Toolbox for Emotional feAture extraction from Physiological signals (TEAP)
- 1Swiss Center for Affective Sciences, University of Geneva, Geneva, Switzerland
- 2CVML Laboratory, Computer Science Department, University of Geneva, Geneva, Switzerland
Physiological response is an important component of an emotional episode. In this paper, we introduce a Toolbox for Emotional feAture Extraction from Physiological signals (TEAP). This open source toolbox can preprocess and calculate emotionally relevant features from multiple physiological signals, namely, electroencephalogram (EEG), galvanic skin response (GSR), electromyogram (EMG), skin temperature, respiration pattern, and blood volume pulse. The features from this toolbox are tested on two publicly available databases, i.e., MAHNOB-HCI and DEAP. We demonstrate that we achieve similar performance to the original work with the features from this toolbox. The toolbox is implemented in MATLAB and is also compatible with Octave. We hope this toolbox to be further developed and accelerate research in affective physiological signal analysis.
1. Introduction
Affective computing thrives to teach machines to understand and express emotions (Picard, 1997). Emotions are multifaceted phenomena with physiological manifestations and bodily expressions (Scherer, 2005). Although the majority of existing methods for automatic emotion recognition are based on audiovisual analysis (D’Mello and Kory, 2015), there is a growing body of research on emotion recognition from peripheral and central nervous system physiological responses (Picard et al., 2001; Lisetti and Nasoz, 2004; Chanel et al., 2009, 2011; Kolodyazhniy et al., 2011; Koelstra et al., 2012; Soleymani et al., 2012b; Mühl et al., 2014). There are certain advantages for using physiological signals for emotion recognition compared to the audiovisual signals; for example, they cannot be easily faked, they do not require a front facing camera, and they can be used in any illumination and in noisy environments. Moreover, they can be combined with audiovisual modalities to construct a more robust and accurate multimodal emotion recognizer (D’Mello and Kory, 2015).
In order to train machines to automatically recognize emotions, we use machine learning techniques. Typically, the procedure includes translating raw signals to a lower dimensional representation space (features) that will be fed into a statistical model. Current physiological feature extraction methods involve calculating statistical moments and time-frequency analysis over segments of physiological signals with the goal of generating emotionally discriminative features. For example, heart rate increase is associated with excitement and activation (arousal) and, as a result, we extract heart rate as a feature for detecting arousal or the level of activation.
To extract features from physiological signals, one can implement methods that translate raw signals into features. Emotional features extracted from physiological signals are often based on the previous work in psychophysiology (Kreibig, 2010). To facilitate research on physiological signal analysis, a number of tools have been developed to the benefit of the community, e.g., EEGLAB (Delorme and Makeig, 2004). To the best of our knowledge, the Augsburg Biosignal Toolobox (AuBT) (Wagner et al., 2005) is the only toolbox available with the goal of extracting emotionally significant physiological features. However, the number of features and the type of signals in AUBT are limited. In this paper, we introduce a Toolbox for Emotional feAture extraction using Physiological signals or TEAP. We aim to create an open source platform that can be further extended by the community with the goal of advancing the field of affective physiological signal analysis. We developed TEAP in MathWorks MATLAB but it also works with Octave, its free alternative. TEAP is able to preprocess and extract features from multiple central and peripheral physiological signals including electroencephalogram (EEG), galvanic skin response (GSR), electrocardiogram (ECG), blood volume pulse (BVP), skin temperature, respiration pattern, and electromyogram (EMG). New physiological channels can be easily added to this toolbox and the implemented statistical and time-frequency analysis functions can be applied on any signal.
TEAP is designed to be useful for both novice and advanced users. A user can simply choose the desired features, define the channels in the input files, and extract physiological features. More advanced users are able to add new functionalities and modules, including support for new signals.
2. Toolboxes for Physiological Signal Processing
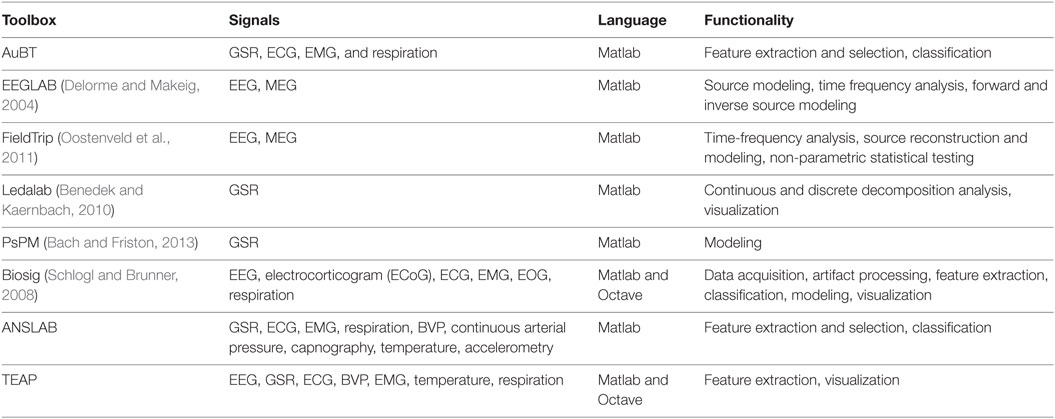
In this section, we review the existing freely available and open source tools for physiological signal analysis. There are a number of toolboxes for processing physiological signals and most of them are tuned for only one type of signal. For instance, EEGLAB (Delorme and Makeig, 2004) and FieldTrip (Oostenveld et al., 2011) are dedicated to brain signal analysis, focusing on signals such as EEG and magnetoencephalogram (MEG). There are also tools for analyzing peripheral physiological signals such as Kubios,1 which analyzes heart rate variability, Ledalab (Benedek and Kaernbach, 2010), and PsPM (Bach and Friston, 2013), which focus on the analysis of galvanic skin responses. There are also tools for analyzing a wide range of physiological signals including Biosig (Schlogl and Brunner, 2008) and the ANSLAB.2 Tools dedicated to one specific physiological signal offer advanced functionalities (e.g., source reconstruction). More general purpose tools handle a diverse range of signals with a more limited set of choices for their analysis. The spirit of TEAP is to allow for the processing of several types of signals, e.g., BVP, ECG, EEG, while maintaining the possibility to extract specific features from each signal. In addition, its architecture and license permits further development in the future.
Most of existing toolboxes requires to have some knowledge about physiological signal processing. In contrast, TEAP can serve researchers who aim at developing data-driven techniques with limited knowledge on the nature of physiological signals. Taking advantage of the BioSig code in its data import interface (Schlogl and Brunner, 2008), TEAP accepts several data formats which are traditionally used to store physiological signals (e.g., the EDF—(European Data Format), while its output is a design matrix representing a set of features for several samples. The Augsburg Biosignal Toolobox (AuBT) (Wagner et al., 2005) has been designed with the same objective in mind. It can perform feature extraction, feature selection, and classification. However, the AuBT only offers the possibility to extract general features based on statistical moments of signal derivatives (e.g., mean of first derivative). In addition, the AuBT input data should be properly formatted and have limited filtering capabilities. This leads to the need to develop file parsers and to gain knowledge in the properties of physiological signals to build proper preprocessing filters. The reviewed characteristics of the existing and proposed toolboxes are summarized in Table 1.
3. Technical Specifications
3.1. Architecture
TEAP has been developed in MATLAB and is compatible with Octave. It is open source and is licensed under the GNU General Public License (GNU GPL). This makes TEAP a completely free and customizable solution. We avoided object oriented programming in MATLAB not to jeopardize Octave compatibility. Although TEAP is programmed without using MATLAB objects, two principal structures are used: signals and bulksigs (hereafter called bulks); bulks are structures containing signals. Functions were created as interfaces to the users to manipulate these two structures. Hence, a user does not need to know how the data are handled by the toolbox in order to use it. However it remains possible to access the content of the structures directly or using the available visualization functions (e.g., Signal_plot()).
TEAP can read many data formats including EDF and EEGLAB. TEAP accepts data recorded from any device as long as the format is supported by Biosig interface, e.g., Biosemi BDF format. Importing EEGLAB data allows performing some preprocessing, which is not available in TEAP. For instance, the raw signals can be segmented using EEGLAB (e.g., according to triggers) and the result can be imported in TEAP. TEAP relies on MATLAB and only requires the signal processing and statistical toolboxes. TEAP can run on the equivalent settings in Octave with its freely available equivalent toolboxes.
The source code and user manuals are available on.3
3.2. Signals and Features
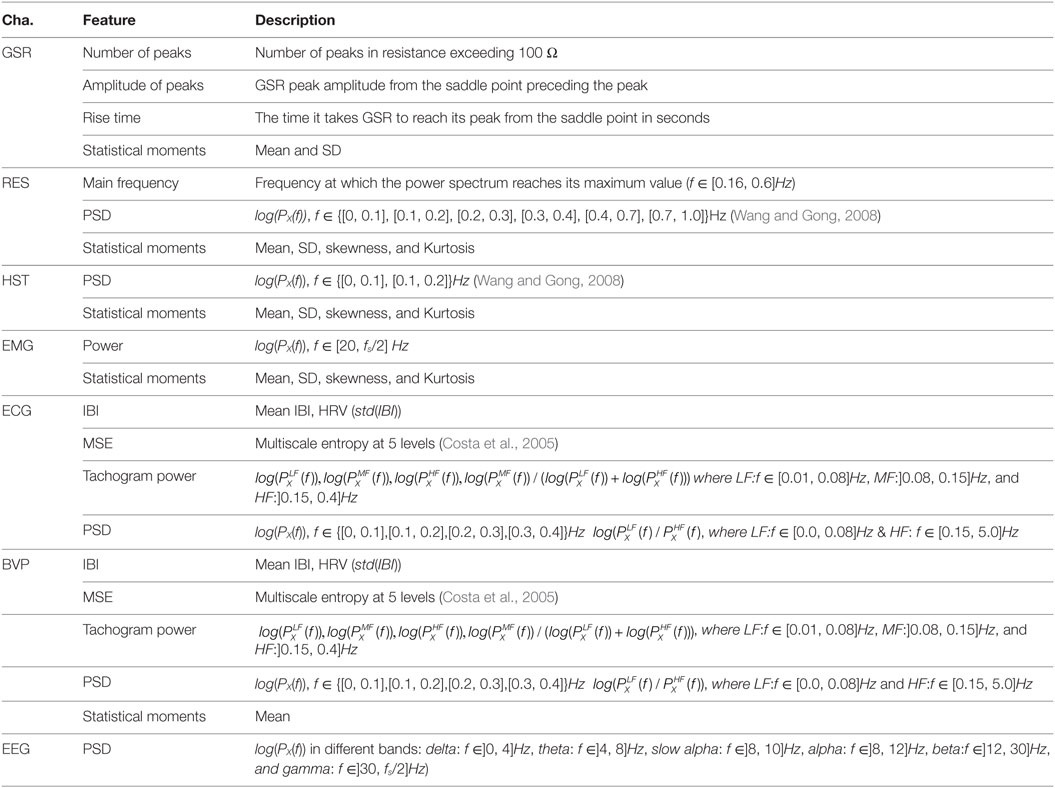
Each signal currently supported by TEAP is presented below. In addition to this list of signals, we added a template dummy signal (DMY), which will facilitate adding new signals and features. The set of features were chosen based on their proven performance in the literature (Kim and André, 2008; Chanel et al., 2009; Koelstra et al., 2012; Soleymani et al., 2012a). The list of existing features and their descriptions are given in Table 2.
3.2.1. Electrocardiogram—ECG
Due to the nature of heart muscles, cardiac activity generates an electrical potential difference, which can be measured by placing electrodes on one’s chest. Electrocardiography (ECG or EKG) is a measurement of this electrical activity. ECG signals can be used to detect heart rate (HR) and heart rate variability (HRV). HR and HRV changes are associated with emotions. For example, pleasant emotions increase heart rate response (Lang et al., 1993), and HRV decreases with fear, sadness, and happiness (Rainville et al., 2006). In TEAP, we analyze ECG signals from one lead (a pair of electrodes) to detect inter-beat-interval (IBI) and HRV features. After calculating the IBI, we can construct a tachogram, which is a signal representing IBIs over time. Tachogram can be further used to extract additional HRV features.
3.2.2. Blood Volume Pulse—BVP
Blood volume pulse (BVP) is the measurement of blood volume in peripheral vessels generally obtained by photoplethysmography. A photoplethysmograph usually consists of a light emitter and detector. The measurement of the reflected light on skin (usually finger) is an indicator of the volume of blood in peripheral vessels. Since blood volume varies by pulse heart rate can be detected from BVP signals. Similar to ECG, TEAP can extract heart rate (HR) and heart rate variability (HRV) related features from BVP.
3.2.3. Galvanic Skin Response (GSR)
Galvanic skin response (GSR) is a measurement of electrical resistance (or conductance) on one’s skin. Skin’s electrical conductance varies with the activity of sweat glands which are controlled by the sympathetic nervous system. GSR responses consists of tonic (slow) and phasic (fast and associated with a stimuli) responses and are related to emotional arousal (Lang et al., 1993; Dawson et al., 2000). TEAP calculates features related to both tonic and phasic responses.
3.2.4. Human Skin Temperature—HST
Skin temperature is a reflection of blood flow and changes in different emotional states (McFarland, 1985). Although temperature changes are slower than other signals, they are associated with emotional responses (Kreibig, 2010). Skin temperature is measured by attaching a temperature sensor on one’s skin. Statistical moments and low frequency power spectral features are extracted from HST by TEAP.
3.2.5. Electromyogram (EMG)
Activity of skeletal muscles generates electromyogenic electrical signals (EMG), which can be recorded by means of electrodes attached to the skin covering those muscles. Typically, a pair of electrodes is attached along the muscle of interest to record the electrical potential between two points. Facial and body expressions associated with emotions activate different muscles. One can record emotionally significant expressions using EMG. For example, smiling activates the zygomaticus major (Ekman, 2006). Spectral power density (in f > 20 Hz) and statistical moments are calculated by TEAP as features from EMG.
3.2.6. Respiration
Respiration pattern can be measured from the expansion of the chest or abdomen circumference. This can be done by a flexible belt with a piezoelectric crystal sensor, which measures the belt’s expansion. Respiration pattern (RES) varies by emotional responses. Slow respiration is linked to relaxation and irregular rhythm, quick variations, and cessation of respiration correspond to high arousal, e.g., anger or fear (Rainville et al., 2006; Kim and André, 2008). Principal frequency, power spectral density, and statistical moments are the features that Teap extract from respiration pattern.
3.2.7. Electroencephalogram—EEG
There is a strong evidence demonstrating the neural activities and circuits engaged in different emotional states (Damasio et al., 2000; Adolphs et al., 2003). Electroencephalogram (EEG) signals are a measurement of electrical neural activity on scalp. EEG signals contain waves in different frequency bands that are associated with different cognitive states. Therefore, power spectral density (PSD) features from different frequency bands are calculated as features in TEAP.
3.3. Usage
The goal of this section is to give an example of TEAP’s usage. Readers are redirected to the user manual (see text footnote 4) for a more detailed documentation.
Suppose a user has an ECG signal and that they want to extract some features. First, a user shall choose which feature to extract; users can use “include” or “exclude” arguments to add or remove features from the available set of features, or simply extract all available features. With TEAP, the process is as follows:
%import probe 1
probe1 = csvread ( ’ ECG example_probe1 . csv ’);
%import probe 2
probe2 = csvread ( ’ ECG example_probe2 . csv ’);
%create the signal from two electrodes and a given sampling
%frequency then display the resulting signal
ECG_sig = ECG_aqn_variable (probe1, probe2, 1,024);
Signal_plot (ECG_sig);
%compute some features (all available features)
[ECG_features, ECG_feats_names]= ECG_feat_extr (ECG_sig);
%compute one feature (for example average inter_beat intervals)
[ECG_features, ECG_feats_names] = ECG_feat_extr (ECG_sig, ’ include ’ , ’ meanIBI ’ );
%compute some features (all available features except HRV)
[ECG_features, ECG_feats_names] = ECG_feat_extr (ECG_sig , ’ exclude ’ , ’HRV ’);
It should be noted that for some specific signals (e.g., HST or GSR), a preprocessing, e.g., filtering, will be automatically applied when the signal is loaded; thus, the user does not have to worry about all the necessary steps.
4. Experimental Validation
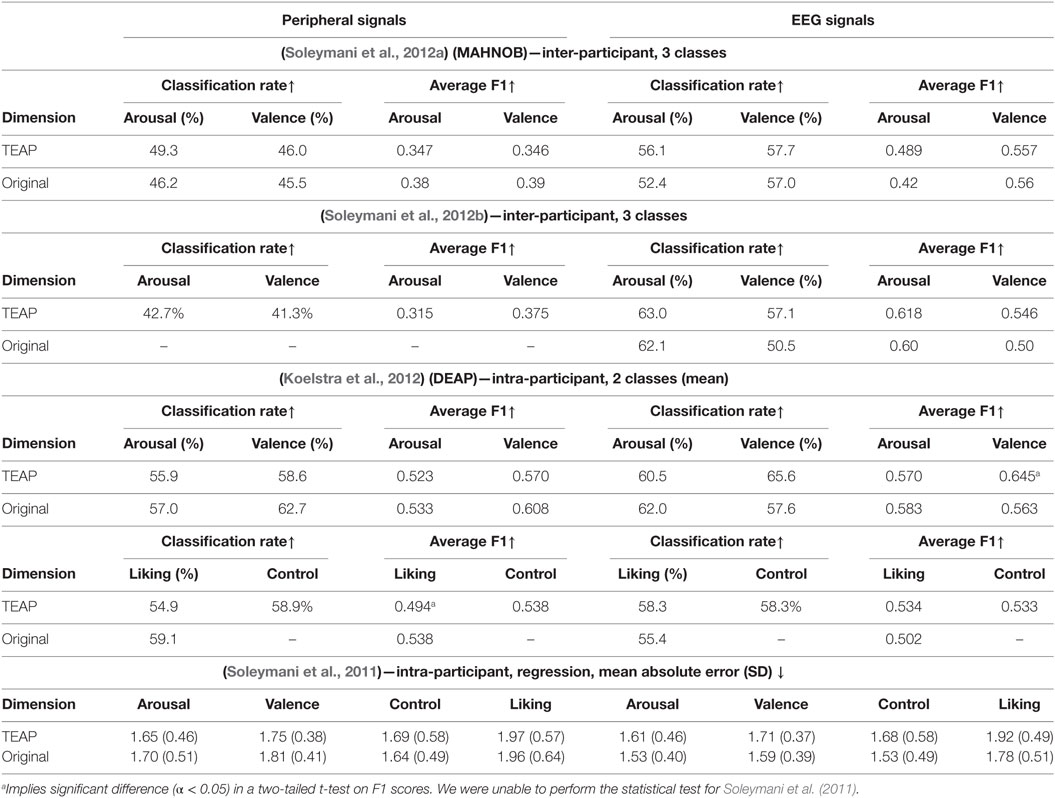
In order to verify TEAP’s performance in emotion recognition, we made use of two publicly available databases to serve as our benchmark, namely MAHNOB-HCI (Soleymani et al., 2012a) and DEAP (Koelstra et al., 2012). We tried to replicate the work presented in four original articles on these databases, i.e., Soleymani et al. (2011, 2012a,b) and Koelstra et al. (2012). In addition to TEAP, reproduction of these results required LIBSVM (Chang and Lin, 2001). We tried to re-implement the same procedure in terms of cross-validation strategy, feature selection, machine learning models, and evaluation metrics. For MAHNOB-HCI database, we only analyzed the emotion experiment whose original results are published in Soleymani et al. (2012a,b). The results of participant-independent cross validation on MAHNOB-HCI database are reported alongside the original results in Table 3. The regression and classification results on DEAP database are also given in Table 3. In most cases, TEAP features are performing similarly with small differences compared to the original work, the reasons behind this difference is twofold. First, we shortened and simplified the set of features in TEAP compared to the original work to make them more generally applicable. For example, EEG asymmetry features are not implemented in TEAP, since it depends on the electrode placement and do not make a large difference. Second, even though we tried to replicate the same machine learning models, our hyper-parameter optimization and feature selection slightly differ from the original work. Nevertheless, the results from TEAP features are, in most cases, not significantly different from the original results and validates its effectiveness in capturing emotionally significant representation of physiological signals.
5. Conclusion
We propose a new open-source toolbox dedicated to extracting and calculating emotionally relevant representations from physiological signals. This toolbox will facilitate and accelerate research in emotional physiological signals analysis. This toolbox is able to import, preprocess, and visualize its signals. It can export features vectors that can be directly fed to the machine learning models. In this paper, we outlined its abilities and demonstrated its effectiveness according to the publicly available benchmarks. Ideal candidates for future feature implementation include synchronization, intermodality features such as pulse ejection time and event-related potentials (ERP). We hope this toolbox can serve the researchers interested in affective physiological signal analysis.
Author Contributions
MS and GC developed and adopted the original features (Chanel et al., 2009, 2011; Soleymani et al., 2009, 2012b; Koelstra et al., 2012). MS performed the experiments and reported the results. FV-D designed the architecture and modules and created the toolbox. TP participated in drafting the article and the discussions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
The work of MS was supported by his Swiss National Science Foundation Ambizione grant. The work of GC and TP was supported by the National Center of Competence in Research (NCCR) Affective sciences financed by the Swiss National Science Foundation (51NF40-104897) and hosted by the University of Geneva.
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/article/10.3389/fict.2017.00001/full#supplementary-material.
Footnotes
References
Adolphs, R., Tranel, D., and Damasio, A. R. (2003). Dissociable neural systems for recognizing emotions. Brain Cogn. 52, 61–69. doi:10.1016/S0278-2626(03)00009-5
Bach, D. R., and Friston, K. J. (2013). Model-based analysis of skin conductance responses: towards causal models in psychophysiology. Psychophysiology 50, 15–22. doi:10.1111/j.1469-8986.2012.01483.x
Benedek, M., and Kaernbach, C. (2010). Decomposition of skin conductance data by means of nonnegative deconvolution. Psychophysiology 47, 647–658. doi:10.1111/j.1469-8986.2009.00972.x
Chanel, G., Kierkels, J. J. M., Soleymani, M., and Pun, T. (2009). Short-term emotion assessment in a recall paradigm. Int. J. Hum. Comput. Stud. 67, 607–627. doi:10.1016/j.ijhcs.2009.03.005
Chanel, G., Rebetez, C., Bétrancourt, M., and Pun, T. (2011). Emotion assessment from physiological signals for adaptation of game difficulty. IEEE Trans. Syst. Man Cybern. A Syst. Humans 41, 1052–1063. doi:10.1109/TSMCA.2011.2116000
Chang, C.-C., and Lin, C.-J. (2011). LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 1–27. doi:10.1145/1961189.1961199
Costa, M., Goldberger, A. L., and Peng, C. K. (2005). Multiscale entropy analysis of biological signals. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 71, 021906. doi:10.1103/PhysRevE.71.021906
Damasio, A. R., Grabowski, T. J., Bechara, A., Damasio, H., Ponto, L. L. B., Parvizi, J., et al. (2000). Subcortical and cortical brain activity during the feeling of self-generated emotions. Nat. Neurosci. 3, 1049–1056. doi:10.1038/79871
Dawson, M. E., Schell, A. M., and Filion, D. L. (2000). “The electrodermal system,” in Handbook of Psychophysiology, 2nd Edn, eds J. T. Cacioppo, L. G. Tassinary and G. G. Berntson (New York, NY: Cambridge University Press), 200–223.
Delorme, A., and Makeig, S. (2004). EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 134, 9–21. doi:10.1016/j.jneumeth.2003.10.009
D’Mello, S. K., and Kory, J. (2015). A review and meta-analysis of multimodal affect detection systems. ACM Comput. Surv. 47, 1–43. doi:10.1145/2682899
Ekman, P. (1990). “Duchenne and facial expression of emotion,” in The Mechanism of Human Facial Expression, Vol. 48. (Cambridge: Cambridge University Press), 270–284.
Kim, J., and André, E. (2008). Emotion recognition based on physiological changes in music listening. IEEE Trans Pattern Anal. Mach. Intell. 30, 2067–2083. doi:10.1109/TPAMI.2008.26
Koelstra, S., Muhl, C., Soleymani, M., Lee, J. S., Yazdani, A., Ebrahimi, T., et al. (2012). DEAP: a database for emotion analysis using physiological signals. IEEE Trans. Affect. Comput. 3, 18–31. doi:10.1109/T-AFFC.2011.15
Kolodyazhniy, V., Kreibig, S. D., Gross, J. J., Roth, W. T., and Wilhelm, F. H. (2011). An affective computing approach to physiological emotion specificity: toward subject-independent and stimulus-independent classification of film-induced emotions. Psychophysiology 48, 908–922. doi:10.1111/j.1469-8986.2010.01170.x
Kreibig, S. D. (2010). Autonomic nervous system activity in emotion: a review. Biol. Psychol. 84, 394–421. doi:10.1016/j.biopsycho.2010.03.010
Lang, P. J., Greenwald, M. K., Bradley, M. M., and Hamm, A. O. (1993). Looking at pictures: affective, facial, visceral, and behavioral reactions. Psychophysiology 30, 261–273. doi:10.1111/j.1469-8986.1993.tb03352.x
Lisetti, C. L., and Nasoz, F. (2004). Using noninvasive wearable computers to recognize human emotions from physiological signals. EURASIP J. Appl. Signal Process. 2004, 1672–1687. doi:10.1155/S1110865704406192
McFarland, R. A. (1985). Relationship of skin temperature changes to the emotions accompanying music. Appl. Psychophysiol. Biofeedback 10, 255–267. doi:10.1007/BF00999346
Mühl, C., Allison, B., Nijholt, A., and Chanel, G. (2014). A survey of affective brain computer interfaces: principles, state-of-the-art, and challenges. Brain Comput. Interfaces 1, 66–84. doi:10.1080/2326263X.2014.912881
Oostenveld, R., Fries, P., Maris, E., and Schoffelen, J.-M. (2011). FieldTrip: open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput. Intell. Neurosci. 2011, 1–9. doi:10.1155/2011/156869
Picard, R. W., Vyzas, E., and Healey, J. (2001). Toward machine emotional intelligence: analysis of affective physiological state. IEEE Trans. Pattern Anal. Mach. Intell. 23, 1175–1191. doi:10.1109/34.954607
Rainville, P., Bechara, A., Naqvi, N., and Damasio, A. R. (2006). Basic emotions are associated with distinct patterns of cardiorespiratory activity. Int. J. Psychophysiol. 61, 5–18. doi:10.1016/j.ijpsycho.2005.10.024
Scherer, K. R. (2005). What are emotions? And how can they be measured? Soc. Sci. Inf. 44, 695–729. doi:10.1177/0539018405058216
Schlogl, A., and Brunner, C. (2008). BioSig: a free and open source software library for BCI research. Computer 41, 44–50. doi:10.1109/MC.2008.407
Soleymani, M., Chanel, G., Kierkels, J. J. M., and Pun, T. (2009). Affective characterization of movie scenes based on content analysis and physiological changes. Int. J. Semantic Comput. 3, 235–254. doi:10.1142/S1793351X09000744
Soleymani, M., Koelstra, S., Patras, I., and Pun, T. (2011). “Continuous emotion detection in response to music videos,” in 2011 IEEE International Conference on Automatic Face Gesture Recognition and Workshops (FG 2011) (Santa Barbara, CA: IEEE), 803–808.
Soleymani, M., Lichtenauer, J., Pun, T., and Pantic, M. (2012a). A multimodal database for affect recognition and implicit tagging. IEEE Trans. Affect. Comput. 3, 42–55. doi:10.1109/T-AFFC.2011.25
Soleymani, M., Pantic, M., and Pun, T. (2012b). Multimodal emotion recognition in response to videos. IEEE Trans. Affect. Comput. 3, 211–223. doi:10.1109/T-AFFC.2011.37
Wagner, J., Jonghwa, K., and Andre, E. (2005). “From physiological signals to emotions: implementing and comparing selected methods for feature extraction and classification,” in IEEE International Conference on Multimedia and Expo (Amsterdam: IEEE), 940–943.
Keywords: physiological signals, emotions, affective computing, electroencephalogram signals, physiological signal processing, code:MATLAB, code:Octave, toolbox
Citation: Soleymani M, Villaro-Dixon F, Pun T and Chanel G (2017) Toolbox for Emotional feAture extraction from Physiological signals (TEAP). Front. ICT 4:1. doi: 10.3389/fict.2017.00001
Received: 08 June 2016; Accepted: 20 January 2017;
Published: 13 February 2017
Edited by:
Nadia Bianchi-Berthouze, University College London, UKReviewed by:
Raymond Robert Bond, Ulster University, UKElisabeth André, Augsburg University, Germany
Copyright: © 2017 Soleymani, Villaro-Dixon, Pun and Chanel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Soleymani, mohammad.soleymani@unige.ch
 Mohammad Soleymani
Mohammad Soleymani Frank Villaro-Dixon
Frank Villaro-Dixon Thierry Pun
Thierry Pun Guillaume Chanel
Guillaume Chanel