Protocol for Heterologous Expression of Insect Odourant Receptors in Drosophila
- 1Chemical Ecology Division, Department of Plant Protection Biology, Swedish University of Agricultural Sciences, Alnarp, Sweden
- 2Department of Neuroethology and Evolution, Max Planck Institute for Chemical Ecology, Jena, Germany
Insect olfactory receptors (ORs) are tuned to volatile chemicals, they are expressed in the membrane of olfactory sensory neurons (OSNs), housed in sensilla on the antenna. The olfactory apparatus is under strong selection and ORs are tuned to vital chemical signals, mediating social communication, feeding and oviposition, and avoidance of predators and pathogens. An emerging technique to reliably and efficiently identify the key ligands of ORs is to express single ORs in heterologous cell systems for subsequent screening. Several in vivo and in vitro platforms have been developed; we here provide a step-by-step protocol for OR expression in Drosophila melanogaster OSNs. Following RNA extraction, molecular cloning of ORs and injection of plasmid vectors into Drosophila embryos to create flies with OR transgenes, single ORs are expressed, via crossing with specific transgene promoters in OSNs of ab3 and T1 antennal sensilla. This approach enables replicable single sensillum electrophysiological recordings (SSR) from readily distinguishable Drosophila sensilla, containing OSNs expressing transgenic ORs. We expect this method to be applicable to ORs across insect orders and to increasingly contribute to chemical ecology research. Heterologous expression enables thorough investigation of single ORs, toward the identification of yet unknown, behaviorally and ecologically relevant chemical signals. It also enables investigations of the functional properties of ORs and their evolutionary diversification, through comparative structure-activity studies across phylogenies.
Introduction
Olfactory communication signals are recruited from countless volatile chemicals filling the air. A foremost goal in insect chemical ecology research is to unambiguously identify behavior-modifying compounds, termed semiochemicals, which convey messages from animals, plants or microbes. Semiochemicals usually are blends of several compounds and it is a sensitive and time-consuming task to discriminate between behaviorally active and inactive compounds found in headspace collections.
In insects, electrophysiological recordings, which employ the antenna as sensor, have been a versatile and widely used tool for selecting candidate compounds (Schneider, 1957; Arn et al., 1975) and facilitate interlacing chemical with behavioral analysis. Recordings from entire antennae are particularly efficient for identification of sex pheromones, used for communication within the same species, and typically elicit a conspicuous response. Knowledge of de-novo produced pheromones also facilitates further identifications, since taxonomically close species use related biosynthetic pathways (Jurenka, 2004). Consequently, hundreds of lepidopteran pheromones have been described (Arn et al., 1992; El-Sayed, 2015).
In comparison, unequivocal identification of kairomones, compounds which guide host plant attraction, in moths and other herbivorous insects is infinitely more difficult. Plants release a wealth of compounds and, in contrast with sex pheromones, there is no producer-receiver correlation—abundance of plant compounds is no criterium for behavioral activity. Plant volatiles that attract herbivores have long been known (Dethier, 1947; El-Sayed, 2015), but we still do not know as to whether, or to what extent these attractants actually correspond to the chemical signatures used by insects to find their host plants. The attractant power of synthetic kairomones is a straightforward criterium, but behavioral assays with kairomones, especially in females, are complex and laborious.
Screening candidate compounds prior to behavioral analysis is therefore paramount. Unfortunately, for the identification of kairomones, conventional antennal electrophysiological recordings fail to deliver. The most abundant compounds in plant headspace invariably produce a response when recording from the entire antenna, disregarding their behavioral relevance. Recordings from single olfactory sensilla, on the other hand, are technically demanding and will only rarely provide exhaustive information. This is exemplified by work on codling moth, where the main apple volatiles produce a strong antennal, but only weak or no behavioral response (Bäckman et al., 2001; Ansebo et al., 2004; Coracini et al., 2004). In contrast, pear ester, a compound which has not been found in the main host apple, is the strongest known adult and larval attractant (Light et al., 2001; Light and Knight, 2005; Light and Beck, 2012).
Following the identification of olfactory receptor (OR) genes from codling moth antennae (Bengtsson et al., 2012), it has recently been shown that CpomOR3, which is highly expressed in male and female antennae, is specifically tuned to pear ester (Bengtsson et al., 2014). This finding corroborates the biological role of pear ester and is supported by intracellular recordings and functional imaging of the codling moth antennal lobe (Trona et al., 2010, 2013). The functional characterization of CpomOR3 also underscores the weight of a reliable screening technique for single ORs—toward a more efficient identification of semiochemicals of plant origin.
In silico identification of putative odourant receptor (OR) genes in Drosophila melanogaster was the starting point for a new era of chemical communication research and opened the door for downstream studies in which ORs are functionally characterized according to the ligands they are tuned to, a process also known as “deorphanization” (Clyne et al., 1999; Gao and Chess, 1999; Vosshall et al., 1999; Hallem et al., 2004; Hallem and Carlson, 2006). Deorphanization of insect ORs is achieved through testing their response spectrum toward odourant compounds, following heterologous expression of these OR proteins in heterospecific cell systems, which facilitates thorough and unambigious screening.
In vitro systems involve the expression of ORs in cell culture platforms, such as human embryonic kidney cells (HEK; Große-Wilde et al., 2006; Syed et al., 2006; Corcoran et al., 2014), Spodoptera frugiperda Sf9 cells (Matarazzo et al., 2005; Kiely et al., 2007; Anderson et al., 2009; Jordan et al., 2009; Xu et al., 2015) and also Xenopus oocytes (Sakurai et al., 2004; Mitsuno et al., 2008; Wanner et al., 2010; Leary et al., 2012; Liu et al., 2013; Zhang and Löfstedt, 2013; Jiang et al., 2014). Recently, a cell-free expression system has been reported (Tegler et al., 2015).
In the case of in vivo systems, heterologous expression is based on the use of mutant, “empty-neuron” lines of D. melanogaster (Dobritsa et al., 2003; Hallem et al., 2004). The antennal basiconic sensilla type 3 (ab3) of the mutant D. melanogaster flies contain an odourant sensory neuron (OSN) that lacks its native OR: expression of the native OR22a/b in ab3A OSNs is disrupted in these mutant flies (Dobritsa et al., 2003). When coupled with the Gal4-UAS transgene expression system (Brand and Perrimon, 1993), using an OR22a Gal4 line, transgenic ORs can be specifically expressed in ab3A empty OSNs, which project their dendrites into large basiconic sensilla (Shanbhag et al., 1999). These OSNs can then be screened for novel responses conferred by the transgenic OR, by means of single sensillum electrophysiological recordings (SSR). This methodology has been successful for the deorphanization of receptors from different subsystems such as antennal ORs as well as maxillary palp ORs (Dobritsa et al., 2003; Goldman et al., 2005). In addition, the empty neuron system has also allowed to deorphanize larval receptors (Kreher et al., 2005, 2008; Mathew et al., 2013).
Deorphanization of putative pheromone receptors (PRs) has proven to be more challenging than OR deorphanization. To provide PRs with a more suitable cellular environment, heterologous expression has instead targeted the trichoid sensillum T1 of D. melanogaster. In wild-type flies, T1 sensilla contain a single neuron expressing a single receptor, OR67d, which is tuned to the male pheromone, 11-cis-vaccenyl acetate (cVA). In knock-in mutant flies, this native receptor is replaced with an OR67d-Gal4 construct (Kurtovic et al., 2007). The T1 system is suitable for the deorphanization of both PRs (Syed et al., 2010; Montagné et al., 2012) and some ORs tuned to plant compounds (Bengtsson et al., 2014; Ronderos et al., 2014).
Heterologous expression in Drosophila has served as a fundamental tool for the deorphanization of insect ORs and PRs across diverse taxa. However, the procedures necessary to produce flies expressing transgenic receptors have not been comprehensively described. Here, we provide a hands-on, step-by-step protocol of how to express and test insect ORs in Drosophila OSNs.
Materials and Equipment
Reagents and materials required for the different steps in producing and testing transgenic fly lines that ectopically express ORs in the empty neuron systems are shown in Table 1.
Procedures
For efficient streamlined cloning of OR genes and generation of transgenic flies, we recommend use of the TOPO/gateway cloning system (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) coupled to PhiC31 integrase-mediated transgenesis system applied to D. melanogaster (Bischof et al., 2007). The TOPO/gateway system facilitates cloning and transfer of DNA inserts from entry to destination plasmid and the Phi3C31 system facilitates highly-efficient, non-random, sequence-directed and irreversible genomic insertion of vector DNA. The following protocols have been formulated specifically for use of these systems. Whether the goal is to express an OR transgene in the ab3A or T1 systems, the molecular cloning procedures in the following section are identical up until the point of embryonic injections, as described below.
An overview of the two main worksteps, molecular cloning (Section Molecular Cloning of Insect ORs) and transgenic expression by fly crossing (Section Transgenic Expression of ORs in Drosophila OSNs) is shown in Figures 1–3. A best-case scenario time plan for the procedures described in the following section is shown in Supplemental Figure 1.
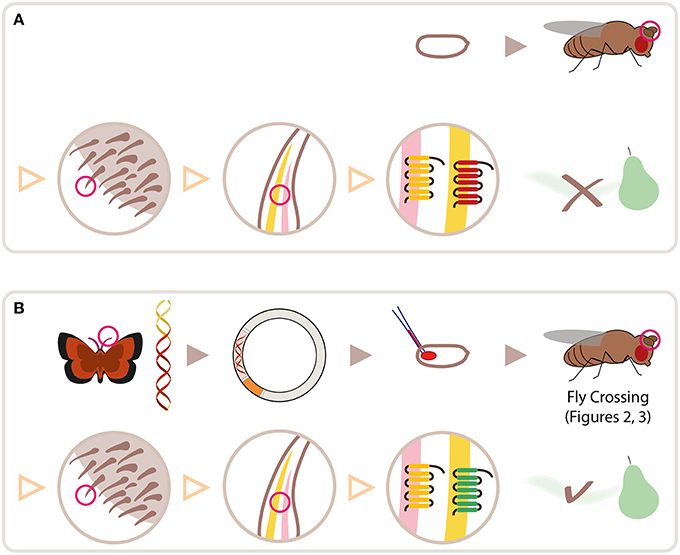
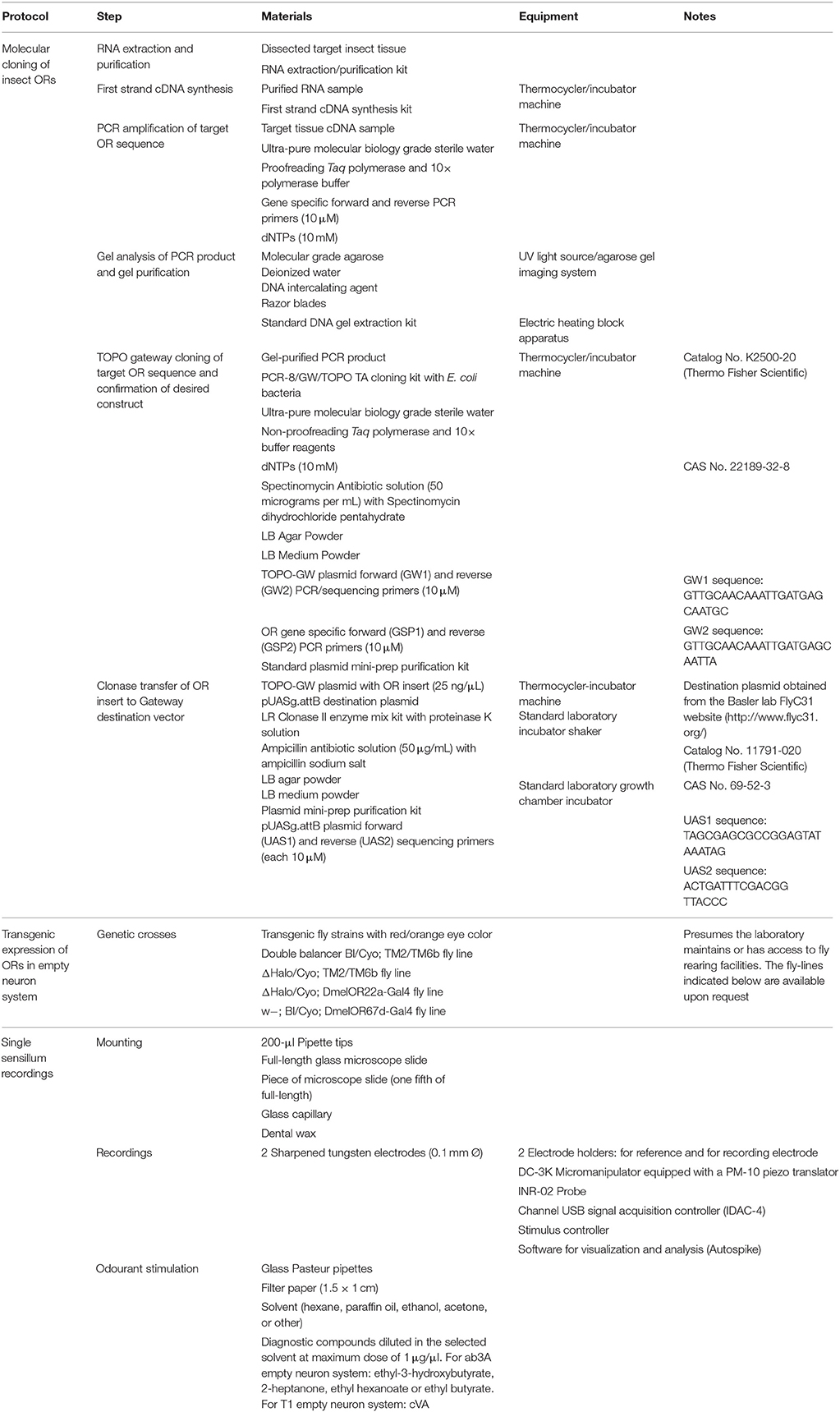
Figure 1. Schematic overview for heterologous expression of insect ORs in Drosophila OSNs. (A) Wild type fly embryo and fly (top row). Red circle highlights antenna, shown in three magnification steps (second row, separated by triangles): sensilla on antenna; 2 olfactory sensory neurons (OSNs) housed in one sensillum; olfactory receptor proteins (ORs) expressed in cell membrane of each OSN. Wild type flies do not smell pear ester. (B) cDNA is synthesized from RNA extracted from lepidopteran antennae; OR gene is cloned into plasmid; plasmid is injected into fly embryo. Following crosses using Gal4/UAS expression system, lepidopteran OR tuned to pear ester is expressed in target OSN on fly antenna, allowing it to detect pear ester. Moth and fly drawing by Katarina Eriksson (www.markadesign.se).
Molecular Cloning of Insect ORs
RNA Extraction and Purification
Dissect antennal (or other target) tissue from a sufficient number of insects into an empty 1.5-mL microcentrifuge tube held in liquid nitrogen, dry ice, or else standard ice. For D. melanogaster, 100 insects are recommended; for moths, 30 insects may be sufficient. Size of the antenna determines the number of specimen required.
Store target tissue in −80°C freezer, or proceed immediately to RNA extraction and purification. Follow standard protocol provided with extraction/purification kit/reagents.
Measure RNA quantity with photospectrometer or equivalent device and store RNA at −80°C or proceed immediately to the next step.
First-Strand cDNA Synthesis
Follow manufacturer's protocol for cDNA synthesis, with maximum quantity of RNA allowed within the volumetric parameters of the enzymatic reaction.
cDNA may commonly be diluted with ultra-pure water (e.g., at 1:1 ratio with cDNA sample) for PCR amplification assays, if necessary. However, dilution of cDNA may not be desirable, when the target genes show relatively low expression patterns compared to other ORs.
Store cDNA at −20°C or proceed immediately to the next step.
PCR Amplification of Target OR Sequence
Generate gene specific primers (GSPs) for PCR amplification of the entire open reading frame (ORF) of the target OR. Utilize forward primers (GSP1) that begin with the start codon and reverse primers (GSP2) that begin with the reverse-complement of the stop codon. If the start to stop codon primers are not ideal for PCR amplification due to mismatched melting temperatures (Tm, greater than 5°C difference) or other factors, it is advisable to design primers upstream or downstream of the ORF, respectively. If positive control primers are not previously available, the Orco gene could serve as a target to control for gene amplification in antennal tissue, since it is always expressed together with ORs and displays high expression in antennal tissue.
Conduct PCR amplification reaction with a DNA polymerase system that includes 3′ to 5′ exonuclease (proofreading) function. At this step, use of a proofreading Taq polymerase is critical; it drastically reduces the likelihood of obtaining unusable plasmid clones that contain OR inserts with incorrect sequence. Set up one PCR reaction per target OR, with positive (e.g., Orco) and negative (e.g., no template) control, according to manufacturers protocol. Run PCR amplification reaction in thermocycler machine according to manufacturers specifications for the Taq polymerase system, with annealing temperature 3°C less than primer melting temperature (Tm) and 30–35 amplification cycles (standard running time, ca. 2 h).
Store PCR overnight at 4°C, for longer periods at −20°C, or proceed immediately to next step.
Gel Analysis of PCR Product and Gel Purification
Run PCR products through 1.5% agarose gel for simultaneous verification of amplification and excision of OR-specific amplicon for purification. Expected band size for ORs is typically around 1200 base-pairs, as compared to fragments of standard DNA ladder.
Use low-intensity UV wavelength so as not to damage/mutate DNA, and minimize exposure time while cutting out the agarose gel that contains OR-specific fragments. Place excised gel in 1.5-mL microcentrifuge tubes and measure the mass of the added gel material. Gel may be frozen at −20°C for later use, or used immediately for the next step.
Purify OR-specific DNA from the gel with standard gel purification/extraction kit according to manufacturers protocol. Elute DNA in ultra pure sterile water or buffer provided with the kit.
Run a small aliquot (e.g., 5 μL) of purified DNA on a 1.5% agarose gel in order to verify success of the procedure and ensure the presence of only OR-specific DNA at the expected size.
Store gel-purified OR DNA at −20°C or proceed immediately to next step.
TOPO/Gateway Cloning of Target OR Sequence and Construct Confirmation
While the use of Taq polymerase with proofreading function is essential to ensure accurate amplification of the target sequence, it results in the removal of adenosine overhang nucleotides at the 5′ and 3′ ends of the DNA amplicon, which is a feature of standard Taq polymerase. These adenosine nucleotides are critical for the function of the TOPO cloning system. Thus, it is necessary, after gel purification, to enzymatically add the adenosine overhangs to the target OR sequence to be cloned.
Use 10 μL of gel-purified DNA, 1.2 μL of 10 × PCR buffer, 1 μL of 10 mM dNTPs (both included in TOPO cloning kit), and 0.5 μL of standard Taq polymerase (without proofreading activity, not included in TOPO cloning kit). It is critical to use only the buffer supplied with the TOPO cloning kit; this buffer is compatible with downstream cloning steps. Mix contents and incubate at 72°C for 10 min. Proceed immediately to next step.
Add 4 μL of previous reaction, 1 μL of salt solution (provided with TOPO cloning kit) and 1 μL of topoisomerase vector mix (provided with TOPO cloning kit). Mix and incubate at room temperature (22–23°C) for more than 5 min, but less than 30 min. For inserts larger than 1 kb in size, the longer incubation time is recommended.
Toward the end of the incubation period, thaw appropriate number of aliquots of One Shot Competent E. coli (provided with TOPO cloning kit) on ice. Mix 2 μL of previous reaction with E. coli and chill on ice for greater than 5 min, but less than 30 min.
Heat shock cell/plasmid mixture at 42°C for 30 s, and place tube promptly on ice. Add 250 μL of SOC media (provided along with E. coli tubes) to cells and grow at 37°C for at least 1 h in incubator shaker. Apply entire contents of cell culture on prepared LB+Spectinomycin (50 μg/ml) bacterial growth plates and incubate overnight at 37°C. Plates may be stored at 4°C for up to 1 month.
To ensure appropriate expression of the OR transgene in D. melanogaster, orientation of the insert from 5′ to 3′ with reference to the attL1 element in the TOPO plasmid is required. To verify correct orientation of the insert, a standard colony PCR protocol is followed, with amplification using one GSP and one TOPO plasmid primer (GW1 or GW2); either combination of GW1 and GSP2 or GW2 and GSP1 will suffice. For either of these combinations, amplification of a PCR product (ca. 1.3 kb) will only occur if the insert is positioned in the plasmid in the desired orientation.
Typically, screening of 4–8 colonies with this assay is sufficient to identify a clone with the insert in the desired orientation. First, select colonies and transfer them each to a 1.5-mL microcentrifuge tube with 50 μL of LB plus spectinomycin (50 μg/ml) growth medium. Incubate culture at 37°C in incubator shaker for at least 1 h. In the meantime, prepare PCR reactions with master mix appropriate to the number of colonies being assayed. Using a Taq polymerase system, without proofreading function, a standard PCR reaction shall be prepared with 2 μL of each colony culture to be added to each PCR reaction tube. Remainder of colony culture is to be stored at 4°C for later use. For the amplification procedure, standard thermocycling parameters shall be followed according to the Taq polymerase system being used, with a 5 min extension period per cycle, and 30–35 amplification cycles. Ensuing gel analysis of PCR amplification products on a 1.5% agarose gel will confirm the presence of amplicon, and thus correct orientation of the insert.
For each TOPO/OR construct, one or more colony cultures with insert may be selected for further processing. After PCR assay and confirmation, the remainder of the colony culture is added to a culture tube with 3 mL of LB plus spectinomycin (50 μg/ml), and this culture is grown overnight at 37°C in a shaker incubator. After overnight growth, the culture may be stored at 4°C for 2–3 weeks or used immediately in the next step.
Using a standard plasmid mini prep purification kit, the culture is to be processed according to manufacturers protocol. Elute plasmid DNA in ultra-pure sterile water or supplied elution buffer and measure concentration of plasmid preparation with photospectrometer or equivalent equipment.
Confirm the sequence of the insert via sequencing reactions with GW1 and GW2 primers supplied with the TOPO cloning kit. This step is critical. Attempts to generate transgenic fly lines without verifying sequence beforehand may lead to otherwise avoidable failure of the experiment.
Store plasmid at −20°C until completion of the sequencing reactions. Discard all plasmids with incorrect sequence or errors otherwise. Select one plasmid with correct sequence for further processing.
Clonase Transfer of OR Insert to Gateway Destination Vector
Using the TOPO/OR and pUASg.attB plasmids diluted to specified concentrations, mix 6 μL of TOPO/OR, 2 μL of pUASg.attB, and 2 μL of the LR clonase enzyme (Thermo Fisher Scientific, USA) and incubate at 25°C for 1 h.
Add 1 μL of proteinase K (supplied with LR Clonase kit) to terminate previous reaction. Mix and incubate at 37°C for 10 min. This step is critical. If omitted, downstream outcomes will not be successful.
During the incubation period, thaw appropriate number of aliquots of One Shot Competent E. coli (provided with TOPO cloning kit) on ice. Mix 2.5 μL of the clonase reaction with E. coli and chill on ice for greater than 5 min, but less than 30 min.
Heat shock cell/plasmid mixture at 42°C for 30 s, and place tubes promptly on ice. Add 250 μL of SOC media (provided with E. coli tubes) to cells and grow at 37°C for at least 1 h in incubator shaker. Apply entire contents of cell culture on previously prepared LB+Ampicilin (50 μg/ml) bacterial growth plates and incubate overnight at 37°C. Plates may be stored at 4°C for up to 1 month.
On account of positive selection of pUASg.attB with OR insert, and negative selection against bacteria with TOPO/OR plasmid (these contain Spectinomycin but not Ampicillin resistance genes) and also those with pUASg.attB lacking OR insert (these contain lethal gene whose gene product results in death of One Shot E. coli), all bacterial colonies on the growth plate will contain the pUASg.attB with OR insert in the correct orientation. Thus, colony PCR is not necessary at this step to confirm presence and orientation of the insert.
For each pUASg.attB/OR construct, transfer one colony to a culture tube with 3 mL of LB plus ampicillin (50 μg/ml), and grow the culture overnight at 37°C in shaker incubator. After overnight growth, culture may be stored at 4°C for up to 2–3 weeks or used immediately in the next step.
Using a standard plasmid mini- or midi-prep purification kit, the culture is to be processed according to manufacturers protocol. Elute plasmid DNA in ultra-pure sterile water and measure concentration of plasmid preparation with photospectrometer or equivalent equipment.
Confirm the sequence of the insert via sequencing reactions with UAS1 and UAS2 sequencing primers (described in Table 1). This step is critical. Attempts to generate transgenic fly lines without verifying sequence beforehand may lead to otherwise avoidable failure of the experiment.
Store plasmid at −20°C until completion of the sequencing reactions. Discard all plasmids with incorrect sequence or errors otherwise. Select one plasmid with correct sequence for injection in fly embryos.
For expression in the ab3A empty neuron system, it is desirable to insert the UAS-OR construct on the 3rd chromosome. Therefore, it is recommended that injections are made into embryos of the following genetic background:
For expression in the T1 neuron system, it is desirable that the UAS-OR construct is inserted on the 2nd chromosome. Therefore, it is recommended that injections are made into embryos of the following genetic background:
Injections are outsourced to a company providing Drosophila embryo injection services. Indeed, fly strains exist that contain landing sites at different locations on the second and third chromosomes. The recommended strains have been selected due to current availability as well as relatively high genomic integration efficiency and transgene expression. Consultation with fly embryo injection companies are advised to determine the best solutions with respect to available fly strains for this purpose.
Transgenic Expression of ORs in Drosophila OSNs
In order to express the OR transgene (UAS-ORx) in OSNs of either ab3 or T1 sensilla, it is necessary to push the transgene through a series of genetic crosses (Figures 2, 3). Injections are made into a fly strain with white-eye mutation (w−) and the UAS-OR construct carries a rescue gene for the white-eye phenotype. Therefore, transgenic flies obtained after injections will have orange/red eyes and a genotype, w−; +; UAS-ORx (w+)/+, for use in ab3 system, or alternatively w−; UAS-ORx(w+)/+, +, for use in T1 system. A series of initial crosses are necessary to screen for the presence of transgene. While it is possible for the end-user to obtain larvae directly from injected embryos and screen for transgenic strains in the laboratory, this is labor intensive and not recommended. Alternatively, these steps are typically offered as service by fly-injection companies for a small fee above and beyond baseline injection costs. For further details on balancer chromosome phenotypes see Greenspan (1997). All stock flylines used for crosses mentioned below are available upon request from our laboratory.
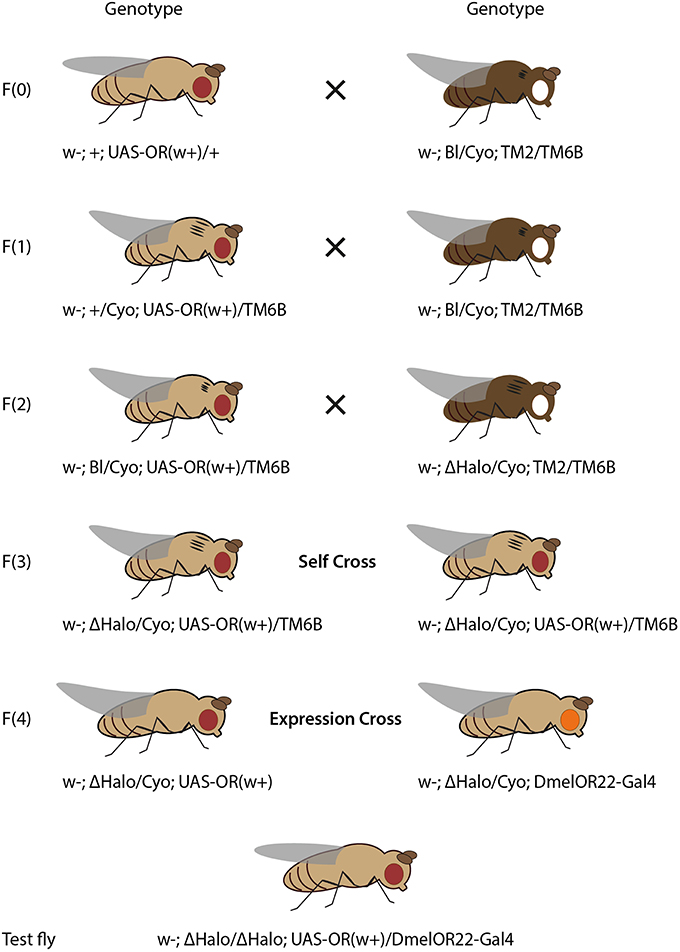
Figure 2. Crossing scheme for heterologous expression of an OR transgene in empty neurons in ab3 sensilla, using ΔHalo mutant background and DmelOR22a-Gal4 driver line. Fly drawing by Katarina Eriksson (www.markadesign.se).
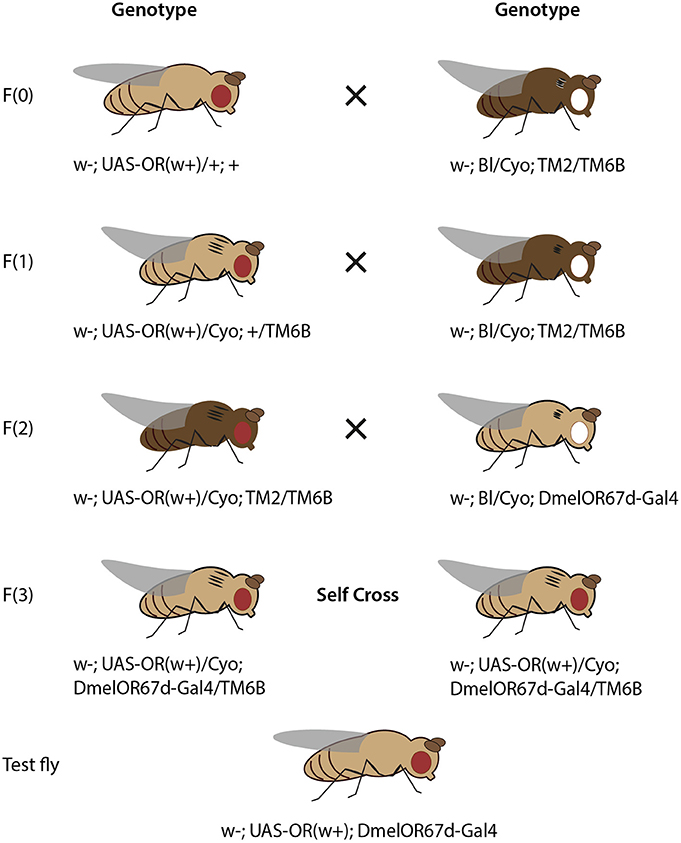
Figure 3. Crossing scheme for heterologous expression of an OR transgene in OSNs in T1 sensilla, using DmelOR67d-Gal4 knock-in with knock-out of DmelOR67d coding sequence. Fly drawing by Katarina Eriksson (www.markadesign.se).
Fly Crossing Scheme for the ab3A Empty Neuron System
The OR transgene must be crossed into the ΔHalo genetic background, which contains a chromosomal deletion spanning the location that includes the OR22a/b locus (Dobritsa et al., 2003; Gross et al., 2003). An outline of the required crosses is shown in Figure 2.
Cross 1. Cross w; +; UAS-ORx(w+)/+ to the double balancer strain, w; Bl/Cyo; TM2/TM6b. Select progeny with red eyes (w+), curly wings (Cyo) and tubby phenotype (with a cluster of bristles on the humerus, TM6b): w; +/Cyo; UAS-ORx(w+)/TM6b. The ebony phenotype features darker pigmentation and presents in flies with both third chromosome balancers (TM2/TM6B). In this schema the OR transgene is present on the third chromosome, its selection is thus mutually exclusive with the ebony phenotype.
Cross 2. Cross selected progeny again to w; Bl/Cyo; TM2/TM6b. Select progeny with red eyes, curly wings, tubby phenotype and short bristles (Bl), with genotype: w−; Bl/Cyo; UAS-ORx(w+)/TM6b. Since the ΔHalo mutation has no phenotypic markers, and is introduced in a genetic background with wild-type (longer) bristles, it is necessary to first pass the OR transgene through a short bristle phenotype in order to be able to discriminate the ΔHalo chromosome from its counterpart wild-type chromosome present in the original transgenic flies received.
Cross 3. Cross selected progeny to w−; ΔHalo/Cyo; TM2/TM6b. Select progeny with red eyes, curly wings, tubby and wild-type bristles (ΔHalo), with genotype, w−; ΔHalo/Cyo; UAS-ORx(w+)/TM6b.
Cross 4. Self-cross selected male and female progeny. Select and breed male and female progeny with red eyes, curly wings, and wild type bristles, without tubby, w−; ΔHalo/Cyo; UAS-ORx, in order to establish a stable stock of fly lines that are ready for the experimental cross and downstream electrophysiological assay. In this stock line, the ΔHalo chromosome is maintained in the presence of the Cyo balancer. While ΔHalo homozygous flies are viable and obtained for downstream assay, they are not fit for reproduction and are relatively sick. It is thus advisable to also maintain a stock of flies with genotype, w−; Bl/Cyo; UAS-ORx(w+)/TM6b, obtained after Cross 2 (above).
Expression Cross. Cross w−; ΔHalo/Cyo; UAS-ORx(w+) to w; ΔHalo/Cyo; DmelOR22a-Gal4(w+). Select female progeny with red eyes and straight wings, w−; ΔHalo/ΔHalo; DmelOR22a-Gal4(w+)/UAS-ORx(w+). These flies are to be used for physiological assay, as described below. Since both the Gal4 and UAS constructs in this system are maintained on the third chromosome, it is not possible to maintain a stable stock of these flies for physiological assays on demand. The expression cross must be made as described above whenever OR assays in the ab3A empty neuron system is required.
Fly Crossing Scheme for the T1 Knock-In Neuron System
The OR transgene must be crossed into the OR67d-knockout/Gal4-knock-in genetic background, which contains a Gal4 transgene in place of the native OR67d gene, and under control of the native OR67d promoter (Kurtovic et al., 2007). An outline of the required crosses is shown in Figure 3.
Cross 1. Cross w; UAS-ORx(w+)/+; + to the double balancer strain, w; Bl/Cyo; TM2/TM6b. Select progeny with red eyes (w+), curly wings (Cyo) and tubby phenotype (TM6b), with genotype: w−; UAS-ORx(w+)/Cyo; +/TM6b.
Cross 2. Cross selected progeny again to w; Bl/Cyo; TM2/TM6b. Select progeny with red eyes, curly wings, tubby phenotype and ebony body color, with genotype: w−; UAS-ORx(w+)/Cyo; TM2/TM6b.
Cross 3. Cross selected progeny to w−; Bl/Cyo; OR67d-Gal4. Select progeny with red eyes, curly wings, wild-type bristles, and tubby phenotype, with genotype w−; UAS-ORx(w+)/Cyo; OR67d-Gal4/TM6b.
Cross 4. Self-cross selected male and female progeny. Select and breed male and female progeny with red eyes, straight wings, and without tubby phenotype, with genotype: w−; UAS-ORx(w+); OR67d-Gal4. In this case, these flies are viable for stock breeding and are also of the correct genotype for experimental testing.
Single Sensillum Recordings
Mounting
Trap a fly inside a 200-μl pipette tip. Horizontally cut the pipette tip close to the head and push the head slightly out of the pipette tip. Place the pipette tip containing the fly facing upwards on dental wax on a microscope slide. Push the antennae on the glass slide fixed with dental wax on the full-length microscope slide.
Fix the glass capillary on dental wax on the microscope slide. Use the glass capillary to push the antenna down by pressing the section between the second and third antennal segment. In the case of transgene ORs expressed in the ab3A OSN, manipulate the glass capillary until exposing the dorso-medial area of the antenna. A cluster of thumb-shaped sensilla (large basiconic sensilla) facing upwards should be visible. For testing transgene ORs that are expressed in trichoid T1 sensilla, manipulate the glass capillary to firmly press the lateral side of the antenna against the microscope slide. The corresponding long pointy sensilla (T1) are then located at the superior side of the antenna from the lateral view. Anatomical maps of the D. melanogaster antenna and sensillum types are found in de Bruyne et al. (2001), Stocker (2001), Dobritsa et al. (2003), and Couto et al. (2005). Guidance to perform electrophysiological recordings can be found in Pellegrino et al. (2010) and in Benton and Dahanukar (2011).
Recording
Place the mounted fly under the microscope and penetrate its right eye with the tungsten reference electrode. At high magnification of the microscope, use the micromanipulator to move the tungsten recording electrode along the antenna. Penetrate either large basiconic sensilla located in the dorso-medial area of the antennae (ab3, empty neuron system), or long trichoid sensilla at the tip of the antennae (T1 knock-in neuron system).
Gently manipulate the recording electrode along the base of the sensilla until a clear pattern of neuronal activity is established (monitored by Autospike; Syntech, Kirchzarten).
Identification of Sensillum Identity
After making contact, confirm sensillum identity prior to testing.
AB3 sensillum. Stable recordings from ab3 sensilla (Dobritsa et al., 2003) will produce spike trains from two OSNs, ab3A and ab3B, with two distinct amplitudes (Figure 4A). A response to stimulation with 2-heptanone, targeting the native Or85b expressed in ab3B, serves as double control: it confirms proper sensillum contact and the identity of the ab3 sensillum.
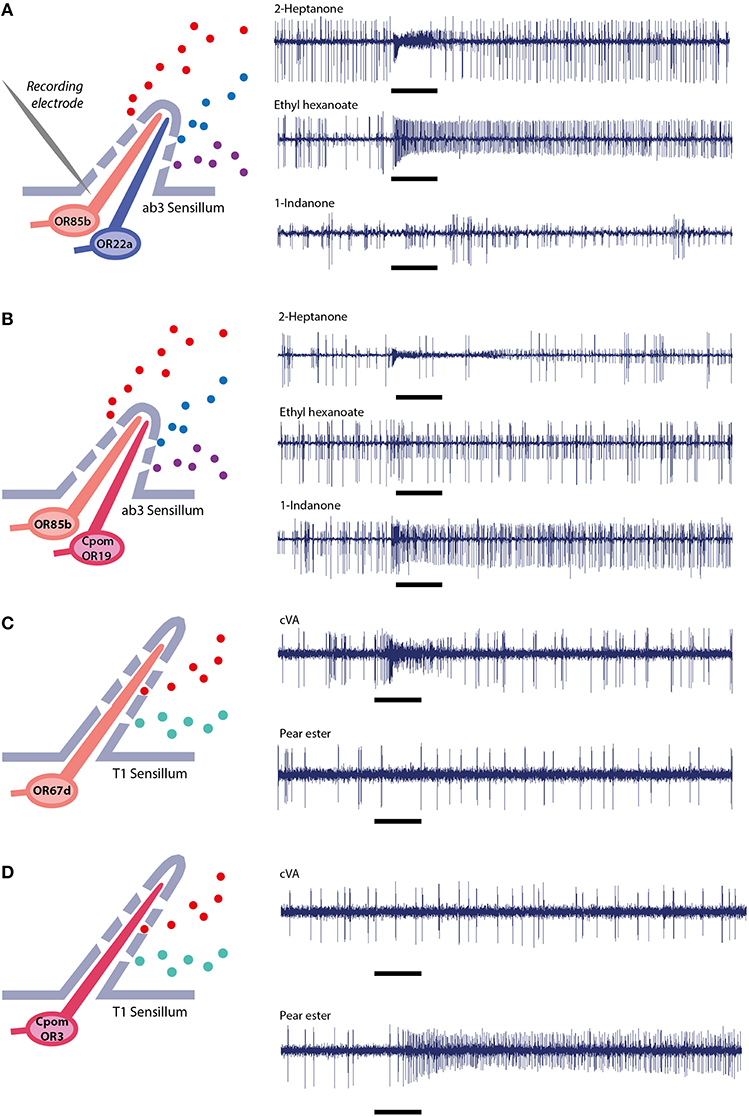
Figure 4. Single sensillum electrophysiological recordings. In ab3 empty neuron system, (A) wild-type flies expressing native ORs, (B) mutant flies expressing native OR85b in the small neuron and transgenic CpomOR19 from C. pomonella in the large neuron (Gonzalez et al., 2015). In T1 empty neuron system, (C) wild-type flies expressing native OR67d, (D) mutant flies expressing transgenic CpomOR3 (Bengtsson et al., 2014).
Regular spontaneous activity of the second neuron ab3A is indicative of a functional transgenic OR. The frequency of spontaneous neuronal activity of OSNs has been shown to be determined by the specific receptor protein that is expressed in the neuron (Hallem et al., 2004), variance of this feature is thus to be expected. However, response to stimulation with the wild type ligands, ethyl hexanoate or ethyl butyrate, is absent or modified and confirms that OR22a is not expressed (Figure 4B).
Last but not least, contact with the wrong large basiconic sensillum subtype (ab1 or ab2) can be ruled out by testing their natural ligands. The ab1 sensillum contains four OSNs (with varying spike amplitudes), one of which is responsive to CO2: breathing gently over the fly will produce a response. For ab2, containing two OSNs, a ethyl-3-hydroxybutyrate stimulus produces a strong response.
T1 sensillum. When recording from T1 sensilla (Kurtovic et al., 2007), only one OSN responds (Figure 4C). The two other types of trichoid sensilla, T2 and T3, contain 2 and 3 neurons, respectively, facilitating discrimination between different trichoid sensilla. The OR insert in T1 is confirmed through lack of a response to cVA (Figure 4D).
Odor Stimulation
Apply 10 μl of test chemical solvent dilutions to filter paper discs inside Pasteur pipettes. Pulses of charcoal-filtered air (2.5 ml) through the pipette are delivered by a stimulus controller (Syntech, Kirchzarten, Germany), lasting at least 0.5 s, into glass tubing delivering air to the fly. Verify sensillum identity before testing. Once contact with correct sensillum subtype is established, sequentially deliver the test panel of compounds. Response magnitude is determined by counting the number of spikes before and after the onset of a response.
Alternatively, test stimuli can be provided by the effluent of a gas chromatograph (GC-SSR). The main advantages of using the GC for stimulation are discussed below (Section Testing Odourants).
Anticipated Results and Troubleshooting
Molecular Cloning and Heterologous Expression
Most attempts to amplify the ORF of a determined OR and clone it into the TOPO entry vector will be successful with little difficulty. Common problems may be remedied after consultation with the troubleshooting section of the TOPO cloning user manual. It must be noted, however, that in some cases, attempts to amplify or obtain clones with the OR construct in the correct orientation can be unsuccessful. In case of amplification issues, it may be necessary to optimize the PCR amplification with gradient PCR or selection of new primer pairs that are more compatible with each other and the target cDNA in question.
Pertaining to issues with identifying TOPO clones with the desired insert in the correct orientation, it may be necessary, during the colony PCR screening step, to assay both combination of plasmid/insert primer pairs, due to primer incompatibility issues. Otherwise, various unknown and unapparent factors may render some OR constructs refractory to plasmid vector propagation. In our experience, this is rare, but may happen; with patience and effort these molecular obstacles can usually be overcome.
In test flies, OR transgenes are expressed in either ab3A or T1 OSNs, which lack their native OR22a/b or OR67d receptors, respectively. This should be verified though PCR assay of transgene OR expression in antennae of progeny obtained through experimental crosses. This can be done by following the procedures described in section 3.1.1 to 3.1.4, using the antennae of 100 test flies as starting material. It is our experience that most ORs will be expressed appropriately in the D. melanogaster antennae, once the appropriate fly crosses have been made. However, in few cases, ORs are refractory to expression in these sensilla, for yet unknown reasons.
Response of OSNs Expressing Novel ORs
The functionality of heterologous expression of ORs in ab3A and T1 OSNs is assessed by SSR. As mentioned above, the ab3 basiconic sensilla house two neurons, ab3A, which natively expresses OR22a (tuned to ethyl hexanoate and ethyl butyrate) and ab3B, which expresses OR85b (tuned to 2-heptanone). Identity and functionality of this sensillum can be verified through stimulation of the ab3B neuron with 2-heptanone.
If ab3A sensilla, expressing a novel OR, should respond to ethyl hexanoate or ethyl butyrate, further testing with other OR22a ligands such as methyl hexanoate, isobutyl acetate and methyl octanoate (Hallem and Carlson, 2006) can help to determine whether the native DmelOR22a or the experimental transgenic OR produce this response. Expression of the transgene OR and lack of expression of the native OR22a receptor can also be confirmed with a PCR assay, as described above. If DmelOR22a is indeed present, it is likely due to erroneous fly-crossings that failed to exclude the wild-type second chromosome. In this case, it will be necessary to carefully perform the fly-crossing schema again to ensure that the ΔHalo chromosome is present in place of a wild-type chromosome containing DmelOR22a.
On the other hand, even if receptors are functionally expressed and confer a background-firing rate on respective ab3A and T1 OSNs, test odourants may not elicit significant responses. The solution is to use a broader test panel of odourants, taking into account a diversity of ecological sources of odourants that are representative of the olfactory environment of the insect being studied. Using volatile collections from natural substrates in combination with GC-SSR is an option.
In a functional transgenic ab3A system, only ligands activating the transgenic OR will produce a response from ab3A neurons. Recently, we have deorphanized CpomOR19 and SlitOR19, of C. pomonella and Spodoptera littoralis, using the ab3A system. A response to 1-indanone was recorded only after expression of CpomOR19 or SlitOR19 in ab3A OSNs, and not from wild-type D. melanogaster (Figures 4A,B; Gonzalez et al., 2015).
Expression of transgenic candidate PRs or other ORs in T1 neurons is characterized by an irregular firing rate (Ronderos et al., 2014). Wild-type flies will show an intense and long-lasting response when stimulated with cVA, while experimental flies will respond with a less intense but more irregular pattern to the ligands of the respective transgene ORs (Figures 4C,D).
CpomOR3 belongs phylogenetically to the clade of moth pheromone receptors and is tuned to the plant volatile pear ester. After functional expression of CpomOR3 in neurons of either ab3 or T1 sensilla, responsiveness and tuning were equally specific and sensitive (Bengtsson et al., 2014).
Sensillum Environment and OR Function
Systematic investigations of the OR repertoires of Drosophila and the malaria mosquito Anopheles gambiae demonstrate that the ab3A empty neuron is a faithful expression system for insect OR genes. OR response profiles in native neurons and in the empty neuron, generally resemble each other, but are not identical (Dobritsa et al., 2003; Hallem et al., 2004; Carey et al., 2010). However, not all receptors will work in ab3A neurons. For example, only 50 out of 72 cloned A. gambiae ORs were functional in the empty neuron (Carey et al., 2010). A similar percentage of D. melanogaster ORs were also not functional in the ab3A empty neuron (Hallem et al., 2004). In cases where transgene ORs are expressed but not functional in ab3A neuron, the background neuronal firing rate phenocopies the ab3A empty neuron condition with regular cluster bursts of multiple action potentials (Dobritsa et al., 2003).
Advances in transcriptomics and the molecular basis of odourant reception in insects will help us to understand what facilitates or impedes correct function of ORs. One explanation is that the cellular environment contributes membrane-bound proteins, such as sensory neuron membrane proteins (SNMPs) and extracellular odourant- or pheromone-binding proteins (OBPs, PBPs), which are known to mediate interactions between ORs or PRs and odourant molecules (Nichols and Vogt, 2008; Leal, 2013; Li et al., 2014; Vogt et al., 2015). Expression patterns of SNMPs and OBPs have been investigated across olfactory organs (Vogt et al., 2002; Shanbhag et al., 2005; Benton et al., 2007), however detailed expression patterns of these genes at the cellular level, with reference to ORs, remain to be fully described.
The combined role of ORs and OBPs, and PRs and PBPs, respectively, in odourant detection and discrimination, has been confirmed by co-expression analysis and by heterologous expression in Xenopus (Schultze et al., 2013; Chang et al., 2015). This is in line with the observation that some ORs are functional only in trichoid sensilla. Presence of the extracellular protein LUSH is necessary for pheromone-sensitive OSNs in Drosophila T1 sensilla (Xu et al., 2005; Laughlin et al., 2008). Similarly, DmelOr83c does not produce a response at all when transgenically expressed in basiconic sensilla OSNs, but requires factors present in trichoid sensilla, including SMNP1 (Ronderos et al., 2014).
Testing Odourants
Odourants used for functional characterization of insect ORs comprise a range of compounds which greatly differ in molecular weight and, accordingly, also in vapor pressure and evaporation rates. In addition, when compounds are formulated onto passive dispensers, their physicochemical affinity to the substrate, including polarity, will modify evaporation rates. Release rates of the odourants included in a test panel will often differ by several orders of magnitude. Yet, these differences in release rates are notoriously ignored or underestimated. For valid structure-activity comparisons, the amounts of test compounds delivered to the antenna need to be corrected for differences in evaporation rates (Bengtsson et al., 1990).
Chemical impurities of test odourants are another serious error source. Standards of natural or synthetic compounds invariably contain impurities, which may be more active than the test compound itself. Even impurities present in trace amounts may elicit strong responses, since ORs are indeed known to be strongly tuned to their key ligands. Last but not least, availability of synthetic standards is often a limiting factor.
Using GC-SSR for stimulation elegantly accounts for these main concerns: release rates, chemical purity and availability of standards. Headspace collections from biological substrates, for example, leaves or fruits of higher plants, will typically contain several dozens of volatiles. Through the GC column, these compounds are delivered at known amounts, independently of vapor pressure. Choice of the column and temperature programme will ensure delivery of pure compound at baseline separation. This includes even geometrical or optical isomers of plant volatiles, which rarely are available as pure standards.
Conclusion
We expect heterologous expression of insect ORs in Drosophila OSNs to make a significant future contribution to the identification of insect semiochemicals, and to investigations of the phylogenetic progression and the functional properties of ORs.
Author Contributions
PW: conceived the idea of the manuscript. FG, WW, and PW designed and prepared the figures. FG, WW, and PW wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge financial support for this study by the Linnaeus initiative “Insect Chemical Ecology, Ethology, and Evolution (IC-E3)” (Formas, SLU; Formas Dnr 2011 1370), the Faculty of Landscape Architecture, Horticulture, and Crop Production Science, SLU Alnarp and Carl Tryggers Stiftelse för Vetenskaplig Forskning. We thank Dr. Bonaventure Aman Omondi for constructive comments on draft versions of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fevo.2016.00024
Supplemental Figure 1. Timeframe for molecular cloning and generation of a transgenic fly line. Multiple ORs can be processed in parallel. Time for molecular work is a best-case scenario, assuming access to DNA sequencing of the generated plasmids. Timeline for obtaining transgenic flies after sending transformation/expression plasmids depends on micro-injection company.
References
Anderson, A. R., Wanner, K. W., Trowell, S. C., Warr, C. G., Jaquin-Joly, E., Zagatti, P., et al. (2009). Molecular basis of female-specific odorant responses in Bombyx mori. Insect Biochem. Mol. Biol. 39, 189–197. doi: 10.1016/j.ibmb.2008.11.002
Ansebo, L., Coracini, M. D. A., Bengtsson, M., Liblikas, I., Ramírez, M., Borg-Karlsson, A.-K., et al. (2004). Antennal and behavioural response of codling moth Cydia pomonella to plant volatiles. J. Appl. Entomol. 128, 488–493. doi: 10.1111/j.1439-0418.2004.00878.488–493
Arn, H., Städler, E., and Rauscher, S. (1975). The electroantennographic detector - a selective and sensitive tool in the gas chromatographic analysis of insect pheromones. Z. Naturforsch. 30c, 722–725. doi: 10.1515/znc-1975-11-1204
Arn, H., Tóth, M., and Priesner, E. (1992). List of Sex Pheromones of Lepidoptera and Related Attractants, 2nd Edn. Montfavet: International Organization for Biological Control.
Bäckman, A.-C., Bengtsson, M., Borg-Karlsson, A.-K., Liblikas, I., and Witzgall, P. (2001). Volatiles from apple eliciting antennal responses in female codling moth Cydia pomonella (L.) (Lepidoptera: Tortricidae): effect of plant injury and sampling technique. Z. Naturforsch. 56c, 262–268. doi: 10.1515/znc-2001-3-415
Bengtsson, J. M., Gonzalez, F., Cattaneo, A. M., Montagné, N., Walker, W. B., Bengtsson, M., et al. (2014). A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2:33. doi: 10.3389/fevo.2014.00033
Bengtsson, J. M., Trona, F., Montagné, N., Anfora, G., Ignell, R., Witzgall, P., et al. (2012). Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS ONE 7:e31620. doi: 10.1371/journal.pone.0031620
Bengtsson, M., Liljefors, T., Hansson, B. S., Löfstedt, C., and Copaja, S. V. (1990). Structure-activity relationships for chain-shortened analogs of (Z)-5-decenyl acetate, a pheromone component of the turnip moth, Agrotis segetum. J. Chem. Ecol. 16, 667–684. doi: 10.1007/BF01016478
Benton, R., and Dahanukar, A. (2011). “Electrophysiological recording from Drosophila taste sensilla,” in Drosophila Neurobiology: A Laboratory Manual, eds B. Zhang, M. R. Freeman, and S. Waddell (New York, NY: Cold Spring Harbor Protocols), 839–850.
Benton, R., Vannice, K. S., and Vosshall, L. B. (2007). An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450, 289–293. doi: 10.1038/nature06328
Bischof, J., Maeda, R. K., Hediger, M., Karch, F., and Basler, K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific ϕC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317. doi: 10.1073/pnas.0611511104
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415.
Carey, A. F., Wang, G., Su, C. Y., Zwiebel, L. J., and Carlson, J. R. (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71. doi: 10.1038/nature08834
Chang, H., Liu, Y., Yang, T., Pelosi, P., Dong, S., and Wang, G. (2015). Pheromone binding proteins enhance the sensitivity of olfactory receptors to sex pheromones in Chilo suppressalis. Sci. Rep. 5:13093. doi: 10.1038/srep13093
Clyne, P. J., Warr, C. G., Freeman, M. R., Lessing, D., Kim, J., and Carlson, J. R. (1999). A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22, 327–338. doi: 10.1016/S0896-6273(00)81093-4
Coracini, M., Bengtsson, M., Liblikas, I., and Witzgall, P. (2004). Attraction of codling moth males to apple volatiles. Ent. Exp. Appl. 110, 1–10. doi: 10.1111/j.0013-8703.2004.00124
Corcoran, J. A., Jordan, M. D., Carraher, C., and Newcomb, R. D. (2014). A novel method to study insect olfactory receptor function using HEK293 cells. Insect Biochem. Mol. Biol. 54, 22–32. doi: 10.1016/j.ibmb.2014.08.005
Couto, A., Alenius, M., and Dickson, B. J. (2005). Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15, 1535–1547. doi: 10.1016/j.cub.2005.07.034
de Bruyne, M., Foster, K., and Carlson, J. R. (2001). Odor coding in the Drosophila antenna. Neuron 30, 537–552. doi: 10.1016/S0896-6273(01)00289-6
Dethier, V. G. (1947). Chemical Insect Attractants and Repellents. Philadelphia, PA: The Blakiston Company.
Dobritsa, A. A., van der Goes van Naters, W., Warr, C. G., Steinbrecht, R. A., and Carlson, J. R. (2003). Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. doi: 10.1016/S0896-6273(03)00094-1
El-Sayed, A. M. (2015). The Pherobase: Database of Insect Pheromones and Semiochemicals. Available online at: http://www.pherobase.com
Gao, Q., and Chess, A. (1999). Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60, 31–39. doi: 10.1006/geno.1999.5894
Goldman, A. L., van der Goes van Naters, W., Lessing, D., Warr, C. G., and Carlson, J. R. (2005). Coexpression of two functional odor receptors in one neuron. Neuron 45, 661–666. doi: 10.1016/j.neuron.2005.01.025
Gonzalez, F., Bengtsson, J. M., Walker, W. B., Rodrigues-Sousa, M. F., Cattaneo, A. M., Montagné, N., et al. (2015). A conserved odorant receptor detects the same 1-indanone analogs in a tortricid and a noctuid moth. Front. Ecol. Evol. 3:131. doi: 10.3389/fevo.2015.00131
Greenspan, R. J. (1997). Fly Pushing: The Theory and Practice of Drosophila Genetics. New York, NY: Cold Springs Harbor Laboratory Press.
Große-Wilde, E., Svatos, A., and Krieger, J. (2006). A pheromone-binding protein mediates the bombykol-induced activation of a pheromone receptor in vitro. Chem. Senses 31, 547–555. doi: 10.1093/chemse/bjj059
Gross, S. P., Guo, Y., Martinez, J. E., and Welte, M. A. (2003). A determinant for directionality of organelle transport in Drosophila embryos. Curr. Biol. 13, 1660–1668. doi: 10.1016/j.cub.2003.08.032
Hallem, E. A., and Carlson, J. R. (2006). Coding of odors by a receptor repertoire. Cell 125, 143–160. doi: 10.1016/j.cell.2006.01.050
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. doi: 10.1016/j.cell.2004.05.012
Jiang, X.-J., Guo, H., Di, C., Yu, S., Zhu, L., Huang, L.-Q., et al. (2014). Sequence similarity and functional comparisons of pheromone receptor orthologs in two closely related Helicoverpa species. Insect Biochem. Mol. Biol. 48, 63–74. doi: 10.1016/j.ibmb.2014.02.010
Jordan, M. D., Anderson, A., Begum, D., Carraher, C., Authier, A., Marshall, S. D., et al. (2009). Odorant receptors from the light brown apple moth (Epiphyas postvittana) recognize important volatile compounds produced by plants. Chem. Senses 34, 383–394. doi: 10.1093/chemse/bjp010
Jurenka, R. (2004). “Insect pheromone biosynthesis,” in The Chemistry of Pheromones and Other Semiochemicals I, ed S. Schulz (Berlin; Heidelberg: Springer), 97–132. doi: 10.1007/b95450
Kiely, A., Authier, A., Kralicek, A. V., Warr, C. G., and Newcomb, R. D. (2007). Functional analysis of a Drosophila melanogaster olfactory receptor expressed in Sf9 cells. J. Neurosci. Methods 159, 189–194. doi: 10.1016/j.jnrumeth.2006.07.005
Kreher, S. A., Kwon, J. Y., and Carlson, J. R. (2005). The molecular basis of odor coding in the Drosophila larva. Neuron 46, 445–456. doi: 10.1016/j.neuron.2005.04.007
Kreher, S. A., Mathew, D., Kim, J., and Carlson, J. R. (2008). Translation of sensory input into behavioral output via an olfactory system. Neuron 59, 110–124. doi: 10.1016/j.neuron.2008.06.010
Kurtovic, A., Widmer, A., and Dickson, B. J. (2007). A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446, 542–546. doi: 10.1038/nature05672
Laughlin, J. D., Ha, T. S., Jones, D. N., and Smith, D. P. (2008). Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 133, 1255–1265. doi: 10.1016/j.cell.2008.04.046
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Leary, G. P., Allen, J. E., Bunger, P. L., Luginbill, J. B., Linn, C. E., Macallister, I. E., et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A. 109, 14081–14086. doi: 10.1073/pnas.1204661109
Li, Z., Ni, J. D., Huang, J., and Montell, C. (2014). Requirement for Drosophila SNMP1 for rapid activation and termination of pheromone-induced activity. PLoS Genet. 10:e1004600. doi: 10.1371/journal.pgen.1004600
Light, D. M., and Beck, J. J. (2012). Behavior of codling moth (Lepidoptera: Tortricidae) neonate larvae on surfaces treated with microencapsulated pear ester. Environ. Entomol. 41, 603–611. doi: 10.1603/EN11273
Light, D. M., and Knight, A. (2005). Specificity of codling moth (Lepidoptera: Tortricidae) for the host plant kairomone, ethyl (2E,4Z)-2,4-decadienoate: field bioassays with pome fruit volatiles, analogue, and isomeric compounds. J. Agric. Food Chem. 53, 4046–4053. doi: 10.1021/jf040431r
Light, D. M., Knight, A. L., Henrick, C. A., Rajapaska, D., Lingren, B., Dickens, J. C., et al. (2001). A pear-derived kairomone with pheromonal potency that attracts male and female codling moth, Cydia pomonella (L.). Naturwiss 88, 333–338. doi: 10.1007/s001140100243
Liu, C., Liu, Y., Walker, W. B., Dong, S., and Wang, G. (2013). Identification and functional characterization of sex pheromone receptors in beet armyworm Spodoptera exigua (Hübner). Insect Biochem. Mol. Biol. 43, 747–754. doi: 10.1016/j.ibjmb.2013.05.009
Matarazzo, V., Clot-Faybesse, O., Marcet, B., Guiraudie-Capraz, G., Atanasova, B., Devauchelle, G., et al. (2005). Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem. Senses 30, 195–207. doi: 10.1093/chemse/bji015
Mathew, D., Martelli, C., Kelley-Swift, E., Brusalis, C., Gershow, M., Samuel, A. D., et al. (2013). Functional diversity among sensory receptors in a Drosophila olfactory circuit. Proc. Natl. Acad. Sci. U.S.A. 110, E2134–E2143. doi: 10.1073/pnas.1306976110
Mitsuno, H., Sakurai, T., Murai, M., Yasuda, T., Kugimiya, S., Ozawa, R., et al. (2008). Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 28, 893–902. doi: 10.1111/j.1460-9568.2008.06429.x
Montagné, N., Chertemps, T., Brigaud, I., François, A., François, M. C., De Fouchier, A., et al. (2012). Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci. 36, 2588–2596. doi: 10.1111/j.1460-9568.2012.08183.x
Nichols, Z., and Vogt, R. G. (2008). The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 398–415. doi: 10.1016/j.ibmb.2007.11.0033
Pellegrino, M., Nakagawa, T., and Vosshall, L. B. (2010). Single sensillum recordings in the insects Drosophila melanogaster and Anopheles gambiae. J. Vis. Exp. 36:e1725. doi: 10.3791/1725
Ronderos, D. S., Lin, C. C., Potter, C. J., and Smith, D. P. (2014). Farnesol-detecting olfactory neurons in Drosophila. J. Neurosci. 34, 3959–3968. doi: 10.1523/JNEUROSCI.4582-13.2014
Sakurai, T., Nakagawa, T., Mitsuno, H., Mori, H., Endo, Y., Tanoue, S., et al. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 101, 16653–16658. doi: 10.1073/pnas.0407596101
Schneider, D. (1957). Elektrophysiologische untersuchungen von chemo- and mechanorezeptoren der Antenne des Seidenspinners Bombyx mori L. Z. Vergl. Physiol. 40, 8–41. doi: 10.1007/BF00298148
Schultze, A., Pregitzer, P., Walter, M. F., Woods, D. F., Marinotti, O., Breer, H., et al. (2013). The co-expression pattern of odorant binding proteins and olfactory receptors identify distinct trichoid sensilla on the antenna of the malaria mosquito Anopheles gambiae. PLoS ONE 8:e69412. doi: 10.1371/journal.pone.0069412
Shanbhag, S. R., Müller, B., and Steinbrecht, R. A. (1999). Atlas of olfactory organs of Drosophila melanogaster: 1. Types, external organization, innervation and distribution of olfactory sensilla. Int. J. Insect Morphol. Embryol. 28, 377–397. doi: 10.1016/S0020-7322(99)00039-2
Shanbhag, S. R., Smith, D. P., and Steinbrecht, R. A. (2005). Three odorant-binding proteins are co-expressed in sensilla trichodea of Drosophila melanogaster. Arthropod Struct. Dev. 34, 153–165. doi: 10.1016/j.asd.2005.01.003
Stocker, R. F. (2001). Drosophila as a focus in olfactory research: mapping of olfactory sensilla by fine structure, odor specificity, odorant receptor expression, and central connectivity. Microsc. Res. Tech. 55, 284–296. doi: 10.1002/jemt.1178
Syed, Z., Ishida, Y., Taylor, K., Kimbrell, D. A., and Leal, W. S. (2006). Pheromone reception in fruit flies expressing a moth's odorant receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 16538–16543. doi: 10.1073/pnas.0607874103
Syed, Z., Kopp, A., Kimbrell, D. A., and Leal, W. S. (2010). Bombykol receptors in the silkworm moth and the fruit fly. Proc. Natl. Acad. Sci. U.S.A. 107, 9436–9439. doi: 10.1073/pnas.1003881107
Tegler, L. T., Corin, K., Hillger, J., Wassie, B., Yu, Y., and Zhang, S. (2015). Cell-free expression, purification, and ligand-binding analysis of Drosophila melanogaster olfactory receptors DmOR67a, DmOR85b and DmORCO. Sci. Rep. 5:7867. doi: 10.1038/srep07867
Trona, F., Anfora, G., Balkenius, A., Bengtsson, M., Tasin, M., Knight, A., et al. (2013). Neural coding merges sex and habitat chemosensory signals in an insect herbivore. Proc. Biol Sci. 280, 20130267. doi: 10.1098/rspb.2013.0267
Trona, F., Anfora, G., Bengtsson, M., Witzgall, P., and Ignell, R. (2010). Coding and interaction of sex pheromone and plant volatile signals in the antennal lobe of the codling moth Cydia pomonella. J. Exp. Biol. 213, 4291–4303. doi: 10.1242/jeb.047365
Vogt, R. G., Große-Wilde, E., and Zhou, J. J. (2015). The lepidoptera odorant binding protein gene family: gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 62, 142–153. doi: 10.1016/j.ibmb.2015.03.003
Vogt, R. G., Rogers, M. E., Franco, M. D., and Sun, M. (2002). A comparative study of odorant binding protein genes: differential expression of the PBP1-GOBP2 gene cluster in Manduca sexta (Lepidoptera) and the organization of OBP genes in Drosophila melanogaster (Diptera). J. Exp. Biol. 205, 719–744.
Vosshall, L. B., Amrein, H., Morozov, P. S., Rzhetsky, A., and Axel, R. (1999). A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96, 725–736. doi: 10.1016/S0092-8674(00)80582-6
Wanner, K. W., Nichols, A. S., Allen, J. E., Bunger, P. L., Garczynski, S. F., Linn, C. E., et al. (2010). Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS ONE 5:e8685. doi: 10.1371/journal.pone.0008685
Xu, P., Atkinson, R., Jones, D. N., and Smith, D. P. (2005). Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 193–200. doi: 10.1016/j.neuron.2004.12.031
Xu, W., Papanicolaou, A., Liu, N. Y., Dong, S. L., and Anderson, A. (2015). Chemosensory receptor genes in the Oriental tobacco budworm Helicoverpa assulta. Insect Mol. Biol. 24, 253–263. doi: 10.1111/imb.12153
Keywords: insect ORs, heterologous expression, empty neuron system, single sensillum recordings, deorphanization
Citation: Gonzalez F, Witzgall P and Walker WB III (2016) Protocol for Heterologous Expression of Insect Odourant Receptors in Drosophila. Front. Ecol. Evol. 4:24. doi: 10.3389/fevo.2016.00024
Received: 21 January 2016; Accepted: 01 March 2016;
Published: 16 March 2016.
Edited by:
Jeffrey A. Riffell, University of Washington, USAReviewed by:
Wynand Van Der Goes Van Naters, Cardiff University, UKZainulabeuddin Syed, University of Notre Dame, USA
Copyright © 2016 Gonzalez, Witzgall and Walker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francisco Gonzalez, francisco.gonzalez@slu.se;
William B. Walker, william.b.walker.iii@slu.se
 Francisco Gonzalez
Francisco Gonzalez Peter Witzgall
Peter Witzgall William B. Walker III
William B. Walker III