- 1School of Sport and Biomedical Sciences, University of Bolton, Bolton, United Kingdom
- 2Extension Division, University of California, Los Angeles, Los Angeles, CA, United States
The Jats represent a large ethnic community that has inhabited the northwest region of India and Pakistan for several thousand years. It is estimated the community has a population of over 123 million people. Many historians and academics have asserted that the Jats are descendants of Aryans, Scythians, or other ancient people that arrived and lived in northern India at one time. Essentially, the specific origin of these people has remained a matter of contention for a long time. This study demonstrated that the origins of Jats can be clarified by identifying their Y-chromosome haplogroups and tracing their genetic markers on the Y-DNA haplogroup tree. A sample of 302 Y-chromosome haplotypes of Jats in India and Pakistan was analyzed. The results showed that the sample population had several different lines of ancestry and emerged from at least nine different geographical regions of the world. It also became evident that the Jats did not have a unique set of genes, but shared an underlying genetic unity with several other ethnic communities in the Indian subcontinent. A startling new assessment of the genetic ancient origins of these people was revealed with DNA science.
Introduction
Population and Demographics
The Jats represent one of the largest ethnic groups that has evolved in the northwest region of the Indian subcontinent—India and Pakistan—over several thousand years. Since the partition of India in 1947, Hindu and Sikh Jats have lived primarily in India, and the Muslim Jats have lived primarily in Pakistan.
In 2012, the Jat population in India—mostly Hindus and Sikhs—was reported to be 82.5 million people (Chatterji, 2012). The last time the population was surveyed according to caste–in the 1931 Census of India–the Jats belonged to three main religions: Hinduism 47%, Islam 33%, and Sikhism 20% (Burdak, 2016). Assuming the ratio among religions has stayed about the same (i.e., 33% for Islam and 67% combined for Hinduism and Sikhism), the population of Muslim Jats in 2012 can be extrapolated to about 40.6 million (82.5 million/67 × 33). On this basis, the total population of all Jats in the Indian subcontinent is estimated to be around 123 million people, roughly equal to the combined population of France, Spain, and Portugal.
Archeological Evidence
The origins of the Indus Valley Civilization—also known as the Harrapan Civilization—can be traced to 7,380–6,201 BCE in northwestern India (Khandekar, 2012). A recent discovery of a large Indus Valley site was made in Rakhighari, about 160 km from New Delhi. Its origins go back to about 5000 BCE (Subramanian and Khan, 2016). This ancient civilization flourished in the third millennium BCE (Harari, 2015), and its people were known as the earliest agriculturists in South Asia (Harris, 1996).
Originally the Jats were pastoralists (Khazanov and Wink, 2001), and gradually became farmers. Although farming settlements emerged in the Indus Valley Civilization about 4,000 BCE (Violatti, 2013), and Jats have been firmly settled as agriculturists in the same geographical region, a connection between the two has not been explored thoroughly. Apparently, this is because there is no conclusive written history of the people of the Indian subcontinent when we look back more than about 2,500 years. As a result, the deep ancestry of the Jat people has remained a mystery for a long time.
Historical Perspectives
Among the earliest available books from India—written in Sanskrit—that provide some glimpses of history are the Rigveda, composed between 1,500 and 500 BCE (Flood, 1996), and the Mahabharata, composed between 400 BCE and 400 CE (Molloy, 2008). This textual evidence contains some references to the existence of agriculture in the area, and mentions people known as the Srinjaya—meaning, sons of the sickle or farmers (Hewitt, 1894).
Some early Greek and Roman historians had acquired fragments of information about India from soldiers and merchants in the Persian Empire. But there is no reliable written history of the Indian subcontinent before Alexander the Great's campaign of India in 327 BCE (Smith, 1921). Although archeology has shed some light about the distant past—and even this record is incomplete—written history of India goes back only about 2,500 years.
More recently, numerous books have been written about Indian history and scholarship has been attempted over the origins of the Jats. Several historians have asserted that Jats were descendants of Indo-Aryans (Risley, 1915; Vaidya, 1921; Singh, 1963; Joon, 1967; Dahiya, 1980; Jindal, 1992; Qanungo, 2003), or Indo-Scythians (Elphinstone, 1841; Cunningham, 1871; Tod, 1920; Mahil, 1955; Marshall, 1960; Dhillon, 1994; Nijjar, 2008). The focus of most historians has been on the Indo-Aryan migrations to north India, which started around 1750 BCE, and the arrival of Indo-Scythians later around 200 BCE. The historical debate between the Aryan and Scythian origins of the Jats has continued (Panwar, 1993). In the scientific community as well, there are varied opinions regarding the Indo-Aryan migrations to India (Wells et al., 2001; Cordaux et al., 2004; Metspalu et al., 2011).
A Pioneer Study Based on Ethnography
In the early days of anthropology, craniometry seemed to offer a solution to the study of antiquity of humans, and attention was directed mainly at the examination of skulls that were excavated. This led to anthropometry, a process of measuring various parts of living humans. Sir Herbert Risley, who was in-charge of the Census of India, introduced anthropometry in India in 1886, and became a pioneer in the application of scientific methods to classify ethnic groups of the country. Based on their tall stature, a long head, fair complexion, and narrow nose, the Jats were classified as Indo-Aryan, and groups with a medium stature, a broad head, fair complexion, and a moderately fine nose, were classified as Scytho-Dravidian (Risley, 1915). The study received criticism, but it opened new fields of enquiry about the people of the subcontinent.
Tracing Deep Ancestry
We can identify our progenitors going back a few hundred years with traditional genealogical methods using records of family history. Beyond that, tracing ancestry is complicated because there is generally no documentation. New methods are now available based on recent developments in DNA science. Because DNA is inherited from our parents, it is possible to track the genes going back thousands of years and determine where our ancestors came from. Genetic tests allow us to trace the origins and paths of ancestors.
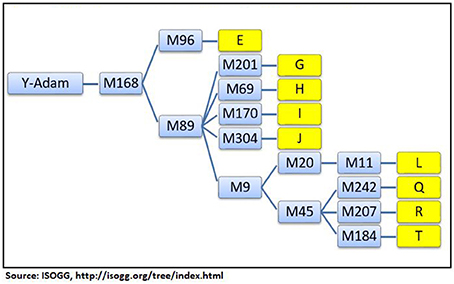
In DNA testing, two kinds of markers on the DNA strand are assessed: short tandem repeats (STRs), and single nucleotide polymorphisms (SNPs). The STRs are found on the Y-chromosome (Y-STRs) and used exclusively for tracing male lines of heredity. The SNPs are found on the Y-chromosome and in MT-DNA. They are used to trace male and female lines of heredity. The result of the test is a set of numbers, referred to as the haplotype, which is used to identify the haplogroup of an individual. Thus, the haplogroup represents a group of people who have inherited common genetic characteristics from the same most recent common ancestor (MRCA). All humans belong to haplogroups which are designated according to their Y-DNA and MT-DNA. The geographic origins of a Y-chromosome haplogroup can be deciphered from the phylogenetic tree of mankind maintained by the International Society of Genetic Genealogy (ISOGG, 2016).
By identifying Y-chromosome haplogroups and their geographic origins, this study has shown that: (a) the genetic origins of the Jats can be traced to at least nine different ancestors and geographical areas of this world, and (b) as a result, this ethnic group did not emerge from a single ancient population such as, the Indo-Aryans or Indo-Scythians.
Materials and Methods
Haplogroups
The nonrecombining portion of the human Y-chromosome is paternally inherited, and passes from father to son essentially unchanged. But occasionally a random change known as a polymorphism or mutation occurs. Such mutations—also called markers—serve as beacons and can be mapped. When geneticists identify a mutation in a DNA test, they try to determine when it first occurred and in which geographic region of the world. Thus, the Y-chromosome haplogroup can be used to trace the paternal line of the individual (Jobling and Tyler-Smith, 2003). The Y-DNA tests are available only for men.
Because Y-DNA haplogroups are closely linked to geography and populations, they serve as important genetic indicators to trace paternal lineages and their ancient origins. This study has relied on the Y-DNA haplogroup as the primary gauge for exploring deep ancestry of the MRCAs of the Jats.
Y-DNA Haplogroup Tree
The Y-DNA haplogroups contain many branches called subhaplogroups or subclades and form a phylogenetic tree of mankind. The branch lengths of the tree are governed by the mutation rates of Y-STRs and Y-SNPs. The markers on the phylogenetic tree provide pieces of evidence regarding the date and geographical origin of the MRCA in the distant past. The top-level haplogroups are identified by letters, A through T. Their subhaplogroups or subclades are expressed as letters and numbers (G2, R1b1, E3b1b, etc.). The tree is updated periodically according to new developments in the field. This study has relied on the Y-DNA haplogroup tree (ISOGG, 2016) to identify the geographical origins of the Jats.
Identifying Haplogroups
Two different methods are incorporated in this study to determine haplogroups. One method examined SNPs on the Y-chromosome in the laboratory with actual DNA samples of men. Another method examined STRs in the Y-chromosome haplotypes found in published literature. For these records with Y-STR profiles, a software program was used to predict the haplogroups.
Several software tools are available that use Y-chromosome haplotypes to identify haplogroups. These software tools are based on mathematical calculations. A study of a software tool, Haplogroup Classifier, developed at the University of Arizona showed that by using machine learning algorithms and data derived from a set of Y-linked STRs, it was possible to assign Y-chromosome haplogroups to individual samples with a high degree of accuracy (Schlecht et al., 2008). The software tool yHaplo was developed at 23andMe (23andme, 2017), a DNA testing company, to enable researchers to identify the Y chromosome haplogroups of males in a genetic sample. The software has been run on more than 600,000 males in the 23andMe database to confirm haplogroup calls for several hundred individuals (Poznik, 2016). In this study, Whit Athey's Haplogroup Predictor software was used (Athey, 2006).
Datasets
Two separate datasets were created for this study, one representing the Jat population, and one representing 38 other ethnic groups of the Indian subcontinent for comparison purposes.
For the Jat population, a dataset of 302 men was compiled consisting of 44 records from the Genographic Project database (Genographic, 2016), with permission of the National Geographic Society, and 258 records from published sources (Henke et al., 2001; Nagy et al., 2007). The haplogroups in the Genographic Project database were already predetermined at source, based on examination of SNPs in the lab with actual Y-DNA samples. The records from published sources contained haplotypes with nine to twelve Y-STR loci (DYS19, DYS385a, DYS385b, DYS389-1, DYS389-2, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, and DYS439) on the Y-chromosome. The haplogroups for these 258 records were identified by processing their haplotypes in the Haplogroup Predictor software. Only the predominant top-level haplogroups were identified (the subclades or subhaplogroups were not used). All haplogroups from the Genographic Project database and published sources were merged and sorted in Excel. This dataset of 302 records represented 294 Jats from India and eight from Pakistan. The Muslim Jats were under represented in the sample.
For other ethnic groups of the Indian subcontinent, a dataset of 1,855 men representing 38 ethnic groups in Bangladesh, India, and Pakistan was compiled from the Genographic Project database (Genographic, 2016), and published sources (Sengupta et al., 2006; Zhao et al., 2009; Giroti and Talwar, 2010; Nair et al., 2011; Chennakrishnaiah et al., 2013; Lee et al., 2014). All haplogroups for these records were predetermined at source, based on examination of SNPs in the lab with actual Y-DNA samples. For this dataset as well, all haplogroups from the Genographic Project database and published sources were merged and sorted in Excel.
Comparison with Foreign Populations
Migrations and invasions have been recurring themes in the history of the Indian subcontinent. To corroborate a nexus with other populations, the genetic relationship of the Jat population was compared with 25 populations of Central Asia and Northern Europe, comprising 21,899 Y-STR haplotypes in the world population database maintained at YHRD.org (see sources of data under Supplementary Information). A sample of 258 Y-STR haplotypes of Jats was used. The calculations were performed with AMOVA and MDS online software tools provided at the YHRD website http://www.yhrd.org.
AMOVA (analysis of molecular variance), is a statistical program for estimating population differentiation directly from molecular distance among DNA haplotypes (Excoffier et al., 1992). This online tool at YHRD.org provides an option to use the Rst or Fst measure—they are analogous—to determine the proportion of variance between populations. The Rst measure was used because it is reported to provide relatively unbiased estimates of population divergence, whereas the Fst measure tends to show too much population similarity (Slatkin, 1995). A multidimensional scaling (MDS) plot based on a non-metric algorithm (Kruskal, 1964) was also created, to provide a visual representation of the pattern of similarities or distances between Jats and 11 foreign populations. The full set of 25 populations from the AMOVA calculation was not used in the MDS plot to avoid overlapping of labels.
Results
Genetic Distance
The results of the AMOVA and MDS tests (Table 1) confirmed that the Jats had genetic affinities with several foreign populations and provided some insights into their genetic makeup. The genetic difference between the Jats and the tested populations ranged from a small distance of 0.0257 in Afghanistan to a larger distance of 0.2128 in FYR Macedonia.
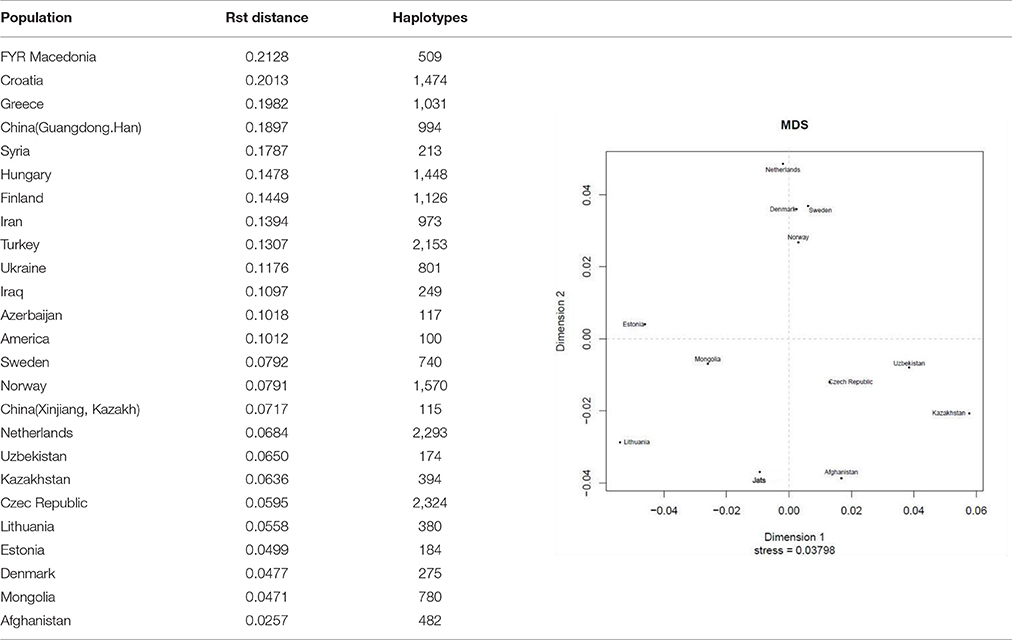
Table 1. Analysis of molecular variance (AMOVA): Pairwise Rst genetic distance between Jats and 25 selected population and a multidimensional scaling (MDS) plot of Jats and 11 closely related populations.
The close genetic affinity between the Jats and the Afghani population was evident because most migrations and invasions into north India have passed through this territory. In the past, several Jat tribes and clans have inhabited parts of Afghanistan (Bellew, 1891).
Haplogroups and Geographic Origins
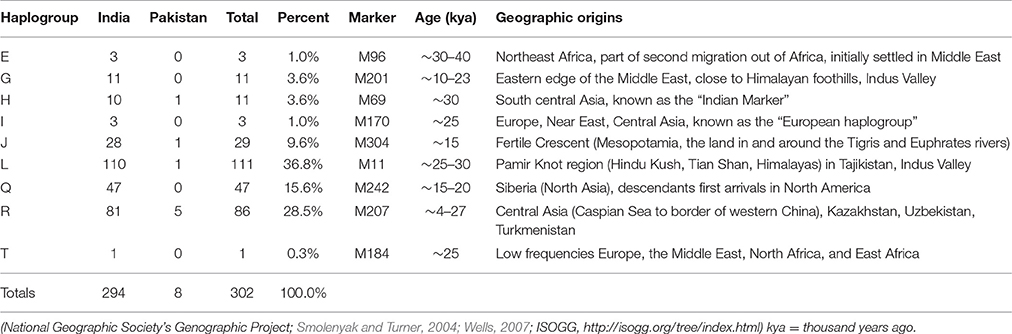
The results of haplogroup analyses revealed that MRCAs of 302 Jats in our dataset belonged to nine different haplogroups—E, G, H, I, J, L, Q, R, and T—with nine different geographic origins.
The same nine haplogroups were used to compare the Jats with other ethnic groups of the Indian subcontinent. The haplogroups of 302 Jats and 1,855 other men in 38 ethnic groups of Bangladesh, India, and Pakistan are displayed in Table 2.
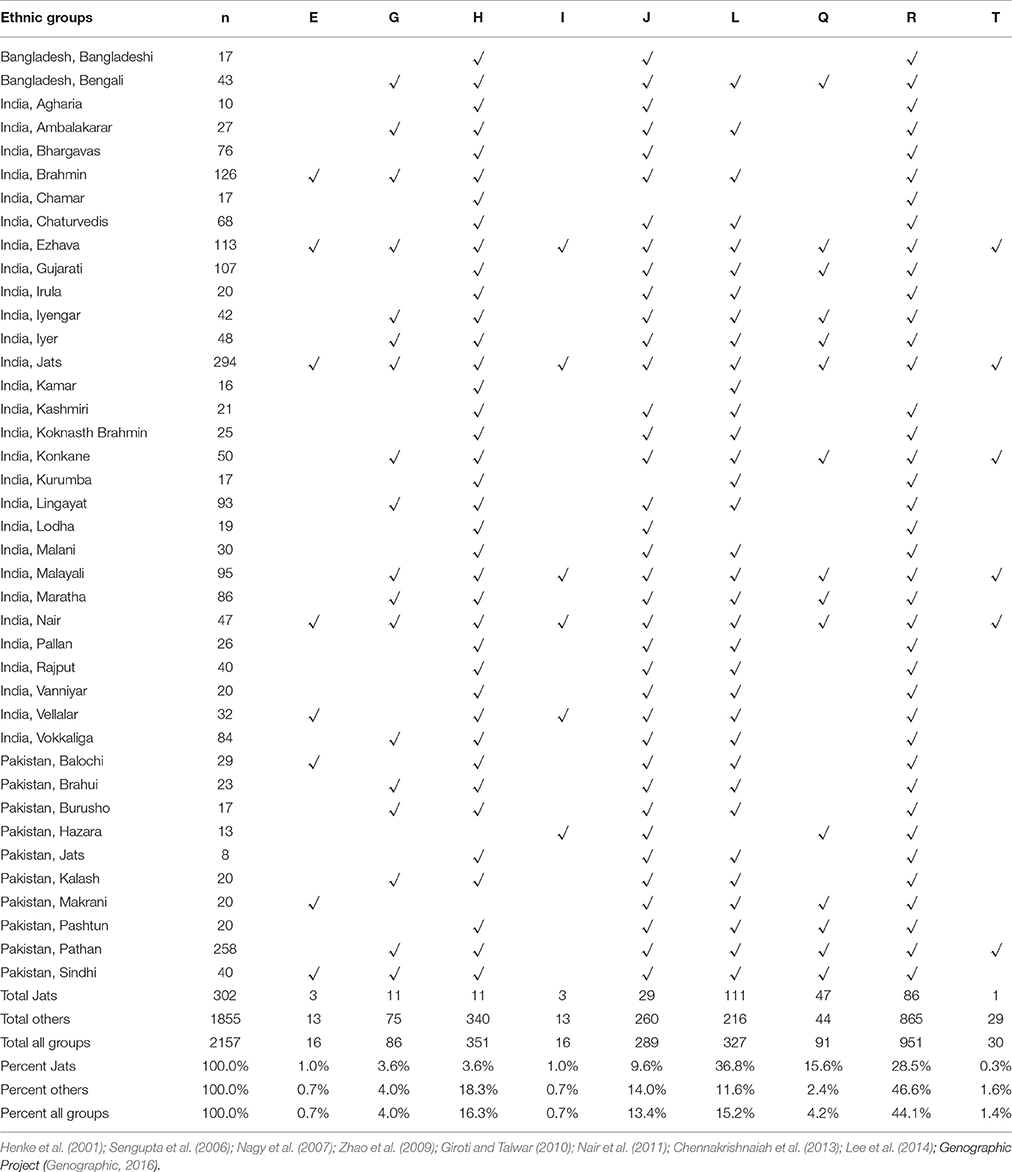
Table 2. Representation in nine Y-Chromosome haplogroups: Jats and thirty-eight ethnic groups of Indian subcontinent.
The results signified that the Jats shared an underlying genetic unity with several other ethnic communities in the Indian subcontinent with the same MRCAs and geographic origins. About 90% of the Jats and about 75% of the other 38 groups in the study belonged to the same four haplogroups J, L, Q, and R.
The geographic origins of the Jats in our study are summarized in Table 3.
A short phylogenetic tree of nine haplogroups of the Jats in this study—with their key top-level markers starting from Y-Adam—appears in Figure 1.
Haplogroup L (36.8%)
This is the largest haplogroup in the Jat sample population. It is present in the Indian population at an overall frequency of about 7–15% (Basu et al., 2003; Cordaux et al., 2004). Genetic studies suggest that this may be one of the original haplogroups of the creators of Indus Valley Civilization (McElreavey and Quintana-Murci, 2005; Sengupta et al., 2006). It has a frequency of about 28% in western Pakistan and Baluchistan, from where the agricultural creators of this civilization emerged (Qamar et al., 2002). The origins of this haplogroup can be traced to the rugged and mountainous Pamir Knot region in Tajikistan (Wells, 2007).
Haplogroup R (28.5%)
This haplogroup originated in north Asia about 27,000 years ago (ISOGG, 2017). It is one of the most common haplogroups in Europe, with its branches reaching 80% of the population in some regions. One branch is believed to have originated in the Kurgan culture, known to be the first speakers of the Indo-European languages and responsible for the domestication of the horse (Smolenyak and Turner, 2004). From somewhere in central Asia, some descendants of the man carrying the M207 mutation on the Y chromosome headed south to arrive in India about 10,000 years ago (Wells, 2007). This is one of the largest haplogroups in India and Pakistan. Of its key subclades, R2 is observed especially in India and central Asia.
Haplogroup Q (15.6%)
With its origins in central Asia, descendants of this group are linked to the Huns, Mongols, and Turkic people. In Europe it is found in southern Sweden, among Ashkenazi Jews, and in central and Eastern Europe such as, the Rhône-Alpes region of France, southern Sicily, southern Croatia, northern Serbia, parts of Poland and Ukraine. A subclade of this haplogroup is associated with Native American populations, and the mutation occurred 8 to 12 thousand years ago during the migration to the Americas through the Bering Strait (Smolenyak and Turner, 2004). It is estimated that as few as twenty people may have founded the initial native population of the Americas (Liu, 2016).
Haplogroup J (9.6%)
The ancestor of this haplogroup was born in the Middle East area known as the Fertile Crescent, comprising Israel, the West Bank, Jordon, Lebanon, Syria, and Iraq. Middle Eastern traders brought this genetic marker to the Indian subcontinent (Kerchner, 2013).
Haplogroups E, G, H, I, T (9.5%)
The ancestors of the remaining five haplogroups E, G, H, I, and T can be traced to different parts of Africa, Middle East, South Central Asia, and Europe (ISOGG, 2016).
Discussion
Sample Size
In statistical analyses, as the population increases in size, the sample size increases at a diminishing rate, and remains relatively constant when it reaches a size of 380 or more. At about 384, the sample is generally representative for a population of one million, or more (Krejcie and Morgan, 1970). Ideally, the sample size should be 380, and preferably larger.
The dataset of 302 Jats used in our research represents a margin of error of 5.7% at a confidence level of 95%. In other words, if a survey is conducted one hundred times among a similar group of people (i.e., 302 × 100; 32,000 people in total), the distribution in haplogroups is expected to be about the same as in this study, with a margin of error of plus or minus 5.7%.
Although the sample of 302 records used in this research revealed key haplogroups for the Jats, the results are not representative of this entire ethnic group of an estimated 123 million people. It is already noted that the Muslim Jats of Pakistan were underrepresented in this study. A larger sample of Muslim Jats is likely to reveal a few additional haplogroups and provide a more complete picture. Therefore, to ascertain a representative distribution of haplogroups for the entire ethnic group of the Jats, the sample size should be at least 380, with a proportional representation of Hindus, Sikhs, and Muslims.
Potential Errors in Haplogroup Prediction
Because of the need for precision in matters relating to criminal and civil laws, the forensic genetics community is generally not in favor of determining haplogroups with STR profiles. It is held that STR haplotypes are not always identical by descent, but also identical by state, and can be rooted in different haplogroups.
A study that used STR profiles of 119 males in Argentina to determine haplogroups with two software programs—Whit Athey's Haplogroup Predictor (used in this study), and a Haplogroup Classifier developed at the University of Arizona—showed that the results were not totally accurate (Muzzio et al., 2011). Another study of 165 males in Nicaragua showed that Athey's Haplogroup Predictor produced accurate results for 95.2% of the sample, but 4.8% of the results were inaccurate (Nunez et al., 2012). For greater reliability in identifying Y chromosomal haplogroups, the forensic community's preferred method is to analyze (SNPs) on the Y chromosome in the lab with actual DNA samples.
Athey has explained that the main drawback of the haplogroup prediction method in his software is the size of the database of some Y-STR haplotypes from which the allele frequencies are calculated. For most haplogroups there is sufficient Y-STR haplotype data. However, for some haplogroups, such as, C, H, L, N, and Q, the database of Y-STR haplotypes is smaller, and the results may be prone to error (Athey, 2006).
Of the 302 records used in this study, 258 were processed through Whit Athey's software. Of these, 169 haplotypes belonged to the potentially error-prone haplogroups H, L, and Q, identified by Athey. Assuming an error rate of 5% for this software, as reported in the Nicaraguan study (Nunez et al., 2012), only 13 haplotypes (5% of 258) may have identified incorrect haplogroups, representing a potential error rate of about 4.3% (13/302) in the total sample used in this study. This suggests an accuracy of about 96% in the haplogroups and geographic origins identified in this study.
Population Mixture Leading to Endogamy
Studies have shown that most ethnic groups of the Indian subcontinent descended from a mixture of two divergent populations. These were the Ancestral North Indians (ANI) who were related to Central Asians, Middle Easterners, Caucasians, and Europeans, and the Ancestral South Indians (ASI) who were not closely related to any groups outside the subcontinent (Reich et al., 2009). These findings explain that admixture was widespread at one time (Moorjani et al., 2013).
The results of the AMOVA and MDS tests in our study confirmed that the Jats had genetic contributions from several populations in the Middle East, Central Asia, and Europe. After the arrival of people called Indo-Aryans—also known as Indo-European speakers—in north India about 2000 BCE, the caste system was introduced, and a stratified social hierarchy evolved. The upper-caste populations started practicing and encouraging endogamy about 70 generations (more than 2,000 years) ago (Basu et al., 2016). Another study suggested that endogamy started much later, about the time of foreign invasions in north India (Vadivelu, 2016).
Consanguinity is another form of endogamy. The word consanguinity comes from the Latin con, meaning shared, and sanguis, meaning blood. Marriage between people who have at least one recent common ancestor is known as consanguineous, and the children are considered inbred. Couples related as second cousins or closer account for an estimated 10.4% of the global population, with the highest rates in West, Central, and South Asia (Bittles and Black, 2010). According to the International Institute for Population Sciences in Mumbai, about 16% of marriages in India are consanguineous (Kuntla et al., 2013). In Pakistan, where first cousin marriages have occurred for generations, the rate is 67% (Yaqoob et al., 1993).
The motivation behind consanguinity is usually to keep bonds, wealth, and property within a family. For this reason, there is a long list of cousin marriages among famous people (e.g., Albert Einstein, Charles Darwin, and others), and in royal families all over the world. Although endogamy has become the general norm in India, and consanguinity is practiced in some parts of the subcontinent, most ethnic groups—including the Jats—carry a blend of genetic components from different populations in the past.
Languages and Genetic Diversity
There are several thousand ethnic and tribal groups in the Indian subcontinent (Papiha, 1996; Xing et al., 2010). Members of these communities share common self-identities that are based on languages, customs, cuisines, and at least six major religions. There are 22 official languages and many dialects in the country (Annamalai, 2006). At least eight different languages—Balochi, Haryanvi, Hindi, Punjabi, Rajasthani, Saraiki, Sindhi, and Urdu—are spoken in the Jat communities, which demonstrates their genetic diversity.
The Aryan-Scythian Conundrum
The estimated population of the Indian subcontinent in 10,000 BCE was about 100,000 people, and stayed at this level until about 5,000 BCE, by when agriculture had spread in the Indus Valley (McEvedy and Jones, 1978). Since then the population has grown exponentially, with about 1.7 billion people in the Indian subcontinent now. Among the several thousand ethnic and tribal groups in the subcontinent, there are no existing population groups known as Indo-Aryan or Indo-Scythian. These appear to be labels that have been loosely applied to people who arrived in north India in a series of waves over a long period in the distant past.
Sir Risley's ethnographic classifications of Indian people did not provide any clues about the origins of the Indo-Aryans and the Scytho-Dravidians. But his studies showed that these two groups were physically different. According to the Imperial Gazetteer of India, the Indo-Scythians were likely pushed toward the south by the Indo-Aryans, mingled with the Dravidian population, and became the ancestors of an entirely different ethnic group known as the Marathas (Gazetteer, 1931).
The Pamir Knot region—from where the MRCA of haplogroup L emerged—is also the home of the Bactria-Margiana Archaeological Complex (BMAC), in a site called Gonur that represents a Bronze Age culture known as the Oxus civilization (Sarianidi, 2007). This BMAC site of around 4000 BCE was discovered and named by the Soviet archeologist Viktor Sarianidi. Among his findings, Sarianidi discovered evidence of sacred alters; traces of ingredients such as, poppy seeds, cannabis, and ephedra, used for a drink called soma; horse sacrifices; four wheeled chariots; and other connections with the Aryans (Sarianidi, 2007; Wood, 2007). Some BMAC materials of this type have been found in the Indus Valley sites. Archaeologist J. P. Mallory from Queens University (Ireland), and Indologist Asko Parpola from the University of Helsinki (Finland), have suggested a connection between the Aryans and BMAC (Mallory, 1989; Parpola, 1999). Because the MRCA of haplogroup L emerged from the same geographical area as the people called the Aryans, there may be a genetic link between the two.
The haplotypes of 26 ancient human specimens from the Krasnoyarsk area in Siberia, dated from between the middle of the second millennium BCE to the fourth century CE (Scythian and Sarmatian timeframe), revealed that nearly all specimens belonged to R1a, a subclade of haplogroup R, which is thought to mark the eastward migration of early Indo-Europeans (Keyser et al., 2009). Another survey of 217 samples from Europe and Asia revealed that R1a1, another subclade of haplogroup R, was spread across Eurasia (Pamjav et al., 2012). Because the origins of haplogroup R can be traced to the same geographical area, there may be a genetic link with the ancient people called Scythians.
Studies have shown that the Hindu Kush area from where these groups migrated to the Indian subcontinent served as a confluence of gene flows from adjoining areas rather than a source of distinctly autochthonous populations (Cristofaro et al., 2013). These people also arrived in north India at different times. As noted earlier, members of haplogroup R arrived about 10,000 years ago, the Indo-Aryan migrations started about 2000 BCE, and the Indo-Scythians arrived much later, around 200 BCE. Because of their physical differences and the large gaps between their arrival times, it can be inferred that these groups were genetically different and not the same people.
This study has shown that the genetic origins of the Jats can be traced to at least nine and possibly more MRCA's, with nine different geographical origins that are spread thousands of miles apart (e.g., from the Fertile Crescent to Serbia). These nine MRCAs were genetically different. Therefore, any assertion that Jats are descendants of a single ancient population such as, the Indo-Aryans or Indo-Scythians cannot be supported. However, certain members of the Jat ethnic group who belong to haplogroups L and R—along with members of several other ethnic groups in the Indian subcontinent who belong to the same two haplogroups—are the most probable candidates to be linked to these ancient populations.
Conclusion
The human Y-chromosome provides a powerful molecular tool for analyzing Y-STR haplotypes and determining their haplogroups which lead to the ancient geographic origins of individuals. For this study, the Jats and 38 other ethnic groups in the Indian subcontinent were analyzed, and their haplogroups were compared. Using genetic markers and available descriptions of haplogroups from the Y-DNA phylogenetic tree, the geographic origins and migratory paths of their ancestors were traced.
The study demonstrated that based on their genetic makeup, the Jats belonged to at least nine specific haplogroups, with nine different lines of ancestry and geographic origins. About 90% of the Jats in our sample belonged to only four different lines of ancestry and geographic origins. Therefore, attributing the origins of this entire ethnic group to loosely defined ancient populations such as, Indo-Aryans or Indo-Scythians represents very broad generalities and cannot be supported. The study also revealed that even with their different languages, religions, nationalities, customs, cuisines, and physical differences, the Jats shared their haplogroups with several other ethnic groups of the Indian subcontinent, and had the same common ancestors and geographic origins in the distant past. Based on recent developments in DNA science, this study provided new insights into the ancient geographic origins of this major ethnic group in the Indian subcontinent. A larger dataset, particularly with more representation of Muslim Jats, is likely to reveal some additional haplogroups and geographical origins for this ethnic group.
Ethics Statement
This study presented in the manuscript does not involve human or animal subjects. All data used in the study are from existing databases and published sources, which are cited.
Author Contributions
DM analyzed data and wrote the paper; IM wrote the paper.
Funding
Research in the laboratory of IM is supported by a Jenkinson TIRI Award and the University of Bolton, UK.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fgene.2017.00121/full#supplementary-material
References
23andme (2017). 23andMe.com Available online at: https://www.23andme.com/ (Accessed January 5, 2017).
Annamalai, E. (2006). “India: language situation,” in Encyclopedia of Language and Linguistics, ed K. Brown (Amsterdam: Elsevier), 610–613.
Athey, T. W. (2006). Haplogroup prediction from Y-STR values using a Bayesian-allele frequency approach. J. Genet. Geneal. 2, 34–39.
Basu, A., Mukherjee, N., Roy, S., Sengupta, S., Banerjee, S., Chakraborty, M., et al. (2003). Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 13, 2277–2290. doi: 10.1101/gr.1413403
Basu, A., Sarkar-Roy, N., and Majumder, P. P. (2016). Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc. Nat. Acad. Sci. U.S.A. 113, 1594–1599. doi: 10.1073/pnas.1513197113
Bellew, H. W. (1891). An Inquiry into the Ethnography of Afghanistan. London: The Oriental University Institute, Woking.
Bittles, A. H., and Black, M. L. (2010). Consanguineous marriage and human evolution. Annu. Rev. Anthropol. 39, 193–207. doi: 10.1146/annurev.anthro.012809.105051
Burdak, L. (2016). Jat Belt: Distribution of Jat Population. Available online at: http://www.jatland.com/home/Jat_Belt (Accessed July 5, 2016).
Chennakrishnaiah, S., Perez, D., Gayden, T., Rivera, L., Regueiro, M., and Herrera, R. J. (2013). Indigenous and foreign Y-chromosomes characterize the Lingayat and Vokkaliga populations of Southwest India. Gene 526, 96–106. doi: 10.1016/j.gene.2013.04.074
Cordaux, R., Aunger, R., Bentley, G., Nasidze, I., Sirajuddin, S. M., and Stoneking, M. (2004). Independent origins of Indian caste and tribal paternal lineages. Curr. Biol. 14, 231–235. doi: 10.1016/j.cub.2004.01.024
Cristofaro, J. D., Pennarun, E., Mazieres, S., Myers, N. M., Lin, A. A., Temori, S. A., et al. (2013). Afghan hindu kush: where eurasian sub-continent gene flows converge. PLoS ONE 8:e76748. doi: 10.1371/journal.pone.0076748
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
Gazetteer (1931). “Possible origin of the scytho-dravidian type,” in The Imperial Gazetteer of India (Oxford: Clarendon Press). Available at Digital South Asia Library, University of Chicago. Published under the authority of His Majesty's secretary of state for India in council. v. 1 (1908–1931), 307.
Genographic (2016). National Geographic Society: The Genographic Project. National Geographic Society. Available online at: https://genographic.nationalgeographic.com (Accessed August 5, 2016).
Giroti, R., and Talwar, I. (2010). The most ancient democracy in the world is a genetic isolate: an autosomal and Y-chromosome study of the hermit village of Malana (Himachal Pradesh, India). Hum. Biol. 82, 123–141. doi: 10.3378/027.082.0201
Harris, D. R. (1996). The Origins and Spread of Agriculture and Pastoralism in Eurasia. New York, NY: Routledge.
Henke, J., Henke, L., Chatthopadhyay, P., Kayser, M., Dulmer, M., Cleef, S., et al. (2001). Application of Y-chromosomal STR haplotypes to forensic genetics. Croat. Med. J. 42, 292–297.
Hewitt, J. F. (1894). The Ruling Races of Prehistoric Times in India, South-Western Asia and Southern Europe. Edinburgh: Archibald Constable & Company.
ISOGG (2016). International Society of Genetic Geology: Y-DNA Haplogroup Tree. Available online at: http://isogg.org/tree/index.html (Accessed December 16, 2016).
ISOGG (2017). Y-DNA Haplogroup Tree 2017. Available online at: http://isogg.org/tree/index.html (Accessed October 10, 2016).
Jindal, M. S. (1992). History of Origins of Some Clans in India, with Special Reference to Jats. New Delhi: Swarup and Sons.
Jobling, M. A., and Tyler-Smith, C. (2003). The human Y chromosome: an evolutionary marker comes of age. Nat. Rev. Genet. 4, 598–612. doi: 10.1038/nrg1124
Kerchner, C. F. (2013). YDNA Haplogroup Descriptions and Information Links. Available online at: http://www.kerchner.com/haplogroups-ydna.htm (Accessed September 2, 2016).
Keyser, C., Bouakaze, C., Crubézy, E., Nikolaev, V. G., Montagnon, D., Reis, T., et al. (2009). Ancient DNA provides new insights into the history of south Siberian Kurgan people. Hum. Genet. 126, 395–410. doi: 10.1007/s00439-009-0683-0
Krejcie, R. V., and Morgan, D. W. (1970). Determining sample size for research activities. Educ. Psychol. Meas. 30, 607–610. doi: 10.1177/001316447003000308
Kruskal, J. B. (1964). Nonmetric multidimensional scaling: a numerical method. Psychometrika 29, 115–129. doi: 10.1007/BF02289694
Kuntla, S., Goli, S., Sekher, T. V., and Doshi, R. (2013). Consanguineous marriages and their effects on pregnancy outcomes in India. Int. J. Soc. Soc. Policy 33, 437–452. doi: 10.1108/IJSSP-11-2012-0103
Lee, E. Y., Shin, K.-J., Rakha, A., Sim, J. E., Park, M. J., Kim, N. Y., et al. (2014). Analysis of 22 Y chromosomal STR haplotypes and Y haplogroup distribution in Pathans of Pakistan. Forensic Sci. Int. Genet. 11, 111–116. doi: 10.1016/j.fsigen.2014.03.004
Liu, D. (2016). Using DNA to Trace Human Migration. Howard Hughes Medical Institute. Available online at: http://www.hhmi.org/biointeractive/using-dna-trace-human-migration (Accessed November 6, 2016).
Mallory, J. P. (1989). In Search of the Indo-Europeans: Language, Archaeology and Myth. London: Thames and Hudson.
McElreavey, K., and Quintana-Murci, L. (2005). A population genetics perspective of the Indus Valley through uniparentally-inherited markers. Ann. Hum. Biol. 32, 154–162. doi: 10.1080/03014460500076223
McEvedy, C., and Jones, R. (1978). Atlas of World Population History. Harmondsworth; Middlesex: Penguin Books Ltd.
Metspalu, M., Romero, I. G., Yunusbayev, B., Chaubey, G., Mallick, C. B., Hudjashov, G., et al. (2011). Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am. J. Hum. Genet. 89, 731–744. doi: 10.1016/j.ajhg.2011.11.010
Molloy, M. (2008). Experiencing the World's Religions: Tradition, Challenge and Change. Columbus, OH: McGraw Hill Education.
Moorjani, P., Thangaraj, K., Patterson, N., Lipson, M., Loh, P.-R., Govindaraj, P., et al. (2013). Genetic evidence for recent population mixture in India. Am. J. Hum. Genet. 93, 422–438. doi: 10.1016/j.ajhg.2013.07.006
Muzzio, M., Santos, M. R., and Bailliet, G. (2011). Software for Y-haplogroup predictions: a word of caution. Int. J. Legal Med. 125, 143–147. doi: 10.1007/s00414-009-0404-1
Nagy, M., Henke, L., Henke, J., Chatthopadhyay, P. K., Völgyi, A., Zalán, A., et al. (2007). Searching for the origin of Romanies: slovakian Romani, Jats of Haryana and Jat Sikhs Y-STR data in comparison with different Romani populations. Forensic Sci. Int. 169, 19–26. doi: 10.1016/j.forsciint.2006.07.020
Nair, P. S., Geetha, A., and Jagannath, C. (2011). Y-short tandem repeat haplotype and paternal lineage of the Ezhava population of Kerala, south India. Croat. Med. J. 52, 344–350. doi: 10.3325/cmj.2011.52.344
Nijjar, B. S. (2008). Origins and History of Jats and Other Allied Nomadic Tribes of India: 900 B.C.-1947 A.D. New Delhi: Atlantic Publishers & Distributors (P) Ltd.
Nunez, C., Geppert, M., Baeta, M., Roewer, L., and Martinez-Jarreta, B. (2012). Y chromosome haplogroup diversity in a Mestizo population of Nicaragua. Forensic Sci. Int. Genet. 6, e192–e195. doi: 10.1016/j.fsigen.2012.06.011
Pamjav, H., Fehér, T., Németh, E., and Pádár, Z. (2012). Brief communication: new Y-chromosome binary markers improve phylogenetic resolution within haplogroup R1a1. Am. J. Phys. Anthropol. 149, 611–615. doi: 10.1002/ajpa.22167
Poznik, G. D. (2016). Identifying Y-chromosome haplogroups in arbitrarily large samples of sequenced or genotyped men. bioRxiv. doi: 10.1101/088716
Qamar, R., Ayub, Q., Mohyuddin, A., Helgason, A., Mazhar, K., Mansoor, A., et al. (2002). Y-Chromosomal DNA variation in Pakistan. Am. J. Hum. Genet. 70, 1107–1124. doi: 10.1086/339929
Qanungo, K. R. (2003). History of the Jats: Upto the death of Mirza Najaf Khan, (1782). Delhi: Books for All.
Reich, D., Thangaraj, K., Patterson, N., Price, A. L., and Singh, L. (2009). Reconstructing Indian population history. Nature 461, 489–494. doi: 10.1038/nature08365
Schlecht, J., Kaplan, M. E., Barnard, K., Karafet, T., Hammer, M. F., and Merchant, N. C. (2008). Machine-learning approaches for classifying haplogroup from Y Chromosome STR data. PLoS Comput. Biol. 4:e1000093. doi: 10.1371/journal.pcbi.1000093
Sengupta, S., Zhivotovsky, L. A., King, R., Mehdi, S. Q., Edmonds, C. A., Chow, C. T., et al. (2006). Polarity and temporality of high-resolution Y-chromosome distributions in india identify both indigenous and exogenous expansions and reveal minor genetic influence of central Asian pastoralists. Am. J. Hum. Genet. 78, 202–221. doi: 10.1086/499411
Slatkin, M. (1995). A measure of population subdividion based on microsatellite allele frequencies. Genetics 139, 457–462.
Smith, V. A. (1921). The Oxford History of India: from the Earliest Times to the End of 1911. London: Oxford at the Clarendon Press.
Smolenyak, M. S., and Turner, A. (2004). Trace Your Roots with DNA: Using Genetic Tests to Explore Your Family Tree. Emmaus, PA: Rodale.
Subramanian, N., and Khan, A. (2016). Who were the people of the Indus Valley Civilisation? The Indian Express. (Accessed January 10,2017).
Tod, J. (1920). Annals and Antiquities of Rajasthan, or the Central and Western Rajput States of India, Vol. 1. London: Humphrey Milford, Oxford University Press.
Vadivelu, M. K. (2016). Emergence of sociocultural norms restricting intermarriage in large social strata (endogamy) coincides with foreign invasions of India. Proc. Natl. Acad. Sci. U.S.A. 113, E2215–E2217. doi: 10.1073/pnas.1602697113
Vaidya, C. V. (1921). History of Mediaeval Hindu India. Vol. 1, Poona City: The Oriental Book-Supplying Agency.
Violatti, C. (2013). Indus Valley Civilization [Online]. Ancient History Encyclopedia. Available online at: http://www.ancient.eu/Indus_Valley_Civilization/ (Accessed January 23, 2017).
Wells, R. S., Yuldasheva, N., Ruzibakiev, R., Underhill, P. A., Evseeva, I., Blue-Smith, J., et al. (2001). The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc. Nat. Acad. Sci. U.S.A. 98, 10244–10249. doi: 10.1073/pnas.171305098
Wells, S. (2007). Deep Ancestry: Inside the Genographic Project. Washington, DC: National Geographic Society.
Xing, J., Watkins, W. S., Hu, Y., Huff, C. D., Sabo, A., Muzny, D. M., et al. (2010). Genetic diversity in India and the inference of Eurasian population expansion. Genome Biol. 11:R113. doi: 10.1186/gb-2010-11-11-r113
Yaqoob, M., Gustavson, K. H., Jalil, F., Karlberg, J., and Iselius, L. (1993). Early child health in Lahore, Pakistan: II inbreeding. Acta Paediatr. Suppl. 82(Suppl. 390), 17–26. doi: 10.1111/j.1651-2227.1993.tb12903.x
Keywords: Y-chromosome, Y-DNA, Y-STR, haplotypes, haplogroups, India, Jats, Pakistan
Citation: Mahal DG and Matsoukas IG (2017) Y-STR Haplogroup Diversity in the Jat Population Reveals Several Different Ancient Origins. Front. Genet. 8:121. doi: 10.3389/fgene.2017.00121
Received: 18 May 2017; Accepted: 30 August 2017;
Published: 20 September 2017.
Edited by:
Mariza De Andrade, Mayo Clinic, United StatesReviewed by:
William C. L. Stewart, Columbia University, United StatesJian Li, Tulane University, United States
Copyright © 2017 Mahal and Matsoukas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David G. Mahal, dgmahal@outlook.com
 David G. Mahal
David G. Mahal Ianis G. Matsoukas
Ianis G. Matsoukas