Revisiting rapid phenotypic evolution in sticklebacks: integrative thinking of standing genetic variation and phenotypic plasticity
- Museum of Zoology, Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, MI, USA
Phenotypic evolution can result from new mutation, standing genetic variation, or phenotypic plasticity. Extensive reviews have focused on either comparing phenotypic evolution by new mutations vs. standing genetic variation (Barrett and Schluter, 2008) or the effect of phenotypic plasticity on generating biodiversity (Moczek, 2010; Pfennig et al., 2010; Moczek et al., 2011). This article focuses on discussing the potential integrative effect of standing genetic variation and phenotypic plasticity on rapid phenotypic evolution. I start with comparing and contrasting how standing genetic variation and plasticity promote phenotypic evolution and conclude by illustrating three examples in threespine sticklebacks—variation in number of lateral plates, the existence of pelvic girdles, and alternative foraging forms—to demonstrate that both mechanisms could be involved in the same pattern of rapid phenotypic evolution, even though in these cases only one mechanism was invoked by the authors to explain the pattern.
Phenotypic Variation in Non-Plastic and Plastic Traits
Standing genetic variation drives rapid evolution (West-Eberhard, 2003; Barrett and Schluter, 2008), and it is a prerequisite for the phenotypic evolution of non-plastic and plastic traits (Figure 1A). For non-plastic traits, selection directly affects the relative frequency of standing genetic variation coding for different phenotypes. However, for plastic traits, genetic variations that lead to different reaction norms between alternative phenotypes are targets of selection (Figure 1). However, the two mechanisms not only interact (e.g., evolution via plasticity not only needs, but also increases the amount of standing genetic variation; Draghi and Whitlock, 2012), but also can be involved in the same process of phenotypic evolution. For example, the ability to digest lactose is a plastic trait in human and the threshold (age) when this ability is turned off can be altered by assimilation (Lomer et al., 2008). The variation in the ability to digest lactose in adulthood among human populations can also result from standing genetic variation (Myles et al., 2005) or as new mutations (Tishkoff et al., 2006) in the lactase gene. Indeed, there are different dominant mechanisms between examples, but the effects of alternative mechanisms have rarely been tested or integrated within one study.
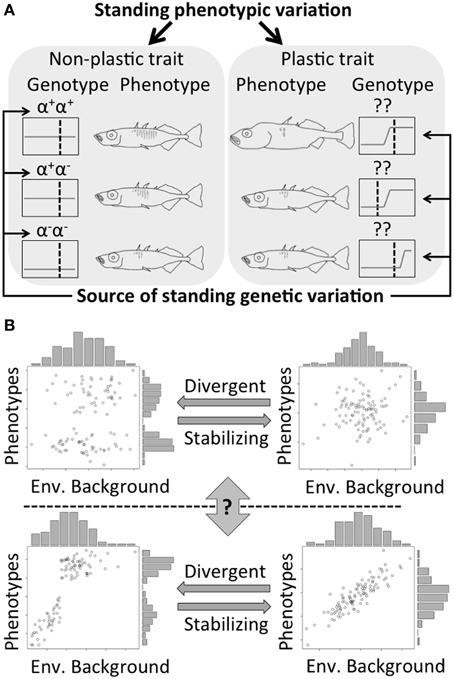
Figure 1. (A) Different types of standing phenotypic variation. Different alleles result in phenotypic difference between individuals for non-plastic traits. Phenotypic difference in plastic trait can result from both environmental variation and changes in the reaction norm. (α and ?: different loci responsible for the focal trait; + and −: the existence and absence of a functional allele for that locus; gray lines in black boxes: the norms of reaction; dash lines: environmental conditions). (B) Transitions between different types of traits (from upper left to lower right: a discrete non-plastic trait, a continuous non-plastic trait, a discrete plastic trait, and a continuous plastic trait). The transition between continuous and discrete phenotypes can result from divergent-stabilizing selections. However, the question of “gene first or environment first?” on morphological evolution is still under intensive debates.
Different mechanisms for rapid phenotypic evolution may be facilitated under different environmental conditions, and the evolutionary outcomes of adopting different mechanisms can be distinct. For example, plastic traits can be advantageous for sessile species or sessile life stages, because adaptation depends on an in situ response to environmental changes (Price et al., 2003). Plastic traits are thus favored when temporal variation occurs on a short timescale such that a given genotype has to cope with multiple environmental conditions. In addition, the effect of environmental filtering on alleles coding for different phenotypes in different habitats can directly lead to genetic differentiation between populations (Schluter and Conte, 2009). Adaptation using phenotypic plasticity, on the other hand, facilitates genetically identical individuals that may colonize different habitats and reduces genetic differentiation between ecologically distinct populations (Wund, 2012). It is important to study how standing variation and plasticity drive evolution simultaneously in different cases.
Identifying Patterns of Rapid Phenotypic Change from Standing Variation
Populations that repeatedly lose and regain an ancestral phenotype are important systems to study evolution from standing genetic variation and phenotypic plasticity (West-Eberhard, 2003; Barrett and Schluter, 2008). Ancestral phenotypes can remain in a population due to standing genetic variation and be resurrected as long as the pertinent alleles are not lost. Change in the threshold between alternative phenotypes of a plastic trait can also resurrect ancestral phenotypes in a descendent population, since genetic materials involved in the genetic module for the ancestral phenotype has never been truly lost (West-Eberhard, 2005). To observe repetitive loss and regain of ancestral phenotypes, the ancestor-descendant relationship between populations must be known or easy to assess. This method is hence limited to lineages with well-reconstructed histories such as the sticklebacks (Bell and Foster, 1994).
Stickleback Examples
Populations of threespine sticklebacks, Gasterosteus aculeatus, have repeatedly colonized freshwater habitats from marine ancestral populations (Bell and Foster, 1994; McKinnon and Rundle, 2002). Freshwater populations have undergone various rapid and parallel phenotypic changes associated with these environments and the genetic and environmental effects on the phenotypes have been extensively investigated. Below I highlight the initial interpretations for phenotypic evolution of the authors and then introduce how an alternative mechanism may help explain the patterns.
Lateral Plate
Evolution in the number of lateral plates between freshwater populations has been demonstrated as evidence for rapid phenotypic adaptation from standing genetic variation (Colosimo et al., 2005). Lateral plates form an antipredator device because they make sticklebacks difficult for predatory fishes to swallow in marine environments. However, plates are not required in freshwater environments that lack large predatory fishes. As a result, they have been lost rapidly and repeatedly in multiple freshwater populations (McKinnon and Rundle, 2002). The alleles for armored (with fully developed plates) and un-armored phenotypes have been identified (Schluter and Conte, 2009). The alleles for lower plate number are found segregating with low frequency in the marine population (Colosimo et al., 2005), while many freshwater descendants are fixed for these alleles.
Instead of loss reflecting standing variation, changes in number of lateral plates can be a result of phenotypic plasticity, as indicated by the effect of diet differences and parasitism severity on the development of lateral plate number in an island population (Reimchen and Nosil, 2001). They can also result from new mutation, as exemplified in a Japanese stickleback population, where a derived allele for lowered plate number, NAKA, at the Eda locus, apparently arose de novo (Colosimo et al., 2005). Furthermore, evolution in the reaction norm could have initiated adaptation in the Japanese freshwater population by decreasing the threshold to develop an un-armored phenotype, and subsequently the NAKA allele emerged as a new mutation and was recruited to stabilize the preferred phenotype (genetic assimilation; West-Eberhard, 2003). However, such argument is still difficult to rigorously test (Laland et al., 2014).
Limnetic and Benthic Forms
Alternation between limnetic and benthic phenotypes in marine stickleback populations is a plastic trait (Day and McPhail, 1996). Plankton feeding induces limnetic forms, while a diet composed of benthic arthropods induces benthic forms. When only one type of food source is found in a local population, adaptation by changing the reaction norm between alternative forms can result in an increased frequency for the better-adapted foraging form (West-Eberhard, 2003). Experiments have demonstrated that an ancestral marine population can seed freshwater populations with distinct phenotypes by feeding the ancestral fish with different food sources (Wund et al., 2008). By altering the type of food source, this process can also be reversed. No loci have decisively been identified for limnetic/benthic forms thus far, and the constant expression of a specific phenotype can be explained by extremely elevated threshold for the alternative one.
Nevertheless, correlation between genetic and phenotypic variations within a foraging form is evident in many freshwater stickleback populations (Jones et al., 2012). Crossing freshwater individuals of different foraging forms produces intermediate forms (Rundle, 2002). Hybrid forms suffer lower fitness because of a significant decrease in foraging efficiency in both limnetic and benthic habitats, but the fitness loss can be rescued by a backcross with a parental form from the corresponding habitats (Rundle, 2002). If a genotype-phenotype correlation for different foraging forms in freshwater stickleback populations exists (e.g., Jones et al., 2012), parallel phenotypic adaptations of foraging forms observed among freshwater populations can result from adaptation via standing genetic variation as a non-plastic trait.
Pelvic Girdle—New Mutation?
A pelvic girdle is an antipreditor device in marine populations, and its development is controlled by the Pitx1 gene (Chan et al., 2010). The pelvic girdle has been repeatedly and independently lost in freshwater populations. An upstream cis-regulatory region of Pitx1 contains different types of deletion among pelvic-absent freshwater populations (Chan et al., 2010), and pelvic formation can be rescued by introducing an intact cis-regulatory region from another population. Independent loss-of-function events in the cis-regulatory region by new mutations (deletions) are hypothesized for parallel pelvic adaptation (Chan et al., 2010).
The evolutionary relationship among the alleles for the Pitx1enhancer (i.e., the pelvic-reduced and wild type alleles) has not yet been reconstructed (cf., Colosimo et al., 2005; Chan et al., 2010). There are also different alleles coding for lower lateral plate number between freshwater populations, but the inference of adaptation by standing genetic variation is made because these alleles share a common ancestry (Colosimo et al., 2005). Although different pelvic-reduced alleles among populations arose from different deletions, they might be derived from an ancestral allele that is prone to deletion events (e.g., alleles that have many tandem/inverted repeats are prone to structural modifications during replications; Gemayel et al., 2010). The excessive number of tandem repeats found in the upstream cis-regulatory region of Pitx1 implies that standing genetic variation as fragile and non-fragile alleles for pelvic girdle formation may be the source for rapid phenotypic evolution. Furthermore, the reduction in genetic diversity occurs specifically in the enhancer region, but not the linked flanking region of Pitx1 (Fig. S1B and S9 A in Chan et al., 2010), which represents a similar pattern to adaptation by standing genetic variation (Figure 1C in Colosimo et al., 2005; Barrett and Schluter, 2008).
Alternatively, the pelvic spine is a developmentally plastic trait (Lescak et al., 2013). Feeding on benthic food sources can induce the separation of the pelvic girdle, which will not occur when feeding on limnetic food sources (Wund et al., 2008). The formation of pelvic girdle can also be affected by the amount of dissolved ions in the environment (Lescak et al., 2013). It is again possible that developmental plasticity may have facilitated the emergence and increased frequency of the pelvic-reduced phenotype by evolution in the reaction norm either because of the absence of predator or to conserve the use of dissolved ion for other body structures (Lescak et al., 2013). New mutations that occurred in the fragile cis-regulatory region were recruited subsequently to stabilize the preferred phenotype in parallel examples.
Identifying the Predominant Mechanism
Although difficult, discriminating patterns driven by different mechanisms is possible. Non-adaptive alleles from loci linked to adaptive alleles and their corresponding non-adaptive phenotypes are not expected to undergo significant frequency changes when phenotypic adaptation occurs from standing genetic variation (Barrett and Schluter, 2008). The rare phenotype of a plastic trait, on the other hand, is predicted to exhibit more phenotypic variants (Moczek, 2010). Because genetic variation in the developmental module for the rare phenotype is less often revealed under selection compared to genetic variation associated with the common phenotype, a higher level of genetic variation should be maintained under selection-mutation balance (Hunt et al., 2011). An example of significantly larger amount of difference in phenotypic variation found between lake populations of sticklebacks—body depth, pelvic spine, and dorsal spines—than those between stream forms (Fig. S2 in Deagle et al., 2012) may arise simply as a by-product of adaptation via phenotypic plasticity, where the giant lake form, which was a rare phenotype, became prevalent under altered environmental conditions (unraveling of cryptic genetic variation; Schlichting, 2008).
Future Perspectives and Concluding Remarks
Although this article discusses discrete phenotypes, there is continuous phenotypic variation in both non-plastic and plastic traits (Figure 1B). Stabilizing and divergent selection can work on both non-plastic and plastic traits to generate discrete and continuous phenotypic variation. Switches between governing by non-plastic or plastic mechanisms for a trait can also occur along an evolutionary lineage (Schwander and Leimar, 2011), although how often and why this switch happens remains elusive. Furthermore, interactions between standing genetic variation and plasticity can exist, where evolution via plastic traits not only requires genetic variation, but also facilitates the accumulation of novel genetic variants during the process (Schlichting, 2008; Draghi and Whitlock, 2012). The fact that a dominant mechanism exists in each example is fully acknowledged in this paper, but the argument is that the alternative mechanism may still play an important role. Before an integrative framework can be established, I hope to encourage integrative thinking about the effects of standing genetic variation and phenotypic plasticity on a common evolutionary pattern.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Sincere thanks are given to L. Lacey Knowles, Mercedes Pascual and Patricia Wittkopp for helpful comments on the previous version of this manuscript. Diego Alvarado-Serrano, Qixin He, Hayley Lanier, Robert T. Massatti, and two reviewers provided critical and constructive suggestions about the contents of this article.
References
Barrett, R. D., and Schluter, D. (2008). Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44. doi: 10.1016/j.tree.2007.09.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bell, M. A., and Foster, S. A. (1994). The Evolutionary Biology of the Threespine Stickback. New York, NY: Oxford University Press.
Chan, Y. F., Marks, M. E., Jones, F. C., Villarreal, G. Jr., Shapiro, M. D., Brady, S. D., et al. (2010). Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a pitx1 enhancer. Science 327, 302–305. doi: 10.1126/science.1182213
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Colosimo, P. F., Hosemann, K. E., Balabhadra, S., Villarrea, G. Jr., Dickson, M., Grimwood, J., et al. (2005). Widespread parallel evolution in sticklebackes by repeated fixation of ectodysplasin alleles. Science 5717, 1928–1933. doi: 10.1126/science.1107239
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Day, T., and McPhail, J. D. (1996). The effect of behavioural and morphological plasticity on foraging efficiency in the threespine stickleback (Gasterosteus sp.). Oecologia 108, 380–388. doi: 10.1007/BF00334665
Deagle, B. E., Jones, F. C., Chan, Y. F., Absher, D. M., Kingsley, D. M., Reimchen, T. E., et al. (2012). Population genomics of parallele phenotypic evolution in stickleback across stream-lake ecological transitions. Proc. R. Soc. Lond. B Biol. Sci. 279, 1277–1286. doi: 10.1098/rspb.2011.1552
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Draghi, J. A., and Whitlock, M. C. (2012). Phenotypic plasticity facilitates mutational variance, genetic variance, and evolvability along the major axis of environmental variation. Evolution 66, 2891–2902. doi: 10.1111/j.1558-5646.2012.01649.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gemayel, R., Vinces, M. D., Legendre, M., and Verstrepen, K. J. (2010). Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477. doi: 10.1146/annurev-genet-072610-155046
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hunt, B. G., Ometto, L., Wurm, Y., Shoemaker, D., Yi, S. V., Keller, L., et al. (2011). Relaxed selection is a precursor to the evolution of phenotypic plasticity. Proc. Natl. Acad. Sci. U.S.A. 108, 15936–15941. doi: 10.1073/pnas.1104825108
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jones, F. C., Chan, Y. F., Schmutz, J., Grimwood, J., Brady, S. D., Southwick, A. M., et al. (2012). A genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr. Biol. 22, 83–90. doi: 10.1016/j.cub.2011.11.045
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Laland, K., Uller, T., Feldman, M., Sterelny, K., Müller, G. B., Moczek, A., et al. (2014). Does evolutionary theory need a rethink? Nature 514, 161–164. doi: 10.1038/514161a
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lescak, E. A., von Hippel, F. A., Bernhardt, R. R., and Bell, M. (2013). Pelvic girdle reduction and asymmetry in threespine stickleback from Wallace Lake, Alaska. Evol. Ecol. Res. 15, 155–170.
Lomer, M. C. E., Parkers, G. C., and Sanderson, J. D. (2008). Lactose intolerance in clinical practice – myths and realities. Aliment. Pharmacol. Ther. 27, 93–103. doi: 10.1111/j.1365-2036.2007.03557.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
McKinnon, J. S., and Rundle, H. D. (2002). Speciation in nature: the threespine stickleback model system. Trends Ecol. Evol. 17, 480–488. doi: 10.1016/S0169-5347(02)02579-X
Moczek, A. P., Sultan, S., Foster, S., Ledón-Rettig, C., Dworkin, I., Nijhout, H. F., et al. (2011). The role of developmental plasticity in evolutionary innovation. Proc. R. Soc. B Lond. Biol. Sci. 278, 2705–2713. doi: 10.1098/rspb.2011.0971
Moczek, A. P. (2010). Phenotypic plasticity and diversity in insects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 593–603. doi: 10.1098/rstb.2009.0263
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Myles, S., Bouzerkri, N., Haverfield, E., Cherkaoui, M., Dugoujon, J.-M., Ward, R., et al. (2005). Genetic evidence in support of a shared Eurasian-North African dairying origin. Hum. Genet. 117, 34–42. doi: 10.1007/s00439-005-1266-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pfennig, D. W., Wund, M. A., Snell-Rood, E. C., Cruickshank, T., Schlishting, C. D., Moczek, A. P., et al. (2010). Phenotypic plasticity's impacts on diversification and speciation. Trends Ecol. Evol. 25, 459–467. doi: 10.1016/j.tree.2010.05.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Price, T. D., Qvarnström, A., and Irwin, D. (2003). The role of phenotypic plasticity in driving genetic evolution. Proc. R. Soc. Lond. B Biol. Sci. 270, 1433–1440. doi: 10.1098/rspb.2003.2372
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reimchen, T. E., and Nosil, P. (2001). Lateral plate asymmetry, diet and parasitism in threespine stickleback. J. Evol. Biol. 14, 632–645. doi: 10.1046/j.1420-9101.2001.00305.x
Rundle, H. D. (2002). A test of ecologically dependent postmating isolation between sympatric sticklebacks. Evolution 56, 322–329. doi: 10.1111/j.0014-3820.2002.tb01342.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schlichting, C. D. (2008). Hidden reaction norms, cryptic genetic variation, and evolvability. Ann. N.Y. Acad. Sci. 1133, 187–203. doi: 10.1196/annals.1438.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schluter, D., and Conte, G. L. (2009). Genetics and ecological speciation. Proc. Natl. Acad. Sci. U.S.A. 106, 9955–9962. doi: 10.1073/pnas.0901264106
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schwander, T., and Leimar, O. (2011). Genes as leaders and followers in evolution. Trends Ecol. Evol. 26, 143–151. doi: 10.1016/j.tree.2010.12.010
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tishkoff, S. A., Reed, F. A., Ranciaro, A., Voight, B. F., Babbitt, C. C., Silverman, J. S., et al. (2006). Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39, 31–40. doi: 10.1038/ng1946
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
West-Eberhard, M. J. (2003). Developmental Plasticity and Evolution. New York, NY: Oxford University Press.
West-Eberhard, M. J. (2005). Phenotypic accommodation: adaptive innovation due to developmental plasticity. J. Exp. Zool. B Mol. Dev. Evol. 304B, 610–618. doi: 10.1002/jez.b.21071
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wund, M. A., Baker, J. A., Clancy, B., Golub, J. I., and Foster, S. A. (2008). A test of the “flexible stem” model of evolution: ancestral plasticity, genetic accommodation, and morphological divergence in the threespine stickleback radiation. Am. Nat. 172, 449–463. doi: 10.1086/590966
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wund, M. A. (2012). Assessing the impacts of phenotypic plasticity on evolution. Integr. Comp. Biol. 52, 5–15. doi: 10.1093/icb/ics050
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: adaptation, colonization, environmental effects, selection
Citation: Huang J-P (2015) Revisiting rapid phenotypic evolution in sticklebacks: integrative thinking of standing genetic variation and phenotypic plasticity. Front. Ecol. Evol. 3:47. doi: 10.3389/fevo.2015.00047
Received: 05 February 2015; Accepted: 20 April 2015;
Published: 08 May 2015.
Edited by:
James J. Cai, Texas A&M University, USAReviewed by:
David Pfennig, University of North Carolina at Chapel Hill, USAGang Li, Texas A&M University, USA
Copyright © 2015 Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jen-Pan Huang, huangjp@umich.edu
 Jen-Pan Huang
Jen-Pan Huang