Characterizing Photosymbiosis Between Fraginae Bivalves and Symbiodinium Using Phylogenetics and Stable Isotopes
- 1Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA, United States
- 2Museum of Natural History, University of Colorado Boulder, Boulder, CO, United States
- 3Department of Ecology and Evolutionary Biology, University of Colorado Boulder, Boulder, CO, United States
- 4Department of Biology, University of Guam, Mangilao, GU, United States
- 5Department of Aquatic Zoology, Western Australian Museum, Perth, WA, Australia
Photosymbiotic associations between heterotrophic hosts and photosynthetic algae play crucial roles in maintaining the trophic and structural integrity of coral reef ecosystems. The marine bivalve subfamily Fraginae contains both non-symbiotic and photosymbiotic lineages, making it an ideal comparative system to study the origin and evolutionary adaptations of photosymbiosis. The symbiotic species exhibit unique morphological adaptations to photosymbiosis. However, the basic biology of these photosymbiotic relationships, such as symbiont diversity and nutritional benefits, has not been thoroughly characterized. In this study, we examined the general morphology of four Fraginae species occupying different depths (0–10 m): Corculum cardissa, Fragum fragum, Fragum scruposum, and Fragum sueziense. Abundant symbionts were found in the mantle, gill, and part of the foot, contained in tubular networks within host tissues. We used molecular phylogenetics to investigate the algal symbiont community of these Fraginae species. Results showed that symbionts from all four species are dinoflagellates belonging to the Symbiodinium clade C and we did not detect any host-specific or geographic-specific genetic structures within the symbionts. We also used stable carbon isotope analyses to examine whether the cockles are directly utilizing photosynthetically derived carbon sources. All species show less depleted 13C compared to filter-feeding bivalves, suggesting at least part of their organic carbon is derived directly from the symbionts. However, 13C depletion of Fragum sueziense collected from deeper habitats are less distinguishable from filter-feeding bivalves. This indicates that species in deeper habitats may rely less on photosymbiosis due to the reduced light availability. Given that the symbiotic fragines exhibit varying morphologies, habitats, and utilization of symbiont photosynthesis, the subfamily represents an ideal model system to study differential adaptations to photosymbiosis.
Introduction
Photosymbiotic associations between heterotrophic hosts and photosynthetic algae play crucial roles in maintaining the trophic and structural integrity of coral reef ecosystems (Cowen, 1988; Trench, 1993; Stanley and Lipps, 2011). The algae provide photosynthetic products to the host and gain shelter and inorganic nutrients in return, allowing the symbiosis to flourish in nutrient deficient habitats. Such associations have evolved in diverse marine lineages, including Foraminifera, Porifera, Cnidaria, and Mollusca (Gast and Caron, 2001; Stanley Jr, 2006; Hill et al., 2011; Vermeij, 2013). Among marine bivalves, photosymbiosis has been documented in at least four groups (Vermeij, 2013; Kirkendale and Paulay, 2017). However, obligate associations are found in only two cockle (Cardiidae) subfamilies, Tridacninae (i.e., giant clams), and Fraginae (i.e., heart cockles and relatives), where the former is relatively well studied. The tridacnids possess dinoflagellates (i.e., zooxanthellae) belonging to the genus Symbiodinium. A combination of photosymbiosis and filter feeding allows them to reach great sizes up to 1.2 m in width (Yonge, 1936; Fitt and Trench, 1981; Hawkins and Klumpp, 1995). The second group, Fraginae cockles, is much less understood despite their remarkable morphological adaptations presumably facilitating photosymbiosis (Kawaguti, 1941; Schneider and Carter, 2001; Kirkendale, 2009).
The subfamily Fraginae is composed of more than fifty morphologically diverse cockle species inhabiting tropical seas worldwide. Unlike the exclusively photosymbiotic Tridacninae giant clams (Fitt and Trench, 1981), Fraginae contains both non-symbiotic and symbiotic species. Seven of the ten modern genera are entirely non-photosymbiotic, while the remaining three are photosymbiotic (Fragum, Corculum, and Lunulicardia; Kirkendale, 2009). Recent molecular phylogenies (Kirkendale, 2009; Herrera et al., 2015) revealed a paraphyletic Fragum giving rise to Corculum and Lunulicardia, suggesting a single origin of Fraginae photosymbiosis, although the possibility of evolutionary loss in the non-symbiotic lineages cannot be rejected (Kirkendale and Paulay, 2017).
Some photosymbiotic fragines exhibit unique morphologies that are hypothesized to be adaptations to photosymbiosis, including large shell surface to volume ratio, highly flattened posterior shells, and transparent shell microstructures to increase light penetration (Watson and Signor, 1986; Ohno et al., 1995; Carter and Schneider, 1997; Schneider and Carter, 2001; Kirkendale, 2005; ter Poorten, 2009). Certain species also show behavioral strategies presumably to maximize sunlight exposure, such as posterior valve gaping (Persselin, 1998; Kirkendale, 2005). To date, the anatomy of symbiont-containing tissues has been studied in a single species, Corculum cardissa (Linnaeus, 1758) (Farmer et al., 2001). The bivalve harbors extracellular symbionts within the lumen of a tertiary tubular network, originating from the digestive system and extending into the mantle and gill tissues. The tubular structure is superficially analogous to that of the giant clams, although giant clams only harbor symbionts in the mantle (for more detailed descriptions of Fraginae morphology, see ter Poorten, 2009).
The diversity of Fraginae-associated symbiont communities are under explored (Kirkendale and Paulay, 2017). The majority of symbiotic algae in marine organisms are dinoflagellates belonging to the genus Symbiodinium, which has been divided into nine distinct clades (A-I) base on molecular phylogenies (Pochon and Gates, 2010). Different lineages in Symbiodinium vary greatly in their geographical distributions (Finney et al., 2010; LaJeunesse et al., 2010), physiological tolerance (Lien et al., 2007), and host specificity (Baker, 2003; Pochon et al., 2006; Yorifuji et al., 2015). Some lineages (e.g., clades A and C) are generalists that occupy a diverse array of hosts from different phyla; others (e.g., clade H) are relatively host-specific (Pochon et al., 2006). Existing literature on Symbiodinium is most comprehensive for coral-associated photosymbioses. Within Mollusca, tridacnid-associated symbionts are relatively well studied, covering 8 out of 12 currently recognized Tridacninae species. Their symbionts are mostly placed into Symbiodinium clade A, C, and occasionally D (Kirkendale and Paulay, 2017; see comprehensive literature list in Supplementary Table 2) and a single host individual can sometimes possess multiple symbiont lineages (DeBoer et al., 2012). Within Fraginae, the symbiont community from four species have been examined to date using molecular markers: Fragum fragum (Linnaeus, 1758), Fragum unedo (Linnaeus, 1758), Corculum cardissa, and Corculum monstrosum (Gmelin, 1791) (Carlos et al., 1999, 2000; Baillie et al., 2000; LaJeunesse, 2001; Dreier et al., 2014). Symbionts freshly isolated from the hosts mostly belong to Symbiodinium clade C, while symbionts cultured from host tissues are from clade A (Carlos et al., 1999). This discrepancy indicates possible selection bias, certain Symbiodinium strains may grow well in culture medium even though they are not the dominant strains within the host (Rowan, 1998). Therefore, to fully capture the symbiont diversity in Fraginae, freshly isolated symbionts from a more comprehensive suite of host species need to be examined.
The nature of Fraginae photosymbiosis has not been thoroughly examined across the subfamily, especially given that different photosymbiotic species show great variations in habitat choice and morphological traits (Kirkendale, 2009). For example, species in the genus Corculum are epifaunal and found in shallow reef flats with their extremely flattened posterior shells fully exposed (although often fouled). In contrast, many other species are shallowly buried in the sand like typical cardiids. Some populations of certain species can even live in deeper, more turbid environments like subtidal lagoons, e.g., Fragum sueziense (Issel, 1869). While some photosymbiotic species have morphologies that are uncommon in extant bivalves, others exhibit shell morphologies that do not differ greatly from non-photosymbiotic fragines. The different forms observed within the photosymbiotic Fraginae are suggestive of a variable reliance on photosymbiotically derived carbon sources. Therefore, it is important to verify whether the bivalve-algal relationship is nutritionally based and truly mutualistic, as research on gastropod-algal associations have suggested possible parasitic interactions between the adult animal and the algal symbionts (Banaszak et al., 2013).
Stable carbon isotope ratios have been useful in assessing trophic interactions in the marine environment (Robinson and Cavanaugh, 1995). The ratios are represented by δ13C values, calculated as (Rsample/Rstd-1) × 1000, where Rsample = 13Csample/12Csample and Rstd refers to the Vienna Pee Dee Belemnite Standard. Marine phytoplankton typically exhibit depleted 13C (i.e., negative δ13C) in their tissues compared to zooplankton, as they use ribulose 1,5-bis-phosphate carboxylase/oxygenase (RubisCO) to catalyze carbon fixation and all forms of RubisCOs fix 12CO2 more rapidly than 13CO2 (Hayes, 2001). This signature is reflected in heterotrophic organisms that consume phytoplankton. For example, common filter-feeding cardiids exhibit δ13C values between −24 ‰ and −22 ‰ (Rossi et al., 2004). Peridinin-containing dinoflagellates, including Symbiodinium, use form II RubisCO that discriminates less against 13C than most other forms of RubisCO (Cavanaugh and Robinson, 1996; Rowan et al., 1996). Thus, organisms relying largely on Symbiodinium-derived carbon sources will be more 13C enriched (i.e., show less negative δ13C values) than those who consume diverse phytoplankton. For instance, the photosymbiotic giant clam Tridacna gigas (Linnaeus, 1758) exhibits δ13C value of −16‰ (Black and Bender 1976). This differential isotope signature may be used to assess the carbons sources of Fragum. If Fragum uses symbiotically derived carbon, their tissues should show less depleted 13C than other filter-feeding cardiids.
The diverse spectrum of habitats, morphologies, and the inclusion of non-symbiotic species in Fraginae suggest that species in this subfamily could represent early evolutionary establishments of photosymbiotic partnerships. Therefore, understanding the origin of photosymbiosis in Fraginae will provide a better picture of how marine metazoans and photosynthetic algae develop symbiotic associations. It is unclear whether the two photosymbiotic cardiid groups, Tridacninae and Fraginae, share a common photosymbiotic ancestor or independently evolved the associations, as the evolutionary relationships between the two groups are currently unresolved (Herrera et al., 2015; Kirkendale and Paulay, 2017). Thus, it is important to examine the morphology and ecology aspects of the two groups to gain more comparative data, especially on the less studied subfamily Fraginae. Here, we characterized photosymbiosis in four representative fragines to deepen our understanding of this subfamily.
Specifically, we identified the phylogenetic relationships of the dinoflagellate symbiont communities from the Fraginae species and synthesized current knowledge on symbiont diversity in Fraginae and Tridacninae to contextualize these findings. We also investigated the stable carbon isotopes of the fragines to evaluate their potential food sources. These results add to our knowledge of the enigmatic bivalve group Fraginae and shed more light on photosymbiotic evolution in the Cardiidae.
Materials and Methods
Specimen Collections
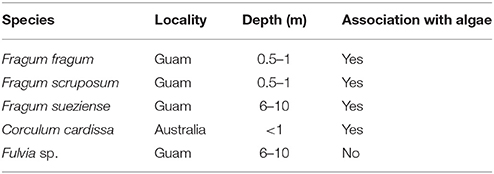
Four photosymbiotic Fraginae species from different habitats were chosen for this study (Table 1). Three of them were collected from Guam: Fragum fragum, Fragum scruposum (Deshayes, 1855), Fragum sueziense; and one from Australia: Corculum cardissa. F. fragum, and F. scruposum were collected from shallow sandy habitats at depth 0.5–1 m at East Agana Bay, Guam, by sieving through sand while snorkeling. F. sueziense were collected from sandy bottoms at depth 6–10 m via scuba diving at Outhouse Beach, Guam. One co-occurring heterotrophic cardiid species (Fulvia sp.) was also collected from Outhouse Beach as a control group. Specimens were processed at the University of Guam Marine Laboratory. Gill, mantle, and foot tissues of the specimens were fixed in 4% paraformaldehyde for histological study or dried for stable isotope analysis (see General morphology and histology and Phylogenetic analysis). Remaining specimens were preserved in 95% ethanol and deposited in the Museum of Natural History, University of Colorado Boulder (Supplementary Table 1). The collecting was done under the banner of the University of Guam Marine Laboratory permits for scientific collection as provided by the Government of Guam Department of Agriculture.
One specimen of Corculum cardissa was collected from intertidal reef habitat during the Western Australia Museum expeditions to Rowley Shoals in October 2014. Mantle and gill tissues were preserved in RNAlater. The rest of the specimen was preserved in 95% ethanol and deposited in the Western Australian Museum.
General Morphology and Histology
The general morphology of Fragum fragum (n = 5), F. scruposom (n = 5), and F. sueziense (n = 2) was examined to identify and characterize symbiont-containing tissues. Slices of gill and mantle tissues were removed and the symbionts were examined in hospite (i.e., within host) using light microscopy.
To examine the organization of the symbionts within the bivalve, gill tissues were embedded in paraffin and sectioned using standard histology protocols. Sections (10 μm) were placed on glass slides and stained with 0.05% toluidine blue solution for 20 min to highlight cell nuclei; rinsed with distilled water and air dried. Slides were then cleared in two changes of xylene and Canada balsam was used to fix the cover slips. Slides were examined using an AxioZoomV16 Microscope (Harvard Center for Biological Imaging).
Phylogenetic Analysis
Total DNA (host and symbiont) was extracted from approximately 3 mm2 of the symbiont-containing bivalve gill tissues using the Omega E.N.Z.A. Mollusc DNA kit. Gill tissues were homogenized in 1.5 ml microcentrifuge tubes using plastic pestles in 200 μL of ML1 buffer provided in the kit. An additional 150 μL of buffer was added to the homogenates and DNA was extracted according to manufacturer's instructions.
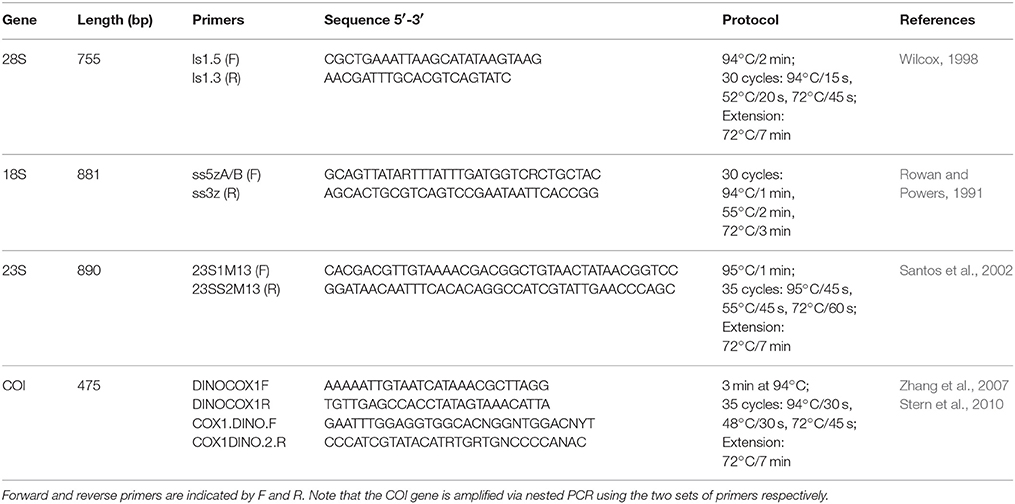
Four different Symbiodinium genetic markers were analyzed in this study: nuclear 18S rRNA, nuclear 28S rRNA, chloroplast 23S rRNA, and mitochondrial COI. Genes were amplified using Symbiodinium-specific primers following published protocols (Table 2). PCR amplifications were performed using Promega GoTaq Green Master Mix. Amplicons were visualized on a 1% agarose gel, purified using the Omega E.N.Z.A. Cycle Pure Kit, and directly sequenced at the Dana-Farber/Harvard Cancer Center DNA resource core. Sequence data were deposited in Genbank (Supplementary Table 1).
Sequences from this study were aligned with Symbiodinium sequences retrieved from Genbank (Supplementary Table 1) in Geneious 8.1.3 (Biomatters) using the built-in Geneious algorithms with default parameter settings. A maximum likelihood phylogeny was constructed for each gene with 100 bootstrap replicates using the RAxML 7.7.1 (Stamatakis et al., 2008) online server hosted at RAxML BlackBox (http://embnet.vital-it.ch/raxml-bb/index.php). The best scoring trees were selected to represent the phylogenies.
Bayesian phylogenies were established for each individual gene and the concatenated dataset using MrBayes v3.2.6 (Ronquist et al., 2012). The concatenated dataset only included Symbiodinium strains that were represented by more than two gene markers. Gene substitution models were selected using PartitionFinder (Lanfear et al., 2012) based on Bayesian information criterion as follows. 23S and 28S: GTR+I+G; 18S: K80+I+G; COI codon 1: HKY+G; COI codon 2, and 3: F81+I. For each dataset, two independent MCMC runs were conducted simultaneously for 10 million iterations and sampled every 1,000 iterations. Convergence was visually examined for each analysis in Tracer V1.6.0 (Drummond et al., 2012). A 50% majority rule consensus phylogeny was generated with 25% burnin. All Bayesian analyses were conducted on the CIPRES science Gateway (http://www.phylo.org). All phylogeny data were deposited in TreeBase (Study ID: 21845).
Stable Carbon Isotopes Analyses
Symbiont-containing mantle and symbiont-free foot tissues of Fragum fragum (N = 5), F. scruposom (N = 9), and F. sueziense (N = 1) were dissected and rinsed in filtered seawater. The mantle and foot tissues of the non-photosymbiotic cardiid (Fulvia sp., N = 6) collected from the same locality as F. sueziense were also harvested. Due to the small sizes of Fulvia specimens, we had to combine foot and mantle tissues together for the analysis. Tissues were placed on pieces of aluminum foil and dried for 48 h at 40°C. Dried tissues were acidified and stable carbon isotopes were measured using IsoPrime isotope ratio mass spectrometer at the Boston University Stable Isotope Laboratory. δ13C values (‰) were reported as (Rsample/Rstd-1) × 1000, where Csample/12Csample and Rstd refers to the Vienna Pee Dee Belemnite Standard. For technical details on isotope analysis, please refer to the Boston University Stable Isotope Laboratory quality assurance webpage: https://www.bu.edu/sil/quality.htm. Stable carbon isotopes were not determined for Corculum cardissa as the specimen available was not preserved for isotope analysis.
A Welch's t-test was used to statistically compare δ13C values between foot and mantle tissues of the Fraginae species, as well as between Fraginae and Fulvia tissues. Values for F. sueziense were not used in the test due to small sample size.
Results
Overall, we found abundant symbionts in the mantle, gill and part of the foot tissues in the photosymbiotic fragines, and the symbionts were extracellular and contained in tubules within host tissues. Symbiont sequences all fell in Symbiodinium clade C based on our molecular phylogenies. All three Fragum species exhibited less depleted C13 compared to filter-feeding cockles, although F. sueziense showed δ13C values closer to those of the filter-feeding bivalves (see details below).
General Observations and Morphology
Abundant single-cellular golden brown algae were found in most of the mantle, the entire gill, and the heel portion of the foot of the three Fragum species (F. fragum, F. scruposum, and F. sueziense) that were observed alive (Figure 1a). The symbiont-containing structures within the tissues were similar among all three species, and images of F. scruposum are shown as representatives. In all symbiont-containing tissues, the algae were extracellular, and tightly packed within the lumen of tubular structures (Figures 1b–d). The tubules appeared to be narrow enough to accommodate only one or two rows of algal cells. When observed in hospite, the algal cells exhibited great flexibility and mostly conformed to the shape of the tubules (Figure 1b). Only when the cells were squeezed out of the tissues after fixation, did they resume the typical spherical shape of the coccoid stage found in other invertebrate hosts.
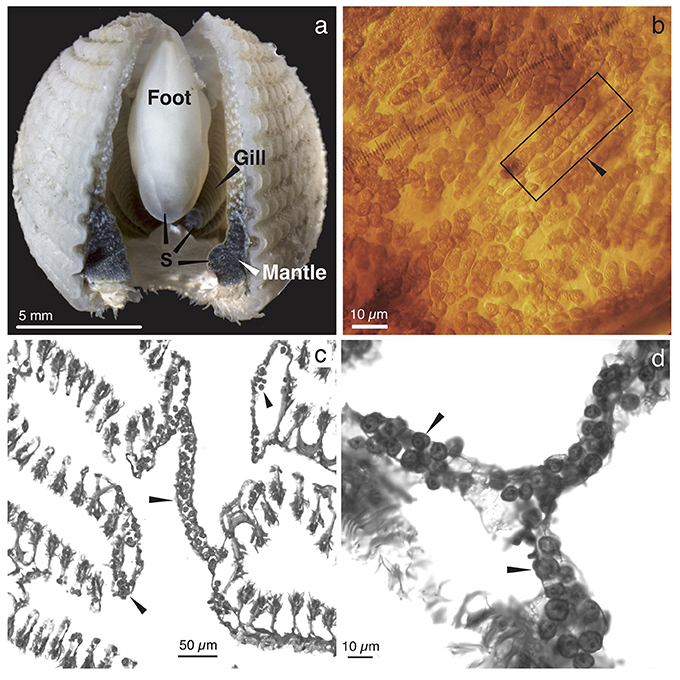
Figure 1. Gross morphology and histology of Fragum scruposum and symbionts. (a). General morphology of the ventral side of an individual. Dark colored regions were occupied by symbiotic algae (S). (b). Image of a piece of bivalve mantle tissue. Box highlights tubules that contain symbionts in hospite. Note that the symbiont cells are tightly packed in the tubules. (c). Light micrograph of a paraffin-embedded cross section through the gill tissue, showing symbionts in tubular network (arrows). (d). Higher magnifications of symbionts within tubules.
Phylogenetic Position of Fraginae Symbionts
All Symbiodinium genetic markers were successfully amplified from tissues of all four Fraginae species: Fragum fragum (N = 7), F. scruposum (N = 5), F. sueziense (N = 1), and Corculum cardissa (N = 1). Both maximum likelihood (ML) and Bayesian analyses recovered well-supported clades (A-H) that are largely congruent with current Symbiodinium phylogenies (Pochon et al., 2012, 2014). For simplicity, only Bayesian topologies are shown here (Figure 2). ML phylogenies are available in TreeBase (ID: 21845).
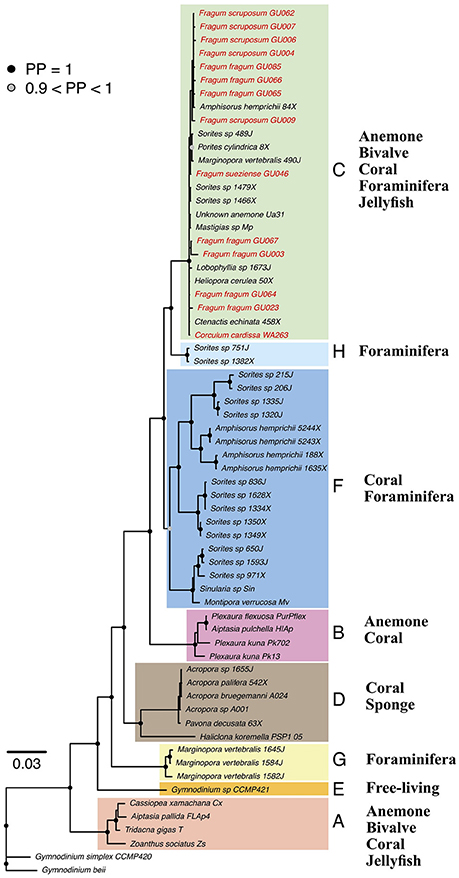
Figure 2. Bayesian phylogeny of Symbiodinium from diverse invertebrate hosts based on concatenated gene dataset. Tip labels indicate host species and symbiont community ID. Node symbols represent posterior probabilities (PP), PP < 90% are not shown. Symbiodinium clades A–H are highlighted by color blocks. Fraginae Symbiodinium lineages are highlighted in red. General host groups are listed for each clade. Detailed concatenated and individual gene trees are deposited in TreeBase (ID: 21845).
Sequences amplified from symbiont communities from all four Fraginae species fell in Symbiodinium clade C. The placement was consistent for the concatenated phylogeny (Figure 2) and all four individual gene trees. Clade A symbionts were not detected in our samples, although their existence is reported from other studies (Carlos et al., 1999, 2000; Baillie et al., 2000; LaJeunesse, 2001). Symbiont sequences showed low variations among different host species for the more conservative genes (18S, 28S, and 23S). The mitochondrial COI gene revealed some substructures among different Fraginae symbionts, but the symbionts did not cluster based on host species or locality. The overall genetic variation in the entire clade C was relatively low compared to other clades such as A, D, and F.
Currently known Fraginae-associated symbiont phylogenetic diversity based on our results and five other molecular studies (Carlos et al., 1999, 2000; Baillie et al., 2000; LaJeunesse, 2001; Dreier et al., 2014) are summarized (Table 3, Supplementary Table 2). All freshly isolated symbiont communities (i.e., non-cultured) were from clade C, except for one individual of Fragum unedo, which harbored clade A symbionts (Baillie et al., 2000). In contrast, all symbiont communities cultured from host tissues belonged exclusively to clade A (refer to Supplementary Table 2 for details on the host species, geographical locations, genetic markers used, and references). To compare symbiont diversity between Fraginae and Tridacninae, a summary of symbiont communities for Tridacninae was also included in Table 3 and Supplementary Table 2.
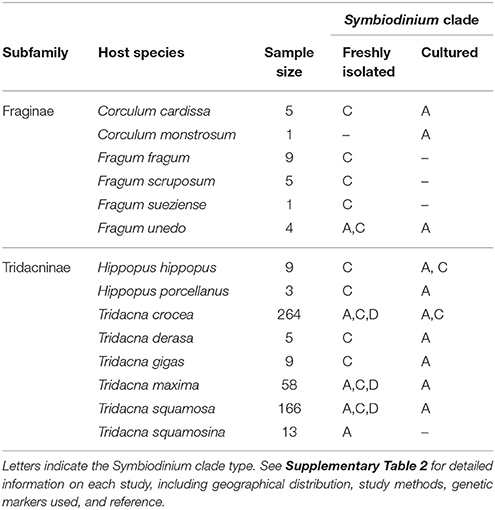
Table 3. Phylogenetic identity of Fraginae and Tridacninae associated Symbiodinium based on this study and literature review.
Stable Isotope Analyses
Stable carbon isotope ratios of the bivalve species studied here are shown in Table 4, and compared with δ13C values for related filter-feeding bivalves in Cardiidae (Figure 3). The stable carbon isotope ratios of the symbiont-containing mantle and symbiont-free tissues of the two shallow water photosymbiotic species, Fragum fragum and F. scruposum, were relatively similar with δ13C = −13.1 to −16.5‰. Both mantle and foot values were statistically different (P < 0.001) from that of the filter-feeding cardiid, Fulvia sp. (δ13C = −20‰). The deeper species, F. sueziense, was more depleted in 13C than the shallower Fragum species, with a mean δ13C = −19‰, closer to that of Fulvia sp. For the three symbiotic Fragum spp., the foot tissue was enriched over the mantle tissues in 13C, averaging 1.4 and 0.5 ‰ for the two shallow and one deep species, respectively. The δ13C differences between foot and mantle tissues were statistically significant (P < 0.01) for F. fragum and F. scruposum (F. sueziense could not be tested due to small sample size). Overall, the δ13C values of all three Fragum species were largely comparable with the photosymbiotic cardiid, the giant clam, Tridacna gigas, but more enriched in 13C than those of the filter-feeding cardiids, Fulvia sp. and Cerastoderma edule (Figure 3).
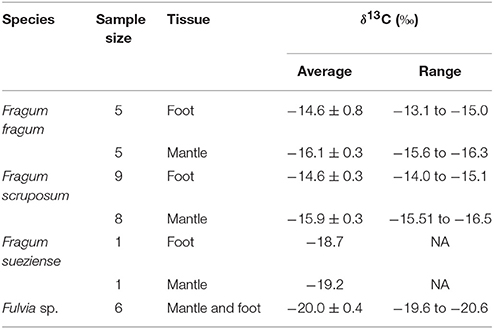
Table 4. Tissue δ13C values of Fragum fragum, F. scruposum, F. sueziense, and the filter-feeing Fulvia sp.
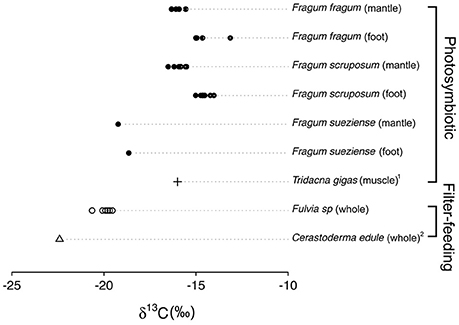
Figure 3. δ13C values of Fraginae species, the giant clam Tridacna gigas, and the filter-feeding cardiids Cerastoderma edule and Fulvia sp. Each point represents one individual sample. Exact values are shown in Table 4. 1Black and Bender (1976). 2Rossi et al. (2004).
Discussion
Our study on representative Fraginae species indicates that they share similarities with the giant clams in terms of symbiont diversity and general symbiont-containing structures. Stable isotope analyses indicate that these bivalves utilize photosymbiosis to obtain at least part of their food sources.
The symbiont-containing morphologies of the three Fragum species are consistent with that of a previously examined Fraginae species Corculum cardissa (Farmer et al., 2001). This is consistent with the preliminary conclusion that fragines may all share similar tubular systems. Farmer et al. (2001) reported that Corculum cardissa contains symbionts in both mantle and gill tissues. We found additional symbionts in foot tissues of all Fragum species tested. The symbionts are mostly concentrated in the heel portion of the foot, which is directly under the siphonal opening of the animal in its natural condition (i.e., exposed to more light); while the rest of the foot is symbiont-free. This is the first published observation of photosymbionts housed in the foot of a bivalve. This new information can be interpreted as evidence that symbiont locations are relatively flexible in Fraginae and the development of symbiont-containing tubules could potentially be influenced by levels of light intensity. The ephemeral nature of zooxanthellal tubules has been observed in Tridacna, with tubules only developing in juveniles in the presence of zooxanthellae (Fitt and Trench, 1981). Tridacnids also exhibit tertiary tubule atrophy following bleaching experiments, suggesting that these structures may be regenerative following bleaching incidents (Norton et al., 1992). Coupling with our observations on Fraginae tubular structures, it is obvious that a research gap exists concerning how this inducible tubular system functions and is maintained at the molecular and cellular levels.
Fraginae symbiont cells that were examined in hospite show surprising shape flexibility. The algae were in full contact with each other and within tubules, exhibiting cylinder shapes due to the tight packing. Symbiodinium in other invertebrate hosts are typically reported as having a spherical (coccoid) shape maintained by a multiple-layered cell wall (Trench et al., 1981; Wakefield et al., 2000). Although we did not examine the Fraginae symbiont cell wall structure in detail, it is likely that the pleomorphic shapes observed here may be a result of reduced cell wall thickness, as it is known that Symbiodinium cell wall development can be somewhat flexible and symbionts cultured outside of their hosts develop significantly thicker cell walls (Palincsar et al., 1988). In addition, artificially removing cell walls can cause Symbiodinium to have pleomorphic shapes (Levin et al., 2017). Therefore, symbiont cell wall construction within host may be suppressed or regulated to facilitate nutrient exchange and/or host communication. It is also possible that reports on symbionts in other hosts (Trench et al., 1981) are affected by tissue fixation artifacts, as even within-tubule symbionts in our samples showed spherical shapes after being fixed in 4% paraformaldehyde (Figures 1c,d).
Phylogenetic analyses recovered exclusively clade C Symbiodinium sequences from all four Fraginae species and the symbiont communities exhibited low levels of genetic variation among different hosts (Figure 2). Although the COI phylogeny showed some structure, support values for the subclades were relatively low. Based on genetic markers used in this study, the symbiont communities do not seem to be Fraginae-specific and do not show any signature of co-diversification with the hosts. Clade C Symbiodinium is the most dominant clade in the Indo-Pacific region and can be found in diverse groups of invertebrate hosts (LaJeunesse, 2005; Pochon et al., 2006). Given that the photosymbiotic Fraginae is a relatively young clade (late Miocene, Vermeij, 2013), existing symbionts in clade C likely established associations with fragines via host expansion. It is worth noting that even though we did not identify any Fraginae-specific Symbiodinium subclades within clade C, a more variable genetic marker (e.g., ITS) may reveal a different picture. Species identification for Symbiodinium requires dense population-level sampling; both morphological examination (e.g., Lee et al., 2015) and more genetic markers are needed to clarify host specificity of the diverse Symbiodinium lineages.
In coral symbioses (LaJeunesse et al., 2004), it is quite common to have multiple Symbiodinium lineages in one host species, as “stocking” multiple ecologically diverse symbiont lineages may facilitate the hosts' ability to cope and adapt to fluctuating environments (Jones et al., 2008; LaJeunesse et al., 2009). Therefore, although we only detected clade C Symbiodinium in our study, symbionts from other clades may coexist within the fragines, but are not the dominant strains. Clade A Symbiodinium have been frequently found in symbiont communities cultured from Fraginae and Tridacninae hosts, but not from freshly isolated symbionts. If this is not a result of the presence of non-intercellular algae (e.g., environmental free-living algae on host body surface), it suggests that a small proportion of Clade A symbionts exist in the host but the numbers are not high enough to be identified by standard PCR methods. To capture these potential rare stains in Fraginae, more sensitive methods that can detect multiple lineages per host (e.g., clade-specific primers, qPCR, cloning, high-throughput sequencing, etc.) will be needed in the future. In addition, our sequences were obtained through standard PCR amplifications from a community of symbionts within each host. Therefore, it is unavoidable that the amplicons are composed of mixed sequences from different symbiont individuals, and these mixed signals could generate artifacts (e.g., chimeric sequences) in the Sanger sequencing process. Therefore, it is important to note that these sequences may represent symbiont community amplicons instead of gene identities of individual symbionts.
Stable carbon isotope ratios suggest that Symbiodinium supplies photosynthates for the Fragum species (Table 4, Figure 3). The tissue carbon isotope ratios of the two shallow water species Fragum fragum and F. scruposum (δ13C = −13.1 to −16.5‰) are consistent with those of other zooxanthellae symbioses, including the giant clam Tridacna gigas, corals, and sea anemones with transfer of symbiont-fixed carbon to host documented in a number of these species (Black and Bender, 1976; Baker et al., 2015). Symbioses with zooxanthellae are typically enriched in 13C relative to filter-feeding invertebrates since Symbiodinium sp. fix CO2 using a form II RubisCO which discriminates against 13C less than form I RubisCOs of most free-living algae (Rowan et al., 1996). Further, the symbiont-free foot tissues of F. fragum and F. scruposum are 13C enriched on average 1.3 to 1.5‰ compared to their symbiont-containing mantle. Such enrichment is typical of the small metabolic fractionations that occur during producer—consumer interactions (DeNiro and Epstein, 1978). δ13C of the symbiont-containing mantle reflects a value combination of both the algae and the host (producer and consumer). δ13C of the foot tissue, on the other hand, only reflects value of the host (consumer). So the difference is partially a result of the isotope composition difference between the symbionts and the bivalve. It is known that animal consumers take on isotopic composition of their food with a small 13C enrichment about 1–2‰ (DeNiro and Epstein, 1978). This enrichment pattern has been confirmed in chemosymbiotic bivalves (Conway et al., 1989). Therefore, the observed 13C enrichment in the bivalve foot over mantle tissues further suggests that the bivalves are utilizing symbiont produced organic carbon.
Tissues of the deeper photosymbiotic Fragum sueziense exhibit an isotopic signature comparable to some photosymbiotic organisms (Baker et al., 2015), with a 0.5‰ 13C enrichment of foot relative to mantle tissue. However, its δ13C values are only slightly less negative than that of the co-occurring filter-feeding cockle Fulvia sp. A probable explanation is that due to the lower light intensity at depth, F. sueziense obtains a smaller proportion of organic carbon from photosymbiosis than F. fragum and F. scruposum, and that this host species likely supplements its nutrition obtained via filter feeding. A similar trend is seen in corals with δ13C values increasing with depth (Muscatine et al., 1989). However, our sample size for F. sueziense is small due to the rarity of this species, hindering our ability to conduct statistical analysis. To further explore this phenomenon, additional sampling of Fraginae species across a broader depth gradient (ter Poorten, 2009), coupled with measurements of photosynthetic activity and conventional feeding, are needed to clarify the host nutritional and the stability/plasticity of the fragine-zooxanthellae trophic interactions.
Our results demonstrated several similarities between the Tridacninae and photosymbiotic Fraginae bivalves. Like Tridacninae, the Fraginae species examined appear to rely on photosynthetically derived carbon sources from the symbionts as a food source. Both can be associated with clades C or A Symbiodinium and harbor them in similar tubule systems. The question remains as to whether these similarities evolved independently or were inherited from a common ancestor. The two groups obviously differ in photosymbiotic ecology and general morphology. The fossil record of tridacnine bivalves can be traced back to Oligocene while symbiotic fragines have only be found extending to late Miocene (Vermeij, 2013). If photosymbiosis was acquired separately by the two subfamilies, mechanisms that generate a convergent evolution of the symbiont harboring tubule systems need to be illustrated. If photosymbiosis is an ancestral trait, losses of symbiotic relationship within the non-symbiotic fragines and possibly other cockle lineages need to be explained (Kirkendale and Paulay, 2017). In addition, given the late appearance of photosymbiotic Fraginae morphology in the fossil record, even if the symbiotic associations are ancestral, the adaptive shell phenotypes must have evolved later and might be driven by habitat shifts and/or expansion to light-abundant environments. Further studies will use comprehensive phylogenetic and functional genomic approaches to illuminate the evolution and origin of symbiotic associations between bivalve hosts and their algal symbionts.
Ethics Statement
Protocols for studying preserved invertebrate animals were exempt from Institutional Animal Care and Use Committee.
Author Contributions
JL and CC designed the study. JL, MV, and LK conducted the fieldwork. JL and MV performed the experiments. JL, LK, and CC wrote the manuscript.
Funding
This work is supported by NSF OCE-PRF 1420967 to JL. LK would like to acknowledge funding from Woodside Energy Ltd to the Western Australian Museum for expedition support to survey Rowley Shoals in October 2014.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Drs. Alexander Kerr, Curt Fiedler, and Dan Lindstrom for supporting the fieldwork of this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00045/full#supplementary-material
Supplementary Table 1. Information of all symbiont sequences used in this study, including host species, genbank ID and voucher ID.
Supplementary Table 2. Current knowledge on Fraginae and Tridacninae-associated symbiont diversity, including host species, geographical distribution, symbiont type, and genetic markers used.
References
Baillie, B. K., Belda-Baillie, C. A., Silvestre, V., Sison, M., Gomez, A. V., Gomez, E. D., et al. (2000). Genetic variation in Symbiodinium isolates from giant clams based on random-amplified-polymorphic DNA (RAPD) patterns. Mar. Biol. 136, 829–836. doi: 10.1007/s002270000290
Baker, A. C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417
Baker, D. M., Freeman, C. J., Knowlton, N., Thacker, R. W., Kim, K., and Fogel, M. L. (2015). Productivity links morphology, symbiont specificity and bleaching in the evolution of Caribbean octocoral symbioses. ISME J. 9, 2620–2629. doi: 10.1038/ismej.2015.71
Banaszak, A. T., Ramos, M. G., and Goulet, T. L. (2013). The symbiosis between the gastropod Strombus gigas and the dinoflagellate Symbiodinium: an ontogenic journey from mutualism to parasitism. J. Exp. Mar. Biol. Ecol. 449, 358–365. doi: 10.1016/j.jembe.2013.10.027
Black, C. C., and Bender, M. M. (1976). δ13C values in marine organisms from the Great Barrier Reef. Funct. Plant Biol. 3, 25–32. doi: 10.1071/PP9760025
Carlos, A. A., Baillie, B. K., Kawachi, M., and Maruyama, T. (1999). Phylogenetic position of Symbiodinium (Dinophyceae) isolates from tridacnids (Bivalvia), cardiids (Bivalvia), a sponge (Porifera), a soft coral (Anthozoa), and a free-living strain. J. Phycol. 35, 1054–1062. doi: 10.1046/j.1529-8817.1999.3551054.x
Carlos, A. A., Baillie, B. K., and Maruyama, T. (2000). Diversity of dinoflagellate symbionts (zooxanthellae) in a host individual. Mar. Ecol. Prog. Ser. 195, 93–100. doi: 10.3354/meps195093
Carter, J. G., and Schneider, J. A. (1997). Condensing lenses and shell microstructure in Corculum (Mollusca: Bivalvia). J. Paleontol. 71, 56–61. doi: 10.1017/S0022336000038956
Cavanaugh, C. M., and Robinson, J. J. (1996). “CO2 fixation in chemoautotroph-invertebrate symbioses: expression of form I and form II RubisCO,” in Microbial Growth on C1 Compounds, eds M. E. Lidstrom and F. R. Tabita (Dordrecht: Kluwer Academic Publishers), 285–292.
Conway, N., Capuzzo, J. M., and Fry, B. (1989). The role of endosymbiotic bacteria in the nutrition of Solemya velum: evidence from a stable isotope analysis of endosymbionts and host. Limnol. Oceanogr. 34, 249–255. doi: 10.4319/lo.1989.34.1.0249
Cowen, R. (1988). The role of algal symbiosis in reefs through time. Palaios 3, 221–227. doi: 10.2307/3514532
DeBoer, T. S., Baker, A. C., Erdmann, M. V., Jones, P. R., and Barber, P. H. (2012). Patterns of Symbiodinium distribution in three giant clam species across the biodiverse Bird's head region of Indonesia. Mar. Ecol. Prog. Ser. 444, 117–132. doi: 10.3354/meps09413
DeNiro, M. J., and Epstein, S. (1978). Influence of diet on the distribution of carbon isotopes in animals. Geochim. Cosmochim. Acta 42, 495–506. doi: 10.1016/0016-7037(78)90199-0
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Dreier, A., Loh, W., Blumenberg, M., Thiel, V., Hause-Reitner, D., and Hoppert, M. (2014). The isotopic biosignatures of photo-vs. thiotrophic bivalves: are they preserved in fossil shells? Geobiology 12, 406–423. doi: 10.1111/gbi.12093
Farmer, M. A., Fitt, W. K., and Trench, R. K. (2001). Morphology of the symbiosis between Corculum cardissa (Mollusca: Bivalvia) and Symbiodinium corculorum (Dinophyceae). Biol. Bull. 200, 336–343. doi: 10.2307/1543514
Finney, J. C., Pettay, D. T., Sampayo, E. M., Warner, M. E., Oxenford, H. A., and LaJeunesse, T. C. (2010). The relative significance of host–habitat, depth, and geography on the ecology, endemism, and speciation of coral endosymbionts in the genus Symbiodinium. Microb. Ecol. 60, 250–263. doi: 10.1007/s00248-010-9681-y
Fitt, W. K., and Trench, R. K. (1981). Spawning, development, and acquisition of zooxanthellae by Tridacna squamosa (Mollusca, Bivalvia). Biol. Bull. 161, 213–235. doi: 10.2307/1540800
Gast, R. J., and Caron, D. A. (2001). Photosymbiotic associations in planktonic foraminifera and radiolaria. Hydrobiologia 461, 1–7. doi: 10.1023/A:1012710909023
Hawkins, A. J. S., and Klumpp, D. W. (1995). Nutrition of the giant clam Tridacna gigas (L.). II. Relative contributions of filter-feeding and the ammonium-nitrogen acquired and recycled by symbiotic alga towards total nitrogen requirements for tissue growth and metabolism. J. Exp. Mar. Biol. Ecol. 190, 263–290. doi: 10.1016/0022-0981(95)00044-R
Hayes, J. M. (2001). “Fractionation of the isotopes of carbon and hydrogen in biosynthetic processes,” in Stable Isotope Geochemistry, Reviews in Mineralogy and Geochemistry, Vol. 43, eds J. W. Valley and D. R. Cole (Washington, DC: Mineralogical Society of America), 225–278.
Herrera, N. D., ter Poorten, J. J., Bieler, R., Mikkelsen, P. M., Strong, E. E., Jablonski, D., et al. (2015). Molecular phylogenetics and historical biogeography amid shifting continents in the cockles and giant clams (Bivalvia: Cardiidae). Mol. Phylogenet. Evol. 93, 94–106. doi: 10.1016/j.ympev.2015.07.013
Hill, M., Allenby, A., Ramsby, B., Schönberg, C., and Hill, A. (2011). Symbiodinium diversity among host clionaid sponges from Caribbean and Pacific reefs: evidence of heteroplasmy and putative host-specific symbiont lineages. Mol. Phylogenet. Evol. 59, 81–88. doi: 10.1016/j.ympev.2011.01.006
Jones, A. M., Berkelmans, R., van Oppen, M. J., Mieog, J. C., and Sinclair, W. (2008). A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc. R. Soc. Lond. B 275, 1359–1365. doi: 10.1098/rspb.2008.0069
Kawaguti, S. (1941). Heart shell Corculum cardissa (L.) and its zooxanthella. Kagaku Nanyo 3, 45–46.
Kirkendale, L. (2005). Evolution of Photosymbiosis in Marine Bivalves (Cardiidae: Fraginae), Ph.D. thesis, University of Florida.
Kirkendale, L. (2009). Their day in the Sun: molecular phylogenetics and origin of photosymbiosis in the other group of photosymbiotic marine bivalves (Cardiidae: Fraginae). Biol. J. Linn. Soc. 97, 448–465. doi: 10.1111/j.1095-8312.2009.01215.x
Kirkendale, L., and Paulay, G. (2017). Part N, Revised, Volume 1, Chapter 9: Photosymbiosis in Bivalvia. Treatise Online 89, 1–39. doi: 10.17161/to.v0i0.6554
Levin, R. A., Suggett, D. J., Nitschke, M. R., Oppen, M. J., and Steinberg, P. D. (2017). Expanding the Symbiodinium (Dinophyceae, Suessiales) toolkit through protoplast technology. J. Eukaryot. Microbiol. 64, 588–597. doi: 10.1111/jeu.12393
LaJeunesse, T. C. (2001). Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: in search of a “species” level marker. J. Phycol. 37, 866–880. doi: 10.1046/j.1529-8817.2001.01031.x
LaJeunesse, T. C., Bhagooli, R., Hidaka, M., DeVantier, L., Done, T., Schmidt, G. W., et al. (2004). Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Mar. Ecol. Prog. Ser. 284, 147–161. doi: 10.3354/meps284147
LaJeunesse, T. C. (2005). “Species” radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition. Mol. Biol. Evol. 22, 570–581. doi: 10.1093/molbev/msi042
LaJeunesse, T. C., Pettay, D. T., Sampayo, E. M., Phongsuwan, N., Brown, B., Obura, D. O., et al. (2010). Long-standing environmental conditions, geographic isolation and host–symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. J. Biogeogr. 37, 785–800. doi: 10.1111/j.1365-2699.2010.02273.x
LaJeunesse, T. C., Smith, R. T., Finney, J., and Oxenford, H. (2009). Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching’ event. Proc. R. Soc. Lond. B 276, 4139–4148. doi: 10.1098/rspb.2009.1405
Lanfear, R., Calcott, B., Ho, S. Y., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Lee, S. Y., Jeong, H. J., Kang, N. S., Jang, T. Y., Jang, S. H., and Lajeunesse, T. C. (2015). Symbiodinium tridacnidorum sp. nov., a dinoflagellate common to Indo-Pacific giant clams, and a revised morphological description of Symbiodinium microadriaticum freudenthal, emended trench & blank. Eur. J. Phycol. 50, 155–172. doi: 10.1080/09670262.2015.1018336
Lien, Y. T., Nakano, Y., Plathong, S., Fukami, H., Wang, J. T., and Chen, C. A. (2007). Occurrence of the putatively heat-tolerant Symbiodinium phylotype D in high-latitudinal outlying coral communities. Coral Reefs 26, 35–44. doi: 10.1007/s00338-006-0185-7
Muscatine, L., Porter, J. W., and Kaplan, I. R. (1989). Resource partitioning by reef corals as determined from stable isotope composition. Mar. Biol. 100, 185–193. doi: 10.1007/BF00391957
Norton, J. H., Shepherd, M. A., Long, H. M., and Fitt, W. K. (1992). The zooxanthellal tubular system in the giant clam. Biol. Bull. 183, 503–506. doi: 10.2307/1542028
Ohno, T., Katoh, T., and Yamasu, T. (1995). The origin of algal-bivalve photo-symbiosis. Palaeontology 38, 1–22.
Palincsar, J. S., Jones, W. R., and Palincsar, E. E. (1988). Effects of isolation of the endosymbiont Symbiodinium microadriaticum (Dinophyceae) from its host Aiptasia pallida (Anthozoa) on cell wall ultrastructure and mitotic rate. Trans. Am. Microsc. Soc. 107, 53–66. doi: 10.2307/3226408
Persselin, S. L. (1998). The Evolution of Shell Windows Within the Fraginae (Bivalvia: Cardiidae) and the Origin of Algal Symbiosis in Cardiids. M.S. thesis, University of Guam.
Pochon, X., and Gates, R. D. (2010). A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai'i. Mol. Phylogenet. Evol. 56, 492–497. doi: 10.1016/j.ympev.2010.03.040
Pochon, X., Montoya-Burgos, J. I., Stadelmann, B., and Pawlowski, J. (2006). Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Mol. Phylogenet. Evol. 38, 20–30. doi: 10.1016/j.ympev.2005.04.028
Pochon, X., Putnam, H. M., Burki, F., and Gates, R. D. (2012). Identifying and characterizing alternative molecular markers for the symbiotic and free-living dinoflagellate genus Symbiodinium. PLoS ONE 7:e29816. doi: 10.1371/journal.pone.0029816
Pochon, X., Putnam, H. M., and Gates, R. D. (2014). Multi-gene analysis of Symbiodinium dinoflagellates: a perspective on rarity, symbiosis, and evolution. PeerJ 2:e394. doi: 10.7717/peerj.394
Robinson, J. J., and Cavanaugh, C. M. (1995). Expression of form I and form II RubisCO in chemoautotrophic symbioses: implications for the interpretation of stable carbon isotope values. Limnol. Oceanogr. 40, 1496–1502. doi: 10.4319/lo.1995.40.8.1496
Ronquist, F., Teslenko, M., Van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rossi, F., Herman, P. M. J., and Middelburg, J. J. (2004). Interspecific and intraspecific variation of δ13C and δ15N in deposit-and suspension-feeding bivalves (Macoma balthica and Cerastoderma edule): evidence of ontogenetic changes in feeding mode of Macoma balthica. Limnol. Oceanogr. 49, 408–414. doi: 10.4319/lo.2004.49.2.0408
Rowan, R. (1998). Diversity and ecology of zooxanthellae on coral reefs. J. Phycol. 34, 407–417. doi: 10.1046/j.1529-8817.1998.340407.x
Rowan, R., and Powers, D. A. (1991). Molecular genetic identification of symbiotic dinoflagellates (zooxanthellae). Mar. Ecol. Prog. Ser. 71, 65–73. doi: 10.3354/meps071065
Rowan, R., Whitney, S. M., Fowler, A., and Yellowlees, D. (1996). RubisCO in marine symbiotic dinoflagellates: form II enzymes in eukaryotic oxygenic phototrophs encoded by a nuclear multigene family. Plant Cell 8, 539–553. doi: 10.1105/tpc.8.3.539
Santos, S. R., Taylor, D. J., Kinzie Iii, R. A., Hidaka, M., Sakai, K., and Coffroth, M. A. (2002). Molecular phylogeny of symbiotic dinoflagellates inferred from partial chloroplast large subunit (23S)-rDNA sequences. Mol. Phylogenet. Evol. 23, 97–111. doi: 10.1016/S1055-7903(02)00010-6
Schneider, J. A., and Carter, J. G. (2001). Evolution and phylogenetic significance of cardioidean shell microstructure (Mollusca, Bivalvia). J. Paleontol. 75, 607–643. doi: 10.1017/S002233600003969X
Stanley, G. D. Jr. (2006). Photosymbiosis and the evolution of modern coral reefs. Science 312, 857–858. doi: 10.1126/science.1123701
Stanley, G. D. Jr., and Lipps, J. H. (2011). Photosymbiosis: the driving force for reef success and failure. Paleontol. Soc. Pap. 17, 33–60. doi: 10.1017/S1089332600002436
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Stern, R. F., Horak, A., Andrew, R. L., Coffroth, M. A., Andersen, R. A., Küpper, F. C., et al. (2010). Environmental barcoding reveals massive dinoflagellate diversity in marine environments. PLoS ONE 5:e13991. doi: 10.1371/journal.pone.0013991
ter Poorten, J. J. (2009). The cardiidae of the panglao marine biodiversity project 2004 and the panglao 2005 deep-sea cruise with descriptions of four new species (Bivalvia). Vita Malacol. 8, 9–96.
Trench, R. K., Wethey, D. S., and Porter, J. W. (1981). Observations on the symbiosis with zooxanthellae among the Tridacnidae (Mollusca, Bivalvia). Biol. Bull. 161, 180–198. doi: 10.2307/1541117
Trench, R. K. (1993). Microalgal-invertebrate symbioses-a review. Endocytobiosis Cell Res. 9, 135–175.
Vermeij, G. J. (2013). The evolution of molluscan photosymbioses: a critical appraisal. Biol. J. Linn. Soc. 109, 497–511. doi: 10.1111/bij.12095
Wakefield, T. S., Farmer, M. A., and Kempf, S. C. (2000). Revised description of the fine structure of in situ “zooxanthellae” genus Symbiodinium. Biol. Bull. 199, 76–84. doi: 10.2307/1542709
Watson, M. E., and Signor, P. W. (1986). How a clam builds windows: shell microstructure in Corculum (Bivalvia: Cardiidae). Veliger 28, 348–355.
Wilcox, T. P. (1998). Large-subunit ribosomal RNA systematics of symbiotic dinoflagellates: morphology does not recapitulate phylogeny. Mol. Phylogenet. Evol. 10, 436–448. doi: 10.1006/mpev.1998.0546
Yonge, C. M. (1936). Mode of Life, Feeding, Digestion and Symbiosis With Zooxanthellae in the Tridacnidae. London: British Museum.
Yorifuji, M., Takeshima, H., Mabuchi, K., Watanabe, T., and Nishida, M. (2015). Comparison of Symbiodinium dinoflagellate flora in sea slug populations of the Pteraeolidia ianthina complex. Mar. Ecol. Prog. Ser. 521, 91–104. doi: 10.3354/meps11155
Keywords: symbiosis, symbiodinium, dinoflagellate, cardiidae, fragum, corculum, tridacna, coral reef
Citation: Li J, Volsteadt M, Kirkendale L and Cavanaugh CM (2018) Characterizing Photosymbiosis Between Fraginae Bivalves and Symbiodinium Using Phylogenetics and Stable Isotopes. Front. Ecol. Evol. 6:45. doi: 10.3389/fevo.2018.00045
Received: 15 January 2018; Accepted: 04 April 2018;
Published: 25 April 2018.
Edited by:
David Michael Baker, University of Hong Kong, Hong KongReviewed by:
Matthew R. Nitschke, University of Aveiro, PortugalShinichiro Maruyama, Tohoku University, Japan
Copyright © 2018 Li, Volsteadt, Kirkendale and Cavanaugh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingchun Li, jingchun.li@colorado.edu
 Jingchun Li
Jingchun Li Megan Volsteadt1,4
Megan Volsteadt1,4  Lisa Kirkendale
Lisa Kirkendale