- 1Institute of Bioinformatics, University Medical Center Göttingen, Göttingen, Germany
- 2Department of Hematology/Medical Oncology, University Medical Center Göttingen, Göttingen, Germany
- 3Department of Medical Statistics, University Medical Center Göttingen, Göttingen, Germany
Transcription factors (TFs) are gene regulatory proteins that are essential for an effective regulation of the transcriptional machinery. Today, it is known that their expression plays an important role in several types of cancer. Computational identification of key players in specific cancer cell lines is still an open challenge in cancer research. In this study, we present a systematic approach which combines colorectal cancer (CRC) cell lines, namely 1638N-T1 and CMT-93, and well-established computational methods in order to compare these cell lines on the level of transcriptional regulation as well as on a pathway level, i.e., the cancer cell-intrinsic pathway repertoire. For this purpose, we firstly applied the Trinity platform to detect signature genes, and then applied analyses of the geneXplain platform to these for detection of upstream transcriptional regulators and their regulatory networks. We created a CRC-specific position weight matrix (PWM) library based on the TRANSFAC database (release 2014.1) to minimize the rate of false predictions in the promoter analyses. Using our proposed workflow, we specifically focused on revealing the similarities and differences in transcriptional regulation between the two CRC cell lines, and report a number of well-known, cancer-associated TFs with significantly enriched binding sites in the promoter regions of the signature genes. We show that, although the signature genes of both cell lines show no overlap, they may still be regulated by common TFs in CRC. Based on our findings, we suggest that canonical Wnt signaling is activated in 1638N-T1, but inhibited in CMT-93 through cross-talks of Wnt signaling with the VDR signaling pathway and/or LXR-related pathways. Furthermore, our findings provide indication of several master regulators being present such as MLK3 and Mapk1 (ERK2) which might be important in cell proliferation, migration, and invasion of 1638N-T1 and CMT-93, respectively. Taken together, we provide new insights into the invasive potential of these cell lines, which can be used for development of effective cancer therapy.
1. Introduction
Cancer undergoes genetic and epigenetic changes through which it acquires cellular and molecular characteristics during invasive tumor growth. These changes allow the tumor cells to evade the immune response, activate the microenvironment, invade surrounding tissues and metastasize to distant sites. The microenvironment plays an important role in this context as it may trigger anti-tumor as well as pro-tumor signals (Gao et al., 2014). Malignant tumor cells stimulate the production and secretion of growth factors, cytokines and enzymes, thereby recruiting the stroma and vasculature, which altogether results in the conversion of a normal tumor-inhibiting into a tumor-promoting microenvironment (Gao et al., 2014). In that respect, tumor aggressiveness can be linked to processes such as cell proliferation, growth, invasion, metastasis, survival as well as inflammation which are regulated by multiple signal transduction pathways. It has been suggested to summarize known signal transduction reactions into about 17 signal transduction pathways (Nebert, 2002). They are usually activated by growth factor signals from the cell surface, and further transmit the signal via transmembrane receptors to their target intracellular effectors. In tumor cells, these pathways are often dysregulated and harbor alterations in key components that can function as driver mutations, i.e., either as activation mutations (Ras, PI3K, Akt) or loss of tumor-suppressor gene function (Pten). Several cancer drivers are important integral parts of these pathways, such as receptor tyrosine kinases, and can be located upstream in signal transduction cascades. Since protein kinases propagate the signals along the cascade, they are considered attractive drug targets for therapeutic intervention using specific protein kinase inhibitors (Zwick et al., 2001; Torkamani et al., 2009; Takeuchi and Ito, 2011; Casaletto and McClatchey, 2012). To this end, many anticancer agents have been used in the context of cancer therapy to account for the number of different pathways (Casaletto and McClatchey, 2012).
The signaling pathways are interconnected and form an elaborate network of pathways that receives signals from a variety of growth factors to tightly regulate processes such as transcription, cell growth, motility, differentiation, apoptosis, and cytoskeletal organization. In addition, the outcome triggered by the integrated signaling may differ between different cell types. Therefore, knowledge on the cell type-specific pathways including their architecture and complexity provides important information on the tumor cell behavior during inhibitor therapy, i.e., the inhibitor may not achieve the desired outcome due to the utilization of alternative bypass pathways in certain tumor cells.
Signal transduction pathways converge on sets of genes with similar key functions which are regulated by upstream transcription factors (TFs). TFs occupy short and specific DNA-sequences denoted as transcription factor binding sites (TFBSs). TFs and their corresponding TFBSs recruit and regulate the transcription machinery, thereby governing selective temporal and spatial activities of their target genes. Moreover, many TFs play important roles as oncogenes and they are usually activated downstream in the signaling cascades. Consequently, their deregulated expression, aberrant activation as well as mutations contribute to tumorigenesis. For example, the TP53 gene which encodes an important transcription factor with tumor suppressor function in cancer, is known to be the most commonly mutated gene in human cancer (Kandoth et al., 2013). Unsurprisingly, TFs are central to cancer and became highly desirable points of interference in cancer gene therapy (Libermann and Zerbini, 2006). In this regard, three major transcription factor families have been considered highly desirable drug targets: (i) the NF-κB and AP-1 families of TFs; (ii) the STAT family members; (iii) the steroid receptors (Libermann and Zerbini, 2006). Although other additional TF families have been implicated in cancer to this day, there is still no comprehensive library on TFs and their specific roles in cancer and, particularly, in different cancer cell types. However, given the tumor heterogeneity and cancer cell plasticity, it can be expected that many more TFs will be associated with potentially important roles in oncogenic pathways of different cancers.
The third most common cancer in the world is colorectal cancer (CRC) which originates in the epithelial cells of the gastrointestinal track and shows a high tendency to metastasize into the liver. CRC is often caused by mutations in two well-studied signal transduction pathways, namely the Wnt and the EGFR pathways (Normanno et al., 2006; Polakis, 2012). Mouse models have been extensively used in cancer studies to directly monitor the metastatic progression in CRC. The ability to study primary tumors as well as distant metastatic sites and to manipulate the spatial and temporal expression levels of certain single genes have proven the animal model technology to be a powerful tool in cancer progression research. Such studies have often made use of APC-deficient mouse models since mutations in the adenomatous polyposis coli (APC), an important component of the Wnt signaling pathway, occur in the majority of human CRC cells (Karim and Huso, 2013). It is estimated that the canonical Wnt/β-catenin signaling pathway is abnormally activated in over 90% of CRCs (Cancer Genome Atlas Network, 2012). Briefly, the canonical Wnt pathway revolves around the intracellular levels of the transcriptional coactivator β-catenin which forms a complex with TCF/LEF, thereby controlling the expression of Wnt signaling targets, such as c-Myc and cyclin D. β-Catenin is degraded by a destruction complex that includes the tumor suppressor APC and other proteins (Stamos and Weis, 2013). Loss of APC leads to a constant activation of WNT signaling, which promotes proliferation of tumor cells.
The bottleneck in cancer research has always been a lack of effective tools to comprehensively study the complex networks of signaling pathways (Kang, 2005; Gupta and Massagué, 2006). Therefore, cancer research has largely taken advantage of the integration of animal models and bioinformatic approaches. Microarrays and nowadays RNA-sequencing techniques (RNA-Seq) are used to infer reliable gene regulatory networks based on the level of all expressed transcripts (transcriptome) (Schena et al., 1995; Mortazavi et al., 2008). The result of a transcriptome profiling experiment can be summarized in a set of expressed genes or transcription units that are meaningful for a certain experimental condition, disease state or developmental process. These technologies have led to paradigm-shifting advances in cancer research. For example, gene expression profiles in combination with supervised clustering approaches were used in breast cancer studies which successfully discriminated between cancer patients with good prognosis from those with poor prognosis, thereby leading to the identification of prognostic cancer genes (van 't Veer et al., 2002; Weigelt et al., 2005). However, solely using genomic profiling of tumor samples only identifies individual genes of a set of signature genes, but does not provide a functional context for these genes, which is important for a mechanistic understanding of cancer-associated processes. Pathway analyses have therefore emerged as powerful tools by benefiting from the statistical power of entire gene sets using the overrepresentation in biologically defined pathways rather than interpreting meaningful functions based on the expression of individual genes.
Despite the presence of a variety of different approaches and rich literature on cancer research as mentioned above, to date, there is still need for comprehensive analyses to detect key regulators in different colorectal cancer cell lines. In this study, we made use of distinct murine cancer cell lines and system biology approaches to identify signature genes and pathways whose activation may specifically affect invasive tumor growth. In addition, we exhaustively covered a broad range of potentially important signaling pathways and focus our discussion selectively on the study of the roles of various classical and novel signaling pathways in CRC. Moreover, we aimed to highlight the meaning of specific TFs in the context of these pathways on the basis of enriched TFBSs in the promoter regions of the signature genes. We provide a comprehensive library on CRC-specific TFs and exemplarily discuss their roles in both CRC cell lines. Taken together, we identified potential discriminators between the two CRC cell lines as well as points of interference for targeted cancer therapy, thus providing further insights into the complexity of cancer.
2. Materials and Methods
2.1. Colorectal Cancer Cell Lines
The CMT-93 cell line, a mouse colorectal polyploid carcinoma cell line, was purchased from the American Type Culture Collection, Manassas, USA (CCL223) and was cultured in DMEM High Glucose Medium (Gibco, Darmstadt, Germany) supplemented with 10% heat inactivated fetal bovine serum (FCS; Sigma, Munich, Germany). The murine colorectal cancer cell line 1638N-T1, derived from Apc1638N adenomas, was kindly provided by Ron Smits (Smits et al., 1997). Remarkably, this cell line harbors a targeted mutation at codon 1638 of the Apc gene, Apc1638T, leading to a truncated Apc protein (Smits et al., 1999). These were cultured in DMEM High Glucose Medium supplemented with 15% not heat inactivated FCS and Insulin/Transferrin/Selenium Solution (Gibco). In contrast to Smits et al., these cells were not cultured on any fibronectin/collagen/albumin-coated plates and were passaged using 0.05% (w/v) trypsin (Biochrom, Berlin, Germany), as long as they did not show any differences in their morphology, viability and proliferation.
2.2. RNA Isolation and Sequencing
Total RNA was isolated using the TRIzol Reagent (Invitrogen, Karlsruhe, Germany) including a DNase I (Roche, Mannheim, Germany) digestion. RNA integrity and quantity was assessed with the Agilent Bioanalyzer 2100 and the NanoDrop DD-1000 UV vis spectrophotometer version 3.2.1. 2 μg of total RNA were used as start material for library preparation (TruSeq Stranded mRNA Sample Prep Kit from Illumina, Cat NRS-122-2101). Accurate quantitation of cDNA libraries was performed by using the QuantiFluor dsDNA System (Promega). The size range of cDNA libraries was determined applying the DNA 1000 chip on the Bioanalyzer 2100 from Agilent (280 bp). cDNA libraries were amplified and sequenced by using the cBot and HiSeq 2000 from Illumina (SR, 1 × 51 bp, 8–9 Gb > 40 M reads per sample). Sequence images were transformed with Illumina software BaseCaller to bcl-files, which were demultiplexed to FASTQ files with CASAVA (version 1.8.2). Quality check was done via FastQC (version 0.10.1, Babraham Bioinformatics).
2.3. Signature Gene Selection
We started our analyses based on 43433 gene annotations from Ensembl (mouse assembly GRCm38.p4), which were retrieved from RNA-seq samples (Section 2.2; three biological replicates for each cell line; GSE78696). Based on these samples, we obtained signature genes as follows:
Using the Trinity platform (Grabherr et al., 2011), we firstly performed a differentially expressed gene (DEG) analysis based on both cell lines. After that, employing the Trinity platform these DEGs were clustered into three main categories using a p-value cutoff for FDR of 0.05 and the default fold change (default: 2 (meaning 22 or 4-fold)): (i) genes which are most significantly upregulated in 1638N-T1 (Supplementary Table S1) and, at the same time, downregulated in CMT-93; (ii) genes which are most significantly upregulated in CMT-93 (Supplementary Table S2) and, at the same time, downregulated in 1638N-T1; (iii) the remaining DEGs which did not fall in the first and second category. In our further analysis, we only considered genes as signature genes which fell into the first or second category.
2.4. Data Processing
For the subsequent analyses we used the geneXplain platform (http://genexplain-platform.com/bioumlweb/), which includes the TRANSFAC and TRANSPATH databases. We used the suggested parameters from this platform if not explicitly stated otherwise.
2.4.1. Enrichment of TFBSs in Promoter Sequences
We applied a conventional enrichment analysis to the previously identified signature gene sets in order to retrieve specific TFs whose binding sites or sequence motifs are particularly enriched in their genomic regions. For the enrichment analysis, we firstly extracted for each signature gene the corresponding promoter sequence covering the −1000 to 100 bp regions relative to transcription start sites. Second, we used position weight matrices (PWMs) from the TRANSFAC database (Wingender, 2008) to predict potential TFBSs in promoters. However, computational TFBS predictions are generally considered as being flooded with high rates of false predictions. The accurate prediction of TFBSs is still a challenging task. To minimize the rate of false predictions in our analysis, we collected a specific PWM library using literature on CRC (Supplementary Table S3). This library contains 229 colorectal cancer-related non-redundant matrices. In our further analysis, this library was used with the minFP profile (cut-offs minimizing false positive rate) that contains the adjusted thresholds for each PWM to minimize the prediction of false positive TFBSs. Using our library, we then employed the F-MATCH program described in Schmid et al. (2006) to determine the enriched TFBSs in promoters of the signature genes (foreground set) in comparison to a background set which contains genes with very small fold changes (~ 0) in both cell lines under study. For this purpose, F-MATCH program applies an iterative process where the initial thresholds in minFP profile are regularly altered until the best possible thresholds are defined which provide most significantly enriched TFBSs. This enrichment analysis yields important key TFs, which may not be mutated themselves, but their altered activation may potentially lead to a persistent expression of their target signature genes, thereby affecting tumorigenesis.
2.4.2. Overrepresented Pathways in Colorectal Cancer
To gain more insights into the functional properties of the signature genes and their transcriptional regulators in CRC, we investigated the overrepresented pathways. For this purpose, we observed the signal transduction and metabolic pathways from TRANSPATH (Krull et al., 2006) database which contains information about genes/molecules and reactions to build complete networks. In this study, we performed two distinct pathway analyses, of which the first one refers to the overrepresented pathways in the signature genes, and the second one is based on the enriched TFBSs found in the promoters of these signature genes.
2.4.3. Identification of Master Regulators with TRANSPATH
Master regulators (MRs) are molecules which are at the very top of regulatory hierarchy and, thus, they are not affected by any of their downstream molecules. Their identification provides important knowledge to display functional relationships of genes. In this study, using the TRANSPATH database, we employed a standard workflow with a maximum radius of 10 steps upstream of TFs to identify their potential MRs.
2.4.4. Transformation of PWMs to Their Corresponding TFs and TF Family/Subfamily Classifications
Multiple PWMs can be assigned to a TF and several TFs belong to a TF family/subfamily. To obtain the correct assignments of the PWMs to their respective TFs and TF family/subfamily, we used the annotations integrated in the geneXplain platform. TF family/subfamily classifications are curated in TFClass (http://tfclass.bioinf.med.uni-goettingen.de/tfclass) which is a classification resource with the aim to catalog TFs based on their DNA-binding characteristics (Wingender et al., 2013). TFClass incorporates a six level classification schema which consists of superclasses, classes, families, subfamilies, genera and factor species of which subfamilies and factor species are optional. At the family level, TFs are primarily grouped on basis of sequence similarities of their DNA-binding domains. The optional subfamily level comprises two more levels which represent genes and gene products, termed genera and species, respectively. TFClass uses a digit-based classification schema which is analogous to the Enzyme Commission numbering system. The schema assigns a four-digit number for the top four classification levels or a six-digit number with respect to the two optional sublevels of the subfamily level.
3. Results
Classical discovery of individual markers usually involves the comparison of normal cells vs. cancer cells, which provides candidates for prognosis as well as individualized treatments. In this study, however, we focused on the in silico comparative analysis of two distinct cancer cell lines which serve as models to describe pathways. The cancer cell-intrinsic pathway repertoire and their activation status may differ between distinct cancer cell lines of the same cancer type, which in turn may have an impact on invasiveness and organ colonization in vivo. Apart from that, it still remains largely unclear as to what extent these processes are promoted or inhibited by the tumor microenvironment. Therefore, it is mandatory to first learn about the cancer cell line-specific pathway repertoire and, further, to test their functional consequences in in vivo models. Above all, the cell lines under study represent suitable models to investigate the molecular mechanisms by which mutations cause predisposition to the formation of multiple colorectal tumors. In addition, they can be used to screen for early disease biomarkers, and to develop therapeutic and preventive strategies.
3.1. Overview of the Analysis Workflow
Our workflow involved four major steps of which the first one was performed using the Trinity platform and all following steps using the geneXplain platform as described below (see also Figure 1):
1. Selection of signature genes (Section 3.2)
a) Analysis of differentially expressed transcripts
b) Clustering of the most differentially expressed transcripts
2. Identification of overrepresented TRANSPATH pathways based on signature genes (Section 3.3)
a) Pathway analysis for 1638N-T1 (Section 3.3.1)
b) Pathway analysis for CMT-93 (Section 3.3.2)
3. Identification of transcription factors (TFs) based on signature genes (Section 3.4)
a) Prediction of enriched TFBSs in promoters using a CRC-specific PWM library
b) Mapping of TFBSs to corresponding TFs as well as TF family/subfamily classifications
c) Grouping of TFs as well as TF family/subfamily into three subsets: 1638NT-1- and CMT-93-intersection-specific TF set; 1638NT-1-specific TF set; CMT-93-specific TF set (Sections 3.4.1, 3.4.3, and 3.4.5)
d) Identification of overrepresented TRANSPATH pathways based on the three TF sets (Sections 3.4.2, 3.4.4, and 3.4.6)
4. Identification of upstream master regulators in pathways based on the three TF sets (Section 3.5)
a) Search for master regulators upstream of TRANSPATH-mapped molecules of each TF set (Sections 3.5.1, 3.5.2, and 3.5.3)
b) Merging of master regulator pathways based on the top three master regulators found for each TF set (Figures 2–4).
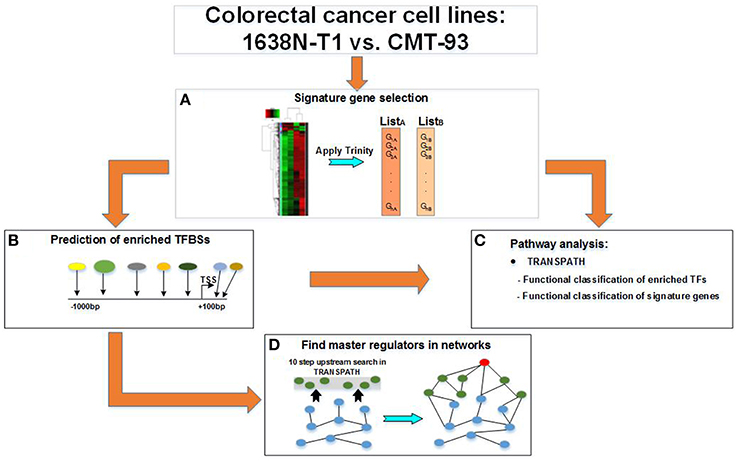
Figure 1. Workflow for the study of distinct colorectal cancer cell lines. A multi-step workflow is outlined for the comparison of the 1638N-T1 and CMT-93. (A) The analysis begins with the identification of signature genes based on RNA-seq samples using the Trinity platform. This step generates two disjunct lists of signature genes which are further applied to different geneXplain analyses. (B) The signature genes are searched for overrepresented TRANSPATH pathways. Enriched transcription factor binding sites (TFBSs) are searched within the −1 kb/+100 bp promoter regions of the signature genes to obtain transcription factors (TFs). (C) The TFs are then searched for overrepresented TRANSPATH pathways. (D) A master regulatory network is generated by searching for a master regulator (red node) up to 10 steps upstream of the TFs (blue nodes) in TRANSPATH. The master regulator is connected via intermediate molecules (green nodes) with the TFs.
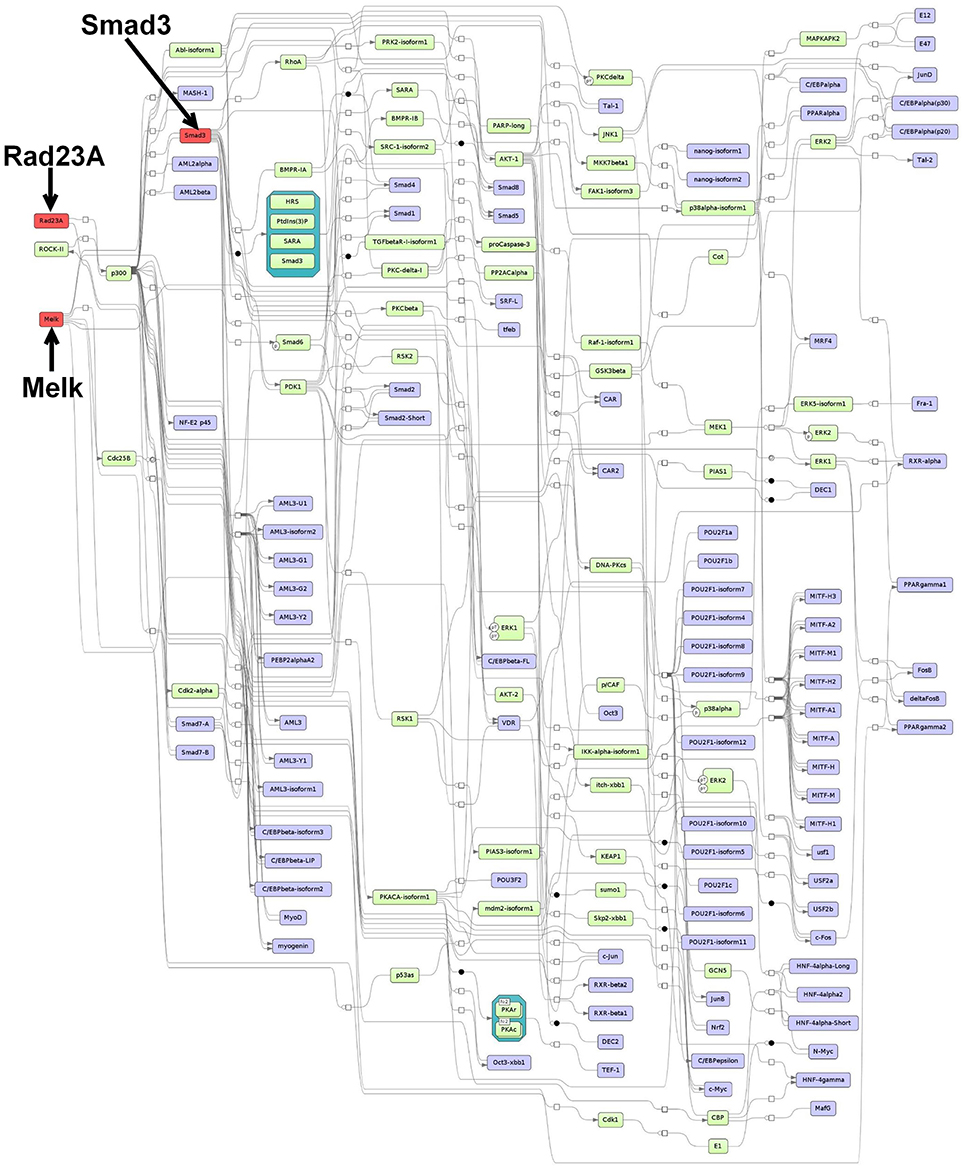
Figure 2. Master regulatory network based on the intersection-specific TF set. The color coding red, blue and green represent nodes for master regulators, regulated transcription factors and connecting molecules, respectively.
3.2. Signature Genes
Tumor initiation, promotion and progression is generally driven by genes whose expression is changed in tumor cells. Comparing gene expression profiles and detection of differentially expressed transcripts between different cancer cell lines can reveal molecular characteristics of the tumor cells under study. Using the Trinity platform we identified signature genes based on their altered transcriptional regulation in the context of CRC. In total, 2296 and 2342 Ensembl gene IDs were identified for 1638N-T1 and CMT-93, respectively. Supplementary Tables S1, S2 provide the full sets of signature genes for 1638N-T1 and CMT-93, respectively.
3.3. Pathway Analyses Based on Signature Gene Sets
The molecular characterization of tumor cells and the molecular mechanisms through which tumor cells acquire the capability to grow progressively, survive and metastasize are numerous and depend on genetic and environmental factors. On the other hand, tumor antigens can be recognized by host T cells, thereby triggering an immune response against the colonization of tumor cells. It is partly the activation of immune system suppressive pathways by the tumor cells which can decide whether cancer evades the anti-tumor immune responses and progresses. Moreover, the expression of various cytokines and chemokines controls the balance between anti-tumor immunity and pro-tumor inflammation. Besides cytokines and chemokines, several TFs and enzymes play critical roles in regulatory functions during tumor development. Therefore, analyzing the tumor-specific expression profiles and detection of these molecules, in particular TFs, are crucial steps in studying the molecular characteristics of tumor cells. Moreover, the knowledge about these molecules and their pathways will provide further information on the molecular mechanisms which may be linked to tumor aggressiveness. In this light, we searched for important pathways for 1638N-T1 and CMT-93 based on their signature genes and exemplarily provided references for their roles in cancer. With the previously defined signature gene sets at hand, we obtained overrepresented TRANSPATH pathways using the geneXplain platform.
3.3.1. Pathway Analysis for 1638N-T1
In total, 30 TRANSPATH pathways were found to be significantly overrepresented based on the signature genes of 1638N-T1 (Table 1). The top four most overrepresented pathways indicated a role for the signature genes Ugt1a1, Ugt1a2, Ugt1a6a, and Ugt1a7c which encode UDP-glucuronosyltransferases (UGTs). These detoxification enzymes are involved in the metabolism of endogenous and xenobiotic compounds (Cooley et al., 1982; Magnanti et al., 2000). Expression of UGTs has been implicated in human urinary bladder and colon cancer (Giuliani et al., 2005; Wang et al., 2012). Furthermore, the first two of the four pathways related to a mechanism for the detoxification of NNAL (the metabolized isoform of NNK) via UGTs-catalyzed glucuronidation pathways (Wiener et al., 2004). NNK is a tobacco agent widely known for promoting tumorigenesis and metastasis through its pro-inflammatory effects (Takahashi et al., 2010). The remaining two pathways related to glucuronidation pathways which are involved in heme degradation in response to oxidative stress. Heme ingestion leads to hyperproliferation and activation of oncogenes as well as the inhibition of the tumor suppressor p53 in response to increased cytotoxicity in the mouse colon (Ijssennagger et al., 2013). The fifth topmost overrepresented pathway corresponded to the activation of Ras-related protein Rap-1A (Rap1A) via interferon gamma (IFNγ). Rap1A is a tumor suppressor which mediates growth inhibitory responses in cancer (Alsayed et al., 2000). The cytokine IFNγ plays an important role in innate and adaptive immune responses and prevents development of primary and transplanted tumors (Ikeda et al., 2002). Further, the pathway analysis found two putative pro-inflammatory metabolic pathways which involve the molecules eicosanoid hepoxilin A3 (hepA3) and platelet activating factor (PAF), respectively. Both molecules have been suggested to play key roles in inflammation-associated cancer (Mrsny et al., 2004; Tsoupras et al., 2009). Furthermore, the results reported a signaling cascade which leads to the activation of mitogen-activated protein kinase 1 (Mapk1/Erk2) via interleukin-8 (IL-8). Several studies have implicated IL-8 in tumor angiogenesis, growth, and metastasis in colon, gastric and pancreatic carcinoma (Li et al., 2001, 2008; Kuai et al., 2012; Sun et al., 2014a). A recent study showed that IL-8 increases the migration in human CRC cells through the integrin alpha-V/beta-6 and chemokine receptors CXCR1/2 involving the activation of Mapk1 and Ets-1 signaling pathway (Sun et al., 2014a). Another reported pathway relates to the interleukin-3 (IL-3)-induced activation of the JAK2/STAT5 pathway. IL-3 expression via the T cell receptor signaling pathway is known to regulate growth and differentiation of hematopoietic stem cells, neutrophils, eosinophils, megakaryocytes, macrophages, lymphoid and erythroid cells (Reddy et al., 2000). Lastly, the results showed overrepresentation for the activation of Wnt signaling which is aberrantly activated in the majority of CRCs (Cancer Genome Atlas Network, 2012).
3.3.2. Pathway Analysis for CMT-93
The pathway analysis resulted in the identification of 28 overrepresented TRANSPATH pathways based on the signature genes of CMT-93 (Table 2). The four topmost overrepresented pathways share 13/14 hit signature genes which are all associated with the assembly of protein complexes called adherens junctions that occur in epithelial and endothelial tissues (Guo et al., 2007). One prominent signature gene amongst these hits was E-cadherin (cadherin-1/CDH1) that belongs to the cadherin superfamily and encodes a calcium-dependent cell adhesion protein. E-cadherin acts as an invasion suppressor and its loss in epithelial carcinomas permits the invasion of adjacent normal tissues. Several studies showed that the level of E-cadherin expression is inversely correlated with tumor malignancy (Vleminckx et al., 1991; Cowin et al., 2005; Junghans et al., 2005). Likewise, protein-protein interactions between E-cadherin and β-catenin result in the formation of a tumor-suppressor system (Müller et al., 1999). The regulation of β-catenin/E-cadherin has been associated with the induction of epithelial-mesenchymal transition (EMT) and metastasis (Morali et al., 2001; Kim et al., 2002; Eger et al., 2004).
The results included further pathways which are related to the phosphorylation and desphosphorylation of the β-catenin/E-cadherin complex. In this regard, it has been reported that phosphorylation of β-catenin, e.g., through the epidermal growth factor receptor (EGFR) or the tyrosine-protein kinase Src, leads to the dissociation of the complex and consequently to the accumulation of free β-catenin. On the contrary, dephosphorylation of β-catenin results in the formation of the complex (Müller et al., 1999). Another overrepresented pathway corresponded to nerve growth factor (NGF) signaling via the tyrosine kinase receptor TrkA. NGF has been associated with cancer cell proliferation as well as apoptosis of colon cancer cells (Molloy et al., 2011; Anagnostopoulou et al., 2013) and with angiogenesis (Romon et al., 2010). Further overrepresented pathways related to the angiopoietin-Tie signaling system which plays a role in the regulation of angiogenesis (Fagiani and Christofori, 2013). In tumors, angiopoietin-2 (Ang2) inhibits the activity of the receptor tyrosine kinase Tie2 and destabilizes blood vessels, thereby facilitating angiogenesis (Holash et al., 1999a,b; Augustin et al., 2009). Moreover, several other overrepresented pathways could be linked to anti-tumor properties. These included two p53-dependent pathways which lead to the induction of the cyclin-dependent kinase inhibitor 1 (p21Cip1) or the p53 upregulated modulator of apoptosis (Puma), respectively. Downregulation of p21Cip1 expression has been associated with poor prognosis and expression of Puma with a rapid apoptosis in CRC (Pasz-Walczak et al., 2001; Yu et al., 2001). Furthermore, the results also included overrepresented pathways which related to vitamin D receptor (VDR) signaling and vitamin D metabolism. VDR signaling is activated upon binding of vitamin D and plays a role in cancer progression as well as cross-talks with multiple other pathways (Slattery, 2007). For example, several studies have suggested interactions of vitamin D or its active vitamin D metabolite, calcitriol, with β-catenin (Deeb et al., 2007; Zheng et al., 2012; Klampfer, 2014). These interactions represent points of convergence between VDR and canonical Wnt signaling in CRC, which has been linked to inhibition of Wnt signaling, tumor growth inhibition, the activation of apoptotic pathways, inhibition of angiogenesis and inhibition of tumor-promoting inflammation (Deeb et al., 2007; Zheng et al., 2012; Klampfer, 2014).
3.4. Promoter Analysis Based on Signature Genes
Altered gene expression is generally a result of the dysregulated activity of TFs that may play central roles as oncogenes and tumor suppressors. These proteins are often potential targets for cancer therapies due to the fact that many oncogenic signaling pathways involve TFs whose aberrant activation and inactivation contributes to tumor development and progression. We applied a promoter analysis to the previously identified signature genes in order to display which TFs are potentially important regulators in the cell lines under study. This analysis was performed using geneXplain which quantifies the enrichment of TFBSs in promoter regions of the signature genes. In total, 135 and 117 TFs were identified for 1638N-T1 and CMT-93, respectively. These numbers include 51 (Supplementary Table S4) and 33 TFs (Supplementary Table S5) that were exclusively enriched in 1638N-T1 or CMT-93, respectively, as well as 84 overlapping TFs in the intersection between both cell lines (Supplementary Table S6). We exemplarily highlighted several TF families/subfamilies which are present for the three TF sets. In a subsequent analysis, we additionally searched for overrepresented pathways on the basis of these sets.
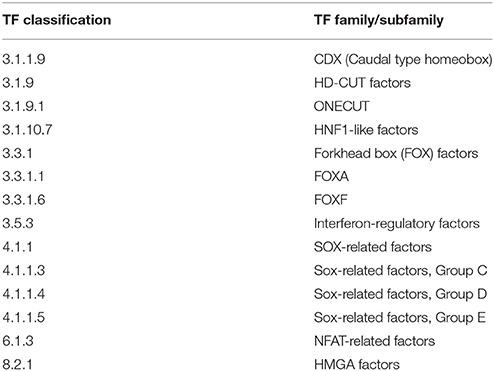
3.4.1. Intersection-Specific TF Families/Subfamilies of 1638N-T1 and CMT-93
The enriched TFBSs were classified into 32 prominent TF families/subfamilies according to TFClass (Table 3). Our analysis detected several members of the SMAD factor family that were found to have enriched binding sites in the promoters. These factors are a major component of TGF-β signaling which is involved in the regulation of cell growth in the normal intestinal epithelium. Alterations in their expression contribute to cancer aggressiveness in CRC (Xie et al., 2003; Xu and Pasche, 2007; Korchynskyi et al., 1999; Fleming et al., 2013). Furthermore, the analysis revealed overrepresentation for members of the Jun-related factors and Fos-related factors. The protein AP-1 is composed of either Jun-Jun homodimers or Jun-Fos heterodimers and plays a role in differentiation, proliferation, and apoptosis (Ameyar et al., 2003). AP-1 is induced by c-Jun N-terminal protein kinases (JNK) and ERK MAPKs pathways or the canonical Wnt signaling pathway in CRC (Licato et al., 1997; Mann et al., 1999), thereby affecting CRC cell proliferation (Suto et al., 2004). Binding site enrichment was also detected for the CCAAT-enhancer binding protein (C/EBP) family of TFs whose expression has been associated with invasiveness of human colorectal cancer (Rask et al., 2000). Likewise, several members of the POU domain factor family, including Oct-4 (Pou5f1), were found in the intersection between both cell lines. It has been reported that Oct-4 promotes metastasis in CRC through EMT (Dai et al., 2013). Furthermore, Oct-4 knockdown leads to decreased Wnt pathway activity and high risk for liver metastases in CRC patients (Dai et al., 2013). Enrichment for binding sites of VDR, which belongs to the Thyroid hormone receptor-related factor (NR1) family, was also detected in the intersection. It has been suggested that vitamin D has no effect on tumor reduction in APC-deficient mice and that VDR expression is lost in the majority of the colon cancer cells (Giardina et al., 2015). Interestingly, the analysis also revealed enrichment for binding sites of β-catenin which interacts as a cofactor with members of the TCF-7-related factor family to activate Wnt target gene expression (see Supplementary Table S6).
3.4.2. Overrepresented TRANSPATH Pathways Based on Intersection-Specific TFs
Based on the 84 overlapping TFs in the intersection of both cell lines, the pathway analysis revealed overrepresentation for 35 TRANSPATH pathways (Table 4). Members of the SMAD factor family were found to be involved in many of the top overrepresented pathways. In this context, the TGF-β pathway was detected among the most overrepresented pathways. Likewise, SMADs were also found to be involved in a pathway which corresponded to the regulation of endothelin-1 (ET-1). ET-1 is a vasoconstrictor peptide, which is known to be produced by CRC cells and stimulates CRC proliferation (Asham et al., 2001; Grant et al., 2007; Knowles et al., 2012). The second most overrepresented pathway corresponded to the transcriptional regulation of ECM components. ECM sustains normal tissue homeostasis and prevents malignant transformation (Gao et al., 2014). Its anti-tumor properties are opposed by chronic inflammation, which may lead to the conversion of a tumor-inhibiting into a tumor-promoting microenvironment (Gao et al., 2014).
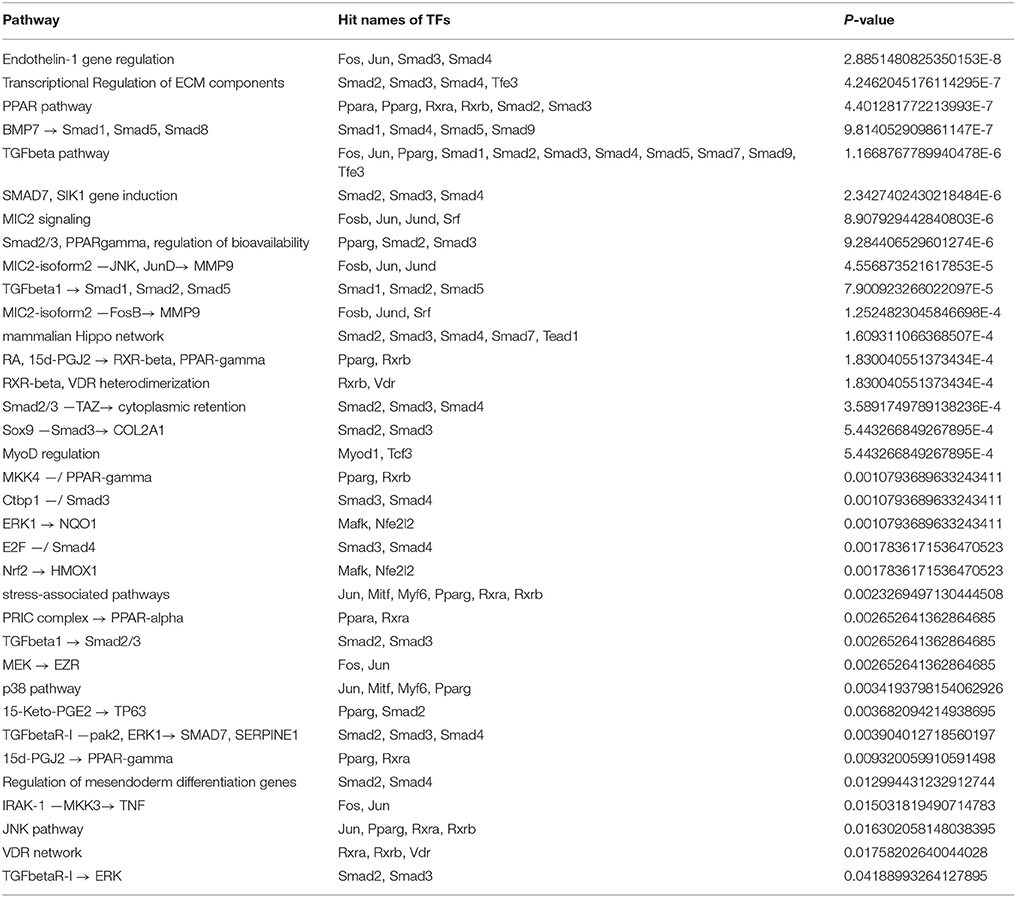
Table 4. Overrepresented TRANSPATH pathways based on the intersection-specific TF set of 1638N-T1 and CMT-93.
Furthermore, the analysis showed overrepresentation for a PPAR-related pathway which comprises the peroxisome proliferator activated receptors PPAR-α, PPAR-γ and Smads. It was shown that activation of PPAR-γ inhibits TGF-β-induced loss of E-cadherin expression, the induction of mesenchymal markers (vimentin, N-cadherin, fibronectin), MMPs and antagonizes Smad3 function, thereby preventing metastasis in lung cancer (Reka et al., 2010). This pathway has also been implicated in the induction of apoptosis as well as inhibition of cyclooxygenase-2 (COX-2) in CRC (Yang and Frucht, 2001). Activation of the PPAR pathway was shown to cause reduction in linear and clonogenic growth and, thus, it has been suggested that PPAR-γ modulates cell growth and differentiation of CRC cells (Sarraf et al., 1998). Moreover, it was shown that PPAR-γ expression is altered in APC-deficient mice, an effect which is thought to be mediated by the Wnt/β-catenin pathway (Jansson et al., 2005). In conformity with the overrepresented pathways, which were found based on the signature genes of CMT-93, a VDR network-related pathway was also found based on the intersection-specific TFs.
3.4.3. 1638N-T1-Specific TF Families/Subfamilies
The enriched TFBSs can be classified into 14 prominent TF families/subfamilies based on the 1638N-T1-specific TFs (Table 5). Amongst these, the factors Onecut1 and Onecut2, which belong to the HD-CUT factors family, were found to be enriched in the signature genes of 1638N-T1. Through targeting of Onecut2, the microRNA miR-429 has been reported to regulate the expression of several EMT-related markers (Sun et al., 2014b). Overall, it has been suggested that Onecut2 is involved in EMT, migration and invasion of CRC cells (Sun et al., 2014b). Onecut1 (Hnf6) expression was found to be positively correlated with the expression of p53 and E-cadherin in human lung cancer. The Onecut1-mediated induction of p53 is thought to inhibit EMT, migration and invasion (Yuan et al., 2013). Moreover, the analysis detected the HOX-related factors Cdx1 and Cdx2, which regulate intestine-specific gene expression and enterocyte differentiation (Suh et al., 1994; Suh and Traber, 1996; Taylor et al., 1997; Freund et al., 1998; Soubeyran et al., 1999; Lynch et al., 2003). In addition, it has been suggested that expression of Cdx1 reduces cancer cell proliferation by reducing cyclin D1 expression (Lynch et al., 2003). Interestingly, Cdx1 and Cdx2 also inhibit proliferation of CRC cells by blocking canonical Wnt signaling activity (Guo et al., 2004). In contrast, another study indicated that Cdx2 can promote expression of Wnt/β-catenin pathway genes (da Costa et al., 1999). Furthermore, the analysis revealed overrepresentation for several members of the interferon regulatory factor (IRF) family. Most IRFs play central roles in immune response, apoptosis and are known to exhibit tumor suppressor properties in cancer (Bouker et al., 2005). For example, anti-tumor function of IRF-1- and IRF-5-associated pathways have been suggested in CRC (Hu and Barnes, 2006; Yuan et al., 2015). The analysis also detected Sox9, a member of the SOX-related factors. Sox9 is a target as well as potential upstream regulator of Wnt signaling (Blache et al., 2004; Bastide et al., 2007).
3.4.4. Overrepresented TRANSPATH Pathways Based on 1638N-T1-Specific TFs
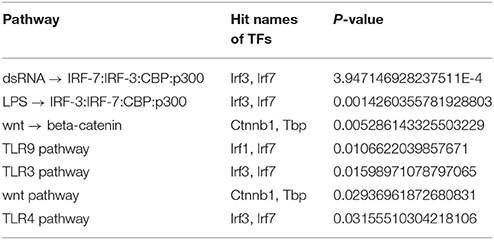
In total, 7 overrepresented pathways were found based on the 51 exclusive TFs for 1638NT-1 (Table 6). The results included overrepresented pathways which corresponded to the TLR (Toll-like receptor) pathways TLR3, TLR4, and TLR9. TLRs are pattern recognition receptors (PRRs) that play key roles in innate and adaptive immune responses. In host defence, TLRs recognize pathogens by pathogen-associated molecular patterns (PAMPs). TLRs are involved in inflammatory reponses, cell proliferation and survival, and have been associated with pro-tumor as well as anti-tumor effects in cancer (Rakoff-Nahoum and Medzhitov, 2009; Basith et al., 2012). TLR signaling pathways promote the production of cytokines and chemokines via interfering with intracellular pathways and activation of TFs, such as IRFs and NF-κB (Li et al., 2014). In particular, activation of the TLR9 pathway promotes the development of anti-tumor T-cell responses (Krieg, 2008). In contrast, it was also shown that this pathway can promote angiogenesis and cancer progression (Belmont et al., 2014; Holldack, 2014). TLR3 activation mediated by dsRNA was shown to trigger apoptosis of human breast cancer cells (Salaun et al., 2006). Additionally, signaling by IRF-3 has been implicated in TLR3-mediated apoptosis in prostate cancer (Gambara et al., 2015). Another overrepresented TRL-related pathway corresponded to the lipopolysaccharide (LPS)-induced activation of the TFs IRF-3, IRF-7, and CBP/p300 via the TLR4/MD2 complex. Moreover, it was shown that metastasis of CRC cells is increased through a signaling cascade involving LPS-induced TLR4 signaling as well as downstream PI3K/Akt signaling and β1 integrin activity (Hsu et al., 2011). LPS also increases phosphorylation of Mapk1 and p38, activation of NF-κB, and promotes cytokine production, such as that of IL-8, vascular endothelial growth factor (VEGF), and TGF-β in human colon cells (Tang and Zhu, 2012). Moreover, the same study has implicated TLR4 in promoting immune escape of the human colon cancer cells by inducing immunosuppressive factors and apoptosis resistance (Tang and Zhu, 2012). Strikingly, two pathways corresponded to the canonical Wnt/β-catenin signaling pathway which is of high relevance in CRC.
3.4.5. CMT-93-Specific TF Families/Subfamilies
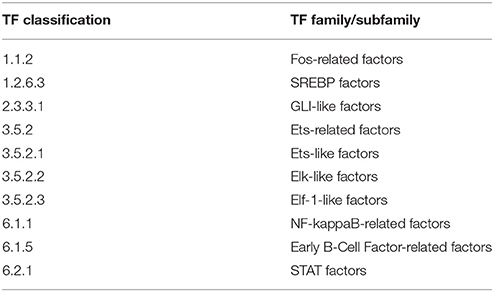
The enriched TFBSs can be classified into 10 prominent TF families/subfamilies for the CMT-93-specific TFs (Table 7). The results included Ebf3 which is a member of the Early B-Cell Factor-related factors family. This family plays a role in differentiation of specific cell types such as B lymphocytes and olfactory cells (Zhao et al., 2006). Expression of Ebf3 was previously shown to promote cell cycle arrest and apoptosis in several tumor cell lines including colon carcinoma (Zhao et al., 2006).
The analysis also reported enriched TFBSs for the NF-κB-related factor family. NF-κB signaling is usually induced by inflammation and also known to be triggered by cancer progression. Many recent findings indicate that NF-κB is constitutively activated in malignant cells of various cancers including CRC (Nakshatri et al., 1997; Wang et al., 1999; Lindholm et al., 2000; Lind et al., 2001; Kojima et al., 2004), thereby promoting, cell proliferation, angiogenesis, metastasis, upregulation of chemokine secretion and other anti-apoptosis proteins (Sakamoto et al., 2009; Wang et al., 2009). Furthermore, enriched binding sites were detected for the signal transducer and activator of transcription (STAT) family which are critical regulators of immune and inflammatory responses (Yu et al., 2009). These factors play an important role in many types of cancer, including colorectal cancer, as they may promote pro-tumor inflammatory pathways such as NF-κB and JAK/STAT pathways, as well as suppress anti-tumor immunity (Wang et al., 2009; Yu et al., 2009; Slattery et al., 2013). The activation of Stat3 and Stat5 has been shown to promote cell proliferation and invasion in cancer (Yu et al., 2009), while Stat3 was also found to be persistently activated and overexpressed in colon cancers (Klampfer, 2008). Our analysis also revealed binding site enrichment for several members of the family of Ets-related factors which are involved in diverse cellular processes, thereby often cooperatively interacting with other TFs and co-factors (Oikawa, 2004). In cancer, this family is known to regulate genes which play a role in angiogenesis, invasion and metastasis. Therefore, their altered expression has been implicated in development and progression of cancer (Bassuk and Leiden, 1997; Graves and Petersen, 1998; Oikawa and Yamada, 2003; Oikawa, 2004). Moreover, it has been suggested to use ETS-related factors as prognostic markers in cytotoxic treatment of metastatic colorectal cancer (Giessen et al., 2013).
3.4.6. Overrepresented TRANSPATH Pathways Based on CMT-93-Specific TFs
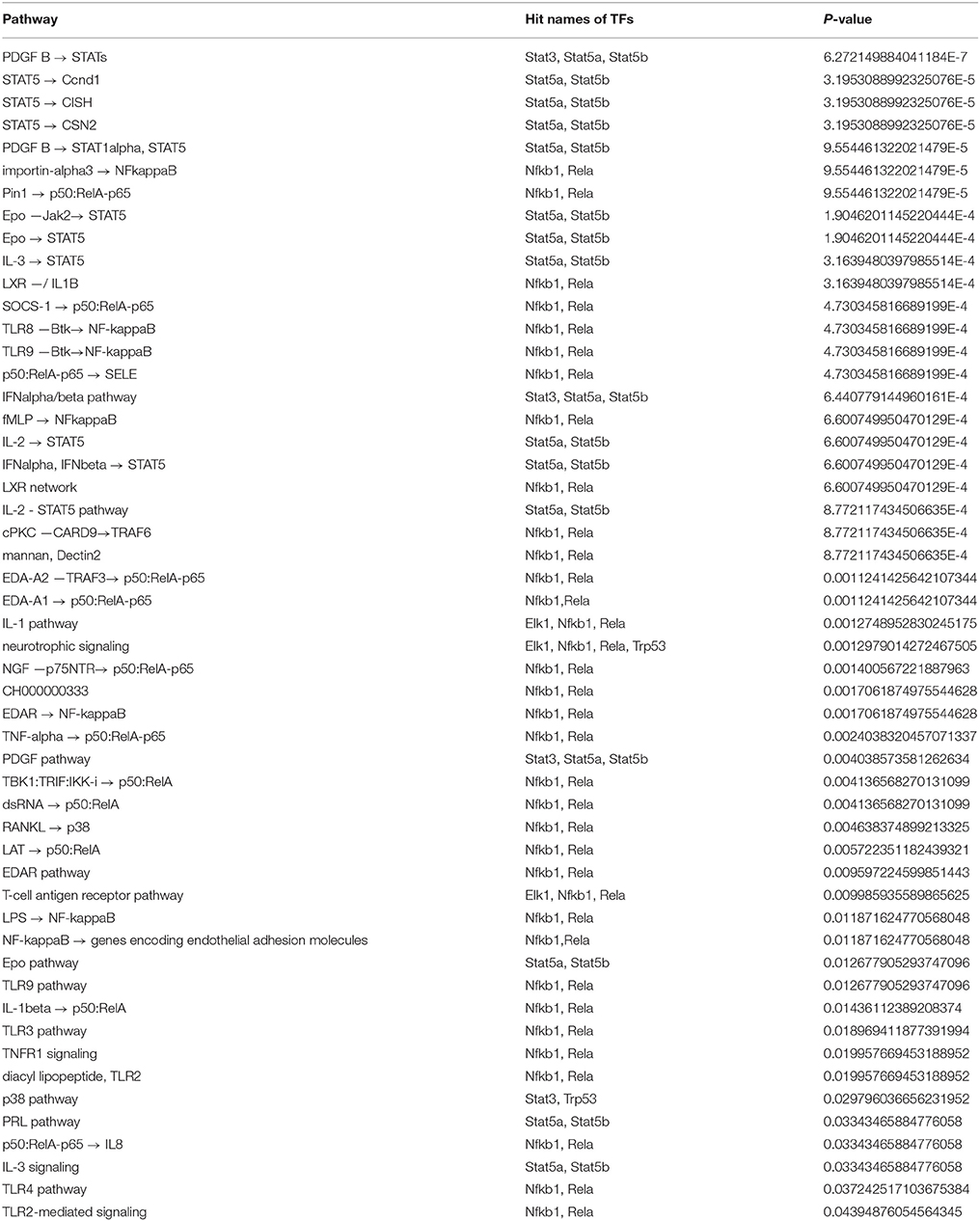
In total, 52 overrepresented pathways were found based on the 33 exclusive TFs for CMT-93 (Table 8). Most of these overrepresented pathways involved NF-κB family members. Further overrepresented pathways involved the tumor necrosis factor-alpha (TNF-α) of which one related to the TNF-α-mediated activation of NFκB. An increase in production of the pro-inflammatory cytokine TNF-α is linked to poor outcome in CRC (Balkwill2005, Mantovani2005, Coussens2002, Balkwill2001). Interestingly, TNF-α was shown to promote Wnt signaling through translocation of β-catenin into the nucleus in gastric tumor cells (Oguma et al., 2008).
In conformity with the results obtained for the 1638N-T1-specific TFs, TLR-related pathways for five different TLRs (TLRs 2,3,4,8,9) were also detected for the TFs of the CMT93-specific set. The results further included several overrepresented STAT factors-related pathways that included an activation of STATs by platelet-derived growth factor (PDGF)-mediated signaling. Signaling via PDGF tyrosine kinase receptors plays an important role in angiogenesis, mesenchymal cell migration, proliferation and the expression and activation of PDGF receptors is particularly associated with invasion and metastasis in CRC (Yu et al., 2003; Kitadai et al., 2006; Steller et al., 2013).
Moreover, the analysis detected overrepresentation for LXR-related pathways that implicate a role for NFκB subunits RELA/p65, NFKB1/p105, NFKB1/p50 as well as interleukin-1 beta (IL-1β). Interestingly, the signature gene set for CMT-93 included the factors Nr1h2 and Nr1h3, two members of the thyroid hormone receptor-related factor (NR1) family. These genes encode liver X receptors (LXRs), of which the oxysterol receptor LXRα (Nr1h3) is thought to increase caspase-dependent apoptosis, slow growth of xenograft tumors in CRC mouse models and may negatively interfere with Wnt signaling through direct binding to β-catenin in CRC (Uno et al., 2009; Sasso et al., 2013). Hence, LXRs have been considered important potential targets in cancer therapeutics on account of their tumor suppressor activities (Sasso et al., 2013; Vedin et al., 2013; Lin and Åke Gustafsson, 2015). With respect to IL-1β, this pro-inflammatory cytokine has been associated with angiogenesis, invasiveness of different tumor cells and increased risk of CRC (Voronov et al., 2003; Andersen et al., 2013).
3.5. Identification of Upstream Master Regulators in Pathways Based on TF Sets
In the previous step, we reported potentially important TFs for the sets of signature genes, on the basis of which we defined sets of TFs for the intersection between the two cell lines as well as for the 1638N-T1-specific and CMT-93-specific TFs. Since signal transduction pathways can modulate the activity of nuclear TFs, activation mutations in these pathways can lead to the altered expression of the TFs and their target genes. These pathways are diverse in both their complexity and the mechanism of signal transduction, and even more complexity is added through cross-talks or transactivation signals between different pathways. Therefore, we were interested in the detection of upstream regulators, called master regulators, for the previously defined TF sets. We additionally aimed to construct the upstream pathways which may regulate activity or inhibition of the TFs.
We applied the master regulator analysis from geneXplain to each of the three TF sets, namely the intersection with overlapping TFs between 1638N-T1 and CMT-93, the 1638N-T1-specific and the CMT-93-specific TFs. This workflow will first map the set-specific TFs to TRANSPATH molecules and then search based on the TRANSPATH knowledge for upstream master regulators. We report the top three master regulators for each TF set (Table 9) and provide references for their roles in cancer. Noteworthy, we only proposed distinct master regulators for each gene set, i.e., different splice variants or isoforms of a master regulator reported by the analysis were counted as the same master regulator.

Table 9. Top three master regulators for three TF sets: Intersection-specific TFs of the two cell lines, 1638N-T1-specific TFs and CMT-93-specific TFs.
The master regulators and their pathways, denoted as master regulator pathways, constitute the set-specific TFs which are either connected to other set-specific TFs or intermediate molecules. These intermediate molecules are not contained within the respective TF sets but function as a bridge between the set-specific TFs and the master regulator(s) in the pathways. Since the pathways of the top ranked master regulators share many of the interacting nodes and, thus, are very similar to each other, we merged the top 3 master regulator pathways for each set into one network.
3.5.1. Prediction of Master Regulators and Construction of a Master Regulatory Network Based on the Intersection-Specific TF Set
For the intersection-specific TF set, we obtained the three master regulators Rad23A, Smad3, and Melk that reach 91, 74, and 93 TFs from the set, respectively. The master regulator Rad23A is involved in DNA damage recognition and nucleotide-excision repair. A recent study has implicated Rad23A in nuclear translocation of the apoptosis-inducing factor (AIF) during induction of cell death (Sudhakar and Chow, 2014). However, not much is known about its specific function in CRC.
As a major component of the TGF-β signaling pathway, the Smad3 master regulator plays a pivotal role in survival, invasion, and metastasis of CRC cells (Xu and Pasche, 2007; Fleming et al., 2013). However, despite the fact that not much is known about the pathogenic role of Smad3, mutations in the gene occur rather rarely in human CRC (Ku et al., 2007). Loss of Smad3 has been associated with metastasis in CRC, an outcome that is thought to be dependent on chronic inflammation, e.g., triggered by bacterial infection (Zhu et al., 1998; Maggio-Price et al., 2006).
The third master regulator maternal embryonic leucine zipper kinase (Melk) is a known embryonic and neural stem cell marker and belongs to the family of serine/threonine kinases (Choi and Ku, 2011). Melk is normally expressed in cells that undergo proliferation during embryonic development, however, elevated expression has been particularly observed in variety of different cancer cell types including colorectal cancer (Gray et al., 2005; Badouel et al., 2010; Ganguly et al., 2015). Moreover, it has been shown that Melk knockdown decreases proliferation and tumor growth in CRC and, thus, it has been proposed to use Melk as a therapeutic target for cancer (Gray et al., 2005).
The merged master regulatory network consisted of 155 nodes (Figure 2, Supplementary Table S7 and Supplementary Figure S7). The master regulators Rad23A and Smad3 were found most upstream in the hierarchy of the network. Rad23A was connected via the nodes p300 and CBP to the other nodes in the network, whereas Smad3 was connected to a variety of nodes which also included important cancer-associated TFs such as c-Myc, Runt-related factors, and Smad factors. Likewise, the master regulator Melk featured cascades through several molecules including Smad factors and p53 (see Figure 2 for more details).
3.5.2. Prediction of Master Regulators and Construction of a Master Regulator Network Based on the 1638N-T1-Specific TF Set
The master regulator analysis detected Mlk3, Tbk1 and Siah2, which reach 28, 22, and 37 TFs from the 1638N-T1-specific set, respectively. The first master regulator MLK3 is a serine/threonine kinase that activates p38 MAP kinase, ERK, and JNK signaling pathways (Velho et al., 2014). MLK3-mediated activation has been shown to promote invasion and metastasis in several cancer types, including breast and gastric cancers (Chen et al., 2010; Mishra et al., 2010; Chen and Gallo, 2012; Cronan et al., 2012). Moreover, it has been proposed that mutant MLK3 is involved in the deregulation of several important CRC-associated signaling pathways such as WNT, MAPK, NOTCH, TGF-β, and P53 (Velho et al., 2014). Concerning Wnt signaling pathways in MLK3 mutant cells, it has been shown that components of the canonical Wnt pathway were found to be downregulated, while components of the non-canonical planar cell polarity (PCP) pathway were found to be upregulated.
The proposed master regulator TBK1 is a member of the non-canonical IκB protein kinases which is involved in the activation of IRF3 and c-Rel and NF-κB in cancer. The role of TBK1 is poorly investigated in CRC. However, several studies associated TBK1 with malignant transformation, cell growth and proliferation (Chien et al., 2006; Kim et al., 2013a,b).
The third master regulator Siah2 is an E3 ubiquitin ligase that regulates the degradation of a variety of substrates such as the nuclear corepressor (N-CoR), TRAF2, 2-oxoglutarate dehydrogenase-complex protein E2 (OGDC-E2), TIEG, and β-catenin (Zhang et al., 1998; Matsuzawa and Reed, 2001; Habelhah et al., 2002, 2004; Johnsen et al., 2002). Siah2 has been implicated in MAPK signaling, mitochondrial dynamics and cell survival (Nakayama et al., 2009; Kim et al., 2011). In addition, several studies have indicated that Siah2 functions as a proto-oncogene, while the Siah1 isoform has been associated with tumor suppressor activity (Wong and Möller, 2013; Gopalsamy et al., 2014). Although its role in CRC remains unclear, Siah2 has been suggested to promote invasion and metastasis in a variety of other cancers, including prostate, breast and liver (Qi et al., 2010, 2013; Behling et al., 2011; Malz et al., 2012; Sarkar et al., 2012; Wong et al., 2012; Gopalsamy et al., 2014).
The merged master regulatory network consisted of 52 nodes (Figure 3, Supplementary Table S8 and Supplementary Figure S8). MLK3 and Siah2 were found most upstream in the hierarchy of the network, whereas TBK1 was found downstream of the network branch which is regulated by Siah2. MLK3 featured cascades through MKK3-isoform1, 4, and 6, and IKK-alpha-isoform1, and -beta. Siah2 was connected via the molecule alpha-synuclein-isoform1, Ubc5A, B, and C. TBK1 was connected via IRF3, 5, and 7, STAT6, and IKK-beta to its downstream nodes.
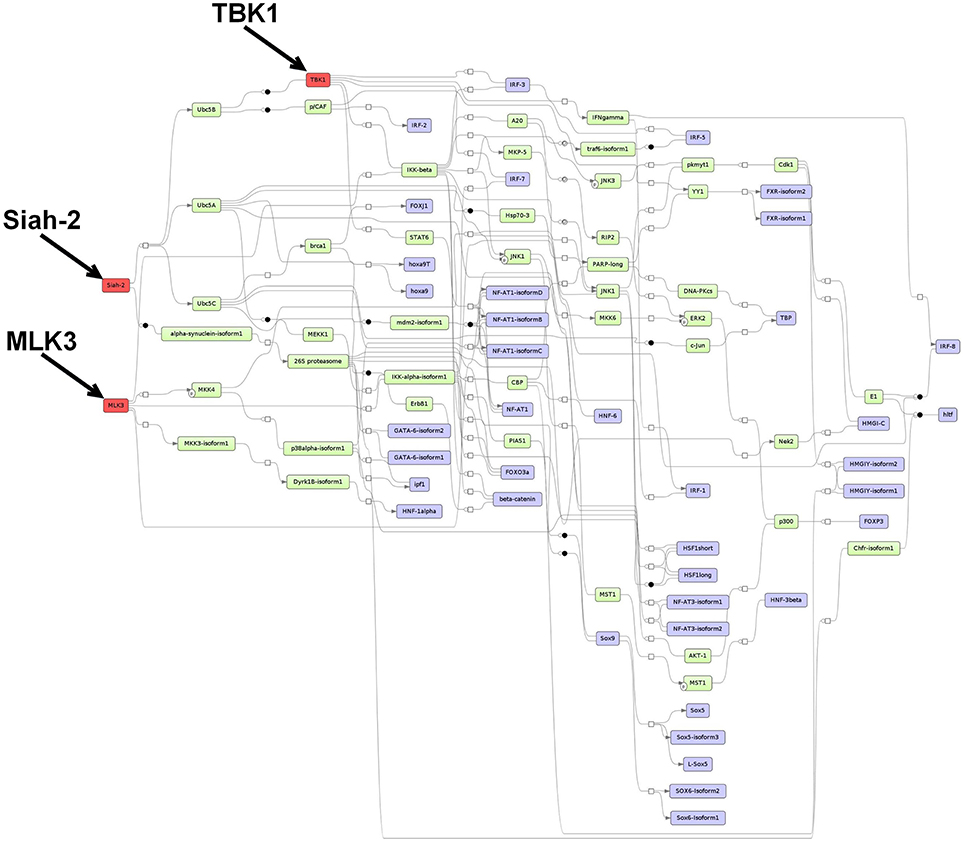
Figure 3. Master regulatory network based on the 1638NT-1-specific TF set. The color coding red, blue and green represent nodes for master regulators, regulated transcription factors and connecting molecules, respectively.
3.5.3. Prediction of Master Regulators and Construction of a Master Regulator Network Based on the CMT-93-Specific TF Set
For the CMT-93-specific TFs, the analysis reported the master regulators Aebp1 (ACLP), Il2rg (gamma-c) and Mapk1 (ERK2), which reach 43, 36, and 31 TFs from the set, respectively. The first proposed master regulator, Aebp1, is known to act as a transcriptional repressor in adipogenesis (Ladha et al., 2012). Aebp1 is upregulated in the majority of the primary glioblastoma multiforme (GBM) and loss of Aebp1 function was shown to result in apoptosis (Ladha et al., 2012). Moreover, Aebp1 induces NF-κB activity which leads to macrophage inflammatory responsiveness and affects tumor cell growth and survival (Majdalawieh et al., 2007). In the context of breast cancer tumorigenesis, Aebp1 has been suggested to be involved in the regulation of the cross-talk between mammary epithelium and stroma (Holloway et al., 2012). To this date, the role of Aebp1 remains largely unclear in CRC.
The second master regulator corresponded to the interleukin 2 receptor subunit gamma (Il2rg/gamma-c) which heterodimerizes with several interleukin receptors, including receptors for the interleukins −2, −4, −7, −9, −15, and −21 (Nata et al., 2015). Interleukins receptor signaling pathways are known to play crucial roles in inflammation-dependent progression and anti-tumor responses in CRC (West et al., 2015).
The last master regulator Mapk1 (ERK2) belongs to the MAP-kinases, which regulate cell growth, differentiation, proliferation, migration, and apoptosis (Santarpia et al., 2012). MAPKs act downstream of several growth-factor receptors such as Egfr, which are often found overexpressed and activated in CRC (Fang and Richardson, 2005). Thus, it has been stated that the ERK MAPK pathway plays a central role in the progression of CRC (Fang and Richardson, 2005). In addition, it has been proposed that this pathway but not the JNK pathway or the p38 MAPK pathway is the key regulator of cell proliferation in CRC (Fang and Richardson, 2005).
The merged master regulatory network was composed of 65 nodes (Figure 4, Supplementary Table S9 and Supplementary Figure S9). ACLP (Aebp1) and Il2rg (gamma-c) were found to be the regulators most upstream in the network. ACLP (Aebp1) was connected via the nodes ERK1 and TNF-alpha to the other nodes in the network. The master regulator Il2rg (gamma-c) featured a cascade through Jak3-isoform1, whereas the master regulator Mapk1 (ERK2) was connected to several molecules and TF families, including SREBP factors, STAT factors and Ets-like factors (see Figure 4 for more details).
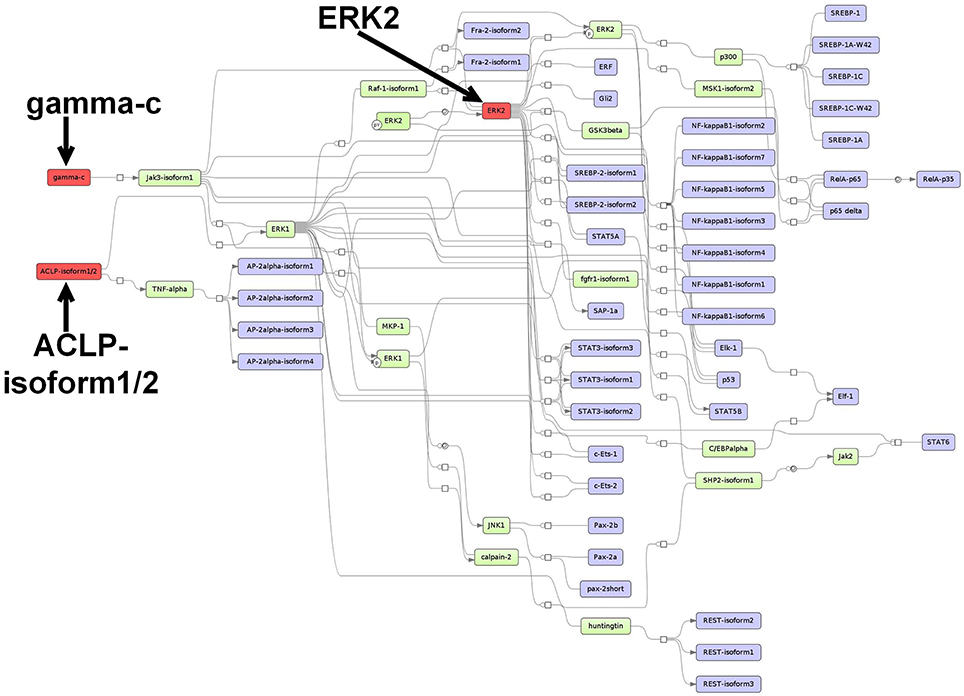
Figure 4. Master regulatory network based on the CMT-93-specific TF set. The color coding red, blue and green represent nodes for master regulators, regulated transcription factors and connecting molecules, respectively.
3.5.4. A Comparison with Randomly Selected Gene Sets
To test the prediction quality of our results and, whether they are specific for CRC, we performed a comparison between our results and those found for randomly drawn gene sets. Thus, we first randomly selected 10 gene sets, each of which had the same sample size as the signature genes analyzed in this study. After that, each random gene set was analyzed in the same way as both signature gene sets. In this regard, we started with TFBS enrichment analyses (see Section 2.4.1) for the detection of enriched TFBSs in the promoter regions of each random gene set. After retrieving the corresponding TFs, we observed that 17 TFs were common to each of the 10 random gene sets. Interestingly, 13 out of these 17 TFs were also detected based on both CRC signature gene sets (see Section 3.4.1). To determine their potential role in the context of our results, we further searched for overrepresented TRANSPATH pathways and master regulators based on these 13 TFs (see Section 2.4.2 and 2.4.3). The results of these analyses showed that there were no overrepresented pathways and, beyond that, the master regulators were completely different from those presented in Section 3.5.1, 3.5.2, and 3.5.3. Finally, we searched for overrepresented TRANSPATH pathways based on each random gene set (see Section 2.3.2). As expected, the overrepresented pathways found for each random gene set were completely different among themselves and, thus, they have no overlap with the pathways presented in the Section 3.3.1 and 3.3.2.
4. Discussion
In this study, we specifically focused on revealing the similarities and differences with respect to the transcriptional regulation as well as the pathway repertoire of two distinct colorectal cancer (CRC) cell lines, namely 1638N-T1 and CMT-93, in a direct comparison. Based on signature genes that are most significantly upregulated in cancer cell type I and cancer cell type II, respectively, our approach aimed to identify the upstream transcriptional regulators and their regulatory networks.
Our results indicated that many of the pathways, which were identified based on the signature genes, can be linked to both pro-tumor as well as anti-tumor properties. In particular, we found pathways for 1638N-T1 which play a role in the detoxification of carcinogens, immune response, and apoptosis. Additionally, we found pathways which can be linked to oxidative stress, inflammation, cell migration, proliferation and survival. Oxidative stress is one important environmental factor in cancer as it is genotoxic and contributes to mutations (Beckman and Ames, 1997). During tumor progression, cells harbor mutations that reduce growth-limiting effects in pathways such as TGF-β signaling which becomes a tumor-promoting pathway due to mutations in later stages of CRC (Jakowlew, 2006; Bellam and Pasche, 2010; Calon et al., 2012). Therefore, it is likely that the results include many putative anti-tumor pathways that contain mutations in the cell lines, which is an important aspect to be addressed in future investigations.
On the level of transcriptional regulation, we identified a number of well-known, cancer-associated TFs with significantly enriched binding sites in the promoter regions of the signature genes. These TFs belong to a variety of TF families/subfamilies and are known to form protein-protein interactions with each other such as Jun factors and Fos factors which form the heterodimeric AP-1 protein (Chen et al., 1996; Shaulian and Karin, 2002; Eferl and Wagner, 2003). Likewise, nuclear receptors (NRs) of the subfamilies vitamin D receptors (NR1I) and retinoid X receptors form the VDR-RXR heterodimer complex (Orlov et al., 2012) that has been implicated in anticancer therapeutics (Friedrich et al., 2002; Sepulveda et al., 2006; Deeb et al., 2007; Matsuda and Kitagishi, 2013). In this light, it is known that TFs do not regulate their target genes in solitude, but interact with other TFs and cofactors in specific combinations for a fine-tuned control of gene expression (Gerstein et al., 2012). In addition, we identified different TF families/subfamilies that have overlapping binding sites and may act in a synergistic, additive, or antagonistic fashion in cancer. Kittler et al. revealed binding redundancy for NRs and their putative cooperating TFs in breast cancer on the basis of 39 factors, whereas non-overlapping binding sites were found to occur rarely (Kittler et al., 2013). Taken together, although the signature genes of both cell lines show no overlap, they may still be regulated by common factors in CRC.
We revealed that 62 and 72% of the TFs for 1638N-T1 and CMT-93, respectively, were found in the intersection of both cell lines. Consequently, only 38 and 28% of the TFs were exclusive for 1638N-T1 and CMT-93, respectively, whose implications in signal transduction pathways might explain phenotypic differences between the two cell lines with regard to tumor growth and metastasis. We deduced cross-talks between several pathways that might have an impact on tumor progression in the cell lines. For the APC-deficient 1638N-T1 cell line, we found overrepresented pathways which related to the activation of the canonical Wnt signaling pathway (Tables 1, 6). Wnt signaling activity is known to contribute to tumor aggressiveness; therefore, it is often targeted in cancer therapy (Anastas and Moon, 2013; Loh et al., 2013). It has also been stated that enhancement of canonical Wnt signaling activity is required for tumor progression and metastasis (Oguma et al., 2008). On the other hand, we showed several pathways for CMT-93 which have been previously associated with an inhibition of Wnt signaling. Two of these pathways related to VDR signaling and LXR-induced signaling (Tables 4, 8). Strikingly, VDR and LXRα (Nr1h3) were included in the signature genes for CMT-93 (see Supplementary Table S2), and VDR also showed significantly enriched binding sites (see Supplementary Table S5). Previous studies have investigated the activation of VDR as well as LXR in APC-deficient mice and observed that the activity of both factors decreased tumor growth (Zheng et al., 2012; Sasso et al., 2013). In addition, LXR expression was found to be downregulated in colon tumors of APC-deficient mice compared with adjacent normal mucosa (Su et al., 1992; Sasso et al., 2013). We also found that CTNNB1, which encodes β-catenin, was not included in the signature genes of any of the two cell lines, but showed significant binding site enrichment (see Supplementary Table S6). With respect to TCF-7-related factors, the genes Tcf7l1 and Lef1, however, were included in the signature genes of 1638N-T1. Interestingly, VDR and LXR can both directly bind to β-catenin, thereby preventing β-catenin from binding to its target sites (Uno et al., 2009; Makoukji et al., 2011; Zheng et al., 2012; Larriba et al., 2013; Shackleford et al., 2013; Lim et al., 2014).
All things considered, supported by the knowledge that 1638N-T1 cells harbor a mutation in the APC gene, which leads to aberrant Wnt pathway activation: we suggest that Wnt signaling is activated in 1638N-T1, but inhibited in CMT-93 through cross-talks of canonical Wnt signaling with VDR signaling pathway and/or LXR-related pathways. Consequently, we suggest that Wnt signaling-driven tumor formation and growth should be increased in mouse models involving 1638N-T1 compared to ones involving CMT-93. Though, many additional factors have to be taken into account when monitoring cell proliferation, invasion, and metastasis in mouse models. Several previous studies indicated synergistic effects between K-Ras and canonical Wnt signaling harboring APC mutations in CRC (Janssen et al., 2006; Luo et al., 2009; Lemieux et al., 2015). Furthermore, during development of effective cancer therapies, tumor cells grown in vitro are transplanted into ectopic sites of immunocompromized mice that do not reject tumor cells (Sharpless and Depinho, 2006; Richmond and Su, 2008; Hung et al., 2010). It has been stated that these xenograft models may fail to recapitulate the heterogeneity of cancer and the microenvironment, i.e., the interaction between tumor cells and supporting stroma (Hung et al., 2010). In the end, regardless of the fact that Wnt signaling may be aberrantly activated in 1638N-T1, a variety of different factors have an impact on the capacity of tumor cells to grow, proliferate, and metastasize in mouse models. We summarized our observations concerning the potential state of canonical Wnt signaling in the cell lines (Figure 5).
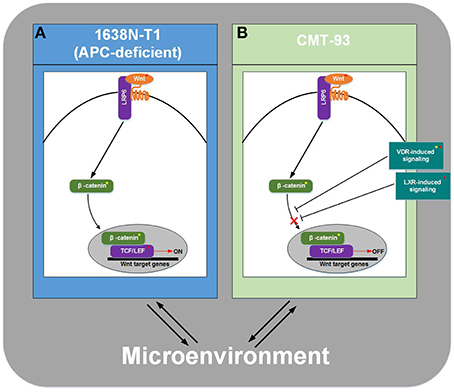
Figure 5. Schema for potential state of canonical Wnt signaling pathway in mouse models. (A) Wnt signaling is activated in 1638N-T1. (B) Wnt signaling is inhibited in CMT-93 through cross-talks with VDR- and/or LXR-induced pathways. Interaction of tumor cells with the microenvironment has an impact on cell proliferation, invasion, and metastasis in mouse models. Signature genes and transcription factors/cofactors, whose binding sites were found to be enriched in promoters, are indicated by a red asterisk or a yellow asterisk, respectively.
The master regulator analyses revealed several potential candidates which might be useful as therapeutic targets for cancer therapy. Master regulators were inferred from a network model that explicitly displayed the regulatory cascades between TFs. Beside several master regulators with yet unknown roles in CRC, we found MLK3 and Mapk1 (ERK2) which might be important in cancer cell proliferation, invasion, and metastasis of 1638N-T1 and CMT-93, respectively. Above all, our master regulatory networks can be used as models to generate testable hypotheses for studying the phenotypic differences between 1638N-T1 and CMT-93.
5. Conclusion
In this study, we have presented a systematic approach which combines colorectal cancer (CRC) cell lines, namely 1638N-T1 and CMT-93, and well-established computational methods in order to compare these cell lines on the level of transcriptional regulation as well as on a pathway level, i.e., the cancer cell-intrinsic pathway repertoire. We used the Trinity platform and the geneXplain platform to identify significantly upregulated genes in each of the cell lines as well as their upstream transcriptional regulators, on the basis of which we generated regulatory networks. Our findings suggested that the Wnt signaling pathway is activated in 1638N-T1, but inhibited in CMT-93 cells through cross-talks with other pathways. Moreover, we identified a number of well-known, cancer-associated TFs for both cell lines and provided indication of several master regulators being present such as MLK3 and Mapk1 (ERK2) which might be important in cell proliferation, migration, and invasion of 1638N-T1 and CMT-93, respectively. Using our systematic approach, we have provided new insights into the invasive potential of individual CRC cell lines, which can be used for development of effective cancer therapy.
Author Contributions
DW and MG participated in the design of the study, conducted computational, and statistical analyses. EW supervised the computational and statistical analyses. MH and TB interpreted the results with DW. JA prepared the colorectal cancer cell lines for this study. AW processed the RNA-Seq data (FASTQ files) and prepared the RNA-Seq count data. AB was involved in the preparation of the cell lines and interpretation of the results in perspective of colorectal cancer biology. DW and MG conceived of and managed the project and wrote the final version of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Ron Smits for providing the murine colorectal cancer cell line 1638N-T1 as well as the Transcriptome and Genome Analysis Laboratory (TAL) in Göttingen for the preparation of the RNA-Seq data (FASTQ files). We would also like to thank Cornelia Meckbach for proofreading the manuscript. This work was supported by the ebio initiative of the German Ministry of Education and Research (BMBF) and DW was funded by the MetastaSys project (0316173A) within the ebio initiative. We acknowledge support by the German Research Foundation and the Open Access Publication Funds of the Göttingen University.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2016.00042
Supplementary Table S1. Signature genes for colorectal cell line 1638N-T1.
Supplementary Table S2. Signature genes for colorectal cell line CMT-93.
Supplementary Table S3. Colorectal cancer-related non-redundant PWM library.
Supplementary Table S4. 1638N-T1-specific TF set.
Supplementary Table S5. CMT-93-specific TF set.
Supplementary Table S6. Intersection-specific TF set between 1638N-T1 and CMT-93.
Supplementary Table S7. Master regulatory network based on the intersection-specific TF set in Pair Graph File format.
Supplementary Figure S7. Master regulatory network based on the intersection-specific TF set as Scalable Vector Graphics (SVG).
Supplementary Table S8. Master regulatory network based on the 1638NT-1-specific TF set in Pair Graph File format.
Supplementary Figure S8. Master regulatory network based on the 1638NT-1-specific TF set as Scalable Vector Graphics (SVG).
Supplementary Table S9. Master regulatory network based on the CMT-93-specific TF set in Pair Graph File format.
Supplementary Figure S9. Master regulatory network based on the CMT-93-specific TF set as Scalable Vector Graphics (SVG).
References
Alsayed, Y., Uddin, S., Ahmad, S., Majchrzak, B., Druker, B. J., Fish, E. N., et al. (2000). IFN-γ activates the C3G/Rap1 signaling pathway. J. Immunol. 164, 1800–1806. doi: 10.4049/jimmunol.164.4.1800
Ameyar, M., Wisniewska, M., and Weitzman, J. B. (2003). A role for AP-1 in apoptosis: the case for and against. Biochimie 85, 747–752. doi: 10.1016/j.biochi.2003.09.006
Anagnostopoulou, V., Pediaditakis, I., Alkahtani, S., Alarifi, S. A., Schmidt, E.-M., Lang, F., et al. (2013). Differential effects of dehydroepiandrosterone and testosterone in prostate and colon cancer cell apoptosis: the role of nerve growth factor (NGF) receptors. Endocrinology 154, 2446–2456. doi: 10.1210/en.2012-2249
Anastas, J. N., and Moon, R. T. (2013). WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 13, 11–26. doi: 10.1038/nrc3419
Andersen, V., Holst, R., Kopp, T. I., Tjønneland, A., and Vogel, U. (2013). Interactions between diet, lifestyle and IL10, IL1B, and PTGS2/COX-2 gene polymorphisms in relation to risk of colorectal cancer in a prospective Danish case-cohort study. PLoS ONE 8:e78366. doi: 10.1371/journal.pone.0078366
Asham, E., Shankar, A., Loizidou, M., Fredericks, S., Miller, K., Boulos, P. B., et al. (2001). Increased endothelin-1 in colorectal cancer and reduction of tumour growth by ET(A) receptor antagonism. Br. J. Cancer 85, 1759–1763. doi: 10.1054/bjoc.2001.2193
Augustin, H. G., Koh, G. Y., Thurston, G., and Alitalo, K. (2009). Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat. Rev. Mol. Cell Biol. 10, 165–177. doi: 10.1038/nrm2639
Badouel, C., Chartrain, I., Blot, J., and Tassan, J.-P. (2010). Maternal embryonic leucine zipper kinase is stabilized in mitosis by phosphorylation and is partially degraded upon mitotic exit. Exp. Cell Res. 316, 2166–2173. doi: 10.1016/j.yexcr.2010.04.019
Basith, S., Manavalan, B., Yoo, T. H., Kim, S. G., and Choi, S. (2012). Roles of toll-like receptors in cancer: a double-edged sword for defense and offense. Arch. Pharm. Res. 35, 1297–1316. doi: 10.1007/s12272-012-0802-7
Bassuk, A. G., and Leiden, J. M. (1997). The role of Ets transcription factors in the development and function of the mammalian immune system. Adv. Immunol. 64, 65–104. doi: 10.1016/S0065-2776(08)60887-1
Bastide, P., Darido, C., Pannequin, J., Kist, R., Robine, S., Marty-Double, C., et al. (2007). Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 178, 635–648. doi: 10.1083/jcb.200704152
Beckman, K. B., and Ames, B. N. (1997). Oxidative decay of DNA. J. Biol. Chem. 272, 19633–19636. doi: 10.1074/jbc.272.32.19633
Behling, K. C., Tang, A., Freydin, B., Chervoneva, I., Kadakia, S., Schwartz, G. F., et al. (2011). Increased SIAH expression predicts ductal carcinoma in situ (DCIS) progression to invasive carcinoma. Breast Cancer Res. Treat. 129, 717–724. doi: 10.1007/s10549-010-1254-8
Bellam, N., and Pasche, B. (2010). TGF-β signaling alterations and colon cancer. Cancer Treat. Res. 155, 85–103. doi: 10.1007/978-1-4419-6033-7_5
Belmont, L., Rabbe, N., Antoine, M., Cathelin, D., Guignabert, C., Kurie, J., et al. (2014). Expression of TLR9 in tumor-infiltrating mononuclear cells enhances angiogenesis and is associated with a worse survival in lung cancer. Int. J. Cancer 134, 765–777. doi: 10.1002/ijc.28413
Blache, P., van de Wetering, M., Duluc, I., Domon, C., Berta, P., Freund, J.-N., et al. (2004). SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 166, 37–47. doi: 10.1083/jcb.200311021
Bouker, K. B., Skaar, T. C., Riggins, R. B., Harburger, D. S., Fernandez, D. R., et al. (2005). Interferon regulatory factor-1 (IRF-1) exhibits tumor suppressor activities in breast cancer associated with caspase activation and induction of apoptosis. Carcinogenesis 26, 1527–1535. doi: 10.1093/carcin/bgi113
Calon, A., Espinet, E., Palomo-Ponce, S., Tauriello, D. V. F., Iglesias, M., Céspedes, M. V., et al. (2012). Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 22, 571–584. doi: 10.1016/j.ccr.2012.08.013
Cancer Genome Atlas Network. (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. doi: 10.1038/nature11252
Casaletto, J. B., and McClatchey, A. I. (2012). Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer 12, 387–400. doi: 10.1038/nrc3277
Chen, J., and Gallo, K. A. (2012). MLK3 regulates paxillin phosphorylation in chemokine-mediated breast cancer cell migration and invasion to drive metastasis. Cancer Res. 72, 4130–4140. doi: 10.1158/0008-5472.CAN-12-0655
Chen, J., Miller, E. M., and Gallo, K. A. (2010). MLK3 is critical for breast cancer cell migration and promotes a malignant phenotype in mammary epithelial cells. Oncogene 29, 4399–4411. doi: 10.1038/onc.2010.198
Chen, T. K., Smith, L. M., Gebhardt, D. K., Birrer, M. J., and Brown, P. H. (1996). Activation and inhibition of the AP-1 complex in human breast cancer cells. Mol. Carcinog. 15, 215–226.
Chien, Y., Kim, S., Bumeister, R., Loo, Y.-M., Kwon, S. W., Johnson, C. L., et al. (2006). RalB GTPase-mediated activation of the IκB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127, 157–170. doi: 10.1016/j.cell.2006.08.034
Choi, S., and Ku, J.-L. (2011). Resistance of colorectal cancer cells to radiation and 5-FU is associated with MELK expression. Biochem. Biophys. Res. Commun. 412, 207–213. doi: 10.1016/j.bbrc.2011.07.060
Cooley, D. A., Frazier, O. H., and Kahan, B. D. (1982). Cardiac transplantation with the use of cyclosporin a for immunologic suppression. Tex Heart Inst. J. 9, 247–251.
Cowin, P., Rowlands, T. M., and Hatsell, S. J. (2005). Cadherins and catenins in breast cancer. Curr. Opin. Cell Biol. 17, 499–508. doi: 10.1016/j.ceb.2005.08.014
Cronan, M. R., Nakamura, K., Johnson, N. L., Granger, D. A., Cuevas, B. D., Wang, J.-G., et al. (2012). Defining MAP3 kinases required for MDA-MB-231 cell tumor growth and metastasis. Oncogene 31, 3889–3900. doi: 10.1038/onc.2011.544
da Costa, L. T., He, T. C., Yu, J., Sparks, A. B., Morin, P. J., Polyak, K., et al. (1999). CDX2 is mutated in a colorectal cancer with normal APC/β-catenin signaling. Oncogene 18, 5010–5014. doi: 10.1038/sj.onc.1202872
Dai, X., Ge, J., Wang, X., Qian, X., Zhang, C., and Li, X. (2013). OCT4 regulates epithelial-mesenchymal transition and its knockdown inhibits colorectal cancer cell migration and invasion. Oncol. Rep. 29, 155–160. doi: 10.3892/or.2012.2086
Deeb, K. K., Trump, D. L., and Johnson, C. S. (2007). Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nat. Rev. Cancer 7, 684–700. doi: 10.1038/nrc2196
Eferl, R., and Wagner, E. F. (2003). AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868. doi: 10.1038/nrc1209
Eger, A., Stockinger, A., Park, J., Langkopf, E., Mikula, M., Gotzmann, J., et al. (2004). β-Catenin and TGFβ signalling cooperate to maintain a mesenchymal phenotype after FosER-induced epithelial to mesenchymal transition. Oncogene 23, 2672–2680. doi: 10.1038/sj.onc.1207416
Fagiani, E., and Christofori, G. (2013). Angiopoietins in angiogenesis. Cancer Lett. 328, 18–26. doi: 10.1016/j.canlet.2012.08.018
Fang, J. Y., and Richardson, B. C. (2005). The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 6, 322–327. doi: 10.1016/S1470-2045(05)70168-6
Fleming, N. I., Jorissen, R. N., Mouradov, D., Christie, M., Sakthianandeswaren, A., Palmieri, M., et al. (2013). SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 73, 725–735. doi: 10.1158/0008-5472.CAN-12-2706
Freund, J. N., Domon-Dell, C., Kedinger, M., and Duluc, I. (1998). The Cdx-1 and Cdx-2 homeobox genes in the intestine. Biochem. Cell Biol. 76, 957–969. doi: 10.1139/bcb-76-6-957
Friedrich, M., Axt-Fliedner, R., Villena-Heinsen, C., Tilgen, W., Schmidt, W., and Reichrath, J. (2002). Analysis of vitamin D-receptor (VDR) and retinoid X-receptor alpha in breast cancer. Histochem. J. 34, 35–40. doi: 10.1023/A:1021343825552
Gambara, G., Desideri, M., Stoppacciaro, A., Padula, F., Cesaris, P. D., Starace, D., et al. (2015). TLR3 engagement induces IRF-3-dependent apoptosis in androgen-sensitive prostate cancer cells and inhibits tumour growth in vivo. J. Cell Mol. Med. 19, 327–339. doi: 10.1111/jcmm.12379
Ganguly, R., Mohyeldin, A., Thiel, J., Kornblum, H. I., Beullens, M., and Nakano, I. (2015). MELK-a conserved kinase: functions, signaling, cancer, and controversy. Clin. Transl. Med. 4:11. doi: 10.1186/s40169-014-0045-y
Gao, F., Liang, B., Reddy, S. T., Farias-Eisner, R., and Su, X. (2014). Role of inflammation-associated microenvironment in tumorigenesis and metastasis. Curr. Cancer Drug Targets 14, 30–45. doi: 10.2174/15680096113136660107
Gerstein, M. B., Kundaje, A., Hariharan, M., Landt, S. G., Yan, K.-K., Cheng, C., et al. (2012). Architecture of the human regulatory network derived from encode data. Nature 489, 91–100. doi: 10.1038/nature11245
Giardina, C., Nakanishi, M., Khan, A., Kuratnik, A., Xu, W., Brenner, B., et al. (2015). Regulation of VDR Expression in Apc-Mutant Mice, Human Colon Cancers and Adenomas. Cancer Prev. Res. 8, 387–399. doi: 10.1158/1940-6207.CAPR-14-0371
Giessen, C., Laubender, R. P., von Weikersthal, L. F., Schalhorn, A., Modest, D. P., Stintzing, S., et al. (2013). Early tumor shrinkage in metastatic colorectal cancer: retrospective analysis from an irinotecan-based randomized first-line trial. Cancer Sci. 104, 718–724. doi: 10.1111/cas.12148
Giuliani, L., Ciotti, M., Stoppacciaro, A., Pasquini, A., Silvestri, I., Matteis, A. D., et al. (2005). Udp-glucuronosyltransferases 1A expression in human urinary bladder and colon cancer by immunohistochemistry. Oncol. Rep. 13, 185–191. doi: 10.3892/or.13.2.185
Gopalsamy, A., Hagen, T., and Swaminathan, K. (2014). Investigating the molecular basis of Siah1 and Siah2 E3 ubiquitin ligase substrate specificity. PLoS ONE 9:e106547. doi: 10.1371/journal.pone.0106547
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Grant, K., Knowles, J., Dawas, K., Burnstock, G., Taylor, I., and Loizidou, M. (2007). Mechanisms of endothelin 1-stimulated proliferation in colorectal cancer cell lines. Br. J. Surg. 94, 106–112. doi: 10.1002/bjs.5536
Graves, B. J., and Petersen, J. M. (1998). Specificity within the ets family of transcription factors. Adv. Cancer Res. 75, 1–55. doi: 10.1016/S0065-230X(08)60738-1
Gray, D., Jubb, A. M., Hogue, D., Dowd, P., Kljavin, N., Yi, S., et al. (2005). Maternal embryonic leucine zipper kinase/murine protein serine-threonine kinase 38 is a promising therapeutic target for multiple cancers. Cancer Res. 65, 9751–9761. doi: 10.1158/0008-5472.CAN-04-4531
Guo, R., Sakamoto, H., Sugiura, S., and Ogawa, M. (2007). Endothelial cell motility is compatible with junctional integrity. J. Cell Physiol. 211, 327–335. doi: 10.1002/jcp.20937
Guo, R.-J., Huang, E., Ezaki, T., Patel, N., Sinclair, K., Wu, J., et al. (2004). Cdx1 inhibits human colon cancer cell proliferation by reducing β-catenin/T-cell factor transcriptional activity. J. Biol. Chem. 279, 36865–36875. doi: 10.1074/jbc.M405213200
Gupta, G. P., and Massagué, J. (2006). Cancer metastasis: building a framework. Cell 127, 679–695. doi: 10.1016/j.cell.2006.11.001
Habelhah, H., Frew, I. J., Laine, A., Janes, P. W., Relaix, F., Sassoon, D., et al. (2002). Stress-induced decrease in TRAF2 stability is mediated by Siah2. EMBO J. 21, 5756–5765. doi: 10.1093/emboj/cdf576
Habelhah, H., Laine, A., Erdjument-Bromage, H., Tempst, P., Gershwin, M. E., Bowtell, D. D. L., et al. (2004). Regulation of 2-oxoglutarate (α-ketoglutarate) dehydrogenase stability by the RING finger ubiquitin ligase Siah. J. Biol. Chem. 279, 53782–53788. doi: 10.1074/jbc.M410315200
Holash, J., Maisonpierre, P. C., Compton, D., Boland, P., Alexander, C. R., Zagzag, D., et al. (1999a). Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF. Science 284, 1994–1998. doi: 10.1126/science.284.5422.1994
Holash, J., Wiegand, S. J., and Yancopoulos, G. D. (1999b). New model of tumor angiogenesis: dynamic balance between vessel regression and growth mediated by angiopoietins and VEGF. Oncogene 18, 5356–5362. doi: 10.1038/sj.onc.1203035
Holldack, J. (2014). Toll-like receptors as therapeutic targets for cancer. Drug Discov. Today 19, 379–382. doi: 10.1016/j.drudis.2013.08.020
Holloway, R. W., Bogachev, O., Bharadwaj, A. G., McCluskey, G. D., Majdalawieh, A. F., Zhang, L., et al. (2012). Stromal adipocyte enhancer-binding protein (AEBP1) promotes mammary epithelial cell hyperplasia via proinflammatory and hedgehog signaling. J. Biol. Chem. 287, 39171–39181. doi: 10.1074/jbc.M112.404293
Hsu, R. Y. C., Chan, C. H. F., Spicer, J. D., Rousseau, M. C., Giannias, B., Rousseau, S., et al. (2011). LPS-induced TLR4 signaling in human colorectal cancer cells increases β1 integrin-mediated cell adhesion and liver metastasis. Cancer Res. 71, 1989–1998. doi: 10.1158/0008-5472.CAN-10-2833
Hu, G., and Barnes, B. J. (2006). Interferon regulatory factor-5-regulated pathways as a target for colorectal cancer therapeutics. Exp. Rev. Anticancer. Ther. 6, 775–784. doi: 10.1586/14737140.6.5.775
Hung, K. E., Maricevich, M. A., Richard, L. G., Chen, W. Y., Richardson, M. P., Kunin, A., et al. (2010). Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc. Natl. Acad. Sci. U.S.A. 107, 1565–1570. doi: 10.1073/pnas.0908682107
Ijssennagger, N., Rijnierse, A., de Wit, N. J. W., Boekschoten, M. V., Dekker, J., Schonewille, A., et al. (2013). Dietary heme induces acute oxidative stress, but delayed cytotoxicity and compensatory hyperproliferation in mouse colon. Carcinogenesis 34, 1628–1635. doi: 10.1093/carcin/bgt084
Ikeda, H., Old, L. J., and Schreiber, R. D. (2002). The roles of IFNγ in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 13, 95–109. doi: 10.1016/S1359-6101(01)00038-7
Jakowlew, S. B. (2006). Transforming growth factor-β in cancer and metastasis. Cancer Metastasis Rev. 25, 435–457. doi: 10.1007/s10555-006-9006-2
Janssen, K.-P., Alberici, P., Fsihi, H., Gaspar, C., Breukel, C., Franken, P., et al. (2006). APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology 131, 1096–1109. doi: 10.1053/j.gastro.2006.08.011
Jansson, E. A., Are, A., Greicius, G., Kuo, I.-C., Kelly, D., Arulampalam, V., et al. (2005). The Wnt/β-catenin signaling pathway targets PPARγ activity in colon cancer cells. Proc. Natl. Acad. Sci. U.S.A. 102, 1460–1465. doi: 10.1073/pnas.0405928102
Johnsen, S. A., Subramaniam, M., Monroe, D. G., Janknecht, R., and Spelsberg, T. C. (2002). Modulation of transforming growth factor β (TGFβ)/Smad transcriptional responses through targeted degradation of TGFβ-inducible early gene-1 by human seven in absentia homologue. J. Biol. Chem. 277, 30754–30759. doi: 10.1074/jbc.M204812200
Junghans, D., Haas, I. G., and Kemler, R. (2005). Mammalian cadherins and protocadherins: about cell death, synapses and processing. Curr. Opin. Cell Biol. 17, 446–452. doi: 10.1016/j.ceb.2005.08.008
Kandoth, C., McLellan, M. D., Vandin, F., Ye, K., Niu, B., Lu, C., et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature 502, 333–339. doi: 10.1038/nature12634
Kang, Y. (2005). Functional genomic analysis of cancer metastasis: biologic insights and clinical implications. Exp. Rev. Mol. Diagn. 5, 385–395. doi: 10.1586/14737159.5.3.385
Karim, B. O., and Huso, D. L. (2013). Mouse models for colorectal cancer. Am. J. Cancer Res. 3, 240–250.
Kim, H., Scimia, M. C., Wilkinson, D., Trelles, R. D., Wood, M. R., Bowtell, D., et al. (2011). Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol. Cell 44, 532–544. doi: 10.1016/j.molcel.2011.08.045
Kim, J.-Y., Beg, A. A., and Haura, E. B. (2013a). Non-canonical IKKs, IKKϵ and TBK1, as novel therapeutic targets in the treatment of non-small cell lung cancer. Exp. Opin. Ther. Targets 17, 1109–1112. doi: 10.1517/14728222.2013.833188
Kim, J.-Y., Welsh, E. A., Oguz, U., Fang, B., Bai, Y., Kinose, F., et al. (2013b). Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc. Natl. Acad. Sci. U.S.A. 110, 12414–12419. doi: 10.1073/pnas.1220674110
Kim, K., Lu, Z., and Hay, E. D. (2002). Direct evidence for a role of β-catenin/LEF-1 signaling pathway in induction of EMT. Cell Biol. Int. 26, 463–476. doi: 10.1006/cbir.2002.0901
Kitadai, Y., Sasaki, T., Kuwai, T., Nakamura, T., Bucana, C. D., Hamilton, S. R., et al. (2006). Expression of activated platelet-derived growth factor receptor in stromal cells of human colon carcinomas is associated with metastatic potential. Int. J. Cancer 119, 2567–2574. doi: 10.1002/ijc.22229
Kittler, R., Zhou, J., Hua, S., Ma, L., Liu, Y., Pendleton, E., et al. (2013). A comprehensive nuclear receptor network for breast cancer cells. Cell Rep. 3, 538–551. doi: 10.1016/j.celrep.2013.01.004
Klampfer, L. (2008). The role of signal transducers and activators of transcription in colon cancer. Front. Biosci. 13, 2888–2899. doi: 10.2741/2893
Klampfer, L. (2014). Vitamin D and colon cancer. World J. Gastrointest. Oncol. 6, 430–437. doi: 10.4251/wjgo.v6.i11.430
Knowles, J. P., Shi-Wen, X., ul Haque, S., Bhalla, A., Dashwood, M. R., Yang, S., et al. (2012). Endothelin-1 stimulates colon cancer adjacent fibroblasts. Int. J. Cancer 130, 1264–1272. doi: 10.1002/ijc.26090
Kojima, M., Morisaki, T., Sasaki, N., Nakano, K., Mibu, R., Tanaka, M., et al. (2004). Increased nuclear factor-kB activation in human colorectal carcinoma and its correlation with tumor progression. Anticancer Res. 24, 675–681.
Korchynskyi, O., Landström, M., Stoika, R., Funa, K., Heldin, C. H., ten Dijke, P., et al. (1999). Expression of Smad proteins in human colorectal cancer. Int. J. Cancer 82, 197–202.
Krieg, A. M. (2008). Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene 27, 161–167. doi: 10.1038/sj.onc.1210911
Krull, M., Pistor, S., Voss, N., Kel, A., Reuter, I., Kronenberg, D., et al. (2006). TRANSPATH®: an information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res. 34, D546–D551. doi: 10.1093/nar/gkj107
Ku, J.-L., Park, S.-H., Yoon, K.-A., Shin, Y.-K., Kim, K.-H., Choi, J.-S., et al. (2007). Genetic alterations of the TGF-β signaling pathway in colorectal cancer cell lines: A novel mutation in Smad3 associated with the inactivation of TGF-β-induced transcriptional activation. Cancer Lett. 247, 283–292. doi: 10.1016/j.canlet.2006.05.008
Kuai, W.-X., Wang, Q., Yang, X.-Z., Zhao, Y., Yu, R., and Tang, X.-J. (2012). Interleukin-8 associates with adhesion, migration, invasion and chemosensitivity of human gastric cancer cells. World J. Gastroenterol. 18, 979–985. doi: 10.3748/wjg.v18.i9.979
Ladha, J., Sinha, S., Bhat, V., Donakonda, S., and Rao, S. M. R. (2012). Identification of genomic targets of transcription factor AEBP1 and its role in survival of glioma cells. Mol. Cancer Res. 10, 1039–1051. doi: 10.1158/1541-7786.mcr-11-0488
Larriba, M. J., González-Sancho, J. M., Barbáchano, A., Niell, N., Ferrer-Mayorga, G., and Muñoz, A. (2013). Vitamin D is a multilevel repressor of Wnt/β-catenin signaling in cancer cells. Cancers 5, 1242–1260. doi: 10.3390/cancers5041242
Lemieux, E., Cagnol, S., Beaudry, K., Carrier, J., and Rivard, N. (2015). Oncogenic KRAS signalling promotes the Wnt/β-catenin pathway through LRP6 in colorectal cancer. Oncogene 34, 4914–4927. doi: 10.1038/onc.2014.416
Li, A., Varney, M. L., and Singh, R. K. (2001). Expression of interleukin 8 and its receptors in human colon carcinoma cells with different metastatic potentials. Clin. Cancer Res. 7, 3298–3304.
Li, M., Zhang, Y., Feurino, L. W., Wang, H., Fisher, W. E., Brunicardi, F. C., et al. (2008). Interleukin-8 increases vascular endothelial growth factor and neuropilin expression and stimulates ERK activation in human pancreatic cancer. Cancer Sci. 99, 733–737. doi: 10.1111/j.1349-7006.2008.00740.x
Li, T.-T., Ogino, S., and Qian, Z. R. (2014). Toll-like receptor signaling in colorectal cancer: Carcinogenesis to cancer therapy. World J. Gastroenterol. 20, 17699–17708. doi: 10.3748/wjg.v20.i47.17699
Libermann, T. A., and Zerbini, L. F. (2006). Targeting transcription factors for cancer gene therapy. Curr. Gene Ther. 6, 17–33. doi: 10.2174/156652306775515501
Licato, L. L., Keku, T. O., Wurzelmann, J. I., Murray, S. C., Woosley, J. T., Sandler, R. S., et al. (1997). In vivo activation of mitogen-activated protein kinases in rat intestinal neoplasia. Gastroenterology 113, 1589–1598. doi: 10.1053/gast.1997.v113.pm9352861
Lim, Y. Y., Kim, S. Y., Kim, H. M., Li, K. S., Kim, M. N., Park, K.-C., et al. (2014). Potential relationship between the canonical Wnt signalling pathway and expression of the vitamin D receptor in alopecia. Clin. Exp. Dermatol. 39, 368–375. doi: 10.1111/ced.12241
Lin, C.-Y., and Åke Gustafsson, J. (2015). Targeting liver x receptors in cancer therapeutics. Nat. Rev. Cancer 15, 216–224. doi: 10.1038/nrc3912
Lind, D. S., Hochwald, S. N., Malaty, J., Rekkas, S., Hebig, P., Mishra, G., et al. (2001). Nuclear factor-κB is upregulated in colorectal cancer. Surgery 130, 363–369. doi: 10.1067/msy.2001.116672
Lindholm, P. F., Bub, J., Kaul, S., Shidham, V. B., and Kajdacsy-Balla, A. (2000). The role of constitutive NF-κB activity in PC-3 human prostate cancer cell invasive behavior. Clin. Exp. Metastasis 18, 471–479. doi: 10.1023/A:1011845725394
Loh, Y. N., Hedditch, E. L., Baker, L. A., Jary, E., Ward, R. L., and Ford, C. E. (2013). The Wnt signalling pathway is upregulated in an in vitro model of acquired tamoxifen resistant breast cancer. BMC Cancer 13:174. doi: 10.1186/1471-2407-13-174
Luo, F., Brooks, D. G., Ye, H., Hamoudi, R., Poulogiannis, G., Patek, C. E., et al. (2009). Mutated K-ras(Asp12) promotes tumourigenesis in Apc(Min) mice more in the large than the small intestines, with synergistic effects between K-ras and Wnt pathways. Int. J. Exp. Pathol. 90, 558–574. doi: 10.1111/j.1365-2613.2009.00667.x
Lynch, J., Keller, M., Guo, R.-J., Yang, D., and Traber, P. (2003). Cdx1 inhibits the proliferation of human colon cancer cells by reducing cyclin D1 gene expression. Oncogene 22, 6395–6407. doi: 10.1038/sj.onc.1206770
Maggio-Price, L., Treuting, P., Zeng, W., Tsang, M., Bielefeldt-Ohmann, H., and Iritani, B. M. (2006). Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res. 66, 828–838. doi: 10.1158/0008-5472.CAN-05-2448
Magnanti, M., Giuliani, L., Gandini, O., Gazzaniga, P., Santiemma, V., Ciotti, M., et al. (2000). Follicle-stimulating hormone, testosterone, and hypoxia differentially regulate UDP-glucuronosyltransferase 1 isoforms expression in rat sertoli and peritubular myoid cells. J. Steroid. Biochem. Mol. Biol. 74, 149–155. doi: 10.1016/S0960-0760(00)00095-9
Majdalawieh, A., Zhang, L., and Ro, H.-S. (2007). Adipocyte Enhancer-binding Protein-1 Promotes Macrophage Inflammatory Responsiveness by Up-Regulating NF-κB via IκBa Negative Regulation. Mol. Biol. Cell 18, 930–942. doi: 10.1091/mbc.E06-03-0217
Makoukji, J., Shackleford, G., Meffre, D., Grenier, J., Liere, P., Lobaccaro, J.-M. A., et al. (2011). Interplay between LXR and Wnt/β-catenin signaling in the negative regulation of peripheral myelin genes by oxysterols. J. Neurosci. 31, 9620–9629. doi: 10.1523/JNEUROSCI.0761-11.2011
Malz, M., Aulmann, A., Samarin, J., Bissinger, M., Longerich, T., Schmitt, S., et al. (2012). Nuclear accumulation of seven in absentia homologue-2 supports motility and proliferation of liver cancer cells. Int. J. Cancer 131, 2016–2026. doi: 10.1002/ijc.27473
Mann, B., Gelos, M., Siedow, A., Hanski, M. L., Gratchev, A., Ilyas, M., et al. (1999). Target genes of β-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc. Natl. Acad. Sci. U.S.A. 96, 1603–1608. doi: 10.1073/pnas.96.4.1603
Matsuda, S., and Kitagishi, Y. (2013). Peroxisome proliferator-activated receptor and vitamin D receptor signaling pathways in cancer cells. Cancers 5, 1261–1270. doi: 10.3390/cancers5041261
Matsuzawa, S. I., and Reed, J. C. (2001). Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 responses. Mol. Cell 7, 915–926. doi: 10.1016/S1097-2765(01)00242-8
Mishra, P., Senthivinayagam, S., Rangasamy, V., Sondarva, G., and Rana, B. (2010). Mixed lineage kinase-3/JNK1 axis promotes migration of human gastric cancer cells following gastrin stimulation. Mol. Endocrinol. 24, 598–607. doi: 10.1210/me.2009-0387
Müller, T., Choidas, A., Reichmann, E., and Ullrich, A. (1999). Phosphorylation and free pool of β-catenin are regulated by tyrosine kinases and tyrosine phosphatases during epithelial cell migration. J. Biol. Chem. 274, 10173–10183. doi: 10.1074/jbc.274.15.10173
Molloy, N. H., Read, D. E., and Gorman, A. M. (2011). Nerve growth factor in cancer cell death and survival. Cancers 3, 510–530. doi: 10.3390/cancers3010510
Morali, O. G., Delmas, V., Moore, R., Jeanney, C., Thiery, J. P., and Larue, L. (2001). IGF-II induces rapid β-catenin relocation to the nucleus during epithelium to mesenchyme transition. Oncogene 20, 4942–4950. doi: 10.1038/sj.onc.1204660
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Mrsny, R. J., Gewirtz, A. T., Siccardi, D., Savidge, T., Hurley, B. P., Madara, J. L., et al. (2004). Identification of hepoxilin A3 in inflammatory events: a required role in neutrophil migration across intestinal epithelia. Proc. Natl. Acad. Sci. U.S.A. 101, 7421–7426. doi: 10.1073/pnas.0400832101
Nakayama, K., Qi, J., and Ronai, Z. (2009). The ubiquitin ligase Siah2 and the hypoxia response. Mol. Cancer Res. 7, 443–451. doi: 10.1158/1541-7786.MCR-08-0458
Nakshatri, H., Bhat-Nakshatri, P., Martin, D. A., Goulet, R. J., and Sledge, G. W. (1997). Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol. Cell Biol. 17, 3629–3639. doi: 10.1128/MCB.17.7.3629
Nata, T., Basheer, A., Cocchi, F., van Besien, R., Massoud, R., Jacobson, S., et al. (2015). Targeting the binding interface on a shared receptor subunit of a cytokine family enables the inhibition of multiple member cytokines with selectable target spectrum. J. Biol. Chem. 290, 22338–22351. doi: 10.1074/jbc.M115.661074
Nebert, D. W. (2002). Transcription factors and cancer: an overview. Toxicology 181–182, 131–141. doi: 10.1016/S0300-483X(02)00269-X
Normanno, N., Luca, A. D., Bianco, C., Strizzi, L., Mancino, M., Maiello, M. R., et al. (2006). Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 366, 2–16. doi: 10.1016/j.gene.2005.10.018
Oguma, K., Oshima, H., Aoki, M., Uchio, R., Naka, K., Nakamura, S., et al. (2008). Activated macrophages promote Wnt signalling through tumour necrosis factor-α in gastric tumour cells. EMBO J. 27, 1671–1681. doi: 10.1038/emboj.2008.105
Oikawa, T. (2004). ETS transcription factors: possible targets for cancer therapy. Cancer Sci. 95, 626–633. doi: 10.1111/j.1349-7006.2004.tb03320.x
Oikawa, T., and Yamada, T. (2003). Molecular biology of the Ets family of transcription factors. Gene 303, 11–34. doi: 10.1016/S0378-1119(02)01156-3
Orlov, I., Rochel, N., Moras, D., and Klaholz, B. P. (2012). Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J. 31, 291–300. doi: 10.1038/emboj.2011.445
Pasz-Walczak, G., Kordek, R., and Faflik, M. (2001). P21 (WAF1) expression in colorectal cancer: correlation with P53 and cyclin D1 expression, clinicopathological parameters and prognosis. Pathol. Res. Pract. 197, 683–689. doi: 10.1078/0344-0338-00146
Polakis, P. (2012). Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol. 4:a008052. doi: 10.1101/cshperspect.a008052
Qi, J., Nakayama, K., Cardiff, R. D., Borowsky, A. D., Kaul, K., Williams, R., et al. (2010). Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 18, 23–38. doi: 10.1016/j.ccr.2010.05.024
Qi, J., Tripathi, M., Mishra, R., Sahgal, N., Fazli, L., Fazil, L., et al. (2013). The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell 23, 332–346. doi: 10.1016/j.ccr.2013.02.016
Rakoff-Nahoum, S., and Medzhitov, R. (2009). Toll-like receptors and cancer. Nat. Rev. Cancer 9, 57–63. doi: 10.1038/nrc2541
Rask, K., Thörn, M., Pontén, F., Kraaz, W., Sundfeldt, K., Hedin, L., et al. (2000). Increased expression of the transcription factors CCAAT-enhancer binding protein-β (C/EBβ) and C/EBPζ (CHOP) correlate with invasiveness of human colorectal cancer. Int. J. Cancer 86, 337–343. doi: 10.1002/(SICI)1097-0215(20000501)86:3<337::AID-IJC6>3.0.CO;2-3
Reddy, E. P., Korapati, A., Chaturvedi, P., and Rane, S. (2000). IL-3 signaling and the role of Src kinases, JAKs and STATs: a covert liaison unveiled. Oncogene 19, 2532–2547. doi: 10.1038/sj.onc.1203594
Reka, A. K., Kurapati, H., Narala, V. R., Bommer, G., Chen, J., Standiford, T. J., et al. (2010). Peroxisome proliferator-activated receptor-γ activation inhibits tumor metastasis by antagonizing Smad3-mediated epithelial-mesenchymal transition. Mol. Cancer Ther. 9, 3221–3232. doi: 10.1158/1535-7163.MCT-10-0570
Richmond, A., and Su, Y. (2008). Mouse xenograft models vs gem models for human cancer therapeutics. Dis. Model. Mech. 1, 78–82. doi: 10.1242/dmm.000976
Romon, R., Adriaenssens, E., Lagadec, C., Germain, E., Hondermarck, H., and Bourhis, X. L. (2010). Nerve growth factor promotes breast cancer angiogenesis by activating multiple pathways. Mol. Cancer 9:157. doi: 10.1186/1476-4598-9-157
Sakamoto, K., Maeda, S., Hikiba, Y., Nakagawa, H., Hayakawa, Y., Shibata, W., et al. (2009). Constitutive NF-κB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth. Clin. Cancer Res. 15, 2248–2258. doi: 10.1158/1078-0432.CCR-08-1383
Salaun, B., Coste, I., Rissoan, M.-C., Lebecque, S. J., and Renno, T. (2006). TLR3 can directly trigger apoptosis in human cancer cells. J. Immunol. 176, 4894–4901. doi: 10.4049/jimmunol.176.8.4894
Santarpia, L., Lippman, S. M., and El-Naggar, A. K. (2012). Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin. Ther. Targets 16, 103–119. doi: 10.1517/14728222.2011.645805
Sarkar, T. R., Sharan, S., Wang, J., Pawar, S. A., Cantwell, C. A., Johnson, P. F., et al. (2012). Identification of a Src tyrosine kinase/SIAH2 E3 ubiquitin ligase pathway that regulates C/EBPδ expression and contributes to transformation of breast tumor cells. Mol. Cell Biol. 32, 320–332. doi: 10.1128/MCB.05790-11
Sarraf, P., Mueller, E., Jones, D., King, F. J., DeAngelo, D. J., Partridge, J. B., et al. (1998). Differentiation and reversal of malignant changes in colon cancer through PPARγ. Nat. Med. 4, 1046–1052. doi: 10.1038/2030
Sasso, G. L., Bovenga, F., Murzilli, S., Salvatore, L., Tullio, G. D., Martelli, N., et al. (2013). Liver X receptors inhibit proliferation of human colorectal cancer cells and growth of intestinal tumors in mice. Gastroenterology 144, 1497–1507, 1507.e1–13. doi: 10.1053/j.gastro.2013.02.005
Schena, M., Shalon, D., Davis, R. W., and Brown, P. O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. doi: 10.1126/science.270.5235.467
Schmid, C. D., Perier, R., Praz, V., and Bucher, P. (2006). EPD in its twentieth year: towards complete promoter coverage of selected model organisms. Nucleic Acids Res 34, D82–D85. doi: 10.1093/nar/gkj146
Sepulveda, V. A. T., Weigel, N. L., and Falzon, M. (2006). Prostate cancer cell type-specific involvement of the VDR and RXR in regulation of the human PTHrP gene via a negative VDRE. Steroids 71, 102–115. doi: 10.1016/j.steroids.2005.08.009
Shackleford, G., Makoukji, J., Grenier, J., Liere, P., Meffre, D., and Massaad, C. (2013). Differential regulation of Wnt/β-catenin signaling by Liver X Receptors in Schwann cells and oligodendrocytes. Biochem. Pharmacol. 86, 106–114. doi: 10.1016/j.bcp.2013.02.036
Sharpless, N. E., and Depinho, R. A. (2006). The mighty mouse: genetically engineered mouse models in cancer drug development. Nat. Rev. Drug Discov. 5, 741–754. doi: 10.1038/nrd2110
Shaulian, E., and Karin, M. (2002). AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4, E131–E136. doi: 10.1038/ncb0502-e131
Slattery, M. L. (2007). Vitamin D receptor gene (VDR) associations with cancer. Nutr. Rev. 65, S102–S104. doi: 10.1301/nr.2007.aug.S102-S104
Slattery, M. L., Lundgreen, A., Kadlubar, S. A., Bondurant, K. L., and Wolff, R. K. (2013). JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol. Carcinog. 52, 155–166. doi: 10.1002/mc.21841
Smits, R., Kartheuser, A., Jagmohan-Changur, S., Leblanc, V., Breukel, C., de Vries, A., et al. (1997). Loss of Apc and the entire chromosome 18 but absence of mutations at the Ras and Tp53 genes in intestinal tumors from Apc1638N, a mouse model for Apc-driven carcinogenesis. Carcinogenesis 18, 321–327. doi: 10.1093/carcin/18.2.321
Smits, R., Kielman, M. F., Breukel, C., Zurcher, C., Neufeld, K., Jagmohan-Changur, S., et al. (1999). Apc1638T: a mouse model delineating critical domains of the adenomatous polyposis coli protein involved in tumorigenesis and development. Genes Dev. 13, 1309–1321. doi: 10.1101/gad.13.10.1309
Soubeyran, P., Andre, F., Lissitzky, J. C., Mallo, G. V., Moucadel, V., Roccabianca, M., et al. (1999). Cdx1 promotes differentiation in a rat intestinal epithelial cell line. Gastroenterology 117, 1326–1338. doi: 10.1016/S0016-5085(99)70283-0
Stamos, J. L., and Weis, W. I. (2013). The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 5:a007898. doi: 10.1101/cshperspect.a007898
Steller, E. J. A., Raats, D. A., Koster, J., Rutten, B., Govaert, K. M., Emmink, B. L., et al. (2013). PDGFRB promotes liver metastasis formation of mesenchymal-like colorectal tumor cells. Neoplasia 15, 204–217. doi: 10.1593/neo.121726
Su, L. K., Kinzler, K. W., Vogelstein, B., Preisinger, A. C., Moser, A. R., Luongo, C., et al. (1992). Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256, 668–670. doi: 10.1126/science.1350108
Sudhakar, J. N., and Chow, K.-C. (2014). Human RAD23 homolog A is required for the nuclear translocation of apoptosis-inducing factor during induction of cell death. Biol. Cell 106, 359–376. doi: 10.1111/boc.201400013
Suh, E., Chen, L., Taylor, J., and Traber, P. G. (1994). A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol. Cell. Biol. 14, 7340–7351. doi: 10.1128/MCB.14.11.7340
Suh, E., and Traber, P. G. (1996). An intestine-specific homeobox gene regulates proliferation and differentiation. Mol. Cell. Biol. 16, 619–625. doi: 10.1128/MCB.16.2.619
Sun, Q., Sun, F., Wang, B., Liu, S., Niu, W., Liu, E., et al. (2014a). Interleukin-8 promotes cell migration through integrin αvβ6 upregulation in colorectal cancer. Cancer Lett. 354, 245–253. doi: 10.1016/j.canlet.2014.08.021
Sun, Y., Shen, S., Liu, X., Tang, H., Wang, Z., Yu, Z., et al. (2014b). MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol. Cell Biochem. 390, 19–30. doi: 10.1007/s11010-013-1950-x
Suto, R., Tominaga, K., Mizuguchi, H., Sasaki, E., Higuchi, K., Kim, S., et al. (2004). Dominant-negative mutant of c-Jun gene transfer: a novel therapeutic strategy for colorectal cancer. Gene Ther. 11, 187–193. doi: 10.1038/sj.gt.3302158
Takahashi, H., Ogata, H., Nishigaki, R., Broide, D. H., and Karin, M. (2010). Tobacco smoke promotes lung tumorigenesis by triggering IKKβ- and JNK1-dependent inflammation. Cancer Cell 17, 89–97. doi: 10.1016/j.ccr.2009.12.008
Takeuchi, K., and Ito, F. (2011). Receptor tyrosine kinases and targeted cancer therapeutics. Biol. Pharm. Bull. 34, 1774–1780. doi: 10.1248/bpb.34.1774
Tang, X., and Zhu, Y. (2012). TLR4 signaling promotes immune escape of human colon cancer cells by inducing immunosuppressive cytokines and apoptosis resistance. Oncol. Res. 20, 15–24. doi: 10.3727/096504012X13425470196092
Taylor, J. K., Boll, W., Levy, T., Suh, E., Siang, S., Mantei, N., et al. (1997). Comparison of intestinal phospholipase A/lysophospholipase and sucrase-isomaltase genes suggest a common structure for enterocyte-specific promoters. DNA Cell Biol. 16, 1419–1428. doi: 10.1089/dna.1997.16.1419
Torkamani, A., Verkhivker, G., and Schork, N. J. (2009). Cancer driver mutations in protein kinase genes. Cancer Lett. 281, 117–127. doi: 10.1016/j.canlet.2008.11.008
Tsoupras, A. B., Iatrou, C., Frangia, C., and Demopoulos, C. A. (2009). The implication of platelet activating factor in cancer growth and metastasis: potent beneficial role of PAF-inhibitors and antioxidants. Infect. Disord. Drug Targets 9, 390–399. doi: 10.2174/187152609788922555
Uno, S., Endo, K., Jeong, Y., Kawana, K., Miyachi, H., Hashimoto, Y., et al. (2009). Suppression of β-catenin signaling by liver X receptor ligands. Biochem. Pharmacol. 77, 186–195. doi: 10.1016/j.bcp.2008.10.007
van 't Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A. M., Mao, M., et al. (2002). Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536. doi: 10.1038/415530a
Vedin, L.-L., Åke Gustafsson, J., and Steffensen, K. R. (2013). The oxysterol receptors LXRα and LXRβ suppress proliferation in the colon. Mol. Carcinog. 52, 835–844. doi: 10.1002/mc.21924
Velho, S., Pinto, A., Licastro, D., Oliveira, M. J., Sousa, F., Stupka, E., et al. (2014). Dissecting the signaling pathways associated with the oncogenic activity of MLK3 P252H mutation. BMC Cancer 14:182. doi: 10.1186/1471-2407-14-182
Vleminckx, K., Vakaet, L., Mareel, M., Fiers, W., and van Roy, F. (1991). Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell 66, 107–119. doi: 10.1016/0092-8674(91)90143-M
Voronov, E., Shouval, D. S., Krelin, Y., Cagnano, E., Benharroch, D., Iwakura, Y., et al. (2003). IL-1 is required for tumor invasiveness and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 2645–2650. doi: 10.1073/pnas.0437939100
Wang, S., Liu, Z., Wang, L., and Zhang, X. (2009). NF-κB signaling pathway, inflammation and colorectal cancer. Cell Mol. Immunol. 6, 327–334. doi: 10.1038/cmi.2009.43
Wang, W., Abbruzzese, J. L., Evans, D. B., Larry, L., Cleary, K. R., and Chiao, P. J. (1999). The nuclear factor-κB RelA transcription factor is constitutively activated in human pancreatic adenocarcinoma cells. Clin. Cancer Res. 5, 119–127.
Wang, Y., Shen, L., Xu, N., Wang, J.-W., Jiao, S.-C., Liu, Z.-Y., et al. (2012). UGT1A1 predicts outcome in colorectal cancer treated with irinotecan and fluorouracil. World J. Gastroenterol. 18, 6635–6644. doi: 10.3748/wjg.v18.i45.6635
Weigelt, B., Peterse, J. L., and van 't Veer, L. J. (2005). Breast cancer metastasis: markers and models. Nat. Rev. Cancer 5, 591–602. doi: 10.1038/nrc1670
West, N. R., McCuaig, S., Franchini, F., and Powrie, F. (2015). Emerging cytokine networks in colorectal cancer. Nat. Rev. Immunol. 15, 615–629. doi: 10.1038/nri3896
Wiener, D., Doerge, D. R., Fang, J.-L., Upadhyaya, P., and Lazarus, P. (2004). Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver: importance of UDP-glucuronosyltransferase 1A4. Drug Metab. Dispos. 32, 72–79. doi: 10.1124/dmd.32.1.72
Wingender, E. (2008). The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 9, 326–332. doi: 10.1093/bib/bbn016
Wingender, E., Schoeps, T., and Dönitz, J. (2013). TFClass: an expandable hierarchical classification of human transcription factors. Nucleic Acids Res. 41, D165–D170. doi: 10.1093/nar/gks1123
Wong, C. S. F., and Möller, A. (2013). Siah: a promising anticancer target. Cancer Res. 73, 2400–2406. doi: 10.1158/0008-5472.CAN-12-4348
Wong, C. S. F., Sceneay, J., House, C. M., Halse, H. M., Liu, M. C. P., George, J., et al. (2012). Vascular normalization by loss of Siah2 results in increased chemotherapeutic efficacy. Cancer Res. 72, 1694–1704. doi: 10.1158/0008-5472.CAN-11-3310
Xie, W., Rimm, D. L., Lin, Y., Shih, W. J., and Reiss, M. (2003). Loss of Smad signaling in human colorectal cancer is associated with advanced disease and poor prognosis. Cancer J. 9, 302–312. doi: 10.1097/00130404-200307000-00013
Xu, Y., and Pasche, B. (2007). TGF-β signaling alterations and susceptibility to colorectal cancer. Hum. Mol. Genet. 16, R14–R20. doi: 10.1093/hmg/ddl486
Yang, W. L., and Frucht, H. (2001). Activation of the PPAR pathway induces apoptosis and COX-2 inhibition in HT-29 human colon cancer cells. Carcinogenesis 22, 1379–1383. doi: 10.1093/carcin/22.9.1379
Yu, H., Pardoll, D., and Jove, R. (2009). STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer 9, 798–809. doi: 10.1038/nrc2734
Yu, J., Ustach, C., and Kim, H.-R. C. (2003). Platelet-derived growth factor signaling and human cancer. J. Biochem. Mol. Biol. 36, 49–59. doi: 10.5483/BMBRep.2003.36.1.049
Yu, J., Zhang, L., Hwang, P. M., Kinzler, K. W., and Vogelstein, B. (2001). PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7, 673–682. doi: 10.1016/S1097-2765(01)00213-1
Yuan, L., Zhou, C., Lu, Y., Hong, M., Zhang, Z., Zhang, Z., et al. (2015). IFN-γ-mediated IRF1/miR-29b feedback loop suppresses colorectal cancer cell growth and metastasis by repressing IGF1. Cancer Lett. 359, 136–147. doi: 10.1016/j.canlet.2015.01.003
Yuan, X.-W., Wang, D.-M., Hu, Y., Tang, Y.-N., Shi, W.-W., Guo, X.-J., et al. (2013). Hepatocyte nuclear factor 6 suppresses the migration and invasive growth of lung cancer cells through p53 and the inhibition of epithelial-mesenchymal transition. J. Biol. Chem. 288, 31206–31216. doi: 10.1074/jbc.M113.480285
Zhang, J., Guenther, M. G., Carthew, R. W., and Lazar, M. A. (1998). Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 12, 1775–1780. doi: 10.1101/gad.12.12.1775
Zhao, L. Y., Niu, Y., Santiago, A., Liu, J., Albert, S. H., Robertson, K. D., et al. (2006). An EBF3-mediated transcriptional program that induces cell cycle arrest and apoptosis. Cancer Res. 66, 9445–9452. doi: 10.1158/0008-5472.CAN-06-1713
Zheng, W., Wong, K. E., Zhang, Z., Dougherty, U., Mustafi, R., Kong, J., et al. (2012). Inactivation of the vitamin D receptor in APC(min/+) mice reveals a critical role for the vitamin D receptor in intestinal tumor growth. Int. J. Cancer 130, 10–19. doi: 10.1002/ijc.25992
Zhu, Y., Richardson, J. A., Parada, L. F., and Graff, J. M. (1998). Smad3 mutant mice develop metastatic colorectal cancer. Cell 94, 703–714. doi: 10.1016/S0092-8674(00)81730-4
Keywords: colorectal cancer, promoter analysis, pathway analysis, master regulator analysis, Wnt pathway
Citation: Wlochowitz D, Haubrock M, Arackal J, Bleckmann A, Wolff A, Beißbarth T, Wingender E and Gültas M (2016) Computational Identification of Key Regulators in Two Different Colorectal Cancer Cell Lines. Front. Genet. 7:42. doi: 10.3389/fgene.2016.00042
Received: 31 October 2015; Accepted: 14 March 2016;
Published: 05 April 2016.
Edited by:
Artemis Georgia Hatzigeorgiou, Biomedical Sciences Research Center Alexander Fleming, GreeceReviewed by:
Nestoras Karathanasis, Diana Lab, GreeceSpyros Tastsoglou, University of Thessaly, Greece
Copyright © 2016 Wlochowitz, Haubrock, Arackal, Bleckmann, Wolff, Beißbarth, Wingender and Gültas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Darius Wlochowitz, darius.wlochowitz@bioinf.med.uni-goettingen.de;
Mehmet Gültas, mehmet.gueltas@bioinf.med.uni-goettingen.de
 Darius Wlochowitz
Darius Wlochowitz Martin Haubrock
Martin Haubrock Jetcy Arackal2
Jetcy Arackal2 Annalen Bleckmann
Annalen Bleckmann Tim Beißbarth
Tim Beißbarth Edgar Wingender
Edgar Wingender Mehmet Gültas
Mehmet Gültas