- 1College of Animal Science and Technology, Northwest A&F University, Yangling, China
- 2Shaanxi Provincial Engineering and Technology Research Center of Cashmere Goats, Yulin University, Yulin, China
- 3Life Science Research Center, Yulin University, Yulin, China
A previous whole-genome association analysis identified lysine demethylase 6A (KDM6A), which encodes a type of histone demethylase, as a candidate gene associated to goat fecundity. KDM6A gene knockout mouse disrupts gametophyte development, suggesting that it has a critical role in reproduction. In this study, goat KDM6A mRNA expression profiles were determined, insertion/deletion (indel) variants in the gene identified, indel variants effect on KDM6A gene expression assessed, and their association with first-born litter size analyzed in 2326 healthy female Shaanbei white cashmere goats. KDM6A mRNA was expressed in all tissues tested (heart, liver, spleen, lung, kidney, muscle, brain, skin and testis); the expression levels in testes at different developmental stages [1-week-old (wk), 2, 3 wk, 1-month-old (mo), 1.5 and 2 mo] indicated a potential association with the mitosis-to-meiosis transition, implying that KDM6A may have an essential role in goat fertility. Meanwhile, two novel intronic indels of 16 bp and 5 bp were identified. Statistical analysis revealed that only the 16 bp indel was associated with first-born litter size (P < 0.01), and the average first-born litter size of individuals with an insertion/insertion genotype higher than that of those with the deletion/deletion genotype (P < 0.05). There was also a significant difference in genotype distributions of the 16 bp indel between mothers of single-lamb and multi-lamb litters in the studied goat population (P = 0.001). Consistently, the 16 bp indel also had a significant effect on KDM6A gene expression. Additionally, there was no significant linkage disequilibrium (LD) between these two indel loci, consistent with the association analysis results. Together, these findings suggest that the 16 bp indel in KDM6A may be useful for marker-assisted selection (MAS) of goats.
Introduction
Improvements in female fertility are of critical importance for the goat industry. As one of the most important factors restricting female fertility, increasing litter size has received much more consideration (Naicy et al., 2016; Yang et al., 2017). However, litter size is a trait with low heritability in many livestock animals, including pigs (Córdoba et al., 2015) and goats (Shaat and Mäki-Tanila, 2009); therefore, traditional direct selection is ineffective. At present, marker-assisted selection (MAS), based on relevant genetic variants, is used extensively to improve traits with low heritability, such as those associated with growth and reproduction (Sharma et al., 2013; An et al., 2015; Tomas et al., 2016). To facilitate MAS application to litter size in the goat industry, critical genetic variants causing phenotypic advantage should be verified.
Currently, whole-genome sequencing and genome-wide association studies (GWAS) are used to explore genetic variants strongly associated with production traits (Lai et al., 2016; Mota et al., 2017; Wu et al., 2018); however, numerous potential genes identified by GWAS have not been fully verified. To address this problem, methods which combine GWAS analysis results and MAS to screen for critical genetic variations in large livestock populations have been developed. Previously, Hubert et al. (2014) used whole-genome re-sequencing to reveal that genomic variations within the bovine transmembrane protein 95 (TMEM95) gene are associated with male reproductive performance. A genome scan in a French dairy goat population also found that variants in diacylglycerol o-acyltransferase 1 (DGAT1) were associated with a notable decrease in milk fat content (Martin et al., 2017). These results demonstrate the feasibility of using combined methods to screen for important genetic variations.
In 2016, a study using whole-genome analysis to compare high and low fecundity groups of the Chinese Laoshan dairy goat identified several genes as potentially critical for fecundity, including lysine demethylase 6A (KDM6A), androgen receptor (AR), and anti-Mullerian hormone receptor type 2 (AMHR2) (Lai et al., 2016). Among these genes, KDM6A encodes a protein that demethylates tri- and dimethylated lysine 27 of histone H3, and can affect gametophyte development. Importantly, numerous studies have verified that the KDM6A gene is vital for animal reproduction. In rodents, knock-out of the KDM6A gene disrupted primordial germ cell development (Mansour et al., 2012). In female mice, the Rhox cluster of genes, which contains reproduction-related homeobox genes, is also regulated by KDM6A (Berletch et al., 2013). Furthermore, KDM6A regulates maturation of the mouse oocyte (Xu et al., 2017). Overall, based on whole-genome analysis and rodent studies, there is strong evidence that KDM6A has crucial roles in modulation of goat fecundity.
To date, goat KDM6A gene expression profiles and DNA polymorphisms are largely unexplored. Therefore, in this study, the tissue expression profiles of the KDM6A gene were investigated, two novel indel variants in this gene identified and the relationship between these loci and first-born litter size evaluated in a large Shaanbei white cashmere goat population. Moreover, the relationship between the identified indel loci and KDM6A expression levels was assessed. Our findings provide a basis for further research about the underlying causal mutation and suggest hypotheses for further study leading to the application of MAS to goat breeding.
Materials and Methods
All experiments in this study involving animals were approved by the Faculty Animal Policy and Welfare Committee of Northwest A&F University (protocol number NWAFAC1008). Moreover, the care and use of experimental animals completely conformed with local animal welfare laws, guidelines, and policies.
Sample Collection
For DNA experiments, a total of 2,326 adult female Shaanbei white cashmere goats were randomly selected from a large population. These goats all received the same diet and were kept under standard conditions after weaning. Among these goats, 1,811 animals had records of first-born litter size data (Wang et al., 2017; Yang et al., 2017). Apart from these female goats, we also collected a total of 18 male goat samples from six different developmental periods for RNA experiments. Nine tissues (heart, liver, spleen, lung, kidney, testis, brain, skin, and muscle) were harvested from 1-week-old (wk) and 2-month-old (mo) male goats (n = 3 per group). Moreover, testes tissues samples were also collected at 2, 3 wk, 1, and 1.5 mo (n = 3 per group). All tissues were immediately frozen in liquid nitrogen and stored at −80°C.
Isolation of DNA
Genomic DNA was isolated from ear tissues using the method published by Lan et al. (2007). The quality of genomic DNA samples was assayed using Nanodrop 2000 Spectrophotometer. DNA samples were each diluted to a working concentration of 10 ng/μL and stored at −20°C.
Primer Design, PCR Amplification, and Indel Genotyping
Five primer pairs for amplification of indel loci in introns were designed using Primer Premier software (version 5.0) based on the goat KDM6A gene sequence (NW_017189516.1) and the NCBI SNP-database (https://www.ncbi.nlm.nih.gov/snp; Table 1). Assays were performed by touch-down PCR in a 13 μL volume, containing 6.5 μL 2 × mix, 0.3 μL each of forward and reverse primers, 0.8 μL genomic DNA (10 ng/μL), and 5.4 μL ddH2O. The PCR protocol was as follows: initial denaturation for 5 min at 95 °C; followed by 18 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 68°C (with a decrease of 1°C per cycle), extension for 30 s at 72°C; another 25 cycles of 30 s at 94°C, 30 s at 50°C, and 2 min at 72°C; and a final extension for 10 min at 72°C, with subsequent cooling to 4°C. The genotyping of indel polymorphisms in goat KDM6A was performed by separation of PCR products (5 μL) by agarose gel electrophoresis.
Total RNA Isolation and Synthesis of cDNA
Total RNA was extracted from tissue samples using TRIzol total RNA extraction reagent (Takara, Dalian, China), according to the manufacturer's instructions. The integrity of total RNA was evaluated by 1% agarose gel electrophoresis in 6 × loading buffer (Takara, Dalian, China). The quantity and quality of total RNA was estimated using a Nanodrop 2000 Spectrophotometer with the OD260 nm/OD280 nm ratio expected to be between 1.8 and 2.0; meanwhile, the OD260 nm/OD230 nm ratio no less than 1.7 (Zhang et al., 2017). Samples were then stored at −80°C. Prime Script™ RT Reagent kit (Takara, Dalian, China) was used to synthesize first strand cDNA, according to the manufacturer's protocol. The resultant cDNA was stored at −20°C.
Analysis of KDM6A mRNA Expression Profiles by Quantitative Real-Time PCR
KDM6A gene expression profiles were analyzed by qPCR using cDNA from 1 wk and 2 mo male goat tissue samples. Expression profiles in testes at different time points (1, 2, 3 wk, 1, 1.5, and 2 mo) were also evaluated. qPCR primers were designed covering different exons in order to assure the amplification of the cDNA (Table 1). qPCR reactions (12 μL) contained 6 μL 2 × SYBR Premix ExTaq (Takara, Dalian, China), 0.5 μL of each primer, and 5 μL cDNA (1/100 dilution). PCR amplification was performed as follows: 95°C for 5 min followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s (Yu et al., 2017). The expression levels of RPL19 (ribosomal protein L19), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and ACTB (β-actin) in all isolated tissues were tested. The reference gene in each tissue was analyzed from the GeNorm program, which based on the M-values (reference gene with the lowest M-value is considered most stable; Vandesompele et al., 2002). After calculation, RPL19 was used as the reference gene in lung, muscle, brain and skin. ACTB was used as the reference gene for evaluation of relative gene expression in heart, liver, spleen, kidney and testis. And previous studies also used ACTB to determine gene expression in goat testis (Yao et al., 2014; Deng et al., 2017b). The results were determined using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Statistical Analysis
To explore the genetic structure of the indel variants in the investigated goat population, genetic diversity indices were calculated. The genotype and allele frequencies reflect the genetic composition of the indel variant in the tested goat population. Nei's methods were used to calculate population genetic diversity indices, including homozygosity (Ho), heterozygosity (He; Ho + He = 1) and polymorphism information content (PIC) (Nei and Roychoudhury, 1974). Ho and He are a measure of genetic variation of a population. PIC is an indicator of polymorphism. Based on PIC values, the genetic variations classified as high genetic diversity (PIC > 0.5), medium genetic diversity (0.25 < PIC < 0.5) and low genetic diversity (PIC < 0.25) (Botstein et al., 1980). The χ2 test using the SHEsis online platform (http://analysis.bio-x.cn) was conducted to evaluate HWE (Li et al., 2009; Chen et al., 2013).
Linkage disequilibrium (LD) is the nonrandom association of alleles at linked loci. In particular, many genetic variations correlated with each other due to LD; thus, LD plays a crucial role for mapping complex disease or trait-associated genes (Pritchard and Przeworski, 2001; Hazelett et al., 2016). Currently, to detect whether there is a linkage between the two indels identified in KDM6A, the LD structure as measured by D' and r2 were performed with the SHEsis online platform (http://analysis.bio-x.cn; Li et al., 2009). The r2-value was used as a pairwise measure of LD (Marty et al., 2010; Huang et al., 2015). The case of r2 = 0 is known as perfect LD, r2 > 0.33 indicates sufficiently strong LD, and r2 = 1 suggests complete LD (Ren et al., 2014).
Associations between indels and first-born litter size, to establish the influence of different parameters on litter size, were analyzed using a general linear model: Yij = μ + HYSi + Gj + eij, where Yij is the phenotypic value of litter size, μ is the overall population mean, HYSi is the fixed effect of the herd-year-season, Gj is the fixed effect of genotype, and eij is the random error (Yang et al., 2017). The litter size data used in this study was first-born litter size; thus, the lambing year and parity were not included in the general linear model. The analysis was performed with SPSS 19.0 software by one-way ANOVA and compared using Tukey multiple test.
Result
mRNA Expression Profile of Goat KDM6A
Goat KDM6A mRNA expression profiles were investigated in different tissues at 1 wk (Figure 1A) and 2 mo (Figure 1B). KDM6A was found to be expressed in all tissues tested at both developmental stages. Notably, the expression levels of KDM6A in heart, liver and spleen tissues were significantly higher at 1 wk than at 2 mo (P < 0.05). In contrast, the expression levels of KDM6A were significantly lower in lung, muscle, brain, and skin at 1 wk than at 2 mo (P < 0.05; Figure 1C).
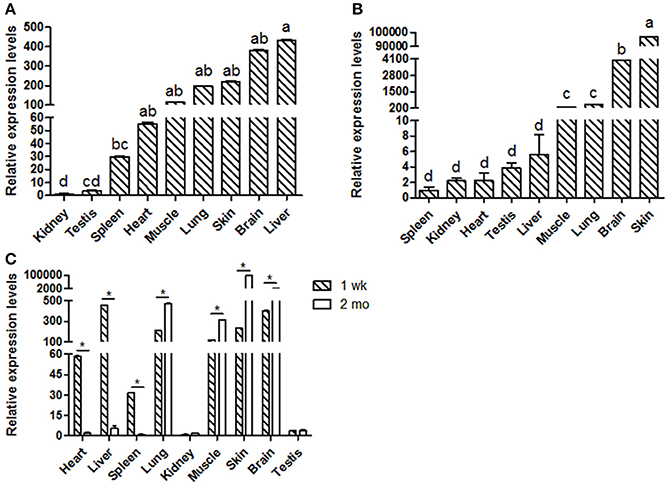
Figure 1. Shaanbei white cashmere goat KDM6A mRNA expression patterns detected by qPCR. (A) Different tissues of 1 wk Shaanbei white cashmere goat; (B) Different tissues of 2 mo Shaanbei white cashmere goat. (C) The comparison of goat KDM6A tissue expression levels between 1 wk and 2 mo. Data represent means ± SE (n = three samples of each tissues). Columns with different letters (a, b) means P < 0.05; *P < 0.05.
Goat KDM6A Gene Expression Profiles in Testis Tissues
This study was focused on the reproductive system, thus the expression levels of KDM6A at different testis developmental stages (1, 2, 3 wk, 1, 1.5, and 2 mo) were explored. In testis tissues, the KDM6A mRNA expression levels at 2 and 3 wk were significantly lower than that at 1, 1.5, and 2 mo (P < 0.05; Figure 2A). In a previous study of Liaoning cashmere goat (the male parents of Shaanbei white cashmere goat) spermatogonia was found to be actively mitotic from the postnatal period, with primary spermatocytes, which result from meiosis, first appearing at 1 mo (Zhan, 2015). Thus, we divided testis development into two phases: birth to 1 mo, referred to as the mitosis period, and 1–2 mo, referred to as the meiosis period. KDM6A mRNA expression levels were significantly increased in meiosis period compared with the mitosis period (P < 0.05; Figure 2B). Together, these findings provide evidence that KDM6A has an important role in fertility. To explore potential DNA markers for improvement of goat fertility, we next focused on the identification of polymorphisms in KDM6A.
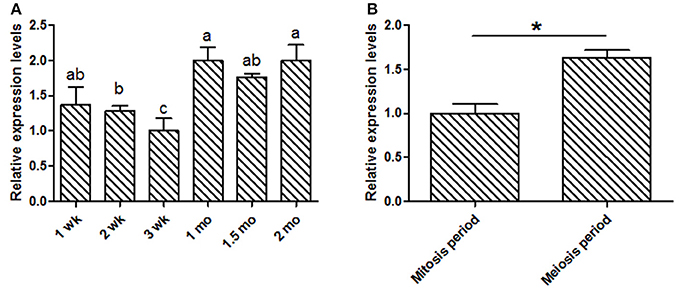
Figure 2. Expression of KDM6A mRNA was detected in goat testes tissues by qPCR. (A) KDM6A mRNA expression profiles at different stages in testis tissue. (B) Expression of KDM6A mRNA at mitosis and meiosis period in testis tissue. Data represent means ± SE (n = three samples per group). Columns with different letters (a, b) means P < 0.05; *P < 0.05.
Identification of Genetic Variants That Regulate KDM6A Expression
In this study, two novel indel variants were detected in goat KDM6A introns; one of 16 bp indel (intron 17) (NW_017189516.1:g.138431_138446delAATGTATAGCTTAAAA; rs636691921) and another of 5 bp indel (intron 17) (NW_017189516.1:g.138708_138712delTTAAT; rs653321281). These indels were detected using primer 3 and 4, respectively. PCR products separated by agarose gel electrophoresis, and sequence diagrams of these two novel indels are presented in Figure 3 and Supplement Figure 1.
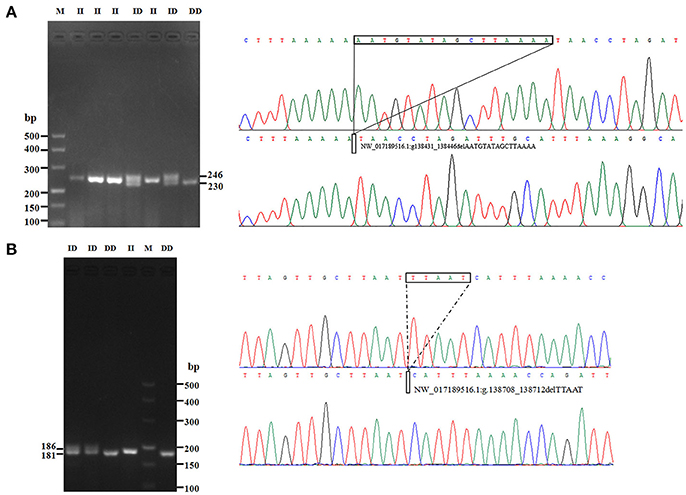
Figure 3. The electrophoresis diagrams and sequence diagrams of goat KDM6A gene indel loci. (A) 16 bp indel locus. (B) 5 bp indel locus.
Several previous studies have reported that variants in intron can affect gene transcription (Ren et al., 2011); therefore, KDM6A expression levels at different developmental periods were conducted in animals with the same genotype. However, in the mitosis period individuals that had DD and ID genotypes were not found at the 16 bp locus; thus, for this locus, we only compared the KDM6A expression levels of II genotype carriers between the mitosis and meiosis periods. The results demonstrated that KDM6A expression levels were significantly higher in the meiosis period of the II genotype at the 16 bp locus (P < 0.01). Furthermore, KDM6A expression was significantly higher during the meiosis period of animals with the II and DD genotypes of the 5 bp locus (P < 0.01; Figure 4). Together, these results indicate that the two indel loci could affect the expression levels of KDM6A and may influence the reproductive phenotype of goats. Therefore, the relationship between these indel loci and goat reproductive traits were further investigated in a large goat population.
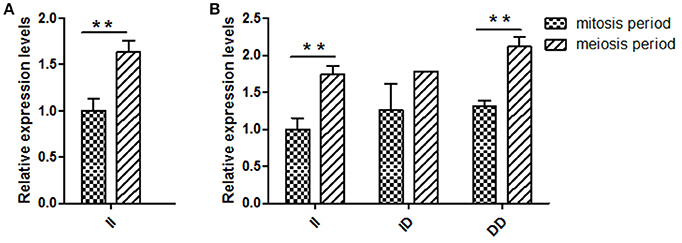
Figure 4. Two indel loci influence KDM6A mRNA expression between two periods. (A) In the 16 bp locus, KDM6A expression levels were significantly higher in the meiosis period of the II genotype. (B) In the 5 bp locus, the II and DD genotype had a significantly difference between two periods. Data represent means ± SE. **P < 0.01.
Genetic Parameters and LD of the Identified Indel Loci
The genotype and allele frequencies, as well as other genetic parameters, associated with the KDM6A indel loci were calculated to determine the genotype distribution among Shaanbei white cashmere goats (Table 2). The data indicated that “I” allele (0.941) of the 16 bp indel was more frequent than “D” allele (0.059). For the 5 bp indel, analysis of 615 individuals indicated that the frequency of the “I” allele was lower than 0.278, with the “D” allele present at a higher frequency (0.722). Additionally, the χ2 test indicated that the 5 bp indel genotype frequency was in agreement with HWE (P > 0.05) in the Shaanbei cashmere goat population; however, the 16 bp indel did not conform to HWE (P < 0.05; Table 2). Based on PIC values, the 16 bp locus had low genetic diversity (PIC = 0.105), and the 5 bp locus had medium genetic diversity (PIC = 0.321). Moreover, we analyzed the LD between these two indel loci; however, no LD was detected between them (r2 = 0.047; Table 3; Figure 5).
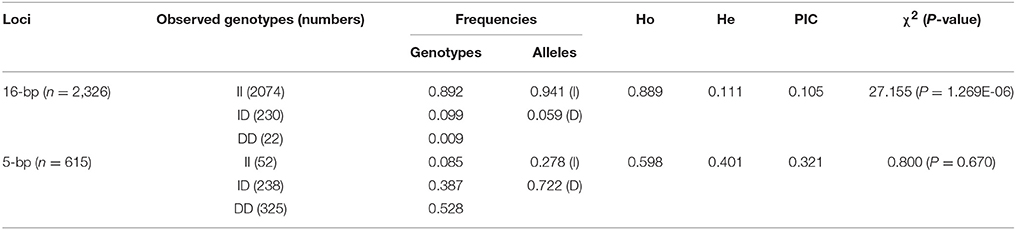
Table 2. Genetic parameters of the 16 and 5 bp loci within KDM6A gene in Shaanbei white cashmere goat.
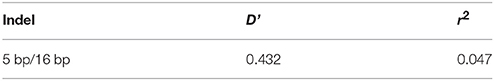
Table 3. Estimated values of linkage equilibrium analysis for two indels in KDM6A gene in studied populations.
Analyses of Associations Between Indel Variations and First-Born Litter Size
Next, the associations between KDM6A indel loci and there productive performance of female goats (first-born litter size) were investigated. The results showed that there was no relationship between the 5 bp indel and first-born litter size in populations of different sizes (n = 300–600 individuals) randomly selected from the whole population (P > 0.05; Table 4). Notably, the 16 bp locus was always associated with first-born litter size (P < 0.01) from 300 to 600 and even reaching 1811 individuals, with animals with the II genotype having larger first-born litter size than those with the DD genotype (Table 4).
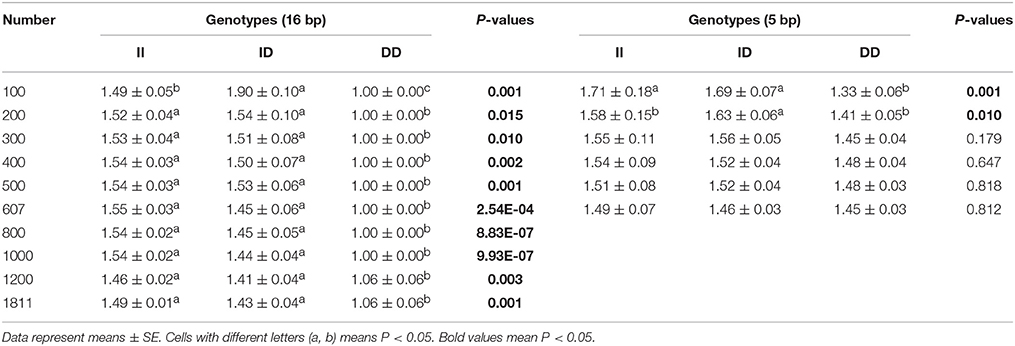
Table 4. Associations of the 16 and 5 bp loci with first-born litter size in detected groups with different numbers.
Furthermore, we investigated the genotype distributions of these two indel loci in groups of goats with first-born single-lamb and multi-lamb litters, using the same test groups described above (n = 100–600 individuals; Tables 5, 6). The results demonstrate that only the 16 bp indel had different genotype distributions between the two groups of goats with different litter types (P < 0.01). These results were consistent with those of association analyses; therefore, we tested the 16 bp indel in a total of 1,811 individuals. The results indicated a significant difference in genotype distributions between groups with first-born single-lamb and multi-lamb litters at the 16 bp indel (P = 0.001; Table 5). There was no LD between the two indel loci, consistent with the results of the association analysis (Figure 5).
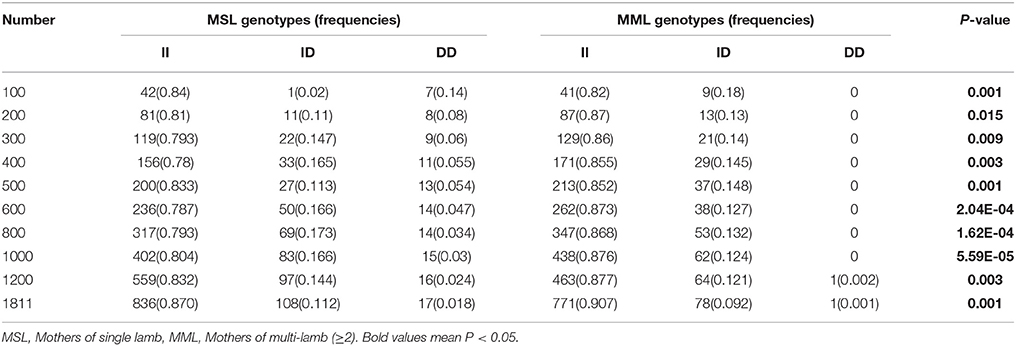
Table 5. The 16 bp locus genotype distribution between mothers of single lamb and multi-lamb litters in Shaanbei white cashmere goats.
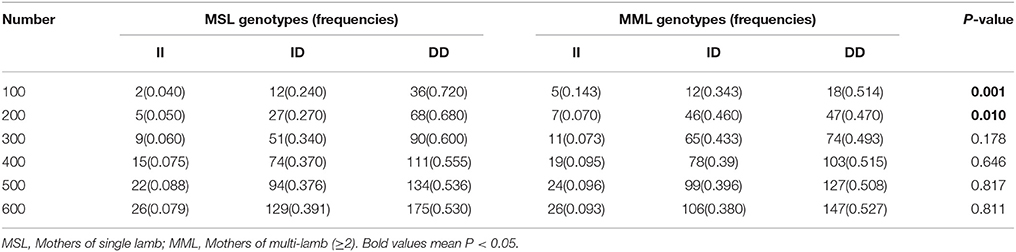
Table 6. The 5 bp locus genotype distribution between mothers of single lamb and multi-lamb litters in Shaanbei white cashmere goats.
Influence of the 16 Bp Indel on KDM6A Expression During the Meiosis Period
Based on the results of the association analyses, we hypothesized that the 16 bp indel can influence goat reproductive phenotype. This phenomenon may be attributable to the effect of genotype at this locus on KDM6A mRNA expression levels. Therefore, we tested KDM6A mRNA expression levels in testis tissue from animals with three genotypes at the two indel loci during the meiosis period. At the 16 bp indel locus, the individuals with II genotype had significantly higher levels of KDM6A mRNA expression than those with the ID and DD genotype (P < 0.05; Figure 6); however, there were no significant differences in KDM6A expression levels among different genotypes at the 5 bp indel locus (P > 0.05; Figure 6).
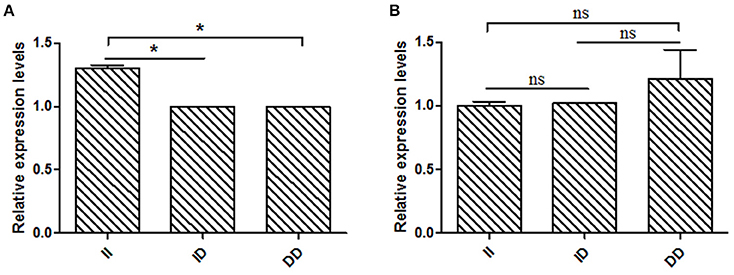
Figure 6. The 16 bp indel locus influence KDM6A expression in meiosis period. (A) In the 16 bp locus, genotype II had a significantly higher KDM6A expression level than genotype ID and DD. (B) In the 5 bp locus, there was non-difference among different genotypes. Data represent means ± SE. *P < 0.05; ns, no significance.
Discussion
Previously, Lai et al. (2016) determined that variants of the KDM6A gene were closely related to fecundity in Laoshan dairy goats using deep sequencing analysis. Several studies have also explored the role of KDM6A in reproductive biology (Yap et al., 2011), and their findings suggest that this gene has an essential role in reproduction. However, there are no previous reports of goat KDM6A tissue expression profiles. The relationship between KDM6A gene variants and first-born litter size in large Shaanbei white cashmere goat population (n = 2,326) required further investigation.
First, we determined the expression profiles of the goat KDM6A gene, and the results demonstrated that it was widely expressed in various organs. As the KDM6A gene is reported to be associated with spermatogenesis (Teperek et al., 2016), we next determined its expression patterns at different developmental stages in testis. Interestingly, the mRNA expression levels of KDM6A at later developmental stages (1, 1.5, and 2 mo) were higher than those at earlier stages (1, 2, and 3 wk). A study of the Liaoning cashmere goat reported that their spermatogonia gradually proliferate via mitotic division from birth, and primary spermatocyte development, which initiate meiosis from 1 mo (Zhan, 2015). Therefore, we combined individuals at 1, 2, and 3 wk classified as the mitosis period; similarly the 1, 1.5, and 2 mo data were considered the meiosis period. Our results demonstrate that KDM6A mRNA expression levels during the mitosis stage were lower than those in the meiosis stage (P < 0.05), suggesting KDM6A may be associated with the mitosis-to-meiosis transition in the Shaanbei white cashmere goat. Additionally, previous reports indicate that KDM6A regulates oocyte meiosis resumption in female mice, and abnormal expression of this gene causes aberrant H3K27me3, leading to disruption of oocyte maturation (Xu et al., 2017). Together, these data indicated that the KDM6A gene may have an essential role in meiosis resumption in both male and female animals.
In addition to the KDM6A gene, deep sequencing analyses of the Laoshan dairy goat have also identified genetic variants in male sex differentiation genes, including AR and AMHR2 that are closely associated with female fecundity (Lai et al., 2016). Furthermore, with the development of modern and intensive breeding condition, the number of male livestock is far less than the female (Wang et al., 2017). We hoped to explore the genetic variation in goat KDM6A, with the aim of implementing the identified polymorphisms as molecular markers to contribute to MAS in goat breeding. Therefore, we performed further analysis of KDM6A genetic effects in the female Shaanbei white cashmere goat population.
Currently, natural genetic variations are divided into three forms: SNPs (single nucleotide polymorphisms), indels and SVs (larger structural variants; Julienne et al., 2010). Unlike other genetic variations, indels can be directly detected by simple PCR amplification and agarose gel electrophoresis, making them convenient and practical (Naicy et al., 2016). Therefore, indel variants in the KDM6A gene were identified and their associations with first-born litter size investigated in a large commercial population of 2,326 Shaanbei white cashmere goats. Two novel indel loci (16 and 5 bp) were identified in putative intron 17 sequences, and each had three genotypes (II, ID, and DD). The 5 bp indel was in HWE (P > 0.05); however, the 16 bp locus was not (P < 0.05), because of the lower number of observed DD genotypes. One possible reason for this is rapid, powerful, and effective selection, which could affect the allelic balance of the indel locus (Zhao et al., 2013; Wang et al., 2015). Therefore, our data indicate that the selection pressure was more powerful on 16 bp than the 5 bp indel locus in the investigated goat population.
To analyze the association between indel loci and first-born litter size, we developed a novel strategy. Initially, analysis of the two indel loci was investigated in the same groups of 100–600 individuals, which were selected randomly from the whole population. When an indel locus in any investigated subset shows significant correlation with phenotype, it can be considered that this site is indeed correlated with the tested trait, especially in large population. This strategy improves the credibility of the test. Using groups of 300–600 individuals, there was no relationship between first-born litter size and the 5 bp indel locus (P > 0.05; Table 4). Interestingly, the 16 bp locus was consistently associated with first-born litter size in the same test groups (P < 0.01). Based on this data, we performed further analysis of the 16 bp indel among all individuals, and found that the association with first-born litter size was retained (P < 0.01), with the II genotype associated with larger litter size relative to the other genotypes, suggesting that the allele “I” of the KDM6A gene positively effects fecundity in this breed. Next, we adopted the same strategy to compare genotype distributions at these two indel loci between females who had first-born single-lamb and multi-lamb litters. The analysis results indicate that the 16 bp indel was very strongly associated with goat first-born litter size. Compared with direct analysis in the tested population (Deng et al., 2017a), this new strategy may provide more detailed and reliable results of association analysis. Additionally, the 16 bp locus had a significant effect on KDM6A gene expression, further implying a huge potential application for analysis of this locus. Moreover, linkage analysis demonstrated no LD between the two analyzed indel loci, consistent with the different results of association analyses.
The association analysis based on the large experimental population revealed that the 16 bp indel located in the 17 intron of KDM6A was strongly associated with litter size in goats, which was consistent with the previous whole-genome analysis for Laoshan dairy goats (Lai et al., 2016). Since Ren et al. (2011) reported that intronic variations could affect the gene expression level, the relationship between the 16 bp indel and the expression of KDM6A was evaluated in the current study. Our results showed that the intronic 16 bp indel significantly associated with the expression of KDM6A. According to previous investigations, the intronic variations could impact the interaction between transcription factors and host genes (Van Laere et al., 2003; Fushan et al., 2009; Soldner et al., 2016). Therefore, the transcription factor binding site on the 16 bp indel sequence was predicted using the online software Genomatix MatInspector (http://www.genomatix.de; Cartharius et al., 2005). The bioinformatics analysis results showed that myocyte-specific enhancer factor 2 (MEF2), as transcription factor, could bind to the sequence in the context of lacking the 16 bp nucleotides (Figure 7). This discovery provided a possibility that MEF2 factor influence goat litter size. However, in mouse, MEF2 was expressed in the testis throughout development but absent in the ovary (Daems et al., 2014), which meant the impact of the 16 bp indel on litter size might not caused by MEF2 factor. In addition, some intronic variations could be in perfect LD with known phenotype-associated mutations (Nakaoka et al., 2016). For example, a 40 bp indel variant residing in the mouse double minute 2 homolog (MDM2) gene promoter is in complete LD with a SNP (rs2279744), and the SNP locus has been demonstrated to be associated with the susceptibility to several cancers. Through linkage with the SNP locus, this indel locus had positive association with risk of colon cancer (Gansmo et al., 2016). Of course, whether the 16 bp intronic indel influences phenotype through linkage with causal mutations need further study to be proven.
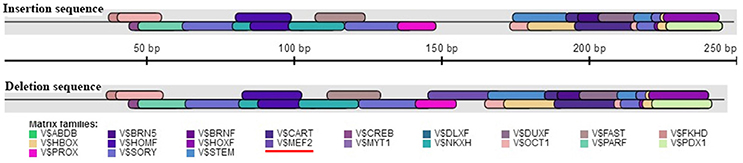
Figure 7. Bioinformatics predict transcription factor binding sites on the 16 bp indel sequences. One potential transcriptional factor MEF2 (myocyte-specific enhancer factor 2) only appear at deletion sequence. Red underline highlights MEF2 factor.
Conclusion
In this study, the results indicated that goat KDM6A mRNA was expressed in all tissues tested (heart, liver, spleen, lung, kidney, muscle, brain, skin, and testis), and the expression levels in testis were significantly increased through mitosis-to-meiosis transition. Meanwhile, two novel intronic indels of 16 and 5 bp were identified, and only the 16 bp indel was significantly associated with first-born litter size (P < 0.01). Additionally, the 16 bp indel had a significant effect on KDM6A gene expression. These findings would provide a basis for further research about the underlying causal mutation and the application of MAS to goat breeding.
Ethics Statement
On the basis of experimental animal management measures in Shaanxi province (016000291szfbgt-2011-000001), all experiment procedures were approved by the Review Committee for the Use of Animal Subjects of Northwest A&F University. Animal experimentation, including sample collection, was performed in agreement with the ethical commission's guidelines.
Author Contributions
YC, XL, and CP came with idea and wrote manuscript. KW, HX, JL, HZ, and LQ collected the goat samples and isolated of genomic DNA. YC, HY, and XZ performed the experiments. YC, HY, and HX analyzed the data. All authors approved the final version of the manuscript for submission.
Funding
This work was funded by the National Natural Science Foundation of China (No.31760650; No.31172184) and Provincial Key Projects of Shaanxi (2014KTDZ02-01).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly thanked the staffs of Shaanbei white cashmere goat breeding farm, Shaanxi province, P.R. China for their collecting samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00091/full#supplementary-material
Supplement Figure 1. The original image of the electrophoresis diagrams. (A) 16 bp indel locus. (B) 5 bp indel locus.
Abbreviations
KDM6A, lysine-specific demethylase 6A; indel, insertion/deletion; SNPs, single nucleotide polymorphisms; LD, linkage disequilibrium; MAS, marker-assisted selection; Ho, homozygosity; He, heterezygosity; PIC, polymorphism information content; HWE, Hardy-Weinberg equilibrium; II, insertion/insertion; ID, insertion/deletion; DD, deletion/deletion; wk, week-old; mo, month-old; LD, linkage disequilibrium; MSL, Mothers of single lamb; MML, Mothers of multi-lamb.
References
An, X., Hou, J., Gao, T., Lei, Y., Li, G., Song, Y., et al. (2015). Single-nucleotide polymorphisms g.151435C> T and g.173057T> C in PRLR gene regulated by bta-miR-302a are associated with litter size in goats. Teriogenology 83, 1477–1483. doi: 10.1016/j.theriogenology.2015.01.030
Berletch, J. B., Deng, X., Nguyen, D. K., and Disteche, C. M. (2013). Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet. 9:e1003489. doi: 10.1371/journal.pgen.1003489
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314–331.
Cartharius, K., Frech, K., Grote, K., Klocke, B., Haltmeier, M., Klingenhoff, A., et al. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942. doi: 10.1093/bioinformatics/bti473
Chen, F., Shi, J., Luo, Y. Q., Sun, S. Y., and Pu, M. (2013). Genetic characterization of the gypsy moth from China (Lepidoptera, Lymantriidae) using inter simple sequence repeats markers. PLoS ONE 8:e73017. doi: 10.1371/journal.pone.0073017
Córdoba, S., Balcells, I., Castelló, A., Ovilo, C., Noguera, J. L., Timoneda, O., et al. (2015). Endometrial gene expression profile of pregnant sows with extreme phenotypes for reproductive efficiency. Sci. Rep. 5:14416. doi: 10.1038/srep14416
Daems, C., Martin, L. J., Brousseau, C., and Tremblay, J. J. (2014). MEF2 is restricted to the male gonad and regulates expression of the orphan nuclear receptor NR4A1. Mol. Endocrinol. 28, 886–898. doi: 10.1210/me.2013-1407
Deng, S. L., Zhang, Y., Yu, K., Wang, X. X., Chen, S. R., Han, D. P., et al. (2017a). Melatonin up-regulates the expression of the GATA-4 transcription factor and increases testosterone secretion from Leydig cells through RORα signaling in an in vitro goat spermatogonial stem cell differentiation culture system. Oncotarget 8, 110592–110605. doi: 10.18632/oncotarget.22855
Deng, T., Pang, C., Ma, X., Duan, A., Liang, S., Lu, X., et al. (2017b). Buffalo SREBP1: molecular cloning, expression and association analysis with milk production traits. Anim. Genet. 48, 720–721. doi: 10.1111/age.12587
Fushan, A. A., Simons, C. T., Slack, J. P., Manichaikul, A., and Drayna, D. (2009). Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose. Curr. Biol. 19, 1288–1293. doi: 10.1016/j.cub.2009.06.015
Gansmo, L. B., Vatten, L., Romundstad, P., Hveem, K., Ryan, B. M., Harris, C. C., et al. (2016). Associations between the MDM2 promoter P1 polymorphism del1518 (rs3730485) and incidence of cancer of the breast, lung, colon and prostate. Oncotarget 7, 28637–28646. doi: 10.18632/oncotarget.8705
Hazelett, D. J., Conti, D. V., Han, Y., Al Olama, A. A., Easton, D., Eeles, R. A., et al. (2016). Reducing GWAS complexity. Cell Cycle 15, 22–24. doi: 10.1080/15384101.2015.1120928
Huang, Y. Z., Jing, Y. J., Sun, Y. J., Lan, X. Y., Zhang, C. L., Song, E. L., et al. (2015). Exploring genotype-phenotype relationships of the LHX3 gene on growth traits in beef cattle. Gene 561, 219–224. doi: 10.1016/j.gene.2015.02.030
Hubert, P., Sabine, K., Christine, W., Hermann, S., Reiner, E., Sandra, J., et al. (2014). A nonsense mutation in TMEM95 encoding a nondescript transmembrane protein causes idiopathic male subfertility in cattle. PLoS Genet. 10:e1004044. doi: 10.1371/journal.pgen.1004044
Julienne, M. M., Ryan, E. M., and Scott, E. D. (2010). Small insertions and deletions (INDELs) in human genomes. Hum. Mol. Genet. 19, R131–R136. doi: 10.1093/hmg/ddq400
Lai, F. N., Zhai, H. L., Cheng, M., Ma, J. Y., Cheng, S. F., Ge, W., et al. (2016). Whole-genome scanning for the litter sizetrait associated genes and SNPs under selection in dairy goat (Capra hircus). Sci. Rep. 6:38096. doi: 10.1038/srep38096
Lan, X. Y., Pan, C. Y., Chen, H., Zhang, C. L., Li, J. Y., Zhao, M., et al. (2007). An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small. Ruminant. Res. 73, 8–12. doi: 10.1016/j.smallrumres.2006.10.009
Li, Z., Zhang, Z., He, Z., Tang, W., Li, T., Zeng, Z., et al. (2009). A partition–ligation–combination–subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis. Cell Res. 19, 519–523. doi: 10.1038/cr.2009.33
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mansour, A. A., Gafni, O., Weinberger, L., Zviran, A., Ayyash, M., Rais, Y., et al. (2012). The H3K27 demethylaseUtx regulates somatic and germ cell epigenetic reprogramming. Nature 488, 409–413. doi: 10.1038/nature11272
Martin, P., Palhière, I., Maroteau, C., Bardou, P., Canale-Tabet, K., Sarry, J., et al. (2017). A genome scan for milk production traits in dairy goats reveals two new mutations in Dgat1 reducing milk fat content. Sci. Rep. 7:1872. doi: 10.1038/s41598-017-02052-0
Marty, A., Amigues, Y., Servin, B., Renand, G., Levéziel, H., and Rocha, D. (2010). Genetic variability and linkage disequilibrium patterns in the bovine DNAJA1 gene. Mol. Biotechnol. 44, 190–197. doi: 10.1007/s12033-009-9228-y
Mota, R. R., Guimarães, S. E., Fortes, M. R., Hayes, B., Silva, F. F., Verardo, L. L., et al. (2017). Genome-wide association study and annotating candidate gene networks affecting age at first calving in Nellore cattle. J. Anim. Breed. Genet. 134, 484–492. doi: 10.1111/jbg.12299
Naicy, T., Venkatachalapathy, R. T., Aravindakshan, T. V., Radhika, G., Raghavan, K. C., Mini, M., et al. (2016). Nerve Growth Factor gene ovarian expression, polymorphism identification, and association with litter size in goats. Theriogenology 86, 2172–2178. doi: 10.1016/j.theriogenology.2016.07.011
Nakaoka, H., Gurumurthy, A., Hayano, T., Ahmadloo, S., Omer, W. H., Yoshihara, K., et al. (2016). Allelic imbalance in regulation of ANRIL through chromatin interaction at 9p21 endometriosis risk locus. PLoS Genet. 12:e1005893. doi: 10.1371/journal.pgen.1005893
Nei, M., and Roychoudhury, A. K. (1974). Sampling variances of heterozygosity and genetic distance. Genetics 76, 379–390.
Pritchard, J. K., and Przeworski, M. (2001). Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69, 1–14. doi: 10.1086/321275
Ren, G., Huang, Y. Z., Wei, T. B., Liu, J. X., Lan, X. Y., Lei, C. Z., et al. (2014). Linkage disequilibrium and haplotype distribution of the bovine LHX4 gene in relation to growth. Gene 538, 354–360. doi: 10.1016/j.gene.2013.12.037
Ren, J., Duan, Y., Qiao, R., Yao, F., Zhang, Z., Yang, B., et al. (2011). A missense mutation in PPARD causes a major QTL effect on ear size in pigs. PLoS Genet. 7:e1002043. doi: 10.1371/journal.pgen.1002043
Shaat, I., and Mäki-Tanila, A. (2009). Variation in direct and maternal genetic effects for meat production traits in Egyptian Zaraibi goats. J. Anim. Breed. Genet. 126, 198–208. doi: 10.1111/j.1439-0388.2008.00784.x
Sharma, R., Ahlawat, S., Maitra, A., Roy, M., Mandakmale, S., and Tantia, M. S. (2013). Polymorphism of BMP4 gene in Indian goat breeds differing in prolifcacy. Gene 532, 140–145. doi: 10.1016/j.gene.2013.08.086
Soldner, F., Stelzer, Y., Shivalila, C. S., Abraham, B. J., Latourelle, J. C., Barrasa, M. I., et al. (2016). Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99. doi: 10.1038/nature17939
Teperek, M., Simeone, A., Gaggioli, V., Miyamoto, K., Allen, G. E., Erkek, S., et al. (2016). Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 26, 1034–1046. doi: 10.1101/gr.201541.115
Tomas, N., Venkatachalapathy, T., Aravindakshan, T., and Raghavan, K. C. (2016). Molecular cloning SNP detection and association analysis of 5′ ?anking region of the goat IGF1 gene with prolifcacy. Anim. Reprod. Sci. 167, 8–15. doi: 10.1016/j.anireprosci.2016.01.016
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome. Biol. 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Van Laere, A. S., Nguyen, M., Braunschweig, M., Nezer, C., Collette, C., Moreau, L., et al. (2003). A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 425, 832–836. doi: 10.1038/nature02064
Wang, C., Zhang, H., Niu, L., Guo, J., Jia, X., Wang, L., et al. (2015). The novel SNPs of leptin gene and their associations with growth traits in Chinese Nanjiang Yellow goat. Gene 572, 35–41. doi: 10.1016/j.gene.2015.06.073
Wang, X. Y., Yang, Q., Wang, K., Zhang, S. H., Pan, C. Y., Chen, H., et al. (2017). A novel 12-bp indel polymorphism within the GDF9 gene is significantly associated with litter size and growth traits in goats. Anim. Genet. 48, 735–736. doi: 10.1111/age.12617
Wu, P., Yang, Q., Wang, K., Zhou, J., Ma, J., Tang, Q., et al. (2018). Single step genome-wide association studies based on genotyping by sequence data reveals novel loci for the litter traits of domestic pigs. Genomics 110, 171–179. doi: 10.1016/j.ygeno.2017.09.009
Xu, K., Chen, X., Yang, H., Xu, Y., He, Y., Wang, C., et al. (2017). Maternal Sall4 Is indispensable for epigenetic maturation of mouse oocytes. J. Biol. Chem. 292, 1798–1807. doi: 10.1074/jbc.M116.767061
Yang, Q., Yan, H., Li, J., Xu, H., Wang, K., Zhu, H., et al. (2017). A novel 14-bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size. Anim. Genet. 48, 499–500. doi: 10.1111/age.12551
Yao, X., Tang, F., Yu, M., Zhu, H., Chu, Z., Li, M., et al. (2014). Expression profile of Nanos2 gene in dairy goat and its inhibitory effect on Stra8 during meiosis. Cell Prolif. 47, 396–405. doi: 10.1111/cpr.12128
Yap, D. B., Walker, D. C., Prentice, L. M., McKinney, S., Turashvili, G., Mooslehner, A. K., et al. (2011). Mll5 is required for normal spermatogenesis. PLoS ONE 6:e27127. doi: 10.1371/journal.pone.0027127
Yu, S., Zhang, P., Dong, W., Zeng, W., and Pan, C. (2017). Identification of stem leydig cells derived from pig testicular interstitium. Stem Cells Int. 2017:2740272. doi: 10.1155/2017/2740272
Zhan, W. (2015). The Study of Spermatogenesis and Seminiferous Performance of Liaoning Cashmere Goat Ram. Master's thesis, Jilin Agricultural University, Changchun.
Zhang, S. H., Xu, H., Liu, X. F., Yang, Q., Pan, C. Y., Lei, C. Z., et al. (2017). The muscle development transcriptome landscape of ovariectomized goat. R. Soc. Open. Sci. 4:171415. doi: 10.1098/rsos.171415
Keywords: cashmere goat, KDM6A gene, meiosis, mitosis, insertion/deletion (indel), litter size
Citation: Cui Y, Yan H, Wang K, Xu H, Zhang X, Zhu H, Liu J, Qu L, Lan X and Pan C (2018) Insertion/Deletion Within the KDM6A Gene Is Significantly Associated With Litter Size in Goat. Front. Genet. 9:91. doi: 10.3389/fgene.2018.00091
Received: 14 November 2017; Accepted: 05 March 2018;
Published: 20 March 2018.
Edited by:
Farai Catherine Muchadeyi, Agricultural Research Council of South Africa (ARC-SA), South AfricaReviewed by:
Kieran G. Meade, Teagasc, The Irish Agriculture and Food Development Authority, IrelandFabyano Fonseca Silva, Universidade Federal de Viçosa, Brazil
Copyright © 2018 Cui, Yan, Wang, Xu, Zhang, Zhu, Liu, Qu, Lan and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianyong Lan, lan342@126.com
Chuanying Pan, panyu1980@126.com
†These authors have contributed equally to this work.
 Yang Cui
Yang Cui Hailong Yan
Hailong Yan Ke Wang
Ke Wang Han Xu
Han Xu Xuelian Zhang
Xuelian Zhang Haijing Zhu2,3
Haijing Zhu2,3 Xianyong Lan
Xianyong Lan Chuanying Pan
Chuanying Pan