), and/or by creating random mutations in core promoter elements (Berg et al., 2013), and this will lead to tight and tuneable control over a metabolic regulatory pathways (Teo and Chang, 2014).
Along with the core promoter elements, upstream elements such as binding sequences for transcription factors can also be modified to increase the strength of the promoters. For example, TetR protein, which is a well-studied and extensively used protein in bacterial molecular biology, retains its binding to DNA control elements in yeast and sensitive to synthetic inducers, such as anhydrotetracycline or doxycycline. So promoter with TetR-binding sites has been used to control different genes in different yeasts (Blount et al., 2012b). Still, non-conventional yeast promoters are poorly characterised, and findings from S. cerevisiae can be applicable to the most the highly efficient non-conventional yeast systems also because most of the yeast promoters are capable of cross recognition (Van Ooyen et al., 2006).
The commercial K. lactis uses inducible promoter PLAC4, which is a lactose inducible promoter for recombinant protein production, and the proteins are produced into the culture fluid, which makes the recombinant protein purification easy. Highly efficient protein secretion machinery and the high biomass attained in submerged culture condition makes K. lactis an attractive cell factory for heterologous protein production (Van den Berg et al., 1990; Gellissen and Hollenberg, 1997). A hybrid promoter approach was established in K. lactis by combining core promoter elements of Trichoderma reesei cellobiohydrolase1 (cbh1) to the β-galactosidase (lac4) promoter of K. lactis for increasing recombinant protein production in the yeast (Madhavan and Sukumaran, 2014). This may open up the possibilities of manipulation of core promoter elements across classes of the organisms by introducing important core promoter sequences to create highly active synthetic promoters.
Pichia pastoris is a widely used protein production platform for enzymes and pharmaceuticals (Vogl et al., 2013). Highly active methanol-inducible promoter, alcohol oxidase 1 gene (pAOX1), is the most widely used and tightly controlled promoter in P. pastoris. This has been well studied in terms of promoter control elements and different transcription factor. Random mutated library of pAOX1 has been constructed and resulted in less glucose repression and increased activity compared to wild pAOX1 (Berg et al., 2013). The constitutive pGAP (glyceraldehyde-3-phosphate dehydrogenase promoter) served as a scaffold, and this resulted in the development of a series of promoters with a range of activity from 0.01- to 19.6-fold of wild-type pGAP activity (Qin et al., 2011). Blazeck et al. (2012) reported that the combination of core promoter elements with synthetic upstream activator sequence could increase the expression level.
CRISPR-Cas
The field of synthetic biology witnessed a transformation by the advent of CRISPR–Cas (Clustered Regularly Interspaced Short Palindromic Repeat) system in several host organisms (Cong et al., 2013). CRISPR-mediated RNA-guided control of gene using nuclease-deficient Cas9 (dCas9) was reported to control and regulate the expression of genes inside the yeast, with synthetic single stranded-guide RNAs (sgRNAs) targeting the genes to silence on a genome. This technique helped to create a series of simultaneous targeted integrations and double- stranded breaks in S. cerevisiae rapidly and efficiently (Bao et al., 2015). The dCas9 was fused to Mxi1, a protein that involved in the attraction of histone deacetylase Sin3p homolog, a component of gene-silencing complex in yeast. Further, the dCas9 along with sgRNAs repressed the TEF promoter of S. cerevisiae about 10-fold, Mxi1 (fusion protein) could downregulate the promoter by 53-fold (Gilbert et al., 2013). This technique is also been applied to an industrially important non-conventional K. lactis strain, where muconic acid production was improved by incorporating six genes at three targeted loci and helped in the introduction of a pathway for the production of muconic acid precursor (Horwitz et al., 2015).
Role of Synthetic Biology in Yeast Metabolic Engineering and Fuel Production
Industrial biofuel production often requires strains that can withstand several stress conditions like extreme pH, high temperature, osmotolerance, shearing forces, organic acids, and other inhibitors. All these properties are complex traits and are encoded by several genes at different loci. Due to the complexity of yeast genome, normal genetic analysis methods, therefore, fail to characterize the underlying genetic regulatory network, and efforts to optimize one of these traits classically depend on adaptive laboratory evolution or other controlled breeding strategies. The phenotypic and genotypic characterization of non-conventional yeast species deserve special attention and will help to identify strains and species with novel and/or improved industrially important properties, and the advent of next-generation sequencing at low cost and in a comparably short time have now opened the way for the use of advanced genome analysis tools like quantitative trait loci analysis to identify, at least under some conditions, all causative genes for a certain trait (Bloom et al., 2013).
Tsai et al. (2015) reviewed about the requirement of genetic circuits in yeast synthetic biology. The timely expression of genes in metabolic pathway could minimize the extra energy and nutrient resources and maximize the production capacity. This emphasizes the need for genetic circuits which include multiple regulatory elements arranged to create different logical gates. The combination of these modules resulted in a complicated network (Elowitz and Leibler, 2000) and genetic switches (Gardner et al., 2000). Recently, Blount et al. (2012a) reviewed the status of Yeast genetic circuits. The genetic circuits in S. cerevisiae are far behind the E. coli. Blount et al. (2012b) also proposed three important principles to create genetic circuits in yeast: (1) the circuit elements should not be related to the host physiologic elements to retain orthogonality, (2) the circuit elements needs to be tuneable and inducible, and (3) self-contained groups should be built to obtain modularity. In order to construct genetic circuits, technologies that create fast prototyping, evaluation, and optimization of metabolic pathways in host species are essential. The current key research area of synthetic biology is how to reduce the time required for making genetic circuits. The finding of new metabolic pathways and its combination can be efficiently addressed by better pathway assembly techniques, like ligation-free assembly (Li and Elledge, 2007; Vroom and Wang, 2008) and BioBricks (Shetty et al., 2008). Another key area of research of synthetic biology is the synthesis of reusable constructs with predictable behavior. The construction of a different array of controllable and tunable expression system is very useful in metabolic engineering. A genetic switch that responds to environmental cues and leads to further downstream regulation is also advantageous for biofuel production (Dueber et al., 2007).
For example, incorporation of FadR-binding sites in GAL1 promoter region, after RNA polymerase binding region and before the transcription start site, represses the promoter activity. But the presence of fatty acid, like myristic acid, changes the conformation of FadR, reduces the affinity to DNA-binding sequences, and deregulates the expression of genes (Blount et al., 2012a). Teo and Chang (2014) incorporated several upstream activator elements which in turn detect copper and phosphate starvation, and then downstream gene will only be activated by the depletion of both fatty acid and copper or fatty acid and phosphate. This synthetic device is useful for sensing the presence of fatty acid and can be included into pathways with the production or depletion of fatty acids. The regulatory responses of yeast to external stimuli have also been engineered for synthetic biology applications. For example, mitogen-activated kinase cascade have genetically engineered to rewire the novel regulatory networks (Kiel and Serrano, 2011).
Non-Conventional Yeast and Biofuel Production
Lignocellulosic biomass is the richest source of sugars for the fermentation to ethanol and other valuable chemicals. For the efficient conversion of lignocellulosic biomass, it is necessary that all the sugars in hydrolyzates should be fermentable to ethanol and other value-added chemicals. The main problem associated with this bioconversion is lack of efficient ethanol-producing yeast, the inefficiency to convert pentoses remains to be topmost challenge in bioconversion of lignocellulosic biomass. Pentose-utilizing yeasts can utilize both glucose and xylose from biomass to ethanol. Their conversion rate is very low due to the presence of inhibitors in the hydrolyzate, less ethanol tolerance, and glucose repression.
Saccharomyces cerevisiae is the best characterized currently used industrial workhorse. In addition to the efforts attempted to genetic engineer S. cerevisiae to overcome a variety of stresses during fermentation and subsequent ethanol production, S. cerevisiae has limitations. Less explored non-conventional yeast species possess better ethanol-producing capabilities, better tolerance to most of these stresses, and could be a better candidate than S. cerevisiae to study the molecular mechanism behind inhibitor tolerance and substrate utilization for second-generation biofuel production.
Saccharomyces cerevisae utilizes glucose by fermentative pathway (Crabtree positive) and some non-conventional yeasts, like K. lactis, P. pastoris, and Y. lipolytica, are predominantly oxidative (Crabtree negative). Several attempts were initiated to convert Crabtree-negative yeasts to Crabtree-positive for improving ethanol fermentation efficiency. Metabolic engineering of K. lactis has been reported by construction of a null mutant in the single gene encoding for a mitochondrial alternative internal dehydrogenase. The mutant showed unaffected rate of oxidation of exogenous NADH, and the mutation also shifted the metabolism to fermentative instead of respiratory. This increased the rate of ethanol production in K. lactis (Gonzalez-Siso et al., 2015).
Dekkera bruxellensis is a crab-positive yeast producing high yield of ethanol. Schifferdecker et al. (2016) developed a metabolically engineered strain by increasing its capability of fermentative metabolism. The encoding for alcohol dehydrogenase was homologously overexpressed under the control of highly active TEF1 promoter, resulted in 1.4–1.7 times more ethanol than the wild-type strain.
The tolerance of yeasts to several stresses like osmolarity and temperature is extremely important for first- and second-generation biofuel production. Zygosaccharomyces rouxii is capable of growing in osmolarity of 3 M NaCl and glucose concentrations of 90% due to the presence of unique transporters in plasma membrane which is higher than S. cerevisiae (Leandro et al., 2011). The temperature tolerance is also essential during the industrial process, which should be up to 50°C. This temperature is different from the normal fermentation temperature of S. cerevisiae, which is between 25 and 37°C (Abdel-Banat et al., 2010). Some non-conventional yeast species-like Ogataea polymorpha have been found to ferment xylose at 45°C (Kurylenko et al., 2014).
Two thermotolerant isolates (withstand above 45°C), NIRE-K1 and NIRE-K3, were screened for fermenting both glucose and xylose and identified as Kluyveromyces marxianus NIRE-K1 and K. marxianus NIRE-K3. The final ethanol yield for both strains was found to be 39.12 and 43.25 g/L (Arora et al., 2015a,b). Another study by Kumar et al. (2014) reported the fermentation of xylose and glucose-rich bagasse hydrolyzates by the thermotolerant (50°C) yeast Kluyveromyces sp. IIPE453 and yielded 0.43 g/g ethanol. The genome sequence information of most of these non-conventional yeasts is available. However, the molecular mechanisms underlying the tolerance of these species to these stress conditions remain poorly investigated for all of them.
Substrate Utilization of Non-Conventional Yeasts
Extensive attempts have been done to increase the range of substrate utilization capabilities of S. cerevisiae through metabolic engineering. A schematic diagram for different routes of substrate assimilation is illustrated in the Figure 2. Efficient conversion of carbon substrates, such as arabinose, xylose, and cellobiose, has been materialized through the introduction of hydrolytic enzymes and transporter genes in yeast. Engineered strains of S. cerevisiae capable of utilizing starch and xylose have been reported (Wei et al., 2013). But all these recombinant strains perform well at a fermentation temperature of 30°C but not at the higher temperatures. But most of the hydrolytic enzymes for lignocellulosic biomass function optimally at 50°C. Majority of the non-conventional yeasts are thermotolerant and ferment sugars at high temperature, but the range of substrate utilization is less (Katahira et al., 2004). Metabolic engineering of these non-conventional yeasts is essential to widen the range of substrate utilization. However, certain non-conventional yeasts utilize a variety of carbon substrates, but they do not possess other important characteristics (ethanol tolerance, inhibitor tolerance, etc.).
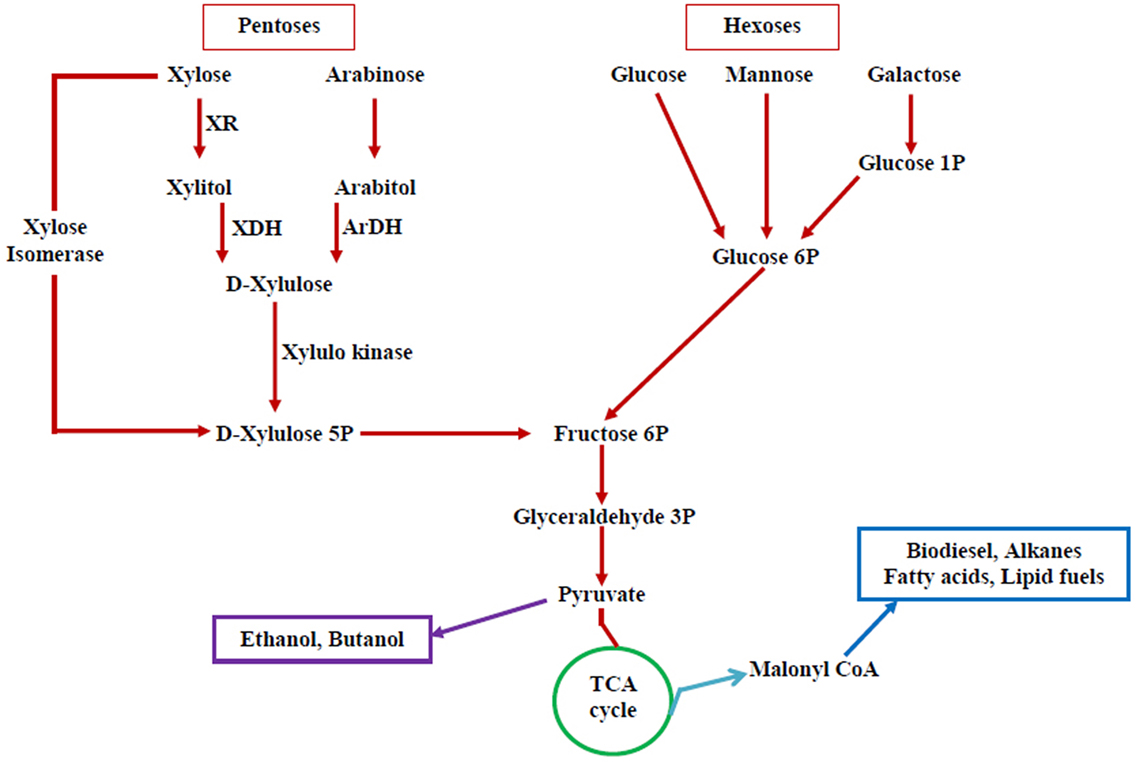
Figure 2. Pentose and hexose utilization pathway in yeasts—possible targets for altering substrate utilization. Pentoses and hexoses, the main components of lignocellulosic biomass that includes xylose, arabinose, glucose, mannose, and galactose and their utilization pathway (XR, xylose reductase; XDH, xylitol dehydrogenase; ArDH, arabitol dehydrogenase). A number of biofuels can be produced from this pathway.
Pichia stipitis are the ascomycetous yeast that can ferment xylose to ethanol at nearly maximum yield without any by-product formation. Few metabolic engineering approaches have been established in P. stipitis to resolve its crabtree effect, low ethanol tolerance, etc. Although P. stipitis is an efficient biofuel producer due to its natural capability for xylose fermentation, further improvement in production host is hampered by a lack of genetic engineering tools. For further improvement of this strain, RNA sequencing was done in search of highly active constitutive promoters. Several promoters like TEF1, Xy11p, ADH1p, and ADH2p have been exploited in S. stipitis for efficient gene expression (Cho and Jeffries, 1999). Few transcriptomic approaches were carried out in S. stipitis under glucose/xylose inducing conditions. Recent transcriptomic study under lignocellulosic biomass inducing condition and oxygen limited condition resulted in the mapping of several important genes (Bullard et al., 2010). Synthetic biology applications in S. stipitis are very less at this moment, and most of them involve the genetic transfer of its efficient biosynthetic genes to S. cerevisiae to make it able to utilize pentose sugars, although the full genome sequencing (Jeffries and Van Vleet, 2009) and the more efficient genetic transformation systems (e.g., plasmid vectors and a loxP/Cre recombination system) and drug resistance markers (Laplaza et al., 2006) might enable better room for genetic modification of this industrial strain.
The thermotolerant yeast H. polymorpha cannot utilize starch and xylose and its ferment glucose and xylose to ethanol at high temperature. For the efficient utilization of xylose and starch, amylolytic and xylanolytic enzymes were heterologously expressed. Genes encoding α amylase and glucoamylase, SWA2 and GAM1, from the yeast Schwanniomyces occidentalis, encoding α-amylase and glucoamylase, were transferred in to H. polymorpha under the well characterized constitutive promoter of the H. polymorpha glyceraldehyde-3-phosphate dehydrogenase gene (Voronovsky et al., 2009). Since the organism is highly thermotolerant, engineering the substrate utilization strategies will help to improve the industrial production.
Kluyveromyces marxianus KY3 is a highly efficient hexose-fermenting yeast and has many other advantages like thermotolerance, high cell density, temperature and pH tolerance, high secretion of heterologous proteins, and efficient substrate utilization (Yanase et al., 2010). The xylose utilization of K. marxianus is weak. To alleviate the xylose utilization, this strain was engineered for the conversion of xylose to xylitol and ethanol at the higher temperature. This was done by replacing native xylose reductase gene of the K. marxianus strain with xylose reductase gene from P. stipitis. This modified strain with both NADPH and NADH as the coenzyme could produce 55 g/L ethanol and 32 g/L xylitol (Zhang et al., 2013). Other engineering approaches include the protein engineering for altering the cofactor requirement of xylose reductase of Candida tenuis (Petschacher and Nidetzky, 2008) and optimized expression levels of Candida shehatae xylose reductase, Candida tropicalis xylose dehydrogenase, and P. pastoris xylose kinase by combinatorial transcriptional engineering (Tsai et al., 2015). Recently, evolutionary adaptation technique has been used to enhance the utilization of xylose in K. marxianus (Sharma et al., 2016).
Development of Consolidated Bioprocessing for Non-Conventional Yeast Strains
Many microorganisms possess biomass-degrading enzymes that can efficiently degrade lignocellulose materials, but their fermentative production of ethanol is less. Recently, Arora et al. (2015a,b) reviewed the importance of highly efficient cellulosomes for the production of biofuels from lignocellulosic biomass. Several attempts have been done to convert enzymatically efficient hosts into ethanol producing host to serve as the cell factory for ethanol production. But still, an efficient ethanol fermenter needs to be developed. To develop yeast for CBP bioethanol production, a synthetic biology technique, called “promoter-based gene assembly and simultaneous overexpression” (PGASO), that can simultaneously insert and express multiple genes into yeast. K. marxianus has a number of advantages, such as heat and toxin tolerance, over the model organisms K. lactis and S. cerevisiae. To formulate an efficient cellulose cocktail, a filter-paper-activity assay for selecting heterologous cellulolytic enzymes was developed in this study and used to select five cellulase degrading genes (two cellobiohydrolases, two endo-β-1,4-glucanases and one beta-glucosidase genes) from different fungi. A fungal cellodextrin transporter gene was selected to transport cellodextrin into the cytoplasm. These six multiple genes were assembled into the genome of the host using PGASO technology. Experimental results indicated that the developed strain KR7 contains five recombinantly expressed heterologous cellulase genes and that strain could transform crystalline cellulose into ethanol (Chang et al., 2013).
Improvement of Tolerance against Inhibitors and Biofuel Products
Despite the engineering of metabolic networks in yeast for biofuel production, yeast could not achieve the high level of biofuel production because of the toxic nature of products. The toxicity is mainly due to the hydrophobicity of the accumulated products inside the membrane, and this will lead to membrane disruption by the inhibition of ATP-generating pumps and conformation changes in the proteins that maintain the fluidity (Jeffries and Jin, 2000; Dunlop, 2011). Several genetic manipulations elicit enhanced tolerance against several advanced biofuels. The success of all these attempts was not up to the mark.
Even though most of the non-conventional yeast species can naturally fight with most of the inhibitors, such as nitroaromatics, aromatics, halogenated organo- phosphates, metals, and alkanes (Zinjarde et al., 2014), still, there is an urgent need for engineering some non-conventional, like Y. lipolytica, against fermentation inhibitors present in biomass hydrolyzates, like acids and phenolics. The inhibitor tolerance is a quantitative trait and is determined by several complex genes. Hence large-scale analysis of gene expression, different expression cassettes construction, and optimization will be essential to impart inhibitor tolerance to yeast. Y. lipolytica has been engineered to express recombinant laccases (Madzak et al., 2005), which could improve its detoxification capacities. Several studies reported the strains of non-conventional yeast species, such as Schizo saccharomycespombe, Pichia kudriavzevii, D. bruxellensis, Torulaspora delbrueckii, and Wickerhamomyces anomala, with promising fermentative features and superior ethanol tolerance levels than that of S. cerevisiae (Mukherjee et al., 2014; Ruyters et al., 2015). D. bruxellensis has been reported as one of the excellent yeast in terms of product tolerance and production. D. bruxellensis and S. cerevisiae possesses an almost similar molecular mechanism for this trait (Piskur et al., 2006). Kwon et al. (2011) reported that P. kudriavzevii is extremely tolerant to 3 g/L furfural and also tolerate acetic acid of up to 10 g/L (Oberoi et al., 2012) and formic acid up to 2 g/L (Dandi et al., 2013). The genetic engineering tools for this yeast are limited, and genome sequence has recently been reported by Chan et al. (2012).
Y. lipolytica Cell Factory for Biofuel Production
Yarrowia lipolytica is a well-studied oleaginous organism and extensively used for industrial biofuel production, and it has been served as a model organism for biofuel research, especially for fatty acid-derived fuels (Beopoulos et al., 2009; Tai and Stephanopoulos, 2013; Blazeck et al., 2014; Zhou et al., 2016). Several metabolic engineering tools are available for Y. lipolytica (Juretzek et al., 2001; Madzak, 2015). The completely annotated genome is available (Dujon et al., 2004), and its metabolism and regulation are also studied in detail (Pan and Hua, 2012). System and synthetic biology approaches have been established in this organism (Morin et al., 2011; Pomraning et al., 2015). Several metabolic engineering studies strengthen the lipid production in this organism. Different target genes were found for overexpression from the metabolic pathway and manipulated to increase the fatty acid accumulation. For example, inhibiting beta-oxidation, by targeted deletion of the six POX genes or the MFE gene (Dulermo and Nicaud, 2011) and overexpression of enzymes leading to TAG production (DGA2) (Beopoulos et al., 2012) and GPD1 (Dulermo and Nicaud, 2011), increased the lipid content. An improved Y. lipolytica strain could produce a very high lipid yield and lipid titers of ~55 g/L under optimized conditions. This further proved the economic feasibility of Y. lipolytica for biofuel production (Qiao et al., 2015). Y. lipolytica is not able to utilize cellulose and starch. Wei et al. (2014) modified the strain by incorporating cellulases for the conversion of cellulosic substrates. In addition, two alpha amylases—starch-degrading enzyme—have been expressed in this host (Park et al., 1997; Celinska et al., 2015).
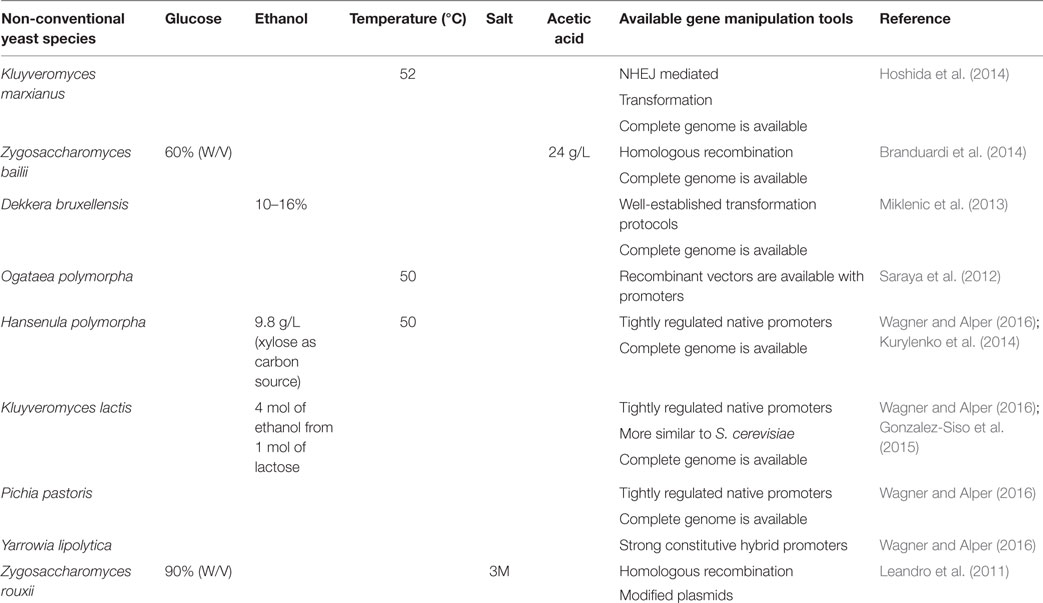
Studies revealed that the use of intron-containing translation elongation factor-1a (TEF) promoter is capable of increasing the oil production Y. lipolytica compared to intron-less TEF promoter. They have exploited this expression system for the overexpression of diacylglycerol acyl transferase (DGA1), the final key enzyme of the triglyceride (TAG) synthesis pathway, which resulted in a fourfold enhancement in lipid yield compared to wild-type, to a lipid yield of 33.8% of DCW. They also proved that the overexpression of acetyl-CoA carboxylase (ACC1), the first key enzyme of fatty acid biosynthesis, increased lipid content twofold over control, or 17.9% lipid content. The co-expression of ACC1 and DGA1 also improved the production of oil content (Tai and Stephanopoulos, 2013). Ledesma-Amaro et al. (2015) performed a strain engineering approach to obtain a consolidated bioprocess for the direct production of lipids from raw starch. Further, they proved that lipid production from starch can be enhanced by both metabolic engineering and culture condition optimization. Blazeck et al. (2014) rewired Y. lipolytica native metabolism for increased lipid titers, by coupling combinatorial multiplexing of lipogenesis targets with phenotypic induction. The tri-level metabolic control results in accumulation of 90% lipid content and 60-fold improvement over wild-type strain. Sheng and Feng (2015) reviewed the metabolic engineering of non-conventional yeast strains to improve the production of fatty acid-derived biofuels, and they discussed the bottlenecks that limit the productivity of biofuels and suggested the appropriate strategies to overcome the current bottlenecks. Some of the non-conventional yeast system currently employed for biofuel production is listed in Table 1.
Optimization of Production Pathways and Host
High yield is very important for biofuel production. The system biology data-driven approaches to synthetic biology for improving microorganisms for producing fuels decreases the cost by reducing the number of expression constructs required for different optimization experiments. The omics data help to identify the rate-limiting steps in biosynthetic pathways. Targeted proteomics approach identified two poorly expressed enzymes in heterologous mevalonate pathway. Codon optimization of the target gene and introduction of a promoter at the 5′ region of the most poorly expressed gene also lead to sesquiterpene production (Redding-Johanson et al., 2011). Some computational prediction tools are involved in changing the metabolic pathway by using knockouts or by adding new catalytic enzymes, and this may help in increasing biofuel production. One important tool for the prediction of metabolic pathway is From Metabolite to Metabolite (FMM)—freely available software (Medema et al., 2012). It compiles the KEGG map and KEGG ligand data to obtain a combined pathway. Recently, a metabolic model with the help of computational tools deleted the NADPH-dependent glutamate dehydrogenase that would result in a 10-fold increase in the production of sesquiterpenes in S. cerevisiae (Asadollahi et al., 2009). These tools can be applied to non-conventional yeasts to expand its desirable traits for biofuel production.
Current Bottlenecks of Synthetic Biology Applications in Non-Conventional Yeast and Future Perspectives
Several non-conventional yeast systems with highly desirable characteristics and genome sequences for biofuel production are available now. The current requirement is to expand the tool box to engineer the non-conventional to make it suitable to digest lignocellulosic biomass and ferment to ethanol. The availability of the complete genome sequences for several non-conventional yeasts will open new opportunity to the system biology and for the enhancement of native production capabilities of these yeasts. Genome-scale metabolic models (GEMs) is tool which can be used to assess the metabolic nature of a cell, its metabolite production capability, species to species relation, identification of genes and transcription factors for metabolic engineering, metabolic flux design, etc. (Zhang and Hua, 2015).
RNA interference is still not implemented in non-conventional yeast despite its success in S. cerevisiae. Optimized RNAi technique in S. cerevisiae has greatly helped for developing tools of metabolic engineering and synthetic biology. Likewise, CRISPR-Cas technology has already been established in S. cerevisiae for biofuel production (Papapetridis et al., 2016). Extension of these technologies to non-conventional yeast strains would enable fastest methods to target many genes and thus by the alteration of metabolic pathway for enhanced biofuel production. CRISPR–Cas technology has been initiated in the non-conventional yeast system K. lactis and S. pombe (Horwitz et al., 2015). The main challenge associated with CRIPR-Cas technology has been the production of guide RNA. In S. cerevisiae, this is achieved by use of RNA polymerase III promoters. But RNA polymerase III promoters are not well in non-conventional yeast. This hinders the progress of CRISPR-Cas technology in non-conventional yeast. Once the CRISPR–Cas technique is a success in the non-conventional yeasts, we will be able to establish transcriptional regulation, genetic circuits, and other metabolic networks to establish a fuel production system and heterologous expression host (Zalatan et al., 2015). The targeted generation of double-stranded breaks also alleviates the challenges associated with homologous recombination in non-conventional yeast.
Concluding Remarks
Current increases in fuels costs have resulted in increased demand in finding an alternative to fossil fuels. This leads to the concept of biofuels by manipulating microbial cellular metabolism for the production of fuels and chemicals. Computational tools are essential for the analysis of high-throughput data generated by gene-sequencing programmes and gene-expression analysis. Analysis of complex metabolic pathway is another challenge in synthetic and metabolic engineering concepts. The field of synthetic biology and metabolic engineering has been growing rapidly in the case of S. cerevisiae. This led to the construction of highly engineered metabolic pathways, strongly regulated metabolic networks, efficiently engineered native and synthetic control elements, and efficient recombinant vectors. Development of mathematical models for studying the complex metabolic pathways is also extremely important. Well-established synthetic biology approaches in S. cerevisiae can be a boost to other non-conventional yeast species by rewiring the metabolic pathways related to biofuel production.
In summary, biofuel production by engineered yeast through synthetic biology requires several synthetic biology pipelines and models. This can be used for predicting metabolically optimal pathways, gene constructs, and metabolite screening. The developments in different “omics” technologies can serve as tool for future synthetic biology of non-conventional yeast.
Author Contributions
AM wrote and reviewed the article. AJ cross checked and modified article and references. PB, RKS, AP, and GC reviewed and modified the article. RS, corresponding author, reviewed and modified the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
The authors are grateful to the Ministry of New and Renewable Energy, Government of India, New Delhi; Department of Science and Technology, Government of India, New Delhi; and Technology Information, Forecasting and Assessment Council, New Delhi, for the financial support provided to the Centre for Biofuels R&D, CSIR-NIIST, Trivandrum. One of the authors RS acknowledges Department of Biotechnology for financial support under DBT Bio-CARe scheme. AM, GC, RS, and PB acknowledge European Commission Seventh Framework Programme, Marie Curie Actions-International Research Staff Exchange Scheme—Contact Number 318931. AM acknowledges Department of Biotechnology for financial support under DBT Research Associateship programme.
References
Abdel-Banat, B. M., Hoshida, H., Ano, A., Nonklang, S., and Akada, R. (2010). High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl. Microbiol. Biotechnol. 85, 861–867. doi: 10.1007/s00253-009-2248-5
Arora, R., Behera, S., Sharma, N. K., and Kumar, S. (2015a). Bioprospecting thermostable cellulosomes for efficient biofuel production from lignocellulosic biomass. Bioresour. Bioprocess 2, 38. doi:10.1186/s40643-015-0066-4
Arora, R., Behera, S., Sharma, N. K., and Kumar, S. (2015b). A new search for thermotolerant yeasts, its characterization and optimization using response surface methodology for ethanol production. Front. Microbiol. 6:889. doi:10.3389/fmicb.2015.00889
Asadollahi, M. A., Maury, J., Patil, K. R., Schalk, M., Clark, A., and Nielsen, J. (2009). Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab. Eng. 11, 328–334. doi:10.1016/j.ymben.2009.07.001
Bao, Z., Xiao, H., Liang, J., Zhang, L., Xiong, X., Sun, N., et al. (2015). Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth. Biol. 4, 585–594. doi:10.1021/sb500255k
Beopoulos, A., Cescut, J., Haddouche, R., Uribelarrea, J. L., Molina-Jouve, C., and Nicaud, J. M. (2009). Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 48, 375–387. doi:10.1016/j.plipres.2009.08.005
Beopoulos, A., Haddouche, R., Kabran, P., Dulermo, T., Chardot, T., and Nicaud, J. M. (2012). Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl. Microbiol. Biotechnol. 93, 1523–1537. doi:10.1007/s00253-011-3506-x
Berg, L., Strand, T. A., Valla, S., and Brautaset, T. (2013). Combinatorial mutagenesis and selection to understand and improve yeast promoters. Biomed Res. Int. 2013, 926985. doi:10.1155/2013/926985
Blazeck, J., Garg, R., Reed, B., and Alper, H. S. (2012). Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol. Bioeng. 109, 2884–2895. doi:10.1002/bit.24552
Blazeck, J., Hill, A., Liu, L., Knight, R., Miller, J., Pan, A., et al. (2014). Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 5, 3131. doi:10.1038/ncomms4131
Bloom, J., Ehrenreich, I., Loo, W., Lite, T., and Kruglyak, L. (2013). Finding the sources of missing heritability in a yeast cross. Nature 494, 234–237. doi:10.1038/nature11867
Blount, B. A., Weenink, T., and Ellis, T. (2012a). Construction of synthetic regulatory networks in yeast. FEBS Lett. 586, 2112–2121. doi:10.1016/j.febslet.2012.01.053
Blount, B. A., Weenink, T., Vasylechko, S., and Ellis, T. (2012b). Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology. PLoS ONE 7:e33279. doi:10.1371/journal.pone.0033279
Branduardi, P., Dato, L., and Porro, D. (2014). Molecular tools and protocols for engineering the acid-tolerant yeast Zygosaccharomyces bailii as a potential cell factory. Methods Mol. Biol. 1152, 63–85. doi:10.1007/978-1-4939-0563-8_4
Bullard, J. H., Purdom, E., Hansen, K. D., and Dudoit, S. (2010). Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11:94. doi:10.1186/1471-2105-11-94
Caspeta, L., Chen, Y., Ghiaci, P., Feizi, A., Buskov, S., Hallström, B. M., et al. (2014). Altered sterol composition renders yeast thermotolerant. Science 346, 75L–78. doi:10.1126/science.1258137
Celinska, E., Bialas, W., Borkowska, M., and Grajek, W. (2015). Cloning, expression, and purification of insect (Sitophilus oryzae) alpha-amylase, able to digest granular starch, in Yarrowia lipolytica host. Appl. Microbiol. Biotechnol. 99, 2727–2739. doi:10.1007/s00253-014-6314-2
Chan, G. F., Gan, H. M., Ling, H. L., and Rashid, N. A. (2012). Genome sequence of Pichia kudriavzevii M12, a potential producer of bioethanol and phytase. Eukaryotic Cell 11, 1300–1301. doi:10.1128/EC.00229-12
Chang, J. J., Ho, F. J., Ho, C. Y., Wu, Y. C., Hou, Y. H., Huang, C. C., et al. (2013). Assembling a cellulase cocktail and a cellodextrin transporter into a yeast host for CBP ethanol production. Biotechnol. Biofuels 6, 19. doi:10.1186/1754-6834-6-19
Cho, J. Y., and Jeffries, T. W. (1999). Transcriptional control of ADH genes in the xylose-fermenting yeast Pichia stipitis. Appl. Environ. Microbiol. 65, 2363–2368.
Chubukov, V., Mukhopadhyay, A., Petzold, C. J., Keasling, J. D., and Martín, H. G. (2016). Synthetic and systems biology for microbial production of commodity chemicals. NPJ Syst. Biol. Appl. 2, 16009. doi:10.1038/npjsba.2016.9
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi:10.1126/science.1231143
Dandi, N. D., Dandi, B. N., and Chaudhari, A. B. (2013). Bioprospecting of thermo- and osmo-tolerant fungi from mango pulp-peel compost for bioethanol production. Antonie Van Leeuwenhoek 103, 723–736. doi:10.1007/s10482-012-9854-4
Dominguez, A., Fermiñán, E., Sánchez, M., González, F. J., Pérez-Campo, F. M., García, S., et al. (1998). Non-conventional yeasts as hosts for heterologous protein production. Int. Microbiol. 1, 131–142.
Dueber, J. E., Mirsky, E. A., and Lim, W. A. (2007). Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat. Biotechnol. 25, 660–662. doi:10.1038/nbt1308
Dujon, B., Sherman, D., Fischer, G., Durrens, P., Casaregola, S., Lafontaine, I., et al. (2004). Genome evolution in yeasts. Nature 430, 35–44. doi:10.1038/nature02579
Dulermo, T., and Nicaud, J. M. (2011). Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 13, 482–491. doi:10.1016/j.ymben.2011.05.002
Dunlop, M. J. (2011). Engineering microbes for tolerance to nextgeneration biofuels. Biotechnol. Biofuels 4, 32. doi:10.1186/1754-6834-4-32
Elowitz, M. B., and Leibler, S. A. (2000). Synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338. doi:10.1038/35002125
Faber, K. N., Haima, P., Harder, W., Veenhuis, M., and Geert, A. B. (1994). Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr. Genet. 25, 305–310. doi:10.1007/BF00351482
Garcia Sanchez, R., Karhumaa, K., Fonseca, C., Sànchez Nogué, V., Almeida, J. R., Larsson, C. U., et al. (2010). Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnol. Biofuels 3, 13. doi:10.1186/1754-6834-3-13
Gardner, T. S., Cantor, C. R., and Collins, J. J. (2000). Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342. doi:10.1038/35002131
Gellissen, G., and Hollenberg, C. P. (1997). Application of yeasts in gene expression studies: a comparison of Saccharomyces cerevisiae, Hansenula polymorpha and Kluyveromyces lactis – a review. Gene 190, 87–97. doi:10.1016/S0378-1119(97)00020-6
Gellissen, G., Kunze, G., Gaillardin, C., Cregg, J. M., Berardi, E., Veenhuis, M., et al. (2005). New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica a comparison. FEMS Yeast Res. 5, 1079–1096. doi:10.1016/j.femsyr.2005.06.004
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. doi:10.1016/j.cell.2013.06.044
Gonzalez-Siso, M. I., Tourino, A., Vizoso, A., Pereira-Rodriguez, A., Rodriguez-Belmonte, E., Becerra, M., et al. (2015). Improved bioethanol production in an engineered Kluyveromyces lactis strain shifted from respiratory to fermentative metabolism by deletion of NDI1. Microb. Biotechnol. 8, 319–330. doi:10.1111/1751-7915.12160
Hartner, F. S., Ruth, C., Langenegger, D., Johnson, S. N., Hyka, P., Lin-Cereghino, G. P., et al. (2008). Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 36, e76. doi:10.1093/nar/gkn369
Horwitz, A. A., Walter, J. M., Schubert, M. G., Kung, S. H., Hawkins, K., Platt, D. M., et al. (2015). Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Syst. 1, 1–9. doi:10.1016/j.cels.2015.02.001
Hoshida, H., Murakami, N., Suzuki, A., Tamura, R., Asakawa, J., Abdel-Banat, B. M. A., et al. (2014). Non-homologous end joining-mediated functional marker selection for DNA cloning in the yeast Kluyveromyces marxianus. Yeast 31, 29–46. doi:10.1002/yea.2993
Jeffries, T. W., and Jin, Y. S. (2000). Ethanol and thermotolerance in the bioconversion of xylose by yeasts. Adv. Appl. Microbiol. 47, 221–268. doi:10.1016/S0065-2164(00)47006-1
Jeffries, T. W., and Van Vleet, J. R. (2009). Pichia stipitis genomics, transcriptomics, and gene clusters. FEMS Yeast Res. 9, 793–807. doi:10.1111/j.1567-1364.2009.00525.x
Juretzek, T., Le Dall, M., Mauersberger, S., Gaillardin, C., Barth, G., and Nicaud, J. (2001). Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast 18, 97–113. doi:10.1002/1097-0061(20010130)18:2<97:AID-YEA652>3.0.CO;2-U
Katahira, S., Fujita, Y., Mizuike, A., Fukuda, H., and Kondo, A. (2004). Construction of a xylan-fermenting yeast strain through co display of xylanolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 70, 5407–5414. doi:10.1128/AEM.70.9.5407-5414.2004
Kiel, C., and Serrano, L. (2011). Challenges ahead in signal transduction: MAPK as an example. Curr. Opin. Biotechnol 23, 305–314. doi:10.1016/j.copbio.2011.10.004
Kooistra, R., Hooykaas, P. J., and Steensma, H. Y. (2004). Efficient gene targeting in Kluyveromyces lactis. Yeast 21, 781–792. doi:10.1002/yea.1131
Kumar, S., Dheeran, P., Singh, S. P., Mishra, I. M., and Adhikari, D. K. (2014). Bioprocessing of bagasse hydrolysate for ethanol and xylitol production using thermotolerant yeast. Bioprocess Biosyst. Eng. 38, 39–47. doi:10.1007/s00449-014-1241-2
Kurylenko, O. O., Ruchala, J., Hryniv, O. B., Abbas, C. A., Dmytruk, K. V., and Sibirny, A. A. (2014). Metabolic engineering and classical selection of the methylotrophic thermotolerant yeast Hansenula polymorpha for improvement of high temperature xylose alcoholic fermentation. Microb. Cell Fact. 13, 122. doi:10.1186/s12934-014-0122-3
Kwon, Y. J., Ma, A. Z., Li, Q., Wang, F., Zhuang, G. Q., and Liu, C. Z. (2011). Effect of lignocellulosic inhibitory compounds on growth and ethanol fermentation of newly-isolated thermotolerant Issatchenkiaorientalis. Bioresource Technol. 102, 8099–8104. doi:10.1016/j.biortech.2011.06.035
Laplaza, J. M., Torres, B. R., Jin, Y. S., and Jeffries, T. W. (2006). Sh ble and Cre adapted for functional genomics and metabolic engineering of Pichia stipitis. Enzyme Microb. Technol. 38, 741–747. doi:10.1016/j.enzmictec.2005.07.024
Leandro, M. J., Sychrová, H., Prista, C., and Loureiro-Dias, M. C. (2011). The osmotolerant fructophilic yeast Zygosaccharomyces rouxii employs two plasmamembrane fructose uptake systems belonging to a new family of yeast sugar transporters. Microbiology 157, 601–608. doi:10.1099/mic.0.044446-0
Ledesma-Amaro, R., Dulermo, T., and Nicaud, J. M. (2015). Engineering Yarrowia lipolytica to produce biodiesel from raw starch. Biotechnol. Biofuels 8, 148. doi:10.1186/s13068-015-0335-7
Li, M. Z., and Elledge, S. J. (2007). Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256. doi:10.1038/nmeth1010
Madhavan, A., and Sukumaran, R. K. (2014). Promoter and signal sequence from filamentous fungus can drive recombinant protein production in the yeast Kluyveromyces lactis. Bioresour. Technol. 165, 302–308. doi:10.1016/j.biortech.2014.03.002
Madzak, C. (2015). Yarrowia lipolytica: recent achievements in heterologous protein expression and pathway engineering. Appl. Microbiol. Biotechnol. 99, 4559–4577. doi:10.1007/s00253-015-6624-z
Madzak, C., Otterbein, L., Chamkha, M., Moukha, S., Asther, M., Gaillardin, C., et al. (2005). Heterologous production of a laccase from the basidiomycete Pycnoporus cinnabarinus in the dimorphic yeast Yarrowia lipolytica. FEMS Yeast Res. 5, 635–646. doi:10.1534/genetics.114.169060
Medema, M. H., Raaphorst, R., Takano, E., and Breitling, R. (2012). Computational tools for the synthetic design of biochemical pathways. Nat. Rev. Microbiol 10, 191–202. doi:10.1038/nrmicro2717
Miklenic, M., Stafa, A., Bajic, A., Zunar, B., Lisnic, B., and Svetec, I.-K. (2013). Genetic transformation of the yeast Dekkera/Brettanomyces bruxellensis with non-homologous DNA. J. Microbiol. Biotechnol. 23, 674–680. doi:10.4014/jmb.1211.11047
Morin, N., Cescut, J., Beopoulos, A., Lelandais, G., Le Berre, V., Uribelarrea, J. L., et al. (2011). Transcriptomic analyses during the transition from biomass production to lipid accumulation in the oleaginous yeast Yarrowia lipolytica. PLoS ONE 6:e27966. doi:10.1371/journal.pone.0027966
Mukherjee, V., Steensels, J., Lievens, B., Van de Voorde, I., Verplaetse, A., Aerts, G., et al. (2014). Phenotypic evaluation of natural and industrial Saccharomyces yeasts for different traits desirable in industrial bioethanol production. Appl Microbiol Biotechnol 98, 9483–9498. doi:10.1007/s00253-014-6090-z
Näätsaari, L., Mistlberger, B., Ruth, C., Hajek, T., Hartner, F. S., and Glieder, A. (2012). Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS ONE 7:e39720. doi:10.1371/journal.pone.0039720
Nielsen, J. (2015). Yeast cell factories on the horizon. Science 349, 1050–1051. doi:10.1371/journal.pone.0039720
Oberoi, H. S., Babbar, N., Sandhu, S. K., Dhaliwal, S. S., Kaur, U., Chadha, B. S., et al. (2012). Ethanol production from alkali-treated rice straw via simultaneous saccharification and fermentation using newly isolated thermotolerant Pichia kudriavzevii HOP-1. J. Ind. Microbiol. Biotechnol. 39, 557–566. doi:10.1007/s10295-011-1060-2
Pan, P., and Hua, Q. (2012). Reconstruction and in silico analysis of metabolic network for an oleaginous yeast, Yarrowia lipolytica. PLoS One 7:e51535. doi:10.1371/journal.pone.0051535
Papapetridis, I., Dijk, M., Dobbe, A., Metz, B., Pronk, J. T., and Maris, A. J. A. (2016). Improving ethanol yield in acetate-reducing Saccharomyces cerevisiae by cofactor engineering of 6-phosphogluconate dehydrogenase and deletion of ALD6. Microb. Cell Fact. 15, 67. doi:10.1186/s12934-016-0465-z
Park, C. S., Chang, C. C., Kim, J. Y., Ogrydziak, D. M., and Ryu, D. D. (1997). Expression, secretion, and processing of rice alpha-amylase in the yeast Yarrowia lipolytica. J. Biol. Chem. 272, 6876–6881. doi:10.1074/jbc.272.11.6876
Peralta-Yahya, P. P., Zhang, F., del Cardayre, S. B., and Keasling, J. D. (2012). Microbial engineering for the production of advanced biofuels. Nature 488, 320–328. doi:10.1038/nature11478
Petschacher, B., and Nidetzky, B. (2008). Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb. Cell Fact. 7, 9. doi:10.1186/1475-2859-7-9
Piskur, J., Rozpedowska, E., Polakova, S., Merico, A., and Compagno, C. (2006). How did Saccharomyces evolve to become a good brewer? Trends Genet. 22, 183–186. doi:10.1016/j.tig.2006.02.002
Pomraning, K. R., Wei, S., Karagiosis, S. A., Kim, Y. M., Dohnalkova, A. C., Arey, B. W., et al. (2015). Comprehensive metabolomic, lipidomic and microscopic profiling of Yarrowia lipolytica during lipid accumulation identifies targets for increased lipogenesis. PLoS ONE 10:e0123188. doi:10.1371/journal.pone.0123188
Qiao, K., Imam Abidi, S. H., Liu, H., Zhang, H., Chakraborty, S., Watson, N., et al. (2015). Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 29, 56–65. doi:10.1016/j.ymben.2015.02.005
Qin, X., Qian, J., Yao, G., Zhuang, Y., Zhang, S., and Chu, J. (2011). GAP promoter library for fine-tuning of gene expression in Pichia pastoris. Appl. Environ. Microbiol. 77, 3600–3608. doi:10.1128/AEM.02843-10
Ramezani-Rad, M., Hollenberg, C. P., Lauber, J., Wedler, H., Griess, E., Wagner, C., et al. (2003). The Hansenula polymorpha (strain CBS4732) genome sequencing and analysis. FEMS Yeast Res. 4, 207–215. doi:10.1016/S1567-1356(03)00125-9
Redding-Johanson, A. M., Batth, T. S., Chan, R., Krupa, R., Szmidt, H. L., Adams, P. D., et al. (2011). Targeted proteomics for metabolic pathway optimization: application to terpene production. Metab. Eng. 13, 194–203. doi:10.1016/j.ymben.2010.12.005
Ruyters, S., Mukherjee, V., Verstrepen, K. J., Thevelein, J. M., Willems, K. A., and Lievens, B. (2015). Assessing the potential of wild yeasts for bioethanol production. J Ind Microbiol Biotechnol 42, 39–48. doi:10.1007/s10295-014-1544-y
Saraya, R., Krikken, A. M., Kiel, J. A., Baerends, R. J., Veenhuis, M., and van der Klei, I. J. (2012). Novel genetic tools for Hansenula polymorpha. FEMS Yeast Res. 12, 271–278. doi:10.1111/j.1567-1364.2011.00772.x
Schifferdecker, A. J., Siurkus, J., Anderson, M. R., Joerck-Ramberg, D., Ling, Z., Zhou, N., et al. (2016). Alcohol dehydrogenase gene ADH3 activates glucose alcoholic fermentation in genetically engineered Dekkera bruxellensis yeast. Appl. Microbiol. Biotechnol. 100, 3219–3231. doi:10.1007/s00253-015-7266-x
Sharma, N. K., Behera, S., Arora, R., and Kumar, S. (2016). Enhancement in xylose utilization using Kluyveromyces marxianus NIRE-K1 through evolutionary adaptation approach. Bioprocess. Biosyst. Eng 39, 835–843. doi:10.1007/s00449-016-1563-3
Sheng, J., and Feng, X. (2015). Metabolic engineering of yeast to produce fatty acid-derived biofuels: bottlenecks and solutions. Front. Microbiol 6:554. doi:10.3389/fmicb.2015.00554
Sherman, D. J., Martin, T., Nikolski, M., Cayla, C., Souciet, J. L, Durrens, P., et al. (2009). Génolevures: protein families and synteny among complete hemi ascomycetous yeast proteomes and genomes. Nucleic Acids Res. 37, D550–D554. doi:10.1093/nar/gkn859
Shetty, R. P., Endy, D., and Knight, T. F. (2008). Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2, 5. doi:10.1186/1754-1611-2-5
Souciet, J. L., Dujon, B., Gaillardin, C., Johnston, M., Baret, P. V., Cliften, P., et al. (2009). Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 19, 1696–1709. doi:10.1101/gr.091546.109
Swinnen, S., Klein, M., Carrillo, M., Mcinnes, J., Nguyen, T., Nevoigt, E., et al. (2013). Re-evaluation of glycerol utilization in Saccharomyces cerevisiae: characterization of an isolate that grows on glycerol without supporting supplements. Biotechnol. Biofuels 6, 157. doi:10.1186/1754-6834-6-157
Tai, M., and Stephanopoulos, G. (2013). Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 15, 1–9. doi:10.1016/j.ymben.2012.08.007
Teo, W. S., and Chang, M. W. (2014). Development and characterization of AND-gate dynamic controllers with a modular synthetic GAL1 core promoter in Saccharomyces cerevisiae. Biotechnol. Bioeng. 111, 144–151. doi:10.1002/bit.25001
Tsai, C. S., Kwak, S., Turner, T. L., and Jin, Y. S. (2015). Yeast synthetic biology toolbox and applications for biofuel production. FEMS Yeast Res. 15, 1–15. doi:10.1111/1567-1364.12206
Unkles, S. E., Valiante, V., Mattern, D. J., and Brakhage, A. A. (2014). Synthetic biology tools for bioprospecting of natural products in eukaryotes. Chem. Biol. 21, 502–508. doi:10.1016/j.chembiol.2014.02.010
Van den Berg, J. A., Van der Laken, K. J., Van Ooyen, A. J., Renniers, T. C., Rietveld, K., Schaap, A., et al. (1990). Kluyveromyces a host for heterologous gene expression: expression and secretion of prochymosin. Biotechnology 8, 135–139. doi:10.1038/nbt0290-135
Van Ooyen, A. J., Dekker, P., Huang, M., Olsthoorn, M. M., Jacobs, D. I., Coluss, P. A., et al. (2006). Heterologous protein production in the yeast Kluyveromyces lactis. FEMS Yeast Res. 6, 381–392. doi:10.1111/j.1567-1364.2006.00049.x
Verbeke, J., Beopoulos, A., and Nicaud, J. M. (2013). Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol. Lett. 35, 571–576. doi:10.1007/s10529-012-1107-0
Vogl, T., Hartner, F. S., and Glieder, A. (2013). New opportunities by synthetic biology for biopharmaceutical production in Pichia pastoris. Curr. Opin. Biotechnol. 24, 1094–1101. doi:10.1016/j.copbio.2013.02.024
Voronovsky, A. Y., Rohulya, O. V., Abbas, C. A., and Sibirny, A. A. (2009). Development of strains of the thermotolerant yeast Hansenula polymorpha capable of alcoholic fermentation of starch and xylan. Metab. Eng. 11, 234–242. doi:10.1016/j.ymben.2009.04.001
Vroom, J. A., and Wang, C. L. (2008). Modular construction of plasmids through ligation-free assembly of vector components with oligonucleotide linkers. BioTechniques 44, 924–926. doi:10.2144/000112808
Wagner, J. M., and Alper, H. S. (2016). Synthetic biology and molecular genetics in non-conventional yeasts: current tools and future advances. Fungal Genet. Biol. 89, 126–136. doi:10.1016/j.fgb.2015.12.001
Wei, H., Wang, W., Alahuhta, M., Vander Wall, T., Baker, J. O., Decker, S. R., et al. (2014). Engineering towards a complete heterologous cellulase secretome in Yarrowia lipolytica reveals its potential for consolidated bioprocessing. Biotechnol. Biofuels 7, 148. doi:10.1186/s13068-014-0148-0
Wei, N., Quarterman, J., Kim, S. R., Cate, J. H. D., and Jin, Y. (2013). Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat. Commun 4, 2580. doi:10.1038/ncomms3580
Wolf, K. (2012). Nonconventional Yeasts in Biotechnology: A Handbook. Seiten: Springer Science & Business Media.
Xuan, Y., Zhou, X., Zhang, W., Zhang, X., Song, Z., and Zhang, Y. (2009). An upstream activation sequence controls the expression of AOX1 gene in Pichia pastoris. FEMS Yeast Res. 9, 1271–1282. doi:10.1111/j.1567-1364.2009.00571.x
Yanase, S., Hasunuma, T., Yamada, R., Tanaka, T., Ogino, C., Fukuda, H., et al. (2010). Direct ethanol production from cellulosic materials at high temperature using the thermotolerant yeast Kluyveromyces marxianus displaying cellulolytic enzymes. Appl. Microbiol. Biotechnol. 88, 381–388. doi:10.1007/s00253-010-2784-z
Zalatan, J. G., Lee, M. E., Almeida, R., Gilbert, L. A., Whitehead, E. H., La Russa, M., et al. (2015). Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160, 339–350. doi:10.1016/j.cell.2014.11.052
Zhang, C., and Hua, H. (2015). Applications of Genome-Scale Metabolic Models in Biotechnology and Systems Medicine. Front. Physiol. 6, 413. doi:10.3389/fphys.2015.00413
Zhang, B., Li, L., Zhang, J., Gao, X., Wang, D., and Hong, J. (2013). Improving ethanol and xylitol fermentation at elevated temperature through substitution of xylose reductase in Kluyveromyces marxianus. J. Ind. Microbiol. Biotechnol 40, 305–316. doi:10.1007/s10295-013-1230-5
Zhou, Y. J., Buijs, N. A., Zhu, Z., Qin, J., Siewers, V., and Nielsen, J. (2016). Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun 7, 11709. doi:10.1038/ncomms11709
Keywords: synthetic biology, yeast, biofuel, metabolic engineering, ethanol
Citation: Madhavan A, Jose AA, Binod P, Sindhu R, Sukumaran RK, Pandey A and Castro GE (2017) Synthetic Biology and Metabolic Engineering Approaches and Its Impact on Non-Conventional Yeast and Biofuel Production. Front. Energy Res. 5:8. doi: 10.3389/fenrg.2017.00008
Received: 30 September 2016; Accepted: 27 March 2017;
Published: 25 April 2017
Edited by:
Rajesh K. Sani, South Dakota School of Mines and Technology, USAReviewed by:
Neha Srivastava, Banaras Hindu University, IndiaSen Li, Chinese Academy of Sciences, China
Navanietha Krishnaraj, National Institute of Technology Durgapur, India
Copyright: © 2017 Madhavan, Jose, Binod, Sindhu, Sukumaran, Pandey and Castro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raveendran Sindhu, sindhurgcb@gmail.com, sindhufax@yahoo.co.in
 Aravind Madhavan
Aravind Madhavan Anju Alphonsa Jose1
Anju Alphonsa Jose1
 Parameswaran Binod
Parameswaran Binod Raveendran Sindhu
Raveendran Sindhu Rajeev K. Sukumaran
Rajeev K. Sukumaran