- 1Department of Microbiology and Botany, Faculty of Science, Zagazig University, Zagazig, Egypt
- 2Department of Molecular Biotechnology, Graduate School of Advanced Sciences of Matter, Hiroshima University, Higashi-Hiroshima, Japan
The integration and excision of various filamentous phage genomes into and out of their host chromosomes occurs by site-specific recombination. The mechanisms proposed for these events include reactions mediated by phage-encoded recombinases and host recombination systems. Site-specific integration of filamentous phages plays a vital role in a variety of biological functions of the host, such as phase variation of certain pathogenic bacterial virulence factors. The importance of these filamentous phages in bacterial evolution is rapidly increasing with the discovery of new phages that are involved in pathogenicity. Studies of the diversity of two different filamentous phages infecting the phytopathogen Ralstonia solanacearum provide us with novel insights into the dynamics of phage genomes, biological roles of prophages, and the regulation and importance of phage–host interactions.
Filamentous Phages and Pathogenic Bacteria
Bacteriophages of the genus Inovirus are filamentous particles containing a circular single-stranded (ss) DNA genome. This kind of phage does not lyse host cells, but it establishes a persistent association with the host, producing and releasing phage particles from the growing and dividing host cells. The genome of inoviruses, represented by the Escherichia coli F-pilus-specific phage Ff (f1, fd, or M13), is generally organized in a modular structure in which functionally related genes are grouped together (Horiuchi, 1997; Rakonjac et al., 2011; Mai-Prochnow et al., 2015). Three functional modules are always present: the replication module (R), the structural module (S), and the assembly and secretion module (A-S; Figure 1A). The R module contains the genes encoding rolling-circle DNA replication and ssDNA-binding proteins pII, pV, and pX (Horiuchi, 1997). The S module contains genes for the major (pVIII) and minor coat proteins (pIII, pVI, pVII, and pIX). The gene gIII encodes the host recognition or adsorption protein pIII (Wang et al., 2006). The A-S module contains the genes for morphogenesis and extrusion of the phage particles (gI and gIV; Marvin, 1998). The gene gIV encodes protein pIV, an aqueous channel (secretin) in the outer membrane, through which phage particles exit from the host cells (Marciano et al., 1999). Although some phages encode their own secretins, others use host products (Davis et al., 2000). For the general infection cycle of inoviruses, see recent reviews (Rakonjac et al., 2011; Mai-Prochnow et al., 2015).
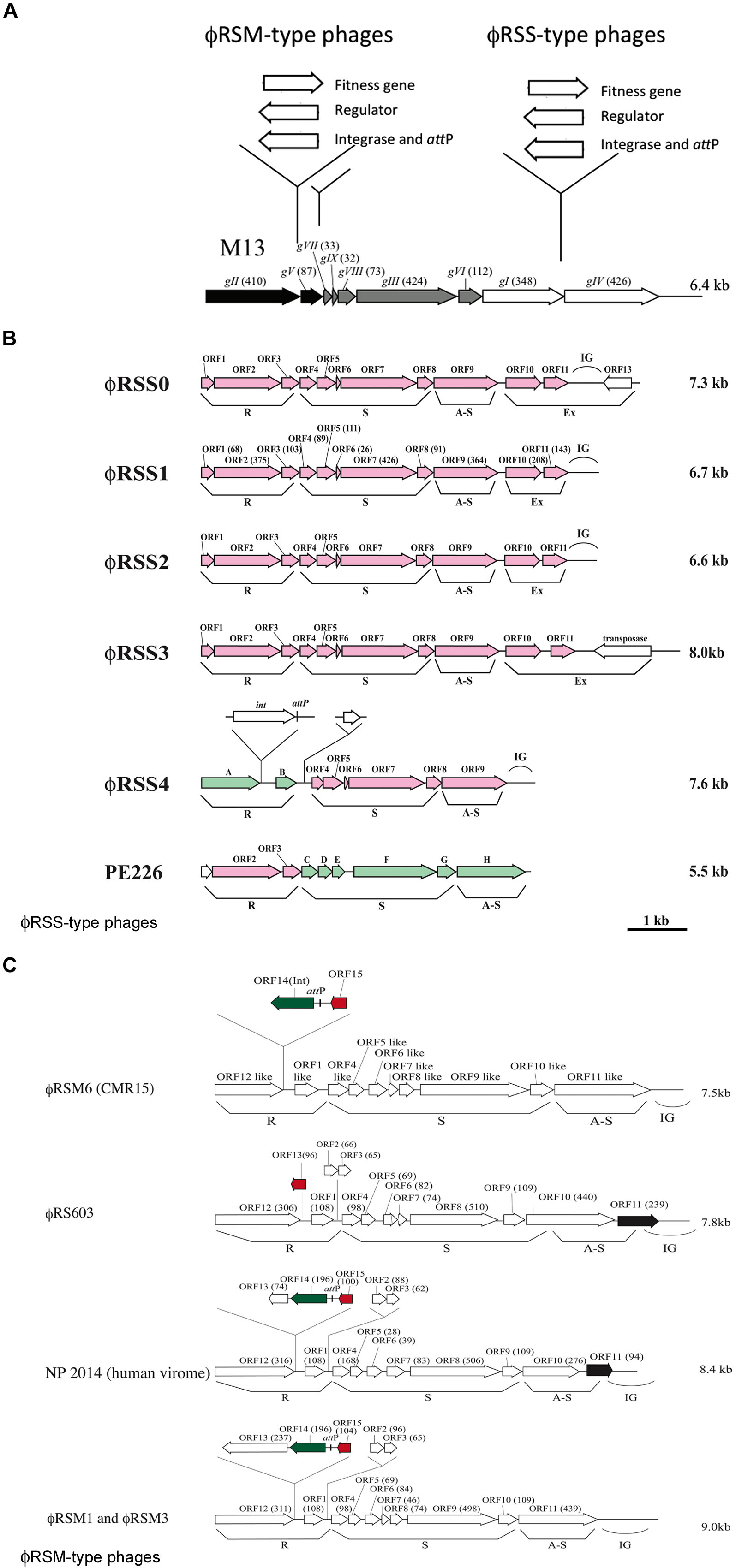
FIGURE 1. Diversity of genomic arrangement in filamentous phages of Ralstonia solanacearum. (A) For ϕRSM-type and ϕRSS-type phages, gene insertion sites are shown along the linear genomic map of Escherichia coli phage M13 (Model and Russel, 1988; Marvin, 1998). Arrows indicate the direction of transcription and represent open reading frames (ORFs) or genes. The functional modules for replication (R), structure (S), and assembly and secretion (A-S) are indicated according to the M13 model. ORF sizes (in amino acids) are in parentheses. IG, intergenic region. (B) Genomic organization of ϕRSS-type phages. According to the E. coli M13-model, ORFs identified in the phage genome are grouped into the R, S, and AS functional modules. IG, large intergenic region. ϕRSS0, ϕRSS2, ϕRSS3, and ϕRSS4 were derived from prophages of strains C319, M4S, MAFF106611, and MAFF211271, respectively. PE226 is a phage of Korean strains of R. solanacearum (Murugaiyan et al., 2010). ORFs shown in pink are homologous to ϕRSS1 ORFs, and those in green are homologous to ϕRSM-type ORFs. (C) Genomic organization of ϕRSM-type phages. ϕRSM1 and ϕRS603 were isolated from soil (Kawasaki et al., 2007; Bich Van et al., 2014). ϕRSM3 and ϕRSM6 are prophages of strains MAFF730139 and CMR15 (phylotype III, Remenant et al., 2010), respectively. NP204 is similar to a phage found in the human virome (Ralstonia phage 1 NP2014, accession no. AHI87735.1). ORFs shown in green, red, and black are genes encoding an integrase (Int), transcriptional repressor, and ϕRSS1-ORF11-like ORF, respectively.
In pathogenic bacteria of either animals or plants, filamentous phage infection has been demonstrated to affect virulence. Examples include (i) enhancing production of virulence factors such as extracellular polysaccharides (EPSs) in Xf- or Lf-infected Xanthomonas campestris (Kamiunten and Wakimoto, 1982; Tseng et al., 1990), (ii) induction of biofilm formation in Pf4-producing Pseudomonas aeruginosa (Webb et al., 2004; Rice et al., 2009), and (iii) reduced twitching motility in ϕRSM-infected Ralstonia solanacearum (Addy et al., 2012a) and in XacF1-infected X. citri (Ahmad et al., 2014). These are likely caused by changes in the host cell surface where phage proteins are secreted and filamentous particles are assembled. More direct involvement of filamentous phages in host virulence is well characterized in Vibrio cholerae. The pathogenicity of this severe diarrheal disease–causing bacterium depends on two key virulence factors, the toxin co-regulated pilus and cholera toxin. Cholera toxin genes are encoded on the filamentous phage CTXϕ and are introduced into bacterial cells by phage integration mediated by the host dif/XerCD recombinase system (Huber and Waldor, 2002; Davis and Waldor, 2003). Also, the filamentous prophage MDA was found at multiple sites in the host chromosome associated with invasive isolates of Neisseria meningitidis (Bille et al., 2005). The prophage Ypfϕ was reported to contribute to the pathogenicity of the plague bacillus, Yersinia pestis (Derbise et al., 2007). The acquisition of the filamentous phage CUS-1 encoding puvA was thought to contribute to the expression of a high-virulence phenotype in Escherichia coli O18:K1:H7 (Gonzalez and Allen, 2003). In these cases, filamentous phages with genes encoding toxins, virulence-enhancing factors, or host fitness factors were integrated into the host genome by various mechanisms. For other examples of filamentous phages infecting pathogenic bacteria, see the recent review by Ilyina (2015).
Different Strategies for Filamentous Phage DNA Integration into the Host Genome
To date, four different integration mechanisms used by filamentous phages have been described (Table 1). Well-characterized filamentous coliphages, such as M13 and fd, typically do not take a lysogenic replication cycle and replicate exclusively as an episome in their host bacteria (Model and Russel, 1988; Rakonjac et al., 2011). Some filamentous phages, including CTXϕ of V. cholerae, accomplish site-specific integration into the dif site of the bacterial chromosome by using the host XerC/D recombination system (Huber and Waldor, 2002). Filamentous phages such as VEJϕ of Vibrio parahaemolyticus (Campos et al., 2010); Cf1c, Cf1t, Cf16v1, and ϕLf of X. campestris (Campos et al., 2003); Xf1c and XacF1 of X. citri (Ahmad et al., 2014); Xfϕf1 of Xylella fastidiosa (de Mello Varani et al., 2008); and Ypfϕ of Yersinia pestis (Lesterlin et al., 2004) also seem to use the host XerC/D recombination for their integration. In contrast, ϕRSM1 and ϕRSM3 of R. solanacearum encode a site-specific integrase (Int) of the resolvase/invertase subfamily of serine recombinases (Askora et al., 2011). This kind of serine recombinase mediates recombination involving the process of double-strand breakage followed by rotation and religation. Both integrative and excisive recombination reactions were catalyzed by ϕRSM-Int (Askora et al., 2011). The phage attP corresponded to the 13 b sequence at the 5′ of serine tRNA (UCG) of the host. The same unit of integration (Int–attP) was also found in a R. pickettii 12J filamentous prophage and in Burkholderia pseudomallei 668 prophage. A different strategy to integrate DNA into the host genome by filamentous phages may be via transposases. Kawai et al. (2005) observed filamentous prophages integrated into the chromosome of Neisseria species. Each prophage copy of the neisserial filamentous phage (Nf) was flanked by a duplication of the 5′-CT and carried an open reading frame (ORF) encoding a transposase homolog (pivNM/irg), suggesting the transposase-mediated integration of Nf DNA into host bacterial chromosomes. Bille et al. (2005) actually showed that the integration of Nf DNA is mediated by its own transposases (pivNM/irg). Meanwhile, Webb et al. (2004) and Mooij et al. (2007) characterized two filamentous prophages, Pf4 and Pf5, in the genome of P. aeruginosa PAO1 and PA14, respectively. Both prophages were integrated into tRNA genes of their host, probably in a reaction mediated by their own Int from the tyrosine-recombinase family. Thus, at least four different strategies for the integration of filamentous bacteriophage DNA into the host chromosome are known (Askora et al., 2012; Table 1).
Structural and Biological Diversity of Two Different Filamentous Phages Infecting Ralstonia solanacearum
Ralstonia solanacearum is a Gram-negative β-proteobacterium that causes bacterial wilt disease in many important crops including tomato, potato, tobacco, eggplant, banana, ginger, and mulberry. Because of its wide geographic distribution and unusually broad host range (more than 50 plant families), it is responsible for significant crop losses worldwide (Hayward, 2000; Denny, 2006). Filamentous phages that were found to infect strains of R. solanacearum were classified into two groups, ϕRSS-type and ϕRSM-type phages. ϕRSS1 is a representative of ϕRSS-type phages and is a relatively small particle (1.1 μm in length) containing an ssDNA genome of 6,662 nt (with a GC content of 62.6%) encoding 11 ORFs (Kawasaki et al., 2007). Genomic DNA of these types of phage was frequently found integrated in the host genome; 23 of 24 strains tested (all isolated in Japan) showed positive hybridization signals in Southern blot analysis (Yamada et al., 2007). Some prophage sequences were determined (Figure 1B). ϕRSS0, ϕRSS2, and ϕRSS3 were derived from prophages of strain C319, M4S, and MAFF106611, respectively (Yamada, 2012). Compared with the M13 gene organization, additional genes are inserted within or next to the A-S module in these ϕRSS genomes (Figure 1A). In the case of ϕRSS0, a putative regulatory gene (with similarity to transcriptional repressors; ORF13) is inserted in the reverse orientation with two unknown ORFs (ORF10 and ORF11). There is an R. solanacearum dif sequence within ORF13 that serves as an attP site for integration into the host genome by host XerC/D recombinases (Yamada, 2013). In the case of ϕRSS3, an additional gene encoding a transposase (IS4 family) was located in the reverse orientation. This may function for integration of the phage DNA in some occasions like the Neisseria cases described above (Kawai et al., 2005). Therefore, these ϕRSS variations represent the possibility of functional equipment at this genomic region with genes for host fitness, integration, and regulatory functions (Figure 1A).
Another type of filamentous phage of R. solanacearum revealed a different story of evolution. ϕRSM1, the first phage to be classified as a member of the ϕRSM-type phages is a longer filamentous particle (1.5 μm in length) containing ssDNA of 9,004 nt (with a GC content of 59.9%) as the genome (Kawasaki et al., 2007; Yamada et al., 2007). A total of 15 ORFs were found on the ϕRSM1 genome including five extra genes in addition to M13-core genes. The extra genes are inserted within the R module or between the R and S modules (Figure 1C). Two of these extra genes (orf14 and orf15) encode a DNA resolvase/invertase-like serine recombinase functioning as an Int (Askora et al., 2011) and a transcriptional repressor (Addy et al., 2012a), respectively. There was an attP site between orf14 and orf15 (Figure 1C). The function of the other extra genes is not known. In contrast to ϕRSS phages, the integration of ϕRSM-type phage DNA into the genome of strains isolated in Japan was not frequent; 6 of 24 strains tested showed positive signals in genomic Southern blot analysis. However, genomic sequences of R. solanacearum strains and related β-proteobacteria in the databases frequently showed ϕRSM-like prophage sequences. A comparison of those sequences revealed the genomic diversity of ϕRSM-type phages as shown in Figure 1C and Supplementary Figure S1. Only one gene encoding a putative repressor (corresponding to ϕRSM1 ORF15) is located within the extra region in the R module of R. solanacearum phage ϕRS603 (Bich Van et al., 2014), whereas ϕRSM6 in strain CMR15 (phylotype III) contained an Int gene (ORF14) in addition to the repressor gene (ORF15; Askora et al., 2014). Like ϕRSM1, ϕRSM3, ϕRSM4 in strain UW551 (phylotype II), ϕRSM5 in strain IPO1609 (phylotype II), and ϕRSM7 in strain Y45 (phylotype IB) contained three genes within this region (ORF13, ORF14, and ORF15) with the same organization (Figure 1C). However, it is noteworthy that there are two different regulatory systems, where the amino acid sequence of ORF15 and its upstream regulatory nucleotide sequence are different in phages infecting different phylotypes (Askora et al., 2014). ϕRSM1, ϕRSM3, and Y45, which infect strains of phylotype I, share similar regulatory systems, whereas ϕRSM5, ϕRSM6, and ϕRSM7, which infect strains of phylotypes II or III, contained another system. Very similar ϕRSM sequences were also found in the genomes of R. syzgii and R. pickettii (Askora et al., 2014). This kind of phage may have an extensive host range in b-proteobacteria. Interestingly, a ϕRSM homolog was found in the human virome (Ralstonia phage 1 NP2014, accession no. AHI87735.1) as shown in Figure 1C. Ralstonia phage 1 NP2014 possesses a circular ssDNA genome that is highly homologous to those of ϕRSM1 and ϕRSM3. Ralstonia phage 1 NP2014 contains a unique ORF11 with high similarity to ϕRSS0 ORF11 (Figure 1B).
As described above, two groups of filamentous phages of R. solanacearum have used different mechanisms for the evolution of genomic arrangements (Figure 1A). However, there may have been some opportunities for them to infect the same host cells by chance, which would have made it possible for the two types of phage to hybridize. Actually, such forms were detected (Figure 1B). A prophage (ϕRSM4) found in strain MAFF211271 showed a gene arrangement with the ϕRSM-type R module containing genes for an Int and regulator and with ϕRSS-type S and A-S modules (Yamada, 2012). A smaller filamentous phage, PE226, was isolated with Korean strains and showed a gene arrangement with a ϕRSS-type R module and ϕRSM-type S and A-S modules (Murugaiyan et al., 2010). Therefore, further genomic diversity by mixing these two types of phage gene arrangement is not surprising.
Filamentous Phage Diversity and Effects on the Host Virulence and Evolution in R. solanacearum
Both ϕRSS-type and ϕRSM-type filamentous phages affect the host physiology including virulence. ϕRSS1-infected cells showed enhanced virulence on tobacco (Yamada et al., 2007) and tomato plants (Addy et al., 2012b). The virulence-enhancing effects by ϕRSS1 infection can be explained as follows: surface-associated ϕRSS1 particles (or phage proteins) may change the surface nature (hydrophobicity) of host cells to generate a high local cell density, resulting in early activation of phcA, the global virulence regulator, or lack of orf13, which is absent from the ϕRSS1 genome (Addy et al., 2012b). The reduced virulence observed for ϕRSS0-infected cells may be caused by the function(s) of ORF13 encoded by ϕRSS0 (Yamada, 2013). Contrasting to the ϕRSS1 effects, upon infection by ϕRSM phages, the host cells showed loss of virulence phenotypes (Addy et al., 2012a). This loss of virulence effect of ϕRSM infection can be explained in three ways: (i) reduced twitching motility and reduced amounts of type IV pili (Tfp), (ii) lower levels of β-1,4-endoglucanase (Egl) activity and EPS production, and (iii) reduced expression of certain virulence/pathogenicity genes (egl, pehC, phcA, phcB, pilT, and hrpB) in the infected cells (Addy et al., 2012a).
Thus, phages sometimes help host bacteria infect plants by enhancing bacterial virulence, and they sometimes interrupt bacterial infection of plants by repressing host genes involved in virulence. Such contradictory effects of these phages largely depend on the phage state, for example, replicating freely in the host, existing as a stable prophage (with Int), or expressing a special transcriptional regulator (Yamada, 2013). In general, the phage-encoded regulator somehow affects the expression of host genes involved in virulence, mostly through repression, in both ϕRSS-type and ϕRSM-type phages. However, integration into the host genome may cause a change in the regulatory function (namely direct effects on the host gene expression may be relaxed). As described above, cell surface changes caused by filamentous phage secretion affect quorum sensing, twitching motility, and biofilm formation. Depending on the lifestyle of host bacterial cells in the environment, phage effects are different, and an advantageous state of cells with phage will be selected under the conditions. Cells whose virulence is enhanced by phage will predominate in the pathogenic stage. Similar types of phage involvement in host virulence regulation may be universal because ϕRSS- or ϕRSM-related sequences are frequently found in various bacterial genomic sequences, including R. picketti (accession no. CP001645), R. syzygii (FR854090), Burkholderia rhizoxinica (FR687359), Pectobacterium wasabiae (CP001790), and Erwinia carotovora (BX950851). The diversity observed in the genome arrangement and biological effects of filamentous phages infecting the phytopathogen R. solanacearum will serve as a good reference to consider interactions between various pathogenic bacteria and their phages.
Hypothesis
Filamentous phages are widely disseminated and exist as prophage states in different strains of pathogenic bacteria. They might evolve rapidly and play roles in the introduction of new genes into their hosts. Therefore, it is highly likely that filamentous phages are mediating the ecological adaptation and virulence of their hosts and thus play significant roles in the evolution of bacterial species.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2015.00217
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported in part by the JSPS Postdoctoral Fellowship for Foreign Researchers (P13086 to AA); the Research and Development Projects for Application in Promoting New Policy of Agriculture, Forestry, and Fisheries (No. 250037B to TY); and JST/BIOTEC Strategic Research Cooperative Program on Biotechnology (A1200357 to TY).
References
Addy, H. S., Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2012a). Loss of virulence of the phytopathogen Ralstonia solanacearum through infection by ϕRSM filamentous phages. Phytopathology 102, 469–477. doi: 10.1094/PHYTO-11-11-0319-R
Addy, H. S., Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2012b). The filamentous phage ϕRSS1 enhances virulence of phytopathogenic Ralstonia solanacearum on tomato. Phytopathology 102, 244–251. doi: 10.1094/PHYTO-10-11-0277
Ahmad, A. A., Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2014). A novel filamentous phage causes loss of virulence to Xanthomonas axonopodis pv citri the causative agent of citrus canker disease. Front. Microbiol. 5:321. doi: 10.3389/fmicb.2014.00321
Askora, A., Abdel-Haliem, M. E. F., and Yamada, T. (2012). Site-specific recombination systems in filamentous phages. Mol. Genet. Genom. 287, 525–530. doi: 10.1007/s00438-012-0700-1
Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2011). Resolvase-like serine recombinase mediates integration/excision in the bacteriophage ϕRSM. J. Biosci. Bioeng. 111, 109–116. doi: 10.1016/j.jbiosc.2010.10.001
Askora, A., Kawasaki, T., Fujie, M., and Yamada, T. (2014). Insight into the diversity of ϕRSM phages infecting strains of the phytopathogen Ralstonia solanacearum complex: regulation and evolution. Mol. Genet. Genomics, 289, 589–598. doi: 10.1007/s00438-014-0835-3
Askora, A., Kawasaki, T., Usami, S., Fujie, M., and Yamada, T. (2009). Host recognition and integration of filamentous phage ϕRSM in the phytopathogen, Ralstonia solanacearum. Virology 384, 69–76. doi: 10.1016/j.virol.2008.11.007
Bich Van, T. T., Yoshida, S., Miki, K., Kondo, A., and Kamei, K. (2014). Genomic characterization of ϕRS603, a filamentous bacteriophage that is infectious to the phytopathogen Ralstonia solanacearum. Microbiol. Immunol. 58, 697–700. doi: 10.1111/1348-0421.12203
Bille, E., Zahar, J. R., Perrin, A., Morelle, S., Kriz, P., Jolley, K. A., et al. (2005). A chromosomally integrated bacteriophageb in invasive meningococci. J. Exp. Med. 201, 1905–1913. doi: 10.1084/jem.20050112
Campos et al., J., Martinez, E., Izquierdo, Y., and Fando, R. (2010). VEJϕ, a novel filamentous phage of Vibrio cholerae able to transduce the cholera toxin genes. Microbiology 156, 108–115. doi: 10.1099/mic.0.032235-0
Campos, J., Martinez, E., Suzarte, E., Rodriguez, B. E., Marrero, K., Silva, Y., et al. (2003). A novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTXϕ. J. Bacteriol. 185, 5685–5696. doi: 10.1128/JB.185.19.5685-5696.2003
Davis, B. M., Lawson, E. H., Sandkvist, M., Sozhamannan, S., Ali, A., and Waldor, M. K. (2000). Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXϕ. Science 288, 333–335. doi: 10.1126/science.288.5464.333
Davis, B. M., and Waldor, M. K. (2003). Filamentous phages linked to virulence of Vibrio cholerae. Curr. Opin. Microbiol. 6, 35–42. doi: 10.1016/S1369-5274(02)00005-X
de Mello Varani, A., Souza, R. C., Nakaya, H. I., de Lima, W. C., de Almeid, P., Kitajima, E. W., et al. (2008). Origins of the Xylella fastidiosa prophage-like regions and their impact in genome differentiation. PLoS ONE 3:e4059. doi: 10.1371/journal.pone.0004059
Denny, T. P. (2006). “Plant pathogenic Ralstonia species,” in Plant-Associated Bacteria, ed. S. S. Gnanamanickam (Amsterdam: Springer) 573–644. doi: 10.1007/978-1-4020-4538-7_16
Derbise, A., Chenal-Francisque, V., Pouillot, F., Fayolle, C., Prevost, M. C., Medigue, C., et al. (2007). A horizontally acquired filamentous phage c ontributes to the pathogenicity of the plague Bacillus. Mol. Microbiol. 63, 1145–1157. doi: 10.1111/j.1365-2958.2006.05570.x
Gonzalez, E. T., and Allen, C. (2003). Characterization of a Ralstonia solanacearum operon required for polygalacturonate degradation and uptake of galacturonic acid. Mol. Plant Microbe Interact. 16, 536–544.
Hayward, A. C. (2000). “Ralstonia solanacearum,” in Encyclopedia of Microbiology, Vol. 4, ed. J. Lederberg (San Diego, CA: Academic Press), 32–42.
Horiuchi, K. (1997). Initiation mechanisms in replication of filamentous phage DNA. Genes Cells 2, 425–432. doi: 10.1046/j.1365-2443.1997.1360334.x
Huber, K. E., and Waldor, M. K. (2002). Filamentous phage integration requires the host recombinases XerC and XerD. Nature 417, 656–659. doi: 10.1038/nature00782
Ilyina, T. S. (2015). Filamentous bacteriophages and their role in the virulence and evolution of pathogenic bacteria. Mol. Genet. Microbiol. Virol. 30, 1–9. doi: 10.3103/S0891416815010036
Kamiunten, H., and Wakimoto, S. (1982). Effect of the infection with filamentous phage Xf-2 on the properties of Xanthomonas campestris var oryzae. Ann. Phytopathol. Soc. Japan 47, 627–636. doi: 10.3186/jjphytopath.47.627
Kawai, M., Uchiyama, I., and Kobayashi, I. (2005). Genome comparison in silico in Neisseria suggests integration of filamentous bacteriophages by their own transposase. DNA Res. 12, 389–401. doi: 10.1093/dnares/dsi021
Kawasaki, T., Nagata, S., Fujiwara, A., Satsuma, H., Fujie, M., Usami, S., et al. (2007). Genomic characterization of the filamentous integrative bacteriophage ϕRSS1 and ϕRSM1, which infect Ralstonia solanacearum. J. Bacteriol. 189, 5792–5802. doi: 10.1128/JB.00540-07
Lesterlin, C., Barre, F. X., and Cornet, F. (2004). Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol. Microbiol. 54, 1151–1160. doi: 10.1111/j.1365-2958.2004.04356.x
Mai-Prochnow, A., Hui, J. G. K., Kjelleberg, S., Rakonjac, J., McDougald, D., and Rice, S. A. (2015). Big things in small packages: the genetics of filamentous phage and effects on fitness of their host. FEMS Microbiol. Rev. doi: org/10.1093/femsre/fuu007 [Epub ahead of print].
Marciano, D. K., Russel, M., and Simon, S. M. (1999). An aqueous channel for filamentous phage export. Science 284, 1516–1519. doi: 10.1126/science.284.5419.1516
Marvin, D. A. (1998). Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 8, 150–158. doi: 10.1016/S0959-440X(98)80032-8
Model, P., and Russel, M. (1988). “Filamentous bacteriophages,” in The Bacteriophages, Vol. 2, ed. R. Calendar (New York, NY: Plenum Press) 375–456.
Mooij, M. J., Drenkard, E., Llamas, M. A., Vandenbroucke-Grauls, C. M. J. E., Savelkoul, P. H. M., Ausubel, F. M., et al. (2007). Characterization of the integrated filamentous phage Pf5 and its involvement in small-colony formation. Microbiology 153, 1790–1798. doi: 10.1099/mic.0.2006/003533-0
Murugaiyan, S., Bae, J. Y., Wu, J., Lee, S. D., Um, H. Y., Choi, H. K., et al. (2010). Characterization of filamentous bacteriophage PE226 infecting Ralstonia solanacearum strains. J. Appl. Microbipl. 110, 296–303. doi: 10.1111/j.1365-2672.2010.04882.x
Rakonjac, J., Bennet, N. J., Spagnuolo, J., Gagic, D., and Russel, M. (2011). Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 13, 51–76.
Remenant, B., Coupat-Goutaland, B., Guidot, A., Cellier, G., Wicker, E., Allen, C., et al. (2010). Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics 11:319. doi: 10.1186/1471-2164-11-379
Rice, S. A., Tan, C. H., Mikkelsen, P. J., Kung, V., Woo, J., Tay, M., et al. (2009). The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. ISME J. 3, 271–282. doi: 10.1038/ismej.2008.109
Tseng, Y. H., Lo, M. C., Lin, K. C., Pan, C. C., and Chang, R. Y. (1990). Characterization of filamentous bacteriophage ϕLf from Xanthomonas campestris pv. campestris. J. Gen. Virol. 71, 1881–1884. doi: 10.1099/0022-1317-71-8-1881
Wang, Y. A., Yu, X., Overman, S., Tsuboi, M., Tomas, G. J. Jr. and Egelman, E. H. (2006). The structure of a filamentous bacteriophage. J. Mol. Biol. 361, 209–215. doi: 10.1016/j.jmb.2006.06.027
Webb, J. S., Lau, M., and Kjelleberg, S. (2004). Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186, 8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004
Yamada, T. (2012). “Bacteriophages of Ralstonia solanacearum: their diversity and utilization as biocontrol agents in agriculture,” in Bacteriophages, ed. I. Kurtboke (Rijeka: InTech-Open Access Publisher), 113–139.
Yamada, T. (2013). Filamentous phages of Ralstonia solanacearum: double-edged swords for pathogenic bacteria. Front. Microbiol. 4:325. doi: 10.3389/fmicb.2013.00325
Keywords: filamentous phage, integration, pathogenic bacteria, virulence change
Citation: Askora A and Yamada T (2015) Two different evolutionary lines of filamentous phages in Ralstonia solanacearum: their effects on bacterial virulence. Front. Genet. 6:217. doi: 10.3389/fgene.2015.00217
Received: 01 April 2015; Accepted: 03 June 2015;
Published: 18 June 2015
Edited by:
Frank T. Robb, University of Maryland, USAReviewed by:
Imke Schroeder, University of California, Los Angeles, USASteven P. T. Hooton, Novolytics, UK
Copyright © 2015 Askora and Yamada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Yamada, Department of Molecular Biotechnology, Graduate School of Advanced Sciences of Matter, Hiroshima University, 1-3-1 Kagamiyama, Higashi-Hiroshima 739-8530, Japan, tayamad@hiroshima-u.ac.jp
 Ahmed Askora
Ahmed Askora Takashi Yamada
Takashi Yamada