- 1Instituto de Biociências, Universidade Federal de Mato Grosso, Cuiabá, Brazil
- 2Instituto de Estudos Costeiros, Universidade Federal do Pará, Bragança, Brazil
- 3Departamento de Biologia Celular, Embriologia e Genética, Universidade Federal de Santa Catarina, Florianópolis, Brazil
The arapaima, Arapaima gigas, is a fish whose populations are threatened by both overfishing and the ongoing destruction of its natural habitats. In the Amazon basin, varying levels of population structure have been found in A. gigas, although no data are available on the genetic diversity or structure of the populations found in the Araguaia-Tocantins basin, which has a topographic profile, hydrological regime, and history of fishing quite distinct from those of the Amazon. In this context, microsatellite markers were used to assess the genetic diversity and connectivity of five wild A. gigas populations in the Araguaia-Tocantins basin. The results of the analysis indicated low levels of genetic diversity in comparison with other A. gigas populations, studied in the Amazon basin. The AMOVA revealed that the Arapaima populations of the Araguaia-Tocantins basin are structured significantly. No correlation was found between pairwise FST values and the geographical distance among populations. The low level of genetic variability and the evidence of restricted gene flow may both be accounted for by overfishing, as well as the other human impacts that these populations have been exposed to over the years. The genetic fragility of these populations demands attention, given that future environmental changes (natural or otherwise) may further reduce these indices and eventually endanger these populations. The results of this study emphasize the need to take the genetic differences among the study populations into account when planning management measures and conservation strategies for the arapaima stocks of the Araguaia-Tocantins basin.
Introduction
Worldwide, the populations of many fish species are declining rapidly (Allan et al., 2005), with the communities occupying lakes, rivers and floodplains being the most affected (Abell et al., 2008). In the Neotropical region, the biodiversity of aquatic ecosystems is under intense pressure, primarily from human activities, such as overfishing, pollution, habitat fragmentation, deforestation and the introduction of exotic species (Agostinho et al., 2005).
The arapaima, Arapaima gigas (Schinz, 1822), is a fish of considerable economic importance in the neotropics, and is one of the species listed in the Convention of International Trade in Endangered Species of Wild Fauna and Flora II. The species is listed as “Data Deficient” by the IUCN (World Conservation Monitoring Centre, 2014). As the arapaima prefers the lentic environments, such as flooded forests, rivers and lakes, of the Amazon, Araguaia-Tocantins and Essequibo basins (Castello and Stewart, 2010; Castello et al., 2013), its populations become increasingly concentrated during the dry season, and the high densities of fish that accumulate during this period greatly increase their vulnerability to capture. Given this vulnerability of the species to fishing and the reduction of its stocks in recent years, researchers have focused increasingly on its genetic diversity and population structure (Farias et al., 2003; Hrbek et al., 2005, 2007; Hrbek and Farias, 2008; Hamoy et al., 2008; Araripe et al., 2013; Vitorino et al., 2015), chromosomal evolution (Marques et al., 2006), and other aspects of its biology (Gomes, 2007; Castello, 2008a,b; Arantes et al., 2010; Fernandes et al., 2012; Farias et al., 2015).
Despite this recent interest, many features of the biology of arapaima are still relatively poorly known, in particular the structure of its natural populations. Up until now, in addition, most studies have focused on the populations of the Amazon basin, and little is known about the genetic diversity of the populations that inhabit the Araguaia-Tocantins basin, even though this system is considered to be a priority area for the conservation of the aquatic biodiversity of the Cerrado biome.
As the characteristics of the Araguaia-Tocantins basin are quite distinct from those of the Amazon basin, the data available for Amazonian Arapaima gigas cannot be extrapolated reliably to the Araguaia-Tocantins. In this way, Latrubesse and Stevaux (2002) identified three principal topographical sectors in the Araguaia basin (the principal focus of the present study): (i) the upper Araguaia River, which extends from the headwaters to Registro do Araguaia, located mainly on Pre-Cambrian rocks, with a V-shaped valley and many rapids, (ii) the middle Araguaia River, between Registro do Araguaia and Conceição do Araguaia, which is characterized by a well-developed alluvial plain located on the lowland Bananal Plain, a prominent geomorphological and sedimentary unit, with some isolated rock formations along the main channel, which form small rapids, and (iii) the lower Araguaia River, which runs downstream from the Bananal Plain, over an area of crystalline rocks until its confluence with Tocantins River, which has no well-defined alluvial plain (Latrubesse and Stevaux, 2002).
Because of these characteristics, the hydrological regime of the Araguaia River is also quite unique, and is determined by well-defined dry and rainy seasons (Aquino et al., 2008). The flood pulse is extremely rapid, with floodplain lakes being connected for only short periods of time. The Araguaia-Tocantins basin also includes portions of the two principal Brazilian biomes, the Amazon, to the north and the Cerrado, to the south (Aquino et al., 2005). However, these important centers of biodiversity have been impacted intensively by industrial-scale farming operations and extensive cattle ranching, the construction of reservoirs for the production of hydroelectric energy (Aquino et al., 2008, 2009; Latrubesse et al., 2009), and the establishment of the Tocantins-Araguaia waterway (Almeida and Peres, 2007). All these impacts have resulted in extensive alterations of the region’s natural environments, in particular the fragmentation of habitats, which reduces gene flow among populations through processes such as siltation and the restriction of aquatic environments (Agostinho et al., 2005).
While arapaima stocks have been reduced by both fishing and habitat loss, few populations have been studied in the Araguaia-Tocantins basin, and there is a pressing need for the expansion of the database to provide a reliable assessment of the conservation status of these populations. The first study to use genetic markers in arapaima from the Araguaia-Tocantins basin was conducted by Vitorino et al. (2015), who found low levels of expected heterozygosity and a distinct structure among the four study populations. The present study provides further advances in the understanding of the genetic characteristics of this important species of Neotropical fish, through the investigation of wild arapaima populations in the Araguaia-Tocantins basin using microsatellite markers. The present study aimed to confirm the findings of Vitorino et al. (2015), and in particular, determine the existence of significant structuring in the arapaima populations of this basin. The prediction that significant population structure exists within the study area is based on the observation that, under natural conditions, arapaima typically form relatively stable family groups, which migrate laterally over short distances (Castello and Stewart, 2010; Castello et al., 2013). In this context, habitat structure is also expected to contribute to genetic diversity, given that marginal lakes increase in number and size moving downstream from the sampling point furthest upstream (point 1, Figure 1), with a larger number of environments been expected to support larger numbers of fish and family groups.
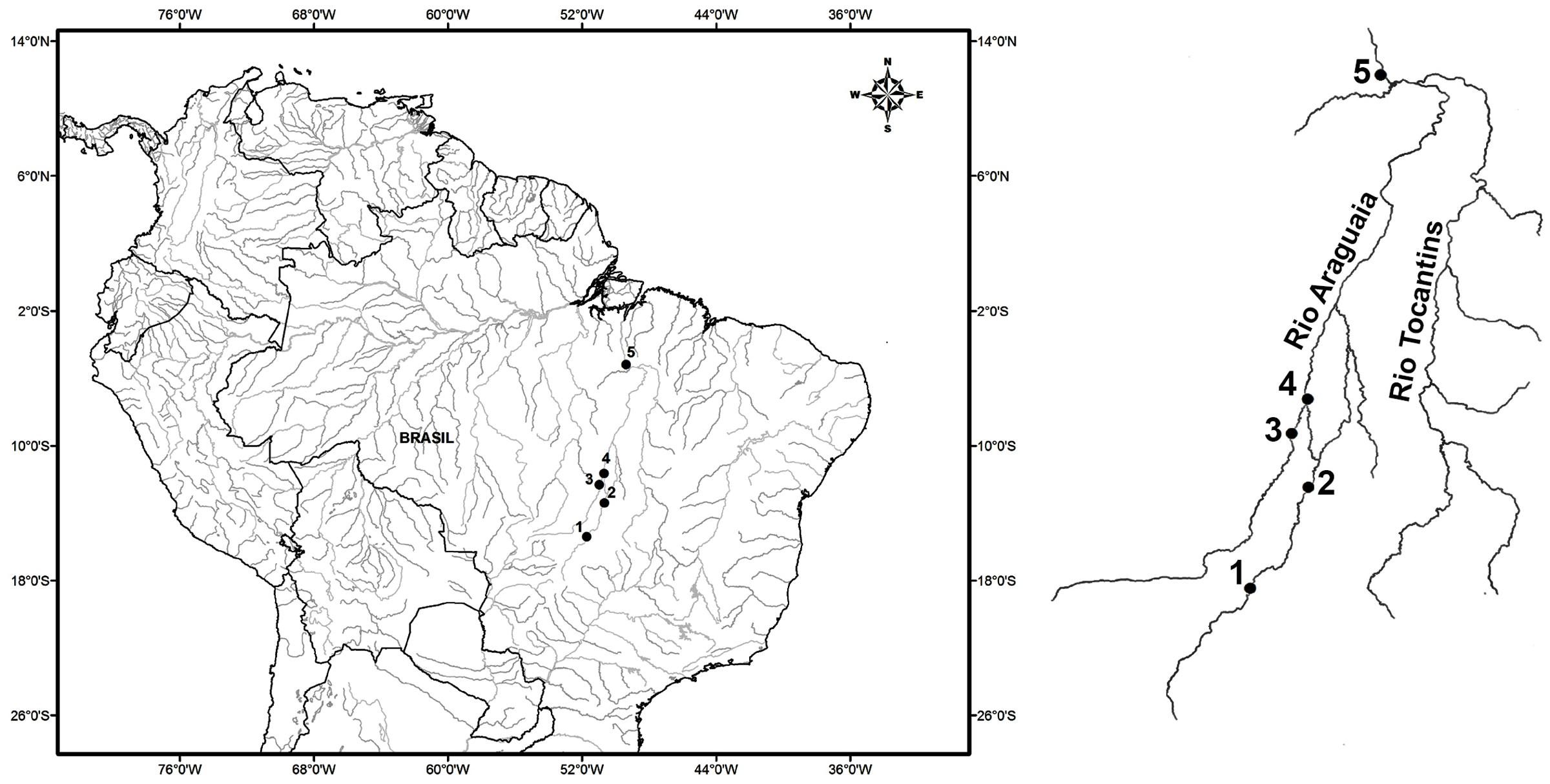
FIGURE 1. Map of Brazil, with emphasis on the Araguaia-Tocantins basin, indicating the collection sites of Arapaima gigas: (1) Araguaiana-MT, (2) Luís Alves, (3) Novo Santo Antônio-MT, (4) São Felix do Araguaia-MT and (5) Itupiranga-PA.
While the study of Vitorino et al. (2015) analyzed the genetic diversity of the arapaima populations of the Araguaia-Tocantins basin using ISSR markers, microsatellites are more appropriate for population analyses, given their co-dominant inheritance (vs. dominant alleles in the ISSR markers) and high degree of polymorphism. Microsatellite markers thus provide a much greater potential for statistical analyses, and were expected to provide a detailed database for the more reliable definition of the genetic diversity of populations. These analyses have important implications for the definition of priority areas for conservation and the implementation of management units for this species.
Materials and Methods
Two hundred and ninety-four samples of the muscle tissue and fins of Arapaima gigas were obtained from five different locations on the Araguaia and Tocantins rivers, three in the state of Mato Grosso (Araguaiana, Novo Santo Antônio, and São Félix do Araguaia), one in Goiás (Luiz Alves), and one in Pará, Itupiranga (Figure 1 and Table 1). The municipality of Araguaiana represents the region furthest upstream of the middle Araguaia River, where the river first forms marginal lakes along its course, appropriate for A. gigas. Araguaiana is also the sector with the smallest number of lakes (Alves and Carvalho, 2007). Luis Alves is just upstream from the bifurcation in the river that creates the Javaés River, and forms Bananal Island. It is important to note that this sector of the river is very popular with sport fisherman, due to the large number of lakes found in this area (Alves and Carvalho, 2007). Novo Santo Antônio, a municipality located between the Mortes and Araguaia rivers, is the region’s principal arapaima fishery center, and the lakes found along the course of the Mortes River provide the bulk of the catch landed on the Araguaia River (Kirsten et al., 2012). The municipality of São Félix do Araguaia is located at the confluence of the Mortes and Araguaia river, on the Bananal plain (de Melo et al., 2007). Most of the arapaima fishermen of Mato Grosso are resident in this municipality (Kirsten et al., 2012). The municipality of Itupiranga is located downstream of the confluence of Araguaia with Tocantins River, 170 km upstream from the Tucurui hydroelectric dam.
The samples collected in Araguaiana were obtained during a rescue operation, which translocated fish from seasonal lakes to a perennial body of water. Small pieces of fin were collected from the live fish during this operation. All the other tissue samples were obtained from specimens caught and marketed by local fishermen at each site, so no specimens were euthanized specifically for the purposes of the study. All the samples (fin and muscle) were preserved in 100% alcohol, and deposited at the Cytogenetics and Animal Genetics Laboratory, of the Federal University of Mato Grosso (LabGen/GEPEMA/UFMT) in Cuiabá, Brazil. When it was necessary to handle animals, all procedures adhered to the recommendations of the Guide for the Care and Use of Laboratory Animals. The collection and transportation of biological specimens by the authors of the present study is authorized by the Brazilian Institute for the Environment and Renewable Resources (IBAMA) through permanent license number 15226-1, issued by the Chico Mendes Institute for the conservation of Biodiversity (ICMBio).
The total DNA was extracted according to the saline extraction protocol of Aljanabi and Martinez (1997), with minor modifications. The amount and quality of the DNA obtained through this procedure were analyzed in a Biophotometer Plus (Eppendorf Hamburg, Hamburg, Germany), and the samples were diluted to a final concentration of 5 ng/μL.
Seven primer pairs (AgCTm4, AgCTm7, AgCAm2, AgCAm15, AgCAm16, AgCAm20, and AgCAm26) were selected based on previous studies and also because they are highly polymorphic for the Arapaima populations of the Amazon Basin (Farias et al., 2003, 2015; Araripe et al., 2013). These primers fluorescently labeled were used for amplification of the microsatellite regions by Polymerase Chain Reaction (PCR), following the conditions described by Farias et al. (2003). For genotyping, the PCR product was mixed with a standard molecular weight marker (MegaBACE ET-550R Size Standard), which was injected into a MegaBace 1000 automatic sequencer (Amersham Biosciences). The alleles were identified using Fragment Profiler 1.2 (Amersham Biosciences).
Once the database was assembled, the existence of possible genotyping errors, null alleles or scoring was verified in MicroChecker (Van Oosterhout et al., 2004). To test whether the populations were in Hardy-Weinberg equilibrium, Genepop (Raymond and Rousset, 1995) was used to estimate the intrapopulation fixation index or coefficient of inbreeding (FIS) for each locus, which was compared with the null hypothesis (FIS = 0). Genepop was also used to determine allele frequencies, and the presence of polymorphic loci, and exclusive and rare alleles. Allele richness was estimated in Fstat 2.9.3.2 (Goudet, 2002). Expected and observed heterozygosity were determined by Arlequin 3.5.1.2 (Excoffier and Lischer, 2010). The significance of the differences between expected and observed heterozygosity was evaluated using a one-way ANOVA, run in PAST 2.17c (Hammer et al., 2001).
Genetic differentiation among populations was evaluated using an Analysis of Molecular Variance (AMOVA), the molecular fixation index (FST), and the divergence parameter (RST), which were all determined by Arlequin 3.5.1.2 (Excoffier and Lischer, 2010). A Mantel test was used to verify possible correlation between genetic differentiation (FST, RST and FST/1-FST) and geographical distance, which was measured following the main channel of the river.
The probability of a given number of stocks was based on a Bayesian approach run in STRUCTURE 2.3.3 (Pritchard et al., 2000). The number of presumed populations (K) was set from 1 to 7. The analyses had a burn-in of 100,000 runs, and a Monte Carlo Markov Chain (MCMC) of 1,000,000, using a model without admixture and allele frequencies. The number of populations was defined by the delta k-value, obtained using the STRUCTURE HARVESTER program (Earl and VonHoldt, 2012).
The estimated effective size (Ne) of each population was derived from the theta values (θ) generated in MIGRATE-n 3.2.6 (Beerli and Felsenstein, 2001), using the formula Ne = θ/4μ, with a microsatellite mutation rate (μ) of 5.56 × 10-4 per locus per generation (Whittaker et al., 2003; Yue et al., 2007).
The MIGRATE-n program was also used to calculate migration rates between pairs of populations by the coalescence method. The number of migrants per generation (Nm) was obtained by multiplying the migration rates obtained by the program (M = m/μ, where m is the fraction of new immigrants from the population per generation) by the θ values of the receptor population in each pairwise comparison. This analysis was performed using the Brownian model, with a uniform distribution and a constant mutation rate between the loci. The standard search strategy values of the MIGRATE program were used, except for the use of 20 short chains and 5 long chains.
The BOTTLENECK program, version 1.2.02 (Cornuet and Luikart, 1996) was used to verify the existence of recent demographic events, such as population bottlenecks. The Wilcoxon test was run using the Two-Phased Mutation (T.P.M) model, established with 30% for the Infinite Allele Mutation (IAM) model and 70% for the Stepwise Mutation Model (SMM).
Results
Genetic Diversity
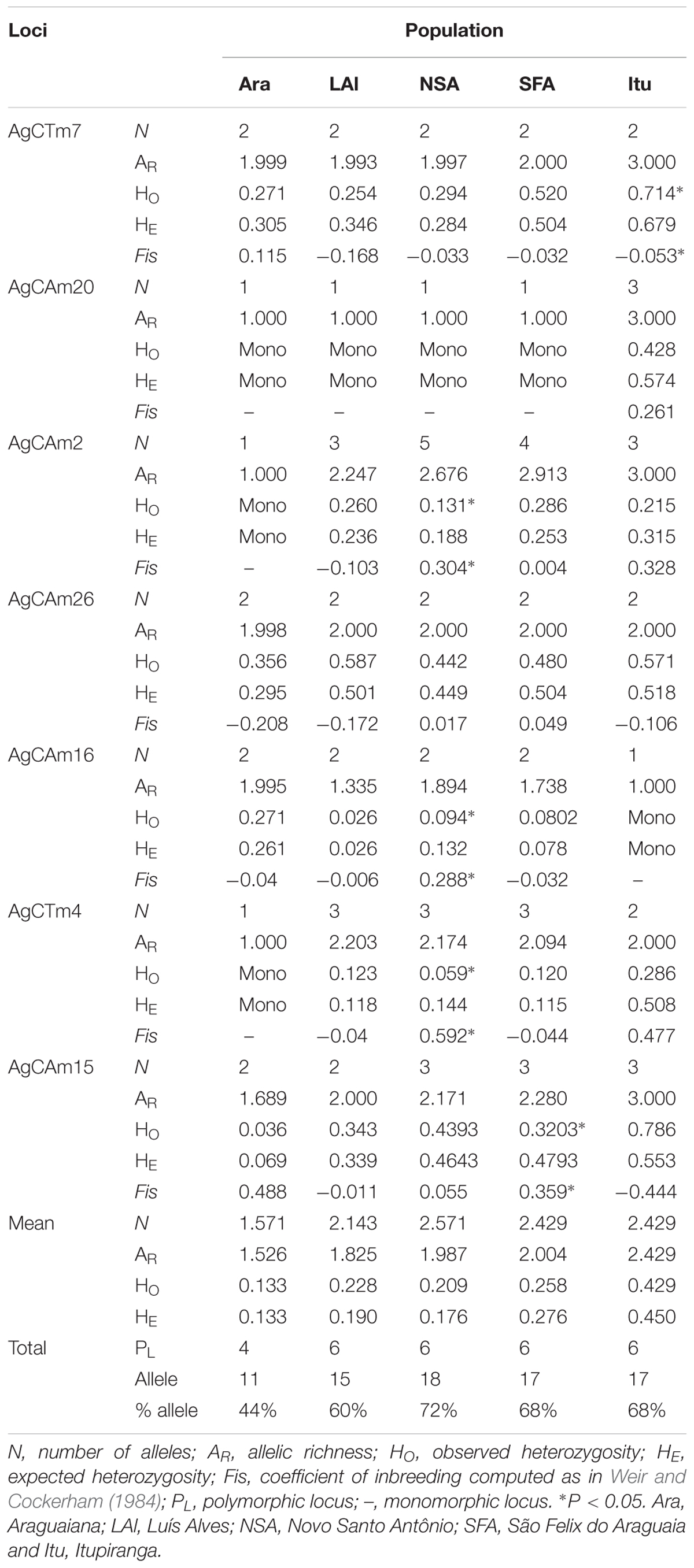
A total of 25 alleles were identified, with an average of 3.57 alleles per locus. While all the loci were polymorphic, four were monomorphic in at least one of the arapaima populations analyzed from the Araguaia-Tocantins basin. The number of alleles, observed and expected heterozygosity, allelic richness, and the fixation index (FIS) were calculated for all seven microsatellite loci analyzed, together with the mean parameters for all loci (Table 2).
The population from Novo Santo Antônio had the highest genetic diversity, with 72% of the identified alleles and an allelic richness of 2.429. At the opposite extreme, the arapaima from Araguaiana were the least diverse (44.0%) with an allelic richness of only 1.521. The specimens from Araguaiana also returned the lowest heterozygosity (Ho = 0.133, He = 0.133), while those from Itupiranga had the highest values, with observed heterozygosity of 0.428 and an expected heterozygosity of 0.449 (Table 2).
Five loci deviated from Hardy-Weinberg equilibrium, with heterozygote frequencies being either lower or higher than expected (Table 2). Only four of the 28 values estimated for the inbreeding coefficient (FIS) were significantly positive, which indicates a deficit of heterozygotes. Three of these loci were recorded in the population from Novo Santo Antônio, while the other was from São Felix do Araguaia. A single locus from Itupiranga was significantly negative, indicating an excess of heterozygotes. When the whole set of loci is considered, a significant FIS value (p < 0.001) was only for the Novo Santo Antônio presented, indicating the occurrence of inbreeding in this population.
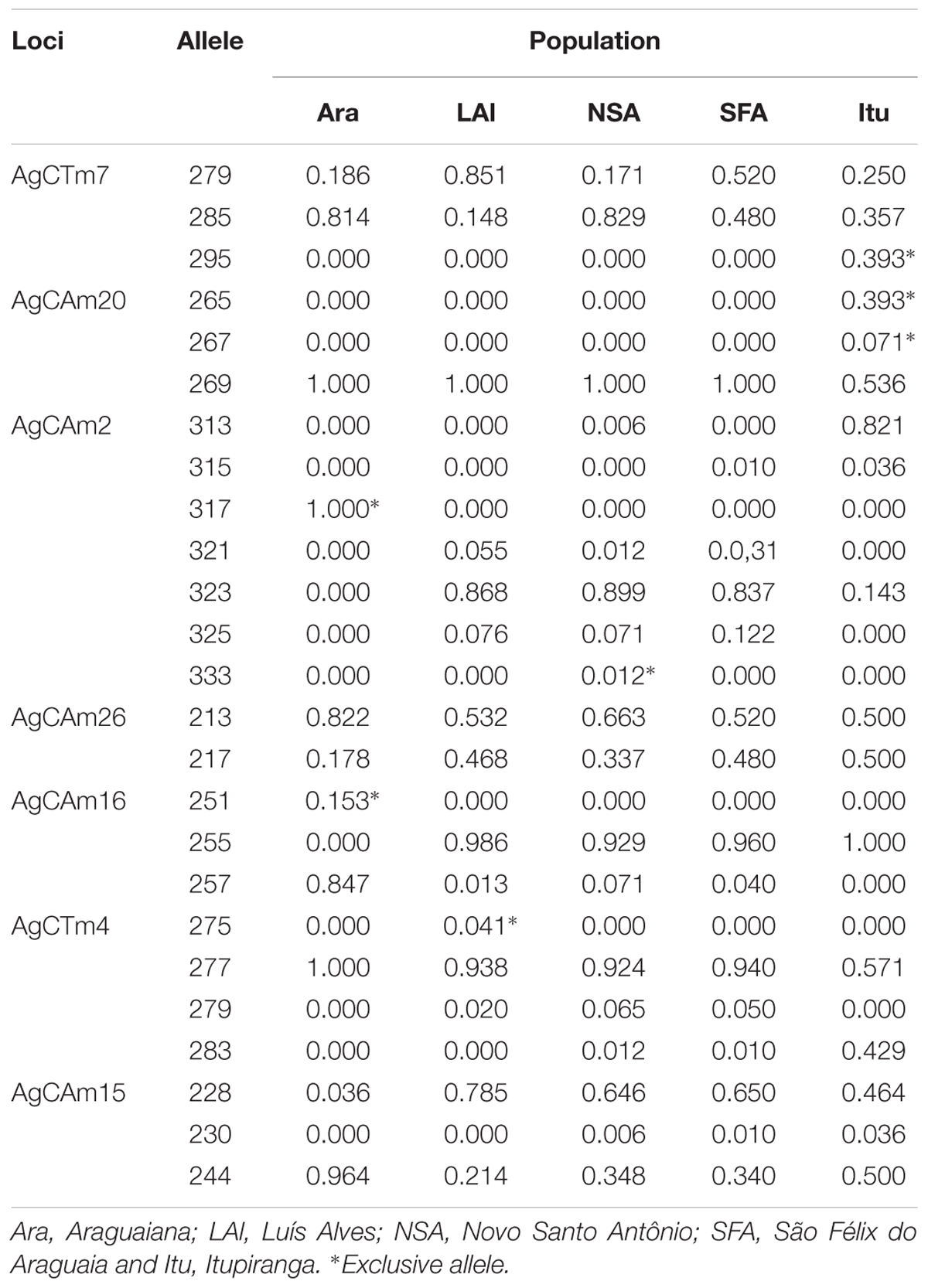
One to three alleles, with high frequencies, were recorded for most loci. All loci except AgCAm15 presented exclusive alleles for the different study populations, at frequencies ranging from 0.012 to 1.00. No exclusive alleles was detected in the population from São Felix do Araguaia (Table 3). No new alleles were detected in this study, given that all the alleles recorded here had been reported previously by Farias et al. (2003) and Araripe (2008).
The MicroChecker analysis (Table 3) indicated the presence of null alleles in the populations from Luís Alves (for loci AgCAm20 and AgCTm4), Novo Santo Antônio (loci AgCAm2, AgCTm4, AgCAm16 and AgCAm20), São Félix do Araguaia (AgCAm15), and Itupiranga (AgCTm4). No systematic pattern was observed in the occurrence of null alleles in the different study populations, however. No evidence was found of either the misidentification of stutters as alleles or dropouts (dominance of small alleles).
Population Structure
The values recorded for Wright index (FST) ranged from 0.061 to 0.669, while those for the Slatkin divergence parameter (RST), varied from 0.025 to 0.688. Both indices were significant for all pairs of locations (Table 4). The results of the Mantel test rejected the hypothesis that genetic differentiation was related to geographical distance, considering the values of either FST (r2 = 0.466115, p = 0.236), RST (r2 = 0.452354, p = 0.240) or FST/1-FST (r2 = 0.219112, p = 0.221000). The results of the Bayesian inference based on the mean likelihood [Ln (K)] of the Δk (Evanno et al., 2005) indicate the presence of two clusters within the Araguaia-Tocantins basin, one formed by the populations of Luís Alves, Novo Santo Antônio, São Félix do Araguaia, and Itupiranga, and the other by that of Araguaiana, which forms a distinct group (Figure 2). These findings were used to establish the clusters for the analysis of molecular variance (AMOVA).
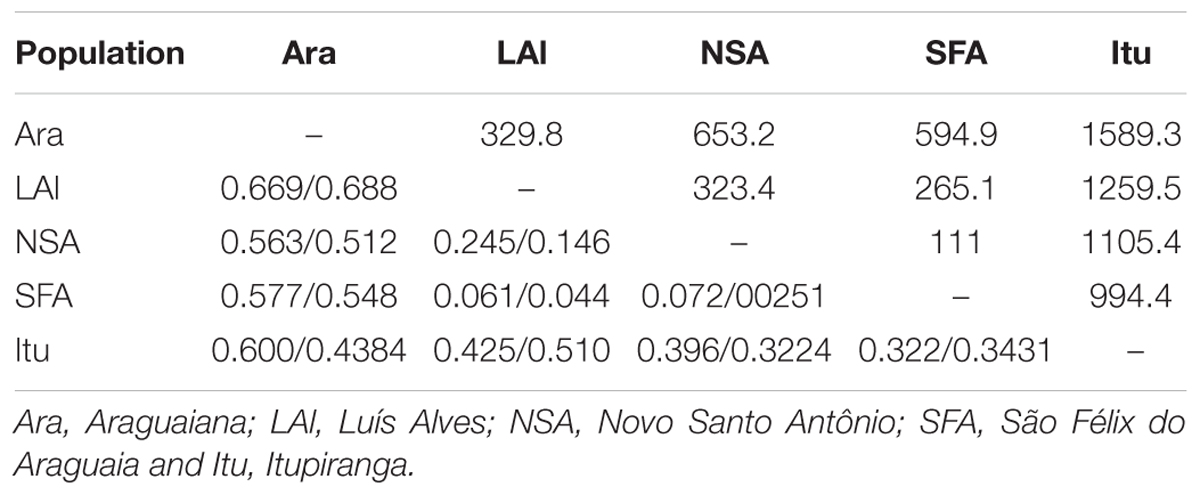
TABLE 4. FST/RST values and geographic distance in kilometers (above the diagonal) between A. gigas populations from Araguaia-Tocantins basin.
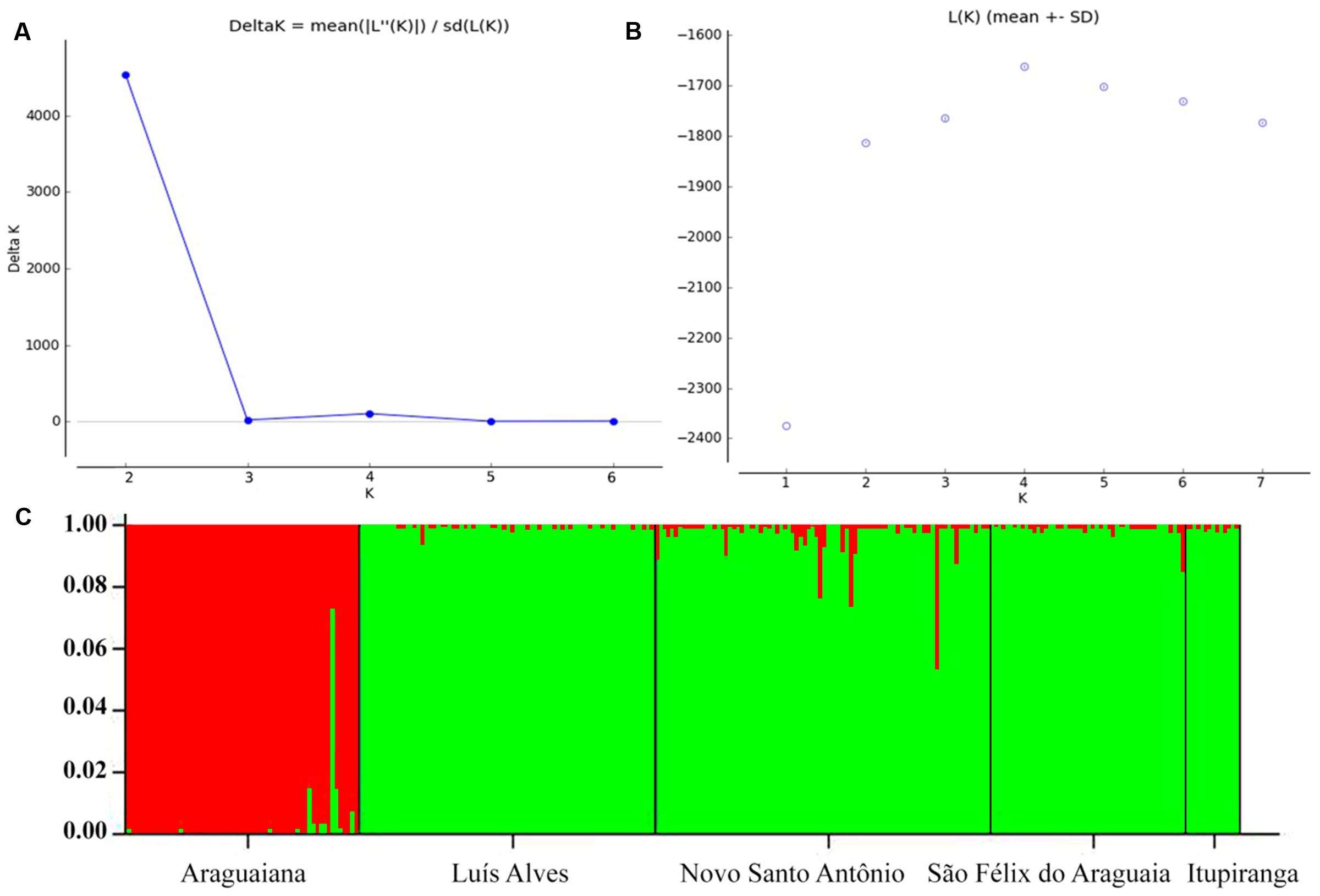
FIGURE 2. Genetic structuring of Arapaima gigas from the Araguaia-Tocantins basin, according to Bayesian analysis. Estimating the number of K groups based on (A) ΔK and (B) the mean maximum likelihood. (C) Each column represents a different individual and the colors represent genetic stocks. The collection sites are separated by black lines.
The results of the AMOVA indicated a lack of significant variation between the two groups (ΦST = 44.15%, RST = 40.07%). The variation among populations within each group contributed only 12.35% (ΦST) and 11.17% (RST) of the variance, while the variance within populations is 43.51% for ΦST and 48.76 for RST. The ΦST (ΦST = 0.564; p = 0.0000) and (RST = 0.512; p = 0.000) were both relatively high among the study populations.
Effective population size ranged from 128 to 351 individuals, with the lowest value being recorded at Araguaiana and the highest at São Félix do Araguaia (Table 5). The MIGRATE-n coalescence analysis (Beerli and Felsenstein, 2001) indicated low levels of gene flow between populations, and only six of the 20 estimates of migrant numbers were higher than one migrant per generation.
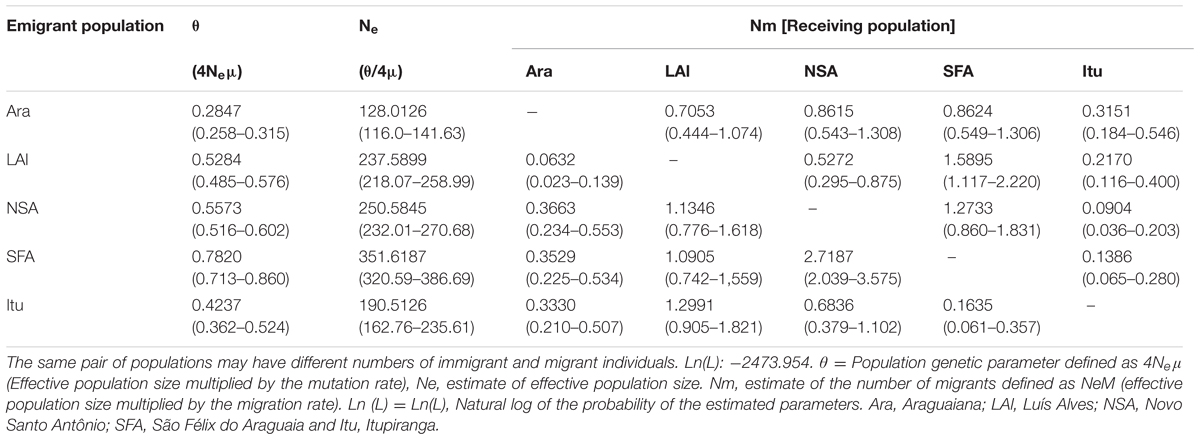
TABLE 5. Analysis of the MIGRATE program showing the estimates of gene flow peer-to-peer among the Arapaima populations of the Araguaia-Tocantins region.
The Bottleneck program did not detect any significant deficit of heterozygotes. However, a significant (p < 0.05) excess of heterozygotes was found in the Itupiranga population using the TPM (Wilcoxon test) model.
Discussion
This study is the first to use microsatellite markers to examine the genetic diversity of the arapaima populations of the Araguaia-Tocantins basin. Overall, the results indicated significantly lower levels of genetic diversity and heterozygosity than those found in previous studies of the same genetic markers in populations from other areas (Farias et al., 2003, 2015; Hamoy et al., 2008; de Alencar Leão, 2009; Araripe et al., 2013).
The arapaima populations that inhabit the study region are affected by natural processes, in particular the hydrological regime, that are quite distinct in comparison with the populations found in the Amazon basin, the region in which the species has been investigated in most detail (Vitorino et al., 2015). The unique features of the Araguaia-Tocantins basin may influence the reduced genetic diversity (number of alleles, heterozygosity, and allelic richness) of its population in comparison with those from the Amazonian basin. This is also true of the allelic richness of the arapaima populations of Tucuruí (Araripe, 2008) and Bananal Island (de Alencar Leão, 2009), two other areas located within the Araguaia-Tocantins basin, which present low values in comparison with population from the Amazon basin. The levels of genetic diversity recorded in the Arapaima populations of the Araguaia-Tocantins basin are lower than expected for the species, but are consistent with the findings of Vitorino et al. (2015), based on ISSR markers. Using mitochondrial markers, da Penha (2014) also found relatively low genetic variability in arapaima populations of the Araguaia-Tocantins basin in comparison with those of the Amazon basin.
The points sampled in the present study are exploited intensively by local fisheries. In fact, most of the arapaima marketed locally are harvested from natural populations (Çiftci and Okumus, 2002), including not only adults, but also the capture of fry as stock for rearing on fish farms. This type of overexploitation often results in a decline in effective population size (Ne), provoking the loss of genetic diversity, which leads to a reduced resilience of populations to environmental stresses and climate change, and a loss of resistance to pathogens (Allendorf and Luikart, 2007; Hughes et al., 2015).
The variation in the Ne values estimated for the different populations analyzed in the present study indicates potential differences in their population dynamics, with distinct patterns of fluctuation over time in the number of individuals reproducing, a process that will have knock-on effects for the sustainability of the population (Carson et al., 2011). In other words, each arapaima population may have a distinct demographic history, based on the differences in variables such as the number of mature individuals, the sex ratio, offspring survival rates, and the availability and quality of habitats. However, the results of the Bottleneck analysis indicate a significant impact on genetic structure only in the case of the population from Itupiranga. This may be related to the construction of the hydroelectric dam at Tucuruí, and the creation of one of the world’s largest reservoirs, which caused considerable impacts on the local fish fauna of the Tocantins River.
The reduced genetic variability found in the arapaima populations of the Araguaia-Tocantins basin may also be related to the anthropogenic impacts that have altered the natural features of the basin extensively throughout most of its length. The hydrological cycle of this basin is more intense than that of the Amazon, and this cycle has been modified by local farming and ranching activities, and the establishment of the Araguaia-Tocantins waterway (Latrubesse and Stevaux, 2006). These modifications have had a major impact, principally on the region’s lacustrine environments, causing a reduction in the size of its lakes, and altering their flood cycle, which affects the lateral migrations typical of this fish (Castello and Stewart, 2010; Castello et al., 2013). These changes may force the arapaima to remain in the same lakes, unable to migrate in search of reproductive partners, eventually creating small, isolated populations susceptible to inbreeding. Genetic drift tends to have a greater impact in smaller populations, which also favor inbreeding, exacerbating the loss of genetic diversity. It seems likely that the combined effects of these processes have reduced the reproductive potential of the local arapaima populations, impacting their effective size. The geomorphology and flood cycle of the Araguaia-Tocantins and Amazon basins are quite distinct, and this appears to be the principal factor determining the variation in the genetic diversity of the stocks analyzed from the Amazon (Farias et al., 2003, 2015; Hamoy et al., 2008; de Alencar Leão, 2009; Araripe et al., 2013) and the Tocantins-Araguaia basin (Vitorino et al., 2015).
However, the possibility that low genetic variability is a natural characteristic of the arapaima populations of the Araguaia-Tocantins basin cannot be ruled out altogether. But whatever the determining factors, this genetic fragility is a cause for concern, given that future environmental impacts (natural or otherwise) may further reduce the diversity of these populations, and threaten their long-term viability.
This fragility is further reinforced by the differences in the genetic variability of each population in the Araguaia-Tocantins basin. Specimens collected at Araguaiana, for example, had the lowest genetic diversity of any population, indicating the smallest effective size of any population, which implies that this population is the most vulnerable to anthropogenic interference from the expansion of the agricultural frontier occurring within the basin (Latrubesse and Stevaux, 2006), as well as being the sector of the basin that has the smallest number of lakes, the preferred habitat of the arapaima (Alves and Carvalho, 2007).
The most likely explanation for the deviations from Hardy-Weinberg equilibrium detected in the arapaima populations of the Araguaia-Tocantins basin is inbreeding, given that significant FIS values (indicating a deficiency of heterozygotes) were found in six of the eight deviations recorded. A number of studies have recorded a deficiency of heterozygotes in fish populations (Castric et al., 2002; Langen et al., 2011; O’Leary et al., 2013; Ferreira et al., 2015), indicating that inbreeding may be relatively common in these vertebrates. In the arapaima this may be related to the relatively sedentary behavior of the species and its parental care, as observed in a number of other species of Neotropical fish (Sofia et al., 2006, 2008; Ferreira et al., 2015).
In addition to these behavioral traits, arapaima is intolerant of lotic environments, so areas of strong rapids represent effective barriers to the dispersal of this species (Castello and Stewart, 2010; Castello et al., 2013). This may lead to the formation of family groups in the different micro-regions of the Araguaia-Tocantins basin. Genetic differences between subpopulations related to geographic distance or the presence of physical barriers, such as waterfalls, have been identified in Amazonian arapaima (Araripe et al., 2013) and in other fish species (Hughes et al., 2015).
The pairwise FST values indicate moderate (0.05–0.15) to extreme (>0.25) genetic differentiation between populations. However, the Bayesian analysis points to the presence of only two genetic stocks in the Araguaia-Tocantins basin, one at Araguaiana, and the other formed by the remaining populations, at Luís Alves, Novo Santo Antônio, Sao Félix do Araguaia and Itupiranga (Figure 2). This arrangement contrasts with that recorded by Vitorino et al. (2015), based on ISSR markers, which indicated that the Araguaiana and Novo Santo Antônio populations shared the same genetic stock, while São Félix do Araguaia and Itupiranga form separate populations (Luís Alves was not included in this analysis).
The structure proposed by the Bayesian analysis is also supported by the estimates of gene flow and the number of migrants between populations, given that the populations that share the same genetic stock had the highest migration rates. It is important to note that the gene flow detected here may reflect ancestral processes, rather than the recent exchange of individuals, because the number of migrants is a somewhat abstract quantity, which cannot be distinguished from the effective population size (Balloux and Lugon-Moulin, 2002).
While the genetic variation found in the present study is not related systematically to geographic distance, the geographically closer populations (Luís Alves, Novo Santo Antônio and São Félix do Araguaia) returned the lowest FST and RST values. Reduced genetic differentiation was expected between the samples from Novo Santo Antônio and São Félix do Araguaia, which are the geographically closest sites (separated by a distance of only 111 km), although the FST values indicate that the populations of São Félix do Araguaia and Luís Alves (265.1 km apart) are the most similar. In addition to the basic differences in comparison with the Amazonian arapaima populations, then, the relationship between population structure and geographic distance is also distinct from that recorded by Araripe et al. (2013).
The present study confirmed the low genetic diversity of the Arapaima gigas populations of the Araguaia-Tocantins basin, which can be linked directly to the environmental fragility of this river system, which reinforces the need for a better understanding of the processes that may further reduce the viability of these populations. The findings of this study also indicated that the genetic diversity of the populations is distributed heterogeneously within the study area, and that the establishment of a single protected area would be insufficient for the preservation of the genetic diversity of the arapaima populations of the Araguaia-Tocantins basin as a whole. In particular, future management measures should consider the population from Araguaiana as an independent unit, distinct from the other Araguaia-Tocantins populations. The Itupiranga region should also be defined as a priority area for conservation, given the high allelic richness found in this population.
Ethics Statement
This study was carried out in strict accordance with the recommendations provided in the Guide for the Care and Use of laboratory Animals. During the development of this work, no animals were sacrificed. Collection was authorized by SEMA (license number 104358/2011/SEMA-MT), Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis - IBAMA and Instituto Chico Mendes de Conservação da Biodiversidade – ICMBio (License number 15226-1).
Author Contributions
CV performed the molecular genetic studies, performed the statistical analyzes and drafted the manuscript. FN performed some molecular genetic studies and contributed to the correction of the text. JA and PV conceived and coordinated the study, participated in its elaboration and helped to draft the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to FAPEMAT (Fundação de Amparo à Pesquisa do Estado de Mato Grosso – Process: 841147/2009/FAPEMAT/PRONEX), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico/INAU), and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) for financial support; Drs. Fabio Porto Foresti and Claudio Oliveira for providing biological samples. We are also very grateful to Mr. Irineu Pirani and Wagner Alves de Santana, for permitting the collection of arapaima tissue samples during rescue operations.
References
Abell, R., Thieme, M. L., Revenga, C., Bryer, M., Kottelat, M., Bogutskaya, N., et al. (2008). Freshwater ecoregions of the world: a new map of biogeographic units for freshwater biodiversity conservation. Bioscience 58, 403–414. doi: 10.1641/B580507
Agostinho, A. A., Thomaz, S. M., and Gomes, L. C. (2005). Conservação da biodiversidade em águas continentais do Brasil. Megadiversidade 1, 70–78.
Aljanabi, S. M., and Martinez, I. (1997). Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25, 4692–4693. doi: 10.1093/nar/25.22.4692
Allan, J. D., Abell, R., Hogan, Z., Revenga, C., Taylor, B. W., Welcomme, R. L., et al. (2005). Overfishing of inland waters. Bioscience 55, 1041–1051. doi: 10.1641/0006-3568(2005)055[1041:OOIW]2.0.CO;2
Allendorf, F. W., and Luikart, G. H. (2007). Conservation an the Genetics of Populations. Oxford: Blackwell Publishing Ltd.
Almeida, A., and Peres, F. C. (2007). Hidrovia Tocantins - Araguaia: importância e impactos econômicos, sociais e ambientais, segundo a percepção dos agentes econômicos locais. Rev. Bras. Recur. Hídricos 12, 169–177. doi: 10.21168/rbrh.v12n2.p169-177
Alves, T. M., and Carvalho, T. M. (2007). Técnicas de sensoriamento remoto para classificação e quantificação do sistema lacustre do Rio Araguaia entre Barra do Garças e foz do Rio Cristalino. Rev. Geogr. Acad. 1, 79–94.
Aquino, S., Latrubesse, E., and Bayer, M. (2009). Assessment of wash load transport in the Araguaia River (Aruanã gauge station), central Brazil. Lat. Am. J. Sedimentol. Basin Anal. 16, 119–128.
Aquino, S., Latrubesse, E. M., and de Souza Filho, E. E. (2008). Relações entre o regime hidrológico e os ecossistemas aquáticos da planície aluvial do Rio Araguaia. Acta Sci. Biol. Sci. 30, 361–369. doi: 10.4025/actascibiolsci.v30i4.5866
Aquino, S., Stevaux, J. C., and Latrubesse, E. M. (2005). Regime hidrológico e aspectos do comportamento morfohidráulico do Rio Araguaia. Rev. Bras. Geomorfol. 6, 29–41. doi: 10.20502/rbg.v6i2.49
Arantes, C. C., Castello, L., Stewart, D. J., Cetra, M., and Queiroz, H. L. (2010). Population density, growth and reproduction of arapaima in an Amazonian river-floodplain. Ecol. Freshw. Fish 19, 455–465. doi: 10.1111/j.1600-0633.2010.00431.x
Araripe, J. (2008). Genética de Populações de Pirarucus (Arapaima gigas) da Reserva de Mamirauá e Considerações sobre a Estrutura Genética para a Espécie. Belém: Universidade Federal do Pará.
Araripe, J., Rêgo, P. S., Queiroz, H., Sampaio, I., and Schneider, H. (2013). Dispersal capacity and genetic structure of Arapaima gigas on different geographic scales using microsatellite markers. PLOS ONE 8:e54470. doi: 10.1371/journal.pone.0054470
Balloux, F., and Lugon-Moulin, N. (2002). The estimation of population differentiation with microsatellite markers. Mol. Ecol. 11, 155–165. doi: 10.1046/j.0962-1083.2001.01436.x
Beerli, P., and Felsenstein, J. (2001). Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl. Acad. Sci. U.S.A. 98, 4563–4568. doi: 10.1073/pnas.081068098
Carson, E. W., Saillant, E., Renshaw, M. A., Cummings, N. J., and Gold, J. R. (2011). Population structure, long-term connectivity, and effective size of mutton snapper (Lutjanus analis) in the Caribbean Sea and Florida keys. Fish. Bull. 109, 416–428.
Castello, L. (2008a). Lateral migration of Arapaima gigas in floodplains of the Amazon. Ecol. Freshw. Fish 17, 38–46. doi: 10.1111/j.1600-0633.2007.00255.x
Castello, L. (2008b). Nesting habitat of Arapaima gigas (Schinz) in Amazonian floodplains. J. Fish Biol. 72, 1520–1528. doi: 10.1111/j.1095-8649.2007.01778.x
Castello, L., and Stewart, D. J. (2010). Assessing CITES non-detriment findings procedures for Arapaima in Brazil. J. Appl. Ichthyol. 26, 49–56. doi: 10.1111/j.1439-0426.2009.01355.x
Castello, L., Stewart, D. J., and Arantes, C. C. (2013). “‘O quê sabemos e precisamos fazer a respeito da conservação do pirarucu (Arapaima spp.) na Amazônia’,” in Biologia, Conservação e Manejo Participativo de Pirarucu na Pan-Amazônia, ed. E. Amaral (Tefé: Instituto de Desenvolvimento Sustentável Mamirauá), 17–31.
Castric, V., Bernatchez, L., Belkhir, K., and Bonhomme, F. (2002). Heterozygote deficiencies in small lacustrine populations of brook Charr Salvelinus fontinalis Mitchill (Pisces, Salmonidae): a test of alternative hypotheses. Heredity 89, 27–35. doi: 10.1038/sj.hdy.6800089
Çiftci, Y., and Okumus, I. (2002). Fish population genetics and applications of molecular markers to fisheries and aquaculture: I- basic principles of fish population genetics. Turk. J. Fish. Aquat. Sci. 2, 145–155.
Cornuet, J. M., and Luikart, G. (1996). Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144, 2001–2014.
da Penha, F. N. (2014). Distintas Linhagens do Gigante Arapaima (Osteoglossiformes: Osteoglossidae) Revelam Diferenças Entre Populações das Bacias Amazônica e Araguaia-Tocantins. Master’s thesis, Universidade Federal do Pará, Pará.
de Alencar Leão, A. S. (2009). Análise da Variabilidade Genética das Populações de Pirarucu (Arapaima gigas, Schinz 1822) dos Principais Tributários do Rio Amazonas Através do uso de Marcadores Microssatélites. Master’s thesis, Universidade Federal do Pará, Pará.
de Melo, T. L., Tejerina-Garro, F. L., and Melo, C. E. (2007). Diversidade biológica da comunidade de peixes no baixo Rio das Mortes, Mato Grosso, Brasil. Rev. Bras. Zool. 24, 657–665. doi: 10.1590/S0101-81752007000300017
Earl, D. A., and VonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Farias, I. P., Hrbek, T., Brinkmann, H., Sampaio, I., and Meyer, A. (2003). Characterization and isolation of DNA microsatellite primers for Arapaima gigas, an economically important but severely over-exploited fish species of the Amazon basin. Mol. Ecol. Resour. 1, 128–130. doi: 10.1046/j.1471-8286
Farias, I. P., Leão, A., Almeida, Y. S., Verba, J. T., Crossa, M. M., Honczaryk, A., et al. (2015). Evidence of polygamy in the socially monogamous Amazonian fish Arapaima gigas (Schinz, 1822) (Osteoglossiformes, Arapaimidae). Neotrop. Ichthyol. 13, 195–204. doi: 10.1590/1982-0224-20140010
Fernandes, M. N., da Cruz, A. L., da Costa, O. T. F., and Perry, S. F. (2012). Morphometric partitioning of the respiratory surface area and diffusion capacity of the gills and swim bladder in juvenile Amazonian air-breathing fish. Arapaima gigas. Micron 43, 961–970. doi: 10.1016/j.micron.2012.03.018
Ferreira, D. G., Galindo, B. A., Frantine-Silva, W., Almeida, F. S., and Sofia, S. H. (2015). Genetic structure of a Neotropical sedentary fish revealed by AFLP, microsatellite and mtDNA markers: a case study. Conserv. Genet. 16, 151–166. doi: 10.1007/s10592-014-0648-2
Gomes, L. D. C. (2007). Physiological responses of pirarucu (Arapaima gigas) to acute handling stress. Acta Amazon. 37, 629–634. doi: 10.1590/S0044-59672007000400019
Goudet, G. J. (2002). FSTAT, A Program for Windows to Estimate and Test Gene Diversities and Fixation Indices (Version 2.9.3.2). Lausanne: Institute of Ecology.
Hammer,Ø., Harper, D. A. T., and Ryan, P. D. (2001). Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 9–18. doi: 10.1016/j.bcp.2008.05.025
Hamoy, I. G., Santos, E. J. M., and Santos, S. E. B. (2008). Rapid and inexpensive analysis of genetic variability in Arapaima gigas by PCR multiplex panel of eight microsatellites. Genet. Mol. Res. 7, 29–32. doi: 10.4238/vol7-1gmr394
Hrbek, T., Crossa, M., and Farias, I. P. (2007). Conservation strategies for Arapaima gigas (Schinz, 1822) and the Amazonian várzea ecosystem. Braz. J. Biol. 67, 909–917. doi: 10.1590/S1519-69842007000500015
Hrbek, T., and Farias, I. P. (2008). The complete mitochondrial genome of the pirarucu. Genet. Mol. Biol. 31, 293–302. doi: 10.1590/S1415-47572008000200024
Hrbek, T., Farias, I. P., Crossa, M., Sampaio, I., Porto, J. I. R., and Meyer, A. (2005). Population genetic analysis of Arapaima gigas, one of the largest freshwater fishes of the Amazon basin: implications for its conservation. Anim. Conserv. 8, 297–308. doi: 10.1017/S1367943005002210
Hughes, J. M., Schmidt, D. J., Huey, J. A., Real, K. M., Espinoza, T., Mcdougall, A., et al. (2015). Extremely low microsatellite diversity but distinct population structure in a long-lived threatened species, the Australian lungfish Neoceratodus forsteri (Dipnoi). PLOS ONE 10:e0121858. doi: 10.1371/journal.pone.0121858
Kirsten, I. F., Puerta, L. R., de Mateus, L. A. F., Catella, A. C., and Lima, I. S. (2012). A pesca do pirarucu (Arapaima sp.) na bacia do Rio Araguaia em Mato Grosso - Brasil. Bol. Inst. Pesca 38, 131–144.
Langen, K., Schwarzer, J., Kullmann, H., Bakker, T. C. M., and Thünken, T. (2011). Microsatellite support for active inbreeding in a cichlid fish. PLOS ONE 6:e24689. doi: 10.1371/journal.pone.0024689
Latrubesse, E. M., Amsler, M. L., de Morais, R. P., and Aquino, S. (2009). The geomorphologic response of a large pristine alluvial river to tremendous deforestation in the South American tropics: the case of the Araguaia River. Geomorphology 113, 239–252. doi: 10.1016/j.geomorph.2009.03.014
Latrubesse, E. M., and Stevaux, J. C. (2002). Geomorphology and environmental aspects of the Araguaia fluvial basin, Brazil. Z. Geomorphol. 129, 109–127.
Latrubesse, E. M., and Stevaux, J. C. (2006). Características físico-bióticas e problemas ambientais associados à planície aluvial do Rio Araguaia. Brasil Central. Rev. Geociências 10, 67–75.
Marques, D. K., Venere, P. C., and Galetti, P. M. Jr. (2006). Chromosomal characterization of the bonytongue Arapaima gigas (Osteoglossiformes: Arapaimidae). Neotrop. Ichthyol. 4, 215–218. doi: 10.1590/S1679-62252006000200007
O’Leary, S. J., Hice, L. A., Feldheim, K. A., Frisk, M. G., McElroy, A. E., Fast, M. D., et al. (2013). Severe inbreeding and small effective number of breeders in a formerly abundant marine fish. PLOS ONE 8:e66126. doi: 10.1371/journal.pone.0066126
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Raymond, M., and Rousset, F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249. doi: 10.1093/oxfordjournals.jhered.a111573
Sofia, S. H., Galindo, B. A., Paula, F. M., Sodré, L. M. K., and Martinez, C. B. R. (2008). Genetic diversity of Hypostomus ancistroides (Teleostei, Loricariidae) from an urban stream. Genet. Mol. Biol. 1, 317–323. doi: 10.1111/jfb.12675
Sofia, S. H., Silva, C. R. M., Galindo, B. A., Almeida, F. S., Sodré, L. M. K., and Martinez, C. B. R. (2006). Population genetic structure of Astyanax scabripinnis (Teleostei, Characidae) from an Urban Stream. Hydrobiologia 553, 245–254. doi: 10.1007/s10750-005-1106-4
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Resour. 4, 535–538. doi: 10.1111/j.1471-8286.2004.00684.x
Vitorino, C. A., Oliveira, R. C. C., Margarido, V. P., and Venere, P. C. (2015). Genetic diversity of Arapaima gigas (Schinz, 1822) (Osteoglossiformes: Arapaimidae) in the Araguaia-Tocantins basin estimated by ISSR marker. Neotrop. Ichthyol. 13, 557–568. doi: 10.1590/1982-0224-20150037
Weir, B. S., and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x
Whittaker, J. C., Harbord, R. M., Boxall, N., Mackay, I., Dawson, G., and Sibly, R. M. (2003). Likelihood-based estimation of microsatellite mutation rates. Genetics 164, 781–787.
World Conservation Monitoring Centre (2014). The IUCN Red List of Threatened Species. Version: 2014.3. Available at: www.iucnredlist.org [accessed May 4, 2015].
Keywords: genetic structure, microsatellite, conservation, habitat fragmentation, genetics population
Citation: Vitorino CA, Nogueira F, Souza IL, Araripe J and Venere PC (2017) Low Genetic Diversity and Structuring of the Arapaima (Osteoglossiformes, Arapaimidae) Population of the Araguaia-Tocantins Basin. Front. Genet. 8:159. doi: 10.3389/fgene.2017.00159
Received: 22 May 2017; Accepted: 10 October 2017;
Published: 24 October 2017.
Edited by:
Rodrigo A. Torres, Universidade Federal de Pernambuco, BrazilReviewed by:
Łukasz Kajtoch, Institute of Systematics and Evolution of Animals (PAN), PolandDaniel Cardoso Carvalho, Pontifícia Universidade Católica de Minas Gerais, Brazil
Copyright © 2017 Vitorino, Nogueira, Souza, Araripe and Venere. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paulo C. Venere, pvenere@uol.com.br
 Carla A. Vitorino
Carla A. Vitorino Fabrícia Nogueira2
Fabrícia Nogueira2 Issakar L. Souza
Issakar L. Souza Juliana Araripe
Juliana Araripe Paulo C. Venere
Paulo C. Venere