Plant Mating Systems Often Vary Widely Among Populations
- 1School of BioSciences, University of Melbourne, Parkville, VIC, Australia
- 2Department of Biological Sciences, University of Wisconsin-Milwaukee, Milwaukee, WI, United States
- 3Ecology and Evolution, Research School of Biology, Australian National University, Canberra, ACT, Australia
- 4Department of Biology, University of Akron, Akron, OH, United States
Most flowering plants are hermaphroditic, yet the proportion of seeds fertilized by self and outcross pollen varies widely among species, ranging from predominant self-fertilization to exclusive outcrossing. A population's rate of outcrossing has important evolutionary outcomes as it influences genetic structure, effective population size, and offspring fitness. Because most mating system studies have quantified outcrossing rates for just one or two populations, past reviews of mating system diversity have not been able to characterize the extent of variation among populations. Here we present a new database of more than 30 years of mating system studies that report outcrossing rates for three or more populations per species. This survey, which includes 741 populations from 105 species, illustrates substantial and prevalent among-population variation in the mating system. Intermediate outcrossing rates (mixed mating) are common; 63% of species had at least one mixed mating population. The variance among populations and within species was not significantly correlated with pollination mode or phylogeny. Our review underscores the need for studies exploring variation in the relative influence of ecological and genetic factors on the mating system, and how this varies among populations. We conclude that estimates of outcrossing rates from single populations are often highly unreliable indicators of the mating system of an entire species.
Introduction
Most flowering plants are hermaphroditic, yet the proportion of seeds fertilized by self and outcross pollen varies widely among species, ranging from predominant self-fertilization to exclusive outcrossing (Schemske and Lande, 1985; Barrett and Eckert, 1990; Vogler and Kalisz, 2001; Goodwillie et al., 2005). The extent to which individuals and populations are outcrossing or selfing (the mating system) can influence the genetic structure of populations, the extent of gene flow, effective population size, and the expression of inbreeding depression (Barrett and Harder, 2017).
Much theoretical work on mating system evolution has focused on conditions favoring the evolution of self-fertilization within populations, such as reproductive assurance when pollen transfer opportunities are limited (Lloyd, 1992; Busch and Delph, 2011), and the genetic transmission advantage of selfing in populations with low inbreeding depression (Fisher, 1941). Conversely, high rates of outcrossing are thought to be favored when the cost of inbreeding depression exceeds the transmission advantage of selfing. These predictions led to the hypothesis that flowering plant populations should be subject to disruptive selection on the rate of outcrossing, causing mixed mating to be rare and transitory (Lande and Schemske, 1985; Barrett, 2003).
Although these predictions of mating system theory largely consider the action of evolution at the population level, many empirical studies investigating mating system variation in both plants and animals have emphasized variation among species, rather than among populations (Vogler and Kalisz, 2001; Goodwillie et al., 2005; Jarne and Auld, 2006; Moeller et al., 2017). For example, Goodwillie et al. (2005) showed that 42% of the 345 species in their survey were mixed-mating (0.2 < t < 0.8). The wide mating system surveys reported in Goodwillie et al. (2005) and Moeller et al. (2017), as well as earlier work with subsets of those data (Schemske and Lande, 1985; Barrett and Eckert, 1990; Barrett and Harder, 1996; Vogler and Kalisz, 2001; Goodwillie et al., 2005) have been important for stimulating research in this field. However, an important limitation of this approach is that most published outcrossing rates report data sampled from just one or two populations (Goodwillie et al., 2005). As noted by Schemske and Lande (1985), a species-level mean cannot adequately characterize highly variable species. Similarly, representing species outcrossing rates by only one or two population estimates potentially overlooks substantial and important variation in outcrossing rate within species. For example, the species mean outcrossing rate of Trillium camtschatcense is t = 0.53, yet population outcrossing estimates are starkly divergent; t > 0.95 for two populations and t < 0.15 in another two populations (Kubota et al., 2008).
We lack quantitative estimates of the prevalence and magnitude of among-population variation in outcrossing rates, despite discussion in the early literature (Schemske and Lande, 1985; Barrett and Eckert, 1990). This broad view is important, because if variation among populations is typically small, then estimates from one or two populations will adequately characterize the mating system of a species. Alternatively, substantial variation among populations provides crucial information needed to explore the evolution of mating-system diversity, and compels researchers to routinely assess multiple populations in mating system studies.
Obtaining a wider view on the distribution of among-population variation in outcrossing rates will also be useful for addressing questions concerning the evolution of such variation. For example, do closely related species tend to have more similar among-population variation than expected by chance? The extent to which mating system variance correlates with phylogenetic distance could help to ascertain the relative influence of environmental vs. heritable genetic factors on the mating system.
Among-population variation in mating system might also be linked to specific ecological influences. For example, biotically-pollinated species more commonly have mixed mating systems, relative to wind-pollinated species (Schemske and Lande, 1985; Aide, 1986; Barrett and Eckert, 1990). It has been suggested that the consistency of abiotic factors (wind pollination) is greater than that of biotic-pollination, as the availability of animal pollinators can vary widely between sites and seasons. Theoretically, this should drive higher variance in outcrossing rate among populations of biotically-pollinated plants (Aide, 1986; Barrett and Eckert, 1990; Waycott and Sampson, 1997), yet this hypothesis has not previously been tested.
Early work on outcrossing rates was largely based on imprecise estimates from single-gene morphological polymorphisms (Harding and Barnes, 1977; Schemske and Lande, 1985), whereas today the availability of molecular markers coupled with maximum likelihood multilocus outcrossing measures (Ritland, 2002) yield more precise estimates of population outcrossing rates. Over the last decade the number of studies reporting molecular marker-based outcrossing rates from three or more populations has nearly doubled, facilitating a detailed, population-level perspective on the distribution of outcrossing rate variation. We therefore surveyed more than 30 years of plant mating system studies in the literature, focusing on those studies presenting multi-population estimates of outcrossing. With these data, we then ask whether variance in the mating system is correlated to phylogeny or pollination mode and discuss the extent of among-population variation in plant mating systems.
Methods
Literature Survey
To measure variability in population-level estimates of outcrossing rate we assembled a database of studies reporting outcrossing rates (tm) for three or more populations per species. We focused exclusively on multilocus estimates of outcrossing rate because these estimates best distinguish true selfing from biparental inbreeding (Ritland, 1985), and are less influenced by selection than are single locus estimates (Ritland, 2002). The maximum likelihood multilocus outcrossing measure tm (Ritland and Jain, 1981) incorporated in software programs MLT (Ritland, 1990b) and MLTR (Ritland, 2002) is the most widely used estimator of the outcrossing rate. We therefore searched for all studies reported in Web of Science through Sept 1, 2016 that cited one of the two software programs (Ritland, 1990b, 2002) or the paper first formalizing tm (Ritland and Jain, 1981). From this set of papers, we extracted a dataset of population-level multilocus outcrossing estimates (tm) for flowering plants. To capture variation among populations our inclusion criteria required each study to have sampled a minimum of three populations for any one species or subspecies. We excluded population tm estimates derived from experimentally manipulated populations, planted crops, and seed orchards. Selfing in hermaphrodite angiosperms can occur within flowers either autogamously or facilitated by pollen vectors. Within-flower selfing does not occur in gymnosperms, and we therefore excluded them from our analysis in order to unite our dataset under a common paradigm of selfing. Similarly, we also excluded data from male-sterile plants in gynodioecious angiosperms.
We collected the reported standard error (s.e.) associated with each population tm, or if the s.e. was not reported we calculated the s.e. using the reported 95% confidence intervals. We set an inclusion threshold for s.e. of tm ≤ 0.12, which excluded 27 studies that either did not report s.e. or 95% confidence intervals, or that reported fewer than three population tm values with s.e. below or equal to this threshold. This quality control threshold was chosen to minimize the frequency of estimates that straddle thresholds between outcrossing/selfing/mixed mating, and to conservatively retain the 80% most reliable tm estimates. We also collected information on the pollination mode for each species by referring to the papers reporting tm, or when not reported, referring elsewhere in the primary literature.
Ten studies reported data for the same population over multiple years and in those cases we used the mean population tm across years. In a few cases, separate papers reported the same data for one or more populations, and we therefore used the supplied population codes to exclude duplicate populations. In 13 studies reporting separate tm for different flower or seed morphs within a population we calculated the mean tm for each population across those categories. Where tm or s.e. was reported in graphical rather than text format, we used DATATHIEF (Tummers, 2006) software to estimate the values. In 16 cases where population tm > 1, we bounded tm to 1.0, the maximum possible outcrossing rate. None of the included studies reported tm < 0.0.
Data Analysis
We performed an analysis testing for phylogenetic signal in both mean and variance of tm. For this, we generated a phylogeny for the species in our dataset using matK accessions obtained from NCBI Genbank, using sequences from the genus when sequences from the species were not available (following Lanfear and Bromham, 2011). We then aligned these sequences using MUSCLE (Edgar, 2004) in Geneious version 8 (Kearse et al., 2012), adjusted the alignment by eye, and removed from the alignment any columns which could not be confidently aligned. We used Modelfinder (Kalyaanamoorthy et al., 2017) to choose an appropriate model of evolution, and IQ-TREE (Nguyen et al., 2015) to estimate a maximum likelihood phylogenetic tree for the aligned sequences. We then used the PHYLOSIGNAL package in R (Keck et al., 2016) to calculate Moran's I (Gittleman and Kot, 1990), an autocorrelation based measure of phylogenetic signal (Münkemüller et al., 2012), varying between zero and one, where higher values indicate correlated traits in neighboring taxa relative to taxa drawn randomly from the tree. Significance in Moran's I was assessed through 999 bootstrap permutations.
We tested for a difference in variance of tm between biotic and abiotically pollinated species using an analysis that accounted for the fact that a 0–1 bounded estimate such as tm will naturally exhibit lower variance at the extreme values. We therefore used a normal approximation to the binomial where for each species, the mean tm (p) and sample variance (σ2) are used to calculate a ratio σ2/p(1 − p). This ratio estimates how well the observed variance matches expected binomial variance under the average tm and when treated as a response variable avoids the influence of intermediate values of tm inflating variance. We calculated 2,000 bootstrapped means for this ratio in both biotic and abiotic pollination classes and tested for significance by calculating the observed difference relative to the bootstrapped 95% confidence interval.
Results
Literature Survey
Our dataset includes tm and s.e. values for 741 populations from 105 species distributed across 44 families and 80 genera (Data Sheet 1). The number of population tm estimates per species ranged from 3 to 37, the mean was 7.06, and the median was 4 (Figure S1). In total, 111 studies met our inclusion criteria, with the number of studies approximately doubling each decade.
Many species in our survey exhibited substantial among-population variation in outcrossing rate (Figure 1). For 43/105 species, the range between minimum and maximum outcrossing rate exceeded 0.24 (twice the maximum population standard error). In the six most variable species, the s.d. of among-population tm was greater than the s.d. of species mean values across all 105 species (0.277). Species with the highest among-population variation in tm include: T. camtschatcense, Geranium carolinianum, Arabis alpina, Eichhornia paniculata, Arabidopsis lyrata, and Amsinckia spectabilis. As expected for a 0–1 bounded variable, among-population variation in tm estimates was lowest for species with extremely high or extremely low mean outcrossing rates (Figure S2). The among-population s.d. for most species was below 0.1 (Figure S2).
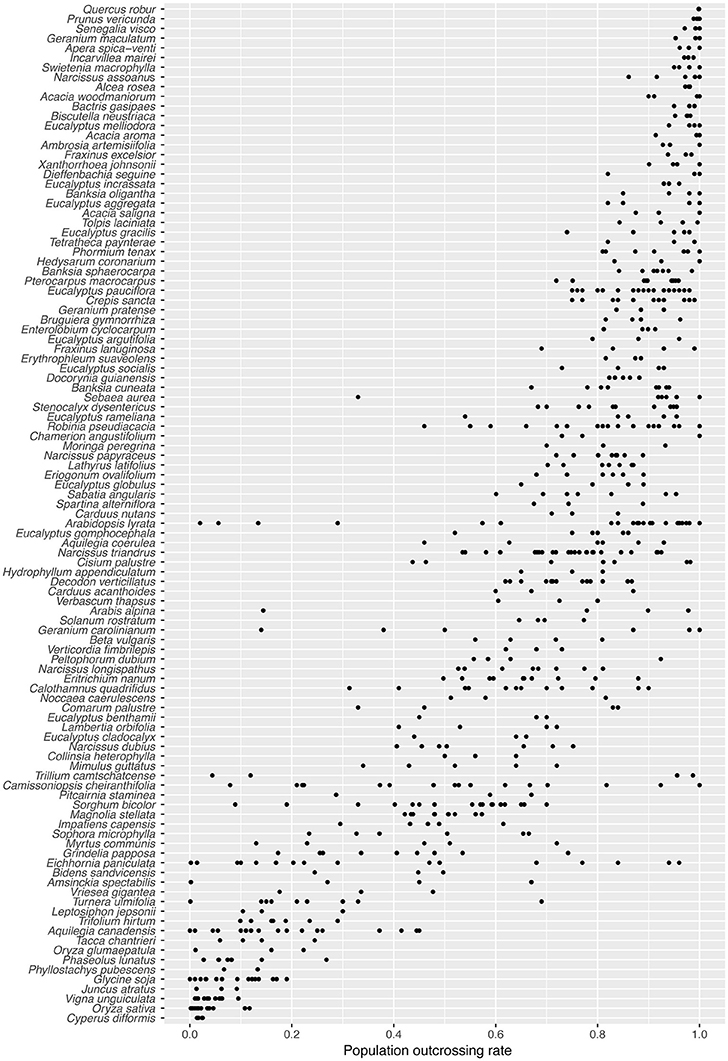
Figure 1. Population-level outcrossing rates (tm) for 105 plant species, arranged in order of decreasing species mean tm.
The distribution of tm estimates in our dataset appears similar to the distribution of species means for 469 species reported by Goodwillie et al. (2005) (Figure S3). A bootstrapped Kolmogorov-Smirnoff test suggests, however, that it is unlikely both datasets are sampling the same underlying distribution (1,000 bootstraps, P = 0.025). While Goodwillie et al. found 42% of species with mixed mating (0.2 < t < 0.8), our population level data shows slightly fewer mixed mating populations (36%), and therefore our dataset is more strongly bimodal. In the majority of species (54/105) the populations do not all fall into a single mating system class, as traditionally defined (tm ≤ 0.2 = selfing; 0.2 < tm < 0.8 = mixed mating; tm > 0.8 = outcrossing; Schemske and Lande, 1985; Figure 1). Most of those cases involve a combination of mixed mating and outcrossing (36/105), or of mixed mating and selfing (10/105). Only one species (T. camtschatcense) had both selfing and outcrossing populations with no mixed mating populations. Mixed mating populations were very common in this dataset, with nearly two-thirds of the species (66/105) having at least one mixed mating population.
Phylogenetic Analysis
Four taxa were not included in the matK phylogeny, as they did not have Genbank accessions (Banksia sphaerocarpa, Banksia oligantha, and Tetratheca paynterae), and one species (Tolpis laciniata) was removed due to its inexplicable placement on an extremely long branch. Moran's I found no phylogenetic correlation in variance (I = 0.003, permuted P = 0.194) and significant but weak phylogenetic correlation in mean tm (I = 0.135, permuted P = 0.001) (Figure S4).
Biotic vs. Abiotic Pollination
We classified species in our database by pollination mode: biotic (n = 87), abiotic (n = 10). Species classified as both biotic and abiotic (n = 4), and those for which pollination mode was unknown (n = 4) were excluded from the analysis. While the highest values of among-population variance were found in biotically pollinated species (Figure 2), the difference in variance between pollination modes was not significant according to our bootstrapped test of binomial variance (0.082, 95% CI: 0.049–0.116).
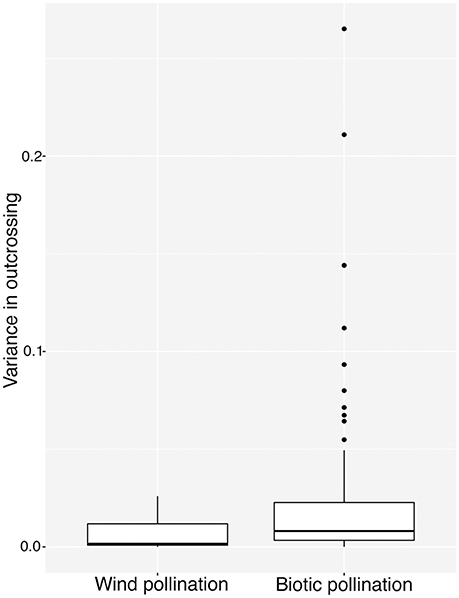
Figure 2. Variance in tm among populations in 87 species of biotically-pollinated and 10 species of abiotically-pollinated flowering plants. Lower and upper hinges correspond to 25 and 75th percentiles, internal horizontal bars indicate the median, vertical whiskers extends up to the largest value and down to the lowest values no further than 1.5 interquartile ranges from the hinge. Points outlying this are represented as dots.
Discussion
Our review of population-level outcrossing reveals prevalent and substantial among-population variation in the mating system of many flowering plant species. This variation cautions against the natural desire to generalize an entire species' mating system (as a “selfing,” “mixed-mating,” or “outcrossing” species) based on estimates from a small number of populations. While previous surveys based on species means or single population estimates concluded that ~40% of species are mixed-mating (0.2 < tm < 0.8) (Vogler and Kalisz, 2001; Goodwillie et al., 2005), 63% of the species in this review have one or more mixed-mating populations. As for previous work, the high frequency of mixed mating is difficult to reconcile with classical models that predict a bimodal distribution of tm (Lande and Schemske, 1985; Charlesworth et al., 1990).
Correlates of Outcrossing Variance: Pollination and Phylogeny
It is difficult to find evidence in our study to support the hypothesis that biotic pollination drives mating system variance, relative to abiotic pollination (Schemske and Lande, 1985; Aide, 1986; Barrett and Eckert, 1990). Although visual inspection of Figure 2 shows many more points with large variation for biotically pollinated species, our statistical analysis suggests that this is an illusion resulting from the necessary relationship between mean and variance for traits constrained to values between 0 and 1. Since many of the abiotically pollinated species had very low or very high outcrossing rates, they necessarily had low variance among populations. In our analysis, we attempted to control for this, and the lack of significant difference leads us to conclude that wind pollinated species are restricted in their variance because of more common obligate selfing or outcrossing, rather than because of any association with the pollen vector itself. This leads to the question of why wind pollinated species might more frequently evolve obligate selfing or outcrossing, which may hinge on important differences in pollen transport dynamics under biotic vs. abiotic pollination.
Our phylogenetic comparative analysis found no support in Moran's I for a correlation of among population variance in outcrossing rates with phylogeny. This implies that mating system variance might be evolving too fast, or too slow, to be reflected by phylogeny. Additionally, a strong intervening influence of environmental or ecological drivers of tm variance might be sufficient to obscure signal in traits that might actually be correlated to phylogeny. Weak but significant phylogenetic correlation in mean tm supports the previous findings of Moeller et al. (2017), and could be driven by correlated tm values in just a few groups (e.g., the grasses/rushes/sedges, and Mimosoideae; Figure S4).
What Are the Causes of Intraspecific Variation in Plant Mating Systems?
Among-population variation in outcrossing rate may reflect the influence of ecological factors that affect the proportion of self pollen deposited on stigmas (Karron et al., 1995, 2012; Devaux et al., 2014b; Sorin et al., 2016), as well as the role of heritable floral traits that influence the amount of outcross-pollen receipt (Barrett and Eckert, 1990; Kalisz et al., 2012) or post-pollination processes (Cruzan and Barrett, 2016; Sorin et al., 2016). It is important to distinguish these two broad causes of among-population variation because they differ in their predicted effects on the evolution of mating systems. Ecological factors exert important short-term effects that can create mating system variance, but by virtue of their stochasticity they will vary in the strength of their influence on adaptation. In contrast, heritable mating system traits dictate long term evolutionary behavior of the mating system (Devaux et al., 2014b). The complex interaction of environmental and heritable mating system traits ultimately directs adaptation in the mating system; for example, pollinator availability strongly influences the fitness benefits of heritable floral traits that promote outcrossing (Eckert et al., 2009).
The wide range of potential drivers of mating system variance is reflected by studies of the most variable species in our database. Considering only the 20% of species with the highest among-population variation in outcrossing rate, 10/21 studies reported among-population differences in floral traits that are likely to have a genetic basis, such as herkogamy, self-incompatibility, heterostyly, and level of autogamy. Environmental and demographic influences such as pollinator service, population size, and density were cited as sources of outcrossing rate variation in 6/21 studies.
Ecological factors influencing the quantity and quality of pollen delivery include pollinator abundance (Knight et al., 2005; Fishman and Willis, 2008); composition of pollinating fauna (Mitchell et al., 2004; Brunet and Sweet, 2006; Karron et al., 2012); plant population density (Murawski and Hamrick, 1991; Karron et al., 1995; Cheptou and Avendaño, 2006); presence and abundance of co-flowering species (Fishman and Wyatt, 1999; Bell et al., 2005; Moeller and Gebre, 2005); population size (Raijmann et al., 1994; Routley et al., 1999; Spigler et al., 2010); and habitat fragmentation (Aguilar et al., 2006; Eckert et al., 2010; Breed et al., 2015). Many of these ecological factors vary temporally as well as spatially, leading to within-year or among-year variation in outcrossing rate (Brunet and Sweet, 2006; Eckert et al., 2009; Kameyama and Kudo, 2009). Ecological factors can, therefore, cause dynamic shifts in mating system over short periods, potentially making single season observations difficult to interpret, and violating assumptions of analyses assuming constant tm (Ritland, 1990a; Routley et al., 1999). Few studies have quantified outcrossing rates of populations over multiple years, and a smaller subset have linked this variation to changes in ecological factors among years (Eckert et al., 2009). Our survey included 10 studies with multi-year outcrossing estimates for the same populations, however most of those did not include more than two time-point estimates for more than three populations. While a statistical analysis of among-year, vs. among-population variance is therefore premature, these studies found populations that varied markedly in outcrossing rate between years (difference in tm between years > 0.2) (Molina-Freamer and Jain, 1992; Herlihy and Eckert, 2002; Brunet and Sweet, 2006; Eckert et al., 2009; Butcher et al., 2011; Coates et al., 2013), supporting findings of Eckert et al. (2009) who found half of studies including multi-year outcrossing estimates reported appreciable temporal variation.
Genetic factors also influence the mating system. Key among these is the magnitude of inbreeding depression (Porcher and Lande, 2005; Devaux et al., 2014a). Among-population variation in outcrossing rates may also reflect the influence of heritable traits that vary within and among populations, such as floral morphology and floral display (Epperson and Clegg, 1988; Lloyd and Schoen, 1992; Harder and Barrett, 2006; Eckert et al., 2009; Karron and Mitchell, 2012), genetic self-incompatibility (Busch, 2005; Tedder et al., 2011; Dart et al., 2012), or stylar discrimination amongst self and outcross pollen (Kruszewski and Galloway, 2006; Cruzan and Barrett, 2016). These traits may not only affect the mating system, they may also affect pollen export to other plants, raising the possibility of “pollen discounting,” a factor which may stabilize mixed mating (Porcher and Lande, 2005; Johnston et al., 2009). Adding further complexity, some traits that influence the mating system, such as flower size, can be under interacting genetic and environmental control (Elle and Hare, 2002; Ivey and Carr, 2012), making it difficult to unambiguously identify the genetic component of variation in mating system (see below).
Designing Future Studies of Mating System Variation
In light of prevalent among-population variation in plant mating systems, future studies should characterize the outcrossing rate of several populations of each species, and should treat the mean outcrossing rate of each species with caution. The most informative studies will also report ecological and genetic axes of variation that correlate with mating system variation. Since ecological drivers of outcrossing rates may vary temporally (Brunet and Sweet, 2006; Eckert et al., 2009), effective studies will also measure population outcrossing rates, pollinator community composition, and co-flowering species over multiple years. Indeed, one hypothesis for the evolution of selfing suggests that high temporal variance in pollinator service might select for selfing as a means of reproductive assurance (Stebbins, 1957; Barrett and Husband, 1990a; Cheptou, 2004; Morgan et al., 2005; Moeller et al., 2017). This needs to be tested with a longitudinal design that explicitly measures variance in pollinator community and visitation alongside measures of outcrossing over multiple seasons.
One challenge when linking ecological factors with mating system variation is that multiple ecological variables are commonly confounded (Barrett and Eckert, 1990). For example, floral display size may correlate with population density (Karron et al., 1995), or pollinator abundance may correlate with presence of co-flowering species (Ghazoul, 2006; Mitchell et al., 2009; Flanagan et al., 2010). Confounding factors are best addressed through factorial experimental designs, where populations represent different combinations of the variables under study. For example, translocation experiments can address how the local pollination environment influences among-population variation in outcrossing rate. Although translocation studies have rarely been used in mating system research (cf. Kelly and Willis, 2002), they have been effective in studies of adaptive divergence in floral morphology (ecotypes) associated with differences in pollinator community (Peter and Johnson, 2014; Sun et al., 2014). In mating system work, individuals could be translocated from a focal population into a different pollination environment. Any associated differences in mating system between translocated and non-translocated plants indicates an environmental influence on the mating system.
Manipulative studies also provide the opportunity to explore how genetic factors influence the mating system. Common garden experiments, for example, are an underused but powerful way to control for the influence of environmental variation. By subjecting plants from several source populations to a common pollination context, and controlling environmental variation that may influence plastic traits such as flower size, these experiments are ideal for understanding the influence of genotype on the mating system (Elle and Hare, 2002; Karron and Mitchell, 2012).
Although seldom assessed, outcrossing rate variation among individuals may be nearly as large as variation among populations (Karron et al., 1997; Nora et al., 2016). This variation in outcrossing rate among individuals is also open to study by many of the correlative and experimental approaches described above. Floral traits, density, phenology, and co-flowering community can all vary on a fine sub-population scale, and the influence of these factors on the emergent mating system could productively be assessed by studies at the sub-population level.
Interpreting This Dataset
There are several characteristics of our database that may influence the distribution of reported outcrossing rates. First, as in previous surveys (Schemske and Lande, 1985; Barrett and Eckert, 1990; Barrett and Harder, 1996; Vogler and Kalisz, 2001; Goodwillie et al., 2005; Moeller et al., 2017), our sample over-represents some taxonomic groups. Our finding of no correlation in tm variance with phylogeny provides evidence that our data are not unduly influenced by these over-represented families. Second, our sample only includes studies reporting tm for three or more populations. If researchers are biased against making replicate outcrossing measures across populations when they do not expect to find variation, our results might also be biased toward over-representing species with tm variance. However, the observed variance in tm in our dataset does not correlate with the number of populations sampled (two-tailed Spearman's r = 0.174, P > 0.05) providing little evidence for this potential bias. Third, when sampling a species, instead of sampling populations randomly, researchers may bias their sample to populations they expect will display wide variation in outcrossing (Schemske and Lande, 1985). If this were occurring, our measures of species' variance in tm would be over-estimated relative to the true variance. Lastly, the proportion of mixed-mating species in our sample may be overestimated because researchers focusing on self-incompatible species may be less likely to measure among-population variation in outcrossing rates creating a bias against the appearance of obligate outcrossers in our dataset (Igic and Kohn, 2006).
Conclusions
Prevalent and substantial among-population variation in outcrossing rates highlights the need for caution in both estimation and interpretation of outcrossing rates in flowering plants. Estimates from single populations should not be used to characterize the mating system of an entire species. In the last several decades of research on the causes and consequences of plant mating system variation, hypotheses, data, and models have proliferated. Nevertheless, a unifying framework explaining the frequency of mixed mating remains elusive. Further insights in future work will require additional multi-population estimates of tm paired with relevant ecological data. In order to move beyond correlative explanations however, careful experimental studies that combine ecological manipulations and sampling over multiple seasons, are needed to distinguish the relative influence of ecology and genetics on the evolutionary enigma of mixed mating.
Author Contributions
MW, RM, and JK jointly conceived this paper, designed the sampling methodology, and drafted the manuscript. MW screened studies, collected, and managed the data. RL carried out phylogenetic analysis, and contributed to writing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer XP and handling Editor declared their shared affiliation.
Acknowledgments
This research was supported by an Endeavor Fellowship and McKenzie Postdoctoral Fellowship to MW, NSF award 1654943 to JK, and NSF 1654951 to RM. We thank Ben Phillips, Filipe Alberto, Emily Latch, Rod Peakall, Karl Duffy, Chris Ivey, Shu-Mei Chang, and Emmanuelle Porcher for discussions and comments on the manuscript. We are also grateful for helpful comments from three reviewers.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00038/full#supplementary-material
Figure S1. Histogram of number of population estimates of tm per species in this study (n = 105).
Figure S2. Standard deviation and mean among estimates of outcrossing rates (tm) for 105 flowering plant species.
Figure S3. Frequency histogram for all reported population estimates of tm pooled across the 105 flowering plant species in this study.
Figure S4. Values for variance and mean in outcrossing rate (tm) for 105 flowering plant species included in this study. The matK phylogeny for all species is on the left, species names on the right. Red points indicate significant correlation according to Moran's Local Indicator of Phylogenetic Association.
Data Sheet S1. Archive of data used in this study.
References
Aguilar, R., Ashworth, L., Galetto, L., and Aizen, M. A. (2006). Plant reproductive susceptibility to habitat fragmentation: review and synthesis through a meta-analysis. Ecol. Lett. 9, 968–980. doi: 10.1111/j.1461-0248.2006.00927.x
Aide, T. M. (1986). The influence of wind and animal pollination on variation in outcrossing rates. Evolution 40, 434–435. doi: 10.1111/j.1558-5646.1986.tb00486.x
Barrett, S. C. (2003). Mating strategies in flowering plants: the outcrossing–selfing paradigm and beyond. Philos. Trans. R. Soc. Biol. Sci. 358, 991–1004. doi: 10.1098/rstb.2003.1301
Barrett, S. C., and Harder, L. D. (1996). Ecology and evolution of plant mating. Trends Ecol. Evol. 11, 73–79. doi: 10.1016/0169-5347(96)81046-9
Barrett, S. C. H., and Harder, L. D. (2017). The ecology of mating and its evolutionary consequences in seed plants. Annu. Rev. Ecol. Evol. Syst. 48:23. doi: 10.1146/annurev-ecolsys-110316-023021
Barrett, S. C., and Husband, B. C. (1990a). Variation in outcrossing rates in Eichhornia paniculata: the role of demographic and reproductive factors. Plant Species Biol. 5, 41–55. doi: 10.1111/j.1442-1984.1990.tb00191.x
Barrett, S. C., and Eckert, C. (1990). “Variation and evolution of mating systems in seed plants,” in Biological Approaches and Evolutionary Trends in Plants ed S Kawano (Tokyo: Academic Press), 229–254.
Bell, J. M., Karron, J. D., and Mitchell, R. J. (2005). Interspecific competition for pollination lowers seed production and outcrossing in Mimulus ringens. Ecology 86, 762–771. doi: 10.1890/04-0694
Breed, M. F., Ottewell, K. M., Gardner, M. G., Marklund, M. H., Dormontt, E. E., and Lowe, A. J. (2015). Mating patterns and pollinator mobility are critical traits in forest fragmentation genetics. Heredity 115, 108–114. doi: 10.1038/hdy.2013.48
Brunet, J., and Sweet, H. R. (2006). Impact of insect pollinator group and floral display size on outcrossing rate. Evolution 60, 234–246. doi: 10.1111/j.0014-3820.2006.tb01102.x
Busch, J. W. (2005). The evolution of self-compatibility in geographically peripheral populations of Leavenworthia alabamica (Brassicaceae). Am. J. Bot. 92, 1503–1512. doi: 10.3732/ajb.92.9.1503
Busch, J. W., and Delph, L. F. (2011). The relative importance of reproductive assurance and automatic selection as hypotheses for the evolution of self-fertilization. Ann. Bot. 109, 553–562. doi: 10.1093/aob/mcr219
Butcher, P. A., Bradbury, D., and Krauss, S. L. (2011). Limited pollen-mediated dispersal and partial self-incompatibility in the rare ironstone endemic Tetratheca paynterae subsp paynterae increase the risks associated with habitat loss. Conserv. Genet. 12, 1603–1618. doi: 10.1007/s10592-011-0258-1
Charlesworth, D., Morgan, M. T., and Charlesworth, B. (1990). Inbreeding depression, genetic load, and the evolution of outcrossing rates in a multilocus system with no linkage. Evolution 44, 1469–1489. doi: 10.1111/j.1558-5646.1990.tb03839.x
Cheptou, P.-O. (2004). Allee effect and self-fertilization in hermaphrodites: reproductive assurance in demographically stable populations. Evolution 58, 2613–2621. doi: 10.1111/j.0014-3820.2004.tb01615.x
Cheptou, P. O., and Avendaño, L. G. (2006). Pollination processes and the Allee effect in highly fragmented populations: consequences for the mating system in urban environments. New Phytol. 172, 774–783. doi: 10.1111/j.1469-8137.2006.01880.x
Coates, D. J., Williams, M. R., and Madden, S. (2013). Temporal and spatial mating-system variation in fragmented populations of Banksia cuneata, a rare bird-pollinated long-lived plant. Aust. J. Bot. 61, 235–242. doi: 10.1071/BT12244
Cruzan, M. B., and Barrett, S. C. (2016). Postpollination discrimination between self and outcross pollen covaries with the mating system of a self-compatible flowering plant. Am. J. Bot. 103, 568–576. doi: 10.3732/ajb.1500139
Dart, S. R., Samis, K. E., Austen, E., and Eckert, C. G. (2012). Broad geographic covariation between floral traits and the mating system in Camissoniopsis cheiranthifolia (Onagraceae): multiple stable mixed mating systems across the species range? Ann. Bot. 109, 599–611. doi: 10.1093/aob/mcr266
Devaux, C., Lande, R., and Porcher, E. (2014a). Pollination ecology and inbreeding depression control individual flowering phenologies and mixed mating. Evolution 68, 3051–3065. doi: 10.1111/evo.12507
Devaux, C., Lepers, C., and Porcher, E. (2014b). Constraints imposed by pollinator behaviour on the ecology and evolution of plant mating systems. J. Evol. Biol. 27, 1413–1430. doi: 10.1111/jeb.12380
Eckert, C. G., Kalisz, S., Geber, M. A., Sargent, R., Elle, E., Cheptou, P.-O., et al. (2010). Plant mating systems in a changing world. Trends Ecol. Evol. 25, 35–43. doi: 10.1016/j.tree.2009.06.013
Eckert, C. G., Ozimec, B., Herlihy, C. R., Griffin, C. A., and Routley, M. B. (2009). Floral morphology mediates temporal variation in the mating system of a self-compatible plant. Ecology 90, 1540–1548. doi: 10.1890/08-1063.1
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Elle, E., and Hare, J. (2002). Environmentally induced variation in floral traits affects the mating system in Datura wrightii. Funct. Ecol. 16, 79–88. doi: 10.1046/j.0269-8463.2001.00599.x
Epperson, B. K., and Clegg, M. T. (1988). Genetics of flower color polymorphism in the common Morning Glory (Ipomoea purpurea). J. Hered. 79, 64–68. doi: 10.1093/oxfordjournals.jhered.a110450
Fisher, R. A. (1941). Average excess and average effect of a gene substitution. Ann. Eugen. 11, 53–63. doi: 10.1111/j.1469-1809.1941.tb02272.x
Fishman, L., and Willis, J. H. (2008). Pollen limitation and natural selection on floral characters in the yellow monkeyflower, Mimulus guttatus. New Phytol. 177, 802–810. doi: 10.1111/j.1469-8137.2007.02265.x
Fishman, L., and Wyatt, R. (1999). Pollinator-mediated competition, reproductive character displacement, and the evolution of selfing in Arenaria uniflora (Caryophyllaceae). Evolution 53, 1723–1733. doi: 10.1111/j.1558-5646.1999.tb04557.x
Flanagan, R. J., Mitchell, R. J., and Karron, J. D. (2010). Increased relative abundance of an invasive competitor for pollination, Lythrum salicaria, reduces seed number in Mimulus ringens. Oecologia 164, 445–454. doi: 10.1007/s00442-010-1693-2
Ghazoul, J. (2006). Floral diversity and the facilitation of pollination. J. Ecol. 94, 295–304. doi: 10.1111/j.1365-2745.2006.01098.x
Gittleman, J. L., and Kot, M. (1990). Adaptation: statistics and a null model for estimating phylogenetic effects. Syst. Biol. 39, 227–241. doi: 10.2307/2992183
Goodwillie, C., Kalisz, S., and Eckert, C. G. (2005). The evolutionary enigma of mixed mating systems in plants: occurrence, theoretical explanations, and empirical evidence. Annu. Rev. Ecol. Evol. Syst.36, 47–79. doi: 10.1146/annurev.ecolsys.36.091704.175539
Harder, L. D., and Barrett, S. C. (2006). Ecology and Evolution of Flowers. Oxford: Oxford University Press.
Harding, J., and Barnes, K. (1977). Genetics of Lupinus. X. Genetic variability, heterozygosity and outcrossing in colonial populations of Lupinus succulentus. Evolution 31, 247–255.
Herlihy, C. R., and Eckert, C. G. (2002). Genetic cost of reproductive assurance in a self-fertilizing plant. Nature 416, 320–323. doi: 10.1038/416320a
Igic, B., and Kohn, J. R. (2006). The distribution of plant mating systems: study bias against obligately outcrossing species. Evolution 60, 1098–1103. doi: 10.1111/j.0014-3820.2006.tb01186.x
Ivey, C. T., and Carr, D. E. (2012). Tests for the joint evolution of mating system and drought escape in Mimulus. Ann. Bot. 109, 583–598. doi: 10.1093/aob/mcr160
Jarne, P., and Auld, J. R. (2006). Animals mix it up too: the distribution of self-fertilization among hermaphroditic animals. Evolution 60, 1816–1824. doi: 10.1111/j.0014-3820.2006.tb00525.x
Johnston, M. O., Porcher, E., Cheptou, P. O., Eckert, C. G., Elle, E., Geber, M. A., et al. (2009). Correlations among fertility components can maintain mixed mating in plants. Am. Nat. 173, 1–11. doi: 10.1086/593705
Kalisz, S., Randle, A., Chaiffetz, D., Faigeles, M., Butera, A., and Beight, C. (2012). Dichogamy correlates with outcrossing rate and defines the selfing syndrome in the mixed-mating genus Collinsia. Ann. Bot. 109, 571–582. doi: 10.1093/aob/mcr237
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14:587. doi: 10.1038/nmeth.4285
Kameyama, Y., and Kudo, G. (2009). Flowering phenology influences seed production and outcrossing rate in populations of an alpine snowbed shrub, Phyllodoce aleutica: effects of pollinators and self-incompatibility. Ann. Bot. 103, 1385–1394. doi: 10.1093/aob/mcp037
Karron, J. D., Ivey, C. T., Mitchell, R. J., Whitehead, M. R., Peakall, R., and Case, A. L. (2012). New perspectives on the evolution of plant mating systems. Ann. Bot. 109, 493–503. doi: 10.1093/aob/mcr319
Karron, J. D., Jackson, R. T., Thumser, N. N., and Schlicht, S. L. (1997). Outcrossing rates of individual Mimulus ringens genets are correlated with anther-stigma separation. Heredity 79, 365–370. doi: 10.1038/hdy.1997.169
Karron, J. D., and Mitchell, R. J. (2012). Effects of floral display size on male and female reproductive success in Mimulus ringens. Ann. Bot. 109, 563–570. doi: 10.1093/aob/mcr193
Karron, J. D., Thumser, N. N., Tucker, R., and Hessenauer, A. J. (1995). The influence of population density on outcrossing rates in Mimulus ringens. Heredity 75, 175–180. doi: 10.1038/hdy.1995.121
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Keck, F., Rimet, F., Bouchez, A., and Franc, A. (2016). phylosignal: an R package to measure, test, and explore the phylogenetic signal. Ecol. Evol. 6, 2774–2780. doi: 10.1002/ece3.2051
Kelly, J. K., and Willis, J. H. (2002). A manipulative experiment to estimate biparental inbreeding in monkeyflowers. Int. J. Plant Sci. 163, 575–579. doi: 10.1086/340735
Knight, T. M., Steets, J. A., Vamosi, J. C., Mazer, S. J., Burd, M., Campbell, D. R., et al. (2005). Pollen limitation of plant reproduction: pattern and process. Annu. Rev. Ecol. Evol. Syst. 36, 467–497. doi: 10.1146/annurev.ecolsys.36.102403.115320
Kruszewski, L. J., and Galloway, L. F. (2006). Explaining outcrossing rate in Campanulastrum americanum (Campanulaceae): geitonogamy and cryptic self-incompatibility. Int. J. Plant Sci. 167, 455–461. doi: 10.1086/501051
Kubota, S., Kameyama, Y., Hirao, A. S., and Ohara, M. (2008). Adaptive significance of self-fertilization in a hermaphroditic perennial, Trillium camschatcense (Melanthiaceae). Am. J. Bot. 95, 482–489. doi: 10.3732/ajb.95.4.482
Lande, R., and Schemske, D. W. (1985). The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution 39, 24–40.
Lanfear, R., and Bromham, L. (2011). Estimating phylogenies for species assemblages: a complete phylogeny for the past and present native birds of New Zealand. Mol. Phylogenet. Evol. 61, 958–963. doi: 10.1016/j.ympev.2011.07.018
Lloyd, D. G. (1992). Self-and cross-fertilization in plants. II. The selection of self-fertilization. Int. J. Plant Sci. 153(3Pt 1), 370–380.
Lloyd, D. G., and Schoen, D. J. (1992). Self-and cross-fertilization in plants. I. Functional dimensions. Int. J. Plant Sci. 153, 358–369. doi: 10.1086/297040
Mitchell, R. J., Flanagan, R. J., Brown, B. J., Waser, N. M., and Karron, J. D. (2009). New frontiers in competition for pollination. Ann. Bot. 103, 1403–1413. doi: 10.1093/aob/mcp062
Mitchell, R., Karron, J., Holmquist, K., and Bell, J. (2004). The influence of Mimulus ringens floral display size on pollinator visitation patterns. Funct. Ecol. 18, 116–124. doi: 10.1111/j.1365-2435.2004.00812.x
Moeller, D. A., Briscoe Runquist, R. D., Moe, A. M., Geber, M. A., Goodwillie, C., Cheptou, P.-O., et al. (2017). Global biogeography of mating system variation in seed plants. Ecol. Lett. 20, 375–384. doi: 10.1111/ele.12738
Moeller, D. A., and Gebre, M. A. (2005). Ecological context of the evolution of self-pollination in Clarkia xantlana: population size, plant communities, and reproductive assurance. Evolution 59, 786–799. doi: 10.1111/j.0014-3820.2005.tb01753.x
Molina-Freamer, F., and Jain, S. K. (1992). Breeding systems of hermaphroditic and gynodioecious populations of the colonizing species Trifolium hirtum all in California. Theor. Appl. Genet. 84, 155–160.
Morgan, M. T., Wilson, W. G., and Knight, T. M. (2005). Plant population dynamics, pollinator foraging, and the selection of self-fertilization. Am. Nat. 166, 169–183. doi: 10.1086/431317
Münkemüller, T., Lavergne, S., Bzeznik, B., Dray, S., Jombart, T., Schiffers, K., et al. (2012). How to measure and test phylogenetic signal. Methods Ecol. Evol. 3, 743–756. doi: 10.1111/j.2041-210X.2012.00196.x
Murawski, D., and Hamrick, J. (1991). The effect of the density of flowering individuals on the mating systems of nine tropical tree species. Heredity 67, 167–174. doi: 10.1038/hdy.1991.76
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nora, S., Aparicio, A., and Albaladejo, R. G. (2016). High Correlated paternity leads to negative effects on progeny performance in two mediterranean shrub species. PLoS ONE 11:e0166023. doi: 10.1371/journal.pone.0166023
Peter, C. I., and Johnson, S. D. (2014). A pollinator shift explains floral divergence in an orchid species complex in South Africa. Ann. Bot. 113, 277–288. doi: 10.1093/aob/mct216
Porcher, E., and Lande, R. (2005). The evolution of self-fertilization and inbreeding depression under pollen discounting and pollen limitation. J. Evol. Biol. 18, 497–508. doi: 10.1111/j.1420-9101.2005.00905.x
Raijmann, L. E., Van Leeuwen, N. C., Kersten, R., Oostermeijer, J. G. B., Den Nijs, H., and Menken, S. B. (1994). Genetic variation and outcrossing rate in relation to population size in Gentiana pneumonanthe L. Conserv. Biol. 8, 1014–1026. doi: 10.1046/j.1523-1739.1994.08041014.x
Ritland, K. (1985). The genetic mating structure of subdivided populations I. Open-mating model. Theor. Popul. Biol. 27, 51–74. doi: 10.1016/0040-5809(85)90015-2
Ritland, K. (1990a). Inferences about inbreeding depression based on changes of the inbreeding coefficient. Evolution 44, 1230–1241.
Ritland, K. (1990b). A series of FORTRAN computer programs for estimating plant mating systems. J. Hered. 81, 236–237.
Ritland, K. (2002). Extensions of models for the estimation of mating systems using n independent loci. Heredity 88, 221–228. doi: 10.1038/sj.hdy.6800029
Ritland, K., and Jain, S. (1981). A model for the estimation of outcrossing rate and gene frequencies using n independent loci. Heredity 47, 35–52. doi: 10.1038/hdy.1981.57
Routley, M. B., Mavraganis, K., and Eckert, C. G. (1999). Effect of population size on the mating system in a self-compatible, autogamous plant, Aquilegia canadensis (Ranunculaceae). Heredity 82, 518–528. doi: 10.1038/sj.hdy.6885220
Schemske, D. W., and Lande, R. (1985). The evolution of self-fertilization and inbreeding depression in plants. II. Empirical observations. Evolution 39, 41–52.
Sorin, Y. B., Mitchell, R. J., Trapnell, D. W., and Karron, J. D. (2016). Effects of pollination and postpollination processes on selfing rate in Mimulus ringens. Am. J. Bot. 103, 1524–1528. doi: 10.3732/ajb.1600145
Spigler, R. B., Hamrick, J. L., and Chang, S. M. (2010). Increased inbreeding but not homozygosity in small populations of Sabatia angularis (Gentianaceae). Plant Syst. Evol. 284, 131–140. doi: 10.1007/s00606-009-0245-x
Stebbins, G. L. (1957). Self fertilization and population variability in the higher plants. Am. Nat. 91, 337–354. doi: 10.1086/281999
Sun, M., Gross, K., and Schiestl, F. P. (2014). Floral adaptation to local pollinator guilds in a terrestrial orchid. Ann. Bot. 113, 289–300. doi: 10.1093/aob/mct219
Tedder, A., Ansell, S. W., Lao, X., Vogel, J. C., and Mable, B. K. (2011). Sporophytic self-incompatibility genes and mating system variation in Arabis alpina. Ann. Bot. 108, 699–713. doi: 10.1093/aob/mcr157
Tummers, B. (2006). DataThief III. Available online at: http://datathief.org/
Vogler, D. W., and Kalisz, S. (2001). Sex among the flowers: the distribution of plant mating systems. Evolution 55, 202–204. doi: 10.1111/j.0014-3820.2001.tb01285.x
Keywords: selfing, outcrossing, breeding system, pollination, self-fertilization, mating system, mixed mating, mating system evolution
Citation: Whitehead MR, Lanfear R, Mitchell RJ and Karron JD (2018) Plant Mating Systems Often Vary Widely Among Populations. Front. Ecol. Evol. 6:38. doi: 10.3389/fevo.2018.00038
Received: 06 October 2017; Accepted: 22 March 2018;
Published: 05 April 2018.
Edited by:
Jordi Figuerola, Estación Biológica de Doñana (CSIC), SpainReviewed by:
Juan Arroyo, Universidad de Sevilla, SpainXavier Pico, Estación Biológica de Doñana (CSIC), Spain
Zhong-Jian Liu, Orchid Conservation and Research Center, China
Copyright © 2018 Whitehead, Lanfear, Mitchell and Karron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael R. Whitehead, michael.r.whitehead@gmail.com
 Michael R. Whitehead
Michael R. Whitehead Robert Lanfear3
Robert Lanfear3  Randall J. Mitchell
Randall J. Mitchell Jeffrey D. Karron
Jeffrey D. Karron