- 1Division of Cardiovascular Diseases, Mayo Clinic, Rochester, MN, USA
- 2Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA
- 3Boston Children's Hospital, Boston, MA, USA
- 4Department of Genetics and Genomic Sciences, Charles R. Bronfman Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, USA
- 5Center for Biomedical Ethics and Society, Department of Pediatrics, Vanderbilt University School of Medicine, Nashville, TN, USA
- 6Biomedical Ethics Program, Mayo Clinic, Rochester, MN, USA
- 7Feinberg School of Medicine, Northwestern University, Chicago, IL, USA
- 8The Sigfried and Janet Weis Center for Research, Geisinger Health System, Danville, PA, USA
- 9Center for Applied Genomics, Abramson Research Center, The Children's Hospital of Philadelphia, Philadelphia, PA, USA
- 10Research Division, Essentia Institute of Rural Health, Duluth, MN, USA
- 11Genomic Medicine Institute, Geisinger Health System, Danville, PA, USA
- 12Departments of Medicine (Medical Genetics) and Genome Sciences, University of Washington, Seattle, WA, USA
The electronic Medical Records and Genomics (eMERGE) (Phase I) network was established in 2007 to further genomic discovery using biorepositories linked to the electronic health record (EHR). In Phase II, which began in 2011, genomic discovery efforts continue and in addition the network is investigating best practices for implementing genomic medicine, in particular, the return of genomic results in the EHR for use by physicians at point-of-care. To develop strategies for addressing the challenges of implementing genomic medicine in the clinical setting, the eMERGE network is conducting studies that return clinically-relevant genomic results to research participants and their health care providers. These genomic medicine pilot studies include returning individual genetic variants associated with disease susceptibility or drug response, as well as genetic risk scores for common “complex” disorders. Additionally, as part of a network-wide pharmacogenomics-related project, targeted resequencing of 84 pharmacogenes is being performed and select genotypes of pharmacogenetic relevance are being placed in the EHR to guide individualized drug therapy. Individual sites within the eMERGE network are exploring mechanisms to address incidental findings generated by resequencing of the 84 pharmacogenes. In this paper, we describe studies being conducted within the eMERGE network to develop best practices for integrating genomic findings into the EHR, and the challenges associated with such work.
Introduction
The availability and reduced costs of high-density genotyping and genome sequencing technologies has accelerated genomic discovery (Green et al., 2011). Genome-wide association studies (GWAS) have revealed numerous common genetic variants that influence susceptibility to disease and adverse drug reactions, as well as inter-individual variation in quantitative traits and drug response (Manolio, 2013). Next-generation sequencing has also enabled discovery of variants associated with rare heritable diseases (Yang et al., 2013). Assessing the utility and identifying best practices for integration of this new genomic knowledge into clinical practice to improve patient care is now a major focus in the area of translational genomics (Kullo et al., 2013; Manolio et al., 2013).
The electronic Medical Records and Genomics (eMERGE) network (see Supplementary Figure) was established in 2007 with support from the National Human Genome Research Institute (NHGRI) to further genomic discovery using biorepositories linked to the electronic health record (EHR). The initial phase (Phase I) of the eMERGE network included five sites: Group Health Cooperative/University of Washington, Marshfield Clinic, Mayo Clinic, Northwestern University, and Vanderbilt University. The network was successful in leveraging the EHR to discover new genetic associations including variants that influence traits such as hematologic traits, lipid levels, Alzheimer disease, and electrocardiographic intervals among many others (Kullo et al., 2010; Naj et al., 2011; Ding et al., 2012, 2013; Rasmussen-Torvik et al., 2012; Crosslin et al., 2013). Phase II of eMERGE began in August 2011 with the addition of Geisinger Health System and Mount Sinai Medical Center to the network. In August 2012, three pediatric sites joined the network: Children's Hospital of Philadelphia, and a joint membership of Cincinnati Children's Hospital Medical Center and Boston Children's Hospital. As was the case for Phase I sites, each new site has its own biorepository linked to clinical phenotypes obtained from the EHR. While continuing efforts at genomic discovery, eMERGE Phase II is also investigating ways of incorporating genomics into the clinical setting (Gottesman et al., 2013), in particular, the return of genomic results in the EHR for use by healthcare providers at the point of care.
Although the focus in Phase I of eMERGE was primarily on genomic discovery efforts, eMERGE investigators also reviewed what research findings should be considered for return to participants (Fullerton et al., 2012). A return of results working group was set up to address the types of genomic findings that might be returned to patients or participants in the future. The genetic data were generated from high-density genotyping arrays and the return of results working group considered two main categories of genomic findings for return to participants. The first related to common variants on these arrays that may have clinical utility, e.g., Factor V Leiden, and hemochromatosis HFE variants. The second related to sex chromosome abnormalities such as Klinefelter and Turner syndromes incidentally discovered on signal intensity analysis of fluorescent data from the genotyping arrays. The investigators agreed that the potential to change immediate medical care was an important criterion for considering return of a result. Based on this criterion, homozygosity of single nucleotide polymorphism (SNP) rs6025 (R506Q, also known as the Factor V Leiden mutation) and rs1800562 (HFE C282Y, associated with hereditary hemochromatosis) were judged to have the highest clinical relevance (Fullerton et al., 2012). Sex chromosome abnormalities were also considered for return to participants, although context, such as the age of the participant, was felt to be an important factor. The working group summarized these recommendations in a position paper (Fullerton et al., 2012).
In Phase II of eMERGE, genomic discovery efforts continue, but the focus has expanded to include clinical implementation of genomic data. In addition to high-density genotype data in a large number of patients (see Supplementary Table) (Gottesman et al., 2013), targeted next-generation sequencing data of 84 ‘very important pharmacogenes’ (VIPs) will be available for nearly 10,000 patients across the network as part of the eMERGE PGx project. Genetic data being considered for return to participants include SNPs of medical or pharmacogenetic relevance, genetic risk scores for common diseases as well as pharmacogenomic variants, and clinically actionable incidental findings related to next-generation sequencing of 84 VIPs (e.g., RYR1 and CACNA1S genes associated with malignant hyperthermia). Candidate variants for possible return to participants and incorporation into EHR, will be validated in a CLIA-certified laboratory. Clinical integration of genomic data is being explored with active examination of the ethical, legal, and social implications of such integration for both patients and clinicians.
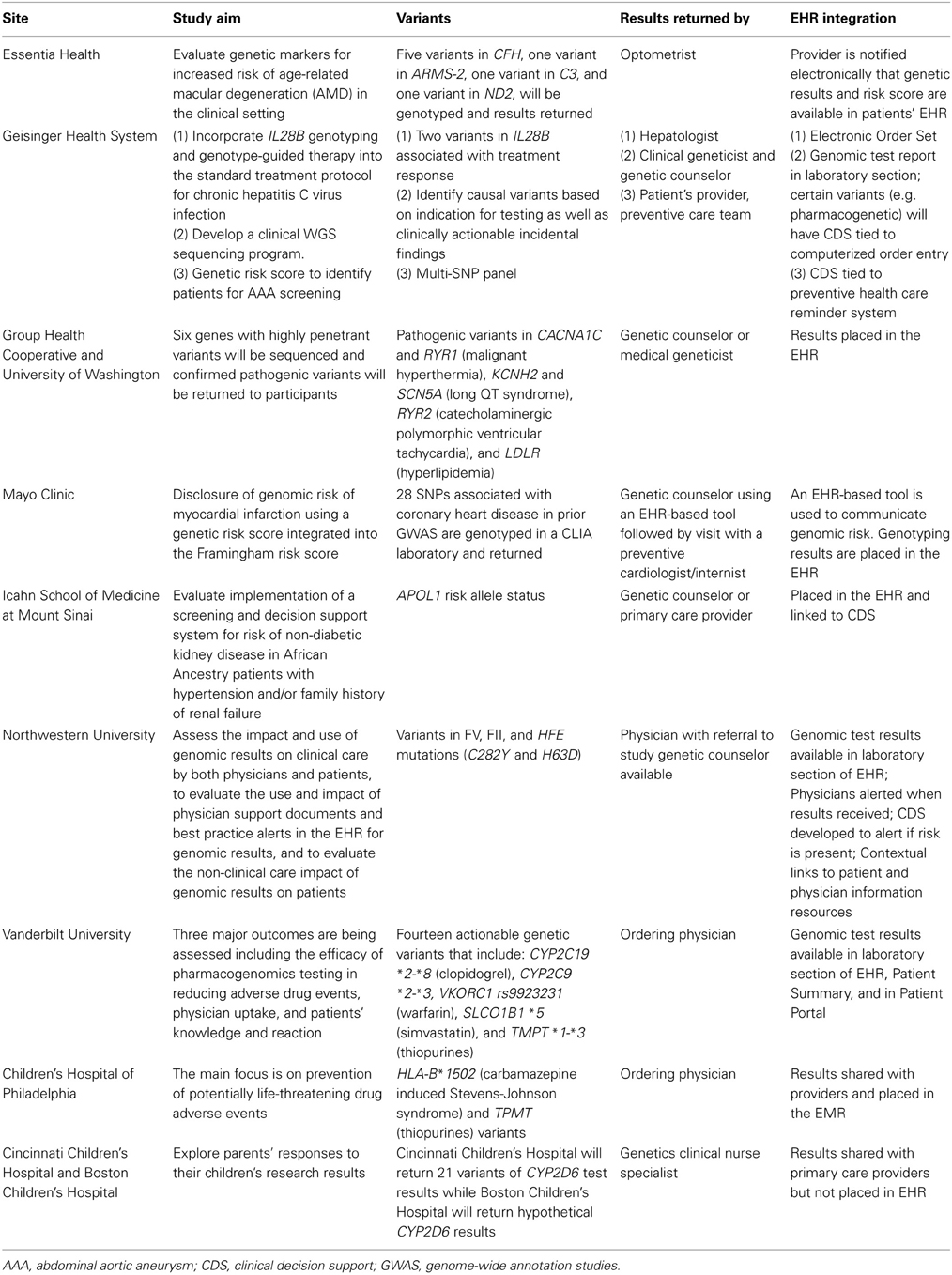
To start addressing the opportunities and challenges in returning genomic results in the clinical setting, each site is conducting genomic medicine pilot projects to return clinically relevant genomic results to participating patients and their health care providers. Additionally, pharmacogenomic information is being placed preemptively in the EHR as part of the network-wide eMERGE PGx project. In this article, we review the current status of the genomic medicine pilot projects at each eMERGE site and the challenges associated with such work. The aim of each project and the genetic variants being considered for return are summarized in Table 1. The categories of genomic results being considered for return to patients in eMERGE II include individual or multiplexed SNPs that influence disease susceptibility or drug responses. The following sections provide an overview of the activities in eMERGE II related to return of results in these pilot genomic medicine implementation projects. A separate manuscript is planned to address return of results in the setting of genomic discovery.
Individual SNPs
Individual SNPs associated with disease risk or of pharmacogenomic relevance are being considered for return of results studies within the eMERGE network, and results of this type are being returned in two eMERGE II pilot studies. A variant in the apolipoprotein L1 (APOL1) gene that is associated with non-diabetic chronic kidney disease (CKD) in patients of African American ancestry (Tzur et al., 2010; Parsa et al., 2013) is being returned in a pilot study at the Mount Sinai Medical Center. To understand processes and the impact of implementing a screening and decision support system for risk of non-diabetic CKD in African American patients with hypertension and/or family history of renal failure, ~40 participants will be genotyped for three risk variants in exon 6 of APOL1. A catalog of current evidence-based guidelines for the management of hypertension and chronic kidney disease will group the participants into three renal care advice message categories including (i) evaluation of CKD, (ii) identification of CKD progression, and (iii) prevention of CKD progression. In-depth, audio-recorded qualitative interviews are being conducted with patients before and after they receive their APOL1 results. Analysis of the transcribed interviews will inform the design of the clinical decision support and patient education materials, and the design of quantitative questionnaires for use in a planned larger study of returning APOL1 results to patients in clinical practice.
At Northwestern University, investigators are following up on the recommendations from the Return of Results workgroup in eMERGE I to include return of potentially actionable findings by reconsenting 150 biobank participants who were genotyped during Phase I of eMERGE. Participants will undergo CLIA-certified genotyping of the Factor V Leiden mutation and the hereditary hemochromatosis HFE mutations (C282Y and H63D). Results will be deposited in the EHR and available to physicians. Study participants will be informed of their results via a letter that will be sent by post or accessible via an online patient web-portal called MyChart). Participants will also complete a baseline survey and follow-up surveys at 1 and 6 months after receiving results. Physicians and participants will also have the opportunity to discuss the results during an appointment or be referred to a study genetic counselor. Semi-structured interviews will be conducted with physicians after results are returned to patients and clinical decision support has been triggered in the EHR. The impact and use of genomic results on both physicians and patients will be assessed, as will the use and impact of physician support documents and best practice alerts in the EHR for genomic results.
Pharmacogenetic Variants
At the Vanderbilt University site, 14 pharmacogenetic variants including: CYP2C19 *2–*8 (clopidogrel), CYP2C9 *2–*3, VKORC1 rs9923231 (warfarin), SLCO1B1 *5 (simvastatin), and TMPT *1–*3 (thiopurines) will be genotyped in their study population. Vanderbilt University has conducted a number of research studies to assess the efficacy of the Pharmacogenomic Resource for Enhanced Decision in Care and Treatment (PREDICT) program in reducing adverse drug events, physician uptake, and patients' knowledge and attitudes toward such testing. Physician surveys and structured interviews have been performed. Interviews have been conducted in the following groups of patients who have been through the interventional cardiology clinic: (i) no medication change following PREDICT, (ii) a change in clopidogrel medication dosing, (iii) change in statin medication dosing, and (iv) not enrolled in PREDICT. Results of these surveys and interviews are awaited.
At the University of Washington-Group Health site, pathogenic variants in six highly penetrant pharmacogenes will be genotyped in ~450 participants. These variants include risk alleles in CACNA1C (malignant hyperthermia), RYR1 (malignant hyperthermia), KCNH2 (long QT syndrome), SCN5A (long QT syndrome), RYR2 (catecholaminergic polymorphic ventricular tachycardia), and LDLR (hyperlipidemia). Results are expected to be returned through the Department of Clinical Genetics with appropriate counseling and subsequent documentation in the EHR. Patients will be surveyed regarding their experience. Healthcare providers will be surveyed to assess their use of genetic information in EHR, ease of use, completeness of information in the EHR, and whether any other resources are needed. Additionally, feasibility of implementing genomic clinical decision support will be assessed by interviews with patients and healthcare providers to guide development and testing of prototype interfaces for the EHR.
In a clinical implementation project at Geisinger Health System, all newly diagnosed patients with chronic HCV are genotyped in a CLIA laboratory for two variants in the interleukin 28B gene (IL28B) that influence treatment response and can influence medication choice. A genotype-guided treatment decision tree was created in consultation with expert clinicians and an electronic order set was implemented in the EHR to insure that all eligible patients underwent testing. Genotype results for any participant who is prescribed interferon alpha and ribavirin for chronic HCV infection are placed in the EHR and are available to hepatologists initiating treatment. The economic impact of use of these variants is being studied.
Investigators at Children's Hospital of Philadelphia are working on developing tools to engage local practitioners in targeted intervention projects trialing in-house web-based software that integrates with the EHR (Fiks et al., 2013). This new tool can be used to query the institutional biobank as well. So far, 515 patients have been genotyped for HLA-B*1502 which is associated with risk of developing Stevens-Johnson syndrome following use of carbamazepine (Chen et al., 2011), in addition to 318 patients genotyped for TPMT in patients that may be treated with thiopurines.
Investigators at Cincinnati Children's Hospital Medical Center are studying the return of CYP2D6 variants in pediatric patients in the content of codeine response, and returning these results to the parents. Variants of CYP2D6 are genotyped using Taqman and long polymerase chain reaction for full gene deletion and duplication. At the time results are returned to parents, they complete a telephone survey about their reactions and plans to share the actual results, and their anticipated preferences regarding receiving hypothetical incidental findings. Follow-up telephone calls are being conducted at 3 and 12 months post-result disclosure to learn how results were used. A subset of parents are also participating in qualitative interviews to further explore their reactions and perceptions to receiving their children's CYP2D6 results. Although results will not be placed in the EHR, the researchers are asking parents for permission to share the results with their child's primary care providers and will evaluate primary care providers' reactions to receiving the results. At the Boston Children's Hospital site, the same study, including the same survey and qualitative interviews, is being carried out but using hypothetical, not actual CYP2D6 results, and without the follow-up telephone calls. Providers will be surveyed about the perceived utility of pharmacogenomics research results in their practice. Between the two sites in Cincinnati and Boston, parents of 400 children will be enrolled. To date, Cincinnati Children's has returned 48 children's CYP2D6 results to parents and has recontacted 21 parents, all of whom have given permission to share the results with their children's primary care providers.
Genetic Risk Scores
Genetic findings of clinical relevance from high-density genotyping arrays include numerous common alleles that influence disease susceptibility. Because most individual variants have modest effect sizes, investigators are exploring the utility of combining the risk variants into genetic risk scores. At the Marshfield/Essentia Health site, a genetic risk score for age-related macular degeneration (AMD) is calculated based on five variants in CFH, one variant in ARMS-2, one variant in C3 and one variant in ND2. Individuals (n = 100) attending optometry clinics are genotyped for these variants (Haines et al., 2005) and a genetic risk score is calculated and incorporated into the EHR by optometrists. Overall risk will be calculated from the genotypes and the patient's smoking status. The results will be shared with the study participants and a telephone survey will be conducted. The participating clinicians will be interviewed after reviewing these results. Recruitment for this study began in June 2013.
At Mayo Clinic, investigators are conducting the Myocardial Infarction Genes (MI-GENES) clinical trial, a pilot study of communicating genetic risk for coronary heart disease (CHD), the leading cause of death in the US. Patients at intermediate risk for CHD based on conventional risk factors will undergo genotyping of ~28 SNPs that are associated with CHD independent of lipid or blood pressure levels, in a CLIA laboratory (Deloukas et al., 2013). Study participants will be randomized to receive a Framingham risk score or a modified Framingham risk score that incorporates a genetic risk score based on the genotyping results. The genetic risk scores will be placed in the EHR and an EHR-based pictogram will be used to assist in discussing genomic risk of CHD. Patients in the study will be followed for at least 6 months to assess the extent to which there are differences in diet, physical activity, and other lifestyle modifications associated with the communication of CHD-related genomic risk information. Participants will also be evaluated for psychosocial and behavioral changes. Recruitment for MI-GENES started in October 2013.
At Geisinger Health System, investigators are combining clinical risk factors and genomic information to develop a risk score for abdominal aortic aneurysm (AAA), a leading cause of death in older men (Kent et al., 2010; Kuivaniemi et al., 2012). The goal is to develop strategies for population screening that combine genetic susceptibility variants with clinical risk factor data mined from EHR. Patients in the Geisinger MyCode biobank (Gerhard et al., 2013; Gottesman et al., 2013) will be genotyped for variants known to be associated with AAA (Kuivaniemi et al., 2013) and this information used to create a risk score that will then be evaluated in the Geisinger patient population after performing abdominal ultrasonography examination.
Targeted Next-Generation Sequencing
The eMERGE pharmacogenomics (PGx) project is using targeted sequencing of 84 pharmacogenes to initiate a multi-site test of the concept that genomic sequence information can be coupled to EHRs for use in the clinical setting (Gottesman et al., 2013). The PGRNseq uses a reagent to “capture” exonic sequences of 84 genes important in pharmacokinetic or pharmacodynamics processes for sequencing on next-generation platforms. Genotypes of established pharmacogenomics utility that influence use of simvastatin, clopidogrel, and warfarin will be confirmed in a CLIA environment and be placed pre-emptively in the EHRs of patients who are “at risk” of receiving these drugs.
Sequence information will also be generated for six genes for which the American College of Medical Genetics and Genomics (ACMG) guidelines suggest returning known or expected pathogenic variants given the association with highly penetrant actionable disorders (Green et al., 2013) (Table 2). These include genes associated with long QT syndrome genes (KCNH2 and SCN5A), malignant hyperthermia (RYR1 and CACNA1S), hypercholesterolemia (LDLR), and catecholaminergic polymorphic ventricular tachycardia (RYR2).
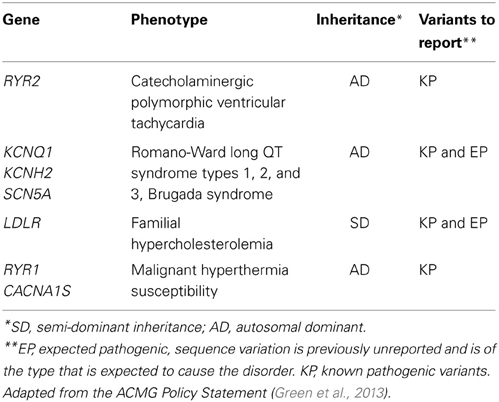
Table 2. Pharmacogenes being sequenced in the eMERGE PGx project and on the ACMG incidental finding list (Green et al., 2013).
ACMG guidelines emphasize return of mutations in these genes that are known to be pathogenic but also suggest returning novel mutations that are “expected” to be pathogenic. The recommendations have stimulated considerable debate (Green et al., 2013), and in particular, whether patient preferences can or should be incorporated into the return of results pipeline has been highlighted (Allyse and Michie, 2013). While the ACMG recommendations do not apply to research participants, members of the eMERGE network's Consent, Education, Regulation and Consultation (CERC) workgroup have weighed in with concerns about the scope of the ACMG position (Burke and Grefenstette, 2013; Ross et al., 2013). The mechanisms for whether and how to return incidental findings are being evaluated at each site. Several sites, in consultation with respective IRBs, are considering confirming pathogenic incidentally found variants for example in RYR1 and CACNA1S, with an orthogonal genotyping method. This would be followed by inviting the study patient to a genetic counseling session during which the option of knowing the research results with confirmation in a CLIA lab will be discussed. Table 3 summarizes the approach of the various sites toward returning incidental findings generated as part of the eMERGE PGx project.
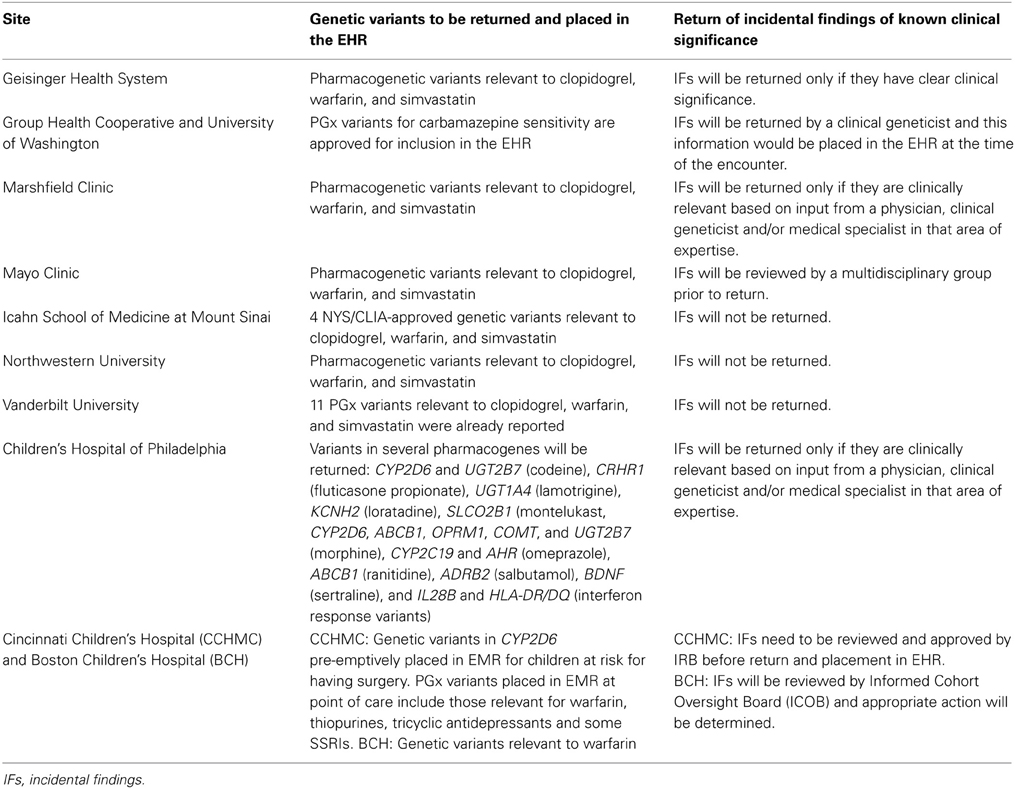
Table 3. Return of results related to the eMERGE pharmacogenomics (PGx) projects at each of the eMERGE network sites.
Whole Genome/Exome Sequencing
Whole genome and exome sequencing are being increasingly utilized in the clinical setting (Yang et al., 2013). Although less than 2% of adults appear to have relevant actionable incidental findings from whole exome sequencing (Dorschner et al., 2013) whether to return such findings is an important topic of debate (Green et al., 2013), particularly in the context of preserving patient autonomy and confidentiality (Klitzman et al., 2013). Institutional committees with diverse expertise will need to adapt guidelines proposed by the ACMG to determine whether and how these can be applied locally, which results will be reported through local EHRs and what format these reports will take. The Clinical Sequencing Exploratory Research (CSER) consortium is also actively investigating return of results in studies utilizing whole genome or whole exome sequencing (https://cser-consortium.org). Several of the eMERGE sites have conducted pilot studies of whole genome/exome sequencing in the clinical setting. For example, at Geisinger Health System, investigators are developing a laboratory report that summarizes results of whole genome sequencing in individuals with intellectual disability and normal chromosomal microarray that have been recruited along with their parents to undergo whole genome sequencing to identify an underlying genetic etiology. Causal variants and incidental findings (from the ACMG list) will be validated using Sanger sequencing. All patients will be informed about the results and undergo counseling. Qualitative data from interviews will be collated and used to improve this process.
Integration of Genomic Findings into the EHR
A recent theme issue of Genetics in Medicine addressed the issues related to integrating genomic findings into the EHR with linkage to clinical decision support at point of care (Kannry and Williams, 2013). Lack of standardized nomenclature for genetic variants is a major hurdle to creating automated decision support. Currently, several groups, including the Health Level 7 Genomics Work Group, are attempting to address this challenge. Additional challenges relate to a number of ethical, legal, and social implications that have been reviewed elsewhere (Hartzler et al., 2013; Hazin et al., 2013). These include uniform provision of genomic CDS to prevent worsening of disparities in healthcare, education of patients and providers, determining which genomic information to include in the EHR, managing incidental findings, privacy and documentation, storage and reinterpretation of genomic data and the results of stakeholder engagement in making these determinations. Education of patients and care providers will be necessary to facilitate the process of return of genomic results. The CERC Work Group in eMERGE has begun to address some of the education issues through jointly developing education materials and a website for patient information (www.myresults.org) and contributing to evaluation of online pharmacogenomics information for physicians. The CERC Working Group also has collaborative interactions with the CSER and Return of Results consortia.
Summary
Genomic technology has advanced greatly through the last decade. As we attempt to implement genomic medicine, many challenges arise, including the need to develop suitable approaches to return results and to deal with incidental findings. We have summarized activities within the eMERGE network that are related to returning genomic results in the EHR setting and also with the return of incidental findings. Initial experiences within the eMERGE network highlight the need for additional studies that address the issues related to disclosure of results from genomic implementation studies. Ongoing work in the eMERGE network will provide important insights into best practices for returning genomic results and dealing with incidental findings using the EHR.
Conflict of Interest Statement
The Review Editor Yiran Guo declares that, despite being affiliated to the same institution as authors John J. Connolly and Brendan J. Keating, the review process was handled objectively and no conflict of interest exists.
Acknowledgments
The eMERGE Network is funded by NHGRI, with additional funding from NIGMS through the following grants: U01HG04599 and U01HG006379 to Mayo Clinic; U01HG004610 and U01HG006375 to Group Health Cooperative; U01HG004608 to Marshfield Clinic; U01HG006389 to Essentia Institute of Rural Health; U01HG004609 and U01HG006388 to Northwestern University; U01HG04603 and U01HG006378 to Vanderbilt University; U01HG006385 to the Coordinating Center; U01HG006382 to Geisinger Clinic; U01HG006380 to Icahn School of Medicine at Mount Sinai; U01HG006830 to The Children's Hospital of Philadelphia; and U01HG006828 to Cincinnati Children's Hospital and Boston Children's Hospital. Additional funding included “Utility of genomic data in population screening for abdominal aortic aneurysm” from The Commonwealth Universal Research Enhancement (CURE) program of the Commonwealth of Pennsylvania (Geisinger).
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fgene.2014.00050/abstract
References
Allyse, M., and Michie, M. (2013). Not-so-incidental findings: the ACMG recommendations on the reporting of incidental findings in clinical whole genome and whole exome sequencing. Trends Biotechnol. 31, 439–441. doi: 10.1016/j.tibtech.2013.04.006
Burke, D. S., and Grefenstette, J. J. (2013). Toward an integrated meta-model of public health dynamics for preparedness decision support. J. Public Health Manag. Pract. 19 (Suppl. 2), S12–S15. doi: 10.1097/PHH.0b013e31828a842f
Chen, P., Lin, J. J., Lu, C. S., Ong, C. T., Hsieh, P. F., Yang, C. C., et al. (2011). Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. N. Engl. J. Med. 364, 1126–1133. doi: 10.1056/NEJMoa1009717
Crosslin, D. R., McDavid, A., Weston, N., Zheng, X., Hart, E., de Andrade, M., et al. (2013). Genetic variation associated with circulating monocyte count in the eMERGE Network. Hum. Mol. Genet. 22, 2119–2127. doi: 10.1093/hmg/ddt010
Deloukas, P., Kanoni, S., Willenborg, C., Farrall, M., Assimes, T. L., Thompson, J. R., et al. (2013). Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 45, 25–33. doi: 10.1038/ng.2480
Ding, K., de Andrade, M., Manolio, T. A., Crawford, D. C., Rasmussen-Torvik, L. J., Ritchie, M. D., et al. (2013). Genetic variants that confer resistance to malaria are associated with red blood cell traits in African-Americans: an electronic medical record-based genome-wide association study. G3 3, 1061–1068. doi: 10.1534/g3.113.006452
Ding, K., Shameer, K., Jouni, H., Masys, D. R., Jarvik, G. P., Kho, A. N., et al. (2012). Genetic Loci implicated in erythroid differentiation and cell cycle regulation are associated with red blood cell traits. Mayo Clin. Proc. 87, 461–474. doi: 10.1016/j.mayocp.2012.01.016
Dorschner, M. O., Amendola, L. M., Turner, E. H., Robertson, P. D., Shirts, B. H., Gallego, C. J., et al. (2013). Actionable, pathogenic incidental findings in 1,000 participants' exomes. Am. J. Hum. Genet. 93, 631–640. doi: 10.1016/j.ajhg.2013.08.006
Fiks, A. G., Grundmeier, R. W., Mayne, S., Song, L., Feemster, K., Karavite, D., et al. (2013). Effectiveness of decision support for families, clinicians, or both on HPV vaccine receipt. Pediatrics 131, 1114–1124. doi: 10.1542/peds.2012-3122
Fullerton, S. M., Wolf, W. A., Brothers, K. B., Clayton, E. W., Crawford, D. C., Denny, J. C., et al. (2012). Return of individual research results from genome-wide association studies: experience of the Electronic Medical Records and Genomics (eMERGE) Network. Genet. Med. 14, 424–431. doi: 10.1038/gim.2012.15
Gerhard, G. S., Carey, D. J., and Steele, G. D. Jr. (2013). “Electronic health records in genomic medicine,” in Genomic and Personalized Medicine, eds. G. S. Ginsburg and H. F. Willard (London: Academic Press) 287–294.
Gottesman, O., Kuivaniemi, H., Tromp, G., Faucett, W. A., Li, R., Manolio, T. A., et al. (2013). The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet. Med. 15, 761–771. doi: 10.1038/gim.2013.72
Green, E. D., Guyer, M. S., Manolio, T. A., and Peterson, J. L. (2011). Charting a course for genomic medicine from base pairs to bedside. Nature 470, 204–213. doi: 10.1038/nature09764
Green, R. C., Berg, J. S., Grody, W. W., Kalia, S. S., Korf, B. R., Martin, C. L., et al. (2013). ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 15, 565–574. doi: 10.1038/gim.2013.73
Haines, J. L., Hauser, M. A., Schmidt, S., Scott, W. K., Olson, L. M., Gallins, P., et al. (2005). Complement factor H variant increases the risk of age-related macular degeneration. Science 308, 419–421. doi: 10.1126/science.1110359
Hartzler, A., McCarty, C. A., Rasmussen, L. V., Williams, M. S., Brilliant, M., Bowton, E. A., et al. (2013). Stakeholder engagement: a key component of integrating genomic information into electronic health records. Genet. Med. 15, 792–801. doi: 10.1038/gim.2013.127
Hazin, R., Brothers, K. B., Malin, B. A., Koenig, B. A., Sanderson, S. C., Rothstein, M. A., et al. (2013). Ethical, legal, and social implications of incorporating genomic information into electronic health records. Genet. Med. 15, 810–816. doi: 10.1038/gim.2013.117
Kannry, J. M., and Williams, M. S. (2013). Integration of genomics into the electronic health record: mapping terra incognita. Genet. Med. 15, 757–760. doi: 10.1038/gim.2013.102
Kent, K. C., Zwolak, R. M., Egorova, N. N., Riles, T. S., Manganaro, A., Moskowitz, A. J., et al. (2010). Analysis of risk factors for abdominal aortic aneurysm in a cohort of more than 3 million individuals. J. Vasc. Surg. 52, 539–548. doi: 10.1016/j.jvs.2010.05.090
Klitzman, R., Appelbaum, P. S., and Chung, W. (2013). Return of secondary genomic findings vs patient autonomy: implications for medical care. JAMA 310, 369–370. doi: 10.1001/jama.2013.41709
Kuivaniemi, H., Ryer, E. J., Yoon, H. R., and Elmore, J. R. (2013). “Genetic risk factors for abdominal aortic aneurysms,” in Aortic Aneurysms: Risk Factors, Diagnosis, Surgery & Repair, eds D. Fischof and F. Hatig (Hauppauge, NY: Nova Science Publishers, Inc.), 1–30. Available online at: https://www.novapublishers.com/catalog/product_info.php?products_id=37952
Kuivaniemi, H., Tromp, G., Carey, D. J., and Elmore, J. R. (2012). “Molecular biology and genetics of aortic aneurysms,” in Molecular and Translational Vascular Medicine, eds M. S. Willis, and J. W. Homeister (NewYork, NY: Springer Science+Business Media, Part 1), 3–33. doi: 10.1007/978-1-61779-906-8_1
Kullo, I. J., Ding, K., Jouni, H., Smith, C. Y., and Chute, C. G. (2010). A genome-wide association study of red blood cell taits using the electronic medical record. PLoS ONE 5:e13011. doi: 10.1371/journal.pone.0013011
Kullo, I. J., Jarvik, G. P., Manolio, T. A., Williams, M. S., and Roden, D. M. (2013). Leveraging the electronic health record to implement genomic medicine. Genet. Med. 15, 270–271. doi: 10.1038/gim.2012.131
Manolio, T. A. (2013). Bringing genome-wide association findings into clinical use. Nat. Rev. Genet. 14, 549–558. doi: 10.1038/nrg3523
Manolio, T. A., Chisholm, R. L., Ozenberger, B., Roden, D. M., Williams, M. S., Wilson, R., et al. (2013). Implementing genomic medicine in the clinic: the future is here. Genet. Med. 15, 258–267. doi: 10.1038/gim.2012.157
Naj, A. C., Jun, G., Beecham, G. W., Wang, L. S., Vardarajan, B. N., Buros, J., et al. (2011). Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 43, 436–441. doi: 10.1038/ng.801
Parsa, A., Kao, W. H. L., Xie, D., Astor, B. C., Li, M., Hsu, C.-Y., et al. (2013). APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 369, 2183–2196. doi: 10.1056/NEJMoa1310345
Rasmussen-Torvik, L. J., Pacheco, J. A., Wilke, R. A., Thompson, W. K., Ritchie, M. D., Kho, A. N., et al. (2012). High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clin. Transl. Sci. 5, 394–399. doi: 10.1111/j.1752-8062.2012.00446.x
Ross, L. F., Rothstein, M. A., and Clayton, E. W. (2013). Mandatory extended searches in all genome sequencing: “incidental findings,” patient autonomy, and shared decision making. JAMA 310, 367–368. doi: 10.1001/jama.2013.41700
Tzur, S., Rosset, S., Shemer, R., Yudkovsky, G., Selig, S., Tarekegn, A., et al. (2010). Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum. Genet. 128, 345–350. doi: 10.1007/s00439-010-0861-0
Keywords: genomics, electronic health records, incidental findings, implementation, genetic counseling, next generation sequencing, pharmacogenetics
Citation: Kullo IJ, Haddad R, Prows CA, Holm I, Sanderson SC, Garrison NA, Sharp RR, Smith ME, Kuivaniemi H, Bottinger EP, Connolly JJ, Keating BJ, McCarty CA, Williams MS and Jarvik GP (2014) Return of results in the genomic medicine projects of the eMERGE network. Front. Genet. 5:50. doi: 10.3389/fgene.2014.00050
Received: 12 December 2013; Paper pending published: 11 January 2014;
Accepted: 18 February 2014; Published online: 26 March 2014.
Edited by:
Marylyn D. Ritchie, The Pennsylvania State University, USAReviewed by:
Yiran Guo, Children's Hospital of Philadelphia, USAJuli Bollinger, Johns Hopkins University, USA
Copyright © 2014 Kullo, Haddad, Prows, Holm, Sanderson, Garrison, Sharp, Smith, Kuivaniemi, Bottinger, Connolly, Keating, McCarty, Williams and Jarvik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iftikhar J. Kullo, Division of Cardiovascular Diseases, Mayo Clinic, 200 First Street Southwest, Rochester, MN 55905, USA e-mail: kullo.iftikhar@mayo.edu;
Gail P. Jarvik, Medical Genetics, University of Washington, Health Sciences Building, K-253B, Box 357720, Seattle, WA 98195-7720, USA e-mail: pair@u.washington.edu
 Iftikhar J. Kullo
Iftikhar J. Kullo Ra'ad Haddad
Ra'ad Haddad Cynthia A. Prows
Cynthia A. Prows Ingrid Holm
Ingrid Holm Saskia C. Sanderson
Saskia C. Sanderson Nanibaa' A. Garrison
Nanibaa' A. Garrison Richard R. Sharp
Richard R. Sharp Maureen E. Smith
Maureen E. Smith Helena Kuivaniemi
Helena Kuivaniemi Erwin P. Bottinger4
Erwin P. Bottinger4 John J. Connolly
John J. Connolly Brendan J. Keating
Brendan J. Keating Catherine A. McCarty
Catherine A. McCarty Marc S. Williams
Marc S. Williams Gail P. Jarvik
Gail P. Jarvik