Diversity of Treegourd (Crescentia cujete) Suggests Introduction and Prehistoric Dispersal Routes into Amazonia
- 1Post–Graduate Program in Botany, Instituto Nacional de Pesquisas da Amazônia, Manaus, Brazil
- 2Centro de Investigaciones en Ecosistemas, Universidad Nacional Autónoma de México, Morelia, Mexico
- 3Institut de Recherche pour le Développement, Université de Montpellier, UMR DIADE, Montpellier, France
- 4Laboratório de Evolução Aplicada, Universidade Federal do Amazonas, Manaus, Brazil
- 5Coordenação de Tecnologia e Inovação, Instituto Nacional de Pesquisas da Amazônia, Manaus, Brazil
The use and dispersal of domesticated plants may reflect patterns of early human diffusion of technologies and lifestyles. Treegourd (Crescentia cujete) has fruits with ancient utilitarian and symbolic value in the Neotropics. We assessed diversity based on chloroplast (SNPs), nuclear (SSR) markers, and fruit shapes of cultivated treegourds and wild relatives across Amazonia and Mesoamerica in order to discuss hypothesis of dispersal routes and diversification of fruits along its distribution. The haplotype network showed three distinct groups: Crescentia amazonica, wild Mesoamerican C. cujete, and cultivated C. cujete from Brazilian Amazonia and Mexico. Mexico and Brazil shared two haplotypes, with slightly different distributions in Amazonia. The most divergent haplotype is well-represented in Eastern Amazonia. Nuclear differentiation between Mesoamerican wild and cultivated C. cujete is relatively low (FST = 0.35), compared with Amazonian cultivated (FST = 0.45–0.61). Differentiation is also higher between wild C. amazonica and cultivated C. cujete (FST = 0.57), but modest within cultivated C. cujete from Amazonia and Mexico (FST = 0.04), with higher genetic similarity in northwestern Amazonia. Mexico and Amazonia showed similar chloroplast nucleotide diversity (4.66 × 10−2 and 5.31 × 10−2, respectively), although sample sizes are very different. Except in Northwestern and Eastern Amazonia, we found ample genetic homogeneity of cultivated C. cujete across Amazonia, but highest morphological diversity in the Northwest, with fruit shapes that are absent in Mexico. We conclude that treegourds introduced into the Amazon Basin and Mexico share a common ancestry with a currently unknown origin. The patterns of genetic diversity across Amazonia allow two hypotheses of the routes of introduction: a northwestern introduction into the Negro and Solimões Rivers, and an eastern introduction from the coastal Guianas into the Amazonas River. The dispersal into Amazonia followed previously proposed routes of human and plant migrations. The contrasting fruit shape diversity suggests different utilitarian demands and cultural preferences for treegourd fruits between Mexico and Amazonia.
Introduction
The use and dispersal of domesticated plants may reflect the patterns of diffusion of human technologies and lifestyles since prehistoric times (Bellwood, 2005; Blench, 2012). Humans greatly expand plants' geographical distributions, which ultimately exerts different ecological and cultural pressures on the evolutionary pathways of plants (Rindos, 1984; Sodero Martins, 2005; Leclerc and Coppensd'Eeckenbrugge, 2012; Meyer and Purugganan, 2013). Various centers of domestication have been proposed in the Americas (Meyer et al., 2012) from where people exchanged plants (Heiser, 1965; Schultes, 1984; Colunga-GarcíaMarín and Zizumbo-Villarreal, 2004). Amazonia is one of them (Clement, 1999), and also encompasses great linguistic diversity (Blench, 2012), ceramic styles (Barreto et al., 2016), and landscape management strategies (Eriksen and Danielsen, 2014), whose geography and chronology are being disentangled (Mayle and Iriarte, 2014; Clement et al., 2015; Neves, 2016; Levis et al., 2017). The Amazonian routes of dispersal of plants and people have been associated with rivers and riparian environments (Schultes, 1984; Godoy et al., 1999; Guix, 2009). However, few studies have demonstrated the genetic signatures of the plants' geographical dispersal mediated by humans in Amazonia (Clement et al., 2010; Shepard and Ramirez, 2011; Thomas et al., 2012; Freitas and Bustamante, 2013), even though they are persistent markers of the long-term use and management of resources (Hanotte et al., 2002; Parker et al., 2010; Armstrong et al., 2017).
Treegourd (Crescentia cujete) is a good case study, since its trees produce fruits with ancient utilitarian and symbolic value widely dispersed across the Neotropics (Gentry, 1980; Arango-Ulloa et al., 2009; Meulenberg, 2011; Aguirre-Dugua et al., 2013; Medeiros and Albuquerque, 2014; Moreira et al., 2017). It currently is one of the most common species in homegardens of the floodplains and adjacent communities of Amazonia (Santos, 1982; Lima and Saragoussi, 2000). Its fruits have different shapes and sizes that are used as bowls, vessels or bottles for drinking or transporting water, bags for provisions, utensils for cooking, and eating, bailing water from canoes, construction of fish traps, manufacture of body ornaments, and musical instruments (Steward, 1948; Patiño, 1967; Morton, 1968; Price, 1982; Bennett, 1992; Heiser, 1993; Meulenberg, 2011). Medicinal uses are also similar across its distribution (Morton, 1968; Duke, 2009), which include neutralization of snake venom and intestinal parasites treatment (Otero et al., 2000; Volpato et al., 2009; Ramos, 2015; Paulo, 2016).
While the great phenotypic variability of cultivated treegourd is a distinctive feature among Crescentia species (Gentry, 1980), its wild populations from Mexican savannahs in the Yucatan Peninsula have smaller, elongated fruits with thinner exocarps (Aguirre-Dugua et al., 2012). The indehiscent and thicker exocarp of cultivated treegourd fruits makes the spontaneous dispersal of seeds impossible (Aguirre-Dugua et al., 2012). Its oldest remains found to date come from a Peruvian archaeological site dating to 5,000–3,800 years BP (Solis, 2006). This pattern contrasts to the bottle gourd, collected from a vine (Lagenaria siceraria), one of the ancient crops similarly used for technological purposes in the Americas (Heiser, 1993). Bottle gourd has been managed at least since the Late Pleistocene (Kistler et al., 2014) and was found in Colombian Amazon by 8,000 BP (Piperno, 2011). The wild progenitor of the cultivated Crescentia cujete remains elusive (Gentry, 1980; Arango-Ulloa et al., 2009; Aguirre-Dugua et al., 2012; Moreira et al., 2017). Gentry (1980) pointed out that C. cujete was certainly native to Mesoamerica, where putative wild populations are found in savannahs and semi-evergreen forests of southern Mexico and northern Central America (Figure 1). However, northern South America cannot be ruled out as part of the original distribution area of wild C. cujete, given the occurrence of apparently spontaneous C. cujete in grazed savannahs of Andean and Caribbean regions of Colombia (Arango-Ulloa et al., 2009). Historical anthropogenic fire management in savannahs (Pinter et al., 2011) may have been advantageous for its early dispersal (Bass, 2004) in these regions. Recently, the wild species native to Amazonian and Orinocan floodplains (Crescentia amazonica) was ruled out as the wild progenitor of cultivated C. cujete (Ducke, 1946; Moreira et al., 2017). Likewise, the wild C. cujete populations found in the southeastern Mexico are not the wild progenitor either (Aguirre-Dugua et al., under revision). In this study, we infer treegourd dispersal and diversification across two pivotal regions of the Neotropics: Amazonia and Mesoamerica. We (1) identify genetic relationships among Mesoamerican and Amazonian cultivated C. cujete; (2) infer routes of introduction into and dispersal within the Amazon Basin; and (3) identify centers of morphological and genetic diversity. We discuss whether this genetic/morphological diversity is linked to (1) introgression with local wild parents, (2) ecological diversification, or (3) cultural diversification, since all three of them are possible along the dispersal routes.
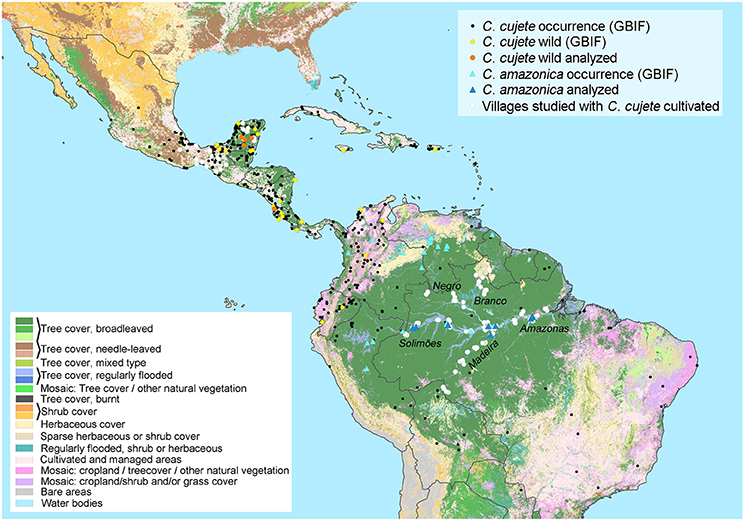
Figure 1. The geographical distribution of cultivated Crescentia cujete, putative wild populations of C. cujete and wild C. amazonica in the Neotropics. Genetic or morphological analyses include samples from Mexico, Costa Rica and five rivers in Brazilian Amazonia (Negro, Branco, Solimões, Madeira, Amazonas). Their distributions were complemented with records from the Global Biodiversity Information Facility (GBIF) and plotted over the vegetation cover (Bartholomé and Belward, 2005). The wild C. cujete distribution was hypothesized based on apparently spontaneous individuals growing in a mosaic of shrub and grass cover, which does not rule out previous human dispersion, since areas might include abandoned or burned croplands.
Materials and Methods
Sampling
We performed molecular analyses using full chloroplast (SNPs) and nuclear (SSR) markers. We also analyzed fruit morphology along the major rivers of Brazilian Amazonia and in parts of Mesoamerica (Supplementary Table S1). We used a previously published genetic and morphological dataset (Moreira et al., 2017) of cultivated C. cujete (N = 372) distributed in 122 localities along the five major rivers of the Brazilian Amazon basin, as well as wild Brazilian treegourds (C. amazonica) (N = 20) distributed in three of the rivers mentioned (Figure 1).
From Mexico, we add new genetic data of cultivated C. cujete from the Yucatan Peninsula, Oaxaca and Chiapas, wild samples from the Yucatan savannahs and a putative wild sample from Costa Rica (Figure 1). We also integrate morphological data from Mesoamerican samples (N = 188), part of which (N = 124) was published previously (Aguirre-Dugua et al., 2013). All Mesoamerican wild samples were identified as C. cujete Linnaeus 1753. In order to depict the putative geographical distribution of wild C. cujete, we searched for individuals of C. cujete described as spontaneous in savannahs on herbarium descriptions found in GBIF (Global Biodiversity Information Facility) (Figure 1).
This research followed the International Society for Ethnobiology's code of ethics (International Society of Ethnobiology, 2006) and was approved by the Committee for Ethics in Research with Human Beings of the National Research Institute for Amazonia (CEP INPA, proc. no. 408.611, 2013). Collection in Brazil was authorized by the Brazilian System for Authorization and Information in Biodiversity, Chico Mendes Institute for Biodiversity Conservation, proc. no. 25052–1, 2012, and transportation by the Brazilian Institute for the Environment and Renewable Natural Resources, proc. no. 14BR015576/DF, 2014. Collection in Mexico and Costa Rica was authorized by proc. no. SGPA/DGGFS/712/3691/10.
Genetic Analysis
We used a previously described protocol for genotyping nuclear microsatellites and the detection of single nucleotide polymorphisms along the entire sequence of the maternally inherited chloroplast genome (Moreira et al., 2016, 2017). In total, 250 samples were genotyped for eight nuclear microsatellites (SSR): 234 from Brazilian Amazonia (215 cultivated C. cujete and 19 wild C. amazonica), and 16 from Mesoamerica (7 cultivated C. cujete from Mexico, 8 wild C. cujete from Mexico, 1 wild C. cujete from Costa Rica). Data from the chloroplast genome was obtained from a total of 215 samples: 191 C. cujete and 16 C. amazonica from Amazonia, 5 cultivated C. cujete from Mexico, 2 wild C. cujete from Mexico, 1 wild C. cujete from Costa Rica. Among the total sample (N = 250), 80 % were genotyped and sequenced for both kinds of markers.
The nuclear SSR dataset was used to assess population structure with a Bayesian approach (Structure 2.3, Pritchard et al., 2000). We applied the admixture model in order to identify ancestral population proportions for each individual and their probable populations of origin. Using total sampling and assuming independent allele frequencies in each population, which reduces the risk of overestimating the number of clusters (Pritchard et al., 2000), we assessed the number of clusters K varying from 1 to 20, with 100,000 burn-in, 100,000 iterations, and five different runs for each K value. To attempt to identify different genetic pools within the cultivated cluster, we performed an additional analysis on a subset including only cultivated C. cujete samples, whose membership probability was higher than 0.6 in the cultivated cluster (N = 200). Using the admixture model, we experimented with two allele frequency assumptions (Pritchard et al., 2000): the independent model as default; and the correlated (assuming lambda = 1), since it is likely that cultivated populations share ancestry due to migration and vegetative propagation. Evanno et al. (2005) ΔK was used to guide our choice of the most likely number of groups. Additionally, we performed a Principal Components Analysis (PCA) with stats R package (R Core Team, 2015) in order to uncover additional genetic structure in our data (Jombart et al., 2009). The PCA was non-centered, but scaled in order to compensate for differences in polymorphism and missing data among the loci analyzed. The spatial interpolation of the clusters obtained in Structure was analyzed using the kriging method in the fields R package (Nychka et al., 2015). Based on geostatistics and maximum likelihood, the krig function estimates the covariance in a grid (we used the scale parameter theta = 50) and infers the fitted surface between geographical coordinates and genetic relationship among samples (Nychka et al., 2015). Nuclear genetic diversity of C. cujete [allelic richness (Ar), private alleles (Ap), observed heterozygosity (Ho), expected heterozygosity (Hs)] was estimated for the five Amazonian rivers considered and the Mexican samples using hierfstat (Goudet, 2005) and poppr (Kamvar et al., 2014) R packages. Pairwise FST between regions were estimated and statistically evaluated using 1,000 bootstraps (Nei, 1987). A neighbor-joining dendrogram of regions was constructed based on Nei's distance and 1,000 bootstraps (Saitou and Nei, 1987). The inbreeding coefficient FIS for each region was estimated and its significance evaluated (considering a Bonferroni corrected p-value of 0.006) using pegas R package (Paradis, 2010).
For the identification of chloroplast SNPs, we used a bioinformatic pipeline previously validated for the sequencing of the entire chloroplast genome (Scarcelli et al., 2016). Briefly, SAMTOOLS 0.1.7 with option-B (Li et al., 2009) was used to generate an mpileup file. VARSCAN 2.3.7 (Koboldt et al., 2012) was used to call SNPs from this mpileup file. The variant call format file (VCF) generated was filtered following Scarcelli et al. (2016) and resulted in a total of 334 cpSNPs detected in our dataset. The final vcf file was exported as a fasta file using VCFtools 1.14 (Danecek et al., 2011) and haplotypes identified with DNAsp 5.10.1 (Librado and Rozas, 2009). An haplotype network was constructed using the median joining algorithm (Bandelt et al., 1999) and samples with up to 6.5% of missing data using POPART 1.7 (Leigh and Bryant, 2015). The geographical distribution of the shared haplotypes of C. cujete samples was plotted using GenGIS 2.5 (Parks et al., 2009). The chloroplast diversity of C. cujete [total number of polymorphic sites (S), number of haplotypes (h), and nucleotide diversity (π)] were estimated according to Nei (1987) using DNAsp 5.10.1. The presence of singleton samples and their contribution with unique alleles were identified by VCFtools 1.14. Paired FST among the Amazonian rivers and Mexico were estimated using the distance method of Tajima and Nei (1984), and their significance was evaluated with 1,000 permutations at a significance level of 0.05 using Arlequin 3.5 (Excoffier and Lischer, 2010).
Morphological Analysis
Fruit shapes of cultivated C. cujete were registered in 286 individuals and fruit diameter was measured in 175 individuals in the Amazon Basin. For Mesoamerican samples, we analyzed 117 cultivated individuals from Mexico, among which 64 were from nine localities in the Yucatan Peninsula (Aguirre-Dugua et al., 2013) and 53 were from 19 localities representing the Gulf of Mexico coast, Tehuacan Valley, and Pacific Ocean coast from the states of Michoacan, Oaxaca, and Chiapas (Figure 1).
The shape of the mature fruits of each individual was classified visually into nine categories: spherical, flattened, oblong, cuneate, elongated, globular, rounded-drop-shaped, oblong-drop-shaped, and kidney-shaped. All of these categories, except spherical, followed the classification created for Colombian fruits (Arango-Ulloa et al., 2009). The spherical fruit was added as a new category, since it is a remarkable shape found in Mexico, which has a higher index of roundness than flattened fruits (Aguirre-Dugua et al., 2012, 2013). For Brazilian samples, the flattened type was sub-divided in order to discriminate these perfectly spherical fruits from flattened ones based on visual comparison of photographs. The Shannon index was adapted to estimate fruit shape diversity using H' = −∑ipilogpi, from Pielou (1975), where pi is the relative frequency of each fruit shape. The Shannon index was calculated for each Amazonian river, and for Amazonia and Mexico.
Results
Geographic Patterns of Nuclear Diversity
Evanno et al. (2005) ΔK suggested that two clusters are the most likely structure in the dataset (K = 2, Figure 2A, Supplementary Figure S1). At K = 2, a clear distinction among wild and cultivated samples was observed (clusters shown in blue and red in Figure 2A, respectively), regardless of their geographical origin. Mexican cultivated C. cujete samples showed an admixed pattern (membership probability to wild cluster from 0.16 to 0.87), as did some of the cultivated C. cujete from the Amazon Basin (membership probability to wild cluster from 0.01 to 0.98). The wild admixture within cultivated C. cujete in the Amazon Basin had higher proportions along the Amazonas River, decreasing values along the Solimões, Madeira, and Negro rivers, and was absent along the Branco River (Figure 2A). The wild Costa Rican sample displayed a membership probability of 0.25 to the cultivated cluster, a larger proportion than the membership shown by the Mexican wild samples (0.01–0.02). In the Principal Component Analysis (PCA), the first two principal components explained 16.7% of the total variance found in the dataset (Figure 2B). Principal component one separated wild from cultivated samples, while principal component two separated the Brazilian wild C. amazonica from the Mesoamerican wild C. cujete samples. The wild sample from Costa Rica was intermediate between wild and cultivated Mexican samples, which agree with its ancestry pattern observed in the clustering analysis performed by Structure. One Brazilian sample from Amazonas River was relatively closer to the Costa Rican sample (Figure 2B). To assess to what extent the intermediate ancestry of cultivated Mexican samples between wild Mesoamerican and Brazilian cultivated samples (Figure 2B) was associated with hybridization or divergence, we performed a Structure analysis among only Mesoamerican samples. This analysis clearly differentiates two groups of wild and cultivated Mesoamerican C. cujete (Supplementary Figure S2). However, we still observed the Costa Rican sample as having intermediate ancestry among these Mesoamerican samples (Supplementary Figure S2). Consequently, the intermediate ancestry detected in cultivated Mesoamerican samples may reflect divergence rather than hybridization. The differentiation between wild C. cujete and cultivated samples was also evident in the neighbor-joining dendrogram (Figure 2C). The level of differentiation between Mesoamerican wild and cultivated C. cujete was relatively low (FST = 0.35, IC95% = 0.13–0.60). The differentiation between Mesoamerican wild and Amazonian cultivated C. cujete samples was lowest with the Negro River (FST = 0.45, IC95% = 0.27–0.65), followed by the Amazonas (FST = 0.50, IC95% = 0.37–0.64), the Solimões (FST = 0.52, IC95% = 0.40–0.68), Madeira (FST = 0.57, IC95% = 0.39–0.81), and Branco (FST = 0.61, IC95% = 0.45–0.82). The wild C. amazonica samples showed high differentiation compared with cultivated C. cujete samples (FST = 0.57, IC95% = 0.37–0.64).
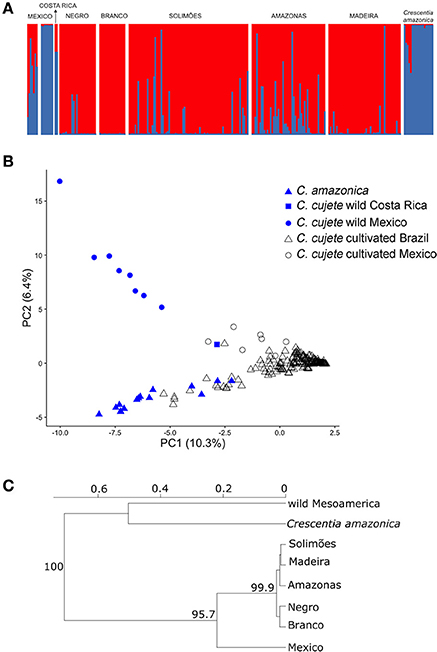
Figure 2. Genetic structure of 250 cultivated and wild treegourds (Crescentia cujete; C. amazonica) from Brazilian Amazonia and Mesoamerica based on 8 nSSR. Wild C. cujete were from the Yucatan Peninsula in Mexico and the Pacific coast of Costa Rica. (A) Structure plots at K = 2. The y-axis shows the proportion of assignment to the cluster and each vertical bar represents a single plant. The geographic locations and river basins are separated by white vertical columns; in Mexico, the first group is cultivated and the second is wild. (B) Principal components analysis (PCA) of nuclear genetic structure. The solid symbols represent the two species in Brazil, while the gray refers to wild and cultivated C. cujete in Mesoamerica. Numbers in parenthesis show the percentage of the allelic variation explained by each axis. (C) Neighbor-joining tree of the geographic relationships between wild and cultivated samples based on Nei's genetic distance with 1,000 bootstraps supports indicated on the nodes.
We performed another Structure analysis with the cultivated samples, using only plants whose membership probability was higher than 0.6 in the cultivated cluster (Figure 2A). The two allele frequency models showed similar patterns, with better defined clusters using the correlated model (Supplementary Figures S3, S4). Again, Evanno et al. (2005) ΔK suggested that two clusters are the most likely structure (K = 2, Supplementary Figure S3); these distinguished Mexican from Brazilian samples, with considerable admixture widely distributed in the Amazon Basin (Figure 3A). Evanno et al.'s ΔK suggested decreasing likelihood of structure up to four clusters (K = 3 and K = 4), although the fourth cluster did not show a pattern that was clearly different from K = 3. At K = 3, Mexican and Brazilian samples showed strong admixture (green and yellow clusters in Figure 3A). The green cluster membership was found in Mexico, but was higher along the Negro River and upper sections of the Branco River, with decreasing membership along the Solimões, Amazonas, and Madeira rivers (Figure 3A). In contrast, the third yellow cluster, also found in Mexico, was predominant along the Amazonas and Madeira rivers, scattered along the Solimões, but also high in the middle Negro River (Figure 3A). The neighbor-joining tree differentiated two groups within Amazonia that are both genetically different from Mexico (Figure 3B). However, the differentiation between Amazonia and Mexico is modest (FST = 0.04, IC 95% = 0.006–0.08). Spatial interpolation of the Structure clusters highlights that, although the admixture between Mexico and Amazonia (Figure 3A), genetic similarity is higher between Mexican samples and northwestern Amazonia (Figure 3C). The spatial interpolation also reveals the wide genetic homogeneity of cultivated C. cujete across Amazonia, except for the genetic differentiation in the Northwest and East, which is free from local wild-admixture effect in this data set (Figure 3C). The Northwestern and Eastern regions are relatively similar (Figure 3C), which agrees with the distribution of the Eastern yellow cluster up to the middle Negro River (Figure 3A). As expected, the Structure clusters in Amazonia without the Mexican samples show similar spatial interpolation pattern (Supplementary Figure S5).
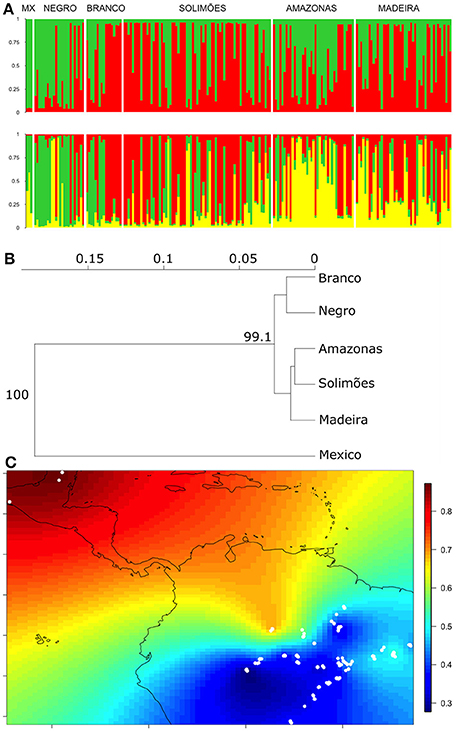
Figure 3. Nuclear genetic differentiation between cultivated C. cujete samples (N = 200) from Mexico and Brazilian Amazonia using 8 nSSR. Only samples with high cultivated membership (>0.6) from Figure 2 were included. (A) Structure analysis based on correlated allele frequency model. Plots show the two likely groupings (K = 2 and K = 3). The y-axis shows the proportion of assignment to the cluster and each vertical bar represents a single plant. Samples were ordered by their geographical location along the main rivers/country: the Negro, Solimões, and Amazonas Rivers are ordered west to east; the Branco River and Mexico are ordered north to south; the Madeira River is ordered south to north. (B) Neighbor-joining tree of the geographic relationships based on Nei's genetic distance with 1000 bootstraps supports indicated on the nodes. (C) Spatial interpolation of the Structure clusters (Q) at K = 2 indicated above (Figure 3A). The colored bar on the right indicates the probability of assignment to the green cluster (Figure 3A) between samples (white dots). Although the admixture between Mexico and Amazonia (Figure 3A), genetic similarity is higher between Mexican samples and northwestern Amazonia. Within Amazonia, cultivated C. cujete is genetically homogeneous, except by the differentiation in the Northwest and in the Eastern, which agrees with K = 3 (Figure 3A).
Geographic Patterns of Chloroplast Diversity
The haplotype network showed three distinct groups: C. amazonica, wild Mesoamerican C. cujete, and cultivated C. cujete from Brazil and Mexico (Figure 4A). The wild Mexican C. cujete lineage is more distant from cultivated C. cujete (55 substitutions + 12 substitutions) than is wild C. amazonica (39 + 12 substitutions). In the cultivated C. cujete group, five common haplotypes were identified, among which four are very close to each other (1 and 2 substitutions) at the core of the cultivated haplogroup (H1, H2, H3, H4). Haplotype H5 is differentiated by at least four substitutions from the core of the network. Divergent cultivated C. cujete samples from the Amazon basin were arranged in the extreme branches of the C. cujete group in the haplotype network (Figure 4A); the highest number of substitutions (36 and 75) was comparable to the differentiation between the wild and cultivated groups.
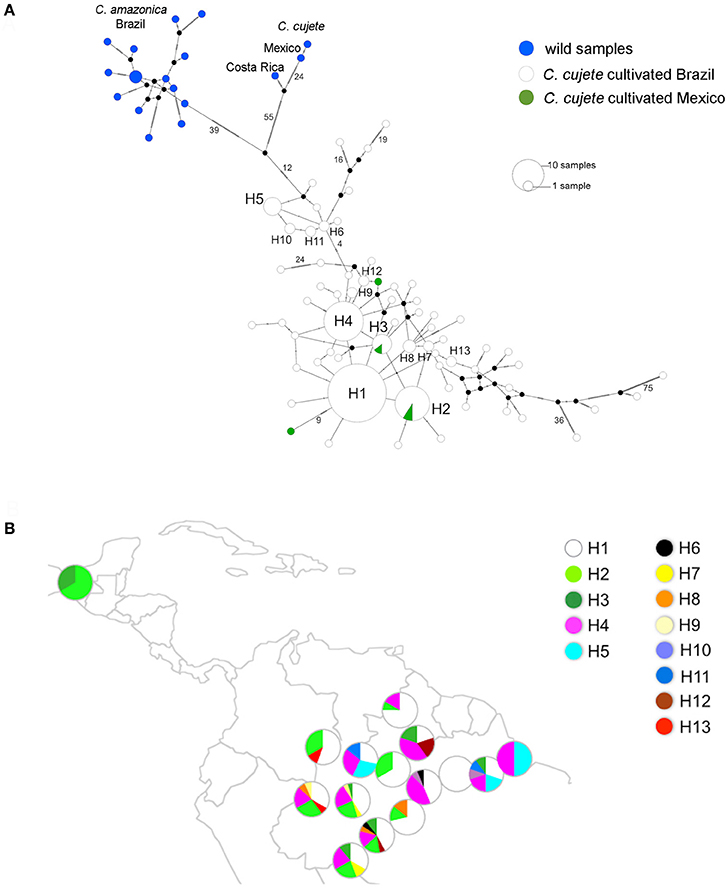
Figure 4. Chloroplast diversity of 215 cultivated and wild treegourds (Crescentia cujete; C. amazonica) from Brazilian Amazonia and Mesoamerica based on 334 SNPs. (A) Haplotype median-joining network of wild samples from Mexico and Costa Rica (C. cujete) and Brazil (C. amazonica), and cultivated C. cujete from Brazil and Mexico. The most abundant haplotypes are H1–H5 and the rare haplotypes, which are shared between at least two samples, are H6–H13. Number of substitutions between haplotypes is indicated on the branches. Black dots represent hypothetical intermediate haplotypes (B) Geographic distribution of 13 haplotypes of cultivated C. cujete depicted in the network (H1–H13). The area of the pie-slice is proportional to relative frequency of each haplotype in the section (upper, middle, lower) of Amazonian rivers and in Mexico.
The most common haplotype in the Amazon basin (H1) was widely dispersed, but not found in Mexico. Mexico and Brazil shared haplotypes H2 and H3, which, although different by only one substitution, showed slightly different distributions in the Amazon Basin (Figure 4B). Haplotype H2, the most common in Mexico, is restricted to the western half of Brazilian Amazonia, with higher frequency in the Northwest. Haplotype H3 is unevenly distributed in the Amazon Basin, but absent in the Northwest. Haplotype H4 is widely distributed, whereas haplotype H5, the most divergent haplotype (Figure 4A), is less abundant and found at low frequencies along the middle Negro River, but is well-represented in Eastern Amazonia. The most divergent rare haplotypes (H6, H10, H11) agree with the geographical distribution of the haplotype H5. The other rare haplotypes (H7, H8, H9) were sparsely distributed along the Solimões and Madeira rivers, except the haplotype H12 shared between Madeira and Branco River and the haplotype H13, restricted to the upper sections of Negro and Solimões Rivers (Figure 4B). None of the Amazonian rivers were significantly divergent from Mexico (Table 1), certainly because of the small sample size from Mexico. Within the Amazon Basin, the Amazonas River is the most differentiated from all other rivers (Table 1).
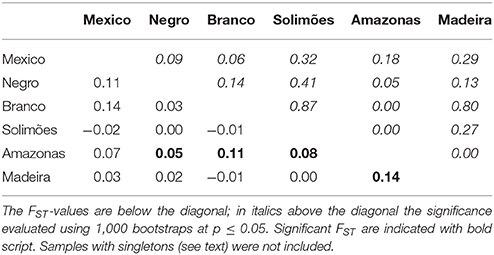
Table 1. Paired FST distance matrix between cultivated C. cujete chloroplast sequences (N = 181) based on 93 SNPs from Mexico and five Amazonian rivers.
Genetic Diversity in Cultivated C. cujete
Based on 8 nSSR of cultivated samples, there were 31 alleles in Mexico and 55 in the Amazon Basin (Table 2), although the sample sizes of the two regions are very different. The number of private alleles among cultivated samples showed that seven alleles were only found in cultivated Mexican samples and 31 alleles in cultivated Amazonian samples (Table 2), among which six are also found in wild Mesoamerican samples. Among Amazonian samples, the Amazonas River concentrated private alleles (5) not found in local wild C. amazonica. The Negro, Solimões and Madeira rivers had fewer private alleles, while none was found in the Branco River (Table 2). Mexico presented the highest expected heterozygosity (Hs). In the Amazon Basin, heterozygosity was highest along the Negro River, followed by the Solimões, Amazonas, Madeira rivers, and was lowest along the Branco River (Table 2). Mexico presented significant inbreeding, while in the Amazon Basin inbreeding was significant along the Branco and Madeira rivers (Table 2).
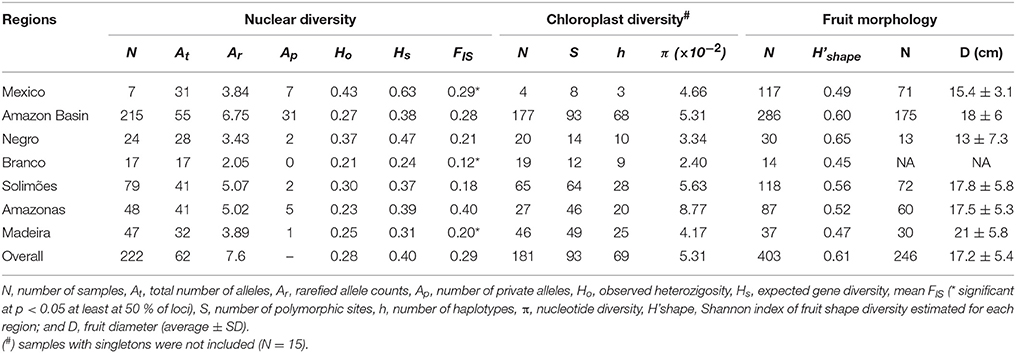
Table 2. Genetic diversity of cultivated Crescentia cujete in Mexico and along major rivers of the Brazilian Amazonia, based on 8 nuclear SSR, 93 chloroplast SNPs and eight fruit shapes.
Among the 334 SNPs found in chloroplast sequences, 206 were found in cultivated C. cujete. Mexico and the Amazon Basin showed similar nucleotide diversity (π), 3.78 × 10−2 and 3.83 × 10−2, respectively, although sample sizes are very different and the Amazon Basin harbors highly divergent samples (Figure 4A). Among cultivated C. cujete, 15 samples produced 119 unique SNP alleles, of which 66 % were from only two samples collected along the Amazonas River, which thus produced an extremely high nucleotide diversity estimate for this river (π = 9.31 × 10−2). When these 15 singleton samples were discarded, there were 93 SNPs and nucleotide diversity in Mexico was still similar to the Amazon Basin (Table 2). The highest nucleotide diversity was still along the Amazonas River, with decreasing values along the Solimões, Madeira, Negro, and Branco rivers (Table 2).
Morphological Diversity of Cultivated C. cujete
We identified a total of eight fruit shapes in the Amazon Basin and five in Mexico (Figure 5A). Fruit shapes shared among these regions were spherical, flattened, oblong, elongated, and cuneate, with higher frequencies of spherical, flattened, and oblong shapes in both regions. Three types (globular, rounded-drop, and oblong-drop) were only recorded in the Amazon Basin. The kidney-shaped fruit found in Colombia was not found in Mexico or Brazilian Amazonia. The absence of drop-shaped fruits in Mexico, which are types clearly distinguished from the others, indicate higher morphological diversity along Amazonian rivers than in Mexico. The Solimões River harbors all the eight fruit shapes described (Figure 5B). The spherical shape, the most frequent in Mexico, is relatively rare in the Amazon Basin, with a higher frequency along the Amazonas River (Figure 5B). The fruit types absent in Mexico were rare in the Amazon Basin as well, except the rounded-drop shape. This fruit type showed relatively high frequency along the Negro River, more than the more common flattened and oblong shapes (Figure 5B). The fruit shape diversity index was higher along the Negro River, with decreasing values along the Solimões and Amazonas rivers, followed by Mexico, and lowest along the Madeira and Branco rivers (Table 2). The fruit shape diversity index was not correlated with any of the genetic estimators (p > 0.05). The fruit diameters showed the lowest average along the Negro River and in Mexico, and the highest along the Madeira River (Table 2). Mexico and the Negro River also showed the extremes of size variation, with Mexico least variable and the Negro most variable (Table 2).
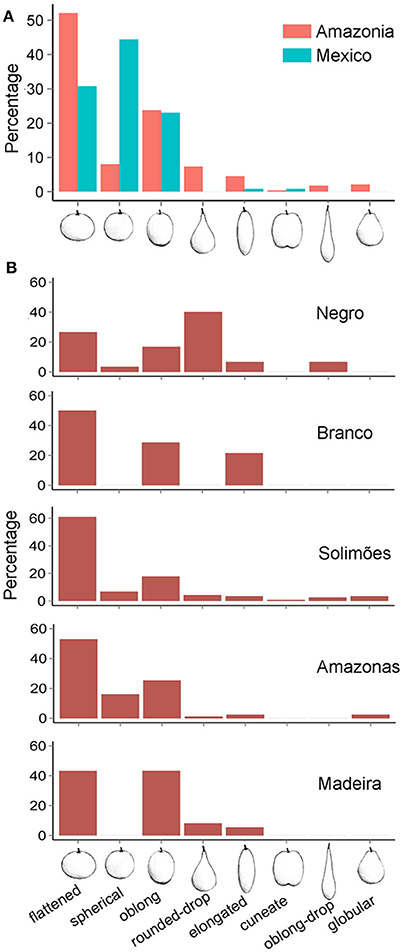
Figure 5. Morphological diversity of cultivated Crescentia cujete in (A) the two domestication centers studied (Mexico N = 117 and Amazonia N = 286) and (B) along Amazonian rivers. Fruit shapes correspond to those described by Arango-Ulloa et al. (2009), except the spherical shape.
Discussion
Cultivated C. cujete are quite similar from Mexico to Brazil, suggesting a common genetic origin. But these cultivated types are strongly differentiated from wild types, both from Mexico and Amazonia, suggesting these wild populations are not the direct ancestors of cultivated C. cujete. The geographical origin of the domestication of this species is still uncertain. However, the high diversity of cultivated C. cujete from Mexico, compared to Amazonia, suggests that its origin may be in Central America. Diversity analyses allowed discussion of the different routes of introduction into Amazonia and subsequent dispersal. More than one route may have been used: a northwestern introduction into the Negro and Solimões Rivers; and an eastern introduction from the coastal Guianas into the Amazonas River. Finally, fruit shape diversity suggests distinct selection pressures across the crop's distribution.
Relationships among Mesoamerican and Amazonian Treegourd Populations
The wild samples from Mexico (taxonomically identified as C. cujete) and the Amazon Basin (identified as C. amazonica) were strongly differentiated from the cultivated samples, given their FST values based on nuclear SSR and number of substitutions in the chloroplast genome. The high number of substitutions in the chloroplast sequences between these wild taxa suggests ancient divergence. The differentiation between wild and cultivated in Mexico (Aguirre-Dugua et al., 2012; under revision) and between wild and cultivated in Amazonia was already noted Moreira et al., 2017). These results suggest that neither of these wild relatives are the direct ancestor of cultivated C. cujete, although Mexican wild samples present clear morphological identification as C. cujete based on Gentry (1980) description.
The Costa Rican sample showed an intermediate admixed nuclear pattern, but high chloroplast differentiation from the cultivated samples (Figures 2, 4A). Consequently, it could be a wild individual pollinated by cultivated C. cujete. However, because ancestry could also reflect divergence, increased sampling in Central America is of interest. Although our results rule out the possibility that cultivated C. cujete was derived from the wild samples from the Yucatan Peninsula, we cannot rule out an origin somewhere between Central America and northern South America, where other potentially wild C. cujete populations occur in savannahs (Figure 1). Nevertheless, our results provide evidence that introduction of domesticated C. cujete in Mexico and Amazonia originated from the same source, given the Mexican relationship with Amazonian samples (Figure 3A, yellow and green clusters) and occurrence of wild Mesoamerican alleles in cultivated Amazonian C. cujete samples.
Hypotheses of Treegourd Introduction into Amazonia
The patterns of treegourd genetic diversity across the Amazon Basin allow two, not mutually exclusive, hypotheses of introduction: a Northwestern route and an Eastern route. A Northwestern route into the upper Negro River is supported by the relatively high levels of heterozygosity and fruit shape diversity (Table 2), higher proportions of Mexican ancestry (Figure 3A, green cluster) and higher frequency of the most common haplotype in Mexico (Figure 4B, haplotype H2). This route into Negro River is possible from the Orinoco River, given the fluvial connections via de Cassiquiare canal. This route was part of an extensive social trading network (Hornborg, 2005), based at least in part on the Arawak network (Eriksen and Danielsen, 2014). This route has also been suggested for various crop dispersals (Schultes, 1984), such as cocona (Solanum sessiliflorum), whose populations were domesticated in the upper Orinoco River (Volpato et al., 2004) and which was widely cultivated in Northwestern Amazonia (Schultes, 1957). Similarly, people from the upper Negro River reported intentional collection of treegourd propagules from the Cassiquiare, where treegourd is considered a spontaneous tree in the floodplains, while along the Negro River cultivation demands more effort (P.A.M., personal observation).
A possible Western route into the upper Solimões River is partially supported by heterozygosity and fruit diversity (Table 2); the presence of all fruit shapes described enhances the possibility (Figure 5B). Moderately high nucleotide diversity with the highest number of haplotypes are the strongest evidence (Table 2), especially because hybridization with wild populations was not reported (Moreira et al., 2017), suggesting that this is C. cujete diversity. This route might reflect introduction from the Pacific coast and crossing of the Andes mountains via the Napo and Putumayo rivers (Schultes, 1984), as might be the case of cacao (Theobroma cacao) (Thomas et al., 2012) and peach palm (Bactris gasipaes) (Rodrigues et al., 2005) demonstrated by molecular evidence. However, it is also possible that this is a continuation of the Negro River route across interfluvial areas, as suggested by the distribution of abundant haplotype H2 and the rare haplotype H13 (Figure 4B).
The Eastern route into the Amazonas River is supported by high heterozygosity and fruit diversity (Table 2), with high Mexican ancestry not found in Western Amazonia (Figure 3A, yellow cluster). The highest levels of nucleotide diversity (Table 2) and the particular distribution of haplotypes not found in Western Amazonia (Figure 4B, haplotype H5), which include one of the Mexican haplotypes (Figure 4B, H3), agree with the nuclear pattern. This route is linked to the coastal Guianas, an ancient area of exchange of Amazonian crops with Mesoamerica (Schultes, 1984). Molecular data of early maize (Zea mays) introduction into South America support dispersal from Mesoamerica through the Caribbean, spreading along the lowlands of the northeastern coast of South America to finally reach Amazon Basin through river systems (Freitas et al., 2003; Bedoya et al., 2017), although the oldest archaeological remains of maize are western (Bush et al., 2016). This route also agrees with pineapple dispersal from the Guianas, where it was domesticated and introduced into Mexico (Coppens D'Eeckenbrugge and Duval, 2009).
The extremely high chloroplast nucleotide diversity along the Amazonas River, almost twice that along the Solimões River (Table 2), is an unexpected result. Such high diversity was also observed with nuclear markers, given the relatively higher number of exclusive cultivated alleles along the Amazonas River (Table 2), which might not be related to local hybridization, since they were not found in C. amazonica (Moreira et al., 2017). While nuclear information is limited by the small number of loci analyzed, the chloroplast pattern is robust and they are in agreement. Therefore, we do not rule out that diversity along the Amazonas River might have been promoted by inter-specific hybridization between Mesoamerica and northern South America, where most diversity of Crescentia species is found (Gentry, 1980) and hybrid samples might have been introduced into Amazonia. Another process that is complementary and also deserves future investigations is the role of seed cultivation to deal with high flooding described along the Amazonas River (Moreira et al., 2017), since seeds might show diversity not found among cuttings as usually practiced (Arango-Ulloa et al., 2009; Aguirre-Dugua et al., 2012; Moreira et al., 2017). This hypothesis follows that of manioc (Manihot esculenta), where cuttings are usually practiced, but seed propagation is important to maintain diversity (Peroni and Sodero Martins, 2000; Elias et al., 2001; Duputié et al., 2009; McKey et al., 2010).
Hypothesis of Fruit Dispersal and Diversification
Domesticated varieties often present greater fruit shape diversity than their wild relatives, as observed in bottle gourd (L. siceraria), whose fruits have similar technological uses (Heiser, 1993; Morimoto et al., 2005). Across its distribution, the pattern of treegourd fruit shape diversity (Figure 5) suggests different cultural preferences affecting diversification. The highest shape diversity was found along the Negro and Solimões rivers (Figure 5B, Table 2). Similar high diversity was also observed in the Orinoco and Caribbean regions of Colombia (Arango-Ulloa et al., 2009), suggesting northwestern South America is an area of treegourd diversification. This pattern of diversity agrees with Amazonian ethnographies that underscore the cultural value of morphotype diversity cultivated for its own sake, such as in manioc (Rival and McKey, 2008) and pequi (Caryocar brasiliense) (Smith and Fausto, 2016). Nevertheless, the greater local frequency of the spherical type in Mexico and rounded-drop shape along the Negro River (Figure 5) suggests distinct selection pressures, as also described for popcorn in Peru (Grobman et al., 2012) and the differential selection of bitter and sweet manioc between Amazonia and the Atlantic Forest in Brazil (Emperaire and Peroni, 2007). Modern Maya people in Mexico and Guatemala have a long history of strong selection of spherical fruits of C. cujete for bowls (jícaras) to use with traditional beverages in rituals and also daily life situations (Ventura, 1996; Aguirre-Dugua et al., 2012, 2013). In Amazonia, the spherical and drop-shaped fruits of C. cujete have different symbolic importance and are recognized with distinct names by Tukano Oriental speakers (Pieter van der Veld, pers. communication), a linguistic family found in Northwestern Amazonia. The spherical fruit is called wahatowê, and is used as bowls to prepare ipadu powder (Erythroxylum coca var. ipadu) in rituals. In contrast, the rounded-drop, called ñahsãwaha, is common in daily life as a spoon and cup for collective food consumption (xibé, a meal of water and manioc flour, and açaí, the juice from Euterpe precatoria). Local people along the upper Negro River reported that the spherical type was also used as an ashtray by healers (pajé) in blessing rituals with tobacco smoke. Ethnographies also reported different treegourd fruits for each type of use, such as cuia-de-tapioca and cuia-de-ipadu (Ribeiro, 1995), although shape differences were not mentioned. In Northwestern South America, these bowls are cultural markers for the traditional use of coca introduced from the Andean foothills (Plowman, 1984). Interestingly, the spherical fruit shape selected in Mexico was the same as the one used in special rituals in Negro River Basin. This suggests that the wide dispersal of plants between South America and Mesoamerica in pre-Columbian times was motivated not essentially by food consumption, as would be expected for agrarian societies, but mainly for recreative and religious purposes (Neves, 2016). Indeed, archaeological remains of C. cujete in Central America and the Antilles were found in ritualistic contexts, such as offerings in funerary rituals (Beaubien, 1993; Conrad et al., 2001). This hypothesis of recreative and religious exchanges is also supported by the ancient dispersal of maize (Zea spp.) for beer preparation and tobacco (Nicotiana spp.) for magic and therapeutic uses, both widely exchanged between these continents (Heiser, 1965; Smalley and Blake, 2003), possibly as sacred gifts (Norton, 2008).
The relatively high morphological diversity found along the Solimões and Amazonas rivers, where most of rare fruit shapes were found (Figure 5B), suggests different demands for fruit shapes since pre-historic times, as expected among plants with technological uses (Blench, 2012). The upper Solimões River and middle Amazonas River were ancient treegourd handicraft centers that were regarded by both Europeans and Native Amazonians as one of the best expressions of their arts and an important article of trade (Rodrigues-Ferreira, 1933; Métraux, 1948). During the colonial period, villages along the Amazonas River produced 5,000–6,000 bowls a year that were exchanged for food (Rodrigues-Ferreira, 1933). This handicraft tradition extends until today, especially for the production of tacacá bowls (a kind of soup), which are made with the rounded (spherical and flattened) fruits (Moreira et al., 2017).
Although there is similarly high biological and cultural complexity in Mesoamerica and Amazonia (Blench, 2012; Clement et al., 2015; Casas et al., 2017), these two plant domestication centers contrast in terms of the morphological diversity of cultivated C. cujete fruits. Curiously, although Mexico pre-history is especially rich in complex societies, such as the Maya (Willey, 1956), morphological fruit diversity is lower and particular fruit shapes are absent, which also reinforces different cultural selection pressures between these regions. It follows that, although the introduction of the cultivated germplasm into both Mexico and Amazonia should lead to a bottleneck (i.e., through founder effect), it might be less severe in Amazonia due to a more diverse array of usages. Moreover, although the spread of a phenotype during dispersal might also be influenced by wild introgression/hybridization (Meyer and Purugganan, 2013), this effect was remarkable only on treegourd fruit size and not on shape diversity in Amazonia (Moreira et al., 2017). Within Mexico, elongated and smaller shapes spontaneously grown in homegardens, resulted possibly from gene flow with wild populations, are not appreciated in Yucatan Peninsula (Aguirre-Dugua et al., 2012), but are selected in the Pacific Coast as spoons (X.A.D, personal observation), although at low frequencies (Figure 5A). Therefore, cultural selection influences the bottleneck during introduction and afterwards the management of hybridization with local wild congeners. Whereas, distribution of shape diversity reflects different culture preferences, size is more influenced by local wild introgression effects.
Conclusions
We demonstrated with molecular evidence that C. cujete introduced into the Amazon Basin and Mexico shares a common ancestry with a currently unknown origin. The dispersal followed previously proposed routes of human and plant migrations into Amazonia. The patterns of genetic diversity across Amazonia allow two, not mutually exclusive, hypotheses of the routes of introduction: a Northwestern introduction into the Negro and Solimões rivers, and an Eastern introduction from the coastal Guianas into the Amazonas River. The fruit shape diversity reveals different ancient utilitarian demands for the fruits. Mesoamerica and Amazonia have contrasting fruit morphological diversity, which suggests different cultural preferences along treegourd's dispersal routes. More comparative studies of its different uses, with a broader genetic and phenotypic distribution, would be useful to better understand the dispersal and diversification of C. cujete in the Americas.
Author Contributions
PM, XA-D, CC, YV, and AC conceived the study. PM and XA-D carried out the field collections and interviews. PM, LZ, MC, CM, and DR performed the molecular work. PM, XA-D, CM, and YV performed the analysis. PM, XA-D, CC, and YV wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-473422/2012-3), the Fundação de Apoio à Pesquisa do Estado do Amazonas (FAPEAM 062.03.137/2012), the Agence Nationale de la Recherche (ANR-13-BVS7-0017), and the ARCAD project funded by the Agropolis Fondation. PM thanks the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior for a scholarship (CAPES-99999.010075/2014-03). We thank the Instituto de Desenvolvimento Agrário do Amazonas for logistical support and farmer families for their support, kindness and consent for this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2017.00150/full#supplementary-material
References
Aguirre-Dugua, X., Eguiarte, L. E., González-Rodríguez, A., and Casas, A. (2012). Round and large: morphological and genetic consequences of artificial selection on the gourd tree Crescentia cujete by the Maya of the Yucatan Peninsula, Mexico. Ann. Bot. 109, 1297–1306. doi: 10.1093/aob/mcs068
Aguirre-Dugua, X., Pérez-Negrón, E., and Casas, A. (2013). Phenotypic differentiation between wild and domesticated varieties of Crescentia cujete L. and culturally relevant uses of their fruits as bowls in the Yucatan peninsula, Mexico. J. Ethnobiol. Ethnomed. 9:76. doi: 10.1186/1746-4269-9-76
Arango-Ulloa, J., Bohorquez, A., Duque, M. C., and Maass, B. L. (2009). Diversity of the calabash tree (Crescentia cujete L.) in Colombia. Agrofor. Syst. 76, 543–553. doi: 10.1007/s10457-009-9207-0
Armstrong, C. G., Shoemaker, A. C., McKechnie, I., Ekblom, A., Szabó, P., Lane, P., et al. (2017). Anthropological contributions to historical ecology: 50 questions, infinite prospects. PLoS ONE 12:e0171883. doi: 10.1371/journal.pone.0171883
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Barreto, C., Lima, H. P., and Betancourt, C. J. (2016). Cerâmicas Arqueológicas da Amazônia: Rumo a Uma Nova Síntese. Belém: IPHAN : Ministério da Cultura.
Bartholomé, E., and Belward, A. S. (2005). GLC2000: a new approach to global land cover mapping from earth observation data. Int. J. Remote Sens. 26, 1959–1977. doi: 10.1080/01431160412331291297
Bass, J. (2004). Incidental agroforestry in Honduras: the jicaro tree (Crescentia spp.) and pasture land use. J. Lat. Am. Geogr. 3, 67–80. doi: 10.1353/lag.2005.0002
Beaubien, H. F. (1993). From codex to calabash: recovery of a painted organic artifact from the archaeological site of Cerén, El Salvador. J. Am. Inst. Conserv. 32, 153–164. doi: 10.2307/3179707
Bedoya, C. A., Dreisigacker, S., Hearne, S., Franco, J., Mir, C., Prasanna, B. M., et al. (2017). Genetic diversity and population structure of native maize populations in Latin America and the Caribbean. PLoS ONE 12:e0173488. doi: 10.1371/journal.pone.0173488
Bellwood, P. (2005). First Farmers: the Origins of Agricultural Societies. Malden, MA: Wiley-Blackwell.
Bennett, B. C. (1992). Plants and people of the amazonian rainforests. Bioscience 42, 599–607. doi: 10.2307/1311925
Blench, R. (2012). “The role of agriculture in explaining the diversity of Amerindian languages,” in The Past Ahead. Language, Culture and Identity in the Neotropics, ed C. Isendahl (Uppsala: ActaUniversitatisUpsaliensis), 13–38.
Bush, M. B., Correa-Metrio, A., McMichael, C. H., Sully, S., Shadik, C. R., Valencia, B. G., et al. (2016). A 6900-year history of landscape modification by humans in lowland Amazonia. Quat. Sci. Rev. 141, 52–64. doi: 10.1016/j.quascirev.2016.03.022
Casas, A., Parra, F., Aguirre-Dugua, X., Rangel-Landa, S., Vázquez, J. B., Vallejo, M., et al. (2017). “Manejo y domesticación de plantas en Mesoamérica: una estrategia de investigación y estado del conocimiento sobre los recursos genéticos,” in Domesticación en el Continente Americano Historia y Perspectivas del Manejo de Recursos Genéticos en el Nuevo Mundo, Vol. 2, eds A. Casas, J. Torres-Guevara and F. Parra-Rondinel (Morelia: UNAM), 69–102.
Clement, C. R. (1999). 1492 and the loss of amazonian crop genetic resources. II. crop biogeography at contact. Econ. Bot. 53, 203–216. doi: 10.1007/BF02866499
Clement, C. R., de Cristo-Araújo, M., D'Eeckenbrugge, G. C., Pereira, A. A., and Picanço-Rodrigues, D. (2010). Origin and domestication of native Amazonian crops. Diversity 2, 72–106. doi: 10.3390/d2010072
Clement, C. R., Denevan, W. M., Heckenberger, M. J., Junqueira, A. B., Neves, E. G., Teixeira, W. G., et al. (2015). The domestication of Amazonia before european conquest. Proc. R. Soc. 282:20150813. doi: 10.1098/rspb.2015.0813
Colunga-GarcíaMarín, P., and Zizumbo-Villarreal, D. (2004). Domestication of plants in Maya lowlands. Econ. Bot. 58, S101–S110. doi: 10.1663/0013-0001(2004)58[S101:DOPIML]2.0.CO;2
Conrad, G. W., Foster, J. W., and Beeker, C. D. (2001). Organic artifacts from the Manantial de la Aleta, Dominican Republic: preliminary observations and interpretations. J. Caribb. Archaeol. 2, 1–20.
Coppens D'Eeckenbrugge, G., and Duval, M. F. (2009). The domestication of pineapple: context and hypotheses. Pineapple News 16, 15–27.
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Ducke, A. (1946). Plantas de cultura precolombiana na Amazônia brasileira. Notas sôbre as espécies ou formas espontâneas que supostamente lhes teriam dado origem. Bol. Técn. Inst. Agron. Norte 8, 1–25.
Duke, J. A. (2009). Duke's handbook of medicinal plants of Latin America. Boca Raton, FL: Taylor & Francis.
Duputié, A., Massol, F., David, P., Haxaire, C., and McKey, D. (2009). Traditional amerindian cultivators combine directional and ideotypic selection for sustainable management of cassava genetic diversity. J. Evol. Biol. 22, 1317–1325. doi: 10.1111/j.1420-9101.2009.01749.x
Elias, M., Penet, L., Vindry, P., McKey, D., Panaud, O., and Robert, T. (2001). Unmanaged sexual reproduction and the dynamics of genetic diversity of a vegetatively propagated crop plant, cassava (Manihot esculenta Crantz), in a traditional farming system. Mol. Ecol. 10, 1895–1907. doi: 10.1046/j.0962-1083.2001.01331.x
Emperaire, L., and Peroni, N. (2007). Traditional management of agrobiodiversity in Brazil: a case study of manioc. Hum. Ecol. 35, 761–768. doi: 10.1007/s10745-007-9121-x
Eriksen, L., and Danielsen, S. (2014). “The Arawakan matrix,” in The Native Languages of South America: Origins, Development, Typology, eds. L. O'Connor and P. Muysken (Cambridge: Cambridge University Press), 152–176. doi: 10.1017/CBO9781107360105.009
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Freitas, F. O., Bendel, G., Allaby, R. G., and Brown, T. A. (2003). DNA from primitive maize landraces and archaeological remains: implications for the domestication of maize and its expansion into South America. J. Archaeol. Sci. 30, 901–908. doi: 10.1016/S0305-4403(02)00269-8
Freitas, F. O., and Bustamante, P. G. (2013). Amazonian maize: diversity, spatial distribution and historical-cultural diffusion. Tipití J. Soc. Anthropol. Lowl. South Am. 11, 60–65. Available online at: http://digitalcommons.trinity.edu/tipiti/vol11/iss2/7
Gentry, A. H. (1980). Bignoniaceae - Part 1 (Crescentieae and Tourrettieae). New York, NY: New York Botanical Garden Press.
Godoy, J. R., Petts, G., and Salo, J. (1999). Riparian flooded forests of the Orinoco and Amazon basins: a comparative review. Biodivers. Conserv. 8, 551–586. doi: 10.1023/A:1008846531941
Goudet, J. (2005). HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Resour. 5, 184–186. doi: 10.1111/j.1471-8286.2004.00828.x
Grobman, A., Bonavia, D., Dillehay, T. D., Piperno, D. R., Iriarte, J., and Holst, I. (2012). Preceramic maize from Paredones and Huaca Prieta, Peru. Proc. Natl. Acad. Sci. U.S.A. 109, 1755–1759. doi: 10.1073/pnas.1120270109
Hanotte, O., Bradley, D. G., Ochieng, J. W., Verjee, Y., Hill, E. W., and Rege, J. E. O. (2002). African pastoralism: genetic imprints of origins and migrations. Science 296, 336–339. doi: 10.1126/science.1069878
Heiser, C. B. J. (1965). Cultivated plants and cultural diffusion in nuclear America. Am. Anthropol. 67, 930–949. doi: 10.1525/aa.1965.67.4.02a00040
Hornborg, A. (2005). Ethnogenesis, regional integration, and ecology in prehistoric Amazonia: toward a system perspective. Curr. Anthropol. 46, 589–620. doi: 10.1086/431530
Jombart, T., Pontier, D., and Dufour, A. B. (2009). Genetic markers in the playground of multivariate analysis. Heredity 102, 330–341. doi: 10.1038/hdy.2008.130
Kamvar, Z. N., Tabima, J. F., and Grünwald, N. J. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. doi: 10.7717/peerj.281
Kistler, L., Montenegro, A., Smith, B. D., Gifford, J. A., Green, R. E., Newsom, L. A., et al. (2014). Transoceanic drift and the domestication of african bottle gourds in the Americas. Proc. Natl. Acad. Sci. U.S.A. 111, 2937–2941. doi: 10.1073/pnas.1318678111
Koboldt, D. C., Zhang, Q., Larson, D. E., Shen, D., McLellan, M. D., Lin, L., et al. (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576. doi: 10.1101/gr.129684.111
Leclerc, C., and Coppensd'Eeckenbrugge, G. (2012). Social organization of crop genetic diversity. The G × E × S interaction model. Diversity 4, 1–32. doi: 10.3390/d4010001
Leigh, J. W., and Bryant, D. (2015). popart: full feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Levis, C., Costa, F. R. C., Bongers, F., Peña-Claros, M., Clement, C. R., Junqueira, A. B., et al. (2017). Persistent effects of pre-Columbian plant domestication on Amazonian forest composition. Science 355, 925–931. doi: 10.1126/science.aal0157
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lima, R., and Saragoussi, M. (2000). “Floodplain home gardens on the central Amazon in Brazil,” in The Central Amazon floodplain: Actual Use and Options for a Sustainable Management, eds W. J. Junk, J. J. Ohly, M. T. F. Piedade, and M. G. M. Soares (Leiden: Backhuys Publishers), 243–268.
Mayle, F. E., and Iriarte, J. (2014). Integrated palaeoecology and archaeology – a powerful approach for understanding pre-columbian Amazonia. J. Archaeol. Sci. 51, 54–64. doi: 10.1016/j.jas.2012.08.038
McKey, D., Elias, M., Pujol, B., and Duputié, A. (2010). The evolutionary ecology of clonally propagated domesticated plants. New Phytol. 186, 318–332. doi: 10.1111/j.1469-8137.2010.03210.x
Medeiros, M. F., and Albuquerque, U. P. (2014). Food flora in 17th century northeast region of Brazil in Historia Naturalis Brasiliae. J. Ethnobiol. Ethnomed. 10:50. doi: 10.1186/1746-4269-10-50
Métraux, A. (1948). “Tribes of the middle and upper Amazon river,” in Handbook of South American Indians: the Tropical Forest Tribes, ed J. H. Steward (Washington, DC: Smithsonian Institution Press), 687–712.
Meulenberg, I. R. M. M. (2011). Calabashes and Bottle Gourds from Suriname: A Comparative Research between Maroons and Amerindians, with a Case-Study in Konomerume, A Kari'na Village. University of Leiden, Netherland.
Meyer, R. S., DuVal, A. E., and Jensen, H. R. (2012). Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 196, 29–48. doi: 10.1111/j.1469-8137.2012.04253.x
Meyer, R. S., and Purugganan, M. D. (2013). Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852. doi: 10.1038/nrg3605
Moreira, P. A., Mariac, C., Scarcelli, N., Couderc, M., Rodrigues, D. P., Clement, C. R., et al. (2016). Chloroplast sequence of treegourd (Crescentia cujete, Bignoniaceae) to study phylogeography and domestication. Appl. Plant Sci. 4:1600048. doi: 10.3732/apps.1600048
Moreira, P. A., Mariac, C., Zekraoui, L., Couderc, M., Rodrigues, D. P., Clement, C. R., et al. (2017). Human management and hybridization shape treegourd fruits in the Brazilian Amazon Basin. Evol. Appl.10, 577–589. doi: 10.1111/eva.12474
Morimoto, Y., Maundu, P., Fujimaki, H., and Morishima, H. (2005). Diversity of landraces of the white-flowered gourd (Lagenaria siceraria) and its wild relatives in Kenya: fruit and seed morphology. Genet. Resour. Crop Evol. 52, 737–747. doi: 10.1007/s10722-004-6119-8
Morton, J. F. (1968). The calabash (Crescentia cujete) in folk medicine. Econ. Bot. 22, 273–280. doi: 10.1007/BF02861961
Neves, E. (2016). “Não existe neolítico ao sul do Equador: as primeiras cerâmicas amazônicas e sua falta de relação com a agricultura,” in Cêramicas Arqueológicas da Amazônia - Rumo a Uma Nova Síntese, eds C. Barreto, H. P. Lima, and C. J. Betancourt (Belém: IPHAN; Ministério da Cultura), 32–39.
Norton, N. (2008). Sacred gifts, Profane Pleasures: a History of Tobacco and Chocolate in the Atlantic World. Ithaca, NY: Cornell University Press.
Otero, R., Núñez, V., Barona, J., Fonnegra, R., Jiménez, S. L., Osorio, R. G., et al. (2000). Snakebites and ethnobotany in the northwest region of Colombia Part III: neutralization of the haemorrhagic effect of Bothrops atrox venom. J. Ethnopharmacol. 73, 233–241. doi: 10.1016/S0378-8741(00)00321-4
Paradis, E. (2010). pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26, 419–420. doi: 10.1093/bioinformatics/btp696
Parker, K. C., Trapnell, D. W., Hamrick, J. L., Hodgson, W. C., and Parker, A. J. (2010). Inferring ancient Agave cultivation practices from contemporary genetic patterns. Mol. Ecol. 19, 1622–1637. doi: 10.1111/j.1365-294X.2010.04593.x
Parks, D. H., Porter, M., Churcher, S., Wang, S., Blouin, C., Whalley, J., et al. (2009). GenGIS: a geospatial information system for genomic data. Genome Res. 19, 1896–1904. doi: 10.1101/gr.095612.109
Patiño, V. M. (1967). Plantas Cultivadas Y Animales Domésticos en América Equinoccial, Tomo III: Fibras, Medicinas Y Misceláneas. Cali: Imprenta Departamental.
Paulo, I. M. M. (2016). Estudo Químico E Biológico de Crescentia Cujete L. (Bignoniaceae). Universidade Federal do Vale do São Francisco, Brazil.
Peroni, N., and Sodero Martins, P. (2000). Influência da dinâmica agrícola itinerante na geração de diversidade de etnovariedades cultivadas vegetativamente. Interciencia 25, 22–29.
Pinter, N., Fiedel, S., and Keeley, J. E. (2011). Fire and vegetation shifts in the Americas at the vanguard of paleoindian migration. Quat. Sci. Rev. 30, 269–272. doi: 10.1016/j.quascirev.2010.12.010
Piperno, D. R. (2011). The origins of plant cultivation and domestication in the New World tropics: patterns, process, and new developments. Curr. Anthropol. 52, S453–S470. doi: 10.1086/659998
Plowman, T. (1984). “The origin, evolution and diffusion of coca, Erythroxylum spp., in south and central America,” in Pre-Columbian Plant Migration. Papers of the Peabody Museum of Archaeology and Ethnology, ed D. Stone (Cambridge: Harvard University Press), 125–163.
Price, S. (1982). When is a calabash not a calabash? New West Indian Guid. 56, 69–82. doi: 10.1163/13822373-90002110
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Ramos, C. S. (2015). Efeitos do Extrato De Coité (Crescentia cujete) Sobre o Reparo Tecidual Em Lesões Cutâneas Não Contaminadas E Contaminadas Induzidas Em Rattus Norvegicus. Universidade Federal de Goiás, Brazil.
R Core Team (2015). R: A Language and Environment for Statistical Computing. Vienna: R Found. Stat. Comput.
Ribeiro, B. G. (1995). Os Índios Das Águas Pretas: Modo De Produção e Equipamento Produtivo, ed. B. G. Ribeiro. São Paulo: Editora da Universidade de São Paulo & Companhia das Letras.
Rindos, D. (1984). The Origins of Agriculture: An Evolutionary Perspective. San Diego, CA: Academic Press.
Rival, L., and McKey, D. (2008). Domestication and diversity in manioc (Manihot esculenta Crantz ssp. esculenta, Euphorbiaceae). Curr. Anthropol. 49, 1119–1128. doi: 10.1086/593119
Rodrigues, D. P., Astolfi Filho, S., and Clement, C. R. (2005). Molecular marker-mediated validation of morphologically defined landraces of pejibaye (Bactris gasipaes) and their phylogenetic relationships. Genet. Resour. Crop Evol. 51, 871–882. doi: 10.1007/s10722-005-0774-2
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Santos, A. M. (1982). Aritapera: uma comunidade de pequenos produtores na várzea amazônica (Santarém-PA). Bol. do Mus. Emílio Goeldi 83, 1–52.
Scarcelli, N., Mariac, C., Couvreur, T. L. P., Faye, A., Richard, D., Sabot, F., et al. (2016). Intra-individual polymorphism in chloroplasts from NGS data: where does it come from and how to handle it? Mol. Ecol. Resour. 16, 434–445. doi: 10.1111/1755-0998.12462
Schultes, R. E. (1957). A little-known cultivated plant from northern South America. Bot. Mus. Lealf. Harv. Univ. 18, 229–244.
Schultes, R. E. (1984). “Amazonian cultigens and their northward and westward migration in pre-Columbian times,” in Pre-Columbian plant migration. Papers of the Peabody Museum of Archaeology and Ethnology, ed D. Stone (Cambridge: Harvard University), 19–38.
Shepard, G., and Ramirez, H. (2011). Made in Brazil : human dispersal of the Brazil nut (Bertholletia excelsa, Lecythidaceae) in ancient Amazonia. Econ. Bot. 65, 44–65. doi: 10.1007/s12231-011-9151-6
Smalley, J., and Blake, M. (2003). Sweet beginnings: stalk sugar and the domestication of maize 1. Curr. Anthropol. 44, 675–703. doi: 10.1086/377664
Smith, M., and Fausto, C. (2016). Sociality and diversity of pequi (Caryocar brasiliense Caryocaraceae) among the Kuikuro of the Upper Xingu river (Brazil). Bol. do Mus. Para. Emílio Goeldi 11, 87–113. doi: 10.1590/1981.81222016000100006
Sodero Martins, P. (2005). Dinâmica evolutiva em roças de caboclos amazônicos. Estud. Avançados 19, 209–220. doi: 10.1590/S0103-40142005000100013
Solis, R. S. (2006). “America's first city? The case of late archaic Caral,” in Andean Archaeology III, eds W. H. Isbell and H. Silverman (New York, NY: Springer), 28–66.
Steward, J. H. (1948). Handbook of South American Indians: The Tropical Forest Tribes. Washington, DC: Smithsonian Institution Press.
Tajima, F., and Nei, M. (1984). Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1, 269–285.
Thomas, E., van Zonneveld, M., Loo, J., Hodgkin, T., Galluzzi, G., and van Etten, J. (2012). Present spatial diversity patterns of Theobroma cacao L. in the neotropics reflect genetic differentiation in Pleistocene refugia followed by human-influenced dispersal. PLoS ONE 7:e47676. doi: 10.1371/journal.pone.0047676
Ventura, C. (1996). The symbolism of Jakaltek Maya tree gourd vessels and corn drinks in Guatemala. J. Ethnobiol. 16, 169–183.
Volpato, G., Godínez, D., Beyra, A., and Barreto, A. (2009). Uses of medicinal plants by haitian immigrants and their descendants in the province of Camagüey, Cuba. J. Ethnobiol. Ethnomed. 5, 16–24. doi: 10.1186/1746-4269-5-16
Volpato, G., Marcucci, R., Tornadore, N., and Paoletti, M. G. (2004). Domestication process of two Solanum section Lasiocarpa species among amerindians in the upper Orinoco, Venezuela, with special focus on Piaroa indians. Econ. Bot. 58, 184–194. doi: 10.1663/0013-0001(2004)058[0184:DPOTSS]2.0.CO;2
Keywords: bignoniaceae, calabash, cuia, domestication, ethnobotany, historical ecology, phylogeography
Citation: Moreira PA, Aguirre-Dugua X, Mariac C, Zekraoui L, Couderc M, Rodrigues DP, Casas A, Clement CR and Vigouroux Y (2017) Diversity of Treegourd (Crescentia cujete) Suggests Introduction and Prehistoric Dispersal Routes into Amazonia. Front. Ecol. Evol. 5:150. doi: 10.3389/fevo.2017.00150
Received: 30 June 2017; Accepted: 14 November 2017;
Published: 29 November 2017.
Edited by:
B. Mohan Kumar, Nalanda University, IndiaReviewed by:
Milton Kanashiro, Embrapa Amazonia Oriental (Embrapa Easter Amazon), BrazilShabir Hussain Wani, Michigan State University, United States
K. S. Rao, University of Delhi, India
Copyright © 2017 Moreira, Aguirre-Dugua, Mariac, Zekraoui, Couderc, Rodrigues, Casas, Clement and Vigouroux. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Priscila A. Moreira, pri.ambrosio@hotmail.com
 Priscila A. Moreira
Priscila A. Moreira Xitlali Aguirre-Dugua
Xitlali Aguirre-Dugua Cédric Mariac
Cédric Mariac Leila Zekraoui
Leila Zekraoui Marie Couderc3
Marie Couderc3  Alejandro Casas
Alejandro Casas Charles R. Clement
Charles R. Clement Yves Vigouroux
Yves Vigouroux