Plant-Mediated Changes in Soil N-Cycling Genes during Revegetation of Copper Mine Tailings
- 1School of Resources and Environmental Engineering, Anhui University, Hefei, China
- 2State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing, China
Nitrogen limitation represents a major bottleneck during the revegetation of mine tailings. However, controls over key nitrogen-cycling genes in rhizospheric soils under differential vegetation management remain poorly understood. The abundance and transcriptional activity of nitrogen-cycling genes and the enzymatic activity of nitrogen transformation are mediated differentially during revegetation of mine tailings by Imperata cylindrica and Chrysopogon zizanioides plants. Results showed that the highest total organic carbon (TOC), total nitrogen (TN), and -N contents were found in the rhizosphere of I. cylindrica. The nifH gene abundances differed between I. cylindrica and C. zizanioides, and were higher in I. cylindrica which demonstrated by 3.39-fold higher mRNA transcript abundance of the nifH gene and a 2.15-fold higher nitrogen fixation rate in the rhizosphere. In addition, C. zizanioides exhibited a 4.94-fold higher transcript abundance of the archaeal amoA gene and the highest nitrification rate (1.706 ± 0.293 μg N- g−1 h−1) in the rhizosphere. In conclusion, I. cylindrica and C. zizanioides stimulated the abundances and activities of nifH gene and archaeal amoA gene, respectively. In addition, I. cylindrica appears to be capable of enhancing nitrogen fixation and exhibited accelerated nitrogen accumulation, which may be particularly useful for the rehabilitation of mine tailings.
Introduction
Mine tailings are typical engineered materials (Paerl et al., 2000) that undergo plant colonization followed by ecosystem development in mine wasteland. Revegetation is an effective and practical strategy for mining wasteland management (Mendez and Maier, 2008). To rehabilitate soil biogeochemical functions in mine tailings using the revegetation strategy, it is necessary to concert changes in physical, mineralogical, geochemical, and biological properties to support sustainable plant communities (Huang et al., 2012; Zornoza et al., 2015). Therein, the development of N-cycling microbial communities and their functions is one of the most important aspects of the ecological development of mine tailings (Nelson et al., 2015). However, research on this subject is greatly lacking. Therefore, it is in urgent need to identify the effect of rehabilitation strategies on the nitrogen cycle in mining wastelands.
The selection of plant species is an important factor in rehabilitating the biogeochemical functions of mine tailings, especially when plant species selection is correlated with N-accumulation (Mendez and Maier, 2008; Parraga-Aguado et al., 2014). Plant species should have a competitive advantage (Hobbie, 2015) to use limiting nutrients more efficiently in the mine tailings, and differences in nitrogen use efficiency of different plant species can lead to positive feedbacks on nitrogen cycling (Knops et al., 2002). It is generally known that cluster roots in environments with low inorganic nutrient availability and nitrogen fixing symbioses in low-N environments increase plant access to nutrients in low supply (Chapman et al., 2006; Lambers et al., 2008). In parallel, plants nutrition is also sensitive to ecosystem nitrogen losses via denitrification and nitrogen assimilation through biological nitrogen fixation (Hai et al., 2009; Töwe et al., 2010) in the mining wastelands with low nitrogen content. Previous research indicated that plants played an important role in shaping the microbial community structure and function in the rhizosphere (Henry et al., 2008; Hai et al., 2009; Hussain et al., 2011). For example, the rhizosphere not only significantly affected the diazotrophs (Zhan and Sun, 2011, 2012) but also influenced nitrate reducers and denitrifiers (Henry et al., 2008). Variation in the abundance and diversity of organic acids or other exudates might explain the influence of plants on the abundance and activity of N-cycling microorganisms (Henry et al., 2008; Weng et al., 2013). Moreover, the rhizosphere can alter the relative abundance and number of functional genes (Hai et al., 2009; Hussain et al., 2011; Zornoza et al., 2015) and influence their transcription (Sharma et al., 2005). The rhizosphere effect on nitrogen cycling genes has been widely investigated in many environments (Yergeau et al., 2014; Concilio et al., 2015; Finzi et al., 2015). However, there is little information concerning the relationship between the rhizosphere and the transcription of functional genes involved in nitrogen cycling in the nitrogen-limited mine tailing environment.
There are many copper mine tailings wastelands in Tongling, China, some of which have been naturally or artificially revegetated. A natural Imperata cylindrica community and an artificial Chrysopogon zizanioides community have been successfully established on the Shuimuchong wasteland of copper mine tailings in Tongling. Both the I. cylindrica community and the C. zizanioides community exhibit a simple community structure, and nitrogen in these ecosystems is primarily derived from diazotrophs living in the copper mine tailings; this provides a good opportunity to hypothesize that the rhizosphere could stimulate the abundance and transcription of functional genes involved in nitrogen cycling. And the major aims of this study are to investigate the quantities and transcription of rhizosphere-associated functional genes in a copper tailings pond and to elucidate the specific rhizosphere effects of distinct plants on the abundance and activity of these genes.
Materials and Methods
Study Areas and Sample Collection
The Shuimuchong copper mine tailing wasteland (30°55′N, 117°50′E) is located in the Tongling copper mine area, Anhui Province, East China, and covers a total surface area of 44 ha. The average annual rainfall in this study district is 1,346 mm, the rainy season is from May to September, the average annual temperature is 16.2°C, and the frost-free period ranges from 237 to 258 days (Sun et al., 2004). On this copper mine tailing wasteland, two types of plant communities were established over 10 years ago, a naturally colonized I. cylindrica community and an artificial seeded C. zizanioides community.
The I. cylindrica and C. zizanioides rhizosphere and bulk samples in the top 20 cm of the mine tailings were collected using the method described by Riley and Barber (1970). In brief, bulk tailings and roots were obtained using a soil corer. The roots plus adhering soil were carefully collected, and the rhizosphere tailings adhering to the roots were removed by shaking. In addition, the remainder portions (mine tailings without roots or adhering soil) in the soil corer were set as the bulk tailings. Twelve samples, including three rhizospheric tailings samples (IC-R) and three bulk tailings samples (IC-B) from I. cylindrica, and three rhizospheric tailings samples (CZ-R) and three bulk tailings samples (CZ-B) from C. zizanioides, were obtained in October of 2013. Each of the 12 samples represented a combination of 3 field tailings samples, and each sample was divided into three portions. The first portion was used to analyze chemical properties, the second was used to extract DNA to measure the abundance of functional genes involved in nitrogen cycling, the third was used to detect the transcription of functional genes involved in nitrogen cycling (i.e., RNA analysis). The first two portions were placed into sterilized self-sealing plastic bags, and the third was stored in RNAlater (1 g of soils: 1 mL of RNAlater) (Ambion, Austin, TX). All samples were stored in a box at a temperature ranging from 0 to 4°C in the field. After returning to the laboratory, the second and third portions were stored at −20°C and the first was dried at room temperature, with the exception of the fractions used to analyze ammonium (-N), nitrate (-N) and nitrite (-N) contents.
Analysis of Chemical Properties
The pH values were measured using a pH meter (soil:water = 1 g:5 mL). The total organic carbon (TOC) was measured as the mass loss of ignition in a muffle furnace at 550 ± 5°C for 6 h to determine total organic matter in mine wastelands. Total nitrogen (TN) was measured using the Kjeldahl method. Ammonium (-N), nitrate (-N) and nitrite (-N) were extracted from fresh samples in 2 mol·L−1 KCl and measured by UV spectrophotometry (Nanjing Institute of Soil Science, 1978).
Nitrogen Fixation Rate, Nitrification Rate and Denitrification Rate
Nitrification rates were measured by culturing fresh tailing samples in 250-mL sterile bottles without any N-amendments. A control (cultured without C2H2) and C2H2 amended bottles were incubated for each sample. C2H2 was added to the headspace at a final partial pressure of 10 Pa (0.01%). Bottles were incubated at 25°C in the dark, and sample analysis was carried out after 2 weeks of incubation. During incubation, bottles were opened and vented every 2 days to maintain aerobic conditions, and C2H2 partial pressure was re-established after resealing. The nitrification rate was calculated as the difference between the nitrite concentration in the control and C2H2-treated samples.
The nitrogen fixation rate was measured in a C2H2 reduction assay in which nitrogenase reduction of C2H2-C2H4 was measured. Denitrification was simultaneously measured by accumulation of N2O because C2H2 inhibits N2O reduction and nitrification (Klemedtsson et al., 1988). Fresh tailings were cultured in 250-mL sterile bottles, and 10% of the headspace was evacuated and replaced with C2H2, and sterile water was used to keep the soil moist. After sealing the bottles for culturing at 25°C, the gas was measured at 4, 8 h, 1, 3, 7, and 14 d (Figure S1). To measure nitrogen fixation, the evolution of C2H2-C2H4 was measured with a gas chromatograph, equipped with a thermal conductivity detector (TCD) and a GDX-501 packed column using N2 as a carrier gas. The temperatures of the detector, injector, and oven were 107, 110, and 60°C, respectively. For denitrification, the N2O emissions were analyzed with an electron capture detector (ECD) connected to a Porapak Q packed column (length: 5 m; O.D.: 3 mm) using N2 as a carrier gas. The temperatures of the detector, injector, and oven were 360, 90, and 50°C, respectively. The denitrification rate was calculated as the difference in N2O produced during the incubation of the control and C2H2-treated samples.
DNA Extraction, RNA Extraction and Inverse Transcription
Total DNA was extracted with a PowerSoil DNA Isolation Kit (MoBio, Carlsbad, USA) according to the manufacturer's instructions. Total RNA was extracted employing the mRNA extraction method described by Mettel et al. (2010). RNA-containing samples in RNAlater were thawed on ice, and supernatants were removed by centrifugation at 20,000 × g for 1 min. Pelleted RNA was shocked at 6 m·s−1 for 35 s with 0.5 g of beads and 700 μL of TPM [50 mmol·L−1 Tris-HCl (pH 8.0); 20 mmol·L−1 MgCl2·6H2O; 2% polyvinylpyrrolidone (PVP)] and then centrifuged at 20,000 × g for 2 min to collect the supernatant. Excess pellet was shocked at 6 m·s−1 for 35 s with 700 μL of PBL (5 mmol·L−1 Tris-HCl (pH 8.0); 50 mmol·L−1 EDTA; 1% SDS; 6% Phenol Water), and centrifuged at 20,000 × g for 2 min to collect the supernatant to mix with the previously obtained supernatant. RNA was extracted with 500 μL of phenol water (pH 4.5), followed by successive treatments with 500 μL of phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1). RNA was precipitated from the supernatant by adding 1 mL of PEG-NaCl (30% PEG-6000; 1.6 mol·L−1 NaCl) and the samples were maintained at room temperature for 2 h. The precipitated RNA was cleaned via the sequential addition of 70% (v/v) ethanol and 40% (v/v) ethanol, and the supernatant was removed following centrifugation at 20,000 × g for 5 min. RNA samples were dissolved in 42.5 μL of water. The DNA was removed with DNase I (TransGen Biotech, Beijing, PRC) and then the total RNA was purified with an RNeasy MinElute cleanup kit (QIAGEN, Hilden, Germany). RNA samples were reverse transcribed into cDNA with TransScript II All-in-one first-strand cDNA supermix for qPCR (TransGen Biotech, Beijing, PRC).
Quantification of DNA and cDNA by Real-Time Quantitative PCR (qPCR)
The PCR primers used to amplify regions of functional genes from nitrogen fixers, nitrifiers, and denitrifiers are listed in Table 1. Amplification was carried out in a total volume of 20 μL containing 2 μL of diluted template DNA, 0.4 μL of each primer (concentration of 10 μM), 0.4 μL of passive reference dye(50×), 6.8 μL of ddH2O, and 10 μL of KAPA SYBR@ FAST qPCR Kit Master Mix (2×) Universal (KAPA Biosystems, Boston, US). All samples and standard reactions were performed in triplicate using a StepOne Real-Time PCR System (ABI, New York, US). Standard curve templates were constructed with 10-fold serial dilutions of linearized recombinant plasmids harboring amplicons amplified from mine tailings samples. Standard curves for the real-time PCR were developed as follows. The genes were amplified from the total genomic DNA using the same primers as in the qPCR. The PCR products were purified by Easypure Quick Gel extraction kit (TransGen Biotech, China) and cloned into a pEASY-T3 clone vector (TransGen Biotech, China). The resulting ligation mixes were transformed into a Trans1-T1 phageresistant chemically competent cell (TransGen Biotech, China). Plasmids were used as the standard from the positive clone, and the standard sequences were deposited in the GenBank Data Library under accession numbers MG256653-MG256661. The recombinant plasmids containing one copy of a gene fragment were diluted from 1:102 to 1:107 copy·μl−1 (list in Table 1) and used as targets for qPCR standard curves. The PCR program for total bacterial quantification was as follows, according to the manufacturer's instructions: 3 min at 95°C, followed by 40 cycles of 3 s at 95°C, and 25 s at 60°C. After each qPCR assay, the amplification specificity was verified by generating melting curves and performing agarose gel electrophoresis. Standard curves were constructed based on triplicate 10-fold dilutions of a DNA standard containing a known copy number. Negative controls without template DNA were included in all experiments to monitor for contamination. The real-time PCR standard curves for all functional genes were repeated three times. Gene copy numbers were calculated by comparing threshold cycles obtained in each PCR run with those of known standard DNA concentrations. Each R2-value of each standard curve for each replicate was determined to be above 0.99.
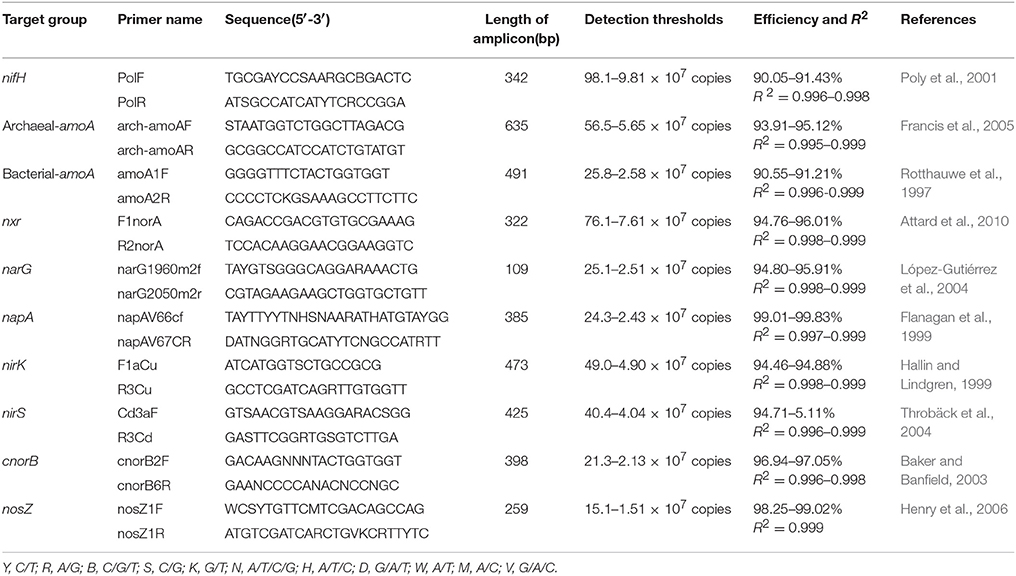
Table 1. Primer pairs used in real-time qPCR analysis for amplification of nitrogenase (nifH), bacterial and archaeal ammonia monooxygenase (amoA), nitrite oxidoreductase (nxr), nitrate reductase (narG and napA), nitrite reductase (nirK and nirS), nitric oxide reductase (cnorB), and nitrous oxide reductase (nosZ) genes.
Data Analysis
Multivariate analysis of variance (MANOVA) was used to analyze the mixed effects of plant species, rhizosphere, and their interaction on chemical properties and the abundances and transcripts of functional genes. One-way analysis of variance (ANOVA, LSD post-hoc test with a significance at P < 0.05) was used to analyze the chemical properties, quantities and transcript abundances of the functional genes in different samples. A Pearson correlation analysis was carried out using SPSS 19.0 to test the relationships between functional gene quantities and chemical properties, the level of significance was set at P < 0.05 (two-tailed), and the false discovery rate (q) was calculated to establish significance of correlations. Regression analysis was performed to structure the linear relationships between the nitrogen transformation rate and the gene and transcript abundances of functional genes. A redundancy analysis (RDA) was performed in the R environment and explanatory variables were chosen with 999 permutations using the “permutest” function of the vegan package, with RDA axes plotted with solid arrows representing chemical variables significantly associated (P < 0.05) with the variation. Variation partitioning was carried out using the “varpart” function in R.
Results
Soil Biogeochemical Properties
The multivariate analysis of variance (MANOVA) of mixed effects of plant species (P), rhizosphere (R) and their interaction on the chemical properties and nitrogen transformation rates (Table 2) showed that plant species significant influenced the soil chemical properties including TOC, TN, C:N ratio, -N, -N, and -N contents, as well as the nitrogen fixation rate and nitrification rate. The rhizosphere significant impacted the inorganic nitrogen (-N, -N, and -N) contents and nitrogen fixation rate. In addition, the interaction effect of plant species and rhizosphere on the chemical properties and nitrogen transformation rates was pointed out in TOC, -N and -N contents as well as the nitrogen fixation rate and nitrification rate.
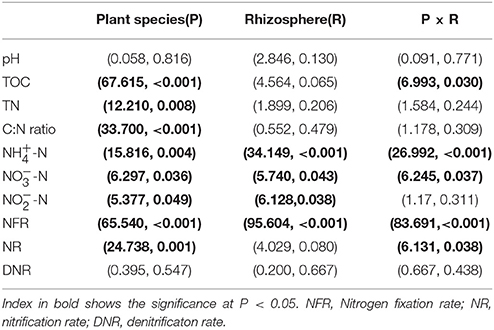
Table 2. Multivariate analysis of variance (MANOVA) (F-value, P-value) of mixed effects of plant species (P), rhizosphere (R) and their interaction on the chemical properties and nitrogen transformation rates.
Variance analysis (ANOVA) showed that TOC was significantly higher under IC than CZ in both the rhizosphere and bulk tailings (Table 3). TN and -N contents were significantly higher in IC than CZ for the rhizospheric samples (Table 3). In addition, the rhizospheric sample of I. cylindrica has the highest nitrogen fixation rate (Table 4). Thus, I. cylindrica was more effective in the formation and accumulation of soil organic matter and nitrogen in copper mine tailings than C. zizanioides.
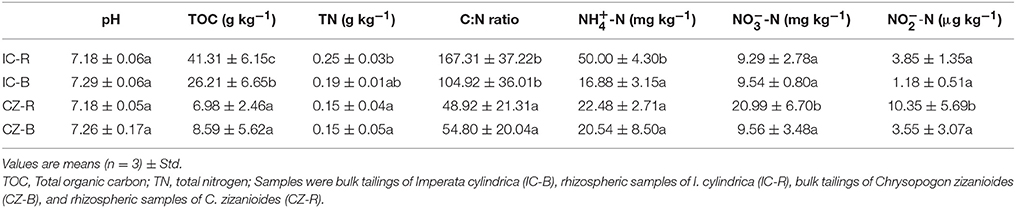
Table 3. Soil basic chemical properties of bulk tailings and rhizospheric samples of Imperata cylindrica and Chrysopogon zizanioides; lowercase letters indicate significant differences in each column (LSD, P < 0.05).
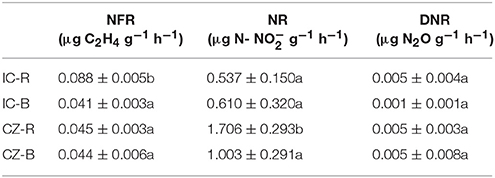
Table 4. The rates of nitrogen fixation (NFR), nitrification (NR) and denitrificaton (DNR) in bulk tailings and rhizospheric samples of plant species; lowercase letters indicate significant differences in each column (LSD, P < 0.05).
When the rhizosphere and bulk tailings of I. cylindrica were compared, the former exhibited higher TOC and -N contents than the latter (Table 3). In contrast to I. cylindrica, the rhizospheric tailings of C. zizanioides contained higher -N and -N contents than the bulk tailings. And the the rhizospheric sample of I. cylindrica has the highest nitrification rate (Table 4).
The effects of plant species on the pH values of the rhizosphere and bulk tailings were not significant (Table 3). However, plant species affected the TOC and TN content, and both the rhizosphere and bulk tailings of I. cylindrica exhibited higher C:N ratios than those of C. zizanioides.
Effects of the Rhizosphere on Functional Genes Involved in the Nitrogen Cycle
The gene and transcript abundances of ten genes involved in the nitrogen cycle, specifically nifH, arch-amoA, bact-amoA, nxr, narG, napA, nirK, nirS, norB, and nosZ, were measured by qPCR (Table 1). The primers used to amplify the functional genes (such as nifH gene) for qPCR do not target all genes, so only a subset would be quantified in the assay. Even so, the plant species merely impacted the abundance of nifH and nxr genes and the transcripts of nifH and amoA (including archaeal amoA and bacterial amoA) genes, and the plant rhizosphere and the interaction of plant species and rhizosphere influenced the transcripts of nifH and archaeal amoA genes (Table 5). In parallel, the abundances of these ten functional genes were not significantly different between the rhizosphere and bulk tailings (Figure 1). However, there were significant differences in transcriptional activities of the nifH gene between the rhizosphere and bulk tailings for I. cylindrica, and for the arch-amoA gene for C. zizanioides (Figure 2). The rhizosphere of I. cylindrica demonstrated a 3.39-fold higher transcript abundance of the nifH gene than in bulk tailings, while the transcript abundance of the archaeal amoA gene in the rhizospheric samples of C. zizanioides were 4.94-fold higher than those in bulk tailings.
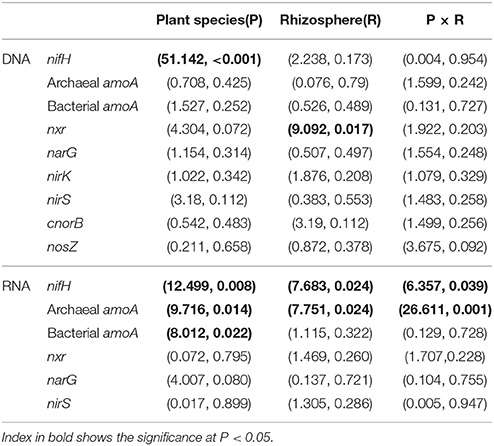
Table 5. Multivariate analysis of variance (MANOVA) (F-value, P-value) of mixed effects of plant species (P), rhizosphere (R) and their interaction on the abundances and transcripts of functional genes.
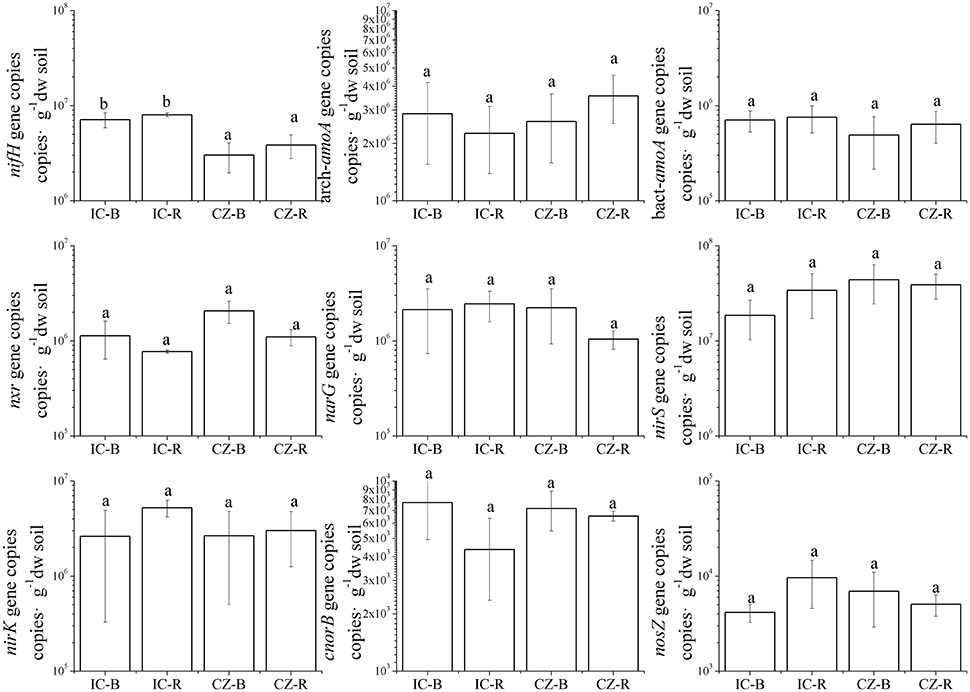
Figure 1. Abundances of genes involved in nitrogen cycling in the rhizosphere and bulk soils of Imperata cylindrica and Chrysopogon zizanioides. Different letters indicate significant differences in samples (LSD, P < 0.05). Samples were bulk tailings of I. cylindrica (IC-B), rhizosphere samples of I. cylindrica (IC-R), bulk tailings of C. zizanioides (CZ-B), and rhizosphere samples of C. zizanioides (CZ-R).
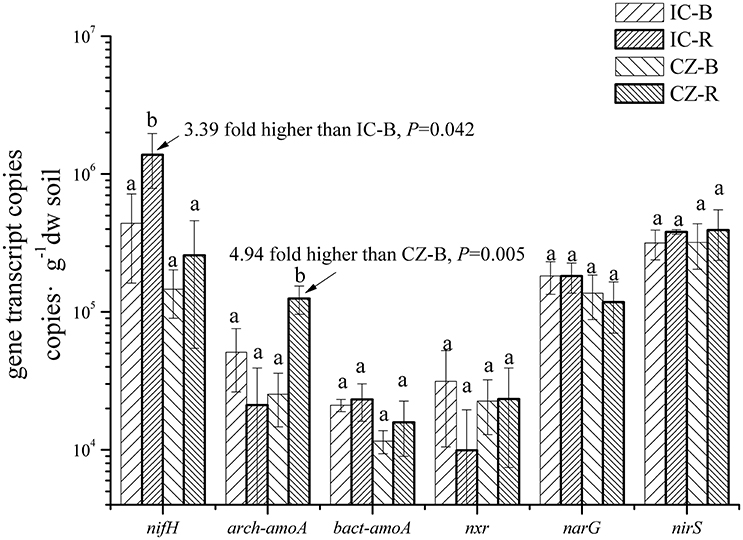
Figure 2. Transcript abundances of genes involved in nitrogen cycling in the rhizosphere and bulk soil of I. cylindrica and C. zizanioides. Different letters indicate significant differences in each gene of samples (LSD, P < 0.05). Samples were bulk talings of I. cylindrica (IC-B), rhizosphere samples of I. cylindrica (IC-R), bulk tailings of C. zizanioides (CZ-B), and rhizosphere samples of C. zizanioides (CZ-R).
Moreover, nifH gene abundance differed significantly between I. cylindrica and C. zizanioides. The nifH gene abundances in both the rhizosphere and bulk tailings of I. cylindrica were significantly higher than those of C. zizanioides (Figure 1).
The quantities of the napA genes, which participate in nitrate reduction, were lower than the detection limit (data not shown). Although the abundance of the nirK gene was >2.0 × 106 copies·g−1 dw soil, its gene transcript abundances were below the detection limit. Similar to the nirK gene, the gene transcript abundance of norB and nosZ were also below the detection limit (data not shown).
Correlations between Gene Abundance, Transcript Abundance and the Chemical Properties of Copper Mine Tailings
The nitrogen fixation rate was significantly enhanced by the increased nifH gene abundance and transcription (Table S1; for gene abundance: YNFR = 0.027 + 4.987 × 10−9 XnifHcopy, F = 5.042, P = 0.049; for transcript abundance: YNFR = 0.038 + 2.921 × 10−8 XnifHtranscript, F = 23.704, P = 0.001). Finally, the nitrification rate was enhanced by the increased transcript abundance of the arch-amoA gene (Table S1; YDNR = 0.031 + 4.475 × 10−7 Xarch−amoAtranscript, F = 8.736, P = 0.014).
A redundancy analysis (RDA, Figures 3A,C) revealed that TOC, TN and the ratio of C:N were the primary environmental factors (according to forwards with a P < 0.05) defining the abundance of microbial communities involved in the nitrogen cycle. TOC, TN, and -N were the primary dimensions affecting the transcript abundance of the microbial communities involved in the nitrogen cycle. Furthermore, variation partitioning indicated that the total N and -N contents explained the most variance in gene abundance (Figure 3B) and transcript abundance (Figure 3D), respectively. In addition, the gene and transcript abundances of the nifH gene were positively correlated with the TOC and -N content, respectively (Table S1). In particular, the nifH transcript was significantly enhanced by increasing the -N content (YnifHtranscript = −2.82 × 105 + 3.02 × 104 Xammonia, F = 13.403, P = 0.004).
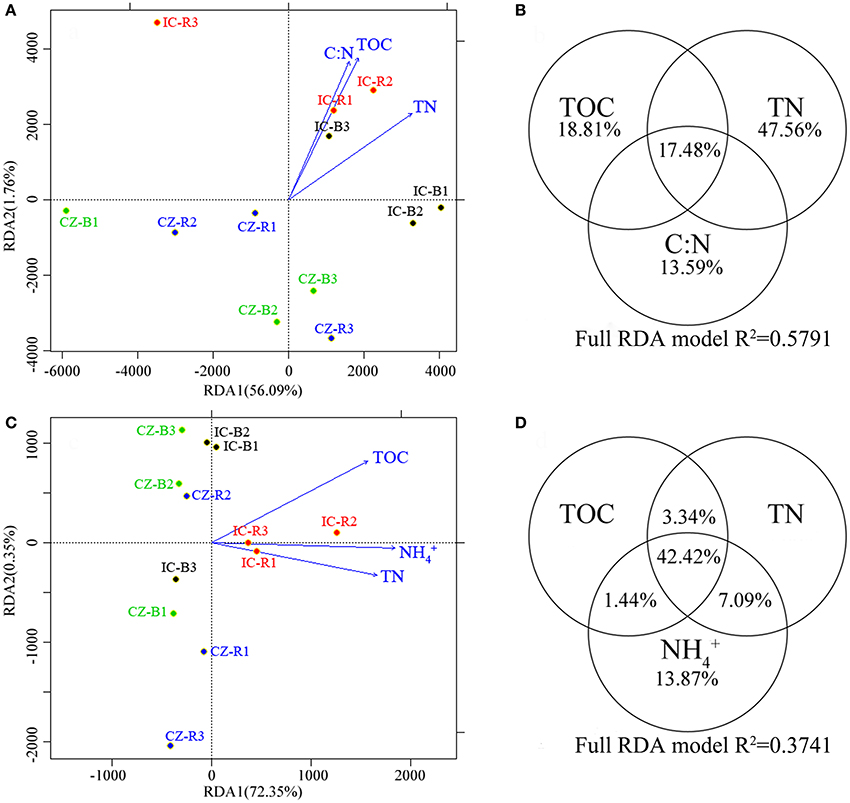
Figure 3. A redundancy analysis (RDA) of the abundances (A) and transcript abundance (C) of microbial functional genes involved in the nitrogen cycle, and variance explained by the most important chemical properties in microbial functional gene abundance (B) and transcript abundance (D). The circled values indicate the contributions of a chemical property to variations in microbial functional gene abundance or transcript abundance. Samples were bulk tailings of I. cylindrica (IC-B), rhizospheric samples of I. cylindrica (IC-R), bulk tailings of C. zizanioides (CZ-B), and rhizospheric samples of C. zizanioides (CZ-R).
Discussion
The major objective was to assess whether different plant communities influenced the N-cycling of functional microbes in the rhizosphere, which were reflected by the abundances and transcripts of N-cycling genes. The comparison between rhizosphere and bulk tailings next to the plant roots in this study could reflect that which genes were most heavily affected. The microbes harboring N-cycling genes in this study included nitrogen-fixing bacteria (with nifH gene), nitrifiers (including ammonia-oxidizing bacteria with the bacterial amoA gene, ammonia-oxidizing archaea with the archaeal amoA gene and nitrate bacteria with the nxr gene) and denitrifiers (microbes participating in one or more reduction reaction with the genes narG, napA, nirS, nirK, norB, and nosZ). In this study, the gene transcript abundance of nirK, norB, and nosZ were below the detection limit, maybe nitrite reduction was not the primary energy source for these denitrifiners in mine tailings. The general knowledge that plant roots can stimulate rhizosphere heterotrophs and activate root-derived N in a soil environment (Thion et al., 2016); formed the basis of our hypothesis that the rhizosphere stimulates the abundances and activities of N-cycling functional microorganisms. Although The primers in the study used to amplify the functional genes for qPCR do not target all genes, the two assayed plants I. cylindrica and C. zizanioides stimulated the abundances and activities of some nifH gene and archaeal amoA gene, respectively. The rhizosphere in this study hardly influenced the abundances of the denitrifiers and the activities of denitrification rate activities. In the paddy and forest soil, the denitrification was unapparent under neutral aerobic (Lan et al., 2015; Pajares and Bohannan, 2016) and/or nitrogen deficiency conditions (Pajares and Bohannan, 2016). In addition, root exudate carbon was previously observed to be effective at decreasing the risk of N loss by increasing the potential for heterotrophic microbial immobilization (Fisk et al., 2015) and plant roots might not provide an appropriate survival environment for denitrifiers (Fisk et al., 2015). Under aerobic conditions, the nitrogen loss was mainly attributable to ammonia oxidizers (Florio et al., 2016), and in this study the stimulating effects of plant roots on ammonia oxidizers still did not aid the accumulation of nitrogen. In the revegetation area, plant litter and root exudate contributed a large amount of organic nitrogen (Baxendale et al., 2014; Thion et al., 2016), and the inorganic nitrogen mineralized by this organic nitrogen was provided to plants and microbes (Thion et al., 2016). In the area revegetated by C. zizanioides, the phytocommunity obtained the inorganic nitrogen in the process of nitrogen transformation and grew well, but the TN in the tailings experienced no improvement compared to the bare wastelands assayed in our previous study (Li et al., 2016a). In contrast, plant-derived C from I. cylindrica stimulated the activity of nitrogen fixing bacteria. The organic carbon released by the root exudation and litter deposition of I. cylindrica increased significantly, causing a higher C:N ratio in the study field. Although no significant correlation between the C:N ratio and nifH gene and transcript abundance or nitrogen fixation rate was exhibited in this study, it has been confirmed that nitrogen-fixing bacteria could increase in abundance with the C:N ratio (Bürgmann et al., 2003).
Plant species selection is a critical issue for the long-term success of phytoremediation in mine tailings (Mendez and Maier, 2008; Parraga-Aguado et al., 2014). Apart from showing tolerance to extreme pH values, high metal concentrations, drought, or salinity, plants must also address nitrogen limitation (Mendez and Maier, 2008; Zornoza et al., 2015). The two selected plant species in this field study exerted significant differential effects on N transformation under the nitrogen-limiting conditions. Plant biomass does not explain this difference, because the below-ground biomass of C. zizanioides was similar to that of I. cylindrica (Li et al., 2016b), whereas the aboveground biomass of C. zizanioides was higher (Shu et al., 2002). There are many causes of this rhizosphere effect, including different types and contents of organic acids or other exudates produced by roots (Henry et al., 2008; Weng et al., 2013). The rhizosphere processes are an important component of the terrestrial nitrogen cycle (Finzi et al., 2015), and that the nitrogen increase by plants such as I. cylindrica exceeds their utilization, particularity with respect to nutrient uptake (Finzi et al., 2015). The TN was the most important factor influencing nitrogen cycle functional genes under neutral conditions in this field study. Plant responses to increased TN content may be critical for microbial adaptation (Huang et al., 2015b), and increased N may lead to higher vegetation density and enhance the competition between plants and microbes (Huang et al., 2015a). In addition, microbial biomass and community structure also influences soil nitrogen cycling by interacting with plants, modulating soil properties (Waring et al., 2013), and immobilizing nutrients (Deng et al., 2016). Therefore, plant-derived C inputs by I. cylindrica stimulated nitrogen fixation and accelerated the nitrogen accumulation, which may be particularly useful for the rehabilitation of mine tailings, and might explain the extensive distribution of this plant in many different habitats (Tabor, 1949; Macdonald, 2004).
Changes in transcription of the nifH and arch-amoA genes were detected, although only a subset of some genes would be quantified in the qPCR assays. And few of the other genes demonstrated a significant response to plant species or rhizosphere, suggesting that nitrogen fixers and archaeal ammonia oxidizers are sensitive to the effects of plant species and the rhizosphere. This sensitivity may be associated with the presence of nitrogen-poor substrates. Nitrogen fixers are a vital source of nitrogen in nitrogen-poor environments, which are widely found in different types of soils (Bürgmann et al., 2003; Coelho et al., 2009; Huang et al., 2011; Chinnadurai et al., 2014) and rhizospheres (Meng et al., 2012). In our previous studies, the application and establishment of certain types of vegetation in mine tailings promotes the presence of important free-living nitrogen fixers in these ecosystems (Zhan and Sun, 2011, 2012). And some studies (Huang et al., 2011; Chinnadurai et al., 2014; Collavino et al., 2014) also indicated that -N improved the abundance and diversity of nitrogen-fixing bacteria. Moreover, stimulation of nitrification in the rhizosphere of C. zizanoides was associated with increases in archaeal ammonium monooxygenase (arch-amoA) transcript abundance. In contrast, no significant differences between the rhizospheres and bulk tailings were identified for bacterial genes involved in nitrification (bact-amoA and nxr) (Figures 1, 2). Archaeal amoA and bacterial amoA genes are known to dominate nitrification activity in different soil niches based on -N availability (Taylor et al., 2012; Sun et al., 2015). Ammonia-oxidizing archaea seem to prefer the low-NH3 substrate conditions in mine tailings, and are more sensitive to changes in soil properties (Di et al., 2010), although no statistical correlation between the gene and transcript abundances of archaeal amoA and -N content was observed (Pearson correlation: r = −0.169, Padjust = 0.829; r = −0.311, Padjust = 0.648, respectively).
In conclusion, our data did not support the hypothesis that the rhizosphere stimulates the abundances and activities of N-cycling functional microorganisms. The nitrogen increased by C. zizanoides was used by plants and microbes as well as reduced by ammonia-oxidizing archaea, which caused no improvement in the TN in mine tailings. In contrast, plant-derived C from I. cylindrica stimulated the nitrogen fixation process in the rhizosphere and further accelerated the nitrogen accumulation in mine tailings. Therefore, we suggest that I. cylindrica exerts much stronger effects on nitrogen accumulation than C. zizanioides, the two selected plant species in this field study.
Author Contributions
YL, ZJ, and QS designed the study; YL, JC, YY, JZ, and DW targeted the research question; YL contributed to writing the manuscript; QS found the funding to carry out the study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (Grant No. 41171418).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2017.00079/full#supplementary-material
References
Attard, E., Poly, F., Commeaux, C., Laurent, F., Terada, A., Smets, B. F., et al. (2010). Shifts between Nitrospira- and Nitrobacter-like nitrite oxidizers underlie the response of soil potential nitrite oxidation to changes in tillage practices. Environ. Microbiol. 12, 315–326. doi: 10.1111/j.1462-2920.2009.02070.x
Baker, B. J., and Banfield, J. F. (2003). Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44, 139–152. doi: 10.1016/S0168-6496(03)00028-X
Baxendale, C., Orwin, K. H., Poly, F., Pommier, T., and Bardgett, R. D. (2014). Are plant–soil feedback responses explained by plant traits? New Phytol. 204, 408–423. doi: 10.1111/nph.12915
Bürgmann, H., Widmer, F., Sigler, W. V., and Zeyer, J. (2003). mRNA extraction and reverse transcription-PCR protocol for detection of nifH gene expression by Azotobacter vinelandii in soil. Appl. Environ. Microbiol. 69, 1928–1935. doi: 10.1128/AEM.69.4.1928-1935.2003
Chapman, S. K., Langley, J. A., Hart, S. C., and Koch, G. W. (2006). Plants actively control nitrogen cycling: uncorking the microbial bottleneck. New Phytol. 169, 27–34. doi: 10.1111/j.1469-8137.2005.01571.x
Chinnadurai, C., Gopalaswamy, G., and Balachandar, D. (2014). Long term effects of nutrient management regimes on abundance of bacterial genes and soil biochemical processes for fertility sustainability in a semi-arid tropical Alfisol. Geoderma 232–234, 563–572. doi: 10.1016/j.geoderma.2014.06.015
Coelho, M. R. R., Marriel, I. E., Jenkins, S. N., Lanyon, C. V., Seldin, L., and O'donnell, A. G. (2009). Molecular detection and quantification of nifH gene sequences in the rhizosphere of sorghum (Sorghum bicolor) sown with two levels of nitrogen fertilizer. Appl. Soil Ecol. 42, 48–53. doi: 10.1016/j.apsoil.2009.01.010
Collavino, M. M., Tripp, H. J., Frank, I. E., Vidoz, M. L., Calderoli, P. A., Donato, M., et al. (2014). nifH pyrosequencing reveals the potential for location-specific soil chemistry to influence N2-fixing community dynamics. Environ. Microbiol. 16, 3211–3223. doi: 10.1111/1462-2920.12423
Concilio, A., Vargas, T., and Cheng, W. (2015). Rhizosphere-mediated effects of the invasive grass Bromus tectorum L. and native Elymus elymoides on nitrogen cycling in Great Basin Desert soils. Plant Soil 393, 245–257. doi: 10.1007/s11104-015-2482-9
Deng, Q., Cheng, X., Hui, D., Zhang, Q., Li, M., and Zhang, Q. (2016). Soil microbial community and its interaction with soil carbon and nitrogen dynamics following afforestation in central China. Sci. Total Environ. 541, 230–237. doi: 10.1016/j.scitotenv.2015.09.080
Di, H. J., Cameron, K. C., Shen, J. P., Winefield, C. S., O'callaghan, M., Bowatte, S., et al. (2010). Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol. Ecol. 72, 386–394. doi: 10.1111/j.1574-6941.2010.00861.x
Finzi, A. C., Abramoff, R. Z., Spiller, K. S., Brzostek, E. R., Darby, B. A., Kramer, M. A., et al. (2015). Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Glob. Chang. Biol. 21, 2082–2094. doi: 10.1111/gcb.12816
Fisk, L. M., Barton, L., Jones, D. L., Glanville, H. C., and Murphy, D. V. (2015). Root exudate carbon mitigates nitrogen loss in a semi-arid soil. Soil Biol. Biochem. 88, 380–389. doi: 10.1016/j.soilbio.2015.06.011
Flanagan, D. A., Gregory, L. G., Carter, J. P., Karakas-Sen, A., Richardson, D. J., and Spiro, S. (1999). Detection of genes for periplasmic nitrate reductase in nitrate respiring bacteria and in community DNA. FEMS Microbiol. Lett. 177, 263–270. doi: 10.1111/j.1574-6968.1999.tb13742.x
Florio, A., Felici, B., Migliore, M., Dell'abate, M. T., and Benedetti, A. (2016). Nitrogen losses, uptake and abundance of ammonia oxidizers in soil under mineral and organo-mineral fertilization regimes. J. Sci. Food Agric. 96, 2440–2450. doi: 10.1002/jsfa.7364
Francis, C. A., Roberts, K. J., Beman, J. M., Santoro, A. E., and Oakley, B. B. (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 102, 14683–14688. doi: 10.1073/pnas.0506625102
Hai, B., Diallo, N. H., Sall, S., Haesler, F., Schauss, K., Bonzi, M., et al. (2009). Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl. Environ. Microbiol. 75, 4993–5000. doi: 10.1128/AEM.02917-08
Hallin, S., and Lindgren, P.-E. (1999). PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 65, 1652–1657.
Henry, S., Bru, D., Stres, B., Hallet, S., and Philippot, L. (2006). Quantitative detection of the nosZ Gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72, 5181–5189. doi: 10.1128/AEM.00231-06
Henry, S., Texier, S., Hallet, S., Bru, D., Dambreville, C., Chèneby, D., et al. (2008). Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ. Microbiol. 10, 3082–3092. doi: 10.1111/j.1462-2920.2008.01599.x
Hobbie, S. E. (2015). Plant species effects on nutrient cycling: revisiting litter feedbacks. Trends Ecol. Evol. 30, 357–363. doi: 10.1016/j.tree.2015.03.015
Huang, G., Li, Y., and Padilla, F. M. (2015a). Ephemeral plants mediate responses of ecosystem carbon exchange to increased precipitation in a temperate desert. Agric. For. Meteorol. 201, 141–152. doi: 10.1016/j.agrformet.2014.11.011
Huang, G., Li, Y., and Su, Y. G. (2015b). Divergent responses of soil microbial communities to water and nitrogen addition in a temperate desert. Geoderma 251–252, 55–64. doi: 10.1016/j.geoderma.2015.03.018
Huang, L., Baumgartl, T., and Mulligan, D. (2012). Is rhizosphere remediation sufficient for sustainable revegetation of mine tailings? Ann. Bot. 110, 223–238. doi: 10.1093/aob/mcs115
Huang, L. N., Tang, F. Z., Song, Y. S., Wan, C. Y., Wang, S. L., Liu, W. Q., et al. (2011). Biodiversity, abundance, and activity of nitrogen-fixing bacteria during primary succession on a copper mine tailings. FEMS Microbiol. Ecol. 78, 439–450. doi: 10.1111/j.1574-6941.2011.01178.x
Hussain, Q., Liu, Y., Jin, Z., Zhang, A., Pan, G., Li, L., et al. (2011). Temporal dynamics of ammonia oxidizer (amoA) and denitrifier (nirK) communities in the rhizosphere of a rice ecosystem from Tai Lake region, China. Appl. Soil Ecol. 48, 210–218. doi: 10.1016/j.apsoil.2011.03.004
Klemedtsson, L., Svensson, B. H., and Rosswall, T. (1988). A method of selective inhibition to distinguish between nitrification and denitrification as sources of nitrous oxide in soil. Biol. Fertil. Soils 6, 112–119. doi: 10.1007/BF00257659
Knops, J. M. H., Bradley, K. L., and Wedin, D. A. (2002). Mechanisms of plant species impacts on ecosystem nitrogen cycling. Ecol. Lett. 5, 454–466. doi: 10.1046/j.1461-0248.2002.00332.x
Lambers, H., Raven, J. A., Shaver, G. R., and Smith, S. E. (2008). Plant nutrient-acquisition strategies change with soil age. Trends Ecol. Evol. 23, 95–103. doi: 10.1016/j.tree.2007.10.008
Lan, T., Han, Y., and Cai, Z. (2015). Denitrification and its product composition in typical Chinese paddy soils. Biol. Fertil. Soils 51, 89–98. doi: 10.1007/s00374-014-0953-4
Li, Y., Jia, Z., Sun, Q., Zhan, J., Yang, Y., and Wang, D. (2016a). Ecological restoration alters microbial communities in mine tailings profiles. Sci. Rep. 6:25193. doi: 10.1038/srep25193
Li, Y., Sun, Q., Zhan, J., Yang, Y., and Wang, D. (2016b). Vegetation successfully prevents oxidization of sulfide minerals in mine tailings. J. Environ. Manage. 177, 153–160. doi: 10.1016/j.jenvman.2016.04.026
López-Gutiérrez, J. C., Henry, S., Hallet, S., Martin-Laurent, F., Catroux, G., and Philippot, L. (2004). Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 57, 399–407. doi: 10.1016/j.mimet.2004.02.009
Macdonald, G. E. (2004). Cogongrass (Imperata cylindrica)-biology, ecology, and management. CRC. Crit. Rev. Plant Sci. 23, 367–380. doi: 10.1080/07352680490505114
Mendez, M. O., and Maier, R. M. (2008). Phytostabilization of mine tailings in arid and semiarid environments-an emerging remediation technology. Environ. Health Perspect. 116, 278–283. doi: 10.1289/ehp.10608
Meng, X., Wang, L., Long, X., Liu, Z., Zhang, Z., and Zed, R. (2012). Influence of nitrogen fertilization on diazotrophic communities in the rhizosphere of the Jerusalem artichoke (Helianthus tuberosus L.). Res. Microbiol. 163, 349–356. doi: 10.1016/j.resmic.2012.03.005
Mettel, C., Kim, Y., Shrestha, P. M., and Liesack, W. (2010). Extraction of mRNA from Soil. Appl. Environ. Microbiol. 76, 5995–6000. doi: 10.1128/AEM.03047-09
Nanjing Institute of Soil Science (1978). Soil Physical and Chemical Analysis. Shanghai: Shanghai Science and Technology Press.
Nelson, K. N., Neilson, J. W., Root, R. A., Chorover, J., and Maier, R. M. (2015). Abundance and activity of 16S rRNA, amoA and nifH bacterial genes during assisted phytostabilization of mine tailings. Int. J. Phytoremediation 17, 493–502. doi: 10.1080/15226514.2014.935284
Paerl, H. W., Pinckney, J. L., and Steppe, T. F. (2000). Cyanobacterial–bacterial mat consortia: examining the functional unit of microbial survival and growth in extreme environments. Environ. Microbiol. 2, 11–26. doi: 10.1046/j.1462-2920.2000.00071.x
Pajares, S., and Bohannan, B. J. M. (2016). Ecology of nitrogen fixing, nitrifying, and denitrifying microorganisms in tropical forest soils. Front. Microbiol. 7:1045. doi: 10.3389/fmicb.2016.01045
Parraga-Aguado, I., González-Alcaraz, M. N., Álvarez-Rogel, J., and Conesa, H. M. (2014). Assessment of the employment of halophyte plant species for the phytomanagement of mine tailings in semiarid areas. Ecol. Eng. 71, 598–604. doi: 10.1016/j.ecoleng.2014.07.061
Poly, F., Monrozier, L. J., and Bally, R. (2001). Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152, 95–103. doi: 10.1016/S0923-2508(00)01172-4
Riley, D., and Barber, S. A. (1970). Salt Accumulation at the Soybean (Glycine Max. (L.) Merr.) root-soil interface. Soil Sci. Soc. Am. J. 34, 154–155. doi: 10.2136/sssaj1970.03615995003400010042x
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712.
Sharma, S., Aneja, M. K., Mayer, J., Munch, J. C., and Schloter, M. (2005). Diversity of transcripts of nitrite reductase genes (nirK and nirS) in rhizospheres of Grain Legumes. Appl. Environ. Microbiol. 71, 2001–2007. doi: 10.1128/AEM.71.4.2001-2007.2005
Shu, W., Xia, H., Zhang, Z., Lan, C., and Wong, M. H. (2002). Use of vetiver and three other grasses for revegetation of Pb/Zn mine tailings: field experiment. Int. J. Phytoremediation 4, 47–57. doi: 10.1080/15226510208500072
Sun, Q., An, S., Yang, L., and Wang, Z. (2004). Chemical properties of the upper tailings beneath biotic crusts. Ecol. Eng. 23, 47–53. doi: 10.1016/j.ecoleng.2004.07.001
Sun, R., Guo, X., Wang, D., and Chu, H. (2015). Effects of long-term application of chemical and organic fertilizers on the abundance of microbial communities involved in the nitrogen cycle. Appl. Soil Ecol. 95, 171–178. doi: 10.1016/j.apsoil.2015.06.010
Tabor, P. (1949). Cogon grass, Imperata Cylindrica (L) Beauv., in the Southeastern United States. Agron. J. 41:270. doi: 10.2134/agronj1949.00021962004100060011x
Taylor, A. E., Zeglin, L. H., Wanzek, T. A., Myrold, D. D., and Bottomley, P. J. (2012). Dynamics of ammonia-oxidizing archaea and bacteria populations and contributions to soil nitrification potentials. ISME J. 6, 2024–2032. doi: 10.1038/ismej.2012.51
Thion, C. E., Poirel, J. D., Cornulier, T., De Vries, F. T., Bardgett, R. D., and Prosser, J. I. (2016). Plant nitrogen-use strategy as a driver of rhizosphere archaeal and bacterial ammonia oxidiser abundance. FEMS Microbiol. Ecol. 92, fiw091–fiw091. doi: 10.1093/femsec/fiw091
Throbäck, I. N., Enwall, K., Jarvis, Å., and Hallin, S. (2004). Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49, 401–417. doi: 10.1016/j.femsec.2004.04.011
Töwe, S., Albert, A., Kleineidam, K., Brankatschk, R., Dümig, A., Welzl, G., et al. (2010). Abundance of microbes involved in nitrogen transformation in the rhizosphere of Leucanthemopsis alpina (L.) Heywood grown in soils from different sites of the Damma glacier forefield. Microb. Ecol. 60, 762–770. doi: 10.1007/s00248-010-9695-5
Waring, B. G., Colin, A., and Hawkes, C. V. (2013). Differences in fungal and bacterial physiology alter soil carbon and nitrogen cycling: insights from meta-analysis and theoretical models. Ecol. Lett. 16, 887–894. doi: 10.1111/ele.12125
Weng, B., Xie, X., Yang, J., Liu, J., Lu, H., and Yan, C. (2013). Research on the nitrogen cycle in rhizosphere of Kandelia obovata under ammonium and nitrate addition. Mar. Pollut. Bull. 76, 227–240. doi: 10.1016/j.marpolbul.2013.08.034
Yergeau, E., Sanschagrin, S., Maynard, C., St-Arnaud, M., and Greer, C. W. (2014). Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J. 8, 344–358. doi: 10.1038/ismej.2013.163
Zhan, J., and Sun, Q. (2011). Diversity of free-living nitrogen-fixing microorganisms in wastelands of copper mine tailings during the process of natural ecological restoration. J. Environ. Sci. 23, 476–487. doi: 10.1016/S1001-0742(10)60433-0
Zhan, J., and Sun, Q. (2012). Diversity of free-living nitrogen-fixing microorganisms in the rhizosphere and non-rhizosphere of pioneer plants growing on wastelands of copper mine tailings. Microbiol. Res. 167, 157–165. doi: 10.1016/j.micres.2011.05.006
Keywords: mine tailings, revegetation, nitrogen cycle, rhizosphere, nifH gene
Citation: Li Y, Jia Z, Sun Q, Cheng J, Yang Y, Zhan J and Wang D (2017) Plant-Mediated Changes in Soil N-Cycling Genes during Revegetation of Copper Mine Tailings. Front. Environ. Sci. 5:79. doi: 10.3389/fenvs.2017.00079
Received: 27 August 2017; Accepted: 09 November 2017;
Published: 21 November 2017.
Edited by:
Luiz Fernando Wurdig Roesch, Federal University of Pampa, BrazilReviewed by:
Roberta Fulthorpe, University of Toronto Scarborough, CanadaLucas William Mendes, University of São Paulo, Brazil
Copyright © 2017 Li, Jia, Sun, Cheng, Yang, Zhan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingye Sun, sunqingye@ahu.edu.cn
 Yang Li1
Yang Li1  Zhongjun Jia
Zhongjun Jia Qingye Sun
Qingye Sun