- Department of Molecular and Cell Biology, University of Connecticut, Storrs, CT, USA
The symbiosis between the squid, Euprymna scolopes, and the bacterium, Vibrio fischeri, serves as a model for understanding interactions between beneficial bacteria and animal hosts. The establishment and maintenance of the association is highly specific and depends on the selection of V. fischeri and exclusion of non-symbiotic bacteria from the environment. Current evidence suggests that the host’s cellular innate immune system, in the form of macrophage-like hemocytes, helps to mediate host tolerance of V. fischeri. To begin to understand the role of hemocytes in this association, we analyzed these cells by high-throughput 454 transcriptomic and liquid chromatography/tandem mass spectrometry (LC-MS/MS) proteomic analyses. 454 high-throughput sequencing produced 650, 686 reads totaling 279.9 Mb while LC-MS/MS analyses of circulating hemocytes putatively identified 702 unique proteins. Several receptors involved with the recognition of microbial-associated molecular patterns were identified. Among these was a complete open reading frame to a putative peptidoglycan recognition protein (EsPGRP5) with conserved residues for amidase activity. Assembly of the hemocyte transcriptome showed EsPGRP5 had high coverage, suggesting it is among the 5% most abundant transcripts in circulating hemocytes. Other transcripts and proteins identified included members of the conserved NF-κB signaling pathway, putative members of the complement pathway, the carbohydrate binding protein galectin, and cephalotoxin. Quantitative Real-Time PCR of complement-like genes, cephalotoxin, EsPGRP5, and a nitric oxide synthase showed differential expression in circulating hemocytes from adult squid with colonized light organs compared to those isolated from hosts where the symbionts were removed. These data suggest that the presence of the symbiont influences gene expression of the cellular innate immune system of E. scolopes.
Introduction
All animals have co-evolved associations with microorganisms. These symbiotic associations have been a major driving force in eukaryotic evolution, including the vastly influential bacterial endosymbiotic origin of essential organelles as championed by the late Lynn Margulis (Sagan, 1967). Given that all metazoans evolved in close and intimate association with microorganisms, it is not surprising that co-evolved microbiota also influenced the evolution of other aspects of animal biology. There has been a long-standing view that the immune system evolved to counter infection by pathogens and to exclude microorganisms from animals. However, recent evidence suggests that the immune system also plays an important role as an interface between hosts and their natural co-evolved benign or beneficial microbiota (Gross et al., 2009; McFall-Ngai et al., 2010; Royet et al., 2011). This interface involves a “dialog” between host and symbiont(s) that ensures that these associations are established and maintained. Growing evidence suggests that these beneficial microorganisms have influenced the evolution of both the innate and adaptive immune systems (McFall-Ngai, 2007; Lee and Mazmanian, 2010; McFall-Ngai et al., 2010). Gnotobiotic studies in mammals have demonstrated that proper development of components of the adaptive immune system require the presence of the host’s normal microbiota. For example the development of the gut-associated lymphoid tissue and differentiation of various T-cell classes are greatly influenced by the bacterial communities in the gut (reviewed in Bauer et al., 2006; Duan and Kasper, 2011; Olszak et al., 2012). It has also been proposed that perhaps the adaptive immune system itself evolved to deal with an increasingly complex gut microbiota (McFall-Ngai, 2007).
Invertebrates, which comprise the vast majority of animals, lack an adaptive immune system and must rely solely on innate immunity to interface with a microbial world. Despite any obvious mechanism to achieve the precision with which the adaptive immune system targets specific pathogens, there are many examples of highly specific associations formed between microorganisms and invertebrates (e.g., as found in insect bacteriocytes, termite hindguts, the trophosomes of hydrothermal vent tubeworms, and the light organs of squids; Nyholm and McFall-Ngai, 2004; Stewart and Cavanaugh, 2006; Moran et al., 2008; Feldhaar and Gross, 2009; Watanabe and Tokuda, 2010). What are the mechanisms for achieving such specificity? Many of these associations exist by creating specialized microenvironments that favor colonization by microorganisms and sequester them to specific cells or tissues (e.g., bacteriocytes; anaerobic, pH, or nutrient-specific environments as in the gut; or specialized tissues as in light organs; Nyholm and McFall-Ngai, 2004; Bäckhed et al., 2005; Stoll et al., 2010). However, increasing evidence suggests that the innate immune system of animal hosts is also important in maintaining these associations (Gross et al., 2009; Nyholm et al., 2009; McFall-Ngai et al., 2010; Krasity et al., 2011; Weiss et al., 2011).
The light organ symbiosis between the Hawaiian bobtail squid, Euprymna scolopes, and the bioluminescent bacterium, Vibrio fischeri, is used as a model system to study the mechanisms by which beneficial bacteria colonize animal hosts (Figure 1; McFall-Ngai, 2002; Nyholm and McFall-Ngai, 2004; Visick and Ruby, 2006). The symbionts reside within the epithelia-lined crypt spaces of a specialized light organ (to densities of 109V. fischeri cells/adult squid). This organ is directly connected to the environment via ciliated ducts that facilitate the environmental transmission of the symbionts. Juvenile squid must select V. fischeri from a background of 106 non-symbiotic bacteria/ml of seawater (Nyholm and McFall-Ngai, 2004). There are many mechanisms by which the partners ensure this specificity and the host’s innate immune system is important in this process. For example, innate immunity effectors like reactive oxygen species (ROS), putative complement members, and phagocytic hemocytes, along with the recognition of microbial-associated molecular patterns (MAMPs) by host pattern recognition receptors (PRRs) have all been implicated in playing a role in this association (Weis et al., 1996; Nyholm and McFall-Ngai, 1998; Davidson et al., 2004; Koropatnick et al., 2004, 2007; Castillo et al., 2009; Nyholm et al., 2009; Troll et al., 2009, 2010; McFall-Ngai et al., 2010, 2012; Heath-Heckman and McFall-Ngai, 2011; Krasity et al., 2011; Schleicher and Nyholm, 2011). Because the symbiosis is binary with a single host and symbiont, it offers the opportunity to ask how such high specificity is established and maintained in the context of interactions with the innate immune system of the host.
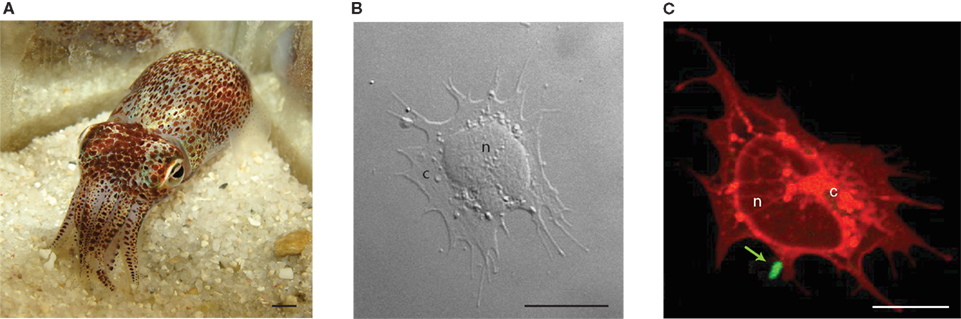
Figure 1. The Hawaiian bobtail squid, Euprymna scolopes, and its hemocytes. (A) Adult E. scolopes. Scale, 3 mm. (B) Hemocyte of the host as viewed by DIC imaging. The macrophage-like hemocyte is the only circulating blood cell type found in the host. These cells can infiltrate the crypt spaces of the light organ where they can interact with the symbiont, V. fischeri. Scale, 10 μm. (C) Host hemocyte stained with CellTracker (red) and viewed by confocal microscopy. A sole V. fischeri cell (green; arrow) can be seen bound to the hemocyte. Scale, 10 μm. n, nucleus; c, cytoplasm.
Euprymna scolopes, like other cephalopods, has a single type of macrophage-like hemocyte (Figure 1). These hemocytes can traverse between tissues and enter into the crypt spaces where the symbionts reside and appear to “sample” these spaces (Nyholm and McFall-Ngai, 1998). Previous studies of hemocytes from E. scolopes have indicated that they behave like macrophages, binding, and phagocytosing bacteria (Nyholm and McFall-Ngai, 1998; Nyholm et al., 2009; Heath-Heckman and McFall-Ngai, 2011). Although the crypts of juvenile symbiotic E. scolopes may contain hemocytes with engulfed bacterial cells, hemocytes in adult crypts have never been observed with phagocytosed bacteria despite being found in a microenvironment with high densities of V. fischeri. These observations suggest that the squid’s hemocyte response may change during development of the association. A study of adult hemocytes revealed that these cells could also differentiate between symbiotic and non-symbiotic bacteria (Nyholm et al., 2009). In vitro binding assays demonstrated that hemocytes differentially bound various species within the Vibrio group. V. fischeri bound poorly to hemocytes from hosts with fully colonized light organs and significantly more to blood cells from hosts from which the symbionts were removed by curing with antibiotics. No significant change was observed for non-symbiotic bacteria. Taken together, these data suggest that hemocytes can differentiate between V. fischeri and other closely related members of the Vibrionaceae and colonization alters the hemocytes’ ability to recognize the symbiont. The phenomenon of hemocytes changing their binding affinity to V. fischeri after colonization may be analogous to vertebrate immune “tolerance”, leading to homeostasis (symbiostasis) of the association. This evidence leads us to ask; how can the innate immune system achieve such specificity and how does the symbiont influence the immune response?
In order to better understand the role of host hemocytes in the squid/Vibrio symbiosis we have used high-throughput sequencing and liquid chromatography/tandem mass spectrometry (LC-MS/MS) to characterize the transcriptome and proteome in circulating blood cells of adult hosts with fully colonized light organs. In addition, we have used quantitative RT-PCR (qRT-PCR) to compare expression of several innate immunity genes from both symbiotic and cured hosts, to understand the molecular mechanisms by which the hemocyte response may change due to colonization.
Materials and Methods
Animal Collections
Adult animals were collected in shallow sand flats off of Oahu, HI by dip net and were either maintained in the laboratory in re-circulating natural seawater aquaria at the Hawaii Institute of Marine Biology or at the University of Connecticut with artificial seawater (ASW, Instant Ocean, IO) at 23°C. All animals were acclimated at least 48 h under laboratory conditions and kept on an approximate 12 h light/12 h dark cycle before sample collection (Schleicher and Nyholm, 2011).
Hemocyte Collection
Squid hemocytes were isolated from adult E. scolopes as previously described (Nyholm et al., 2009; Collins and Nyholm, 2010). Animals were first anesthetized in a 2% solution of ethanol in seawater. Hemolymph was extracted from the cephalic artery, located between the eyes, using a sterile 1-ml syringe with a 28-gauge needle. An average of 50–100 μl of hemolymph (∼5,000 hemocytes/μl) was obtained per animal using this method. Freshly collected hemocytes were washed and resuspended in Squid Ringer’s solution and hemocyte concentrations were determined by hemocytometer.
Curing Experiments
To generate naïve hemocytes, adult E. scolopes were cured of their population of V. fischeri symbionts (Nyholm et al., 2009). Briefly, adult squid were maintained individually in 5-gal tanks containing IO ASW (Aquarium Systems). For one set of animals, chloramphenicol and gentamicin was added to the seawater to a final concentration of 20 μg/m1 of each. The concentration of antibiotics used in these experiments effectively eliminates V. fischeri symbionts from the light organ without compromising the health of the host in any detectable way (Doino and McFall-Ngai, 1995; Lamarcq and McFall-Ngai, 1998; Foster et al., 2000; Nyholm et al., 2002, 2009; Koropatnick et al., 2007). The animals were transferred daily into fresh IO, either with or without antibiotics, for 5 days, and samples of hemolymph were collected on day 5. The resulting two sets of hemocytes were designated either “normal” (untreated/symbiotic) or “naïve” (treated/cured). The effectiveness of the antibiotics was determined by dissecting and homogenizing the central core of the light organ, and plating an aliquot of the homogenate on seawater tryptone agar (Nyholm et al., 2009).
Transcriptome Sequencing
RNA was extracted from the hemocytes of 11 adult squid using the RNAqueous-Micro kit (Ambion, Life Technologies) and treated with Turbo DNAse (Ambion, Life Technologies). Ribosomal RNA was removed using a eukaryote RiboMinus kit (Invitrogen) and quantified using an Agilent Bioanalyzer PicoRNA chip. Two-hundred nanograms of treated and pooled RNA (approximately 18 ng from the hemocytes of each of the 11 animals) was used to make a 454 cDNA library according to standard protocol (Roche). The library was titrated with an SVemPCR kit and sequenced on one-half of a 454 picotiter plate.
454 Bioinformatics
454 sequencing reads were assembled using the CLC Genomic Workbench with standard de novo assembly parameters (mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.5, similarity = 0.8, conflicts resolved by voting, minimum contig length = 200 bases). Contigs were used to query the UniProt and NCBI nr protein databases using blastx with a maximum e-value of 0.01. Significant alignments were further analyzed using SignalP (Petersen et al., 2011), InterProScan (Zdobnov and Apweiler, 2001), and ClustalX (Jeanmougin et al., 1998) using default parameters. The output from the blastx searches were imported into the program BLAST2GO (Conesa et al., 2005) and were used to assign Gene Ontology categories. Level 2 hierarchies were used for comparison between proteomic and transcriptomic sequencing. BLAST results were also used to make annotated protein databases to be searched with MS/MS peptide data.
Protein Extraction
Hemocytes collected from an adult symbiotic squid were homogenized in 64 mM Tris pH 8, 1% SDS, 1× protease inhibitor cocktail (Sigma Aldrich, P2714). After the homogenate was centrifuged (Eppendorf 5810 R, 14,000 rpm, 4°C, 30 min), the protein concentration of the supernatant was quantified spectrophotometrically using the RC DC protein assay (Bio-Rad, Hercules, CA, USA). Alternatively, hemocyte proteins from adult symbiotic squid were extracted using the Ready Prep Protein Extraction Kit (Membrane I) as described by the manufacturer’s protocol (Bio-Rad, Hercules, CA, USA). Hemocyte proteins from two adult squid were separated into either a hydrophilic or a hydrophobic layer based on temperature dependent phase partitioning. The protein concentration of each layer was determined with the RC DC protein assay as described above (Bio-Rad, Hercules, CA, USA).
Protein Digestion
Proteins were digested as previously described (Schleicher and Nyholm, 2011). Hemocyte proteins were precipitated with 10% (w/v) trichloroacetic acid (Fisher Scientific) at 4°C overnight. The precipitates were collected by centrifugation (Eppendorf 5810 R, 11,000 × g, 30 min, 4°C) and washed twice with ice-cold acetone. Protein pellets were briefly air-dried and then resuspended in 25 μl of 8 M urea, 0.4 M ammonium bicarbonate, pH 8.0. Samples were reduced and alkylated with 5 μl of 45 mM dithiothreitol (DTT; Acros Organics) at 37°C for 20 min and 5 μl of 100 mM iodoacetamide (Acros Organics) at room temperature in the dark for 20 additional minutes. Sequencing grade trypsin was added 1:15 (w/w enzyme to protein; Promega, V5111). The solutions were diluted in water to 100 μl (2 M urea final concentration). Both samples were digested at 37°C for 18–24 h and then stored at −80°C until further analysis.
Protein Identification Using NanoLC-MS/MS
Protein identifications were completed at the W.M. Keck Biotechnology Resource Laboratory at Yale University as previously described (Schleicher and Nyholm, 2011). Briefly, 5 μl (0.375 μg) of the hydrophilic and hydrophobic fraction of hemocyte protein extraction from the Ready Prep Protein Extraction Kit I were analyzed on a LTQ Orbitrap equipped with a Waters nanoAcquity UPLC system. Samples were concentrated at 15 μl/min with 99% Buffer A (100% water, 0.1% formic acid) for 1 min on a trap column (Waters Symmetry® C18 180 μm × 20 mm). Peptides were separated at 300 nl/min with Buffer A and Buffer B (100% CH3CN, 0.075% formic acid) using a 1.7-μm, 75 μm × 250 mm nanoAcquity™ UPLC™ column (35°C). A 51-min linear gradient was established using increasing amounts of Buffer B (95% Buffer A, 5% Buffer B at initial conditions, 50% A, 50% B, at 50 min, and 15% A, 85% B at 51 min). MS was acquired in the Orbitrap using one microscan and an inject time of 900 followed by four data dependent MS/MS acquisitions in the ion trap.
Hemocyte proteins from an adult symbiotic squid were also identified using a 5600 TripleTOF (AB SCIEX, Concord, ON, Canada) equipped with a Waters nanoAcquity UPLC system. Briefly, 5 μl (3 μg) of sample were loaded onto a Waters Symmetry® C18 180 μm × 20 mm trap column at 15 μl/min with 99% Buffer A for 1 min. Peptides were separated with a 1.7-μm, 75-μm × 150-mm nanoAcquity™ UPLC™ column (45°C) using a 161-min linear gradient at 500 nl/min. The gradient included 95% Buffer A, 5% Buffer B at initial conditions, 60% A, 40% B at 160 min, and 15% A, 85% B at 161 min. MS was acquired from 400 to 1250 Da for 0.25 s. While in high sensitivity mode, 20 data dependent MS/MS were acquired for 0.05 s each.
Proteomic Data Analysis
MS/MS spectra were converted to Mascot compatible files using either the Mascot Distiller (Orbitrap MS/MS) or the AB Sciex MGF Converter (5600 TripleTOF MS/MS) and searched with the Mascot algorithm (Version 2.3.01; Hirosawa et al., 1993; Perkins et al., 1999) using an E. scolopes protein sequence database (25,745 sequences) generated from available transcriptomic data: juvenile light organ ESTs (Chun et al., 2006) and hemocyte transcripts (this study). Searching parameters included peptide charge states of +2 or +3, partial methionine oxidation, carboxamidomethylated cysteine, a peptide tolerance of ±20 ppm, and MS/MS fragment tolerance of ±0.6 Da. A decoy database search was also performed by Mascot.
Scaffold (version Scaffold_3.2.0, Proteome Software Inc., Portland, OR, USA) was used to validate MS/MS based peptide and protein identifications by Mascot. In addition, Scaffold applied X! Tandem (Craig and Beavis, 2004) [The GPM, thegpm.org; version CYCLONE (2010.12.01.1)] as a supplementary search to further confirm protein and peptide identifications. X! Tandem applied the same search parameters as Mascot. Peptide and protein identifications were accepted at greater than 95.0% probabilities each. Only proteins that met the necessary criteria and contained two or more peptides were considered as positive identifications. For positive identifications, the false discovery rates (FDRs) were 0% at both the protein and peptide level as calculated by Scaffold. Putative protein identifications included proteins with only one matching peptide (protein FDR, 0.4%).
Polymerase Chain Reaction
Total RNA from adult symbiotic or naïve hemocytes was extracted using TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. Approximately 1–2 μg of RNA per animal was isolated. Subsequent PCR and qRT-PCR was performed using cDNA template created from 200 ng of total RNA using the iScript cDNA synthesis kit (Bio-Rad) and using the manufacturer’s instructions for random primers. End-point PCR was performed on a MyCycler Thermal Cycler (Bio-Rad, Hercules, CA, USA) using the following program: 3 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 55°C, and 45 s at 72°C; with a final extension of 10 min at 72°C. Products were separated on a 1% agarose gel, stained with SYBR safe DNA Gel Stain (Life Technologies, Carlsbad, CA, USA) and visualized using a Molecular Image Gel Doc (Bio-Rad, Hercules, CA, USA).
qRT-PCR reactions were performed on a Cycle iQ Multicolor Real-Time Detection System (Bio-Rad, Hercules, CA). qRT-PCR was then performed using gene specific primers, 10 μl Sso Fast Evagreen Supermix (Bio-Rad, Hercules, CA) and distilled water to a total volume of 20 μl. Amplification was performed under the following conditions: 3 min at 95°C, followed by 40 cycles at 95°C for 5 s, 55°C for 15 s, 72°C for 30 s, followed by a melt curve program, performed to confirm the presence of one optimal dissociation temperature for the resulting amplicons. Standard curves were created using a five-fold cDNA dilution series with each primer set. All efficiencies were between 90 and 105%. Each gene was assayed in three technical triplicates for each of three biological replicates, and data was analyzed using the ΔΔCT relative quantification method with the 40S and L21 ribosomal subunits as control genes. Primers for both end-point and qRT-PCR are seen in Table A1 in Appendix.
Results
We analyzed circulating adult hemocytes by high-throughput 454 transcriptomic and LC-MS/MS proteomic analyses. 454 high-throughput sequencing produced 650,686 reads totaling 279.9 Mb (Table 1; Dataset S1 in Supplementary Material) while LC-MS/MS analyses of circulating hemocytes putatively identified 702 unique proteins (Table 2; Datasets S2 and S3 in Supplementary Material). Analysis of gene ontologies (GO) from these datasets revealed that both the transcriptome and proteome yielded similar types of genes/proteins involved with biological processes, molecular function, and cell components (Figure 2).
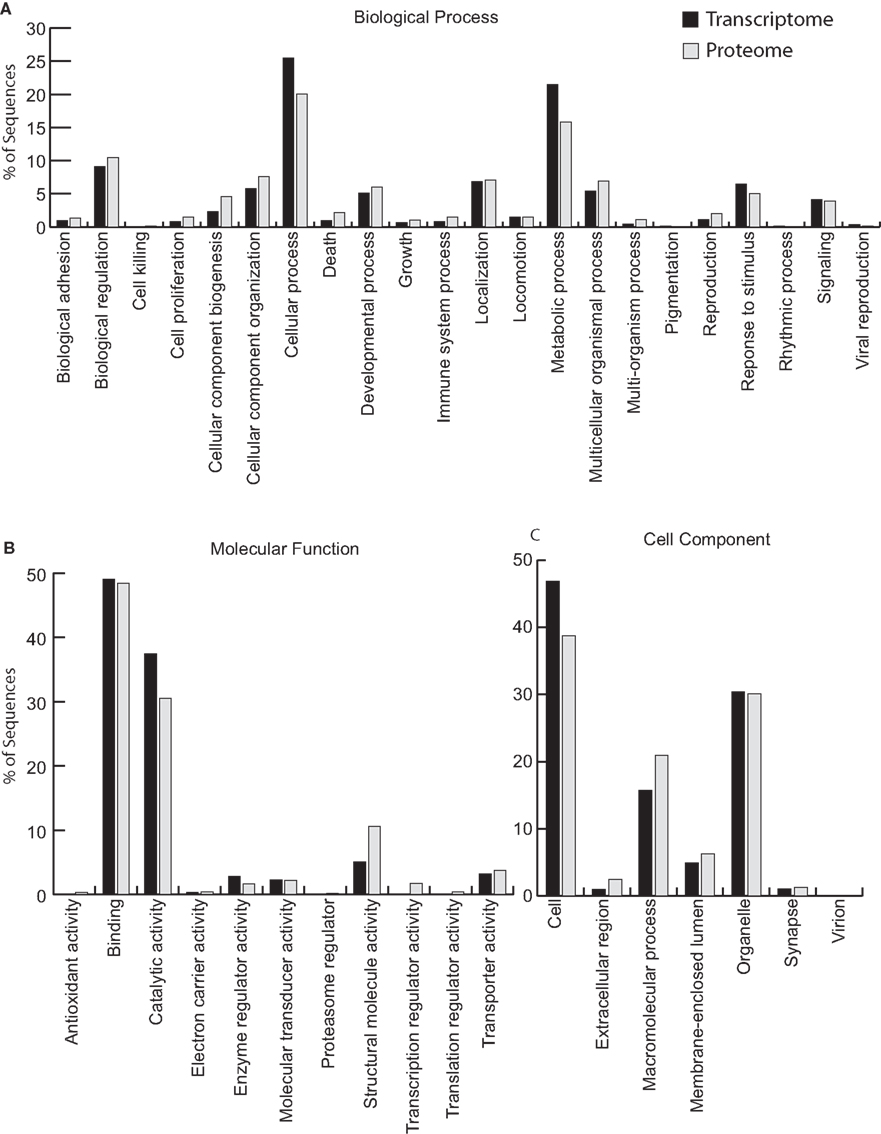
Figure 2. Gene ontology (GO) analysis of the hemocyte transcriptome and proteome. Annotation by GO revealed similar proportions of GO terms in both the transcriptome and proteome (Datasets 1 and 2). Sequences were annotated using previously described BLAST parameters (see Materials and Methods), and level 2 GO terms in (A) biological process, (B) molecular function and (C) cell component were assigned by BLAST2GO. Black bars represent transcriptome sequences; gray bars represent proteome sequences.
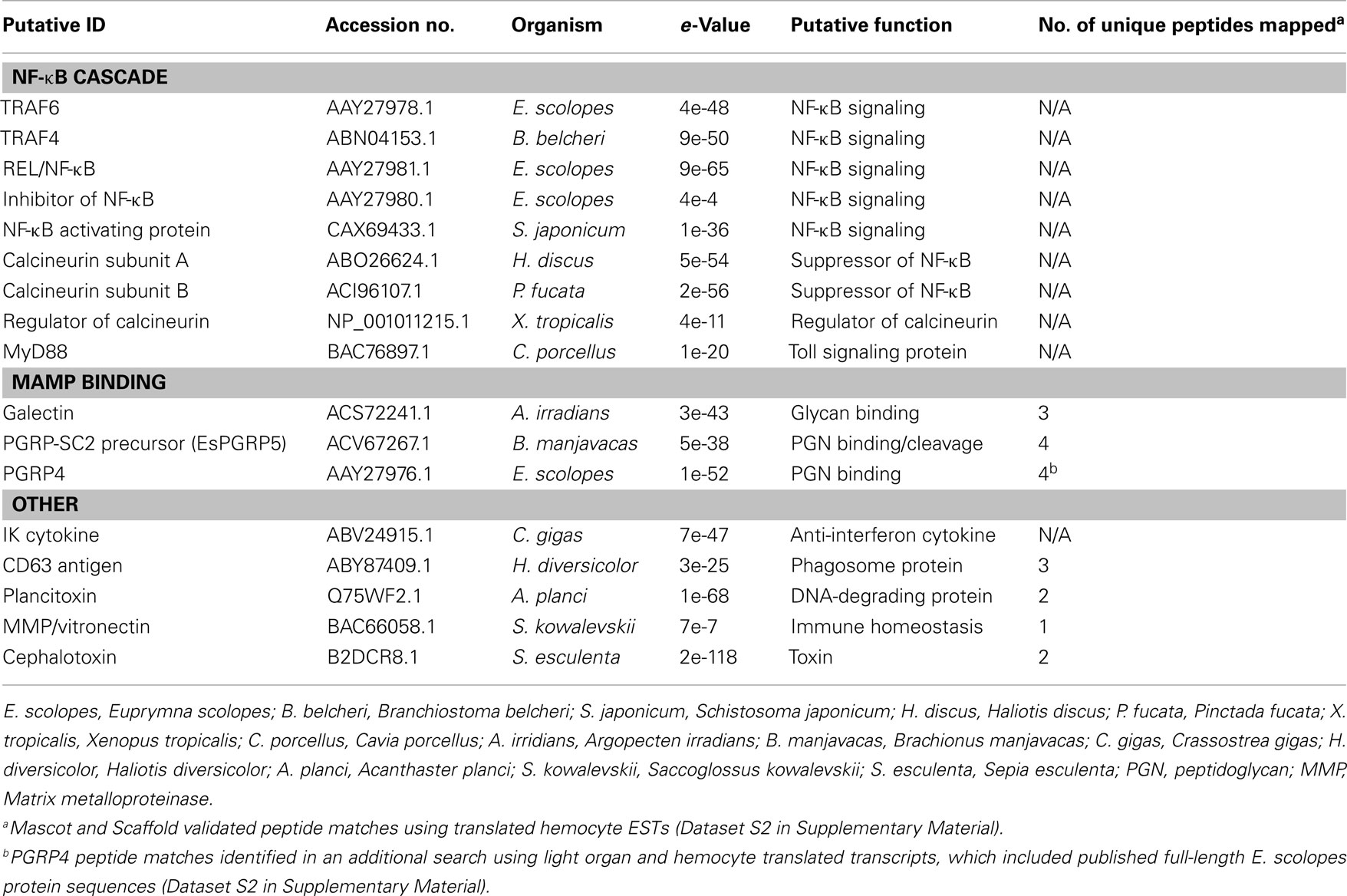
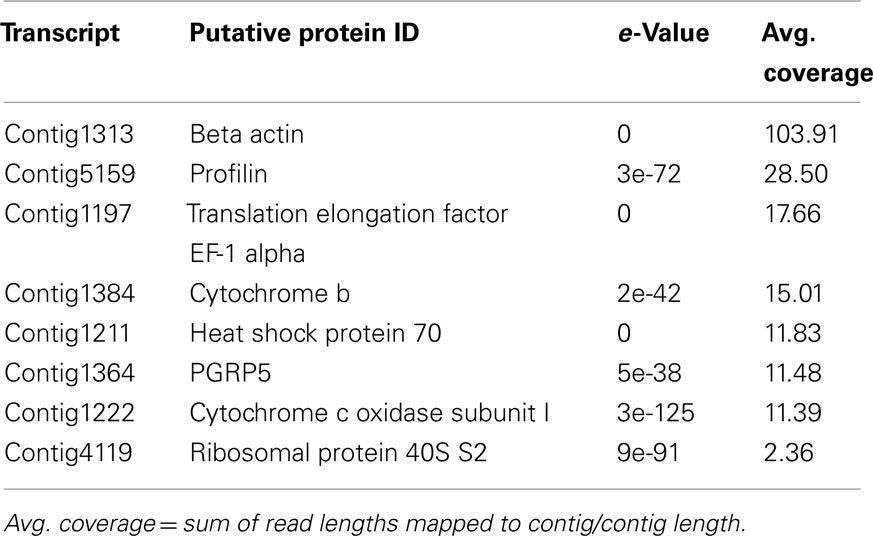
Despite a large number of reads associated with ribosomal genes, assembly of the transcriptome resulted in 5,586 contigs having an N50 of 521 bp, of which 1,995 were annotated (Table 1). Many of these were predicted to be involved with cytoskeletal structure (Figure 2; Dataset S1 in Supplementary Material). Among innate immunity-related genes, several members, and/or putative regulators of the NF-κB signaling pathway were identified including TRAF4, TRAF6, REL/NF-κB, inhibitor of NF-κB, NF-κB activating protein, both calcineurin subunits, a regulator of calcineurin, and MyD88 (Table 3). Several PRRs were identified including galectin and a previously described peptidoglycan recognition protein (EsPGRP4) (Table 3). An undescribed PGRP, most closely related to PGRP-SC2 from the rotifer Brachionus manjavacas, was also found (Table 3). Further analysis revealed a complete open reading frame (ORF) consisting of a 759bp transcript encoding a predicted protein containing 253 amino acids (Figure 3). SignalP software predicted that this newly described PGRP (EsPGRP5) has a signal peptide and is therefore likely secreted (Figure 3). In addition, alignment with four other PGRPs from E. scolopes showed that EsPGRP5 has conserved residues for amidase activity, suggesting that it is capable of degrading bacterial peptidoglycan and its derivatives (Figure 3). The EsPGRP5 gene was one of the most abundant annotated transcripts (95/1995, Dataset S1 in Supplementary Material) and it had significant coverage compared to other highly expressed housekeeping genes (Table 4). Peptides from all three PRRs found in the transcriptome were also detected in the proteome (Figure 4; Table 5; Datasets S1 and S2 in Supplementary Material). Other transcripts found included IK cytokine, CD63 antigen, two putative toxins, plancitoxin and cephalotoxin, and a matrix metalloproteinase that shares similarity with vitronectin, a member of the complement cascade (Table 5).

Figure 3. Alignment of E. scolopes peptidoglycan recognition proteins. Alignment of EsPGRP5 with the previously described EsPGRPs 1–4, revealed a predicted 253 aa protein with a predicted signal peptide sequence (dark shade) as well as amidase activity (light shade). (*) Conserved residues among all EsPGRPs; (.) substitutions with weak similarity <0.5 Gonnet PAM 250 matrix; (:) substitutions with strong similarity, >0.5 Gonnet PAM 250 matrix.
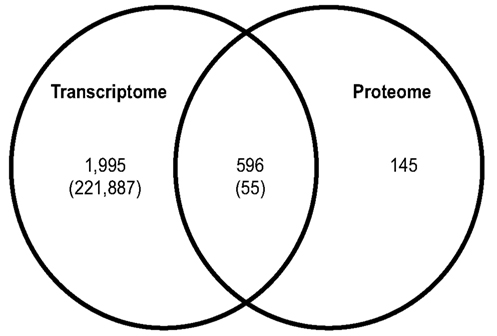
Figure 4. Comparison of the hemocyte transcriptome and proteome. Protein sequences identified by hemocyte peptides were compared to the hemocyte transcriptome using tblastn. 596 sequences were found to be in common between the transcriptome and proteome while 145 protein sequences were identified solely using proteomics. Numbers in parentheses represent total annotated singletons (Transcriptome) and proteome matches to singletons (Shared).
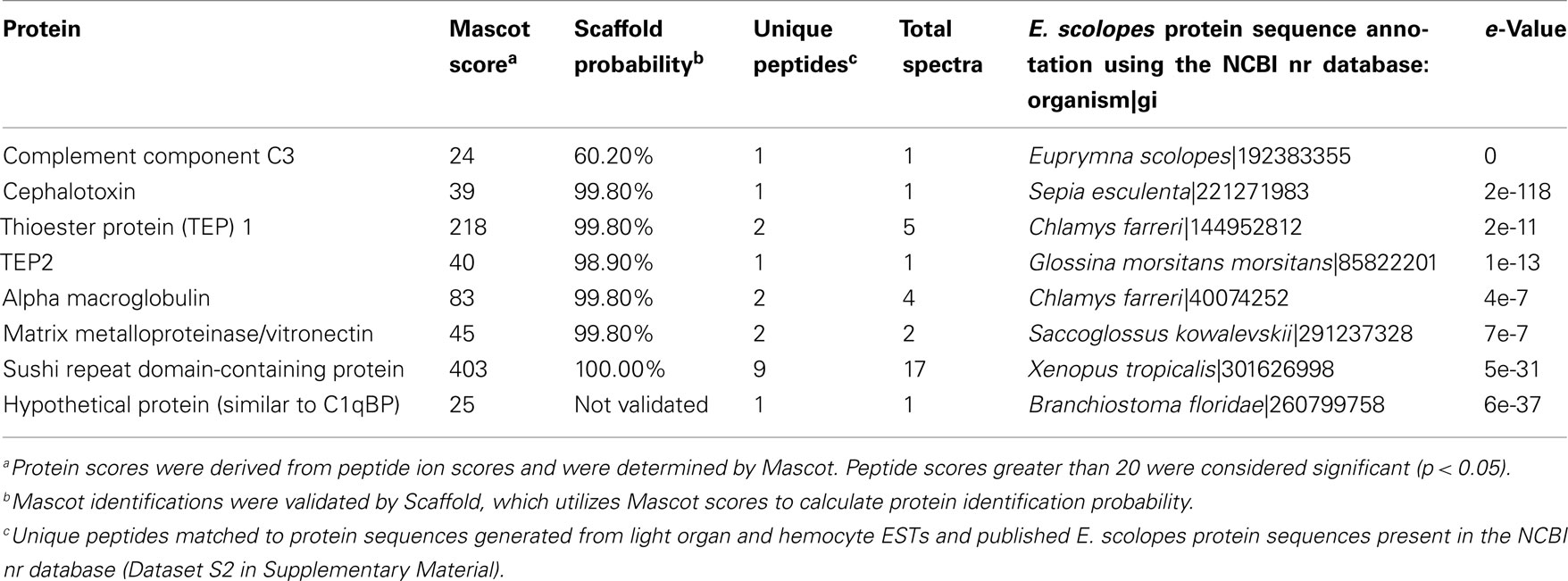
Table 5. Putative complement pathway and other innate immunity-related proteins identified in E. scolope s hemocytes.
Analyses of the hemocyte proteome revealed a number of proteins involved with innate immune responses, and some of these were also found in the transcriptome (Figure 4; Tables 3 and 5, Dataset S3 in Supplementary Material). Comparing the MS/MS peptide data solely to the hemocyte transcriptome from this study revealed 596 genes/proteins in common and 145 proteins identified only from the proteome (Figure 4; Dataset S3 in Supplementary Material).
Several putative proteins with similarity to members of the complement cascade and other innate immunity functions were identified from the LC-MS/MS analyses, including the lynchpin component EsC3, a cephalotoxin protein, two thioester proteins (TEPs), an alpha macroglobulin, and a matrix metalloproteinase similar to vitronectin (Table 5). A 398 aa residue protein was detected having six sushi repeat domains that are common to complement and cell adhesion proteins like CR1, CR2, and selectins. In addition there was a putative protein with similarity to the complement member C1qBP.
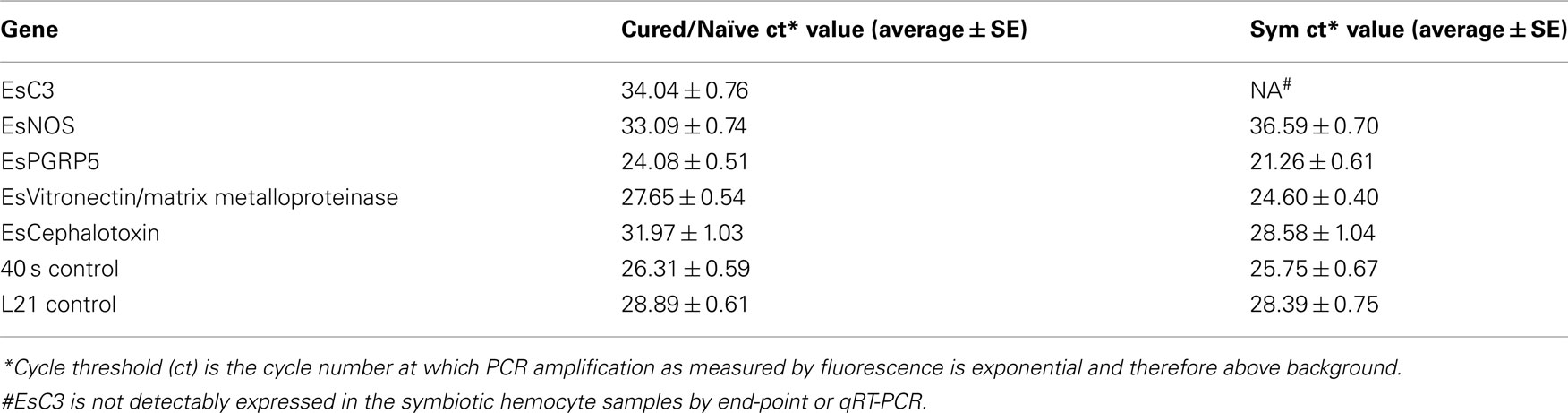
Both end-point RT- and qRT-PCR showed that members of the complement cascade including EsC3 and a putative matrix metalloproteinase/vitronectin along with cephalotoxin were differentially expressed in hemocytes isolated from hosts with fully colonized light organs compared to those hosts that had been cured (naïve; Figure 5). EsC3 transcripts were detected in naïve hemocytes but not in hemocytes from symbiotic hosts. The matrix metalloproteinase/vitronectin and cephalotoxin were both down-regulated in naïve hemocytes (−5.6 and −7.1 fold respectively). EsPGRP5 decreased in expression in naïve hemocytes compared to those from symbiotic hosts (−4.7 fold). EsPGRP5 also had the lowest CT value of all the genes analyzed, including the 40S and L21 ribosomal controls, adding further evidence that this gene is abundantly expressed in hemocytes (Table A2 in Appendix). In addition, nitric oxide synthase (NOS), which has previously been shown to be differentially expressed in symbiotic and aposymbiotic (uncolonized) juvenile light organs (Davidson et al., 2004), was up-regulated in hemocytes from cured hosts (+16.8 fold).
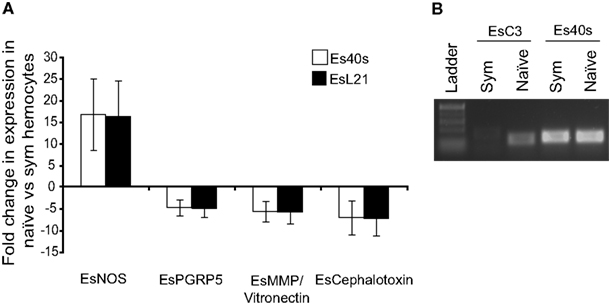
Figure 5. Differential expression of putative innate immunity genes. qRT-PCR (A) and end-point PCR (B) of select immune genes in adult symbiotic and naïve hemocytes. For qRT-PCR transcription of indicated genes was normalized to the 40S and L21 ribosomal subunit genes and represented as fold difference of the naïve relative to the symbiotic state. Error bars represent SD of three triplicate samples. qRT-PCR and end-point PCR results are representative of three separate RNA extractions.
Discussion
This study represents the first transcriptomic and proteomic analyses of the circulating hemocytes from E. scolopes. As might be expected, in addition to factors involved with general biological processes (Figure 2), hemocytes express a suite of innate immunity genes under a normal symbiotic state (Figure 6). Our data have revealed a number of genes and proteins that have previously been shown to be important in the squid/Vibrio association. In addition we have described a fifth PGRP (EsPGRP5), which was among the most abundant transcripts in hemocytes. We also identified novel genes/proteins, for example a putative cephalotoxin and plancitoxin that have not been previously described in invertebrate hemocytes. Finally, we demonstrate that some of these genes are differentially expressed in hemocytes from symbiotic and cured hosts, suggesting that colonization influences gene expression of the cellular innate immune system of E. scolopes. The cellular localization and function of many of these genes and proteins have yet to be characterized in detail but we discuss below the putative function of some of these innate immunity effectors in the context of host hemocytes (Figure 6).
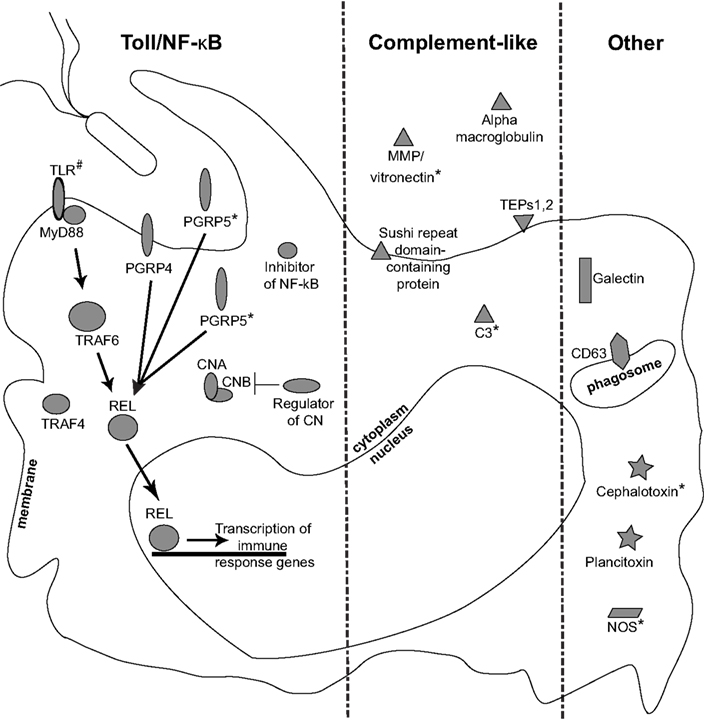
Figure 6. Identification of transcripts and proteins associated with hemocytes. Representation of putative immune components present in circulating squid hemocytes. Interactions with symbiotic and/or non-symbiotic bacteria likely involve recognition by host PRRs, like PGRPs and TLR. Down stream signaling via the NF-κB pathway may modulate immune effector response. Putative members of a complement cascade as well as other immune-related genes were also expressed in circulating hemocytes. Colonization state influenced expression of a subset of these genes (*; Figure 4), suggesting that V. fischeri can modulate the host’s cellular innate immune response. (#), immune gene not identified in this study (Goodson et al., 2005; Rader et al., unpublished data). Localizations are inferred from protein domain predictions as well as similarity to other systems.
Among the transcripts and identified peptides, many were predicted to be involved in cytoskeletal structure (Figure 2; Datasets S1–S3 in Supplementary Material). This is not surprising considering that these macrophage-like hemocytes can switch within minutes from a round spheroid to an activated cell with amoeboid characters. These activated cells are then capable of migrating through tissues and have been shown to enter the crypts of the light organ where they can then interact with V. fischeri (Nyholm and McFall-Ngai, 1998; Heath-Heckman and McFall-Ngai, 2011).
Among those genes involved with innate immunity, several putative PRRs were found including a galectin and two PGRPs (EsPGRP4 and EsPGRP5). Galectins are proteins that can bind carbohydrates and have been implicated as PRRs during the immune response (reviewed in Vasta, 2009). In other mollusks, galectins have been shown to bind microorganisms and are up-regulated in the presence of bacteria (Tasumi and Vasta, 2007; Song et al., 2011; Zhang et al., 2011). The role of galectin in this symbiosis has yet to be characterized, but efforts are underway to compare gene expression and protein levels in hemocytes of colonized and uncolonized hosts.
Whether an association is pathogenic or beneficial the immune system must first recognize the microorganism(s). These encounters are mediated by the initial recognition of MAMPs by PRRs (Koropatnick et al., 2004; Medzhitov, 2007; McFall-Ngai et al., 2012). PGRPs are one of a suite of PRRs that recognize MAMPs and are evolutionarily conserved (Royet et al., 2011). Five PGRPs have now been described from E. scolopes (Goodson et al., 2005; Troll et al., 2009, 2010; this study) and prior work has demonstrated that some of these are important during the onset of the symbiosis (Troll et al., 2009, 2010). Analyses of EsPGRP1 and EsPGRP2 protein expression in the juvenile light organ have shown that they have different cellular localizations within the crypt epithelia and nascent light organ tissues. V. fischeri can influence this expression with specific MAMPS. For example, tracheal cytotoxin (TCT), a derivative of peptidoglycan that along with lipopolysaccharide signals early development of the host light organ (Foster et al., 2000; Koropatnick et al., 2004), influences the nuclear loss of EsPGRP1 in host epithelial cells (Troll et al., 2009). EsPGRP2 which is secreted into the light organ crypts has TCT-degrading amidase activity and may ameliorate the effect of this toxin, especially during times when the symbiont numbers, and presumably TCT concentrations, are high (Troll et al., 2010). The ability to have a diverse repertoire of PGRPs may give the host greater ability to respond to peptidoglycan and TCT depending on the MAMP microenvironment that it encounters. The results of this study describe a fifth PGRP (Table 3; Figure 3) and suggest that EsPGRP5 is one of the most abundant transcripts expressed in host hemocytes (Table 4; Figure 5; Dataset S1 in Supplementary Material). Furthermore, qRT-PCR showed that this gene is down-regulated 4.7 fold in hemocytes from cured hosts suggesting that colonization of the light organ may directly influence PRRs in hemocytes. Currently, a polyclonal antibody to EsPGRP5 was generated and is being used to localize this protein in hemocytes at different stages of the association and under challenge by symbiotic and non-symbiotic bacteria.
The Toll/NF-κB pathway is an evolutionarily conserved signaling pathway often involved in regulating innate immune genes (Leulier and Lemaitre, 2008). In E. scolopes, homologs of this pathway have been identified (Goodson et al., 2005) and microarray data demonstrated different expression levels in colonized vs. uncolonized juvenile light organs (Chun et al., 2006). Our transcriptomic data revealed several members of this pathway, including MyD88, the universal adaptor protein for the TLRs as well as other IL-1 receptor proteins that plays an important and well-studied role in inflammation and host defense (Kenny and O’Neill, 2008). MyD88 is also involved with interactions of epithelial surfaces with microbial communities. For example in mice, production of certain antimicrobial peptides by paneth cells is dependent on MyD88, and is sufficient to limit bacterial translocation to the mesenteric lymph nodes without altering the number of luminal bacteria (reviewed in Wells et al., 2011). Two TNF receptor-associated factors TRAF4 and TRAF6 were also identified. TRAF4 is a signal transducer of the Toll/IL-1 family. In mouse macrophages, TRAF4 has been shown to negatively regulate the NF-κB signaling cascade in response to the peptidoglycan byproduct muramyl dipeptide (Marinis et al., 2011) while TRAF6 is involved with regulating ROS via the NF-κB pathway in mammalian macrophages (West et al., 2011). Both the A and B subunits of calcineurin were also detected in the transcriptome. Calcineurin is known for modulating immune responses in macrophages and this protein can inhibit both IL-12 and TNFα production. Furthermore, inhibiting calcineurin in vitro increases expression of effector genes, including NOS, and NF-κB as much as eight-fold (Conboy et al., 1999). Calcineurin may also be involved in responses to LPS since mice lacking a subunit of calcineurin show a tolerance to LPS, with much lower TNFα production than wild type animals (Jennings et al., 2009). Recent transcriptome profiling of other molluscan hemocytes from an oyster and mussel challenged with either pathogenic vibrios or lipopolysaccharide also revealed a number of expressed pattern recognition and signaling molecules related to the Toll/NF-κB pathway, thus highlighting the importance of this evolutionarily conserved pathway in this cell type (de Lorgeril et al., 2011; Philipp et al., 2012).
Recent analyses of the squid/Vibrio system have identified a number of putative members of the complement pathway (Castillo et al., 2009; Schleicher and Nyholm, 2011). C3 is the central protein to all three pathways of the complement system and is involved in opsonization and phagocytosis (Dunkelberger and Song, 2010). A C3 ortholog was recently discovered in E. scolopes and shown to be located at the apical surfaces of crypt epithelial cells (Castillo et al., 2009). In this study, C3 was found in the hemocyte proteome (Table 5). End-point RT-PCR and qRT-PCR showed C3 expression only in hemocytes from cured hosts. We do not yet know the turnover rate of the EsC3 protein in hemocytes but removing the symbionts led to an increase in gene expression of this main component of the complement pathway. Also identified were two TEPs. TEPs are similar to complement-like components and alpha macroglobulins, which have basic functions as proteases and protease inhibitors respectively (Bou Aoun et al., 2011). Among invertebrates, TEPs play an important role in the innate immune response as members of the complement-like system (Blandin and Levashina, 2004; Bou Aoun et al., 2011). A recent analysis of the light organ proteome found several TEPs (Schleicher and Nyholm, 2011). Also identified was an alpha macroglobulin belonging to a family of protease inhibitors and has been shown to be an important component of the innate immune response of arthropods (Sottrup-Jensen, 1989).
Among the other proteins identified was a matrix metalloproteinase similar to vitronectin. Vitronectin has been described as a serum glycoprotein involved in regulating innate immunity homeostasis (Schvartz et al., 1999). In humans, vitronectin inhibits the membrane attack complex of the complement pathway. Bacteria, including Haemophilus influenzae and Neisseria meningitides can interact with vitronectin and evade the complement response (Singh et al., 2010). Recent evidence also suggests that the parasite Plasmodium falciparum may use vitronectin-like domains to evade the immune system (Ludin et al., 2011). LC-MS/MS analyses identified a putative vitronectin in circulating hemocytes and results of qRT-PCR showed that MMP/vitronectin was down-regulated 5.6 fold in the absence of V. fischeri (Figure 5). The putative MMP/vitronectin in this study contains a hemopexin-like domain (data not shown) but whether it serves a role similar to vitronectin in the squid host remains to be determined. Among other proteins identified was one containing six sushi repeat domains in a partial ORF (Table 5). Sushi domains are commonly found among complement receptors (e.g., CR1, CR2, C4b binding protein, and factor H) as well as other proteins involved with innate immunity and self/non-self recognition (Kirkitadze and Barlow, 2001; Nyholm et al., 2006). This protein has yet to be characterized in the squid/Vibrio association but offers an enticing future target for analysis. A putative peptide was also found that had similarity to C1qBP which is a component of the complement protein cascade but will have to be validated further.
Both the transcriptome and proteome identified a protein with high similarity to cephalotoxin (Tables 3 and 5; Ueda et al., 2008). In octopus and cuttlefish, cephalotoxins are normally expressed in the salivary glands and are involved with immobilizing prey. This is the first report of this protein in another cell type. A search of the UniProt database identified a thrombospondin domain similar to ones described in properdin, a proenzyme in the complement cascade (Kemper and Hourcade, 2008; Kemper et al., 2010). Whether cephalotoxin functions as a toxin, a protease, or in another capacity in the hemocytes of E. scolopes remains to be determined but qRT-PCR showed a 7.1 fold downturn in expression in naïve hemocytes, suggesting that colonization also influences expression of this gene (Figure 5). A second toxin was identified with similarity to plancitoxins found in the crown-of-thorns starfish Acanthaster planci (Shiomi et al., 2004). These proteins are predicted to be deoxyribonucleases and future studies should focus on characterizing any potential DNAse activity in squid hemocytes.
Reactive oxygen species are common effectors used by the innate immune system to combat pathogens. In symbiotic associations, ROS have also been shown to be important, often in contributing to microenvironments that may promote a microbial member (Weis et al., 1996; Davidson et al., 2004; Weis, 2008; Ryu et al., 2010). The enzyme nitric oxide synthase (NOS) is required for generating antimicrobial reactive nitrogen species (RNS) such as nitric oxide (NO). Both NOS and NO are active during colonization of the host and NO was detected in vesicles found within mucus secretions that interact with V. fischeri during initiation of the association (Davidson et al., 2004). Colonization leads to an attenuation of NOS and NO in the crypts suggesting that the RNS response may change in order to accommodate the symbiont (Davidson et al., 2004). Although not detected in the transcriptome or proteome, we decided to analyze NOS expression in symbiotic and naïve hemocytes because of its likely importance in the host immune response (Figure 5). Naïve hemocytes showed a 16.8 fold increase in expression suggesting that colonization leads to a suppression of NOS in hemocytes, similar to what’s been described for juvenile light organs.
Another condition that may influence hemocyte gene expression is a changing symbiont population over a 24-h day/night cycle. During the evening when the host is active, there is a full complement of V. fischeri, but at dawn, the host expels 95% of these symbionts into the surrounding seawater (Boettcher et al., 1996; Nyholm and McFall-Ngai, 1998). We do not yet know how gene expression of host hemocytes might change during the day/night cycle and in the context of the microenvironment encountered in the crypt spaces of the light organ, but prior studies have reflected changes in host and symbiont transcription in adults light organ tissues over a 24-h period (Wier et al., 2010). Light organ gene expression is also altered by colonization in juvenile hosts (Chun et al., 2006). Our transcriptome was derived from circulating hemocytes from a variety of time points to understand the overall background gene expression in colonized adults. Growing evidence suggests that the immune response of hosts can fluctuate on a daily rhythm (Shirasu-Hiza et al., 2007; Binuramesh and Michael, 2011) and future studies should also consider how innate immunity effectors and pathways might vary over the day/night cycle in the squid/Vibrio association.
We still have much to understand concerning the role of host hemocytes in the squid/Vibrio symbiosis, but future research into this cell type will likely yield interesting results. Macrophage-like phagocytes serve at the interface between hosts and symbionts. Besides combating pathogens, invertebrate hemocytes have recently been found to play important roles in mediating bacterial symbioses as in the medicinal leech (Silver et al., 2007), linked to a primed immune response to specific bacteria in Drosophila (Pham et al., 2007) and promoting symbiont tolerance in the squid/Vibrio association (Nyholm et al., 2009). This is the first report to apply both transcriptomics and proteomics to analyze hemocytes in a beneficial symbiosis. One advantage of combining both techniques in future studies is that genes/proteins not identified by one method may be detected by the other. For example, both TEPs and alpha macroglobulin were found in the proteome but not in the transcriptome (Figure 4; Dataset S3 in Supplementary Material). Future studies will also use quantitative transcriptomic and proteomic methods to compare gene and protein expression between symbiotic and naïve hemocytes and those challenged with V. fischeri and non-symbiotic bacteria. This study lays the groundwork for exploring the molecular mechanisms of hemocyte/symbiont specificity in the squid/Vibrio symbiosis and adds further evidence that interactions between beneficial bacteria and the innate immune system are important in mediating these associations.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank C. Obergfell and R. O’Neill and the Center for Applied Genetics and Technology at the University of Connecticut for assistance with 454 analyses. We thank T. Abbott, K. Stone, and the Mass spectrometry and Protein Chemistry Facility of the W.M. Keck Biotechnology Resource Laboratory at Yale University for assistance with proteomic data. We thank Kewalo Marine Laboratory and the Hawaii Institute of Marine Biology for assistance with collections. We thank P. Lapierre for assisting us with bioinformatic analyses. The UConn Bioinformatics Facility provided computing resources for BLAST searches and ClustalX alignments. This work was supported by the National Science Foundation IOS-0958006 and the University of Connecticut Research Foundation to Spencer V. Nyholm.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Molecular_Innate_Immunity/10.3389/fimmu.2012.00091/abstract
References
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920.
Bauer, E., Williams, B. A., Smidt, H., Verstegen, M. W., and Mosenthin, R. (2006). Influence of the gastrointestinal microbiota on development of the immune system in young animals. Curr. Issues Intest. Microbiol. 7, 35–51.
Binuramesh, C., and Michael, R. D. (2011). Diel variations in the selected serum immune parameters in Oreochromis mossambicus. Fish Shellfish Immunol. 3, 824–829.
Blandin, S., and Levashina, E. A. (2004). Thioester-containing proteins and insect immunity. Mol. Immunol. 40, 903–908.
Boettcher, K. J., Ruby, E. G., and McFall-Ngai, M. J. (1996). Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J. Comp. Physiol. A 179, 65–73.
Bou Aoun, R., Hetru, C., Troxler, L., Doucet, D., Ferrandon, D., and Matt, N. (2011). Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster. J. Innate Immun. 3, 52–64.
Castillo, M. G., Goodson, M. S., and McFall-Ngai, M. J. (2009). Identification and molecular characterization of a complement C3 molecule in a lophotrochozoan, the Hawaiian bobtail squid Euprymna scolopes. Dev. Comp. Immunol. 33, 69–76.
Chun, C. K., Scheetz, T. E., Bonaldo, Mde. F., Brown, B., Clemens, A., Crookes-Goodson, W. J., Crouch, K., DeMartini, T., Eyestone, M., Goodson, M. S., Janssens, B., Kimbell, J. L., Koropatnick, T. A., Kucaba, T., Smith, C., Stewart, J. J., Tong, D., Troll, J. V., Webster, S., Winhall-Rice, J., Yap, C., Casavant, T. L., McFall-Ngai, M. J., and Soares, M. B. (2006). An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics 7, 154. doi: 10.1186/1471-2164-7-154
Collins, A. J., and Nyholm, S. V. (2010). Obtaining hemocytes from the Hawaiian bobtail squid Euprymna scolopes and observing their adherence to symbiotic and non-symbiotic bacteria. J. Vis. Exp. 36, 1714.
Conboy, I. M., Manoli, D., Mhaiskar, V., and Jones, P. (1999). Calcineurin and vacuolar-type H+-ATPase modulate macrophage effector functions. Proc. Natl. Acad. Sci. U.S.A. 96, 6324–6329.
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676.
Craig, R., and Beavis, R. C. (2004). TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467.
Davidson, S. K., Koropatnick, T. A., Kossmehl, R., Sycuro, L., and McFall-Ngai, M. J. (2004). NO means ‘yes’ in the squid-Vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell. Microbiol. 6, 1139–1151.
de Lorgeril, J., Zenagui, R., Rosa, R. D., Piquemal, D., and Bachere, E. (2011). Whole transcriptome profiling of successful immune response to Vibrio infections in the oyster Crassostrea gigas by digital gene expression analysis. PLoS ONE 6, e23142. doi: 10.1371/journal.pone.0023142
Doino, J. A., and McFall-Ngai, M. J. (1995). A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol. Bull. 189, 347–355.
Duan, J., and Kasper, D. L. (2011). Regulation of T cells by gut commensal microbiota. Curr. Opin. Rheumatol. 23, 372–376.
Dunkelberger, J. R., and Song, W. (2010). Complement and its role in innate and adaptive immune responses. Cell Res. 20, 34–50.
Feldhaar, H., and Gross, R. (2009). Insects as hosts for mutualistic bacteria. Int. J. Med. Microbiol. 299, 1–8.
Foster, J. S., Apicella, M. A., and McFall-Ngai, M. J. (2000). Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev. Biol. 226, 242–254.
Goodson, M. S., Kojadinovic, M., Troll, J. V., Scheetz, T. E., Casavant, T. L., Soares, M. B., and McFall-Ngai, M. J. (2005). Identifying components of the NF-κB pathway in the beneficial Euprymna scolopes-Vibrio fischeri light organ symbiosis. Appl. Environ. Microbiol. 71, 6934–6946.
Gross, R., Vavre, F., Heddi, A., Hurst, G. D., Zchori-Fein, E., and Bourtzis, K. (2009). Immunity and symbiosis. Mol. Microbiol. 73, 751–759.
Heath-Heckman, E. A., and McFall-Ngai, M. J. (2011). The occurrence of chitin in the hemocytes of invertebrates. Zoology 114, 191–198.
Hirosawa, M., Hoshida, M., Ishikawa, M., and Toya, T. (1993). MASCOT: multiple alignment system for protein sequences based on three-way dynamic programming. Comput. Appl. Biosci. 9, 161–167.
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., and Gibson, T. J. (1998). Multiple sequence alignment with ClustalX. Trends Biochem. Sci. 23, 403–405.
Jennings, C., Kusler, B., and Jones, P. (2009). Calcineurin inactivation leads to decreased responsiveness to LPS in macrophages and dendritic cells and protects against LPS-induced toxicity in vivo. Innate Immun. 15, 109–200.
Kemper, C., Atkinson, J. P., and Hourcade, D. E. (2010). Properdin: emerging roles of a pattern-recognition molecule. Annu. Rev. Immunol. 28, 131–155.
Kemper, C., and Hourcade, D. E. (2008). Properdin: new roles in pattern recognition and target clearance. Mol. Immunol. 45, 4048–4056.
Kenny, E. F., and O’Neill, L. A. (2008). Signalling adaptors used by Toll-like receptors: an update. Cytokine 43, 342–349.
Kirkitadze, M. D., and Barlow, P. N. (2001). Structure and flexibility of multiple domain proteins that regulate complement activation. Immunol. Rev. 180, 146–161.
Koropatnick, T. A., Engle, J. T., Apicella, M. A., Stabb, E. V., Goldman, W. E., and McFall-Ngai, M. J. (2004). Microbial factor-mediated development in a host-bacterial mutualism. Science 306, 1186–1188.
Koropatnick, T. A., Kimbell, J. R., and McFall-Ngai, M. J. (2007). Responses of host hemocytes during the initiation of the squid-Vibrio symbiosis. Biol. Bull. 212, 29–39.
Krasity, B. C., Troll, J. V., Weiss, J. P., and McFall-Ngai, M. J. (2011). LBP/BPI proteins and their relatives: conservation over evolution and roles in mutualism. Biochem. Soc. Trans. 39, 1039–1044.
Lamarcq, L. H., and McFall-Ngai, M. J. (1998). Induction of a gradual, reversible morphogenesis of its host’s epithelial brush border by Vibrio fischeri. Infect. Immun. 66, 777–785.
Lee, Y. K., and Mazmanian, S. K. (2010). Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768–1773.
Leulier, F., and Lemaitre, B. (2008). Toll-like receptors–taking an evolutionary approach. Nat. Rev. Genet. 9, 165–178.
Ludin, P., Nilsson, D., and Mäser, P. (2011). Genome-wide identification of molecular mimicry candidates in parasites. PLoS ONE 6, e17546. doi: 10.1371/journal.pone.0017546
Marinis, J. M., Homer, C. R., McDonald, C., and Abbot, D. W. (2011). A novel motif in the Crohn’s disease susceptibility protein, NOD2, Allows TRAF4 to down-regulate innate immune responses. J. Biol. Chem. 286, 1938–1950.
McFall-Ngai, M. J. (2002). Unseen forces: the influence of bacteria on animal development. Dev. Biol. 242, 1–14.
McFall-Ngai, M. J., Heath-Heckman, E. A., Gillette, A. A., Peyer, S. M., and Harvie, E. A. (2012). The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin. Immunol. 24, 3–8.
McFall-Ngai, M. J., Nyholm, S. V., and Castillo, M. G. (2010). The role of the immune system in the initiation and persistence of the Euprymna scolopes – Vibrio fischeri symbiosis. Semin. Immunol. 22, 48–53.
Medzhitov, R. (2007). Recognition of microorganisms and activation of the immune response. Nature 449, 819–826.
Moran, N. A., McCutcheon, J. P., and Nakabachi, A. (2008). Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190.
Nyholm, S. V., Deplancke, B., Gaskins, H. R., Apicella, M. A., and McFall-Ngai, M. J. (2002). Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 68, 5113–5122.
Nyholm, S. V., and McFall-Ngai, M. J. (1998). Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol. Bull. 195, 89–97.
Nyholm, S. V., and McFall-Ngai, M. J. (2004). The winnowing: establishing the squid-Vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642.
Nyholm, S. V., Passegue, E., Ludington, W. B., Voskoboynik, A., Mitchel, K., Weissman, I. L., and DeTomaso, A. W. (2006). Fester, a candidate allorecognition receptor from a primitive chordate. Immunity 25, 163–173.
Nyholm, S. V., Stewart, J. J., Ruby, E. G., and McFall-Ngai, M. J. (2009). Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ. Microbiol. 11, 483–493.
Olszak, T., An, D., Zeissig, S., Vera, M. P., Richter, J., Fanke, A., Glickman, J. N., Siebert, R., Baron, R. M., Kasper, D. L., and Blumberg, R. S. (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science (in press).
Perkins, D. N., Pappin, D. J., Creasy, D. M., and Cottrell, J. S. (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567.
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786.
Pham, L. N., Dionne, M. S., Shirasu-Hiza, M., and Schneider, D. S. (2007). A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 3, e26. doi: 10.1371/journal.ppat.0030026
Philipp, E. E. R., Kraemer, L., Melzner, F., Poustka, A. J., Thieme, S., Findeisen, U., Schreiber, S., and Rosentiel, P. (2012). Massive parallel RNA sequencing identifies a complex immune gene repertoire in the lophotrochozoan Mytilus edulis. PLoS ONE 7, e33091. doi: 10.1371/journal.pone.0033091
Royet, J., Gupta, D., and Dziarski, R. (2011). Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nat. Rev. Immunol. 11, 837–851.
Ryu, J. H., Ha, E. M., and Lee, W. J. (2010). Innate immunity and gut-microbe mutualism in Drosophila. Dev. Comp. Immunol. 34, 369–376.
Schleicher, T. R., and Nyholm, S. V. (2011). Characterizing the host and symbiont proteomes in the association between the bobtail squid, Euprymna scolopes, and the bacterium, Vibrio fischeri. PLoS ONE 6, e25649. doi: 10.1371/journal.pone.0025649
Schvartz, I., Seger, D., and Shaltiel, S. (1999). Vitronectin. Int. J. Biochem. Cell Biol. 31, 539–544.
Shiomi, K., Midorikawa, S., Ishida, M., Nagashima, Y., and Nagai, H. (2004). Plancitoxins, lethal factors from the crown-of-thorns starfish Acanthaster planci, are deoxyribonucleases II. Toxicon 44, 499–506.
Shirasu-Hiza, M. M., Dionne, M. S., Pham, L. N., Ayres, J. S., and Schneider, D. S. (2007) Interactions between circadian rhythm and immunity in Drosophila melanogaster. Curr. Biol. 17, R353–R355.
Silver, A. C., Kikuchi, Y., Fadl, A. A., Sha, J., Chopra, A. K., and Graf, J. (2007). Interaction between innate immune cells and a bacterial type III secretion system in a mutualistic and pathogenic associations. Proc. Natl. Acad. Sci. U.S.A. 104, 9481–9486.
Singh, B., Su, Y., and Riesbeck, K. (2010). Vitronectin in bacterial pathogenesis: a host protein used in complement escape and cellular invasion. Mol. Microbiol. 78, 545–560.
Song, X., Zhang, H., Wang, L., Zhao, J., Mu, C., Song, L., Qiu, L., and Liu, X. (2011). A galectin with quadruple-domain from bay scallop Argopecten irradians is involved in the innate immune response. Dev. Comp. Immunol. 35, 592–602.
Sottrup-Jensen, L. (1989). α-Macroglobulins: structure, shape, and mechanism of proteinase complex formation. J. Biol. Chem. 264, 11539–11542.
Stewart, F. J., and Cavanaugh, C. M. (2006). Symbiosis of thioautotrophic bacteria with Riftia pachyptila. Prog. Mol. Subcell. Biol. 41, 197–225.
Stoll, S., Feldhaar, H., Fraunholz, M. J., and Gross, R. (2010). Bacteriocyte dynamics during development of a holometabolous insect, the carpenter ant Camponotus floridanus. BMC Microbiol. 10, 308. doi: 10.1186/1471-2180-10-308
Tasumi, S., and Vasta, G. R. (2007). A galectin of unique domain organization from hemocytes of the eastern oyster (Crassostrea virginica) is a receptor for the protistan parasite Perkinsus marinus. J. Immunol. 179, 3086–3098.
Troll, J. V., Adin, D. M., Wier, A. M., Paquette, N., Silverman, N., Goldman, W. E., Stadermann, F. J., Stabb, E. V., and McFall-Ngai, M. J. (2009). Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell. Microbiol. 11, 1114–1127.
Troll, J. V., Bent, E. H., Pacquette, N., Wier, A. M., Goldman, W. E., Silverman, N., and McFall-Ngai, M. J. (2010). Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ. Microbiol. 12, 2190–2203.
Ueda, A., Nagai, H., Ishida, M., Nagashima, Y., and Shiomi, K. (2008). Purification and molecular cloning of SE-cephalotoxin, a novel proteinaceous toxin from the posterior salivary gland of the cuttlefish Sepia esculenta. Toxicon 52, 574–581.
Visick, K. L., and Ruby, E. G. (2006). Vibrio fischeri and its host: it takes two to tango. Curr. Opin. Microbiol. 9, 632–638.
Watanabe, H., and Tokuda, G. (2010). Cellulolytic systems in insects. Annu. Rev. Entomol. 55, 609–632.
Weis, V. M. (2008). Cellular mechanisms of cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211(Pt 19), 3059–3066.
Weis, V. M., Small, A. L., and McFall-Ngai, M. J. (1996). A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc. Natl. Acad. Sci. U.S.A. 93, 13683–13688.
Weiss, B. L., Wang, J., and Aksoy, S. (2011). Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 9, e1000619. doi: 10.1371/journal.pbio.1000619
Wells, J. M., Rossi, O., Meijerink, M., and van Baarlen, P. (2011). Epithelial crosstalk at the microbiota-mucosal interface. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4607–4614.
West, A. P., Brodsky, I. E., Rahner, C., Woo, D. K., Bromage, H., Tempst, P., Walsh, M. C., Choi, Y., Shadel, G. S., and Ghosh, S. (2011). TLR signaling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480.
Wier, A. M., Nyholm, S. V., Mandel, M. J., Massengo-Tiassé, R. P., Schaefer, A. L., Koroleva, I., Splinter-Bondurant, S., Brown, B., Manzella, L., Snir, E., Almabrazi, H., Scheetz, T. E., Bonaldo, Mde. F., Casavant, T. L., Soares, M. B., Cronan, J. E., Reed, J. L., Ruby, E. G., and McFall-Ngai, M. J. (2010). Transcriptional patterns in both host and bacterium underlie a daily rhythmof anatomical and metabolic change in a beneficial symbiosis. Proc. Natl. Acad. Sci. U.S.A. 107, 2259–2264.
Zdobnov, E. M., and Apweiler, R. (2001). InterProScan- an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848.
Zhang, D., Jiang, S., Hu, Y., Cui, S., Guo, H., Wu, K., Li, Y., and Su, T. (2011). A multidomain galectin involved in innate immune response of pearl oyster Pinctada fucata. Dev. Comp. Immunol. 35, 1–6.
Appendix
Keywords: innate immunity, symbiosis, Euprymna scolopes, Vibrio fischeri, hemocytes, transcriptomics, proteomics
Citation: Collins AJ, Schleicher TR, Rader BA and Nyholm SV (2012) Understanding the role of host hemocytes in a squid/Vibrio symbiosis using transcriptomics and proteomics. Front. Immun. 3:91. doi: 10.3389/fimmu.2012.00091
Received: 19 January 2012; Accepted: 08 April 2012;
Published online: 10 May 2012.
Edited by:
Larry J. Dishaw, University of South Florida, USAReviewed by:
Ioannis Eleftherianos, The George Washington University, USAVirginia Weis, Oregon State University, USA
Copyright: © 2012 Collins, Schleicher, Rader and Nyholm. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Spencer V. Nyholm, Department of Molecular and Cell Biology, University of Connecticut, 91 N. Eagleville Road, Unit 3125, Storrs, CT 06269, USA. e-mail: spencer.nyholm@uconn.edu
†Andrew J. Collins and Tyler R. Schleicher have contributed equally to this work.