- 1Molecular Epidemiology, German Cancer Research Center, Heidelberg, Germany
- 2Molecular Biology of Breast Cancer, Department of Gynecology and Obstetrics, University Clinic, Heidelberg, Heidelberg, Germany
In the recent years, circulating microRNAs (miRNAs) have garnered a lot of attention and interest in the field of disease biomarkers. With characteristics such as high stability, low cost, possibility of repeated sampling and minimal invasiveness, circulating miRNAs are ideal for development into diagnostic tests. There have been many studies reported on the potential of circulating miRNAs as early detection, prognostic, and predictive biomarkers in cancer. Here, we have reviewed the application of plasma and serum miRNAs as biomarkers for cancer focusing on epithelial carcinomas [prostate, breast, lung, colorectal, and gastric cancer (GC)] and hematological malignancies (leukemia and lymphoma). We have also addressed the common challenges that need to be overcome to achieve a successful bench to bedside transition.
Introduction
Cancer accounts for the highest mortality in developed countries, and second highest in developing countries, making it a worldwide health problem (Ferlay et al., 2008; Siegel et al., 2012). One method of disease management is the development of biomarkers, which could enhance early cancer detection, improve patient stratification and therapy response prediction. This in turn would result in a more favorable disease outcome. Blood-based protein biomarkers, such as carcinoembryonic antigen (CEA), carbohydrate antigen (CA), or prostate specific antigen (PSA), have gained a lot of recognition. Nevertheless, they suffer from low sensitivity, especially with respect to their use in screening for early stages or inability to distinguish aggressive from indolent tumors (Lumachi et al., 2000; Hanash et al., 2011). This has prompted the investigation for novel and more sensitive biomarkers, which can supplement or complement existing detection methods and aid in disease monitoring.
Since their discovery, microRNAs (miRNAs) have proven to be an essential and indispensable layer in gene regulation, mainly, post-transcriptional regulation. Involved in many physiological processes and also found to be deregulated in many disease mechanisms, miRNAs have also gained importance as they carry information about the patho-physiological state of a person, and thus can serve as biomarkers. miRNAs present in the cell-free body fluids such as plasma, serum, urine, saliva, etc., termed as circulating miRNAs, have been exploited as biomarkers in many diseases in the past five years. Placental miRNAs were the first class of circulating miRNAs to be detected in maternal plasma during pregnancy (Chim et al., 2008). Parallel to this discovery, miRNAs were found to be elevated in the serum of lymphoma patients compared to healthy individuals (Lawrie et al., 2008). Since then circulating miRNAs have attracted a great deal of attention as novel, minimally invasive biomarkers for various diseases. Features such as ease of access and remarkable stability increase their potential as disease biomarkers (Mitchell et al., 2008; Turchinovich et al., 2011). Since they are bound to proteins like argonaute (AGO) or high density lipoprotein, they are highly resistant to RNase activity, unlike mRNAs measured in gene signature markers (Turchinovich and Burwinkel, 2012). Detection of miRNAs by quantitative polymerase chain reaction (qPCR) has the advantages of being relatively inexpensive, sensitive to even low amounts due to amplification of signal, and robust. In these terms, they seem to have an edge over protein markers, which currently represent the most common circulating biomarker type. A further testament to their diverse and dynamic uses in a clinical settings is the various reports outlining use of circulating miRNAs as a diagnostic, prognostic and predictive marker (Figure 1). The impact of circulating miRNAs in the field of biomarkers has been unequivocal with more than 250 studies probing their use in cancer diagnosis and prognosis.
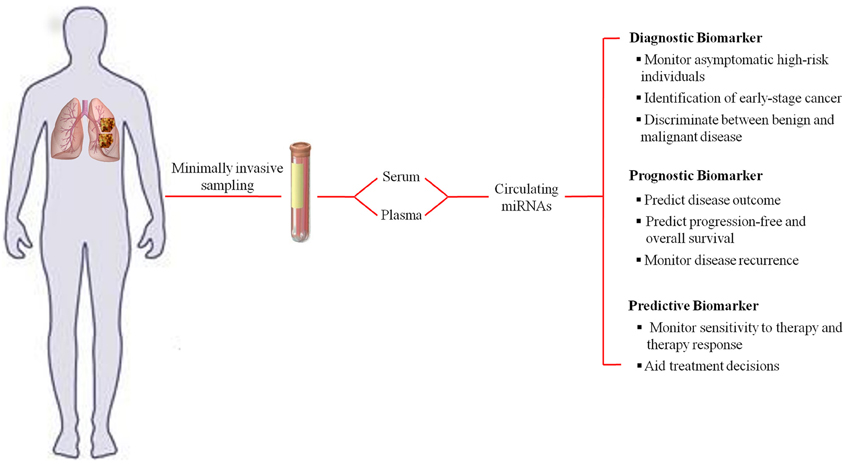
Figure 1. Representation of the diagnostic, prognostic, and predictive uses of circulating miRNAs as a biomarker using lung cancer as an example.
In this review, we have summarized the recent-findings in cancer related circulating miRNAs and presented them also in a tabular form, with a focus on epithelial cancers such as prostate (Table 1), breast (Table 2), lung (Table 3), colorectal (Table 4), and gastric (Table 5) cancer, and also touched upon lymphomas and leukemia (Table 6). We present a comprehensive summary of circulating miRNAs present in plasma or serum which are associated with diagnosis and/or prognosis of both primary and metastatic cancers, along with an appraisal of the translational implications of this field.
Prostate Cancer
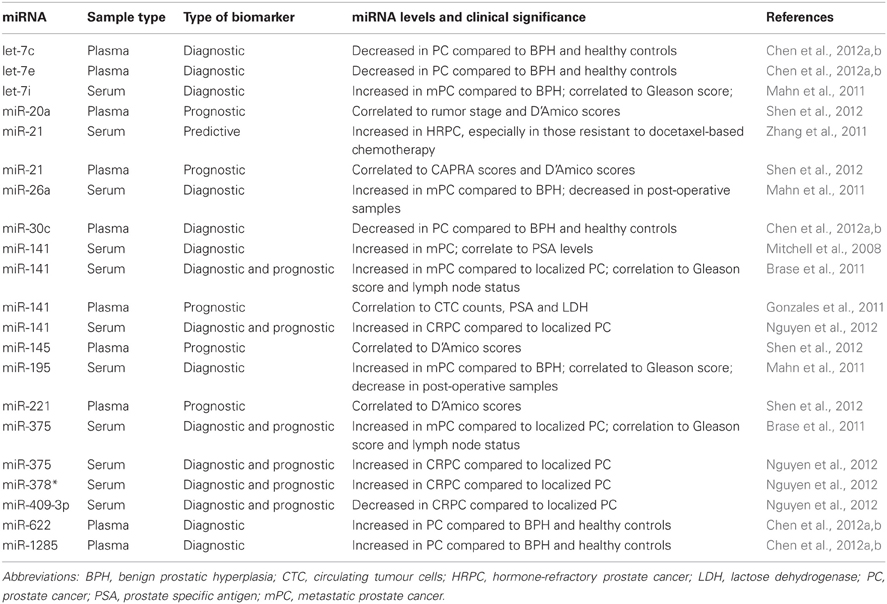
Prostate cancer (PC) was the first cancer type to be used as a disease model for the establishment of circulating miRNAs' potential as blood-based biomarkers. The pioneer work by Mitchell and colleagues have provided very important insights and laid the foundation for future investigations on circulating miRNAs and cancers. This study demonstrated that the tumor-derived miRNAs from the donor could reach the circulation of the recipient PC xenograft mouse, and could be subsequently measured in the recipients' plasma. Additionally, Mitchell et al. also investigated a few miRNA candidates which were chosen based on their expression in human prostate specimens and absence in plasma of healthy controls. This investigation performed with serum of metastatic PC (mPC) patients and healthy control individuals found increased levels of miR-141, a miRNA involved in epithelial-mesenchymal transition, in the mPC group (Mitchell et al., 2008). In another study, pre-selected oncogenic miR-26a, miR-195, and let-7i were first confirmed to be upregulated in tissue, and then found to be significantly elevated in serum of PC patients compared to those with benign prostate hyperplasia (BPH). However, this study was not able to reproduce the significant difference in miR-141, and claimed that the miR-141 levels were too low for reliable testing (Mahn et al., 2011). To achieve the same end-result of identifying miRNAs that could discriminate PC from BPH and healthy controls, global miRNAs levels were screened for and top hits validated. A five miRNA panel consisting of let-7c, let-7e, miR-30c, miR-622, and miR-1285 was proposed (Chen et al., 2012a,b). The miRNAs described in the above studies, due to their ability to discriminate PC from even BPH, are ideal candidates for early detection of PC, especially those at high-risk.
Apart from detection, circulating miRNAs have also been connected to prognosis of PC. miR-141, miR-200b, and miR-375 were the first miRNAs identified in this context due to correlations to Gleason score and lymph node affliction (Brase et al., 2011). The prognostic promise of miR-141 and miR-375 has been confirmed by two separate studies, in which their amounts were found to increase from low-risk through high-risk localized disease toward metastatic cancer (Bryant et al., 2012; Nguyen et al., 2012). Circulating miRNAs have also been found to be linked to various PC risk scores such as tumor stage, CAPRA (Cancer of the Prostate Risk Assessment) and D'Amico scores (miR-20a, miR-21, miR-145, and miR-221) (Shen et al., 2012) or have predictive abilities similar to that of circulating tumour cell (CTC) counts, PSA, and lactate dehydrogenase (LDH) levels of PC patients (miR-21, miR-141) (Gonzales et al., 2011).
Breast Cancer
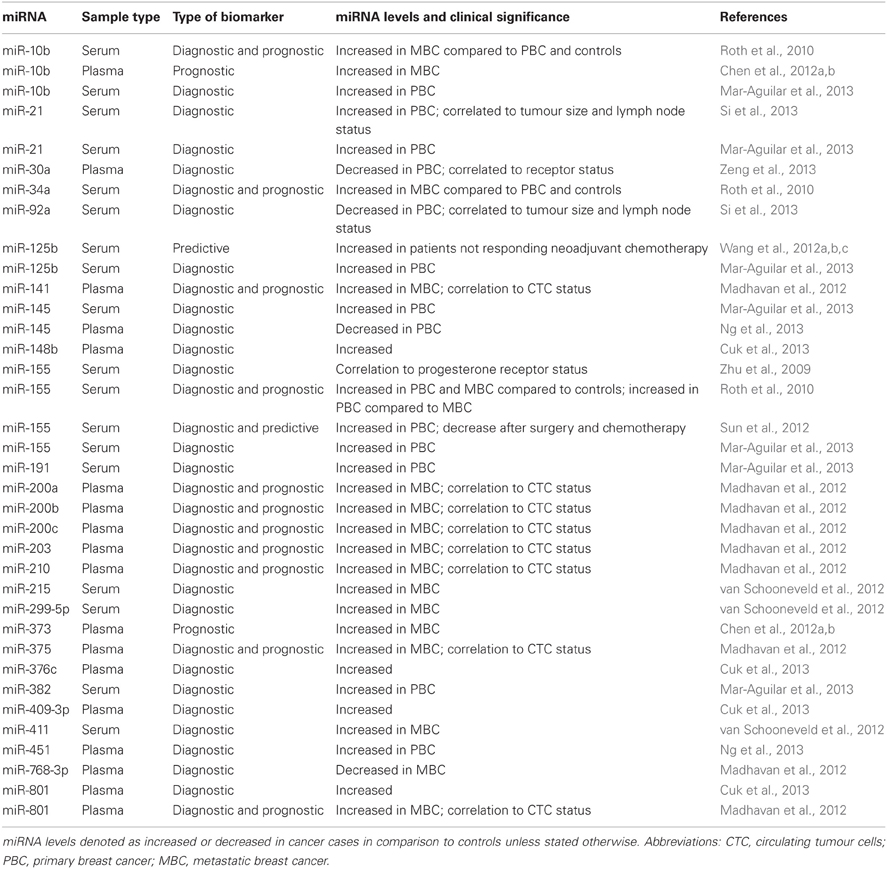
Currently, there are no early detection markers for primary breast cancer (PBC) in routine clinical use. Zhu and colleagues performed the first study to understand the association between serum miR-16, miR-145, miR-155 levels, and breast cancer risk, and found a correlation of miR-155 to progesterone receptor status (Zhu et al., 2009). In one of our studies, which was the first investigation based on genome-wide approach in PBC, miR-148b, miR-376c, miR-409-3p, and miR-801 were identified as potential early stage breast cancer biomarkers in plasma. Interestingly, miR-148b, miR-376c, and miR-409-3p were present at lower levels in malignant tissue compared to benign breast tissue, whereas no difference could be detected in miR-801 (Cuk et al., 2013). An alternate approach of identifying circulating miRNAs capable of early detection, combed for miRNAs commonly deregulated in tissue and plasma. This led to the identification of decreased miR-145 and increased miR-451 levels in plasma of breast cancer patients in relation to controls, and their plasma levels reflected those of tissue (Ng et al., 2013). Contrarily, miR-145 was reported to be upregulated in serum of PBC in another study, which could be due to the analysis of different sample types by the two studies (Mar-Aguilar et al., 2013). Recently, analysing circulation miRNAs that are known to be aberrantly expressed in breast tissue or have functional role in tumorigenesis has led to the addition of miR-21 and miR-92a (Si et al., 2013), miR-10b, miR-125b, miR-155, miR-191, and miR-382 (Mar-Aguilar et al., 2013), and miR-30a (Zeng et al., 2013) to this growing list of miRNAs for early detection of PBC. Interestingly, while exploring the difference in plasma miRNA levels between two distinct ethnic groups (Caucasian and African-American), different miRNAs were found to be deregulated in the Caucasian American and the African-American patients in comparison to their ethnically matched controls, with an overlap of only two miRNAs (miR-181a and miR-1304) between them, thus showing that circulating miRNA profile could depend on ethnicity to some degree (Zhao et al., 2010).
With respect to the metastatic setting, many reports outline the identification of circulating miRNAs which have the ability to discriminate metastatic breast cancer (MBC) from PBC and/or controls. One such study focusing on both PBC and MBC uncovered a significant difference in serum total RNA and miR-155 between PBC and healthy controls, while miR-10b, miR-34a, and miR-155 correlated with overt metastasis (Roth et al., 2010). In contrast to this candidate gene-approach, van Schooneveld et al. found miR-215, miR-299-5p, and miR-411 to be increased in tissue and also in serum of patients with MBC compared to healthy individuals by Taqman low density array cards (van Schooneveld et al., 2012). In a genome-wide array based approach, focusing not only on the identification of miRNAs for detection of MBC, but also those which could serve as prognostic markers, we investigated plasma from patients and controls, and correlated them to CTC status. We detected nine miRNAs (miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-210, miR-375, and miR-801) which were able to discriminate between MBC patients based on the presence or absence of CTCs, as well as MBC patients from healthy individuals. Combinations of these circulating miRNAs or even miR-200b alone were a better marker of progression-free survival and overall survival (OS) than CTC status, which is an FDA-approved prognostic marker for MBC (Madhavan et al., 2012). Another recent study interrogated plasma miR-10b and miR-373 levels, which are known mediators of metastasis in breast cancer, wherein they uncovered association of these miRNAs with lymph node metastasis, thereby showing potential as prognostic biomarkers (Chen et al., 2012a,b).
Apart from detection and disease prognosis, circulating miRNAs have also been associated with response to breast cancer treatment and thus possess predictive capabilities. Serum miR-125b (Wang et al., 2012a,b,c) and miR-155 (Sun et al., 2012) have been linked to chemotherapy response. Furthermore, miR-30a and miR-155 have been demonstrated as more accurate than the predictive markers CEA and CA 15-3, which are currently in use for monitoring treatment in MBC (Harris et al., 2007; Uehara et al., 2008; Sun et al., 2012; Zeng et al., 2013).
Lung Cancer
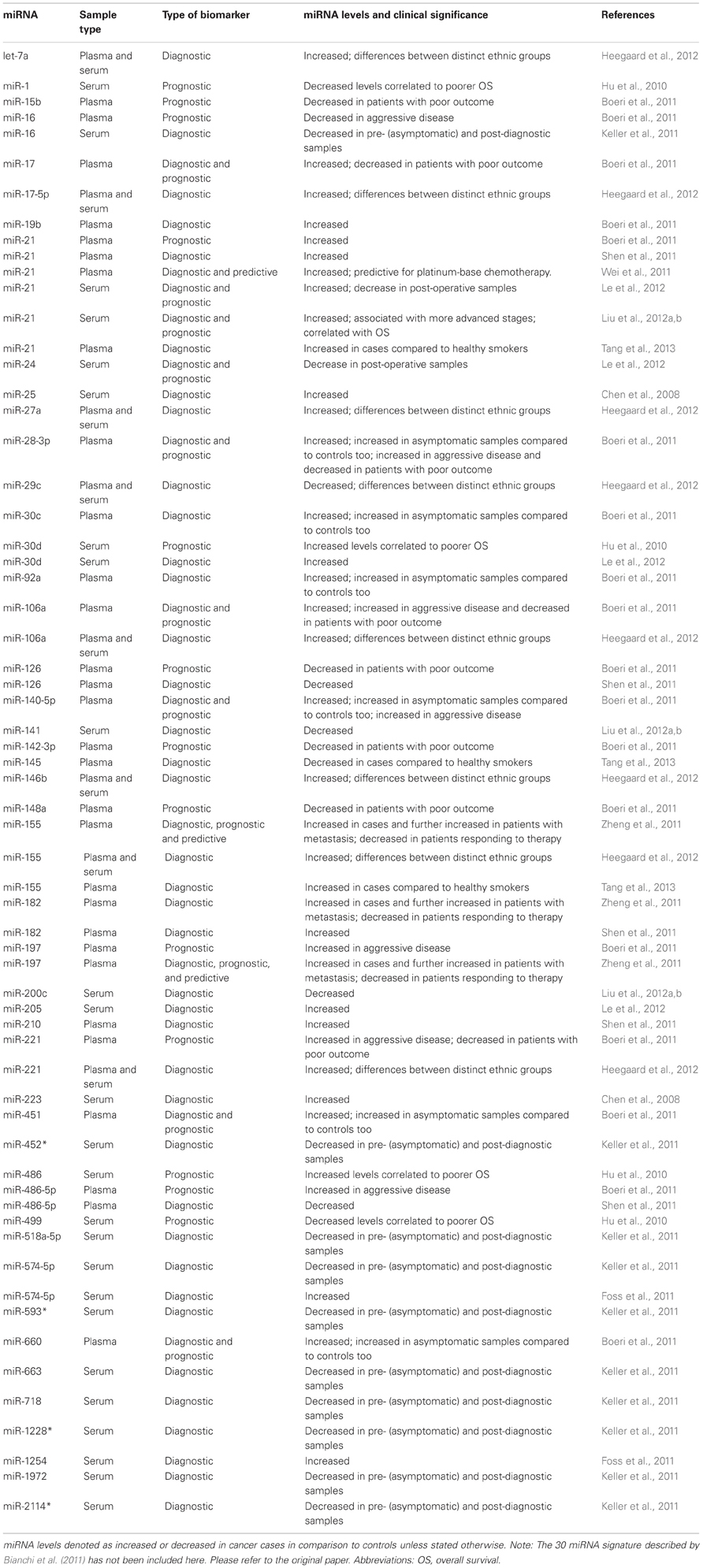
Lung cancer has been extensively studied with respect to its circulating miRNA profile and majority of studies reviewed here have concentrated on non-small-cell lung cancer (NSCLC), the more common type. The first study to systematically characterize miRNAs in serum by Solexa sequencing identified sixty three circulating miRNAs in NSCLC patients that were not detected in control samples, and ultimately miR-25 and miR-223 were highlighted as the most promising diagnostic markers (Chen et al., 2008). Another study adopting a global miRNA screening recommended miR-574-5p and miR-1254 as detection markers (Foss et al., 2011). As opposed to this global approach, Shen et al. verified miRNAs in plasma of NSCLC patients and controls, which were previously reported as differentially expressed in sputum. A panel of four miRNAs (miR-21, miR-126, miR-486-5p, and miR-210) was declared as having the highest predictive power for all stages of NSCLC, with a higher diagnostic sensitivity for more advanced stages (Shen et al., 2011). The diagnostic value of miR-21, once again, along with miR-145 and miR-155 was confirmed by validating their significant deregulation between malignant NSCLC and healthy smokers (Tang et al., 2013).
While the above studies aimed to identify circulating miRNAs that could identify NSCLC patients, Boeri et al. and Bianchi et al. attempted to distinguish miRNAs that could also be useful in the diagnosis of asymptomatic patients in plasma and serum, respectively (Bianchi et al., 2011; Boeri et al., 2011). The first study conducted a comprehensive analysis, and put forth a miRNA signature in plasma capable of detecting lung cancer in asymptomatic patients before clinical presentation of disease. miR-28-3p, miR-30c, miR-92a, miR-140-5p, miR-451, and miR-660 were the most robust miRNAs to predict the development of lung cancer, as they were deregulated in samples collected from patients even 1–2 years prior to diagnosis when compared to control individuals. In case of samples collected at the time-point of diagnosis, miR-17, miR-19b, miR-92a, miR-106a, and miR-660 were also found to be informative (Boeri et al., 2011). Bianchi and co-workers analysed serum samples from asymptomatic NSCLC patients and healthy smokers, and identified a signature of 34 miRNAs with the capacity to (1) identify early stage NSCLC patients and, (2) discriminate between benign and malignant lesions, thus detecting the onset of a malignant disease (Bianchi et al., 2011). A retrospective study, which profiled serum miRNAs in pre- and post-diagnostic NSCLC samples, and compared them to normal controls validated miR-16, miR-452*, miR-518a-5p, miR-574-5p, miR-593*, miR-663, miR-718, miR-1228*, miR-1972, and miR-2114* to be significantly deregulated in both categories of cases, thus making them ideal markers for early detection (Keller et al., 2011). These studies concur on their inference that dramatic changes in circulating miRNA profile occur at time points closer to cancer diagnosis, thus supporting the idea that miRNAs may predict cancer development years prior to clinically symptomatic disease.
With a very poor 5-year survival rate in lung cancer patients, markers to assess prognosis or predict therapy response are equally important. When pre-diagnostic samples of patients with an unfavorable clinical outcome were investigated, two sets of miRNAs were developed to help identify patients at risk for aggressive disease and prognosticate outcome (Boeri et al., 2011). By genome-wide sequencing of serum of patients with longer and those with shorter survival, Hu et al. determined the levels of serum miR-1, miR-30d, miR-486, and miR-499 to be linked to survival in NSCLC patients. Specifically, patients having two or more of these “high-risk” miRNAs deregulated were found to have a higher probability of shortened survival compared to patients with none or just one deregulated miRNA. The four-miRNA signature was propounded as an independent predictor of OS (Hu et al., 2010). Although in this study miR-21 was one of the top three hits of sequencing, it could not be successfully validated in a larger cohort. Nevertheless, circulating miR-21 has been repeatedly reported as being deregulated in lung cancer patients and possessing prognostic value, and has specifically been associated with (1) lymph node metastasis and advanced cancer stage, (2) patient survival prognosis, (3) sensitivity to platinum-based chemotherapy, and (4) post-operative disease monitoring (Wei et al., 2011; Le et al., 2012; Liu et al., 2012a,b). A recent study found miR-142-3p to be associated with a high risk of recurrence in early-stage lung adenocarcinoma patients, and thus proposed it as a putative serum marker for risk assessment (Kaduthanam et al., 2013). With respect to the predictive value, plasma levels of miR-155, miR-182, and miR-197 which were shown to be increased in patients, and especially those with metastasis, exhibited a significant decrease in lung cancer patients who respond to chemotherapy (Zheng et al., 2011).
Heegaard et al., in addition to identifying circulating miRNAs for diagnostic purposes in NSCLC, compared plasma and serum miRNA profile, and miRNA profile of different ethnic groups. Notably, the miRNA levels in paired plasma and serum from the early stage NSCLC patients showed no significant correlation. Reduced levels of miR-17-5p, miR-27a, miR-106a, miR-146b, miR-155, miR-221, and let-7a, and increased levels of miR-29c were detected only in the serum of NSCLC patients and not in plasma (Heegaard et al., 2012). Analogous to the findings in a breast cancer study, the miRNA profiles also displayed differences between the African American and European American ethnic groups (Zhao et al., 2010; Heegaard et al., 2012).
Colorectal Cancer
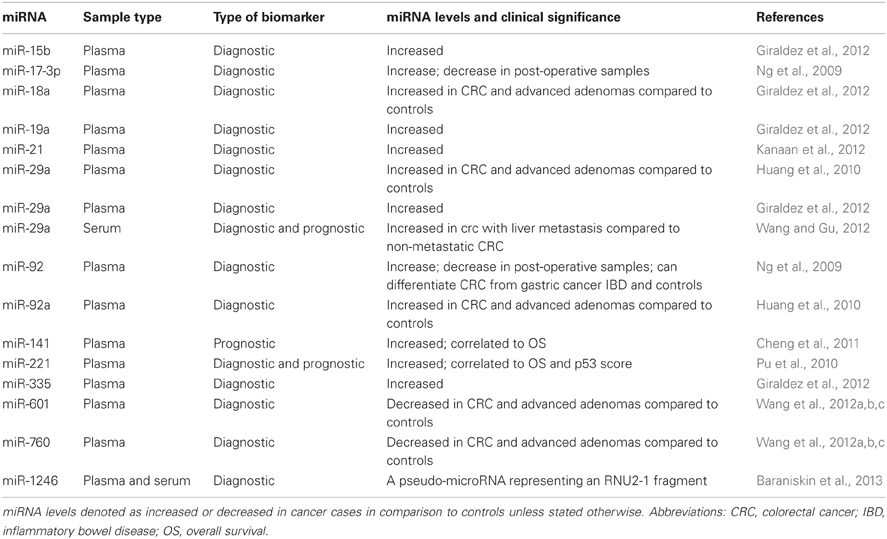
In colorectal cancer (CRC) many studies have evaluated the feasibility of circulating miRNAs for detecting early stage cancer and also as prognostic/predictive marker (Pfütze et al., 2013). Ng et al. achieved this by profiling miRNAs in tissue and plasma, and short-listing miRNAs that were differentially expressed in both. Ultimately, miR-17-3p and miR-92, both belonging to the same miRNA gene cluster and classified as oncogenic, were validated as elevated in CRC plasma and CRC tissue, in comparison to their normal counterparts. The two miRNAs were also proposed as responsive to surgery as lower levels were detected in post-operative plasma compared to pre-operative plasma, thus expanding their role as biomarkers. Interestingly, miR-92 alone had the potential to discriminate CRC samples from other diseases, such as gastric cancer (GC) and inflammatory bowel disease samples, apart from controls (Ng et al., 2009). The diagnostic potential of miR-92a was further confirmed by another study in plasma, which in addition also identified miR-29a as being increased in CRC, and similar to the previous study the miRNAs showed reduction following surgery. However, this study was not able to validate miR-17-3p as it could not be measured due to very low levels in plasma. miR-29a and miR-92a could also differentiate adenomas (pre-malignant lesions) from control samples, however their sensitivity to differentiate adenomas from carcinomas was not mentioned, which might influence their diagnostic utility (Huang et al., 2010). By miRNA profiling and subsequent validation, miR-601 and miR-760 were also suggested as potential diagnostic biomarkers of adenomas and CRC by the same group. Combining miR-29a, miR-92a, and miR-760, the detection sensitivity of early stages of CRC was further improved, especially when complemented with CEA (Wang et al., 2012a,b,c). miR-29a amounts in serum has also been reported to discriminate metastatic and non-metastatic CRC patients, specifically liver metastasis (Wang and Gu, 2012). Another study which undertook a genome-wide miRNA profiling of plasma, identified miR-15b, miR-19a, miR-19b, miR-29a, and miR-335 as being able to differentiate CRC patients from healthy individuals, while miR-18a could do so also between advanced adenomas and healthy individuals (Giraldez et al., 2012). miR-21 was identified as an early detection marker, but here, rather than profiling plasma, the strategy followed was to profile CRC and normal tissue, and validate the top hits from this discovery round in plasma (Kanaan et al., 2012). miR-1246, in spite of being designated as a pseudo-miRNA, was discerned as a diagnostic marker for pancreatic ductal adenocarcinoma and CRC in serum and plasma, and predictive for early therapy response prediction of chemo- and radiation therapy due to its decline following surgical resection. The measured signal, which was discriminating cancer and control samples, was proposed to have come from the U2 small nuclear RNA (snRNA) fragment RNU2-1f (Baraniskin et al., 2013).
Although many studies have outlined the diagnostic potential of circulating miRNAs in CRC, not many have explored their prognostic potential. One such study measured oncogenic miRNAs and identified plasma miR-221 to have prognostic prowess in addition to being diagnostic for CRC. miR-221 levels significantly correlated to OS and p53 expression (Pu et al., 2010). The only other report in this direction showed miR-141 to be highly correlated to cancer stage, complemented CEA in identifying stage IV CRC, and associated with poorer OS (Cheng et al., 2011).
Gastric Cancer
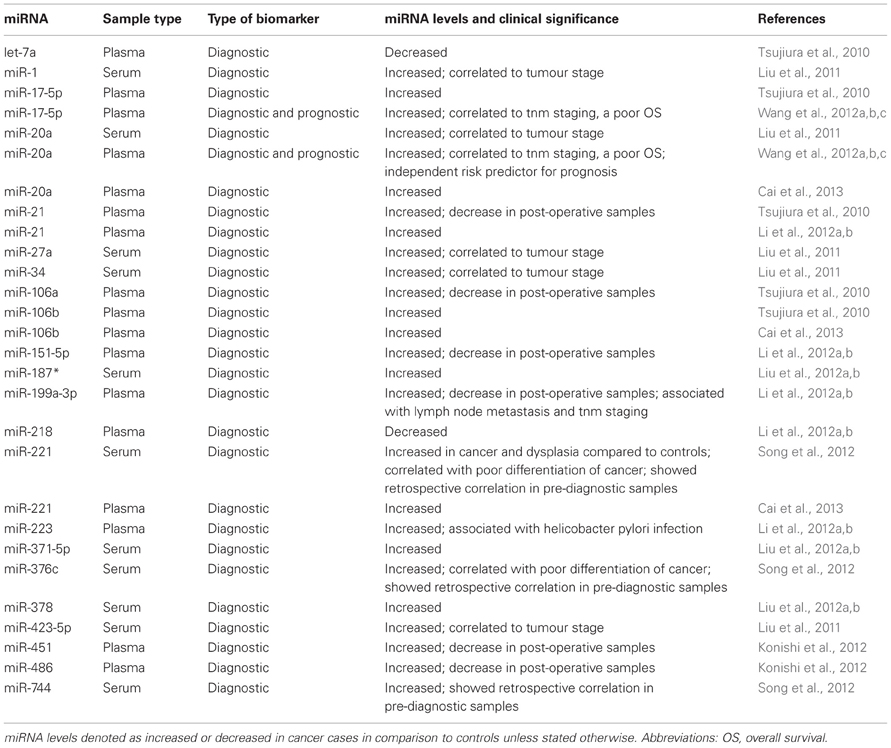
GC has one of the poorest survival rates, which makes the discovery of suitable biomarkers for early detection or prognosis all the more important (Siegel et al., 2012). Based on previous findings in GC tissue samples, miR-17-5p, miR-21, miR-106a, miR-106b, and let-7a were chosen for analysis in plasma samples. While the first four were demonstrated to be present at significantly higher levels, let-7a was lower in GC patients than in healthy individuals. In addition to this, a significant drop in the levels of miR-21 and miR-106b in post-operative paired samples, and a concordance between tissue and plasma miRNAs were also observed. The authors interpreted from these results that the levels of plasma miRNAs reflected the corresponding expression level in tumour (Tsujiura et al., 2010). Unlike this study, Liu et al. carried out a global miRNA screening in GC and healthy control serum samples, independent of miRNA profile in tissue. A five miRNA signature in serum was identified to be markedly upregulated in GC patients, and within GC samples, additionally correlated to tumour stage, thereby an advanced clinical stage. Its discriminatory power was claimed to be even higher than currently established markers, CEA and CA 19-9. This was attributed to the use of multiple miRNAs which were functionally associated to various tumorigenic processes (immune response-related: miR-20a and miR-423-5p; tissue-specific: miR-1; tumour cell growth/cycle-related: miR-27a and miR-34). This, they claimed, could prove to be more comprehensive than a conventional single protein-based biomarker (Liu et al., 2011). Another study using miRNA profiling of serum uncovered 16 upregulated miRNAs in the discovery round. These included miR-17, miR-20a, miR-21, and miR-106a/b among others, which eventually could not be validated unlike in the previous studies mentioned here. After validation, miR-221, miR-376c, and miR-744 were delineated as markers for GC detection, capable of identifying GC even 5 years prior to clinical diagnosis (Song et al., 2012). By comparing pre- and post-operative plasma samples, miR-451 and miR-486 (both lowered in post-operation) were selected as candidates and confirmed to be present in higher amounts in GC as opposed to controls (Konishi et al., 2012). One more study dealing with GC plasma samples demonstrated miR-21 and miR-223 to be significantly higher in GC patients than in healthy controls, while miR-218 was significantly lower. Interestingly, miR-223 was also specifically correlated to Helicobacter pylori infection, an etiological agent for GC (Li et al., 2012a,b). Other miRNAs proposed as diagnostic markers for GC include miR-151-5p and miR-199a-3p (Li et al., 2012a,b), and miR-221 (Cai et al., 2013) which were identified in plasma. The latter study also re-affirmed the potential of miR-20a and miR-106b to differentiate GC from controls (Cai et al., 2013). Of these, miR-199a-3p was also associated with lymph node metastasis and TNM staging, thus increasing its diagnostic importance (Li et al., 2012a,b). With respect to serum, miR-187*, miR-371-5p, and miR-378 were found to be significantly elevated in GC patients, with miR-378 being singled out as the most informative (Liu et al., 2012a,b).
Of the above mentioned miRNAs, miR-17-5p, and miR-21 were tested in an independent study to not only be elevated in GC, but also significantly correlated to TNM staging, and a poor OS, and miR-21 alone was defined as an independent prognostic marker (Wang et al., 2012a,b,c). Other than this one report, not many studies focusing on the prognostic role of circulating miRNAs in GC have been published.
Haematological Cancers
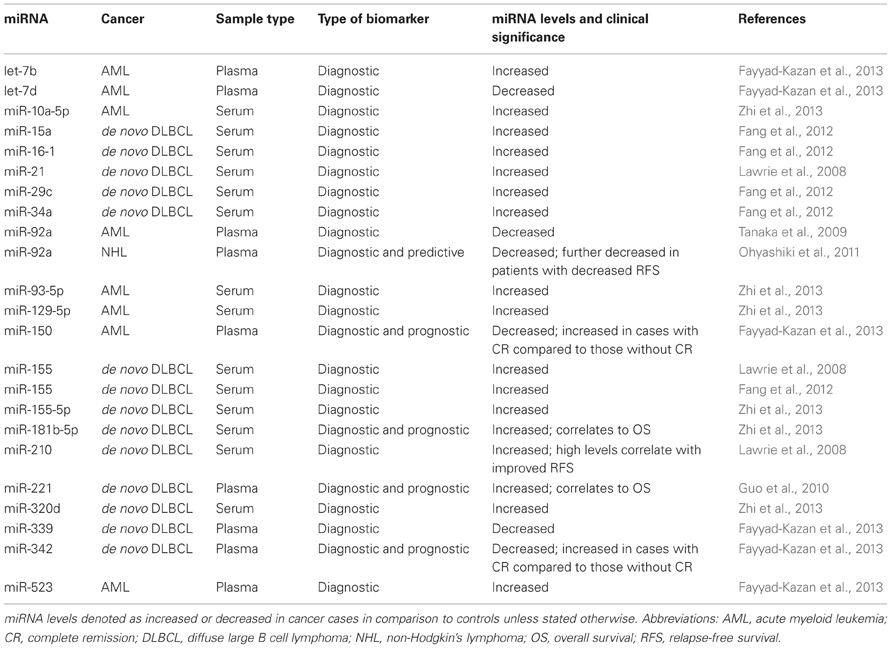
Although circulating miRNAs from plasma or serum are in close contact with their cellular counterpart and could easily reflect any abnormality of blood cells, there has been very little research in this direction. The first foray into the field of circulating miRNAs in cancer was with diffuse large B cell lymphoma (DLBCL) where tumour-associated miR-21, miR-155, and miR-210 were shown to be significantly higher in the serum of cancer patient serum than in healthy controls. In addition to its diagnostic proficiency, miR-21 also showed correlation to relapse-free survival (Lawrie et al., 2008). miR-15a, miR-16-1, miR-29c, miR-34a, and miR-155, which are known tumour-associated miRNAs, were later also defined as early detection markers for DLBCL, with miR-155 being the only common miRNA in these two studies (Lawrie et al., 2008; Fang et al., 2012). Since the miR-17-92 polycistronic miRNA cluster plays a crucial role in lymphomagenesis and affects neo-angiogenesis, one study explored the diagnostic and prognostic value of plasma miR-92a in non-Hodgkin's lymphoma (NHL). miR-92a was found to be extremely low in patients, and additionally evinced a strong correlation to relapse rates among patients (Ohyashiki et al., 2011). Another study, extrapolating the functional role of miRNAs in cancer to its diagnostic potential, screened for miR-221. Based on the hypothesis that miRNAs functionally involved in resistance to a particular chemotherapeutic drug would harbor information regarding treatment response, Guo et al. measured miR-221 (increased in adriamycin resistant cancer cells) in extra-nodal natural killer T-Cell (NK/T-cell) lymphoma, which is a very aggressive form with very poor prognosis. Plasma miR-221 was able to distinguish patients from controls, and also demonstrated prognostic value by being correlated to OS (Guo et al., 2010).
Leukemia is another type of hematological cancer where circulating miRNAs have been probed. In addition to NHL, miR-92a has also been observed to be decreased in plasma of acute myeloid leukemia (AML) patients. This study reported the ratio of miR-92a/miR-638 as having the strongest detection value for AML, where miR-638 was considered an internal reference gene (Tanaka et al., 2009). As opposed to this knowledge-based approach, a recent article outlined the identification of candidate miRNAs from microarray experiments in plasma. let-7d, miR-150, miR-339, and miR-342 were identified to be depressed, whilst let-7b, and miR-523 to be increased in AML patients at diagnosis when compared to controls. The combination of miR-150 and miR-342 was found to be the most accurate model for diagnostic purposes, and their prognostic value was also contemplated due to their elevation in patients with complete remission (Fayyad-Kazan et al., 2013). Another study on AML carried out whole miRNome sequencing to identify candidate miRNAs in serum differentially regulated from controls, and was able to successfully validate six miRNAs; miR-10a-5p, miR-93-5p, miR-129-5p, miR-155-5p, miR-181b-5p, and miR-320d. More importantly, miR-181b-5p levels in serum showed significant association with OS (Zhi et al., 2013).
Conclusion and Future Perspectives
We have seen a tremendous growth of interest in the area of circulating miRNAs that could be developed as cancer biomarkers. From the body of work presented here some interesting observations and interpretations can be made. A few miRNAs seem to be prevalent and are associated with different cancer, for e.g., oncogenic miR-17-92 cluster which shows involvement in both solid and blood cancers, hence their use as diagnostic markers could be a double edged sword for example, while they could cover a broad spectrum of cancers, they would lack specificity. Circulating miRNAs that can identify cancer development years prior to disease onset or in asymptomatic individuals, as shown in lung and GC, could be used for the screening of high-risk groups (Song et al., 2012). Different approaches have been adopted to identify these circulating miRNA biomarkers such as genome-wide profiling, knowledge-based candidate approach, analysing for those miRNAs known to be deregulated in tissue, etc., with each approach having its own advantages and disadvantages. From the conflicting results regarding concordance between miRNAs in tumour tissue and those in circulation between studies, one can infer that the circulating miRNAs have a heterogeneous origin, which has also been corroborated by other studies (Turchinovich et al., 2012). Finally, most studies propose combination of miRNAs rather than a single miRNA for discriminating cancer patients from controls, or for determining prognosis of patients, implying the advantage of using miRNAs involved in different physio-pathological pathways to obtain a complete picture.
Crucial to the translational value of any biomarker is its robustness and reproducibility. As with all upcoming fields, there are a few technical glitches, such as sample preparation for plasma or serum and data normalization, that still need to be dealt with. Establishing a standardized protocol which should be preferably followed across all studies is very important and desirable. This would facilitate comparison and (re-)affirmation of results from different cohorts and enhance detection of only those circulating miRNAs that give us information about the biological state of samples rather than confounding us with physiological or systemic differences. Most studies, with very few exceptions, deal with relatively small sample sizes and lack validation in independent cohort(s). To achieve true translational relevance and bring circulating miRNAs into routine diagnostics, we need to invest in larger study cohorts and reproducibly validate the results. Nevertheless, all the current data underlines the enormous prospective for circulating miRNAs in the field of cancer as (1) diagnostic biomarkers for early detection of cancer; (2) prognostic biomarkers helping to predict outcome and (3) predictive biomarkers which are associated with response or resistance to a particular therapy option, and thus aid in treatment planning.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Our work was supported by the Dietmar–Hopp Foundation, the Helmholtz Society, and the German Cancer Research Center (DKFZ). We apologize to those whose work was not cited owing to space constraints.
References
Baraniskin, A., Nopel-Dunnebacke, S., Ahrens, M., Jensen, S. G., Zollner, H., Maghnouj, A., et al. (2013). Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int. J. Cancer 132, e48–e57. doi: 10.1002/ijc.27791
Bianchi, F., Nicassio, F., Marzi, M., Belloni, E., Dall'olio, V., Bernard, L., et al. (2011). A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol. Med. 3, 495–503. doi: 10.1002/emmm.201100154
Boeri, M., Verri, C., Conte, D., Roz, L., Modena, P., Facchinetti, F., et al. (2011). MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 3713–3718. doi: 10.1073/pnas.1100048108
Brase, J. C., Johannes, M., Schlomm, T., Falth, M., Haese, A., Steuber, T., et al. (2011). Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 128, 608–616. doi: 10.1002/ijc.25376
Bryant, R. J., Pawlowski, T., Catto, J. W., Marsden, G., Vessella, R. L., Rhees, B., et al. (2012). Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 106, 768–774. doi: 10.1038/bjc.2011.595
Cai, H., Yuan, Y., Hao, Y. F., Guo, T. K., Wei, X., and Zhang, Y. M. (2013). Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med. Oncol. 30, 452. doi: 10.1007/s12032-012-0452-0
Chen, W., Cai, F., Zhang, B., Barekati, Z., and Zhong, X. Y. (2012a). The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumour Biol. 34, 455–462. doi: 10.1007/s13277-012-0570-5
Chen, Z. H., Zhang, G. L., Li, H. R., Luo, J. D., Li, Z. X., Chen, G. M., et al. (2012b). A panel of five circulating microRNAs as potential biomarkers for prostate cancer. Prostate 72, 1443–1452. doi: 10.1002/pros.22495
Chen, X., Ba, Y., Ma, L., Cai, X., Yin, Y., Wang, K., et al. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006. doi: 10.1038/cr.2008.282
Cheng, H., Zhang, L., Cogdell, D. E., Zheng, H., Schetter, A. J., Nykter, M., et al. (2011). Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE 6:e17745. doi: 10.1371/journal.pone.0017745
Chim, S. S., Shing, T. K., Hung, E. C., Leung, T. Y., Lau, T. K., Chiu, R. W., et al. (2008). Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 54, 482–490. doi: 10.1373/clinchem.2007.097972
Cuk, K., Zucknick, M., Heil, J., Madhavan, D., Schott, S., Turchinovich, A., et al. (2013). Circulating microRNAs in plasma as early detection markers for breast cancer. Int. J. Cancer 32, 1602–1612. doi: 10.1002/ijc.27799
Fang, C., Zhu, D. X., Dong, H. J., Zhou, Z. J., Wang, Y. H., Liu, L., et al. (2012). Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann. Hematol. 91, 553–559. doi: 10.1007/s00277-011-1350-9
Fayyad-Kazan, H., Bitar, N., Najar, M., Lewalle, P., Fayyad-Kazan, M., Badran, R., et al. (2013). Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J. Transl. Med. 11, 31. doi: 10.1186/1479-5876-11-31
Ferlay, J., Shin, H. R., Bray, F., Forman, D., Mathers, C, and Parkin, D. M. (2008). Cancer Incidence and Mortality Worldwide. GLOBOCAN (2008). v2.0. IARC CancerBase No. 10 [Internet]. Available online at: http://globocan.iarc.fr/
Foss, K. M., Sima, C., Ugolini, D., Neri, M., Allen, K. E., and Weiss, G. J. (2011). miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J. Thorac. Oncol. 6, 482–488. doi: 10.1097/JTO.0b013e318208c785
Giraldez, M. D., Lozano, J. J., Ramirez, G., Hijona, E., Bujanda, L., Castells, A., et al. (2012). Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin. Gastroenterol. Hepatol. 11, 681–688. doi: 10.1016/j.cgh.2012.12.009
Gonzales, J. C., Fink, L. M., Goodman, O. B. Jr., Symanowski, J. T., Vogelzang, N. J., and Ward, D. C. (2011). Comparison of circulating MicroRNA 141 to circulating tumor cells, lactate dehydrogenase, and prostate-specific antigen for determining treatment response in patients with metastatic prostate cancer. Clin. Genitourin. Cancer 9, 39–45. doi: 10.1016/j.clgc.2011.05.008
Guo, H. Q., Huang, G. L., Guo, C. C., Pu, X. X., and Lin, T. Y. (2010). Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis. Markers 29, 251–258. doi: 10.3233/DMA-2010-0755
Hanash, S. M., Baik, C. S., and Kallioniemi, O. (2011). Emerging molecular biomarkers–blood-based strategies to detect and monitor cancer. Nat. Rev. Clin. Oncol. 8, 142–150. doi: 10.1038/nrclinonc.2010.220
Harris, L., Fritsche, H., Mennel, R., Norton, L., Ravdin, P., Taube, S., et al. (2007). American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J. Clin. Oncol. 25, 5287–5312. doi: 10.1200/JCO.2007.14.2364
Heegaard, N. H., Schetter, A. J., Welsh, J. A., Yoneda, M., Bowman, E. D., and Harris, C. C. (2012). Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int. J. Cancer 130, 1378–1386. doi: 10.1002/ijc.26153
Hu, Z., Chen, X., Zhao, Y., Tian, T., Jin, G., Shu, Y., et al. (2010). Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 28, 1721–1726. doi: 10.1200/JCO.2009.24.9342
Huang, Z., Huang, D., Ni, S., Peng, Z., Sheng, W., and Du, X. (2010). Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 127, 118–126. doi: 10.1002/ijc.25007
Kaduthanam, S., Gade, S., Meister, M., Brase, J. C., Johannes, M., Dienemann, H., et al. (2013). Serum miR-142-3p is associated with early relapse in operable lung adenocarcinoma patients. Lung Cancer 80, 223–227. doi: 10.1016/j.lungcan.2013.01.013
Kanaan, Z., Rai, S. N., Eichenberger, M. R., Roberts, H., Keskey, B., Pan, J., et al. (2012). Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann. Surg. 256, 544–551. doi: 10.1097/SLA.0b013e318265bd6f
Keller, A., Leidinger, P., Gislefoss, R., Haugen, A., Langseth, H., Staehler, P., et al. (2011). Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol. 8, 506–516. doi: 10.4161/rna.8.3.14994
Konishi, H., Ichikawa, D., Komatsu, S., Shiozaki, A., Tsujiura, M., Takeshita, H., et al. (2012). Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br. J. Cancer 106, 740–747. doi: 10.1038/bjc.2011.588
Lawrie, C. H., Gal, S., Dunlop, H. M., Pushkaran, B., Liggins, A. P., Pulford, K., et al. (2008). Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 141, 672–675. doi: 10.1111/j.1365-2141.2008.07077.x
Le, H. B., Zhu, W. Y., Chen, D. D., He, J. Y., Huang, Y. Y., Liu, X. G., et al. (2012). Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med. Oncol. 29, 3190–3197. doi: 10.1007/s12032-012-0303-z
Li, B. S., Zhao, Y. L., Guo, G., Li, W., Zhu, E. D., Luo, X., et al. (2012a). Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS ONE 7:e41629. doi: 10.1371/journal.pone.0041629
Li, C., Li, J. F., Cai, Q., Qiu, Q. Q., Yan, M., Liu, B. Y., et al. (2012b). MiRNA-199a-3p in Plasma as a potential diagnostic biomarker for gastric cancer. Ann. Surg. Oncol. doi: 10.1245/s10434-012-2600-3. [Epub ahead of print].
Liu, H., Zhu, L., Liu, B., Yang, L., Meng, X., Zhang, W., et al. (2012a). Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 316, 196–203. doi: 10.1016/j.canlet.2011.10.034
Liu, X. G., Zhu, W. Y., Huang, Y. Y., Ma, L. N., Zhou, S. Q., Wang, Y. K., et al. (2012b). High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med. Oncol. 29, 618–626. doi: 10.1007/s12032-011-9923-y
Liu, R., Zhang, C., Hu, Z., Li, G., Wang, C., Yang, C., et al. (2011). A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur. J. Cancer 47, 784–791. doi: 10.1016/j.ejca.2010.10.025
Lumachi, F., Brandes, A. A., Ermani, M., Bruno, G., and Boccagni, P. (2000). Sensitivity of serum tumor markers CEA and CA 15-3 in breast cancer recurrences and correlation with different prognostic factors. Anticancer Res. 20, 4751–4755.
Madhavan, D., Zucknick, M., Wallwiener, M., Cuk, K., Modugno, C., Scharpff, M., et al. (2012). Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin. Cancer Res. 18, 5972–5982. doi: 10.1158/1078-0432.CCR-12-1407
Mahn, R., Heukamp, L. C., Rogenhofer, S., von Ruecker, A., Muller, S. C., and Ellinger, J. (2011). Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology 77, 1265.e9–1265.e16. doi: 10.1016/j.urology.2011.01.020
Mar-Aguilar, F., Mendoza-Ramirez, J. A., Malagon-Santiago, I., Espino-Silva, P. K., Santuario-Facio, S. K., Ruiz-Flores, P., et al. (2013). Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis. Markers 34, 163–169. doi: 10.3233/DMA-120957
Mitchell, P. S., Parkin, R. K., Kroh, E. M., Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518. doi: 10.1073/pnas.0804549105
Ng, E. K., Chong, W. W., Jin, H., Lam, E. K., Shin, V. Y., Yu, J., et al. (2009). Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58, 1375–1381. doi: 10.1136/gut.2008.167817
Ng, E. K., Li, R., Shin, V. Y., Jin, H. C., Leung, C. P., Ma, E. S., et al. (2013). Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS ONE 8:e53141. doi: 10.1371/journal.pone.0053141
Nguyen, H. C., Xie, W., Yang, M., Hsieh, C. L., Drouin, S., Lee, G. S., et al. (2012). Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate 73, 346–354. doi: 10.1002/pros.22572
Ohyashiki, K., Umezu, T., Yoshizawa, S., Ito, Y., Ohyashiki, M., Kawashima, H., et al. (2011). Clinical impact of down-regulated plasma miR-92a levels in non-Hodgkin's lymphoma. PLoS ONE 6:e16408. doi: 10.1371/journal.pone.0016408
Pfütze, K., Lou, X., and Burwinkel, B. (2013). MicroRNA Signatures as Biomarkers of Colorectal Cancer. New Jersey, NJ: Blackwell Publisher.
Pu, X. X., Huang, G. L., Guo, H. Q., Guo, C. C., Li, H., Ye, S., et al. (2010). Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J. Gastroenterol. Hepatol. 25, 1674–1680. doi: 10.1111/j.1440-1746.2010.06417.x
Roth, C., Rack, B., Muller, V., Janni, W., Pantel, K., and Schwarzenbach, H. (2010). Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 12, R90. doi: 10.1186/bcr2766
Shen, J., Hruby, G. W., McKiernan, J. M., Gurvich, I., Lipsky, M. J., Benson, M. C., et al. (2012). Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 72, 1469–1477. doi: 10.1002/pros.22499
Shen, J., Todd, N. W., Zhang, H., Yu, L., Lingxiao, X., Mei, Y., et al. (2011). Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab. Invest. 91, 579–587. doi: 10.1038/labinvest.2010.194
Si, H., Sun, X., Chen, Y., Cao, Y., Chen, S., Wang, H., et al. (2013). Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J. Cancer Res. Clin. Oncol. 139, 223–229. doi: 10.1007/s00432-012-1315-y
Siegel, R., Naishadham, D., and Jemal, A. (2012). Cancer statistics, 2012. CA Cancer J. Clin. 62, 10–29. doi: 10.3322/caac.20138
Song, M. Y., Pan, K. F., Su, H. J., Zhang, L., Ma, J. L., Li, J. Y., et al. (2012). Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS ONE. 7:e33608. doi: 10.1371/journal.pone.0033608
Sun, Y., Wang, M., Lin, G., Sun, S., Li, X., Qi, J., et al. (2012). Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS ONE 7:e47003. doi: 10.1371/journal.pone.0047003
Tanaka, M., Oikawa, K., Takanashi, M., Kudo, M., Ohyashiki, J., Ohyashiki, K., et al. (2009). Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS ONE 4:e5532. doi: 10.1371/journal.pone.0005532
Tang, D., Shen, Y., Wang, M., Yang, R., Wang, Z., Sui, A., et al. (2013). Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur. J. Cancer Prev. doi: 10.1097/CEJ.0b013e32835f3be9. [Epub ahead of print].
Tsujiura, M., Ichikawa, D., Komatsu, S., Shiozaki, A., Takeshita, H., Kosuga, T., et al. (2010). Circulating microRNAs in plasma of patients with gastric cancers. Br. J. Cancer 102, 1174–1179. doi: 10.1038/sj.bjc.6605608
Turchinovich, A., and Burwinkel, B. (2012). Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 9, 1066–1075. doi: 10.4161/rna.21083
Turchinovich, A., Weiz, L., and Burwinkel, B. (2012). Extracellular miRNAs: the mystery of their origin and function. Trends Biochem. Sci. 37, 460–465. doi: 10.1016/j.tibs.2012.08.003
Turchinovich, A., Weiz, L., Langheinz, A., and Burwinkel, B. (2011). Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233. doi: 10.1093/nar/gkr254
Uehara, M., Kinoshita, T., Hojo, T., Akashi-Tanaka, S., Iwamoto, E., and Fukutomi, T. (2008). Long-term prognostic study of carcinoembryonic antigen (CEA) and carbohydrate antigen 15-3 (CA 15-3) in breast cancer. Int. J. Clin. Oncol. 13, 447–451. doi: 10.1007/s10147-008-0773-3
van Schooneveld, E., Wouters, M. C., Van der Auwera, I., Peeters, D. J., Wildiers, H., Van Dam, P. A., et al. (2012). Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 14, R34. doi: 10.1186/bcr3127
Wang, H., Tan, G., Dong, L., Cheng, L., Li, K., Wang, Z., et al. (2012a). Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS ONE 7:e34210. doi: 10.1371/journal.pone.0034210
Wang, M., Gu, H., Wang, S., Qian, H., Zhu, W., Zhang, L., et al. (2012b). Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol. Med. Rep. 5, 1514–1520. doi: 10.3892/mmr.2012.828
Wang, Q., Huang, Z., Ni, S., Xiao, X., Xu, Q., Wang, L., et al. (2012c). Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS ONE 7:e44398. doi: 10.1371/journal.pone.0044398
Wang, L. G., and Gu, J. (2012). Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 36, e61–e67. doi: 10.1016/j.canep.2011.05.002
Wei, J., Gao, W., Zhu, C. J., Liu, Y. Q., Mei, Z., Cheng, T., et al. (2011). Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin. J. Cancer 30, 407–414. doi: 10.5732/cjc.010.10522
Zeng, R. C., Zhang, W., Yan, X. Q., Ye, Z. Q., Chen, E. D., Huang, D. P., et al. (2013). Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med. Oncol. 30, 477. doi: 10.1007/s12032-013-0477-z
Zhang, H. L., Yang, L. F., Zhu, Y., Yao, X. D., Zhang, S. L., Dai, B., et al. (2011). Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 71, 326–331. doi: 10.1002/pros.21246
Zhao, H., Shen, J., Medico, L., Wang, D., Ambrosone, C. B., and Liu, S. (2010). A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS ONE 5:e13735. doi: 10.1371/journal.pone.0013735
Zheng, D., Haddadin, S., Wang, Y., Gu, L. Q., Perry, M. C., Freter, C. E., et al. (2011). Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int. J. Clin. Exp. Pathol. 4, 575–586.
Zhi, F., Cao, X., Xie, X., Wang, B., Dong, W., Gu, W., et al. (2013). Identification of circulating micrornas as potential biomarkers for detecting acute myeloid leukemia. PLoS ONE 8:e56718. doi: 10.1371/journal.pone.0056718
Keywords: circulating miRNA, cancer, biomarkers, prognostic marker, detection marker
Citation: Madhavan D, Cuk K, Burwinkel B and Yang R (2013) Cancer diagnosis and prognosis decoded by blood-based circulating microRNA signatures. Front. Genet. 4:116. doi: 10.3389/fgene.2013.00116
Received: 22 April 2013; Accepted: 01 June 2013;
Published online: 21 June 2013.
Edited by:
Arun Bhardwaj, University of South Alabama Mitchell Cancer Institute, USAReviewed by:
Sumit Arora, University of south Alabama, USATarun K. Sharma, Translational Health Science and Technology Institute, India
Copyright © 2013 Madhavan, Cuk, Burwinkel and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Dharanija Madhavan, Molecular Epidemiology, C080, German Cancer Research Center (DKFZ), Heidelberg, Germany; Molecular Biology of Breast Cancer, Department of Gynecology and Obstetrics, University Clinic, Heidelberg, Vossstrasse 9, D-69115 Heidelberg, Germany e-mail: d.madhavan@dkfz.de
†These authors have contributed equally to this work.