Phylogenetic Structure of Plant Communities: Are Polyploids Distantly Related to Co-occurring Diploids?
- 1Department of Biology, University of Central Florida, Orlando, FL, United States
- 2Department of Ecology and Evolutionary Biology, University of Colorado Boulder, Boulder, CO, United States
Polyploidy is widely acknowledged to have played an important role in the evolution and diversification of vascular plants. However, the influence of genome duplication on population-level dynamics and its cascading effects at the community level remain unclear. In part, this is due to persistent uncertainties over the extent of polyploid phenotypic variation, and the interactions between polyploids and co-occurring species, and highlights the need to integrate polyploid research at the population and community level. Here, we investigate how community-level patterns of phylogenetic relatedness might influence escape from minority cytotype exclusion, a classic population genetics hypothesis about polyploid establishment, and population-level species interactions. Focusing on two plant families in which polyploidy has evolved multiple times, Brassicaceae and Rosaceae, we build upon the hypothesis that the greater allelic and phenotypic diversity of polyploids allow them to successfully inhabit a different geographic range compared to their diploid progenitor and close relatives. Using a phylogenetic framework, we specifically test (1) whether polyploid species are more distantly related to diploids within the same community than co-occurring diploids are to one another, and (2) if polyploid species tend to exhibit greater ecological success than diploids, using species abundance in communities as an indicator of successful establishment. Overall, our results suggest that the effects of genome duplication on community structure are not clear-cut. We find that polyploid species tend to be more distantly related to co-occurring diploids than diploids are to each other. However, we do not find a consistent pattern of polyploid species being more abundant than diploid species, suggesting polyploids are not uniformly more ecologically successful than diploids. While polyploidy appears to have some important influences on species co-occurrence in Brassicaceae and Rosaceae communities, our study highlights the paucity of available geographically explicit data on intraspecific ploidal variation. The increased use of high-throughput methods to identify ploidal variation, such as flow cytometry and whole genome sequencing, will greatly aid our understanding of how such a widespread, radical genomic mutation influences the evolution of species and those around them.
Introduction
Polyploidy, or whole genome duplication, has been an important force shaping the evolutionary history of vascular plants (Adams and Wendel, 2005; Rieseberg and Willis, 2007; Soltis et al., 2009; Ramsey and Ramsey, 2014). Not only is polyploidy considered an important mechanism of speciation (Coyne and Orr, 2004; Soltis et al., 2014; Zhan et al., 2016), it is also often associated with major phenotypic shifts such as in size, flower color, water use, reproductive system, pollinator specialization, herbivore resistance, and phenology (Levin, 1983; Masterson, 1994; Segraves and Thompson, 1999; Husband et al., 2007; Maherali et al., 2009; Balao et al., 2011; Ramsey and Ramsey, 2014). Genome duplication has also been associated with novel alterations to genomic architecture and regulation that may affect adaptation (Comai, 2005; Madlung, 2013). However, despite the prevalence of polyploid events, the biodiversity implications of genome duplication, and the phenotypic differences often observed between diploids and polyploids, much remains unknown about how far reaching the impact of whole genome duplication is on interactions with other species and at the community level (Laport and Ng, 2017; Segraves, 2017).
Renewed interest in studying polyploidy over the last several decades has bent recent opinion toward acknowledging the significance of genome duplication on patterns of biodiversity (Coyne and Orr, 2004; Soltis et al., 2007; Ramsey and Ramsey, 2014; Laport and Ng, 2017; Segraves, 2017). Yet, the influence of genome duplication on population- and community-level dynamics remains unclear, in part because the evolutionary origin of polyploids may strongly influence the extent of polyploid phenotypic variation. Polyploids formed via the hybridization of two closely related species with partially diverged genomes (allopolyploidy) often exhibit phenotypes that are intermediate to, or outside the range of (i.e., transgressive), the parental species. In contrast, polyploids formed via the union of unreduced gametes within a population (autopolyploidy) often exhibit more subtle phenotypic differences when compared to their diploid progenitors. Historically, the more pronounced phenotypic variation among allopolyploids was considered as being important for interspecific interactions and patterns of biodiversity (Soltis et al., 2007; Ramsey and Ramsey, 2014). Research over the last few decades has shown, however, that the phenotypic, and underlying genetic, variation associated with both allo- and autopolyploids has the potential to influence ecological affinities, and play an important role in facilitating the establishment of new cytotypes, their expansion into a broader range of environmental conditions, and consequently their interactions with other species.
From the extensive body of empirical and theoretical work on the ecology and evolution of polyploids, whole genome duplication can be expected to have cascading effects on interspecific interactions and community-level dynamics (Ramsey and Ramsey, 2014; Čertner et al., 2017; Laport and Ng, 2017; Segraves, 2017). Although the direction and strength of the effect remains unclear, a number of predictions can be made about how species interactions and co-occurrence may be shaped by whole genome duplication based on previous species- and population-level work. For example, when considering first generation polyploids (i.e., tetraploids), it is thought that these neopolyploids must immediately compete with their co-occurring diploid progenitor upon formation while suffering a distinct frequency-dependent reproductive disadvantage. Because the relatively rare tetraploids are most likely to mate with more abundant diploids, this disadvantage, known as minority cytotype exclusion, arises from the lower fitness realized through the production of inviable or infertile triploid hybrid offspring (Hagberg and Ellerström, 1959; Levin, 1975). With few or no potential mates with which to reproduce, the neopolyploid is effectively “bred to death.” However, even slight differences between polyploids and diploids, such as phenological shifts in the timing of reproduction, reproductive strategy (e.g., sexual vs. asexual), and ecological differences, may satisfy theoretical requirements for successful escape from minority cytotype exclusion (Husband, 2000). By easing direct ecological competition and promoting assortative mating, phenotypic differences may allow neopolyploids to persist within the range of their diploid progenitors. Present day communities would therefore reflect signatures of these historic events, whereby polyploids will often co-occur with their close diploid relatives.
Alternatively, polyploids may overcome minority cytotype exclusion by dispersing to new, unexploited habitats and maintaining the exclusion of their progenitors. Theoretical work predicts that the likelihood of neopolyploids becoming established at their site of origin is very low (Fowler and Levin, 2016), while dispersal and exploitation of novel habitat due to phenotypic differences either accompanying or arising rapidly after genome duplication greatly increases the probability of persistence (Lewis, 1962; Kay, 1969; Leitch and Leitch, 2008; Levin and Soltis, 2017). Indeed, the phenotypic differences associated with polyploidy may be great enough to facilitate the establishment of new cytotypes and their expansion into a broader range of ecological and environmental conditions. For example, polyploids have been documented to differ in ecological niche affinities and adaptive traits (Ramsey, 2011; McIntyre, 2012; Laport et al., 2013; Glennon et al., 2014; Marchant et al., 2016), experience morphological and physiological differences affecting phenological and physiological rates (Beaulieu et al., 2008; Manzaneda et al., 2012; Laport et al., 2016; Rey et al., 2017), have unique interactions with herbivores and pollinators (Thompson et al., 1997; Kennedy et al., 2006; Arvanitis et al., 2007; Halverson et al., 2008; Thompson and Merg, 2008; Roccaforte et al., 2015), and exhibit unique water relations (Maherali et al., 2009) and mycorrhizal associations (Těšitelová et al., 2013). Shifts from sexual to asexual reproduction, or a breakdown of self-incompatibility systems (Comai, 2005; Otto, 2007), could further promote the establishment of polyploids in geographic areas isolated from their diploid progenitors by providing a means of reproduction and population increase. If strong ecological differentiation between cytotypes and establishment in geographically isolated areas is the predominant mode of neopolyploid success and persistence, polyploids should more often occur in different communities than their close diploid relatives.
In addition to phenotypic differences between polyploids and diploids, variation at the molecular level also likely bears strongly on community assembly. In particular, genetic changes associated with whole genome duplication could increase the ecological success of polyploids in novel communities. Doubled nuclear DNA content on its own can have cellular phenotypic consequences that alter intracellular stoichiometric relationships and physiological rates, causing shifts to growth rate, gas exchange, and flowering time (Comai, 2005; Beaulieu et al., 2008; Madlung, 2013; Bilinski et al., 2018), which may allow polyploids to outcompete co-occurring diploids. The novel genetic architecture and regulatory environment of duplicated genomes may also lead to greater adaptability, and the larger genome size may be a larger target for functional mutations that could influence adaptation (Comai, 2005; Madlung, 2013; Soltis et al., 2015; Song and Chen, 2015; Mei et al., 2018). For example, the increased genomic content of polyploids presents potential opportunities for rapid paralog subfunctionalization or neofunctionalization that could lead to greater competitive ability or ecological success, and even invasiveness, relative to diploid progenitors (Thompson and Lumaret, 1992; Schlaepfer et al., 2010; te Beest et al., 2011; Green et al., 2013; Pyšek et al., 2013; Nagy et al., 2017). Indeed, the increased genomic/allelic diversity of larger genomes, decreased inbreeding depression, multisomic inheritance, intergenomic recombination, and accelerated epigenetic processes of polyploids have been identified as major factors that may predispose polyploid populations to rapidly exploit novel ecological niches (Comai, 2005; Soltis et al., 2009; Parisod et al., 2010; Green et al., 2013; Madlung, 2013). Thus, while the genetic changes associated with whole genome duplication and their influence over ecologically relevant phenotypic shifts may be used as a basis to make predictions about the ecological success of polyploids within communities, it remains relatively unexplored whether polyploids are indeed better competitors in a community context.
One way to investigate the influence of genome duplication on community structure is by analyzing diploid and polyploid co-occurrence within multiple communities using a comparative phylogenetic framework. Although there has been an increase in studies integrating phylogenetic data with questions about community ecology over the last decade (Webb et al., 2002; Cavender-Bares et al., 2006; Emerson and Gillespie, 2008; Vamosi et al., 2009), no studies have explicitly included ploidal information to assess the influence of genome duplication (and associated phenotypes) on community structure. Here, we use a novel approach to examine how polyploids influence phylogenetic community structure by combining ploidal information with phylogenetic analyses of plant communities across the United States. Specifically, we focus on two large plant families that are well represented across North American biomes and in which polyploidy has evolved multiple times, Brassicaceae and Rosaceae, to test (1) whether polyploid species are more distantly related to diploids within the same community than co-occurring diploids are to one another. We expect this phylogenetic pattern if polyploids escaped minority cytotype exclusion by inhabiting a different geographic range compared to their diploid progenitor and close relatives. We also test (2) whether polyploid species tend to exhibit greater ecological success than diploid species, using the relative abundance of polyploids vs. diploids as an indicator of successful establishment within communities. We further compare the abundance of native and non-native species to examine whether species experiencing recent ecological range expansions (i.e., non-native species) also tend to be polyploid.
Materials and Methods
Community Data Collection
We obtained species composition and abundance data for Brassicaceae and Rosaceae communities across the United States from the National Ecological Observatory Network (NEON; https://www.neonscience.org; Keller et al., 2008). NEON has established sites across the United States and conducted plant surveys of replicated 400 m2 plots across each site. We specifically focused on Brassicaceae and Rosaceae communities because they are polyploid-rich, broadly represent contrasting life histories, and were present in a large number of NEON communities. We focused on 16 communities (Figure 1), each of which had three or more representatives from the respective family for which we could obtain ploidal data (6 Brassicaceae communities, 11 Rosaceae communities; Figure 2). For each species, we determined its ploidal level based on scientific literature and online databases (Kew C-value database, http://data.kew.org/cvalues/; Chromosome Count Database, https://ccdb.tau.ac.il; Table S1), as well as its native status following the designation assigned in the United States Department of Agriculture (USDA) PLANTS database (https://plants.usda.gov) (Figure 2). While mode of polyploid origin is likely important for interspecific interactions and ecological success, we were unable to consider differences in origin for this study as we could not consistently determine whether a species was an allo- or autopolyploid. As geographic variation in ploidy can be common (Baack, 2005; Kolár et al., 2009; Ståhlberg, 2009; Trávníček et al., 2011; Castro et al., 2012; Laport et al., 2012; Ramsey and Ramsey, 2014; Zozomová-Lihová et al., 2015; Wefferling et al., 2017), we aimed to determine the community-specific ploidal level of each species. When species were reported to comprise multiple ploidal levels for the region around a NEON site (Figure 2), we repeated analyses with each ploidy. When assigning native status to each species, we considered species to be either native or non-native to the lower 48 states.
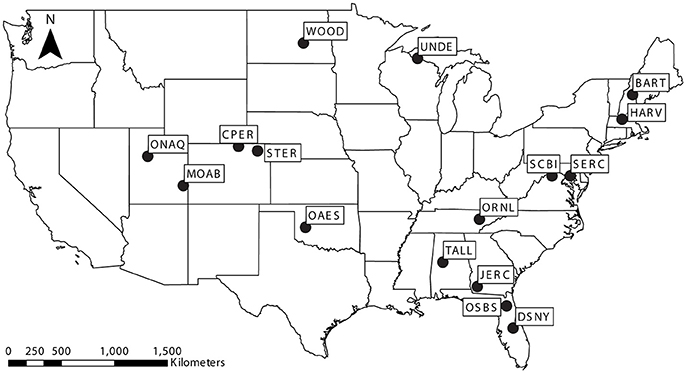
Figure 1. Map of the United States showing study communities collected from established National Ecological Observatory Network sites: Bartlett Experimental Forest (BART), Central Plains Experimental Range (CPER), Disney Wilderness Preserve (DSNY), Harvard Forest (HARV), Jones Ecological Research Center (JERC), Moab (MOAB), Klemme Range Research Station (OAES), Onaqui-Ault (ONAQ), Oak Ridge National Laboratory (ORNL), Ordway-Swisher Biological Station (OSBS), Smithsonian Environmental Research Center (SERC), Smithsonian Conservation Biology Institute (SCBI), North Sterling (STER), Talladega National Forest (TALL), Woodworth (WOOD), and University of Notre Dame Environmental Research Center (UNDE).
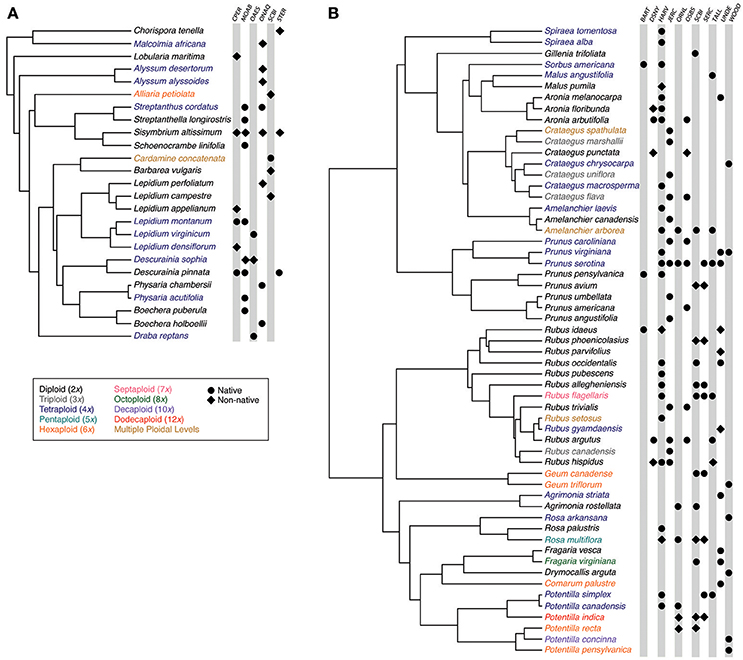
Figure 2. Phylogenetic trees showing (A) Brassicaceae and (B) Rosaceae community members and their occurrence in each study community. Species names are colored to indicate ploidal level. Symbols indicate the species' presence within a study community, with circles and diamonds indicating whether the species is native or non-native, respectively. Site abbreviations follow Figure 1.
Phylogenetic Reconstruction
As published phylogenies of Brassicaceae and Rosaceae did not include all members of our study communities (Huang et al., 2016; Zhang et al., 2017), we reconstructed phylogenies for each family using sequence data from GenBank and newly generated sequence data for species that did not have publicly available sequence data for our target genetic loci. We focused on one nuclear locus, ITS (internal transcribed spacer), and two chloroplast loci, rbcL (ribulose bisphosphate carboxylase large chain) and matK (maturase K). To generate our own sequences, leaf tissue was obtained from the Rocky Mountain Herbarium (RM) and the Missouri Botanical Gardens Herbarium (MO). DNA was extracted using the Qiagen Plant Mini Kit or CTAB DNA extraction method (Doyle and Doyle, 1987). We amplified the gene regions using previously published primers (Table S2) and following PCR protocols available in the Supplementary Materials (Supplementary Material 1). As we had difficulty amplifying ITS for Rosaceae due to polymorphisms in binding sites, we designed a new primer using Primer 3 (Koressaar and Remm, 2007; Untergasser et al., 2012) based on previously sequenced Rosaceae species: ITS_SGR (5′-AGG TTT GAC AAC CAC CGA TT-3′). We sent PCR products to Genewiz (Cambridge, Massachusetts) for purification and sequencing, and checked sequence quality in Geneious v6.0.5 (Biomatters Ltd., Auckland, NZ).
To ensure that the evolutionary relationships among members of the community were consistent with known relationships, we reconstructed phylogenies that included all species in this study, as well as any other available sequences from GenBank for each family. The inclusion of additional species not occurring within the communities of focus in phylogenetic reconstruction has been shown to reduce error in node age estimates, and consequently in calculations of community phylogenetic diversity metrics (Park et al., 2018). High quality sequence data for the targeted genetic loci were downloaded from GenBank using the PHLAWD pipeline (Smith et al., 2009). We combined GenBank sequences with newly generated sequences, aligned them in Mafft v7 (Katoh et al., 2002) and concatenated the gene regions in Mesquite v3.10 (Maddison and Maddison, 2017). The final data set included 1,912 species for Brassicaceae including five outgroup members (Cleome lutea Hook., Cleome viscosa L., Cleome rutidosperma DC., Moringa oleifera Lam., Polanisia dodecandra (L.) DC.). For Rosaceae, the final data set included 1,450 species including four outgroup members (Rhamnus cathartica L., Ceanothus verrucosus Nutt., Pisum sativum L., Astragalus membranaceus (Fisch.) Bunge).
We used Bayesian inference to reconstruct a time-calibrated phylogeny for each family using BEAST2 v2.4.5 (Bouckaert et al., 2014) on the CIPRES Science Gateway (www.phylo.org). For the phylogenetic reconstruction of Brassicaceae, the stem and crown nodes were constrained with a lognormal offset of 59.5 and 42.0 million years ago (Ma) (mean 0.01, standard deviation 1.0), respectively, following Huang et al. (2016). For the phylogenetic reconstruction of Rosaceae, the stem and crown nodes were constrained with a lognormal offset of 106.50 and 95.09 Ma (mean 0.01, standard deviation 1.0), respectively, following Zhang et al. (2017). We conducted two runs of 120 million generations and sampled trees every 12,000 generations. We used Tracer v1.6 (Rambaut et al., 2014) to verify that both runs reached stationarity and converged on the posterior distribution of trees. As identified in Tracer, we discarded 10% of the trees from each run as burn-in, then combined and summarized trees as a maximum clade credibility (MCC) tree using LogCombiner and TreeAnnotator (included as part of the BEAST2 package). We pruned all species that were not included in each of our study communities from the trees prior to site-specific analyses.
Diploid and Polyploid Phylogenetic Relationships
We used two approaches to determine whether polyploid species are more distantly related to diploids within the same community than co-occurring diploids are to one another. First, we used a broad-scale approach to investigate patterns of phylogenetic relatedness across all sites by calculating the phylogenetic distance between diploids and their closest diploid relative within the same community (nearest taxon distance; NTD2x−2x), and comparing these distances to the phylogenetic distance between polyploids and their most closely related, co-occurring diploid species (NTDpolyploid−2x). We pooled these values across sites and compared NTD2x−2x to NTDpolyploid−2x by conducting a Mann-Whitney U test using the wilcox.test function in R. We also evaluated our hypothesis of closer relationships between co-occurring diploids than among co-occurring diploids and polyploids by comparing the proportion of NTD2x−2x and NTDpolyploid−2x comparisons that fell below a threshold of the mean nearest taxon distance (MNTD) of the family-level phylogeny. MNTD was calculated using the cophenetic.phylo function in the ape R package (Paradis et al., 2004).
Second, we examined patterns of phylogenetic relatedness within each site by testing whether the MNTD between polyploids and diploids (MNTDpolyploid−2x) was significantly greater than MNTD2x−2x than expected by chance. We employed a simulation approach by comparing the observed metric MNTDpolyploid−2x / MNTD2x−2x within each community to a null distribution generated by replacing polyploid community members with randomly drawn species from a pool of polyploids from all sites, and recalculating the MNTDpolyploid−2x / MNTD2x−2x metric for the new community. Our null distribution comprised 1,000 random communities per site. We considered polyploids to be more distantly related to diploids than expected by chance if the MNTDpolyploid−2x / MNTD2x−2x metric was greater than 1 and was greater than 95% of the null distribution (P < 0.05). Any communities that did not have both diploid and polyploid species (Rosaceae: DSNY; Brassicaceae: STER, OAES) or only had one diploid or polyploid representative (Rosaceae: WOOD) were excluded from these analyses. All MNTD calculations were performed using the ses.mntd function in the picante R package (Kembel et al., 2010).
Tests of Polyploid Ecological Success
We identified whether polyploid species showed patterns consistent with having greater ecological success than diploids by using species abundance as an indicator of successful establishment within a community (Levin, 1975; Callaway and Aschehoug, 2000; Cleland et al., 2004). Specifically, we tested whether polyploids occurred at greater total relative abundance than diploids within each community by conducting a Mann-Whitney U-test with the wilcox.test function in R. We further assessed whether differences in abundance could be attributed to non-native species, reflecting ecological success of recent range expansions, by testing whether the total relative abundance of diploid and polyploid species significantly differed between natives and non-natives. We tested significance using a Kruskal-Wallis test and when appropriate, followed the analysis with Dunn's post-hoc test. This was performed using the kruskal.test and the dunnTest (FSA package) functions, respectively, in R.
Results
Community Data Collection
Our six Brassicaceae communities comprised 3–8 Brassicaceae species while our eleven Rosaceae communities comprised 3–24 Rosaceae species (Figure 2). Most Brassicaceae in our communities were 2x, 4x, or 6x, with the exception of one species where the ploidy ranged from 20x to 30x [Cardamine concatenata (Michx.) O. Schwarz.; Kreiner et al., 2017; Table S1]. In Brassicaceae communities, 33–100% of the species were polyploid, and 22–75% of the species were non-native (Figure 2). In Rosaceae communities, species ranged in ploidal level from 2x to 12x, with 33–86% of the species being polyploid. These communities also ranged from not having any non-native species to 44% of the species being non-native.
Phylogenetic Reconstruction
We generated 63 new sequences for species missing sequence data for our target loci (GenBank accessions KY427264-KY427326; Table S3). The final Brassicaceae alignment comprised 1,912 species and was 8,242 basepairs (bp) in length, while the final Rosaceae alignment comprised 1,450 species and was 12,007 bp in length (TreeBASE accession: S22405). All study species within the communities were represented in our time-calibrated phylogenetic trees, and both phylogenies for Brassicaceae and Rosaceae community members were congruent in topology to previously published phylogenies (Huang et al., 2016; Zhang et al., 2017; Figure 2).
Diploid and Polyploid Phylogenetic Relationships
Our broad-scale analysis examining phylogenetic patterns of relatedness between co-occurring polyploids and diploids vs. co-occurring diploids found that across all sites, NTDpolyploid−2x was significantly greater than NTD2x−2x for both Brassicaceae (P < 0.05) and Rosaceae (P < < 0.01; Figure 3). Further supporting this result for both families was that a larger proportion of NTD2x−2x comparisons fell below the MNTD threshold compared to NTDpolyploid−2x. For Brassicaceae communities, 76.9% of diploid-diploid comparisons and 34.1% of polyploid-diploid comparisons fell below the Brassicaceae MNTD, while for Rosaceae, 84.1% of diploid-diploid comparisons and 53.8% of polyploid-diploid comparisons fell below the Rosaceae MNTD (Figure 3). This pattern suggests that fewer polyploids co-occur with a close diploid relative compared to diploids.
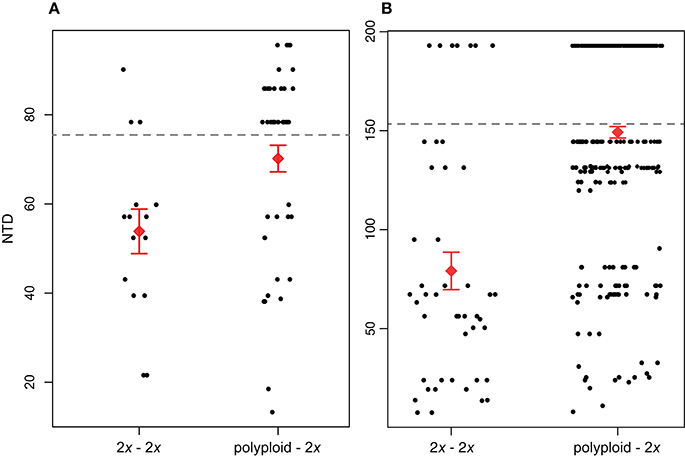
Figure 3. Phylogenetic distance between diploids and their closest diploid relative (nearest taxon distance; NTD) within the same community (2x – 2x) and NTD between polyploids and diploids within the same community (polyploid – 2x). NTD differences between the two groups are significant for both (A) Brassicaceae (P < 0.05) and (B) Rosaceae (P < < 0.01). The red diamond and error bars show the mean of the distribution ±1 standard error. The mean NTD (MNTD) for each family-level phylogeny is indicated by the dashed horizontal line.
When examining each site, MNTDpolyploid−2x was greater than MNTD2x−2x for three of the four Brassicaceae communities (MNTDpolyploid−2x / MNTD2x−2x > 1; Figure 4). However, MNTDpolyploid−2x / MNTD2x−2x was only significantly greater than expected by chance at one site (ONAQ; P < 0.05) in our simulation analyses. At MOAB, although MNTDpolyploid−2x was greater than MNTD2x−2x, the phylogenetic distance was smaller than expected by chance (lower 2.5% of the null distribution). Within Rosaceae communities, MNTDpolyploid−2x was greater than MNTD2x−2x for seven of the nine communities, but none of these differences were significantly different from the random expectation in our simulation analyses (Figure 4). For one community (JERC), we considered Crataegus spathulata Michx. to either be a diploid or a triploid. When analyzed as a diploid, we found that MNTDpolyploid−2x was smaller than expected by chance, although overall, MNTDpolyploid−2x was still greater than MNTD2x−2x (MNTDpolyploid−2x / MNTD2x−2x > 1). However, we did not find any significant patterns when C. spathulata was treated as a triploid in the analyses. At another community (HARV), we performed analyses with Rubus setosus Bigelow as either diploid or triploid, however there was no effect on the overall results.
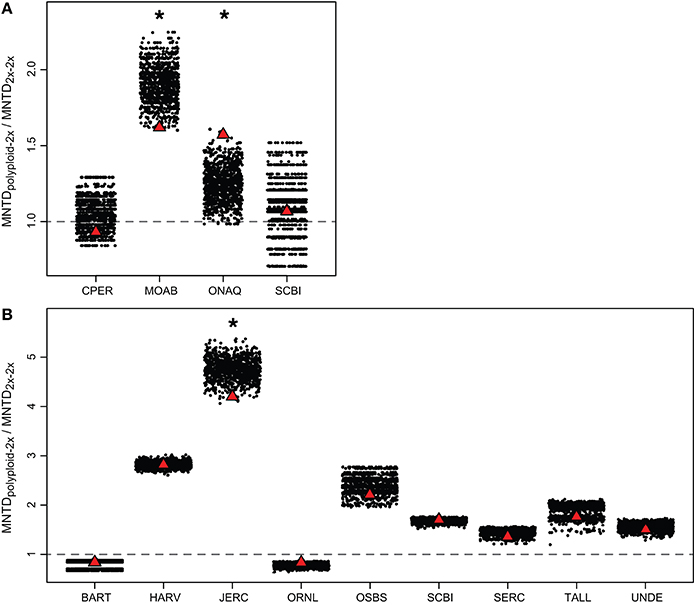
Figure 4. Comparison of observed MNTDpolyploid−2x / MNTD2x−2x at each site (red triangle) to simulated random communities (black dots) for (A) Brassicaceae and (B) Rosaceae. The random expectation was generated by randomly replacing polyploid species from a pool of polyploids from all study communities. MNTDpolyploid−2x / MNTD2x−2x > 1 (above dashed line) indicates that the mean phylogenetic distance between polyploids and the closest diploid relative in the same community is greater than that between co-occurring diploids. Asterisks indicate that the observed MNTDpolyploid−2x / MNTD2x−2x is significantly different from random (P < 0.05). If the observed value is significantly higher than the random distribution, MNTDpolyploid−2x / MNTD2x−2x is significantly greater than expected by chance. Alternatively, if the observed value is significantly lower than the random distribution, MNTDpolyploid−2x / MNTD2x−2x is significantly less than expected by chance. The significant difference at JERC for Rosaceae (B) reflects the analysis with Crataegus spathulata as a diploid (vs. triploid). Site name abbreviations follow Figure 1.
Tests of Polyploid Ecological Success
In Brassicaceae communities, polyploids tended to be more abundant than diploids (P > 0.05; Figure 5A). Though not a significant pattern, in all communities that included both diploid and polyploid species, ≥70% of the individuals were polyploid. The greater abundance of polyploids appears to be driven by non-native polyploids, which tended to be greater in number than native polyploids (Figure 5C). However, we did not find a significant difference in the abundance of non-native and native diploid or polyploid individuals within any of the communities (P > 0.05). This may be due to the small number of Brassicaceae communities included in the analysis, or could suggest that the ecological success of polyploid species is not the result of non-native species experiencing recent range expansions.
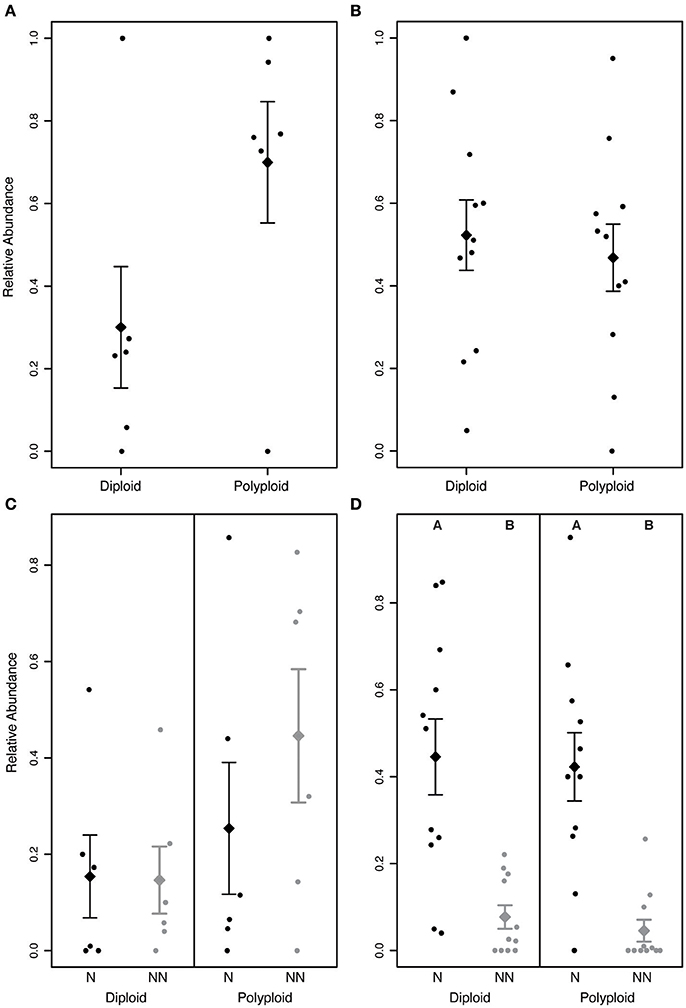
Figure 5. Relative abundance of diploids and polyploids in (A,C) Brassicaceae and (B,D) Rosaceae communities. (A) In Brassicaceae communities, polyploids tend to be more abundant than diploids, though the difference was not significant (P > 0.05). (B) In Rosaceae communities, diploids and polyploids do not significantly differ in abundance (P > 0.05). (C) In Brassicaceae communities, non-native (NN) polyploids tend to occur at a greater abundance than the other groups, but the difference is not significant (P > 0.05). (D) In Rosaceae communities, native species (N) are significantly more abundant than non-native species for both diploid and polyploid species (P < 0.05). Letters above the distributions in (D) indicate significantly different groups. The diamond and error bars indicate the mean of the distribution ± 1 standard error.
In Rosaceae communities, we found no significant difference between diploid and polyploid abundance (P > 0.05; Figure 5B). When polyploids and diploids were categorized as native or non-native, however, we found that native species were significantly more abundant than co-occurring non-native species for both diploids and polyploids (P < 0.05; Figure 5D) suggesting that ecological success is not necessarily associated with genome duplication.
Discussion
Polyploidy is now widely accepted as a mechanism of reproductive isolation and plant speciation, but much remains to be clarified about the influence of genome duplication on population- and community-level dynamics. In this study, we draw upon the extensive body of work conducted on the ecology and evolution of polyploids to predict and test how genome duplication may affect phylogenetic community structure. By examining two large flowering plant families with high incidences of polyploidy using phylogenetic data and cytogeographic information from a diversity of sources, we found that communities may be shaped in diverse ways by genome duplication and that the impacts of polyploidy are far from clear-cut. Polyploidy appears to influence patterns of phylogenetic relationships and species co-occurrence in Brassicaceae and Rosaceae communities, but these patterns appear to be lineage-specific rather than due to properties intrinsic to all genome duplication events. These results reflect the complexities and multifaceted consequences of polyploidy (Soltis et al., 2016), but our study also highlights the current paucity of information on ploidal variation at fine spatial scales (especially at cytotype contact zones), which may have contributed, in part, to some inconsistencies in our results.
Patterns of Polyploid Community Structure Are Lineage-Specific
For both Brassicaceae and Rosaceae, we found that ploidal variation is a common feature of communities across the United States. We especially observed a higher diversity of ploidal levels, and higher overall ploidies, among the Rosaceae. Of the 11 Rosaceae communities, all but one comprised both diploid and polyploid species, while two of the six Brassicacae communities were either composed of only polyploid species or of only diploid species. It is not immediately clear why Rosaceae species would exhibit a greater diversity of ploidies and higher ploidal complements, or why Rosaceae communities almost always included polyploids. This pattern may simply be due to the greater number of Rosaceae species present in the included communities, or that Rosaceae is an older family than Brassicaceae (~95 vs. ~42 million years old, respectively; Huang et al., 2016; Zhang et al., 2017) allowing more time for the evolution of greater ploidal diversity. However, it is notable that, compared to Brassicaceae, Rosaceae species tend to have perennial life histories. The longer-lived life histories of perennial species may satisfy conditions that promote unreduced gamete and polyploid formation, or polyploid phenotypes may best experience higher fitnesses when they have longer-lived life histories. Previous studies suggest that polyploid populations may arise more regularly in herbaceous species, but not necessarily in short-lived or annual species (Stebbins, 1938; Grant, 1981; Ramsey and Schemske, 2002; Zenil-Ferguson et al., 2017). It is possible that, on average, the Rosaceae species included in our analyses fall into a “sweet spot” of non-woody perennial life-history traits favoring genome duplication.
Our phylogenetic analyses of Brassicaceae and Rosaceae community structure indicate that in both families, polyploid species tend to be more distantly related to co-occurring diploids than diploids are to each other. Indeed, the proportion of diploid-diploid relationships falling below the MNTD of the family-level phylogeny was greater than that for the proportion of polyploid-diploid relationships (Figure 3). This suggests that the polyploid members of these communities may not have arisen in situ, but rather these polyploids are likely to have arisen in disjunct communities, or from interspecific hybridizations (i.e., allopolyploidy; Symonds et al., 2010), before dispersing to the surveyed communities. This is consistent with polyploids escaping minority cytotype exclusion by inhabiting differing geographic or ecological areas compared to their close relatives (Levin, 1975; Husband, 2000; Čertner et al., 2017). Alternatively, this phylogenetic pattern could have arisen if polyploids did establish within the same community as their diploid ancestors, but interploidal competition resulted in the local extinction of the diploid. Further studies incorporating a temporal aspect to community structure to capture interspecific interactions through time would allow us to distinguish between these two alternatives.
When considering each site separately, we did not find a consistent pattern in our simulation analyses. Although one Brassicaceae community showed polyploids to be more distantly related to diploids than expected by chance, at all other sites, we did not find a significant pattern, or found that polyploids were more closely related to diploids than expected, despite phylogenetic distances between polyploids and diploids being larger than the distances between diploids to one another. Together, the results from our broad-scale analyses and site-specific simulation analyses suggest that polyploidy can play an important role in shaping community structure but that the effect is species-specific. For example, the extent to which polyploids differ in phenotype and genetic composition could influence interactions with co-occurring species and the mode of escape from minority cytotype exclusion. Polyploids can exhibit wider variation in phenotypes compared to diploids, ranging from striking to subtle, which may depend in part upon the mode of polyploid formation. While allopolyploids often exhibit phenotypes that are intermediate to the parental species, the combination of two evolutionarily differentiated genomes, and their attendant regulatory elements, can sometimes produce transgressive phenotypes outside the range of variation harbored by either parental species (McCarthy et al., 2015, 2017). In contrast, autopolyploids often exhibit more subtle phenotypic differences when compared to their diploid progenitors (Maherali et al., 2009; Thompson et al., 2015). Therefore, our lack of a consistent result could be due, at least in part, to the inherent genetic differences between autopolyploids and allopolyploids, and further investigations examining how these two modes of polyploid formation may differ in their influence on community structure would go far toward illuminating interspecific interactions involving polyploids.
The apparent lineage-specific effect of polyploidy on phylogenetic community structure may also be due to varying ecological niche affinities and/or differences in life history. The hypothesized association between greater ploidal diversity and perennial life history (Müntzing, 1936; Stebbins, 1938; Grant, 1981) may mean that genome duplication shapes communities dominated by perennial species more strongly than communities comprising mostly annual species (Stebbins, 1938; Leitch and Leitch, 2012; Zenil-Ferguson et al., 2017). Though the incidence of polyploidy among woody species (which also tend to be perennial) is lower than among herbaceous species, this may consequently mean that the ecoregions or habitats dominated by perennial species (e.g., forests, woodlands, shrublands) are influenced more strongly by genome duplication than habitats where annual and herbaceous species dominate (e.g., grasslands, meadows). Our findings clearly provide motivation for broader investigations of the differences in impact on community structure between polyploid plant species with differing life histories.
Polyploids Not Consistently More Ecologically Successful Than Diploids
Polyploidy has classically been argued to be an important enabler of plant invasions and the exploitation of novel ecological niches (Pandit et al., 2014; Ramsey and Ramsey, 2014). Indeed, chromosome number has been identified as a correlate of invasiveness (Pyšek et al., 2013), and non-native polyploids in some flora are more likely to successfully become naturalized than diploid species (Nagy et al., 2017). As a measure of ecological success, a greater relative abundance of non-native species within a community should reflect their ability to successfully occupy and exploit novel habitat or outcompete and displace resident species (Levin, 1975; Callaway and Aschehoug, 2000; Cleland et al., 2004; te Beest et al., 2011). In our study, we found opposing patterns within Brassicaceae and Rosaceae communities. Brassicaceae polyploids showed patterns of abundance consistent with being more ecologically successful than diploids, which may have been driven by non-native species. Although there was not a significant difference between diploid and polyploid abundance, perhaps due to the relatively small sample of Brassicaceae sites analyzed, it is striking that in all communities that had both diploid and polyploid species, polyploids made up over 70% of the total relative abundance. On the other hand, Rosaceae diploid species were just as abundant as polyploids, and native species appeared to be more ecologically successful, with higher abundances, than the non-native species regardless of ploidy, suggesting that ecological success is not always a correlate of non-native and/or polyploid species.
The lack of a clear pattern for greater non-native polyploid abundance relative to diploids in Brassicaceae and Rosaceae communities is consistent with the varying findings of prior studies on invasive polyploids. For example, although many polyploids are invasive (Thompson, 1991; Pandit et al., 2006), species with smaller genome sizes have also been found to occur at higher species abundance, especially among annual species (Herben et al., 2012), and are more likely to be invasive (Grotkopp et al., 2002; Pandit et al., 2006, 2014; Kubešová et al., 2010; Lavergne et al., 2010; Herben and Goldberg, 2014; Schmidt et al., 2017). These counterintuitive findings may also reflect species-specific effects where a polyploid's potential for successful establishment and population expansion within a community may be highly dependent upon species-specific attributes, life histories, source locations, or the local environment of the community. For example, in anthropogenically disturbed habitats, non-native or invasive species are often polyploid (Lumaret and Borrill, 1988; Ramsey and Schemske, 1998). The importance of source locations and the ecology of the non-native range can also be seen in English Ivy (Hedera spp.), where the observation that diploids are invasive on the east coast of North America and tetraploids are invasive on the west coast of North America is thought to be due to adaptation that has occurred within the native European range, followed by subsequent exploitation of similar habitat within the invasive range (Green et al., 2013). Moreover, different cytotypes can also vary in ecological attributes and fitness across their range (McIntyre and Strauss, 2017), further nuancing the probability of establishment success within a community.
Conflicting observations of polyploid ecological success relative to diploids may also be due to the eco-evolutionary dynamics that occur over ecological timescales that affect interspecific competition and adaptation (Yoshida et al., 2003; Hairston et al., 2005; Reznick, 2013; DeLong et al., 2016). It is possible that when considered over time, the polyploid species observed in Brassicaceae and Roseaceae communities may be superior competitors that are in the process of displacing resident diploid species (or other ploidies). Alternatively, the polyploid species may be transient or ephemeral community members, documented at the present moment in time, and will eventually be displaced by the resident diploid species (Čertner et al., 2017). Additional studies incorporating phenotypic traits, and temporal data on species occurrence and abundance are needed to parse these alternatives and identify the underlying drivers of community structure. NEON's mission to repeatedly survey these sites over the next 30 years may provide an avenue to examine how community structure changes temporally, and offer insight into how polyploids and diploids interact within communities.
Observations that polyploids are not always ecologically superior suggest that polyploidy per se may have limited influence on the successful establishment of a population, or that the effects of genome duplication may not be uniformly predictable after polyploidy “primes the pump.” This can be seen in studies explicitly examining ecological differences between diploids and polyploids that show variable patterns of ecological niche divergence for both auto- and allopolyploids (Glennon et al., 2014; Marchant et al., 2016). Studies involving synthetically generated polyploids have further demonstrated that interploidal trait differences only partially arise as a direct consequence of polyploidy, and similar studies in established polyploids are consistent with genome duplication either representing or generating intra-population variation that can be elaborated upon by natural selection (e.g., Husband and Schemske, 2000; Raabová et al., 2008; Ramsey, 2011; Laport et al., 2016). Additional studies incorporating ecological data (i.e., climate, soil, water availability, pollinators, etc.) would likely provide greater detail about diploid and polyploid differences at the community level in both native and non-native systems, and should be undertaken for a broader range of species (Kolár et al., 2017). Yet, additional comparative studies examining multiple diploid-polyploid pairs would go far in disentangling the influence of lineage- or cytotype-specific life history attributes, functional traits, and genomic contributions on the adaptive potential of genome duplication for range expansion and the establishment of non-native species within communities.
The Need for Greater Documentation of Geographic Ploidal Variation
Our study highlights the need for better documentation of intraspecific ploidal variation in a geographical context to better understand the role of genome duplication on plant community structure. Our characterization of members within a community was reliant upon local-scale documentation of ploidal variation, but we often found a paucity of available geographically explicit intraspecific ploidy data. Despite the known prevalence of geographic variation in ploidy within species (e.g., Baack, 2005; Kolár et al., 2009; Ståhlberg, 2009; Trávníček et al., 2011; Castro et al., 2012; Laport et al., 2012; Zozomová-Lihová et al., 2015; Wefferling et al., 2017; reviewed in Ramsey and Ramsey, 2014), species harboring populations differing in ploidy have historically been geographically under-sampled. Modern technologies, such as high throughput flow cytometry screening for DNA content (Kron et al., 2007), have improved our ability to identify intraspecific ploidal variation, representing potential cryptic biodiversity, and can facilitate tying phenotypic variation to different ploidies within polyploid complexes. Furthermore, new genomic tools and the ever-increasing trove of genomic data for non-model organisms could be used in post-hoc analyses to further reveal novel cytotype variation (e.g., modifications to genotype-by-sequencing approaches; Gompert and Mock, 2017). The implementation of these approaches, paired with broader usage of electronic databases (e.g., Kew C-value database, Chromosome Count Database) and inclusion of ploidy or genome size information on herbarium specimens will facilitate the documentation of polyploid complexes and further aid explorations of polyploid biodiversity and its influence on community structure.
Conclusion and Perspective
This is an exciting time to study the ecological and evolutionary implications of polyploidy at the population and community level. The growing body of work on polyploid evolution and population-level dynamics suggests that polyploidy may potentially have cascading effects on communities, yet few studies have explicitly tested the effect genome duplication has on community structure. Our novel study on Brassicaceae and Rosaceae communities suggests that the effects of genome duplication on community structure may often be lineage-specific, but polyploidy should still be considered as a potentially important driver of biodiversity patterns given the pervasiveness of genome duplication among vascular plants. Our findings contribute to the increasing number of studies highlighting the complexity and multifaceted consequences of whole genome duplication (reviewed in Ramsey and Ramsey, 2014; Soltis et al., 2016). Although explicitly population-level studies may reveal the processes underlying the pattern (e.g., inter-trophic-level interactions such as with herbivores, pollinators, mycorrhiza, and other microbial symbionts; reviewed in Segraves, 2017), macro-scale studies such as ours complement the many population-level studies of polyploids by providing a “zoomed out” perspective on general patterns, a comparative evaluation of a greater diversity of plant species and life histories, and offer nuance into how different evolutionary lineages may interact within communities comprising multiple ploidies.
At the same time, our understanding of the effect of polyploidy on community structure may have been hindered by the paucity of available geographically meaningful data on intraspecific ploidal variation, and the difficulty in compiling existing data from scattered literature reports. Alongside the recognized need to characterize intraspecific genetic and trait variation to understand their subsequent effects on community structure (Hughes et al., 2008; Bolnick et al., 2011), we urge continued emphasis on the characterization and documentation of ploidal variation across species' ranges. Such information will greatly aid comparative studies at the population and community level, and help shed light on how such a common, but profound, mutation influences the evolution of species and those around them.
Author Contributions
All authors contributed to the design of the research and writing the paper. MG collected the data, and MG and JN performed statistical analyses.
Funding
This research was supported by a National Science Foundation grant to JN and RL (NSF-EF 1550813), an REU supplement to that grant, and a National Science Foundation grant to RL (NSF-DEB-1556371).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Vivianna Sanchez and William Weaver for their assistance in data collection and interpretation, and Chelsea Pretz for fruitful discussions in planning this study. We also thank Stacey D. Smith for generously sharing laboratory space to conduct this research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2018.00052/full#supplementary-material
References
Adams, K. L., and Wendel, J. F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141. doi: 10.1016/j.pbi.2005.01.001
Arvanitis, L., Wiklund, C., and Ehrlén, J. (2007). Butterfly seed predation: effects of landscape characteristics, plant ploidy level and population structure. Oecologia 152, 275–285. doi: 10.1007/s00442-007-0659-5
Baack, E. J. (2005). To succeed globally, disperse locally: effects of local pollen and seed dispersal on tetraploid establishment. Heredity 94, 538–546. doi: 10.1038/sj.hdy.6800656
Balao, F., Herrera, J., and Talavera, S. (2011). Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: a multivariate morphological approach. New Phytol. 192, 256–265. doi: 10.1111/j.1469-8137.2011.03787.x
Beaulieu, J. M., Leitch, I. J., Patel, S., Pendharkar, A., and Knight, C. A. (2008). Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 179, 975–986. doi: 10.1111/j.1469-8137.2008.02528.x
te Beest, M., Le Roux, J. J., Richardson, D. M., Brysting, A. K., Suda, J., Kubešová, M., et al. (2011). The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 109, 19–45. doi: 10.1093/aob/mcr277
Bilinski, P., Albert, P. S., Berg, J. J., Birchler, J., Grote, M., Lorant, A., et al. (2018). Parallel altitudinal clines reveal adaptive evolution of genome size in Zea mays. bioRxiv. doi: 10.1101/134528
Bolnick, D. I., Amarasekare, P., Araújo, M. S., Bürger, R., Levine, J. M., Novak, M., et al. (2011). Why intraspecific trait variation matters in community ecology. Trends Ecol. Evol. 26, 183–192. doi: 10.1016/j.tree.2011.01.009
Bouckaert, R., Heled, J., Kühnert, D., Vaughan, T., Wu, C.-H., Xie, D., et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Callaway, R. M., and Aschehoug, E. T. (2000). Invasive plants versus their new and old neighbors: a mechanism for exotic invasion. Science 290, 521–523. doi: 10.1126/science.290.5491.521
Castro, S., Loureiro, J., Procházka, T., and Münzbergová, Z. (2012). Cytotype distribution at a diploid–hexaploid contact zone in Aster amellus (Asteraceae). Ann. Bot. 110, 1047–1055. doi: 10.1093/aob/mcs177
Cavender-Bares, J., Keen, A., and Miles, B. (2006). Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology 87, S109–S122. doi: 10.1890/0012-9658(2006)87[109:PSOFPC]2.0.CO;2
Cleland, E. E., Smith, M. D., Andelman, S. J., Bowles, C., Carney, K. M., Horner-Devine, C. M., et al. (2004). Invasion in space and time: non-native species richness and relative abundance respond to interannual variation in productivity and diversity. Ecol. Lett. 7, 947–957. doi: 10.1111/j.1461-0248.2004.00655.x
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Čertner, M., Fenclová, E., Kúr, P., Kolár, F., Koutecký, P., Krahulcová, A., et al. (2017). Evolutionary dynamics of mixed-ploidy populations in an annual herb: dispersal, local persistence and recurrent origins of polyploids. Ann. Bot. 120, 303–315. doi: 10.1093/aob/mcx032
DeLong, J. P., Forbes, V. E., Galic, N., Gibert, J. P., Laport, R. G., Phillips, J. S., et al. (2016). How fast is fast? Eco-evolutionary dynamics and rates of change in populations and phenotypes. Ecol. Evol. 6, 573–581. doi: 10.1002/ece3.1899
Doyle, J. D., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Emerson, B. C., and Gillespie, R. G. (2008). Phylogenetic analysis of community assembly and structure over space and time. Trends Ecol. Evol. 23, 619–630. doi: 10.1016/j.tree.2008.07.005
Fowler, N. L., and Levin, D. A. (2016). Critical factors in the establishment of allopolyploids. Am. J. Bot. 103, 1139–1145. doi: 10.3732/ajb.1500407
Glennon, K. L., Ritchie, M. E., and Segraves, K. A. (2014). Evidence for shared broad-scale climatic niches of diploid and polyploid plants. Ecol. Lett. 17, 574–582. doi: 10.1111/ele.12259
Gompert, Z., and Mock, K. E. (2017). Detection of individual ploidy levels with genotyping-by-sequencing (GBS) analysis. Mol. Ecol. Resour. 17, 1156–1167. doi: 10.1111/1755-0998.12657
Green, A. F., Ramsey, T. S., and Ramsey, J. (2013). Polyploidy and invasion of English ivy (Hedera spp., Araliaceae) in North American forests. Biol. Invasions 15, 2219–2241. doi: 10.1007/s10530-013-0446-7
Grotkopp, E., Rejmánek, M., and Rost, T. L. (2002). Toward a causal explanation of plant invasiveness: seedling growth and life-history strategies of 29 pine (Pinus) species. Am. Nat. 159, 396–419. doi: 10.1086/338995
Hagberg, A., and Ellerström, S. (1959). The competition between diploid, tetraploid and aneuploid rye. Hereditas 45, 369–416. doi: 10.1111/j.1601-5223.1959.tb03058.x
Hairston, N. G., Ellner, S. P., Geber, M. A., Yoshida, T., and Fox, J. A. (2005). Rapid evolution and the convergence of ecological and evolutionary time. Ecol. Lett. 8, 1114–1127. doi: 10.1111/j.1461-0248.2005.00812.x
Halverson, K., Heard, S. B., Nason, J. D., and Stireman, J. O. (2008). Differential attack on diploid, tetraploid, and hexaploid Solidago altissima L. by five insect gallmakers. Oecologia 154, 755–761. doi: 10.1007/s00442-007-0863-3
Herben, T., and Goldberg, D. E. (2014). Community assembly by limiting similarity vs. competitive hierarchies: testing the consequences of dispersion of individual traits. J. Ecol. 102, 156–166. doi: 10.1111/1365-2745.12181
Herben, T., Suda, J., Klimešová, J., Mihulka, S., Ríha, P., and Šímová, I. (2012). Ecological effects of cell-level processes: genome size, functional traits and regional abundance of herbaceous plant species. Ann. Bot. 110, 1357–1367. doi: 10.1093/aob/mcs099
Huang, C.-H., Sun, R., Hu, Y., Zeng, L., Zhang, N., Cai, L., et al. (2016). Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 33, 394–412. doi: 10.1093/molbev/msv226
Hughes, A. R., Inouye, B. D., Johnson, M. T. J., Underwood, N., and Vellend, M. (2008). Ecological consequences of genetic diversity. Ecol. Lett. 11, 609–623. doi: 10.1111/j.1461-0248.2008.01179.x
Husband, B. C. (2000). Constraints on polyploid evolution: a test of the minority cytotype exclusion principle. Proc. R. Soc. Lond. B Biol. Sci. 267, 217–223. doi: 10.1098/rspb.2000.0990
Husband, B. C., Ozimec, B., Martin, S. L., and Pollock, L. (2007). Mating consequences of polyploid evolution in flowering plants: current trends and insights from synthetic polyploids. Int. J. Plant Sci. 169, 195–206. doi: 10.1086/523367
Husband, B. C., and Schemske, D. W. (2000). Ecological mechanisms of reproductive isolation between diploid and tetraploid Chamerion angustifolium. J. Ecol. 88, 689–701. doi: 10.1046/j.1365-2745.2000.00481.x
Katoh, K., Misawa, K., Kuma, K. i., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kay, Q. O. N. (1969). The origin and distribution of diploid and tetraploid Tripleurospermum inodorum (L.) Schultz Bip. Watsonia 7, 130–141.
Keller, M., Schimel, D. S., Hargrove, W. W., and Hoffman, F. M. (2008). A continental strategy for the national ecological observatory network. Front. Ecol. Environ. 6, 282–284. doi: 10.1890/1540-9295(2008)6[282:ACSFTN]2.0.CO;2
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: r tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. doi: 10.1093/bioinformatics/btq166
Kennedy, B. F., Sabara, H. A., Haydon, D., and Husband, B. C. (2006). Pollinator-mediated assortative mating in mixed ploidy populations of Chamerion angustifolium (Onagraceae). Oecologia 150, 398–408. doi: 10.1007/s00442-006-0536-7
Kolár, F., Certner, M., Suda, J., Schönswetter, P., and Husband, B.C. (2017). Mixed-ploidy species: progress and opportunities in polyploid research. Trends Plant Sci. 22, 1041–1055. doi: 10.1016/j.tplants.2017.09.011
Kolár, F., Štech, M., Trávníček, P., Rauchová, J., Urfus, T., Vít, P., et al. (2009). Towards resolving the Knautia arvensis agg. (Dipsacaceae) puzzle: primary and secondary contact zones and ploidy segregation at landscape and microgeographic scales. Ann. Bot. 103, 963–974. doi: 10.1093/aob/mcp016
Koressaar, T., and Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291. doi: 10.1093/bioinformatics/btm091
Kreiner, J. M., Kron, P., and Husband, B. C. (2017). Frequency and maintenance of unreduced gametes in natural plant populations: associations with reproductive mode, life history and genome size. New Phytol. 214, 879–889. doi: 10.1111/nph.14423
Kron, P., Suda, J., and Husband, B. C. (2007). Applications of flow cytometry to evolutionary and population biology. Ann. Rev. Ecol. Evol. Syst. 38, 847–876. doi: 10.1146/annurev.ecolsys.38.091206.095504
Kubešová, M., Moravcova, L., Suda, J., Jarošík, V., and Pyšek, P. (2010). Naturalized plants have smaller genomes than their non-invading relatives: a flow cytometric analysis of the Czech alien flora. Preslia 82, 81–96.
Laport, R. G., Hatem, L., Minckley, R. L., and Ramsey, J. (2013). Ecological niche modeling implicates climatic adaptation, competitive exclusion, and niche conservatism among Larrea tridentata cytotypes in North American deserts. J. Torrey Bot. Soc. 140, 349–363. doi: 10.3159/TORREY-D-13-00009.1
Laport, R. G., Minckley, R. L., and Ramsey, J. (2012). Phylogeny and cytogeography of the North American creosote bush (Larrea tridentata, Zygophyllaceae). Syst. Bot. 37, 153–164. doi: 10.1600/036364412X616738
Laport, R. G., Minckley, R. L., and Ramsey, J. (2016). Ecological distributions, phenological isolation, and genetic structure in sympatric and parapatric populations of the Larrea tridentata polyploid complex. Am. J. Bot. 103, 1358–1374. doi: 10.3732/ajb.1600105
Laport, R. G., and Ng, J. (2017). Out of one, many: the biodiversity considerations of polyploidy. Am. J. Bot. 104, 1119–1121. doi: 10.3732/ajb.1700190
Lavergne, S., Mouquet, N., Thuiller, W., and Ronce, O. (2010). Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Annu. Rev. Ecol. Evol. Syst. 41, 321–350. doi: 10.1146/annurev-ecolsys-102209-144628
Leitch, A. R., and Leitch, I. J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320, 481–483. doi: 10.1126/science.1153585
Leitch, A. R., and Leitch, I. J. (2012). Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytol. 194, 629–646. doi: 10.1111/j.1469-8137.2012.04105.x
Levin, D. A. (1975). Minority cytotype exclusion in local plant populations. Taxon 24, 35–43. doi: 10.2307/1218997
Levin, D. A. (1983). Polyploidy and novelty in flowering plants. Am. Nat. 122, 1–25. doi: 10.1086/284115
Levin, D. A., and Soltis, D. E. (2017). Factors promoting polyploid persistence and diversification and limiting diploid speciation during the K-Pg interlude. Curr. Opin. Plant Biol. 42, 1–7. doi: 10.1016/j.pbi.2017.09.010
Lewis, W. H. (1962). Phylogenetic study of Hedyotis (Rubiaceae) in North America. Am. J. Bot. 49, 855–865. doi: 10.1002/j.1537-2197.1962.tb15020.x
Lumaret, R., and Borrill, M. (1988). Cytology, genetics, and evolution in the genus dactylis. Crit. Rev. Plant Sci. 7, 55–91. doi: 10.1080/07352688809382259
Maddison, W. P., and Maddison, D. R. (2017). Mesquite: a modular system for evolutionary analysis.Version 3.31. Available online at: http://mesquiteproject.org
Madlung, A. (2013). Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110, 99–104. doi: 10.1038/hdy.2012.79
Maherali, H., Walden, A. E., and Husband, B. C. (2009). Genome duplication and the evolution of physiological responses to water stress. New Phytologist 184, 721–731. doi: 10.1111/j.1469-8137.2009.02997.x
Manzaneda, A. J., Rey, P. J., Bastida, J. M., Weiss-Lehman, C., Raskin, E., and Mitchell-Olds, T. (2012). Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae). New Phytol. 193, 797–805. doi: 10.1111/j.1469-8137.2011.03988.x
Marchant, D. B., Soltis, D. E., and Soltis, P. S. (2016). Patterns of abiotic niche shifts in allopolyploids relative to their progenitors. New Phytol. 212, 708–718. doi: 10.1111/nph.14069
Masterson, J. (1994). Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264, 421–424. doi: 10.1126/science.264.5157.421
McCarthy, E. W., Arnold, S. E., Chittka, L., Le Comber, S. C., Verity, R., Dodsworth, S., et al. (2015). The effect of polyploidy and hybridization on the evolution of floral colour in Nicotiana (Solanaceae). Ann. Bot. 115, 1117–1131. doi: 10.1093/aob/mcv048
McCarthy, E. W., Berardi, A. E., Smith, S. D., and Litt, A. (2017). Related allopolyploids display distinct floral pigment profiles and transgressive pigments. Am. J. Bot. 104, 92–101. doi: 10.3732/ajb.1600350
McIntyre, P. J. (2012). Polyploidy associated with altered and broader ecological niches in the Claytonia perfoliata (Portulacaceae) species complex. Am. J. Bot. 99, 655–662. doi: 10.3732/ajb.1100466
McIntyre, P. J., and Strauss, S. (2017). An experimental test of local adaptation among cytotypes within a polyploid complex. Evolution 71, 1960–1969. doi: 10.1111/evo.13288
Mei, W., Stetter, M. G., Gates, D. J., Stitzer, M., and Ross-Ibarra, J. (2018). Adaptation in plant genomes: bigger is different. Am. J. Bot. 105, 16–19. doi: 10.1002/ajb2.1002
Müntzing, A. (1936). The evolutionary significance of autopolyploidy. Hereditas 21, 363–378. doi: 10.1111/j.1601-5223.1936.tb03204.x
Nagy, D. U., Stranczinger, S., Godi, A., Weisz, A., Rosche, C., Suda, J., et al. (2017). Does higher ploidy level increase the risk of invasion? A case study with two geo-cytotypes of Solidago gigantea Aiton (Asteraceae). J. Plant Ecol. 11, 317–327. doi: 10.1093/jpe/rtx005
Otto, S. P. (2007). The evolutionary consequences of polyploidy. Cell 131, 452–462. doi: 10.1016/j.cell.2007.10.022
Pandit, M. K., Tan, H. T. W., and Bisht, M. S. (2006). Polyploidy in invasive plant species of Singapore. Bot. J. Linnean Soc. 151, 395–403. doi: 10.1111/j.1095-8339.2006.00515.x
Pandit, M. K., White, S. M., and Pocock, M. J. O. (2014). The contrasting effects of genome size, chromosome number and ploidy level on plant invasiveness: a global analysis. New Phytol. 203, 697–703. doi: 10.1111/nph.12799
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Parisod, C., Holderegger, R., and Brochmann, C. (2010). Evolutionary consequences of autopolyploidy. New Phytol. 186, 5–17. doi: 10.1111/j.1469-8137.2009.03142.x
Park, D. S., Worthington, S., and Xi, Z. (2018). Taxon sampling effects on the quantification and comparison of community phylogenetic diversity. Mol. Ecol. 27, 1296–1308. doi: 10.1111/mec.14520
Pyšek, P., Hulme, P. E., Meyerson, L. A., Smith, G. F., Boatwright, J. S., Crouch, N. R., et al. (2013). Hitting the right target: taxonomic challenges for, and of, plant invasions. AoB Plants 5:plt042. doi: 10.1093/aobpla/plt042
Raabová, J., Fischer, M., and Münzbergová, Z. (2008). Niche differentiation between diploid and hexaploid Aster amellus. Oecologia 158, 463–472. doi: 10.1007/s00442-008-1156-1
Rambaut, A., Suchard, M., Xie, D., and Drummond, A. (2014). Tracer v1. 6 Available online at: http://beast.bio.ed.ac.uk
Ramsey, J. (2011). Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. U.S.A. 108, 7096–7101. doi: 10.1073/pnas.1016631108
Ramsey, J., and Ramsey, T. S. (2014). Ecological studies of polyploidy in the 100 years following its discovery. Philos. Trans. R. Soc. B 369:20130352. doi: 10.1098/rstb.2013.0352
Ramsey, J., and Schemske, D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29, 467–501. doi: 10.1146/annurev.ecolsys.29.1.467
Ramsey, J., and Schemske, D. W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. Syst. 33, 589–639. doi: 10.1146/annurev.ecolsys.33.010802.150437
Rey, P. J., Manzaneda, A. J., and Alcantara, J. M. (2017). The interplay between aridity and competition determines colonization ability, exclusion, and ecological segregation in the heteroploid Brachypodium distachyon species complex. New Phytol. 215, 85–96. doi: 10.1111/nph.14574
Reznick, D. N. (2013). A critical look at reciprocity in ecology and evolution: introduction to the symposium. Am. Nat. 181, S1–S8. doi: 10.1086/670030
Rieseberg, L. H., and Willis, J. H. (2007). Plant speciation. Science 317, 910–914. doi: 10.1126/science.1137729
Roccaforte, K., Russo, S. E., and Pilson, D. (2015). Hybridization and reproductive isolation between diploid Erythronium mesochoreum and its tetraploid congener E. albidum (Liliaceae). Evolution 69, 1375–1389. doi: 10.1111/evo.12666
Schlaepfer, D. R., Edwards, P. J., and Billeter, R. (2010). Why only tetraploid Solidago gigantea (Asteraceae) became invasive: a common garden comparison of ploidy levels. Oecologia 163, 661–673. doi: 10.1007/s00442-010-1595-3
Schmidt, J. P., Drake, J. M., and Stephens, P. (2017). Residence time, native range size, and genome size predict naturalization among angiosperms introduced to Australia. Ecol. Evol. 7, 10289–10300. doi: 10.1002/ece3.3505
Segraves, K. A. (2017). The effects of genome duplications in a community context. New Phytol. 215, 57–69. doi: 10.1111/nph.14564
Segraves, K., and Thompson, J. (1999). Plant polyploidy and pollination: floral traits and insect visits to diploid and tetraploid Heuchera grossulariifolia. Evolution 53, 1114–1127. doi: 10.1111/j.1558-5646.1999.tb04526.x
Smith, S. A., Beaulieu, J. M., and Donoghue, M. J. (2009). Mega-phylogeny approach for comparative biology: an alternative to supertree and supermatrix approaches. BMC Evol. Biol. 9:37. doi: 10.1186/1471-2148-9-37
Soltis, D. E., Albert, V. A., Leebens-Mack, J., Bell, C. D., Paterson, A. H., Zheng, C., et al. (2009). Polyploidy and angiosperm diversification. Am. J. Bot. 96, 336–348. doi: 10.3732/ajb.0800079
Soltis, D. E., Soltis, P. S., Schemske, D. W., Hancock, J. F., Thompson, J. N., Husband, B. C., et al. (2007). Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56, 13–30. doi: 10.2307/25065732
Soltis, D. E., Visger, C. J., Marchant, D. B., and Soltis, P. S. (2016). Polyploidy: pitfalls and paths to a paradigm. Am. J. Bot. 103, 1146–1166. doi: 10.3732/ajb.1500501
Soltis, P. S., Liu, X., Marchant, D. B., Visger, C. J., and Soltis, D. E. (2014). Polyploidy and novelty: Gottlieb's legacy. Philos. Trans. R. Soc. B 369:20130351. doi: 10.1098/rstb.2013.0351
Soltis, P. S., Marchant, D. B., Van de Peer, Y., and Soltis, D. E. (2015). Polyploid and genome evolution in plants. Curr. Opin. Genet. Dev. 35, 119–125. doi: 10.1016/j.gde.2015.11.003
Song, Q., and Chen, Z. J. (2015). Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 24, 101–109. doi: 10.1016/j.pbi.2015.02.007
Ståhlberg, D. (2009). Habitat differentiation, hybridization and gene flow patterns in mixed populations of diploid and autotetraploid Dactylorhiza maculata s.l. (Orchidaceae). Evol. Ecol. 23, 295–328. doi: 10.1007/s10682-007-9228-y
Stebbins, G. L. (1938). Cytological characteristics associated with the different growth habits in the Dicotyledons. Am. J. Bot. 25, 189–198. doi: 10.1002/j.1537-2197.1938.tb09203.x
Symonds, V. V., Soltis, P. S., and Soltis, D. E. (2010). Dynamics of polyploid formation in Tragopogon (Asteraceae): recurrent formation, gene flow, and population structure. Evolution 64, 1984–2003. doi: 10.1111/j.1558-5646.2010.00978.x
Těšitelová, T., Jersáková, J., Roy, M., Kubátová, B., Těšitel, J., Urfus, T., et al. (2013). Ploidy-specific symbiotic interactions: divergence of mycorrhizal fungi between cytotypes of the Gymnadenia conopsea group (Orchidaceae). New Phytol. 199, 1022–1033. doi: 10.1111/nph.12348
Thompson, J. D. (1991). The biology of an invasive plant. BioScience 41, 393–401. doi: 10.2307/1311746
Thompson, J. D., and Lumaret, R. (1992). The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends Ecol. Evol. 7, 302–307. doi: 10.1016/0169-5347(92)90228-4
Thompson, J. N., Cunningham, B. M., Segraves, K. A., Althoff, D. M., and Wagner, D. (1997). Plant polyploidy and insect/plant interactions. Am. Nat. 150, 730–743. doi: 10.1086/286091
Thompson, J. N., and Merg, K. F. (2008). Evolution of polyploidy and the diversification of plant-pollinator interactions. Ecology 89, 2197–2206. doi: 10.1890/07-1432.1
Thompson, K. A., Husband, B. C., and Maherali, H. (2015). No influence of water limitation on the outcome of competition between diploid and tetraploid Chamerion angustifolium (Onagraceae). J. Ecol. 103, 733–741. doi: 10.1111/1365-2745.12384
Trávníček, P., Kubátová, B., Curn, V., Rauchová, J., Krajníková, E., Jersáková, J., et al. (2011). Remarkable coexistence of multiple cytotypes of the Gymnadenia conopsea aggregate (the fragrant orchid): evidence from flow cytometry. Ann. Bot. 107, 77–87. doi: 10.1093/aob/mcq217
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res. 40, e115–e115. doi: 10.1093/nar/gks596
Vamosi, S. M., Heard, B., Vamosi, J. C., and Webb, C. O. (2009). Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 18, 572–592. doi: 10.1111/j.1365-294X.2008.04001.x
Webb, C. O., Ackerly, D. D., McPeek, M. A., and Donoghue, M. J. (2002). Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448
Wefferling, K. M., Castro, S., Loureiro, J., Castro, M., Tavares, D., and Hoot, S. B. (2017). Cytogeography of the subalpine marsh marigold polyploid complex (Caltha leptosepala s.l., Ranunculaceae). Am. J. Bot. 104, 271–285. doi: 10.3732/ajb.1600365
Yoshida, T., Jones, L. E., Ellner, S. P., Fussmann, G. F., and Hairston, N. G. Jr. (2003). Rapid evolution drives ecological dynamics in a predator–prey system. Nature 424, 303–306. doi: 10.1038/nature01767
Zenil-Ferguson, R., Ponciano, J. M., and Burleigh, J. G. (2017). Testing the association of phenotypes with polyploidy: an example using herbaceous and woody eudicots. Evolution 71, 1138–1148. doi: 10.1111/evo.13226
Zhan, S. H., Drori, M., Goldberg, E. E., Otto, S. P., and Mayrose, I. (2016). Phylogenetic evidence for cladogenetic polyploidization in land plants. Am. J. Bot. 103, 1252–1258. doi: 10.3732/ajb.1600108
Zhang, S.-D., Jin, J.-J., Chen, S.-Y., Chase, M. W., Soltis, D. E., Li, H.-T., et al. (2017). Diversification of Rosaceae since the late cretaceous based on plastid phylogenomics. New Phytol. 214, 1355–1367. doi: 10.1111/nph.14461
Keywords: Brassicaceae, genome duplication, non-native species, phylogenetic community ecology, polyploidy, Rosaceae
Citation: Gaynor ML, Ng J and Laport RG (2018) Phylogenetic Structure of Plant Communities: Are Polyploids Distantly Related to Co-occurring Diploids? Front. Ecol. Evol. 6:52. doi: 10.3389/fevo.2018.00052
Received: 22 January 2018; Accepted: 11 April 2018;
Published: 30 April 2018.
Edited by:
Hans D. Daetwyler, La Trobe University, AustraliaReviewed by:
Jacob A. Tennessen, Oregon State University, United StatesRubén Torices, Université de Lausanne, Switzerland
Copyright © 2018 Gaynor, Ng and Laport. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert G. Laport, rob.laport@gmail.com
 Michelle L. Gaynor
Michelle L. Gaynor Julienne Ng
Julienne Ng Robert G. Laport
Robert G. Laport