Mechanism of Resistance to Glyphosate in Lolium perenne from Argentina
- 1Chacra Experimental Integrada Barrow (MAIBA—INTA), National Scientific and Technical Research Council, Tres Arroyos, Argentina
- 2Institute of Plant Physiology (INFIVE), National Scientific and Technical Research Council, La Plata, Argentina
- 3Chacra Experimental Integrada Barrow (MAIBA—INTA), Ministry of Agro-Industry and National Institute of Agricultural Technology, Tres Arroyos, Argentina
- 4Plant Proteins Research Center (CIPROVE), Faculty of Exact Sciences, National University of La Plata, La Plata, Argentina
- 5Institute of Plant Physiology (INFIVE), National University of La Plata, La Plata, Argentina
- 6Centro de Investigaciones en Sanidad Vegetal Faculty of Agronomy and Forestry, National University of La Plata, La Plata, Argentina
In Argentina, glyphosate resistance was reported in a Lolium perenne population after 12 years of successful herbicide use. The aim of the current paper was to put in evidence for the mechanism of glyphosate resistance of this weed. Susceptible leaves treated with different doses of glyphosate and incubated in vitro showed an accumulation of shikimic acid of around three to five times the basal level, while no changes were detected in leaves of glyphosate-resistant plants. The resistance mechanism prevents shikimate accumulation in leaves, even under such tissue-isolation conditions. The activity of the glyphosate target enzyme (EPSPS: 5-enolpyruvylshikimate-3-phosphate synthase) was quantified at different herbicide concentrations. EPSPS from resistant plants showed no difference in glyphosate-sensitivity compared to EPSPS from susceptible plants, and, accordingly, no amino acid substitution causing mutations associated with resistance were found. While the glyphosate target enzymes were equally sensitive, the basal EPSPS activity in glyphosate resistant plants was approximately 3-fold higher than the EPSPS activity in susceptible plants. This increased EPSPS activity in glyphosate resistant plants was associated with a 15-fold higher expression of EPSPS compared with susceptible plants. Therefore, the over-expression of EPSPS appears to be the main mechanism responsible for resistance to glyphosate. This mechanism has a constitutive character and has important effects on plant fitness, as recently reported.
Introduction
Glyphosate-resistant weeds put at risk the efficacy of glyphosate, an important herbicide used worldwide in current cropping systems. Since the first report of glyphosate resistance in 1998, the evolution of glyphosate resistant weed populations has rapidly escalated (Powles et al., 1998; Duke and Powles, 2008). At present, glyphosate resistance has been documented in 36 species (Heap, 2017). Herbicide-resistant weeds have several negative effects on farms and surrounding regions. They would increase the costs and difficulties for weed control (Norsworthy et al., 2012). Accordingly, the evolution of glyphosate-resistant weeds threatens the long-term efficacy of the world's most important herbicide resource (Duke and Powles, 2008).
The target enzyme of glyphosate is 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS; EC. 2.5.1.19) that catalyses the reaction of shikimate-3-phosphate and phosphoenolpyruvate to yield 5-enolpyruvylshikimate-3-phosphate (Franz et al., 1997). This is the sixth enzyme on the shikimate pathway, which is essential for the synthesis of aromatic amino acids (Schönbrunn et al., 2001). The inhibition of EPSPS triggers the accumulation of shikimate, a substrate of the enzyme. Therefore, the effect of glyphosate on plants can be easily measured by monitoring the accumulation of this substrate of EPSPS (Dayan et al., 2015).
After EPSPS inhibition, several physiological processes are affected by glyphosate. The effect of the herbicide is not restricted to the inhibition of aromatic amino acid biosynthesis alone. It also disrupts the de novo biosynthesis of most non-aromatic amino acids, coupled with decreased protein synthesis (Maroli et al., 2016). In consequence, growth is inhibited (Pline et al., 2002; Orcaray et al., 2012), the transport of assimilates is reduced (Yanniccari et al., 2012a) and photosynthetic CO2 assimilation decreases significantly (Olesen and Cedergreen, 2010). Several of these effects are limited or absent in glyphosate-resistant weed plants treated with the herbicide (Yanniccari et al., 2012a,b,c; Maroli et al., 2016).
Although more than 200 weed populations have shown glyphosate resistance in 26 countries (Heap, 2017), mechanism of plant survival was determined in a few cases. These mechanisms can be grouped into target-site and non-target-site based mechanisms (Powles and Yu, 2010). In the first category, the resistance can occur due to a gene mutation conferring an amino acid change that prevents herbicide binding. EPSPS target site mutations have been reported in six species, most frequently in the genus Lolium (Sammons and Gaines, 2014). Amino acid substitutions at Pro-106 have been the most common target site mutations in glyphosate-resistant Lolium spp. (Wakelin and Preston, 2006; Perez-Jones et al., 2007; Jasieniuk et al., 2008; Simarmata and Penner, 2008; Kaundun et al., 2011; Collavo and Sattin, 2012; Gonzalez-Torralva et al., 2012). However, Pro-106 substitutions confer only a modest degree of glyphosate resistance (Powles and Yu, 2010). Mutations in the EPSPS gene at Gly-101, Thr-102, Gly-144, and Ala-192 have been detected as sources of glyphosate resistance when expressed in Escherichia coli, but have not yet been reported in glyphosate-resistant weeds (Sammons and Gaines, 2014).
In crop fields under glyphosate selection, the evolution of a double amino acid substitution at Thr-102 and Pro-106 in the EPSPS in Eleusine indica conferring a high level of glyphosate resistance was recently communicated (Yu et al., 2015). In this species, other mechanisms of glyphosate resistance have also been found, associated with the over-expression of EPSPS (Chen et al., 2015). Target-gene duplication was previously reported in Amaranthus palmeri and Lolium perenne ssp. multiflorum (Gaines et al., 2010; Salas et al., 2012) and today it is a common and robust glyphosate resistance mechanism (Sammons and Gaines, 2014). On the other hand, non-target-site mechanisms of glyphosate resistance based on exclusion systems such as decreased herbicide retention and foliar uptake (Michitte et al., 2007), or reduced glyphosate translocation and vacuolar sequestration (Lorraine-Colwill et al., 2003; Perez-Jones et al., 2007; Ge et al., 2010; Vila-Aiub et al., 2012; Ghanizadeh et al., 2016) are well documented.
In the genus Lolium, allogamous species have demonstrated an unprecedented capacity to evolve resistance to multiple herbicides on five continents (Heap, 2017). In Argentina, glyphosate resistance was reported in a L. perenne population after 12 years of successful use (Yanniccari et al., 2012d). In this population, a 10.8-fold greater dose of glyphosate was necessary to match the control efficiency of a susceptible population (Yanniccari et al., 2012d). Nonetheless, the underlying mechanism of herbicide resistance has not been studied. The aim of the current paper was to determine the mechanism of glyphosate resistance in L. perenne from Argentina.
Materials and Methods
Plant Material
Perennial ryegrass seeds from a glyphosate-resistant population were collected in a field in the south of Buenos Aires province (37°S, 62°W) in 2009. The fields had a history of 12 years of no-tillage agriculture, with weed control based on three applications of glyphosate per year, at doses ranging from 360 to 720 g ae ha−1 (Yanniccari et al., 2012d). One hundred field-collected seeds from 50 randomly chosen plants were germinated in Petri dishes containing filter paper with distilled water. The germination occurred in a growth chamber with 75 μmol m−2 s−1 of photosynthetically active radiation, in a regime of 12/12 h of light/darkness and temperatures of 25/15°C day/night. After 7 d, 25 seedlings taken at random were transferred to 250 cm3 pots (contained in a plastic tray) filled with soil. The plants were grown in a greenhouse and the pots were randomized and subirrigated daily maintaining a water level of about 10 mm in the irrigation tray throughout the assay. Fertilizer (12:10:20, Nitrofoska, Compo Argentina) (2 g L−1) was added every 15 d. Plants were grown for 8 weeks and vegetative clones of individual plants were propagated by tiller partition in order to obtain four ramets per plant. When individual ramets reached the BBCH 23–24 scale, each one was treated with glyphosate (isopropylamine salt of glyphosate, Roundup, 360 g L−1, Monsanto Argentina) at 0, 500, or 2,000 g acid equivalent (ae) ha−1. To this end, a backpack sprayer equipped with flat-fan nozzles (Teejet 11002) was used, delivering 150 L ha−1. At 21 d post-application, plants were characterized as glyphosate susceptible (with no survivors at 500 g ae ha−1 and higher doses) or glyphosate resistant (surviving plants at 2,000 g ha−1 and lower doses).
Shikimate Accumulation Test
Six plants of each susceptible and glyphosate-resistant phenotype were employed to evaluate the shikimate accumulation in glyphosate-treated leaves incubated under in vitro conditions. Four last full-expanded leaves were cut at 0.5 cm above the ligule from tillers of each plant. Subsequently, the leaves of each plant were randomly placed vertically on metal lab racks and treated with different doses of glyphosate (0, 500, 1,500 and 3,000 g ae ha−1) using a manual sprayer calibrated to deliver 200 L ha−1. After 10 min of application, each leaf was placed in a 20 cm3 glass test tube, where the base of the leaf was immersed in with 1 mL of Hoagland nutrient solution diluted 1:3 with distilled water. The tubes were closed with flexible thermoplastic (Parafilm®) in which a 1 mm-diameter hole was made to promote gas exchange between the confined atmosphere and the exterior. Immediately, the tubes were incubated for 72 h under continuous light (300 μmol m−2 s−1 of photosynthetically active radiation) at 20°C (tubes were randomly placed every 24 h). After that, 0.05 g of fresh weight from the middle third of the each leaf was used for shikimic acid extraction, following the methodology of Singh and Shaner (1998) with the modifications of Perez-Jones et al. (2007). Shikimic acid was quantified with a double beam spectrophotometer (Shimadzu UV-160A, Shimadzu Corporation) at 382 ηm. The determination of the concentration of shikimic acid was based on a shikimate (3α,4α,5β-Trihydroxy-1-cyclohexene-1-carboxylic acid, 99%. Sigma Aldrich, Inc.) standard curve. The experiment was conducted as a completely randomized design and was repeated twice.
ANOVA was performed to evaluate the differences between experiment replicates, phenotypes, herbicide treatments, and interactions. Residual plots indicated that the variances were normally distributed and homogeneous. Means were compared using Fisher's LSD test (p < 0.05) when there were significant differences.
Enzyme Assay
The activity of EPSPS from glyphosate-susceptible and -resistant plants was evaluated under increasing glyphosate concentrations. The EPSPS extraction and assay protocols were modified from procedures described by Sammons et al. (2007). Forty grams of fresh unexpanded leaves were harvested from non-glyphosate treated clones of both phenotypes. Using liquid nitrogen, the samples were ground with 8 g of polyvinyl polypyrrolidone. Subsequently, 200 mL of extraction buffer (100 mM Tris-HCl, 5 mM EDTA, 10% (v/v) glycerol, 50 mM KCl, 10 mM 2-mercaptoethanol, 2 mM benzamidine, 0.5 mM phenymethysulphonyl fluoride and 10 μM leupeptine, pH 7.0) was added. Then, the crude extract was homogenized at 1,000 rpm for 5 min using a vertical axis micromotor with steel blades. After this, the extract was centrifuged at 800 × g for 45 min at 4°C and was subsequently filtered using a muslin cloth. Finally, the supernatant was fractionated with ammonium sulfate, and the 45–70% fraction was collected by centrifugation at 20,000 × g for 45 min. The pellet obtained was resuspended in a minimal volume of extraction buffer (2–3 mL) and desalted on a Sephadex G-25 column (PD-10, GE®) pre-equilibrated with extraction buffer according to the manufacturer's instructions. The eluate obtained was stored at −20°C until use in EPSPS activity determinations. For every fraction obtained, protein quantifications were carried out using the method of Bradford (1976) with bovine serum albumin as a reference standard.
EPSPS activity was assayed in 100 μL of 100 mM HEPES 100 mM, pH 7.0; 2 mM (NH4)6Mo7O24, 10 mM KF, 1 mM MgCl2, 10% v/v glycerol, 0.5 mM shikimate-3-phosphate, 1.25 mM phosphoenolpyruvate (using ultrapure water (Milli-Q®) at 25°C by determining the amount of inorganic phosphate produced in the reaction following Eschenburg et al. (2002). EPSPS activity was expressed in enzyme unit (U) per milligram of protein (1 U stands for 1 ηmol of phosphate produced per minute in the assayed reaction). The activities of glyphosate-susceptible and -resistant plants were determined at 0, 1, 5, 25, 50, 500, 1,000, 10,000 μg L−1 glyphosate [96% N-(phosphomethyl) glycine, Sigma Aldrich] concentrations.
EPSPS activity data was used to build dose response curves with a non-linear log-logistic regression model of standard slope:
where y represents the EPSPS activity at the glyphosate concentration x; C is the lower asymptote, D is the upper asymptote and I50 is the herbicide concentration required to achieve 50% of the maximum response. To assess the accuracy of the model, an F-test for model significance, residual variance analysis and coefficient of determination (R2) were calculated. Finally, the parameters from glyphosate-susceptible and -resistant models were contrasted using an F-test (P < 0.05).
EPSPS Partial Sequencing
Total RNA was extracted from leaf tissue of both glyphosate-susceptible and -resistant plants (S1, S2, S3 and R1, R2, R3; respectively) using the Trizol® reagent (Invitrogen) method according to the manufacturer's protocol. DNA in the RNA samples was degraded using DNase (RQ1 RNase-Free DNase kit, Promega) for 30 min at 37°C. Subsequently, the RNA was quantified spectrophotometrically at 260 ηm.
First strand complementary DNA (cDNA) was synthesized from 2 μg of total RNA. Reverse transcription was carried out in a 25 μL reaction mixture containing 1 μg oligo d(T)15 primer, M-MLV reverse transcriptase (200 U), RNasin (25 U), 8 mM dNTPs and M-MLV reverse transcriptase reaction buffer (250 mM Tris-HCl, pH 8.3, 375 mM KCl, 15 mM MgCl2 and 50 mM DTT) at 42°C for 1 h.
The resulting cDNA was used as a template for amplification of the EPSPS sequence. Forward primer (5′-AAAGGATGCCAAGGAGGAAGTAA-3′) and reverse primer (5′-GTACTTCTGTCCTCCTTTAATG-3′) were employed to amplify a highly conserved region in which point mutations conferring glyphosate resistance have been reported (Sammons and Gaines, 2014). A 466-bp fragment was obtained in PCR reactions (initial denaturation at 94°C for 2 min and 30 cycles of 94°C for 1 min, 62°C for 1 min, 72°C for 1 min and final extension at 72°C for 10 min) containing: 250 ηg cDNA template, 0.4 μM of each primer, 0.2 mM dNTPs, 0.5 mM MgCl2, 1X reaction buffer (Inbio Highway), and 0.5 U Taq polymerase (Inbio Highway) in a 25 μL reaction mix. PCR products were purified by precipitation at 20°C with three parts of absolute ethanol and 0.1 parts of sodium acetate (3 M) for each PCR product. The samples were centrifuged at 32,000 g for 20 min at 4°C and then washed three times with 70% ethanol. Each DNA pellet was diluted in ultrapure water and sequenced from both ends. DNA sequences obtained were cleaned, aligned, translated and compared at the 101, 102, 106, 144, and 192 codons (numbers based on the plant EPSPS numbering system used by Padgette et al., 1996).
EPSPS Expression
The cDNAs obtained from three plants of both phenotypes were employed as a template for two step qPCR reactions using an StepOnePlus™ Real-Time PCR System (Life Technologies) and FastStart Universal SYBR Green Master (Roche). Cinnamoyl-CoA reductase (CCR, AY061888.1) was used for normalization (forward primer: 5′-AGCAGCCATACAAGATGT-3′ and reverse primer: 5′- AGCTAGGGTTTCCTTGTC-3′). Primers specific to EPSPS were designed (forward: 5′-CTTGAGTTCCTTGCTGATG-3′ and reverse: 5′-GTACTTCTGTCCTCCTTTAATG-3′) and the following program was used: one cycle at 95°C for 10 min, then 40 cycles of 95°C for 15 s and 60°C for 1 min. Real time fluorescence data were captured at the end of each cycle of 15 s. Melt-curve analysis was conducted by holding the samples at 95°C for 15 s, then reducing the temperature to 60°C for 1 min, and then increasing the temperature to 95°C for 15 s. During this change of temperature fluorescence was measured continuously. Negative controls consisting of primers with no template were included. Each measurement was performed in triplicate. Results were expressed as fold increases in EPSPS expression relative to CCR and data were analyzed using ANOVA. Residual plots corroborated that the variances were normally distributed and homogeneous. The means were compared using the LSD test (P < 0.05).
Results
Shikimate Accumulation Test
Shikimate concentration in leaves was strongly affected by the herbicide treatment depending on the phenotype studied. The interaction between the phenotype and the treatment showed highly significant differences (Table 1). Both experiments carried out had similar results (P > 0.05, Table 1) and data from them were pooled.
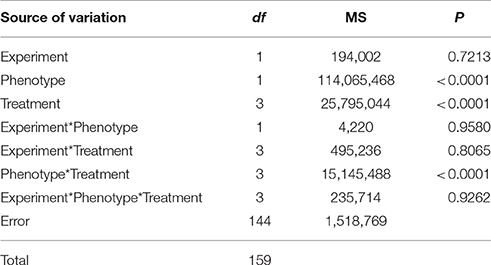
Table 1. Summary of ANOVA: mean square (MS), degrees of freedom (df), and probability values (P) for the effect of replication of the experiment comparing the effect of Lolium perenne phenotype (glyphosate-susceptible and -resistant phenotypes), treatment (dose of glyphosate: 0, 500, 1,500, and 3,000 g ae ha−1) and their interaction on shikimate concentration.
Shikimic acid levels quantified in glyphosate treated leaves were significantly different between samples from glyphosate-susceptible and -resistant plants (Figure 1). Leaves from susceptible plants increased their shikimate concentration by around 3.5-fold in response to glyphosate application at 500 or 1,500 g ae ha−1 (Figure 1). When 3,000 g ae ha−1 was sprayed the susceptible leaves increased their shikimate levels 5-fold compared with controls without glyphosate (Figure 1). Neither of the glyphosate doses applied significantly affected the shikimic acid concentration of leaves from glyphosate-resistant plants (Figure 1).
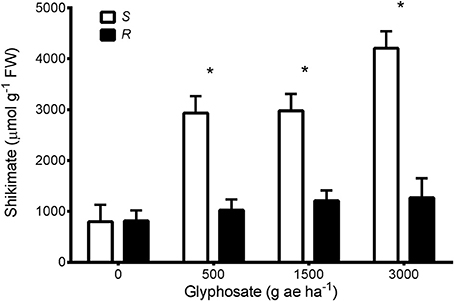
Figure 1. Shikimate accumulation in foliar tissues of glyphosate-susceptible (S) and -resistant phenotypes (R) after glyphosate treatments. Vertical bars represent ±1 standard error of the mean. Asterisks indicate significant differences between phenotypes within each dose.
EPSPS Activity
Based on the EPSPS activity data, a model was fitted for each biotype (P < 0.001 in both cases) and the parameters of glyphosate-susceptible and -resistant models were contrasted (Table 2, Figure 2). Comparison of the lower asymptotes (C), i.e., the level of EPSPS activity at infinitely high concentrations of the inhibitor, showed no significant differences between phenotypes. The upper asymptote (D), which is associated with EPSPS activity without glyphosate in the reaction medium, was significantly different between phenotypes (P = 0.0018). In this sense, basal EPSPS activity from glyphosate-resistant plants was approximately 3-fold higher than the EPSPS activity from susceptible plants. The half-maximal inhibitory concentration (I50) was not significantly different between the glyphosate-susceptible and -resistant models (P = 0.93) (Table 2, Figure 2).
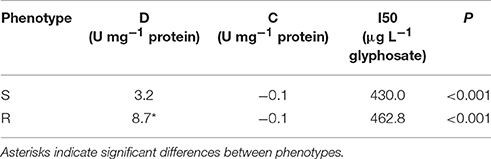
Table 2. Non-linear regression parameters for the activity of EPSPS from glyphosate-susceptible (S) and -resistant (R) phenotypes and P-values for model accuracy.
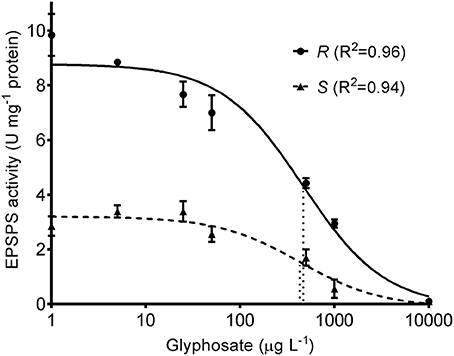
Figure 2. EPSPS activity of glyphosate-susceptible (S) and -resistant phenotypes (R) subjected to different glyphosate concentrations. Symbols represent mean values and the vertical bars are ±1 standard error of the mean. The curves represent predicted responses and dotted lines indicate I50 for each model.
EPSPS Partial Sequencing
A cDNA fragment of the EPSPS gene was sequenced and no nucleotide differences were identified between the sequences from glyphosate-resistant and -susceptible individuals. Therefore, no substitutions were recorded at Gly-101, Thr-102, Pro-106, Gly-144, or Ala-192 residues, which have been associated with a mechanism of resistance to glyphosate (Sammons and Gaines, 2014; Figure 3).

Figure 3. Sequence of EPSPS cDNAs isolated from the glyphosate-resistant phenotype and conceptual translation of the amino acid sequence. Resistance conferring codons are show in boxes (numbers refer to amino acid positions in the EPSPS plant sequence according to Padgette et al. (1996). No mutations were detected in positions reported as sources of glyphosate resistance.
EPSPS Expression
EPSPS transcript abundance was measured relative to cinnamoyl-CoA reductase. The relative expression levels of EPSPS were significantly different between phenotypes (P < 0.01) wherein glyphosate-resistant plants showed around 15-fold higher expression than susceptible plants (Figure 4).
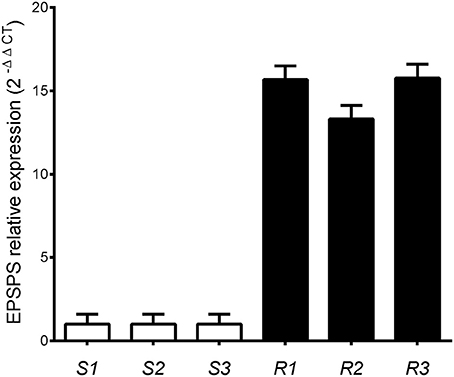
Figure 4. Relative EPSPS expression in glyphosate-susceptible (S) and -resistant (R) plants of Lolium perenne plants, using the cinnamoyl-CoA reductase gene for normalization. Vertical bars represent ±1 standard error of the mean.
Discussion
The quantification of shikimic acid in glyphosate-treated plants has been used as metabolic marker of glyphosate sensitivity (Dayan et al., 2015). The shikimate pathway consumes ≥30% of carbon fixed through the Calvin cycle (Maeda and Dudareva, 2012) and the inhibition of EPSPS causes a non-regulated carbon flux into the shikimate pathway through induction of the expression of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase, the first enzyme of this metabolic pathway (Zabalza et al., 2017). These alterations are reflected in a massive carbon flow to shikimate-3-phosphate, which is converted into high levels of shikimate (Duke and Powles, 2008).
The shikimic acid concentration measured in susceptible L. perenne plants sprayed with glyphosate was approximately three to five times the basal level while no changes were detected in glyphosate-resistant counterparts (Yanniccari et al., 2012d). In the current results, susceptible leaves treated with different doses of glyphosate and incubated in vitro showed shikimic acid accumulation, but leaves from glyphosate-resistant plants showed no change in shikimate concentration. This could be considered as a semi-destructive method in order to evaluate the glyphosate sensitivity of plant materials, where susceptible genotypes could be tested without killing them.
While in vitro methods based on shikimate quantification have been proposed for identifying glyphosate-resistant plants, in most methods leaf fragments are immersed in a medium with glyphosate and thus incubated (Shaner et al., 2005). This methodology has been effective to quickly detect differences between glyphosate-susceptible and -resistant plants (Perez-Jones et al., 2005). However, the doses used in these methodologies cannot be compared to the doses sprayed under field conditions. Moreover, the process of herbicide absorption in a system where the leaves are immersed could be very different to the foliar uptake that occurs in treated plants under field conditions. In this regard, the shikimate accumulation test in the current work successfully addressed these limitations.
The glyphosate-resistant and -susceptible plants showed differential glyphosate sensitivity when the whole plant was sprayed with the herbicide (Yanniccari et al., 2012d), as when a sample of leaf was treated and shikimate concentration was recorded. In both cases, the mechanism of resistance put into play would prevent shikimate accumulation in glyphosate-resistant plants.
The activity of herbicide target enzymes at different herbicide concentrations has been used to determine if a target site mechanism provokes low sensitivity in resistant plants (Baerson et al., 2002; Burke et al., 2006; Yu et al., 2010; Cruz-Hipolito et al., 2011). Following this approach, glyphosate-susceptible and -resistant models of EPSPS activity showed no significant differences in half maximal inhibitory concentration (I50) (Table 2). As a consequence, EPSPS from glyphosate-resistant plants was not different in glyphosate sensitivity compared to EPSPS from susceptible plants. Accordingly, no changes in the nucleotide sequences associated with Gly-101, Thr-102, Pro-106, Gly-144, and Ala-192 were found in either case. The affinity between the glyphosate molecule and EPSPS from resistant plants was not affected, unlike in other cases of glyphosate resistance (Ng et al., 2004; Yu et al., 2007; Simarmata and Penner, 2008; Kaundun et al., 2011).
While the glyphosate target-site was equally sensitive, differential levels of EPSPS activity was detected when comparing the plant materials. At a glyphosate concentration of 1,000 μg L−1, when the EPSPS activity of susceptible plants was inhibited by ~90%, the level of EPSPS activity measured in the crude extract from glyphosate-resistant plants was not different to the EPSPS activity of the susceptible material without glyphosate (Figure 2). This higher EPSPS activity likely ensures maintenance of a functional shikimate pathway after glyphosate treatment in the glyphosate-resistant plants.
The increased EPSPS activity may be due to increased transcription of the EPSPS gene or greater stability of the EPSPS enzyme at a similar transcription rate compared with susceptible plants. In the current work, the glyphosate-resistant plants showed 15-fold higher expression of EPSPS than the susceptible plants and this explains the differential glyphosate sensitivity between individuals. In this sense, the glyphosate effect on net carbon assimilation, chlorophyll fluorescence, and production and transportation of assimilated carbon would be prevented by the high activity of the target enzyme (Yanniccari et al., 2012a,b).
This mechanism of resistance is constitutive in character, i.e., it does not depend of a glyphosate treatment to be induced. In consequence, the high level of expression of EPSPS likely has important effects on plant fitness, as have recently been detected on the inhibition of growth and seed production (Yanniccari et al., 2016). Considering that molecules from the shikimate pathway can account for up to 60% of the total plant dry weight (Haslam, 1993), the over-expression of EPSPS likely affects the regulation of the shikimate pathway and would therefore provoke pleiotropic effects on several physiological processes associated with growth and seed production.
Although is not possible to rule out that other resistance mechanisms also operate, the over-expression of EPSPS appears to be the main mechanism responsible for resistance to glyphosate. This mechanism would be controlled by a single locus with incomplete dominance (Yanniccari et al., 2015) and would provoke low glyphosate sensitivity in plants treated in the field as observed for foliar tissues sprayed with glyphosate and incubated in vitro.
Author Contributions
MY, CI, CN, DG, and AC conceived and designed the experiments; MY and MG performed the experiments; MY, DG, and AC analyzed the data and MY wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was partially supported by Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, National Research Council of Argentina—PIP 0165), the Instituto Nacional de Tecnología Agropecuaria (INTA—PNPV113034, BASUR-1272409 PRET) and the Universidad Nacional de La Plata (UNLP—A180). Thanks are also due to Dr. R. D. Sammons for helpful tips on carrying out the EPSPS activity assays.
References
Baerson, S. R., Rodriguez, D. J., Tran, M., Feng, Y., Biest, N. A., and Dill, G. M. (2002). Glyphosate-resistant goosegrass. identification of a mutation in the target enzyme 5-enolpyruvylshikimate-3- phosphate synthase. Plant Physiol. 129, 1265–1275. doi: 10.1104/pp.001560
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Burke, E. I., Burton, J., York, A., and Wilcut, J. (2006). Mechanism of resistance to clethodim in a johnsongrass (Sorghum halepense) biotype Weed Sci. 54, 401–406. doi: 10.1614/WS-05-153R.1
Chen, J., Huang, H., and Zhang, C. (2015). Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 242, 859–868. doi: 10.1007/s00425-015-2324-2
Collavo, A., and Sattin, M. (2012). Resistance to glyphosate in Lolium rigidum selected in Italian perennial crops: bioevaluation, management and molecular bases of target-site resistance. Weed Res. 52, 16–24. doi: 10.1111/j.1365-3180.2011.00883.x
Cruz-Hipolito, H., Osuna, M., Domínguez-Valenzuela, J., Espinoza, N., and De Prado, R. (2011). Mechanism of resistance to ACCase-inhibiting herbicides in wild oat (Avena fatua) from Latin America. J. Agric. Food Chem. 59, 7261–7267. doi: 10.1021/jf201074k
Dayan, F. E., Owens, D. K., Corniani, N., Lima Silva, F. M., Watson, S. B., Howell, J., et al. (2015). Biochemical markers and enzyme assays for herbicide mode of action and resistance studies. Weed Sci. 63, 23–63. doi: 10.1614/WS-D-13-00063.1
Duke, S. O., and Powles, S. B. (2008). Glyphosate : a once-in-a-century herbicide. Pest Manage Sci. 325, 319–325. doi: 10.1002/ps.1518
Eschenburg, S., Healy, M. L., Priestman, M. A., Lushington, G. H., and Schönbrunn, E. (2002). How the mutation glycine96 to alanine confers glyphosate insensitivity to 5-enolpyruvyl shikimate-3-phosphate synthase from Escherichia coli. Planta 216, 129–135. doi: 10.1007/s00425-002-0908-0
Franz, J. E., Mao, M. K., and Sikorski, J. A. (1997). Glyphosate: A Unique Global Herbicide. Washington, DC: American Chemical Society.
Gaines, T. A., Zhang, W., Wang, D., Bukun, B., Chisholm, S. T., Shaner, D. L., et al. (2010). Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. U.S.A. 107, 1029–1034. doi: 10.1073/pnas.0906649107
Ge, X., d'Avignon, D. A., Ackerman, J. J. H., and Sammons, R. D. (2010). Rapid vacuolar sequestration: the horseweed glyphosate resistance mechanism. Pest Manage Sci. 66, 345–348. doi: 10.1002/ps.1911
Ghanizadeh, H., Harrington, K. C., and James, T. K. (2016). Genetic inheritance of restricted herbicide translocation in a glyphosate-resistant Lolium perenne population. New Zeal. J. Agric. Res. 59, 269–279. doi: 10.1080/00288233.2016.1189438
Gonzalez-Torralva, F., Gil-Humanes, J., Barro, F., Brants, I., and De Prado, R. (2012). Target-site mutation and reduced translocation are present in a glyphosate-resistant Lolium multiflorum Lam. biotype from Spain. Plant Physiol. Biochem. 58, 16–22. doi: 10.1016/j.plaphy.2012.06.001
Heap, I. (2017). The International Survey of Herbicide Resistant Weeds. Available online at: http://weedscience.org/ (Accessed January 22, 2017).
Jasieniuk, M., Riaz Ahmad, A., Sherwood, A., Firestone, J., Perez-Jones, A., Lanini, W., et al. (2008). Glyphosate-resistant Italian ryegrass (Lolium multiflorum) in California: distribution, response to glyphosate, and molecular evidence for an altered target enzyme. Weed Sci. 56, 496–502. doi: 10.1614/WS-08-020.1
Kaundun, S., Dale, R., Zelaya, I., Dinelli, G., Marotti, I., McIndoe, E., et al. (2011). A novel P106L mutation in EPSPS and an unknown mechanism(s) act additively to confer resistance to glyphosate in a South African Lolium rigidum population. J. Agric. Food Chem. 59, 3227–3233. doi: 10.1021/jf104934j
Lorraine-Colwill, D. F., Powles, S. B., Hawkes, T. R., Hollinshead, P. H., Warner, S. A. J., and Preston, C. (2003). Investigations into the mechanism of glyphosate resistance in Lolium rigidum. Pest. Biochem. Physiol. 74, 62–72. doi: 10.1016/S0048-3575(03)00007-5
Maeda, H., and Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Maroli, A. S., Nandula, V. K., Duke, S. O., and Tharayil, N. (2016). Stable isotope resolved metabolomics reveals the role of anabolic and catabolic processes in glyphosate-induced amino acid accumulation in Amaranthus palmeri biotypes. J. Agr. Food Chem. 64, 7040–7048. doi: 10.1021/acs.jafc.6b02196
Michitte, P., De Prado, R., Espinoza, N., Pedro Ruiz-Santaella, J., and Gauvrit, C. (2007). Mechanisms of resistance to glyphosate in a ryegrass (Lolium multiflorum) biotype from Chile. Weed Sci. 55, 435–440. doi: 10.1614/WS-06-167.1
Ng, C. H., Wickneswary, R., Salmijah, S., Teng, Y. T., and Ismail, B. S. (2004). Glyphosate resistance in Eleusine indica (L.) Gaertn. from different origins and polymerase chain reaction amplification of specific alleles. Aust. J. Agric. Res. 55, 407–414. doi: 10.1071/AR03155
Norsworthy, J. K., Ward, S. M., Shaw, D. R., Llewellyn, R. S., Nichols, R. L., Webster, T. M., et al. (2012). Reducing the risks of herbicide resistance: best management practices and recommendations. Weed Sci. 60, 31–62. doi: 10.1614/WS-D-11-00155.1
Olesen, C. F., and Cedergreen, N. (2010). Glyphosate uncouples gas exchange and chlorophyll fluorescence. Pest Manage Sci. 66, 536–542. doi: 10.1002/ps.1904
Orcaray, L., Zulet, A., Zabalza, A., and Royuela, M. (2012). Impairment of carbon metabolism induced by the herbicide glyphosate. J. Plant Physiol. 169, 27–33. doi: 10.1016/j.jplph.2011.08.009
Padgette, S. R., Re, R. D., Barry, G. F., Eichholtz, D. E., Delannay, X., Fuchs, R. L., et al. (1996). “New weed control opportunities: development of soybeans with a Roundup Ready gene,” in Herbicide-Resistant Crops: Agricultural, Economic, Environmental, Regulatory, and Technological Aspects, ed S. O. Duke (Boca Raton, FL: CRC Press), 53–84.
Perez-Jones, A., Park, K., Polge, N., Colquhoun, J., and Mallory-Smith, C. A. (2007). Investigating the mechanisms of glyphosate resistance in Lolium multiflorum. Planta 226, 395–404. doi: 10.1007/s00425-007-0490-6
Perez-Jones, A., Park, K. W., Colquhoun, J., Mallory-Smith, C., and Shaner, D. (2005). Identification of glyphosate-resistant Italian ryegrass (Lolium multiflorum) in Oregon. Weed Sci. 53, 775–779. doi: 10.1614/WS-04-200R.1
Pline, W. A., Wilcut, J. W., Edmisten, K. L., and Wells, R. (2002). Physiological and morphological response of glyphosate-resistant and non-glyphosate-resistant cotton seedlings to root-absorbed glyphosate. Pest. Biochem. Physiol. 73, 48–58. doi: 10.1016/S0048-3575(02)00014-7
Powles, S. B., Lorraine-Colwill, D. F., Dellow, J. J., and Preston, C. (1998). Evolved resistance to glyphosate in rigid ryegrass (Lolium rigidum) in Australia. Weed Sci. 46, 604–607.
Powles, S. B., and Yu, Q. (2010). Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. doi: 10.1146/annurev-arplant-042809-112119
Salas, R. A., Dayan, F. E., Pan, Z., Watson, S. B., Dickson, J. W., Scott, R. C., et al. (2012). EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manage Sci. 68, 1223–1230. doi: 10.1002/ps.3342
Sammons, R. D., and Gaines, T. A. (2014). Glyphosate resistance: state of knowledge. Pest Manage Sci. 70, 1367–1377. doi: 10.1002/ps.3743
Sammons, R. D., Meyer, J., Hall, E., Ostrander, E., and Schrader, S. (2007). “A simple continuous assay for EPSP synthase in plant tissue,” in Annual Meeting of the North Central Weed Science Society 2007 (St. Louis, MO).
Schönbrunn, E., Eschenburg, S., Shuttleworth, W. A., Schloss, J. V., Amrhein, N., Evans, J. N. S., et al. (2001). Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. U.S.A. 98, 1376–1380. doi: 10.1073/pnas.98.4.1376
Shaner, D. L., Nadler-Hassar, T., Henry, W. B., and Koger, C. H. (2005). A rapid in vivo shikimate accumulation assay with excised leaf discs. Weed Sci. 53, 769–774. doi: 10.1614/WS-05-009R.1
Simarmata, M., and Penner, D. (2008). The basis for glyphosate resistance in rigid ryegrass (Lolium rigidum) from California. Weed Sci. 56, 181–188. doi: 10.1614/WS-07-057.1
Singh, B. K., and Shaner, D. (1998). Rapid determination of glyphosate injury to plants and identification of glyphosate resistant plants. Weed Technol. 12, 527–530.
Vila-Aiub, M. M., Balbi, M. C., Distefano, J., Fernández, L., Hopp, E., and Powles, S. B. (2012). Glyphosate resistance in perennial Sorghum halepense (johnsongrass), endowed by reduced glyphosate translocation and leaf. Pest Manage Sci. 68, 430–436. doi: 10.1002/ps.2286
Wakelin, A. M., and Preston, C. (2006). A target-site mutation is present in a glyphosate- resistant Lolium rigidum population. Weed Res. 54, 432–440. doi: 10.1111/j.1365-3180.2006.00527.x
Yanniccari, M., Istilart, C., Giménez, D. O., and Castro, A. M. (2012a). Effects of glyphosate on the movement of assimilates of two Lolium perenne L. populations with differential herbicide sensitivity. Environ. Exp. Bot. 82, 14–19. doi: 10.1016/j.envexpbot.2012.03.006
Yanniccari, M., Istilart, C., Giménez, D. O., Acciaresi, H., and Castro, A. M. (2012c). Efecto del glifosato sobre el crecimiento y acumulación de azúcares libres en dos biotipos de Lolium perenne de distinta sensibilidad al herbicida. Planta Daninha 30, 155–164. doi: 10.1590/S0100-83582012000100018
Yanniccari, M., Istilart, C., Giménez, D. O., and Castro, A. M. (2012d). Glyphosate resistance in perennial ryegrass (Lolium perenne L.) from Argentina. Crop Prot. 32, 12–16. doi: 10.1016/j.cropro.2011.09.021
Yanniccari, M., Istilart, C., Giménez, D. O., and Castro, A. M. (2015). Inheritance of glyphosate resistance in Lolium perenne and hybrids with Lolium multiflorum. Crop Prot. 71, 72–78. doi: 10.1016/j.cropro.2015.01.024
Yanniccari, M., Tambussi, E., Istilart, C., and Castro, A. M. (2012b). Glyphosate effects on gas exchange and chlorophyll fluorescence responses of two Lolium perenne L. biotypes with differential herbicide sensitivity. Plant Physiol. Biochem. 57, 210–217. doi: 10.1016/j.plaphy.2012.05.027
Yanniccari, M., Vila-Aiub, M., Istilart, C., Acciaresi, H., and Castro, A. M. (2016). Glyphosate resistance in perennial ryegrass (Lolium perenne L.) is associated with a fitness penalty. Weed Sci. 64, 71–79. doi: 10.1614/WS-D-15-00065.1
Yu, L. P. C., Kim, Y. S., and Tong, L. (2010). Mechanism for the inhibition of the carboxyltransferase domain of acetyl-coenzyme A carboxylase by pinoxaden. Proc. Natl. Acad. Sci. U.S.A. 107, 22072–22077. doi: 10.1073/pnas.1012039107
Yu, Q., Cairns, A., and Powles, S. (2007). Glyphosate, paraquat and ACCase multiple herbicide resistance in a Lolium rigidum biotype. Planta 225, 499–513. doi: 10.1007/s00425-006-0364-3
Yu, Q., Jalaludin, A., Han, H., Chen, M., Sammons, R. D., and Powles, S. B. (2015). Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance. Plant Physiol. 167, 1440–1447. doi: 10.1104/pp.15.00146
Keywords: glyphosate resistance, shikimate assay, EPSPS activity, EPSPS overexpression, perennial ryegrass
Citation: Yanniccari M, Gómez-Lobato ME, Istilart C, Natalucci C, Giménez DO and Castro AM (2017) Mechanism of Resistance to Glyphosate in Lolium perenne from Argentina. Front. Ecol. Evol. 5:123. doi: 10.3389/fevo.2017.00123
Received: 27 July 2017; Accepted: 22 September 2017;
Published: 09 October 2017.
Edited by:
Ilias Travlos, Agricultural University of Athens, GreeceReviewed by:
Pei Wang, Jiangsu University, ChinaLeonardo Bianco de Carvalho, São Paulo State University, Brazil
Copyright © 2017 Yanniccari, Gómez-Lobato, Istilart, Natalucci, Giménez and Castro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcos Yanniccari, marcosyanniccari@conicet.gov.ar
 Marcos Yanniccari
Marcos Yanniccari María E. Gómez-Lobato
María E. Gómez-Lobato Carolina Istilart3
Carolina Istilart3